Abstract
Two commercial PRRSV ELISA kits (IDEXX and Bionote) were evaluated for their sensitivity and specificity using 476 PRRS-positive serum samples collected from 7 animal challenge experiments and 1,000 PRRS-negative sera. Both ELISA kits exhibited 100% sensitivity with sera collected 14 to 42 days post-infection, and the results from the kits were highly correlated (R2=0.9207). The specificity of IDEXX or Bionote kit was 99.9% or 99.7%, respectively. In addition, the Bionote ELISA kit was used to examine 100 sera that were determined to be falsely positive either by IDEXX 2XR or 3XR ELISA, and only 7 of these samples were found to be positive. These results indicate that both ELISA kits exhibited similar levels of sensitivity and specificity and would complement one another for the verification of false-positive samples.
Keywords: antibodies detection, Bionote ELISA, IDEXX ELISA, nucleocapsid protein, PRRSV
Porcine reproductive and respiratory syndrome virus (PRRSV) is a cause of major economic loss to the pig industry worldwide [1]. PRRSV primarily affects the respiratory tract and reproductive organs [11]. PRRSV was first isolated in North America in 1987 and in Europe in 1990 [2, 3]. The PRRSV genome is approximately 15 kb in length and contains at least ten open reading frames (ORFs: OPF1a/1b, 2a/b, 3, 4, 5a/5b, 6 and 7) [13, 14, 19]. Nucleocapsid protein (N) encoded by ORF7 is a 15-kDa, multi-functional protein and the most abundant component of the virion [15, 21, 22]. The N protein is conserved among various PRRSV strains and is highly immunogenic [7, 10, 16, 18, 21]. Previous studies demonstrated that the development of immunoassays using the N protein for the detection of anti-PRRSV antibodies is sensitive, specific and repeatable [4, 12]. Therefore, the N protein is a suitable candidate for diagnosis of the disease using virus-specific antibody detection [4]. Various serological tests, such as indirect immunofluorescent antibody (IFA) assays, immunoperoxidase monolayer antibody (IPMA) assays, virus neutralization and enzyme-linked immunosorbent assays (ELISAs), have been used for the routine diagnosis of PRRS [5, 8, 17, 20, 23,24,25,26], and recombinant N protein-based ELISAs are most commonly used for PRRSV diagnosis [6, 12]. Among these serological methods, the IDEXX PRRS ELISA kit in particular has become an industry standard for the diagnosis of PRRSV due to its simple protocols and high sensitivity, specificity and reproducibility [4, 9]. However, control of this virus requires increasingly reliable detection tools, import controls, outbreak investigations and follow-up studies. In this study, the sensitivity and specificity of two commercial PRRSV antibody (Ab) ELISA kits, IDEXX PRRS 3XR Ab ELISA (Westbrook, ME, U.S.A.) and Bionote PRRS Ab ELISA 4.0 (Hwasung, South Korea), are compared using samples collected from animal challenge studies with various PRRSV strains as well as field samples from Korean PRRS-negative swine farms.
Four hundred seventy-six serum samples were selected from 7 different animal challenge studies that were conducted with 7 different North America (type II) PRRSV strains [VR2332 (GenBank accession number: JF430265), JA142 (AY424271), SDSU73 (JN654458), MN184 (EF488739), K07-2273 (JQ656251), K08-1053 (JQ656266) or Ingelvac PRRS MLV (hereafter “MLV”, AF159149)] that share 88–99% nucleotide sequence homology based on ORF5 sequences. In short, ten 4-week-old, PRRS-negative pigs were purchased for each virus strain, challenged intramuscularly with each of the 7 virus strains at 103 TCID50/ml and bled at 0, 7, 14, 21, 28, 35 and 42 days post-challenge (dpc). Fourteen samples from two pigs challenged with MN184 or MLV were excluded from the study due to insufficient sample quantities. These collected samples were used as known positive samples to determine the sensitivities of the ELISA kits. The animal experiment protocol was approved by the Chonbuk National University Institutional Animal Care and Use Committee (approval number: 2012-0025). Forty swine farms that have maintained PRRS-negative status over the past year were confirmed to be negative by real-time reverse transcription-polymerase chain reaction (RT-PCR) and IDEXX PRRS 3XR Ab ELISA and were selected for the study. Information regarding the primers and probe for the real-time RT-PCR is as follows: forward primer: TGTCAGATTCAGGGAGRATAAGTTAC; probe: TTTTGCACCACMGCCAGCCC; and reverse primer: ATCARGCGCACAGTRTGATGC. RT-PCR was conducted with the AgPath-IDTM One-Step RT-PCR Kit (Ambion, Austin, TX, U.S.A.) in a 25 µl reaction volume using 5 µl of extracted template. PCR amplification included (a) reverse transcription for 10 min at 45°C; (b) a 10 min activation step at 95°C; and (c) 40 cycles of 15 sec at 95°C and 45 sec at 60°C. Samples demonstrating a threshold cycle (Ct) of 35 cycles or less were considered positive. One thousand sera samples collected from the 40 PRRS-negative farms were used to determine the specificity of the Bionote PRRS Ab ELISA 4.0 kit. One hundred sera samples that yielded false-positive results by either IDEXX 2XR (n=23) or 3XR ELISA (n=81) but were confirmed negative by IFA were evaluated using the Bionote PRRS Ab ELISA. IFAs were conducted in 96-well plates prepared by inoculating MARC-145 cell monolayers with VR2332 at the titer of 104 TCID50/ml. The presence of antibody was confirmed by immunofluorescence microscopy using a PRRS virus-specific monoclonal antibody, SDOW17, labeled with FITC (Rural Technologies, Brookings, SD, U.S.A.). The PRRS virus-specific IFA antibody titer was determined as the reciprocal of the highest dilution in which specific fluorescence was detected. IDEXX PRRS Ab ELISA and Bionote Ab ELISA kit assays were performed according to the manufacturers’ specifications, and the presence of anti-PRRSV antibodies was measured by calculating the sample-to-positive (S/P) cut-off value of 0.4 to distinguish between positive and negative samples. All of the field serum samples were collected with consent from the producers who submitted the samples to Chonbuk National University-Veterinary Diagnostic Center. All false-positive or false-negative samples were determined either by IDEXX PRRS Ab ELISA or Bionote PRRS Ab ELISA and were confirmed by IFA. After two weeks, this process was repeated again on serum samples collected from the same pigs for final confirmation.
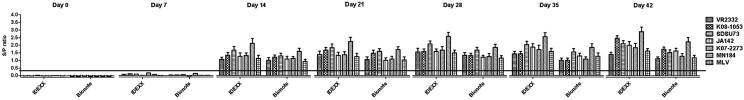
Four hundred seventy-six samples collected weekly from the pigs challenged with 7 different PRRSV strains were evaluated by IDEXX ELISA and Bionote ELISA. Both kits were capable of detecting PRRSV-specific antibodies at 14 to 42 dpc, although neither method could detect PRRSV-specific antibodies at 7 dpc (Fig. 1). Good correlation (R2=0.9207) of the S/P ratios from both ELISA kits was observed, whereas overall S/P values from the IDEXX ELISA were consistently higher than those from the Bionote ELISA (Fig. S1). Based on this comparison, it was concluded that both ELISA kits could be used as effective complementary methods given that the ELISA kits exhibited 100% sensitivity with a high correlation. To determine the specificity of the Bionote ELISA, 1,000 samples collected from 40 PRRS-negative farms were also evaluated. Three samples with S/P ratios ranging from 0.4389 to 0.6097 (considered positive) were later determined to be false-positives, because those samples were negative by IFA and no evidence of PRRS outbreak was observed in those pigs. The three samples that returned false-positive signals with the Bionote ELISA were evaluated using the IDEXX ELISA, and two were determined to be negative. Thus, the IDEXX and Bionote ELISA kits detected only 1 and 3 false-positives out of 1,000 negative samples, resulting in 99.9 and 99.7% specificity rates, respectively (Table 1 and Fig. S2). Recently, IDEXX 3XR ELISA was also compared to another commercial PRRS antibody ELISA kit (HIPRA ELISA) as well as fluorescent microbead immunoassays (FMIA) [10]. IDEXX ELISA detected positive animals earlier than HIPRA ELISA, and IDEXX ELISA also exhibited 100% sensitivity with sera collected at 14 dpc and presented the highest efficiency rate (100%), while HIPRA ELISA exhibited a lower efficiency (80%) when applied to the same set of samples. The FMIA assay exhibited the highest false-positive rate for known negative field samples when compared with the commercial ELISAs (IDEXX and HIPRA ELISA kits) for the detection of PRRSV antibodies.
Fig. 1.
Test results from two commercial PRRS ELISA kits applied to 476 serum samples sequentially collected from PRRS-negative pigs challenged with 7 different PRRSV strains. *The horizontal line indicates the cut-off value (0.4) for positivity.
Table 1. Test results from IDEXX PRRS 3XR ELISA for samples that yielded false-positive results based on the Bionote PRRS Ab ELISA.
| Case No. | Bionote | IDEXX | ||
|---|---|---|---|---|
| S/P ratio | Result | S/P ratio | Result | |
| 23-31-1 | 0.4389 | F-Posa) | –0.049 | Negb) |
| 100-2 | 0.6097 | F-Pos | 0.0142 | Neg |
| 8-63 | 0.5646 | F-Pos | 0.4286 | F-Pos |
a) F-Pos; False-positive, b) Neg; Negative.
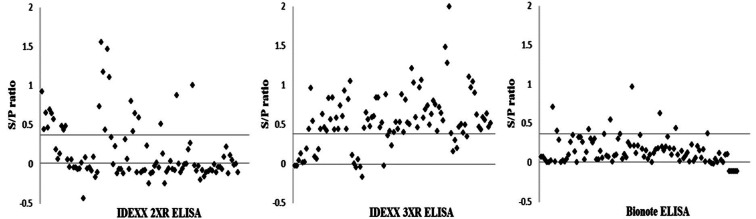
Bionote ELISA was performed on 100 serum samples that were judged to be falsely positive by either IDEXX 2XR or 3XR ELISA, and only 7 samples were determined to actually be positive (Fig. 2). S/P ratios ranged from 0.41 to 1.55 with an average of 0.7401 for IDEXX 2XR. For IDEXX 3XR, an S/P ratio range of 0.41 to 1.99 with an average of 0.6834 was noted. S/P ratios ranged from 0.41 to 0.97 S/P with an average of 0.5889 for Bionote false-positives (Table 2). These results indicate that Bionote ELISA or IDEXX ELISAs could complement one another for the verification of false-positive samples, rather than suggesting an interpretation that the Bionote ELISA is superior to IDEXX 2XR or 3XR ELISA, because the falsely positive samples were collected from routine PRRSV serological testing with IDEXX 3XR ELISA on field samples submitted from PRRS-negative farms between the years 2012 and 2014. Although commercial ELISA kits are the most reliable tests to detect PRRSV infection at the herd level, the specificity of the kits has been frequently challenged by unexpected false-positive results at various rates. In a previous study [9], a blocking ELISA (bELISA) was developed and applied to 133 serum samples that yielded unexpected positive IDEXX ELISA results from 4,038 diagnostic samples submitted by farms from which PRRS-negative results were expected. The bELISA was developed using a recombinant PRRSV N protein and a biotinylated monoclonal antibody. The sensitivity and specificity of this technique were 97.8% and 100%, respectively, based on the evaluation of 686 sera collected from PRRSV-infected or uninfected pigs. In the study, the bELISA identified 97% of the samples as negative. Therefore, the data indicate that a significantly increased percentage of unexpected positive results could be observed with the IDEXX ELISA, despite the fact that it is one of the most widely accepted assays [9]. In this regard, the Bionote ELISA appears to be a good alternative method to address singleton reactors detected by IDEXX ELISA, given that the Bionote ELISA only detected 7 samples as false-positives among 100 singleton samples determined to be positive by IDEXX 2XR or 3XR ELISAs.
Fig. 2.
Test results from the Bionote PRRS ELISA kit using 100 samples that returned false-positive results based on either IDEXXX PRRS 2XR or 3XR ELISA. *The horizontal line indicates the cut-off value (0.4) for positivity.
Table 2. Test results from the Bionote PRRS Ab ELISA on samples identified as false positives either by IDEXX PRRS 2XR or 3XR ELISA.
| Case No. | Sample No. | 2XR | 3XR | Bionote | IFA | |||
|---|---|---|---|---|---|---|---|---|
| S/P ratio | Result | S/P ratio | Result | SP ratio | Result | |||
| 12-0099 | 1 | 0.92 | F-Posa | -0.03 | Negb | 0.07 | Neg | Neg |
| 2 | 0.44 | F-Pos | -0.03 | Neg | 0.07 | Neg | Neg | |
| 12-0909 | 3 | 0.65 | F-Pos | 0.04 | Neg | 0.04 | Neg | Neg |
| 4 | 0.46 | F-Pos | 0.13 | Neg | 0 | Neg | Neg | |
| 12-0948 | 5 | 0.69 | F-Pos | 0.01 | Neg | 0 | Neg | Neg |
| 12-0966 | 6 | 0.64 | F-Pos | 0.02 | Neg | 0.03 | Neg | Neg |
| 13-2694 | 7 | 0.57 | F-Pos | 0.19 | Neg | 0.71 | F-Pos | Neg |
| 13-3294 | 8 | 0.18 | Neg | 0.44 | F-Pos | 0.01 | Neg | Neg |
| 13-4519 | 9 | 0.06 | Neg | 0.96 | F-Pos | 0.41 | F-Pos | Neg |
| 13-4509 | 10 | 0.13 | Neg | 0.54 | F-Pos | 0.29 | Neg | Neg |
| 12-0796 | 11 | 0.48 | F-Pos | 0.09 | Neg | 0 | Neg | Neg |
| 12 | 0.43 | F-Pos | 0.06 | Neg | 0.02 | Neg | Neg | |
| 12-0959 | 13 | 0.48 | F-Pos | 0.18 | Neg | 0.01 | Neg | Neg |
| 13-1999 | 14 | 0.05 | Neg | 0.44 | F-Pos | 0.05 | Neg | Neg |
| 13-2413 | 15 | –0.04 | Neg | 0.63 | F-Pos | 0.1 | Neg | Neg |
| 13-2862 | 16 | 0.05 | Neg | 0.46 | F-Pos | 0.26 | Neg | Neg |
| 17 | –0.05 | Neg | 0.42 | F-Pos | 0.35 | Neg | Neg | |
| 18 | –0.05 | Neg | 0.83 | F-Pos | 0.14 | Neg | Neg | |
| 13-3168 | 19 | –0.07 | Neg | 0.56 | F-Pos | 0 | Neg | Neg |
| 13-3318 | 20 | –0.06 | Neg | 0.84 | F-Pos | 0.32 | Neg | Neg |
| 21 | 0.01 | Neg | 0.43 | F-Pos | 0.33 | Neg | Neg | |
| 13-3399 | 22 | –0.44 | Neg | 0.58 | F-Pos | 0.26 | Neg | Neg |
| 13-3609 | 23 | 0.08 | Neg | 0.44 | F-Pos | 0.04 | Neg | Neg |
| 13-4129 | 24 | –0.06 | Neg | 0.74 | F-Pos | 0.13 | Neg | Neg |
| 13-4200 | 25 | –0.05 | Neg | 0.6 | F-Pos | 0.43 | F-Pos | Neg |
| 13-4372 | 26 | –0.09 | Neg | 0.93 | F-Pos | 0.3 | Neg | Neg |
| 13-4451 | 27 | 0.09 | Neg | 0.44 | F-Pos | 0.26 | Neg | Neg |
| 28 | –0.17 | Neg | 0.82 | F-Pos | 0.31 | Neg | Neg | |
| 13-4997 | 29 | –0.11 | Neg | 1.05 | F-Pos | 0.04 | Neg | Neg |
| 12-0651 | 30 | 0.73 | F-Pos | 0.11 | Neg | 0.04 | Neg | Neg |
| 12-0741 | 31 | 1.55 | F-Pos | 0 | Neg | 0.15 | Neg | Neg |
| 32 | 1.17 | F-Pos | -0.05 | Neg | 0.06 | Neg | Neg | |
| 33 | 0.43 | F-Pos | 0.05 | Neg | 0.37 | Neg | Neg | |
| 12-0757 | 34 | 1.46 | F-Pos | -0.04 | Neg | 0.09 | Neg | Neg |
| 35 | 1.1 | F-Pos | -0.17 | Neg | 0.07 | Neg | Neg | |
| 12-0855 | 36 | 0.34 | Neg | 0.45 | F-Pos | 0.54 | F-Pos | Neg |
| 12-1344 | 37 | –0.01 | Neg | 0.65 | F-Pos | 0.01 | Neg | Neg |
| 12-2084 | 38 | 0.22 | Neg | 0.56 | F-Pos | 0.08 | Neg | Neg |
| 12-2621 | 39 | –0.12 | Neg | 0.47 | F-Pos | 0.1 | Neg | Neg |
| 13-1828 | 40 | –0.07 | Neg | 0.59 | F-Pos | 0.3 | Neg | Neg |
| 12-1692 | 41 | –0.07 | Neg | 0.6 | F-Pos | 0.37 | Neg | Neg |
| 12-3616 | 42 | –0.12 | Neg | 0.84 | F-Pos | 0.04 | Neg | Neg |
| 13-1852 | 43 | 0.31 | Neg | 0.84 | F-Pos | 0.11 | Neg | Neg |
| 13-2904 | 44 | 0.06 | Neg | 0.46 | F-Pos | 0.06 | Neg | Neg |
| 13-4680 | 45 | –0.07 | Neg | 0.52 | F-Pos | 0.26 | Neg | Neg |
| 12-2294 | 46 | 0.8 | F-Pos | -0.03 | Neg | 0.21 | Neg | Neg |
| 12-3260 | 47 | 0.41 | F-Pos | 0.88 | F-Pos | 0.97 | F-Pos | Neg |
| 13-1829 | 48 | 0.64 | F-Pos | 0.36 | Neg | 0.21 | Neg | Neg |
| 49 | –0.11 | Neg | 0.41 | F-Pos | 0.12 | Neg | Neg | |
| 13-1871 | 50 | 0.59 | F-Pos | 0.23 | Neg | 0.22 | Neg | Neg |
| 51 | –0.11 | Neg | 0.4 | F-Pos | 0.35 | Neg | Neg | |
| 13-2136 | 52 | –0.09 | Neg | 0.53 | F-Pos | 0.17 | Neg | Neg |
| 13-0930 | 53 | –0.08 | Neg | 0.44 | F-Pos | 0.07 | Neg | Neg |
| 13-1741 | 54 | 0.23 | Neg | 0.53 | F-Pos | 0.16 | Neg | Neg |
| 13-2262 | 55 | –0.25 | Neg | 0.88 | F-Pos | 0.05 | Neg | Neg |
| 13-3412 | 56 | –0.12 | Neg | 0.4 | F-Pos | 0.1 | Neg | Neg |
| 12-2994 | 57 | –0.11 | Neg | 0.81 | F-Pos | 0.01 | Neg | Neg |
| 58 | –0.03 | Neg | 0.52 | F-Pos | 0.12 | Neg | Neg | |
| 12-3031 | 59 | 0.01 | Neg | 0.5 | F-Pos | 0.17 | Neg | Neg |
| 60 | –0.05 | Neg | 1.21 | F-Pos | 0.18 | Neg | Neg | |
| 13-1509 | 61 | 0.51 | F-Pos | 1.03 | F-Pos | 0.63 | F-Pos | Neg |
| 13-1676 | 62 | 0.13 | Neg | 0.59 | F-Pos | 0.2 | Neg | Neg |
| 63 | –0.25 | Neg | 0.46 | F-Pos | 0.16 | Neg | Neg | |
| 13-1987 | 64 | –0.1 | Neg | 0.97 | F-Pos | 0.2 | Neg | Neg |
| 13-2836 | 65 | –0.06 | Neg | 1.06 | F-Pos | 0.33 | Neg | Neg |
