Abstract
A novel Gram-stain negative, aerobic, non-flagellated, rod-shaped gliding bacterial strain, designated HFJT, was isolated from a skin lesion of a diseased Atlantic salmon (Salmo salar L.) in Finnmark, Norway. Colonies were observed to be yellow pigmented with entire and/or undulating margins and did not adhere to the agar. The 16S rRNA gene sequence showed that the strain belongs to the genus Tenacibaculum (family Flavobacteriaceae, phylum ‘Bacteroidetes’). Strain HFJT exhibits high 16S rRNA gene sequence similarity values to Tenacibaculum dicentrarchi NCIMB 14598T (97.2 %). The strain was found to grow at 2–20 °C and only in the presence of sea salts. The respiratory quinone was identified as menaquinone 6 and the major fatty acids were identified as summed feature 3 (comprising C16:1 ω7c and/or iso-C15:0 2-OH), iso-C15:0, anteiso-C15:0, iso-C15:1 and iso-C15:0 3-OH. The DNA G+C content was determined to be 34.1 mol%. DNA–DNA hybridization and comparative phenotypic and genetic tests were performed with the phylogenetically closely related type strains, T. dicentrarchi NCIMB 14598T and Tenacibaculumovolyticum NCIMB 13127T. These data, as well as phylogenetic analyses, suggest that strain HFJT should be classified as a representative of a novel species in the genus Tenacibaculum, for which the name Tenacibaculum finnmarkense sp. nov. is proposed; the type strain is HFJ T = (DSM 28541T = NCIMB 42386T).
Keywords: Norway, Polyphasic taxonomy, Salmon farming, Skin lesions, Ulcerative disease, Winter ulcers
Introduction
During an outbreak of an ulcerative disease in Atlantic salmon at a seawater site in Finnmark, Norway, long rod shaped bacteria were found to predominate in the skin lesions. One strain of these isolates designated HFJT is described in the present study. The 16S rRNA gene sequence showed that it belongs to the genus Tenacibaculum (family Flavobacteriaceae, phylum ