Abstract
In our previous study, we genetically analyzed bovine viral diarrhea viruses (BVDVs) isolated from 2000 to 2006 in Japan and reported that subgenotype 1b viruses were predominant. In the present study, 766 BVDVs isolated from 2006 to 2014 in Hokkaido, Japan, were genetically analyzed to understand recent epidemics. Phylogenetic analysis based on nucleotide sequences of the 5′-untranslated region of viral genome revealed that 766 isolates were classified as genotype 1 (BVDV-1; 544 isolates) and genotype 2 (BVDV-2; 222). BVDV-1 isolates were further divided into BVDV-1a (93), 1b (371) and 1c (80) subgenotypes, and all BVDV-2 isolates were grouped into BVDV-2a subgenotype (222). Further comparative analysis was performed with BVDV-1a, 1b and 2a viruses isolated from 2001 to 2014. Phylogenetic analysis based on nucleotide sequences of the viral glycoprotein E2 gene, a major target of neutralizing antibodies, revealed that BVDV-1a, 1b and 2a isolates were further classified into several clusters. Cross-neutralization tests showed that BVDV-1b isolates were antigenically different from BVDV-1a isolates, and almost BVDV-1a, 1b and 2a isolates were antigenically similar among each subgenotype and each E2 cluster. Taken together, BVDV-1b viruses are still predominant, and BVDV-2a viruses have increased recently in Hokkaido, Japan. Field isolates of BVDV-1a, 1b and 2a show genetic diversity on the E2 gene with antigenic conservation among each subgenotype during the last 14 years.
Keywords: antigenicity, bovine viral diarrhea virus, E2, genetic diversity
Bovine viral diarrhea virus (BVDV) is the causative agent of bovine viral diarrhea (BVD) [15], leading to economic losses for the livestock industry and is distributed worldwide [16, 17]. BVDVs are distinguished into two biotypes, cytopathic (cp) and noncytopathic (ncp), depending on their cytopathic effect on cell culture [15, 27]. Acute infection with BVDV in cattle results in temporary fever, respiratory symptoms and/or diarrhea, and most cattle develop an effective immune status [8, 27]. Only ncp BVDVs can cause births of persistently infected (PI) calves through infection of pregnant cattle during approximately 40–120 days of gestation [9, 26]. The PI calves shed BVDVs during their entire lifespan and may succumb to fatal mucosal disease [10]. Therefore, prompt identification and stamping out of the PI calves are necessary for the control of BVD.
BVDV belongs to the genus Pestivirus of the family Flaviviridae, together with classical swine fever virus and border disease virus [24]. It possesses a single-stranded positive-sense RNA genome of approximately 12.3 kb with one large open reading frame flanked by a 5′ and 3′ untranslated region (UTR). The open reading frame encodes approximately 4,000 amino acids that yield at least 12 cleavage products, Npro, Core, Erns, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B, through co- and post-translational processing of the polyprotein by cellular and viral proteases [22]. The 5′-UTR is a conserved region among BVDVs and used to detect viral genome for diagnosis [5]. The glycoprotein E2 is a major target of neutralizing antibodies [12, 13], and the E2 gene is used to comparative genomic analysis among pestiviruses [6]. The Npro is a unique nonstructural protein of pestiviruses and also used for comparative analysis of viral genome [6].
BVDVs are genetically divided into genotype 1 (BVDV-1) and genotype 2 (BVDV-2), recognized as separate virus species [33, 35]. HoBi-like pestivirus, tentatively designated as BVDV genotype 3 (BVDV-3), has been recognized as an atypical pestivirus, while infection with this virus results in BVD-like symptoms in cattle [4, 39]. BVDVs are further divided genetically into at least 17 subgenotypes (1a–1q) for BVDV-1 and 3 subgenotypes (2a–2c) for BVDV-2 [11, 34]. Different BVDV subgenotypes predominate in different geographic locations [36, 45]. Antigenic diversity of inter-subgenotypes has been well reported [1, 3, 28, 31, 36].
In Japan, outbreaks of BVD have been reported since 1960s, and genetic and antigenic analyses of BVDVs have been carried out [25, 28,29,30,31, 38, 40]. BVDV-1a, 1b, 1c, 1j, 1n, 1o and 2a strains were isolated, and BVDV-1b viruses were predominant from 2000 to 2006 in Japan [25]. Different subgenotype strains isolated in Japan showed antigenic differences [28, 31]. To investigate recent epidemics in Hokkaido, Japan, we genetically and antigenically analyzed BVDVs isolated from 2001 to 2014 in the present study.
MATERIALS AND METHODS
Viruses and cells: The 766 BVDV isolates were kindly provided from 14 livestock hygiene service centers in Hokkaido Prefecture, Japan, from 2006 to 2014. These viruses were isolated from serum, buffy coat, nasal discharge, pleural effusion, fecal specimen or emulsion of an organ (lung, liver, kidney, spleen, heart or brain) of cattle or aborted fetuses infected with BVDVs. Seventeen of the 766 isolates were cp BVDVs, and the other 749 isolates were ncp BVDVs. Isolates were designated as “BVDV/municipalities of isolation/isolate number/year of isolation”, for example, BVDV/Nakashibetsu/881/10 strain was isolated in 2010 at Nakashibetsu, Hokkaido, Japan. BVDVs isolated from 2001 to 2006 in Hokkaido were described in our previous report [25] and also analyzed genetically and antigenically in the present study. Nose strain (BVDV-1a), IS27CP/01 strain (BVDV-1b) and KZ-91-NCP strain (BVDV-2a) [31] were selected as reference strains. Hokudai-Lab/09 strain (BVDV-2b) was isolated from a commercial fetal bovine serum batch derived from North America. GBK_E− strain (BVDV-2a) was kindly provided from Takashi Kozasa (National Veterinary Assay Laboratory, Kokubunji, Japan) [21]. These viruses were also used as reference strains. The bovine kidney cell line MDBK-HS [20] was propagated in Eagle’s minimum essential medium (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 0.295% tryptose phosphate broth (Becton Dickinson, San Jose, CA, U.S.A.), 10 mM N,N-bis (2-hydroxyethyl)-2-aminoethanesulfonic acid (Sigma-Aldrich, St. Louis, MO, U.S.A.), 0.3 mg/ml L-glutamine (MEM-BES) and 10% horse serum (Life Technologies, Carlsbad, CA, U.S.A.). Bovine fetal muscle (BFM) cell cultures within 20 passages were propagated in MEM-BES with 5% horse serum and 5% fetal bovine serum which is free from both BVDV antigens and antibodies (Japan Bio Serum, Hiroshima, Japan). MDBK-HS cells were used for virus preparation, and BFM cells were used for cross-neutralization tests.
Virus genome sequencing and phylogenetic analysis: Viral RNA was extracted by TRIzol LS Reagent (Life Technologies) from the supernatants of MDBK-HS cells inoculated with viruses. The part of 5′-UTR genome was amplified by reverse transcription (RT) and PCR using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Life Technologies) and the primer set, 324 and 326 [44]. For amplification of the entire E2 gene, extracted RNA was reverse-transcribed with the Random Primer (N)9 (Takara Bio, Otsu, Japan) using the M-MLV Reverse Transcriptase (Life Technologies). The entire E2 gene of BVDVs was amplified using primers in Supplementary Table 1 and Ex-Taq DNA Polymerase (Takara Bio). Nucleotide sequences of PCR fragments were determined using the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies) and the 3500 Genetic Analyzer (Life Technologies) according to the manufacturer’s protocol. Sequencing data were analyzed using the GENETYX Network version 12.0.1 software (GENETYX, Tokyo, Japan). The entire E2 sequences of field isolates used for antigenic analysis were deposited in the DDBJ/EMBL/GenBank databases under accession numbers described in Supplementary Table 2. For phylogenetic analysis, multiple sequences were aligned with the CLUSTAL W algorithm using the default parameters [43]. Phylogenetic trees based on nucleotide sequences of the 5′-UTR and the E2 gene were constructed by the maximum-likelihood method and bootstrap analysis (n=1,000) using the MEGA 6.0 software with default parameters [42]. The sequence data of reference strains were obtained from the DDBJ/EMBL/GenBank databases. The sequence data of No.12 strain were kindly provided from Dr. Hiroshi Aoki (Nippon Veterinary and Life Science University, Musashino, Japan).
Statistical analysis: Pearson’s chi-square test was performed in order to estimate association between viral genotypes and incidences of clinical signs in cattle infected with BVDVs. Fisher’s exact test was performed in order to identify the categories responsible for a significant chi-square statistic. All tests were performed at the 0.05 level of significance.
Antisera: Antisera against IS27CP/01, BVDV/Nakashibetsu/881/10, BVDV/Nakasatsunai/719/09-CP, BVDV/Setana/1103/13, BVDV/Hamanaka/646/08, GBK_E−, Hokudai-Lab/09 and BVDV/Hamanaka/843/10 strains were prepared using Slc: JW/CSK rabbits (Japan SLC, Hamamatsu, Japan). These viruses were injected four times intravenously and intramuscularly with each virus suspension containing approximately 107.0 TCID50 at 3, 5 and 7 weeks after the first injection. Eight weeks later, sera were obtained and inactivated for 30 min at 56°C to be used for cross-neutralization tests. Rabbit antisera against the Nose strain (BVDV-1a) and the KZ-91-NCP strain (BVDV-2a) were also produced by four times injection and kindly provided from Dr. Hiroshi Aoki.
Cross-neutralization tests: Cross-neutralization tests were performed in BFM cell cultures grown in 96-well microplates as described previously [37]. Briefly, serial twofold dilutions of sera were prepared in a volume of 25 µl in a microplate with cell culture media, mixed with an equal volume of virus suspension containing 200 TCID50 and incubated at 37°C for 1 hr. Then, 100 µl of BFM cell suspension (approximately 2 × 104 cells) were added into all the wells of the microplate and incubated at 37°C under 5% CO2 for 5 days. The plates were air dried and fixed at 80°C for 1 hr. The fixed cells were stained using an immunoperoxidase method with anti-NS3 MAbs [20]. Each assay was performed in quadruplicate, and neutralization titers were expressed as the reciprocal of the highest dilution that inhibited 50% viral growth. Antigenic similarity (R) values were calculated according to the following formula of Archetti and Horsfall [2].
 |
RESULTS
Genotyping of the isolates based on the 5′-UTR nucleotide sequences: The 766 BVDVs were isolated from 2006 to 2014 from PI cattle in Hokkaido, Japan. Phylogenetic analysis of the 5′-UTR nucleotide sequences revealed that these isolates were classified as BVDV-1 (544 isolates) and BVDV-2 (222). BVDV-1 were further divided into BVDV-1a (93 isolates), 1b (371) and 1c (80), and all BVDV-2 isolates were grouped into BVDV-2a (222) (Table 1). Proportion of subgenotypes 1a, 1b, 1c and 2a was 12.1%, 48.4%, 10.5% and 29.0%, respectively. The sequence identity of the 5′-UTR was 72–75% between BVDV-1 and BVDV-2 isolates, and almost equal to those of the previous report [35]. The sequence identities among BVDV-1 and BVDV-2 isolates were 87–100% and 93–100%, respectively.
Table 1. Genotyping of BVDVs isolated in Hokkaido from 2006 to 2014.
| Year of isolation | No. of isolate | Genotype | |||
|---|---|---|---|---|---|
| BVDV-1 | BVDV-2 | ||||
| 1a | 1b | 1c | 2a | ||
| 2006a) | 66 | 2 | 43 | 19 | 2 |
| 2007 | 123 | 14 | 82 | 12 | 15 |
| 2008 | 85 | 17 | 40 | 4 | 24 |
| 2009 | 78 | 7 | 44 | 10 | 17 |
| 2010 | 83 | 11 | 35 | 15 | 22 |
| 2011 | 133 | 10 | 50 | 11 | 62 |
| 2012 | 102 | 17 | 42 | 4 | 39 |
| 2013 | 61 | 13 | 19 | 2 | 27 |
| 2014b) | 35 | 2 | 16 | 4 | 13 |
| Total | 766 | 93 | 371 | 80 | 222 |
a) From April to December. b) From January to July.
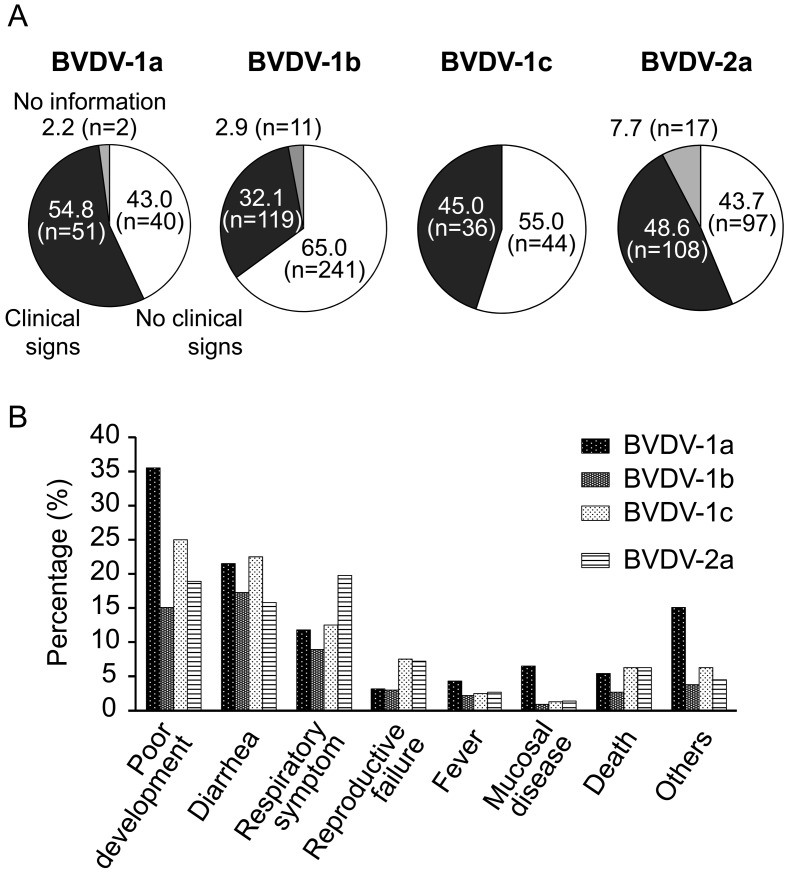
Association between viral subgenotypes and their pathogenicity in cattle: To investigate association between viral subgenotypes and their pathogenicity in cattle, the present isolates were classified according to viral subgenotypes and clinical signs of cattle. Between 43.0–65.0% of cattle infected with each subgenotype virus showed no clinical signs, and the others showed typical clinical signs of BVD except for no information (Fig. 1A). In statistical analyses, Pearson’s chi-squared test was performed in order to estimate association between viral genotypes and incidences of clinical signs in cattle infected with BVDVs. Difference of viral genotypes significantly relates to incidence of clinical signs in cattle (P<0.01). The percentages of clinical sings of BVDV-1b vs. other subgenotypes were compared with Fisher’s exact test. This analysis revealed that cattle infected with BVDV-1b viruses significantly showed less clinical signs compared with other subgenotypes (P<0.01). Clinical signs frequently observed in cattle were poor development, diarrhea, respiratory symptoms, reproductive failure, fever, mucosal disease and death (Fig. 1B).
Fig. 1.
Viral subgenotypes and clinical signs in cattle persistently infected with BVDVs. (A) The pie charts show percentage incidence of clinical signs in cattle infected with each subgenotype virus. Statistical analysis using Pearson’s chi-square test and Fisher’s exact test showed that cattle infected with BVDV-1b viruses showed significantly less clinical signs compared with other subgenotypes (P<0.01). (B) Clinical signs frequently observed in cattle infected with BVDVs of each subgenotype. “Others” include rough fur, stomatitis and neurologic symptoms. Complications of clinical signs in the same cattle were dually counted for each sign.
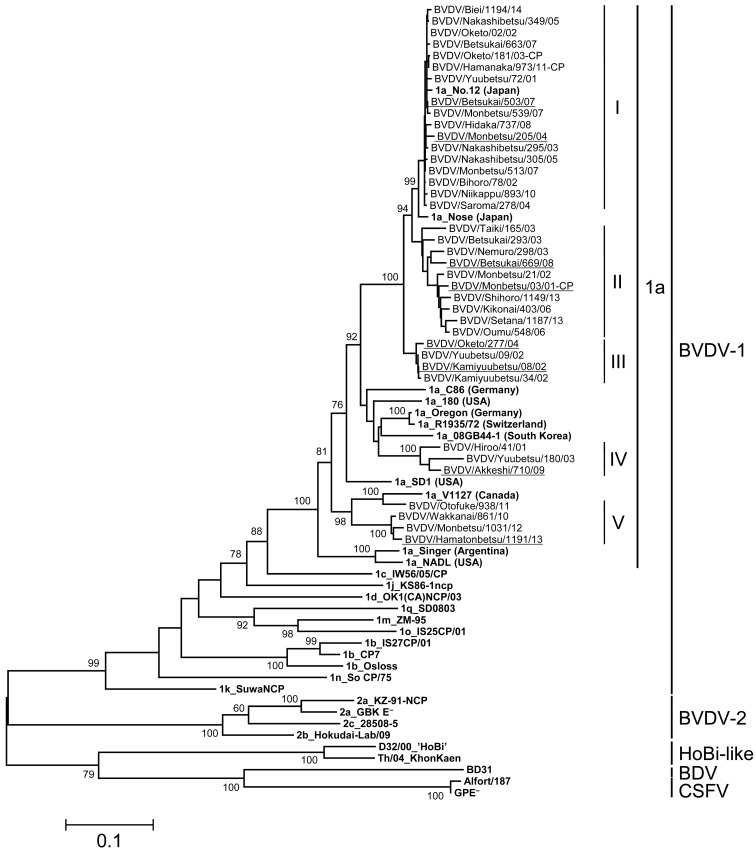
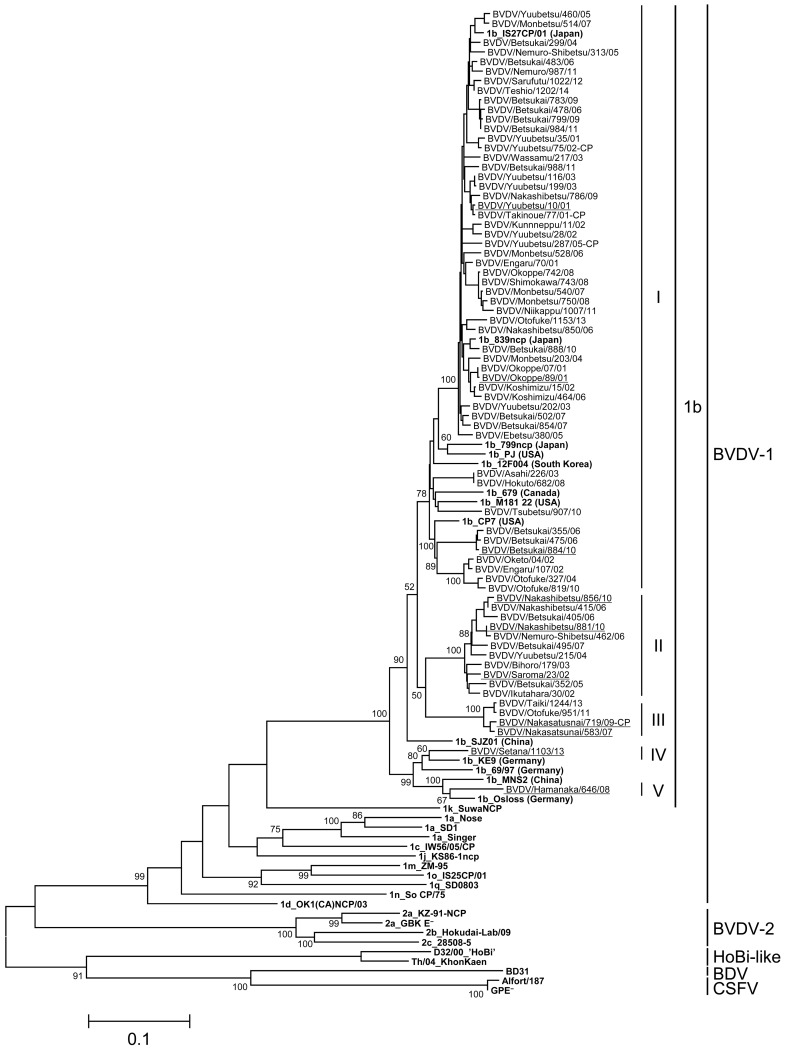
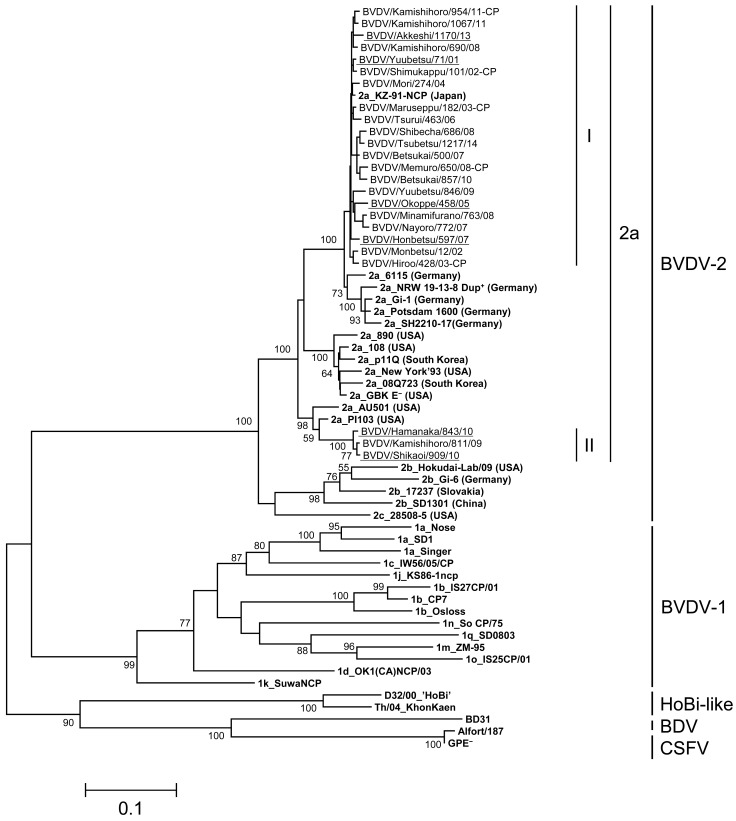
Phylogenetic analysis on the basis of nucleotide sequences of the viral glycoprotein E2 gene: The entire E2 genes of BVDV-1a, 1b and 2a isolates were analyzed for more comparative genetic investigation. Thirty-eight isolates of BVDV-1a, 70 isolates of BVDV-1b and 24 isolates of BVDV-2a from 2001 to 2014 in Hokkaido were randomly selected for phylogenetic analysis on the basis of the entire E2 array, avoiding biases by year and geographical localization of viral isolation. The BVDV-1a, 1b and 2a isolates in Hokkaido were blanched into 5, 5 and 2 clusters by phylogenetic analysis supported with bootstrap values more than 50, respectively (Figs. 2, 3, 4). On phylogenetic analysis of the E2 gene, all isolates were classified as the same subgenotypes based on the 5′-UTR. The classification of each E2 cluster of BVDV-1a and 1b was not biased by year and geographical localization of viral isolation. Only isolates of 2009 and 2010 years were classified as BVDV-2a cluster II.
Fig. 2.
Phylogenetic tree of BVDV-1a field isolates based on nucleotide sequences of the entire E2 gene. Nucleotides 1,122 bp of the entire E2 gene were used for phylogenic analysis. The phylogenetic tree of BVDV-1a field isolates was constructed using the maximum-likelihood method and bootstrap analysis (n=1,000) by the MEGA 6.0 software with 31 sequences obtained from the DDBJ/EMBL/GenBank databases and through personal communication. The 31 reference strains were described in bold. The 38 field isolates in Hokkaido were classified as 5 clusters (I–V) followed by bootstrap values of phylogenetic analysis. The 8 field isolates used for antigenic analysis were underlined.
Fig. 3.
Phylogenetic tree of BVDV-1b field isolates based on nucleotide sequences of the entire E2 gene: Nucleotides 1,122 bp of the entire E2 gene were used for phylogenic analysis. The phylogenetic tree of BVDV-1b field isolates was constructed using the maximum-likelihood method and bootstrap analysis (n=1,000) by the MEGA 6.0 software with 33 sequences obtained from the DDBJ/EMBL/GenBank databases and through personal communication. The 33 reference strains were described in bold. The 70 field isolates in Hokkaido were classified as 5 clusters (I–V) followed by bootstrap values of phylogenetic analysis. The 10 field isolates used for antigenic analysis were underlined.
Fig. 4.
Phylogenetic tree of BVDV-2a field isolates based on nucleotide sequences of the entire E2 gene: Nucleotides 1,116 bp of the entire E2 gene were used for phylogenic analysis. The phylogenetic tree of BVDV-2a field isolates was constructed using the maximum-likelihood method and bootstrap analysis (n=1,000) by the MEGA 6.0 software with 38 sequences obtained from the DDBJ/EMBL/GenBank databases and through personal communication. The 38 reference strains were described in bold. The 24 field isolates in Hokkaido were classified as 2 clusters (I–II) followed by bootstrap values of the phylogenetic analysis. The 6 field isolates used for antigenic analysis were underlined.
Antigenic analysis of BVDV-1a and 1b field isolates by cross-neutralization tests: To compare the phylogenetic E2 classification and the antigenicity, 8 isolates of BVDV-1a and 10 isolates of BVDV-1b were randomly selected from each E2 cluster for cross-neutralization tests, avoiding biases by year and geographical localization of viral isolation. Nose (BVDV-1a) and IS27CP/01 (BVDV-1b) strains were also analyzed as representative strains of each subgenotype. Rabbit antisera against BVDV-1b field isolates, BVDV/Nakashibetsu/881/10, BVDV/Nakasatsunai/719/09-CP, BVDV/Setana/1103/13 and BVDV/Hamanaka/646/08 strains were prepared to analyze antigenic property of each BVDV-1b E2 cluster. The cross-neutralization tests showed 4 to 64-fold antigenic differences between BVDV-1a and 1b isolates in Japan (Table 2). Antigenic similarity (R) values of BVDV-1a and 1b strains were 6.3 to 17.7, which suggested significant antigenic differences between BVDV-1a and 1b subgenotypes. The cross-neutralization tests of BVDV-1a field isolates showed 1 to 2-fold antigenic differences against BVDV-1a Nose strain, and this result indicated that antigenicity of BVDV-1a isolates was similar regardless of further classification by E2 array. BVDV-1b field isolates of clusters I, II, III and IV showed 8-fold or less antigenic differences, and R values were 25.0 to 50.0. These results indicated that antigenicity of these BVDV-1b isolates was almost similar among E2 clusters. The BVDV/Hamanaka/646/08 strain of BVDV-1b cluster V showed lowest neutralization titers compared with other BVDV-1b isolates, and R values compared with other E2 clusters were 12.5 to 25.0. These results indicated that the antigenicity of this isolate was slightly different from other BVDV-1b isolates.
Table 2. Cross-neutralization titers of antisera raised against different strains of BVDV-1a and 1b.
| Subgenotype | Cluster of E2 gene | Virusb) | Antisera against BVDVsa) | |||||
|---|---|---|---|---|---|---|---|---|
| 1a | 1b | |||||||
| Nose | Cluster I | Cluster II | Cluster III | Cluster IV | Cluster V | |||
| IS27CP/01 | Nakashibetsu /881/10 | Nakasatsunai /719/09-CP | Setana/1103/12 | Hamanaka /646/08 | ||||
| 1a | Cluster I | Nose | 2,048 | 64 | 64 | 32 | 128 | 64 |
| Betsukai/503/07 | 2,048 | 64 | 64 | 32 | 128 | 32 | ||
| Monbetsu/205/04 | 1,024 | 64 | 32 | 32 | 64 | 32 | ||
| Cluster II | Monbetsu/03/01-CP | 1,024 | 64 | 64 | 16 | 32 | 32 | |
| Betsukai/669/08 | 1,024 | 64 | 32 | 32 | 64 | 64 | ||
| Cluster III | Oketo/277/04 | 2,048 | 64 | 64 | 32 | 128 | 64 | |
| Kamiyuubetsu/08/02 | 2,048 | 64 | 32 | 32 | 128 | 64 | ||
| Cluster IV | Akkeshi/710/09 | 1,024 | 64 | 32 | 16 | 32 | 32 | |
| Cluster V | Hamatonbetsu/1191/13 | 1,024 | 128 | 16 | 16 | 128 | 128 | |
| 1b | Cluster I | IS27CP/01 | 256 | 512 | 256 | 256 | 128 | 128 |
| Yuubetsu/10/01 | 256 | 512 | 256 | 256 | 512 | 256 | ||
| Okoppe/89/01 | 256 | 256 | 256 | 256 | 256 | 128 | ||
| Betsukai/884/10 | 128 | 512 | 256 | 128 | 512 | 256 | ||
| Cluster II | Saroma/23/02 | 256 | 256 | 1,024 | 256 | 512 | 256 | |
| Nakashibetsu/856/10 | 256 | 512 | 1,024 | 512 | 256 | 256 | ||
| Nakashibetsu/881/10 | 128 | 512 | 1,024 | 1,024 | 256 | 128 | ||
| Cluster III | Nakasatsunai/583/07 | 256 | 128 | 256 | 512 | 256 | 256 | |
| Nakasatsunai/719/09-CP | 256 | 128 | 128 | 512 | 256 | 128 | ||
| Cluster IV | Setana/1103/12 | 256 | 128 | 256 | 256 | 512 | 64 | |
| Cluster V | Hamanaka/646/08 | 64 | 128 | 64 | 64 | 128 | 512 | |
a) Neutralization titers are expressed as the reciprocal of the highest dilution that inhibits 50% viral growth, and homologous titers are described in bold. b) “BVDV/” is omitted from the name of field isolates.
Antigenic analysis of BVDV-2a field isolates by cross-neutralization tests: The 6 isolates of BVDV-2a were also selected for antigenic analysis by cross-neutralization tests based on the same criteria. KZ-91-NCP (BVDV-2a), GBK_E− (BVDV-2a) and Hokudai-Lab/09 (BVDV-2b) strains were also analyzed as representative strains of each subgenotype. Rabbit antisera against the BVDV/Hamanaka/843/10 strain were prepared to analyze antigenic property of BVDV-2a cluster II. The cross-neutralization tests showed 2 to 16-fold antigenic differences, comparing with BVDV-2a and 2b (Table 3). The R values among subgenotypes were 12.5 to 35.4, and significant antigenic differences were not observed among BVDV-2 subgenotypes. The cross-neutralization tests of BVDV-2a field isolates showed 1 to 4-fold antigenic differences, and antigenicity of BVDV-2a field isolates was almost similar among E2 clusters.
Table 3. Cross-neutralization titers of antisera raised against different strains of BVDV-2.
| Subgenotype | Cluster of E2 gene | Virusb) | Antisera against BVDVsa) | |||
|---|---|---|---|---|---|---|
| 2a | 2b | |||||
| KZ-91-NCP | Hamanaka /843/10 | GBK_E− | Hokudai-Lab/09 | |||
| 2a | Cluster I | KZ-91-NCP | 512 | 64 | 16 | 32 |
| Yuubetsu/71/01 | 512 | 128 | 64 | 64 | ||
| Okoppe/458/05 | 256 | 128 | 32 | 64 | ||
| Honbetsu/597/07 | 256 | 64 | 16 | 64 | ||
| Akkeshi/1170/13 | 256 | 64 | 16 | 32 | ||
| Cluster II | Hamanaka/843/10 | 128 | 128 | 16 | 16 | |
| Shikaoi/909/10 | 128 | 128 | 8 | 16 | ||
| GBK_E− | 128 | 128 | 128 | 8 | ||
| 2b | Hokudai-Lab/09 | 256 | 64 | 32 | 128 | |
a) Neutralization titers are expressed as the reciprocal of the highest dilution that inhibits 50% viral growth, and homologous titers are described in bold. b) “BVDV/” is omitted from the name of field isolates.