| 13-3315 | 66 | 0.03 | Neg | 0.58 | F-Pos | 0.18 | Neg | Neg |
| 67 | –0.07 | Neg | 0.69 | F-Pos | 0.1 | Neg | Neg | |
| 13-2134 | 68 | –0.08 | Neg | 0.74 | F-Pos | 0.17 | Neg | Neg |
| 13-2462 | 69 | 0.87 | F-Pos | 0.49 | F-Pos | 0.44 | F-Pos | Neg |
| 13-3278 | 70 | 0 | Neg | 0.63 | F-Pos | 0.1 | Neg | Neg |
| 13-3513 | 71 | –0.11 | Neg | 0.8 | F-Pos | 0.02 | Neg | Neg |
| 13-3707 | 72 | –0.06 | Neg | 0.75 | F-Pos | 0.15 | Neg | Neg |
| 13-2330 | 73 | –0.01 | Neg | 0.41 | F-Pos | 0.05 | Neg | Neg |
| 13-3481 | 74 | 0 | Neg | 0.72 | F-Pos | 0.08 | Neg | Neg |
| 13-4622 | 75 | 0.18 | Neg | 0.64 | F-Pos | 0.12 | Neg | Neg |
| 76 | 0.26 | Neg | 0.55 | F-Pos | 0.01 | Neg | Neg | |
| 13-4931 | 77 | 1 | F-Pos | 1.48 | F-Pos | 0.23 | Neg | Neg |
| 14-1370 | 78 | –0.02 | Neg | 1.28 | F-Pos | 0.02 | Neg | Neg |
| 14-1342 | 79 | –0.09 | Neg | 2 | F-Pos | 0.06 | Neg | Neg |
| 14-1335 | 80 | –0.03 | Neg | 0.38 | Neg | 0.21 | Neg | Neg |
| 81 | –0.2 | Neg | 0.16 | Neg | 0.16 | Neg | Neg | |
| 82 | –0.08 | Neg | 0.3 | Neg | 0.06 | Neg | Neg | |
| 83 | –0.16 | Neg | 0.19 | Neg | 0.17 | Neg | Neg | |
| 14-1324 | 84 | –0.11 | Neg | 0.47 | F-Pos | 0.02 | Neg | Neg |
| 14-1328 | 85 | –0.1 | Neg | 0.5 | F-Pos | 0.37 | Neg | Neg |
| 14-1234 | 86 | –0.08 | Neg | 0.39 | Neg | 0.01 | Neg | Neg |
| 14-1206 | 87 | –0.09 | Neg | 0.49 | F-Pos | –0.01 | Neg | Neg |
| 14-1052 | 88 | –0.01 | Neg | 0.34 | Neg | –0.02 | Neg | Neg |
| 89 | –0.11 | Neg | 1.1 | F-Pos | 0.04 | Neg | Neg | |
| 14-1018 | 90 | –0.04 | Neg | 0.96 | F-Pos | 0.01 | Neg | Neg |
| 91 | –0.04 | Neg | 1.05 | F-Pos | 0.12 | Neg | Neg | |
| 92 | –0.07 | Neg | 0.9 | F-Pos | 0.03 | Neg | Neg | |
| 14-1002 | 93 | 0.09 | Neg | 0.62 | F-Pos | 0.01 | Neg | Neg |
| 14-0961 | 94 | 0.21 | Neg | 0.46 | F-Pos | 0.1 | Neg | Neg |
| 14-0929 | 95 | –0.12 | Neg | 0.43 | F-Pos | 0.11 | Neg | Neg |
| 96 | 0.11 | Neg | 0.59 | F-Pos | –0.11 | Neg | Neg | |
| 14-0837 | 97 | 0.05 | Neg | 0.56 | F-Pos | –0.11 | Neg | Neg |
| 98 | –0.02 | Neg | 0.63 | F-Pos | –0.11 | Neg | Neg | |
| 14-0700 | 99 | 0 | Neg | 0.47 | F-Pos | –0.11 | Neg | Neg |
| 100 | –0.11 | Neg | 0.52 | F-Pos | –0.11 | Neg | Neg | |
a) F-Pos; False-positive, b) Neg; Negative.
In this study, the results obtained indicate that these two commercial kits exhibit similar sensitivities and specificities. Therefore, both PRRSV ELISA kits can complement one another for the verification of false-positive samples.
Supplementary Material
Acknowledgments
The study was supported by the “Cooperative Research Program for Agriculture Science & Technology Development (Development of Monitoring and Diagnostic Methods for Environmental Animal Disease, PJ010530)” of the Rural Development Administration, Republic of Korea.
REFERENCES
- 1.Albina E.1997. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet. Microbiol. 55: 309–316. doi: 10.1016/S0378-1135(96)01322-3 [DOI] [PubMed] [Google Scholar]
- 2.Andreyev V. G., Wesley R. D., Mengeling W. L., Vorwald A. C., Lager K. M.1997. Genetic variation and phylogenetic relationships of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch. Virol. 142: 993–1001. doi: 10.1007/s007050050134 [DOI] [PubMed] [Google Scholar]
- 3.Benfield D. A., Nelson E. A., Collins J. E., Harris L., Goyal S. M., Robison D., Cristianson W. T., Morrison R. B., Gorcyca D., Chladek D. W.1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Invest. 4: 127–133. doi: 10.1177/104063879200400202 [DOI] [PubMed] [Google Scholar]
- 4.Chen C., Fan W., Jia X., Li J., Bi Y., Liu W.2013. Development of a recombinant N-Gp5c fusion protein-based ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus. J. Virol. Methods 189: 213–220. doi: 10.1016/j.jviromet.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 5.Cho S. H., Freese W. R., Yoon I. J., Trigo A. V., Joo H. S.1993. Seroprevalence of indirect fluorescent antibody to porcine reproductive and respiratory syndrome virus in selected swine herds. J. Vet. Diagn. Invest. 5: 259–260. doi: 10.1177/104063879300500220 [DOI] [PubMed] [Google Scholar]
- 6.Collins J., Dee S., Halbur P., Keffaber K., Lautner B., McCaw M., Rodibaugh M., Sanford E., Yeske P.1996. Laboratory diagnosis of porcine reproductive and respiratory syndrome (PPRS) virus infection. Swine Health and Prod. 4: 33–35. [Google Scholar]
- 7.Dea S., Wilson L., Therrien D., Cornaglia E.2000. Competitive ELISA for detection of antibodies to porcine reproductive and respiratory syndrome virus using recombinant E. coli-expressed nucleocapid protein as antigen. J. Virol. Methods 87: 109–122. doi: 10.1016/S0166-0934(00)00158-0 [DOI] [PubMed] [Google Scholar]
- 8.Denac H., Moser C., Tratschin J. D., Hofmann M. A.1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65: 169–181. doi: 10.1016/S0166-0934(97)02186-1 [DOI] [PubMed] [Google Scholar]
- 9.Ferrin N. H., Fang Y., Johnson C. R., Murtaugh M. P., Polson D. D., Torremorell M., Gramer M. L., Nelson E. A.2004. Validation of a blocking enzyme-linked immunosorbent assay for detection of antibodies against porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 11: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber P. F., Gimenez-Lirola L. G., Halbur P. G., Zhou L., Meng X. J., Opriessnig T.2014. Comparison of commercial enzyme-linked immunosorbent assays and fluorescent microbead immunoassays for detection of antibodies against porcine reproductive and respiratory syndrome virus in boars. J. Virol. Methods 197: 63–66. doi: 10.1016/j.jviromet.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 11.Goyal S. M.1993. Porcine reproductive and respiratory syndrome. J. Vet. Diagn. Invest. 5: 656–664. doi: 10.1177/104063879300500435 [DOI] [PubMed] [Google Scholar]
- 12.Janková J., Celer V.2012. Expression and serological reactivity of Nsp7 protein of PRRS genotype I virus. Res. Vet. Sci. 93: 1537–1542. doi: 10.1016/j.rvsc.2012.06.007 [DOI] [PubMed] [Google Scholar]
