Abstract
Feline morbillivirus (FmoPV) is an emerging virus in domestic cats and considered to be one of the causes of chronic renal failure in cats. In this study, we established a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of FmoPV. The results indicated that the detection limit of the assay was 10 50% tissue culture infective dose (TCID50)/ml in the original sample, and sensitivity of the assay was calculated as 0.12 TCID50 per one RT-LAMP reaction. We also detected FmoPV in clinical urine samples from cats infected with FmoPV. The FmoPV RT-LAMP assay is rapid, simple and highly specific for the detection of FmoPV, and thus, it would be a reliable detection method for FmoPV.
Keywords: detection, feline, feline morbillivirus, reverse transcription loop-mediated isothermal amplification
Many pathogenic morbilliviruses, such as measles virus, canine distemper virus (CDV) and rinderpest virus, are present in various mammals including humans [5]. Feline morbillivirus (FmoPV) is a new member of morbillivirus family, which was first identified in domestic cats in Hong Kong in 2012 [11]. This emerging virus is considered to be associated with tubulointerstitial nephritis [11], most common diagnosis of chronic renal failure in cats [1]. FmoPV RNAs were detected in 12% of the stray cats in Hong Kong and mainland China [11]. Subsequently, the existence of FmoPV infection was also reported in Japan [2, 8, 10], and FmoPV RNAs were detected in 13.7% of the cat urine specimens [8] and 40% of the kidney tissues of cats with nephritis [2]. Chronic renal failure is commonly seen in cats over 7 years old [1, 6]; therefore, FmoPV infection represents an important disease for cats, if the virus causes tubulointerstitial nephritis. Because of the absence of effective vaccines or antiviral drugs, rapid laboratory diagnostic techniques are essential to reduce the risk of FmoPV transmission from FmoPV-infected cats to uninfected cats.
Laboratory diagnostic techniques including virus isolation, anti-FmoPV antibody test by immunoblot analysis and viral genome detection by reverse transcription polymerase chain reaction (RT-PCR) have been used for detection of FmoPV [2, 8, 10, 11]. In general, virus isolation is a gold standard and most reliable method for virus detection. However, virus isolation from clinical specimens requires multiple passages, and cytopathic effect induced by FmoPV is relatively small and unclear [3]. Moreover, the virus isolation rates from FmoPV-infected cats are still unknown at present. Antibody test is a rapid and suitable diagnostic method in laboratory, but we found that there are potential cross-reactions between FmoPV and CDV, and the humoral immune responses against FmoPV in cats are weak in immunoblot analysis [10]. Therefore, at the moment, RT-PCR test is the most reliable detection method in diagnosis of FmoPV infection. However, RT-PCR test requires multiple steps of amplification and the use of thermal cyclers. Thus, an alternative method is required for practical diagnosis of FmoPV.
Loop-mediated isothermal amplification (LAMP) is an innovative technique for the amplification of nucleic acids [7]. LAMP has advantages over other techniques in its rapid detection and high specificity [7]. The principle is based on strand displacement by Bst DNA polymerase and formation of stem-loop structure with four specific primers in six regions. The reaction is conducted under isothermal conditions at 60–65°C, without the need for expensive and complex equipments, such as thermal cyclers [7]. This simplicity of the methods makes the LAMP assay suitable for use in clinic or in field studies. With these advantages, LAMP has been widely applied to detect various pathogenic organisms [4, 9]. In this study, we aimed to establish a rapid, simple and highly specific assay for the diagnosis of FmoPV with a one-step RT-LAMP method.
Crandell-Rees feline kidney (CRFK) cells (ATCC, CCL-94) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, penicillin (100 IU/ml) and streptomycin (100 µg/ml) (Invitrogen, Carlsbad, CA, U.S.A.) at 37°C in 5% CO2. To prepare stock virus of FmoPV, we inoculated FmoPV strain SS1 [10] into CRFK cells. Two weeks after inoculation, culture supernatants were harvested, filtered through a 450-nm membrane filter (PALL, Port Washington, NY, U.S.A.) and stored at −80°C as stock viruses. The infectious titer of the stock virus was determined as described previously [3] and expressed as the 50% tissue culture infective dose (TCID50). The titer of the stock virus of FmoPV strain SS1 was 1.0 × 106 TCID50/ml. Clinical urine samples were obtained from household cats from which FmoPV strains, SS1, SS2 and SS3, were isolated [10].