DISCUSSION
BVDVs have been detected since 1969 in Japan, and BVD has not been eradicated as yet. Approximately 60% of dairy cattle and 20% of beef cattle in a total of Japan are raised in the Hokkaido Prefecture, and thus, it is important to control BVD in this area. The 766 BVDVs were isolated from cattle or aborted fetuses by the 14 livestock hygiene service centers of Hokkaido Prefecture from 2006 to 2014, and now, we have a total of 1,155 field isolates since 2001 [25]. To confirm a diagnosis of persistent infection with BVDVs, cattle should be retested after an interval of at least 3 weeks. In Japan, most of cattle were not retested, because of economic losses by detention of ‘viral pollutant’ in herds. These infected cattle were sporadically identified in herds, not in a group, and thus, almost of these cattle can be considered as PI cattle.
To investigate recent epidemics in Hokkaido for the control of BVD, we firstly analyzed the 766 BVDVs in the present study. Phylogenetic analysis on the basis of the 5′-UTR revealed that field isolates were divided into BVDV-1a, 1b, 1c and 2a. These subgenotype viruses have been circulating at least since 2001 in Hokkaido, Japan [25], but HoBi-like viruses (BVDV-3) were not detected from field isolates by the present phylogenetic analysis. On classification of BVDV-2 subgenotypes, some criteria were reported in the previous studies [19, 34, 41]. In the present study, the classification of BVDV-2 isolates was followed using that of Peterhans et al. [34], and all Japanese BVDV-2 strains were classified as BVDV-2a. In Japan, no epidemic has been reported, that was caused by highly pathogenic BVDV-2 viruses, such as the 890 strain isolated in North America [7] or the NRW 19-13-8 strain recently isolated in Germany [19]. Taken together, BVDV-1b viruses are still predominant, and BVDV-2a has increased since 2011 in Hokkaido, Japan. Interestingly, statistical analysis showed that BVDV-1b isolates in Hokkaido tended to result in less clinical signs in cattle. This result may indicate that the predominance of BVDV-1b is due to difficulty of diagnosis by clinical signs.
Inactivated vaccines containing BVDV-1a and 2a antigens and live-attenuated vaccines containing BVDV-1a antigen had been used widely in Japan until 2014, whereas BVDV-1b and 2a viruses were predominant recently in Hokkaido. Therefore, we analyzed nucleotide sequences of the E2 of BVDV-1a, 1b and 2a field isolates from 2001 to 2014, since the E2 is a main target of immune responses in hosts and important for viral antigenicity [12, 13]. These field isolates were further classified, supported with bootstrap values in the phylogenetic trees. Regardless of the classification of E2 clusters, all BVDV-1a and 2a field isolates were antigenically similar. Antigenicity of BVDV-1b isolates was almost similar among E2 clusters, while only one isolate of BVDV-1b cluster V, BVDV/Hamanaka/646/08 strain, showed antigenic difference. The BVDV/Hamanaka/646/08 strain was blanched into the most distant cluster from other BVDV-1b clusters and showed the lowest identity of the E2 gene (84–88%) with other BVDV-1b isolates. These results indicate that genetic differences on the E2 gene correlate with differences in viral antigenicity, and thus, phylogenetic analysis on the E2 array improves understanding of viral antigenicity in detail. Additionally, the isolates of BVDV-1b clusters IV and V were blanched into the same cluster with Osloss strain, which was classified as ‘BVDV-1b1’ in the previous report [41]. Almost Japanese BVDV-1b isolates were closely related with CP7 strain of ‘BVDV-1b2’, and it is the first report of isolation of BVDV-1b1 viruses in Japan. BVDV/Setana/1103/13 strain of BVDV-1b cluster IV was antigenically similar with BVDV-1b2 viruses, and antigenicity could not be clearly distinguished between BVDV-1b1 and 1b2 viruses. Taken together, it has been confirmed that the BVDV-1a, 1b and 2a isolates are likely to be antigenically conserved among each subgenotype during the last 14 years in Hokkaido, Japan.
Antigenic diversity among subgenotypes of BVDV-1 has been well demonstrated [1, 3, 28, 31, 36]. Japanese BVDV-1a and 1b viruses showed significant antigenic differences in the previous studies [28, 31], and the present study also showed antigenic differences between BVDV-1a and 1b isolates in Hokkaido, supported by R values (6.3 to 17.7) lower than 25.0 [2]. In contrast, BVDV-1a and 1b isolates in other countries, such as Switzerland, U.S.A. and Argentina, showed higher R values (24.2 to 50.0) [1, 3, 36]. In such countries, the several reports also showed that BVDV-1a vaccines can induce only lower antibody titers against BVDV-1b viruses [14, 32, 36], and some authors discussed that this antigenic difference may lead to predominance of BVDV-1b viruses over the other subgenotypes [14, 36]. In Japan, there are no complete data of the frequency of vaccination in cattle, and thus, it is still unclear whether the predominant situation of BVDV-1b viruses was due to BVDV-1a vaccine pressure or not. Recently, a commercial inactivated vaccine containing BVDV-1b and 2a strains and a live attenuated vaccine containing BVDV-1a and 2a strains have become available in Japan, and further studies are needed to consider the epidemiological impact of these vaccines on the predominant BVDVs.
In summary, we revealed that BVDV-1b and 2a viruses were recently predominant in Hokkaido, Japan. BVDV-1a, 1b and 2a isolates show genetic diversity on the E2 gene with antigenic conservation among each subgenotype during the last 14 years. Based on the present study, it is required to use effective vaccine against predominant field isolates for the control of BVD in Japan. Vaccination as a stand-alone control measure is not adequate in preventing viral circulation [18, 23], and thus, it is important to both stamp out PI cattle and to use effective vaccines for reduction of PI cattle to control BVD.
Supplementary Material
Acknowledgments
We are grateful to the 14 livestock hygiene service centers of Hokkaido Prefecture for providing BVDVs. We thank Dr. Takashi Kozasa (National Veterinary Assay Laboratory, Japan) and Dr. Hiroshi Aoki (Nippon Veterinary and Life Science University, Musashino, Japan) for providing BVDVs, sequence data of viral genome and antisera. We also thank Drs. Keita Matsuno and Norikazu Isoda for an excellent support on this research.
REFERENCES
- 1.Alpay G., Yeşilbağ K.2015. Serological relationships among subgroups in bovine viral diarrhea virus genotype 1 (BVDV-1). Vet. Microbiol. 175: 1–6. doi: 10.1016/j.vetmic.2014.10.034 [DOI] [PubMed] [Google Scholar]
- 2.Archetti I., Horsfall F. L.1950. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 92: 441–462. doi: 10.1084/jem.92.5.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachofen C., Stalder H., Braun U., Hilbe M., Ehrensperger F., Peterhans E.2008. Co-existence of genetically and antigenically diverse bovine viral diarrhoea viruses in an endemic situation. Vet. Microbiol. 131: 93–102. doi: 10.1016/j.vetmic.2008.02.023 [DOI] [PubMed] [Google Scholar]
- 4.Bauermann F. V., Ridpath J. F., Weiblen R., Flores E. F.2013. HoBi-like viruses: an emerging group of pestiviruses. J. Vet. Diagn. Invest. 25: 6–15. doi: 10.1177/1040638712473103 [DOI] [PubMed] [Google Scholar]
- 5.Becher P., Orlich M., Shannon A. D., Horner G., König M., Thiel H. J.1997. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 78: 1357–1366. doi: 10.1099/0022-1317-78-6-1357 [DOI] [PubMed] [Google Scholar]
- 6.Becher P., Orlich M., Kosmidou A., König M., Baroth M., Thiel H. J.1999. Genetic diversity of pestiviruses: identification of novel groups and implications for classification. Virology 262: 64–71. doi: 10.1006/viro.1999.9872 [DOI] [PubMed] [Google Scholar]
- 7.Bolin S. R., Ridpath J. F.1992. Differences in virulence between two noncytopathic bovine viral diarrhea viruses in calves. Am. J. Vet. Res. 53: 2157–2163. [PubMed] [Google Scholar]
- 8.Brownlie J., Clarke M. C., Howard C. J.1984. Experimental production of fatal mucosal disease in cattle. Vet. Rec. 114: 535–536. doi: 10.1136/vr.114.22.535 [DOI] [PubMed] [Google Scholar]
- 9.Brownlie J., Clarke M. C., Howard C. J.1989. Experimental infection of cattle in early pregnancy with a cytopathic strain of bovine virus diarrhoea virus. Res. Vet. Sci. 46: 307–311. [PubMed] [Google Scholar]
- 10.Brownlie J.1991. The pathways for bovine virus diarrhoea virus biotypes in the pathogenesis of disease. Arch. Virol. Suppl. 3: 79–96. doi: 10.1007/978-3-7091-9153-8_10 [DOI] [PubMed] [Google Scholar]
- 11.Deng Y., Sun C. Q., Cao S. J., Lin T., Yuan S. S., Zhang H. B., Zhai S. L., Huang L., Shan T. L., Zheng H., Wen X. T., Tong G. Z.2012. High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet. Microbiol. 159: 490–493. doi: 10.1016/j.vetmic.2012.04.023 [DOI] [PubMed] [Google Scholar]
- 12.Deregt D., Bolin S. R., van den Hurk J., Ridpath J. F., Gilbert S. A.1998. Mapping of a type 1-specific and a type-common epitope on the E2 (gp53) protein of bovine viral diarrhea virus with neutralization escape mutants. Virus Res. 53: 81–90. doi: 10.1016/S0168-1702(97)00129-9 [DOI] [PubMed] [Google Scholar]
- 13.Donis R. O.1995. Molecular biology of bovine viral diarrhea virus and its interactions with the host. Vet. Clin. North Am. Food Anim. Pract. 11: 393–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulton R. W., Ridpath J. F., Confer A. W., Saliki J. T., Burge L. J., Payton M. E.2003. Bovine viral diarrhoea virus antigenic diversity: impact on disease and vaccination programmes. Biologicals 31: 89–95. doi: 10.1016/S1045-1056(03)00021-6 [DOI] [PubMed] [Google Scholar]
- 15.Gillespie J. H., Baker J. A., McEntee K.1960. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 50: 73–79. [PubMed] [Google Scholar]
- 16.Hessman B. E., Fulton R. W., Sjeklocha D. B., Murphy T. A., Ridpath J. F., Payton M. E.2009. Evaluation of economic effects and the health and performance of the general cattle population after exposure to cattle persistently infected with bovine viral diarrhea virus in a starter feedlot. Am. J. Vet. Res. 70: 73–85. doi: 10.2460/ajvr.70.1.73 [DOI] [PubMed] [Google Scholar]
- 17.Houe H.1999. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet. Microbiol. 64: 89–107. doi: 10.1016/S0378-1135(98)00262-4 [DOI] [PubMed] [Google Scholar]
- 18.Houe H., Lindberg A., Moennig V.2006. Test strategies in bovine viral diarrhea virus control and eradication campaigns in Europe. J. Vet. Diagn. Invest. 18: 427–436. doi: 10.1177/104063870601800501 [DOI] [PubMed] [Google Scholar]
- 19.Jenckel M., Höper D., Schirrmeier H., Reimann I., Goller K. V., Hoffmann B., Beer M.2014. Mixed triple: allied viruses in unique recent isolates of highly virulent type 2 bovine viral diarrhea virus detected by deep sequencing. J. Virol. 88: 6983–6992. doi: 10.1128/JVI.00620-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kameyama K., Sakoda Y., Tamai K., Igarashi H., Tajima M., Mochizuki T., Namba Y., Kida H.2006. Development of an immunochromatographic test kit for rapid detection of bovine viral diarrhea virus antigen. J. Virol. Methods 138: 140–146. doi: 10.1016/j.jviromet.2006.08.005 [DOI] [PubMed] [Google Scholar]
- 21.Kozasa T., Abe Y., Mitsuhashi K., Tamura T., Aoki H., Ishimaru M., Nakamura S., Okamatsu M., Kida H., Sakoda Y.2015. Analysis of a pair of END+ and END− viruses derived from the same bovine viral diarrhea virus stock reveals the amino acid determinants in Npro responsible for inhibition of type I interferon production. J. Vet. Med. Sci. 77: 511–518. doi: 10.1292/jvms.14-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamp B., Riedel C., Roman-Sosa G., Heimann M., Jacobi S., Becher P., Thiel H. J., Rümenapf T.2011. Biosynthesis of classical swine fever virus nonstructural proteins. J. Virol. 85: 3607–3620. doi: 10.1128/JVI.02206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg A., Houe H.2005. Characteristics in the epidemiology of bovine viral diarrhea virus (BVDV) of relevance to control. Prev. Vet. Med. 72: 55–73. doi: 10.1016/j.prevetmed.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach B. D., Murray C. L., Thiel H. J., Rice C. M.2013. Flaviviridae. Fields Virology, 6th ed. 1: 712–794. [Google Scholar]