- 13.Johnson C. R., Griggs T. F., Gnanandarajah J., Murtaugh M. P.2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 92: 1107–1116. doi: 10.1099/vir.0.030213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mounir S., Mardassi H., Dea S.1995. Identification and characterization of the porcine reproductive and respiratory virus ORFs 7, 5 and 4 products. Adv. Exp. Med. Biol. 380: 317–320. doi: 10.1007/978-1-4615-1899-0_51 [DOI] [PubMed] [Google Scholar]
- 15.Nelson E. A., Christopher-Hennings J., Benfield D. A.1994. Serum immune responses to the proteins of porcine reproductive and respiratory syndrome (PRRS) virus. J. Vet. Diagn. Invest. 6: 410–415. doi: 10.1177/104063879400600402 [DOI] [PubMed] [Google Scholar]
- 16.Nelson E. A., Christopher-Hennings J., Drew T., Wensvoort G., Collins J. E., Benfield D. A.1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31: 3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okinaga T., Yoshii M., Suzuki T., Miyazaki A., Takagi M., Tsunemitsu H., Yamagishi T.2009. Evaluation of unexpected positive results from a commercial ELISA for antibodies to PRRSV. Vet. Rec. 164: 455–459. doi: 10.1136/vr.164.15.455 [DOI] [PubMed] [Google Scholar]
- 18.Seuberlich T., Tratschin J. D., Thur B., Hofmann M. A.2002. Nucleocapsid protein-based enzyme-linked immunosorbent assay for detection and differentiation of antibodies against European and North American porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 9: 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadejek T., Stankevicius A., Storgaard T., Oleksiewicz M. B., Belak S., Drew T. W., Pejsak Z.2002. Identification of radically different variants of porcine reproductive and respiratory syndrome virus in Eastern Europe: towards a common ancestor for European and American viruses. J. Gen. Virol. 83: 1861–1873. [DOI] [PubMed] [Google Scholar]
- 20.Takikawa N., Kobayashi S., Ide S., Yamane Y., Tanaka Y., Yamagishi H.1996. Detection of antibodies against porcine reproductive and respiratory syndrome (PRRS) virus in swine sera by enzyme-linked immunosorbent assay. J. Vet. Med. Sci. 58: 355–357. doi: 10.1292/jvms.58.355 [DOI] [PubMed] [Google Scholar]
- 21.Venteo A., Rebollo B., Sarraseca J., Rodriguez M. J., Sanz A.2012. A novel double recognition enzyme-linked immunosorbent assay based on the nucleocapsid protein for early detection of European porcine reproductive and respiratory syndrome virus infection. J. Virol. Methods 181: 109–113. doi: 10.1016/j.jviromet.2012.01.024 [DOI] [PubMed] [Google Scholar]
- 22.Wootton S. K., Nelson E. A., Yoo D.1998. Antigenic structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 5: 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon I. J., Joo H. S., Christianson W. T., Kim H. S., Collins J. E., Carlson J. H., Dee S. A.1992a. Isolation of a cytopathic virus from weak pigs on farms with a history of swine infertility and respiratory syndrome. J. Vet. Diagn. Invest. 4: 139–143. doi: 10.1177/104063879200400204 [DOI] [PubMed] [Google Scholar]
- 24.Yoon I. J., Joo H. S., Christianson W. T., Kim H. S., Collins J. E., Morrison R. B., Dial G. D.1992b. An indirect fluorescent antibody test for the detection of antibody to swine infertility and respiratory syndrome virus in swine sera. J. Vet. Diagn. Invest. 4: 144–147. doi: 10.1177/104063879200400205 [DOI] [PubMed] [Google Scholar]
- 25.Yoon I. J., Joo H. S., Goyal S. M., Molitor T. W.1994. A modified serum neutralization test for the detection of antibody to porcine reproductive and respiratory syndrome virus in swine sera. J. Vet. Diagn. Invest. 6: 289–292. [DOI] [PubMed] [Google Scholar]
- 26.Yoon K. J., Zimmerman J. J., Swenson S. L., McGinley M. J., Eernisse K. A., Brevik A., Rhinehart L. L., Frey M. L., Hill H. T., Platt K. B.1995. Characterization of the humoral immune response to porcine reproductive and respiratory syndrome (PRRS) virus infection. J. Vet. Diagn. Invest. 7: 305–312. doi: 10.1177/104063879500700302 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.