RNAs from cell lysates were extracted using RNeasy Mini Kit (Qiagen, Venlo, Limburg, the Netherlands) according to the manufacturer’s instructions. Viral RNAs from the 10-fold serially diluted stock virus or clinical samples were extracted using QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instruction.
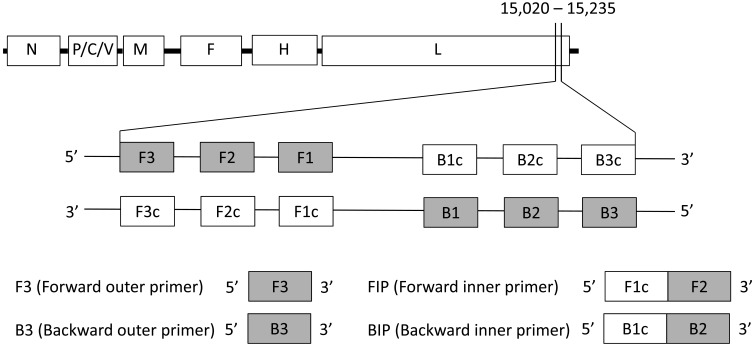
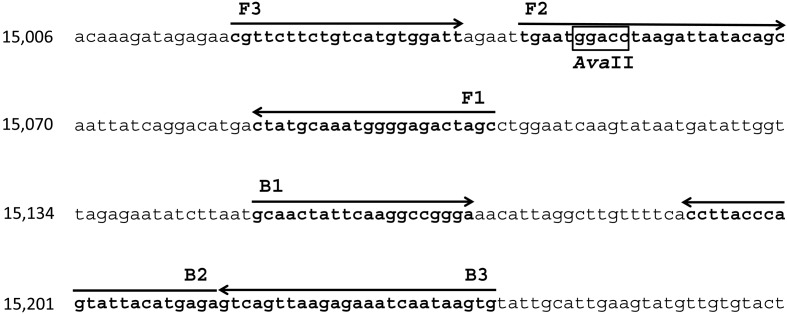
RT-LAMP primers were designed to target the L gene, a highly conserved region among FmoPV strains [10]. Four primers, including two outer primers (F3 and B3) and two inner primers (FIP and BIP), were generated using PrimerExplorer V4 software from Eiken Chemical Co., Ltd., Tokyo, Japan (http://primerexplorer.jp/e/). These primers recognize six distinct regions in the target sequence, consisting of F1, F2 and F3 regions at the 5′ end and B1c, B2c and B3c at the 3′ end (Fig. 1). The primer sequences and nucleotide positions are shown in Table 1 and Fig. 2. The nucleotide sequences of FmoPV corresponding to these primers are identical among FmoPV strains, SS1, SS2 and SS3 (Fig. 2).
Fig. 1.
Primer design of the FmoPV RT-LAMP assay. A part of L gene (nucleotide position 15,020-15,235 based on the FmoPV strain SS1 [GenBank Accession no. AB910309.1]), which is a highly conserved region among FmoPV isolates, was used for the target region of the RT-LAMP. F1c, F2c, F3c, B1c, B2c and B3c are complementary to F1, F2, F3, B1, B2 and B3, respectively. FIP consists of the region complementary to F1c linked with F2 region. BIP consists of B1c region linked with the region complementary to B2.
Table 1. List of RT-LAMP primers used in this study.
| Primer | Sequence (5′-3′) | Genome position |
|---|---|---|
| F3 | CGTTCTTCTGTCATGTGGATT | 15,020–15,040 |
| B3 | CACTTATTGATTTCTCTTAACTGAC | 15,211–15,235 |
| FIP (F1c+F2) | GCTAGTCTCCCCATTTGCATAG | 15,086–15,107; |
| TGAATGGACCTAAGATTATACAGC | 15,046–15,069 | |
| BIP (B1c+B2) | GCAACTATTCAAGGCCGGGA | 15,150–15,169; |
| TCTCATGTAATACTGGGTAAGG | 15,189–15,210 |
Fig. 2.
Nucleotide sequence of the target gene for the FmoPV RT-LAMP assay. The primer binding sites are identical among FmoPV strains, SS1, SS2 and SS3 [GenBank Accession nos. AB910309.1; LC036586.1; and LC036587.1]. The arrows show the six distinct regions in the target sequence and the direction of DNA synthesis. F3 and B3 regions at the 5′ end were used as outer primers. AvaII restriction site is boxed.
The RT-LAMP reaction was carried out with a Loopamp RNA amplification kit (Eiken Chemical Co., Ltd.), following the manufacturer’s instructions. A total of 25 µl reaction mixture containing 40 pmol each of primers FIP and BIP, 5 pmol each of the outer primers F3 and B3, 12.5 µl of 2 × reaction mix, 1 µl of enzyme mix and 5 µl of sample RNA was incubated at 63°C for 60 min, and then inactivated the enzyme and terminated the reaction by heating at 95°C for 2 min. After the amplification reaction, the RT-LAMP products were analyzed on a 1.5% agarose gel stained with ethidium bromide. The specificity of the RT-LAMP products was confirmed with a restriction enzyme digestion [7]. After ethanol precipitation of the amplified RT-LAMP products, precipitates were dissolved in distilled water and digested with AvaII at 37°C for 2 hr and subjected to electrophoresis on a 1.5% agarose gel.
RT-PCR was performed using the specific primers (5′-GGAACATGGCCTCCTGTAGA-3′ and 5′-CTCCATTGGCAATCAGGTTT-3′) targeting a 487 base pair fragment of the L gene of FmoPV [2]. Five µl of viral RNA was reverse-transcribed using Verso cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, U.S.A.) with random primers, followed by PCR amplification of cDNA. The PCR mixture consisted of 1 µl of cDNA, PCR buffer (10 nmol/l Tris-HCl, pH 8.3, 50 mmol/l KCl, 3 mmol/l MgCl2 and 0.01% gelatine), 200 µM of each dNTP and 1.0 unit of ExTaq polymerase (TaKaRa, Otsu, Japan). Thermal-cycling condition for PCR was as follows: 35 cycles of 94°C for 30 sec, 48°C for 30 sec and 72°C for 30 sec.
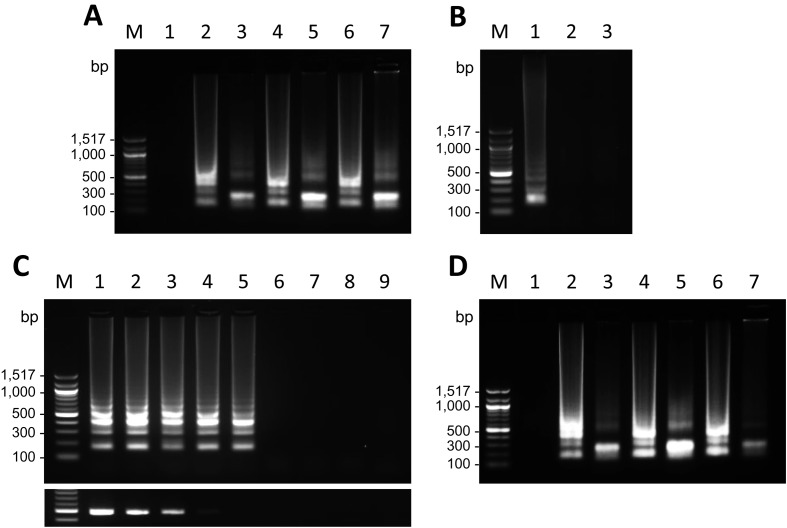
Firstly, FmoPV RNAs extracted from lysates of CRFK cells infected with FmoPV strains, SS1, SS2 and SS3, were used to evaluate the RT-LAMP assay. In all specimens, the amplification by RT-LAMP showed a ladder-like pattern on agarose gel electrophoresis (Fig. 3A), which is consistent with the presence of various sized DNA fragments consisting of alternately inverted repeats of the target sequence on the same strand. The exact nucleotide sequence of the RT-LAMP products is predicted from the sequence of the target DNA and the primers. Therefore, it is possible to predict the size of the digested products with a restriction enzyme that cleaves the DNA fragments in specific recognition sites [7]. To confirm that the products were amplified from the target sequence, the RT-LAMP products were digested with AvaII whose cleavage site is present in the target sequence (Fig. 2). By digestion with AvaII, a band sized approximately 200 bp became evident on agarose gel electrophoresis in each sample (Fig. 3A), indicating that the ladder-like fragments were RT-LAMP products amplified from the targeted FmoPV L gene.
Fig. 3.
Specificity and sensitivity of the RT-LAMP products and clinical application. (A) Specificity of the FmoPV RT-LAMP assay. RT-LAMP products amplified from RNAs extracted from CRFK infected with FmoPV strains, SS1, SS2 and SS3 (lanes 2, 4 and 6, respectively), and RT-LAMP products digested with AvaII (lanes 3, 5 and 7, respectively) are shown. An RT-LAMP product amplified from RNA extracted from uninfected CRFK cells is shown as a negative control (lane 1). Lane M: 100 bp DNA ladder. (B) Potential cross-reactions of the FmoPV RT-LAMP assay with CDV. RT-LAMP products amplified from RNA extracted from CRFK cells infected with FmoPV strain SS1 and uninfected Vero cells are shown as positive (lane 1) and negative (lane 2) controls, respectively. An RT-LAMP product amplified from RNA extracted from Vero cells infected with CDV strain Snyder Hill is shown in lane 3. Lane M: 100 bp DNA ladder. (C) Sensitivity comparison of the RT-LAMP assay with conventional RT-PCR. RT-LAMP (upper panel) and RT-PCR products (lower panel) from 10-fold serial dilutions of stock virus of FmoPV strain SS1 were electrophoresed. Amplicons from 10−1(lane 1), 10−2(lane 2), 10−3(lane 3), 10−4(lane 4), 10−5(lane 5), 10−6(lane 6), 10−7(lane 7) and 10−8 dilutions (lane 8) are shown. RT-LAMP and RT-PCR products amplified from RNA extracted from uninfected CRFK cells are shown as a negative control (lane 9). Lane M: 100 bp DNA ladder. (D) Detection of FmoPV RNAs in clinical urine samples. RT-LAMP products amplified from RNAs extracted from urine samples of cats infected with FmoPV strains, SS1, SS2 and SS3 (lanes 2, 4 and 6, respectively), and the RT-LAMP products digested with AvaII (lanes 3, 5 and 7, respectively) are shown. An RT-LAMP product amplified from RNA extracted from an uninfected cat is shown as a negative control (lane 1). Lane M: 100 bp DNA ladder.
Next, the potential cross-reactions of the RT-LAMP assay for FmoPV were examined with CDV, the closely related morbillivirus species in dogs and large felids [2]. The RT-LAMP assay was performed using RNA extracted from lysates of Vero cells (ATCC, CCL-81) infected with CDV strain Snyder Hill (Gene Accession no. JN896987.1). As shown in Fig. 3B, CDV was found to be negative by the FmoPV RT-LAMP assay, indicating that there is no cross-reactive amplification with CDV.
To evaluate the sensitivity of the FmoPV RT-LAMP assay, both RT-LAMP and RT-PCR were carried out, and their detection limits were compared using 10-fold serial dilutions of stock virus of FmoPV strain SS1. Viral RNAs were extracted from 140 µl per dilution, and 5 µl of each RNA extract was then subjected to RT-LAMP and RT-PCR. The results showed that the RT-LAMP assay can detect FmoPV RNA up to 10−5 dilution, which is approximately 100-time more sensitive than conventional RT-PCR (Fig. 3C).
Finally, we tried to detect FmoPV RNAs in urine samples of cats infected with FmoPV strains, SS1, SS2 and SS3 [10]. Viral RNAs were extracted from urine of the cats and then subjected to the FmoPV RT-LAMP assay. As a result, we successfully detected FmoPV RNA in all urine samples (Fig. 3D). The specificity of RT-LAMP products was also confirmed by AvaII digestion (Fig. 3D).
In this study, we developed an RT-LAMP assay specific for FmoPV using primers targeting the L gene of FmoPV. The RT-LAMP assay produced the characteristic ladder-like patterns in agarose gel electrophoresis, and the specificity of the products was confirmed by AvaII digestion (Fig. 3A). Furthermore, the RT-LAMP assay did not show any cross-reactivity between FmoPV and CDV (Fig. 3B). The use of four primers recognizing six regions of target sequence gave a high degree of specificity for the RT-LAMP assay.
The results demonstrated that the RT-LAMP assay has 100-fold higher sensitivity compared to conventional RT-PCR. The detection limit of the assay was 10 TCID50/ml in the original sample (Fig. 3C). Sensitivity of the FmoPV RT-LAMP assay was calculated as 0.12 TCID50 of FmoPV, because we used 5 µl out of 60 µl RNA solution extracted from 140 µl of the stock virus in one reaction of FmoPV RT-LAMP. These results indicate that sensitivity of the established RT-LAMP assay is sufficiently high to detect low concentrations of FmoPV.
The RT-LAMP assay was also efficient in specifically detecting FmoPV RNA from all three urine samples of infected cats (Fig. 3D). This finding reveals the potential clinical feasibility of the RT-LAMP assay as a useful diagnostic tool to detect FmoPV.
The RT-LAMP assay is simple and rapid compared to conventional RT-PCR; reverse transcription reaction and DNA amplification proceed in a single step as the reaction mixture is incubated at 63°C for 1 hr. Detection of RT-LAMP requires only water baths or heat blocks to keep a constant temperature [7].
A critical factor for the RT-LAMP assay is to design specific primers in a highly conserved region. In the present study, 3 sequences of FmoPV strains, SS1, SS2 and SS3, were aligned, and a highly conserved region of the L gene was chosen as a target sequence (Fig. 1). Because FmoPV has genetic diversity [8, 10], for the practical use of the FmoPV RT-LAMP assay in diagnosis, it is essential to accumulate the genetic data of FmoPV isolates.
In conclusion, the RT-LAMP assay was successfully established for the rapid detection of FmoPV RNA in clinical specimens. The assay was shown to be highly specific and sensitive for the detection of FmoPV. Although further studies are necessary for the application, the RT-LAMP assay reported here has great potential for diagnosis of FmoPV RNAs in clinic or in field studies.
Acknowledgments
Shoichi Sakaguchi was supported by a fellowship of the Japan Society for the Promotion of Science.
REFERENCES
- 1.DiBartola S. P., Rutgers H. C., Zack P. M., Tarr M. J.1987. Clinicopathologic findings associated with chronic renal disease in cats: 74 cases (1973–1984). J. Am. Vet. Med. Assoc. 190: 1196–1202. [PubMed] [Google Scholar]
- 2.Furuya T., Sassa Y., Omatsu T., Nagai M., Fukushima R., Shibutani M., Yamaguchi T., Uematsu Y., Shirota K., Mizutani T.2014. Existence of feline morbillivirus infection in Japanese cat populations. Arch. Virol. 159: 371–373. doi: 10.1007/s00705-013-1813-5 [DOI] [PubMed] [Google Scholar]
- 3.Koide R., Sakaguchi S., Miyazawa T.2015. Basic biological characterization of feline morbillivirus. J. Vet. Med. Sci. 77: 565–569. doi: 10.1292/jvms.14-0623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurosaki Y., Takada A., Ebihara H., Grolla A., Kamo N., Feldmann H., Kawaoka Y., Yasuda J.2007. Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplification. J. Virol. Methods 141: 78–83. doi: 10.1016/j.jviromet.2006.11.031 [DOI] [PubMed] [Google Scholar]
- 5.Lamb R. A., Parks G. D.2013. Paramyxoviridae. pp. 957–995. In: Fields Virology, 6th ed. (Knipe, D. M. and Howley, P. eds.), Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 6.Lulich J. P., Osborne C. A., O’Brien T. D., Polzin D. J.1992. Feline renal failure: questions, answers, questions. Compend. Contin. Educ. Pract. Vet. 14: 127–152. [Google Scholar]
- 7.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T.2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28: E63. doi: 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E. S., Suzuki M., Kimura M., Maruyama K., Mizutani H., Saito R., Kubota N., Furuya T., Mizutani T., Imaoka K., Morikawa S.2014. Identification of a natural recombination in the F and H genes of feline morbillivirus. Virology 468–470: 524–531. doi: 10.1016/j.virol.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 9.Poon L. L., Leung C. S., Chan K. H., Lee J. H., Yuen K. Y., Guan Y., Peiris J. S.2005. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 43: 427–430. doi: 10.1128/JCM.43.1.427-430.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi S., Nakagawa S., Yoshikawa R., Kuwahara C., Hagiwara H., Asai K. I., Kawakami K., Yamamoto Y., Ogawa M., Miyazawa T.2014. Genetic diversity of feline morbilliviruses isolated in Japan. J. Gen. Virol. 95: 1464–1468. doi: 10.1099/vir.0.065029-0 [DOI] [PubMed] [Google Scholar]
- 11.Woo P. C. Y., Lau S. K. P., Wong B. H. L., Fan R. Y. Y., Wong A. Y. P., Zhang A. J. X., Wu Y., Choi G. K. Y., Li K. S. M., Hui J., Wang M., Zheng B.J., Chan K. H., Yuen K.Y.2012. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc. Natl. Acad. Sci. U.S.A. 109: 5435–5440. doi: 10.1073/pnas.1119972109 [DOI] [PMC free article] [PubMed] [Google Scholar]