- 25.Matsuno K., Sakoda Y., Kameyama K., Tamai K., Ito A., Kida H.2007. Genetic and pathobiological characterization of bovine viral diarrhea viruses recently isolated from cattle in Japan. J. Vet. Med. Sci. 69: 515–520. doi: 10.1292/jvms.69.515 [DOI] [PubMed] [Google Scholar]
- 26.McClurkin A. W., Littledike E. T., Cutlip R. C., Frank G. H., Coria M. F., Bolin S. R.1984. Production of cattle immunotolerant to bovine viral diarrhea virus. Can. J. Comp. Med. 48: 156–161. [PMC free article] [PubMed] [Google Scholar]
- 27.McClurkin A. W., Bolin S. R., Coria M. F.1985. Isolation of cytopathic and noncytopathic bovine viral diarrhea virus from the spleen of cattle acutely and chronically affected with bovine viral diarrhea. J. Am. Vet. Med. Assoc. 186: 568–569. [PubMed] [Google Scholar]
- 28.Minami F., Nagai M., Ito M., Matsuda T., Takai H., Jinkawa Y., Shimano T., Hayashi M., Seki Y., Sakoda Y., Sugiura K., Akashi H.2011. Reactivity and prevalence of neutralizing antibodies against Japanese strains of bovine viral diarrhea virus subgenotypes. Comp. Immunol. Microbiol. Infect. Dis. 34: 35–39. doi: 10.1016/j.cimid.2009.10.007 [DOI] [PubMed] [Google Scholar]
- 29.Nagai M., Sato M., Nagano H., Pang H., Kong X., Murakami T., Ozawa T., Akashi H.1998. Nucleotide sequence homology to bovine viral diarrhea virus 2 (BVDV 2) in the 5′ untranslated region of BVDVs from cattle with mucosal disease or persistent infection in Japan. Vet. Microbiol. 60: 271–276. doi: 10.1016/S0378-1135(98)00158-8 [DOI] [PubMed] [Google Scholar]
- 30.Nagai M., Ito T., Sugita S., Genno A., Takeuchi K., Ozawa T., Sakoda Y., Nishimori T., Takamura K., Akashi H.2001. Genomic and serological diversity of bovine viral diarrhea virus in Japan. Arch. Virol. 146: 685–696. doi: 10.1007/s007050170139 [DOI] [PubMed] [Google Scholar]
- 31.Nagai M., Hayashi M., Itou M., Fukutomi T., Akashi H., Kida H., Sakoda Y.2008. Identification of new genetic subtypes of bovine viral diarrhea virus genotype 1 isolated in Japan. Virus Genes 36: 135–139. doi: 10.1007/s11262-007-0190-0 [DOI] [PubMed] [Google Scholar]
- 32.Pecora A., Malacari D. A., Ridpath J. F., Perez Aguirreburualde M. S., Combessies G., Odeón A. C., Romera S. A., Golemba M. D., Wigdorovitz A.2014. First finding of genetic and antigenic diversity in 1b-BVDV isolates from Argentina. Res. Vet. Sci. 96: 204–212. doi: 10.1016/j.rvsc.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 33.Pellerin C., van den Hurk J., Lecomte J., Tijssen P.1994. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology 203: 260–268. doi: 10.1006/viro.1994.1483 [DOI] [PubMed] [Google Scholar]
- 34.Peterhans E., Bachofen C., Stalder H., Schweizer M.2010. Cytopathic bovine viral diarrhea viruses (BVDV): emerging pestiviruses doomed to extinction. Vet. Res. 41: 44. doi: 10.1051/vetres/2010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridpath J. F., Bolin S. R., Dubovi E. J.1994. Segregation of bovine viral diarrhea virus into genotypes. Virology 205: 66–74. doi: 10.1006/viro.1994.1620 [DOI] [PubMed] [Google Scholar]
- 36.Ridpath J. F., Fulton R. W., Kirkland P. D., Neill J. D.2010. Prevalence and antigenic differences observed between Bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the southwestern United States. J. Vet. Diagn. Invest. 22: 184–191. doi: 10.1177/104063871002200203 [DOI] [PubMed] [Google Scholar]
- 37.Sakoda Y., Hikawa M., Tamura T., Fukusho A.1998. Establishment of a serum-free culture cell line, CPK-NS, which is useful for assays of classical swine fever virus. J. Virol. Methods 75: 59–68. doi: 10.1016/S0166-0934(98)00098-6 [DOI] [PubMed] [Google Scholar]
- 38.Sakoda Y., Ozawa S., Damrongwatanapokin S., Sato M., Ishikawa K., Fukusho A.1999. Genetic heterogeneity of porcine and ruminant pestiviruses mainly isolated in Japan. Vet. Microbiol. 65: 75–86. doi: 10.1016/S0378-1135(98)00284-3 [DOI] [PubMed] [Google Scholar]
- 39.Schirrmeier H., Strebelow G., Depner K., Hoffmann B., Beer M.2004. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J. Gen. Virol. 85: 3647–3652. doi: 10.1099/vir.0.80238-0 [DOI] [PubMed] [Google Scholar]
- 40.Shimizu M., Satou K.1987. Frequency of persistent infection of cattle with bovine viral diarrhea-mucosal disease virus in epidemic areas. Jpn. J. Vet. Sci. 49: 1045–1051. doi: 10.1292/jvms1939.49.1045 [DOI] [PubMed] [Google Scholar]
- 41.Tajima M., Frey H. R., Yamato O., Maede Y., Moennig V., Scholz H., Greiser-Wilke I.2001. Prevalence of genotypes 1 and 2 of bovine viral diarrhea virus in Lower Saxony, Germany. Virus Res. 76: 31–42. doi: 10.1016/S0168-1702(01)00244-1 [DOI] [PubMed] [Google Scholar]
- 42.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilcek S., Herring A. J., Herring J. A., Nettleton P. F., Lowings J. P., Paton D. J.1994. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 136: 309–323. doi: 10.1007/BF01321060 [DOI] [PubMed] [Google Scholar]
- 45.Vilcek S., Durkovic B., Kolesarova M., Paton D. J.2005. Genetic diversity of BVDV: consequences for classification and molecular epidemiology. Prev. Vet. Med. 72: 31–35. doi: 10.1016/j.prevetmed.2005.08.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.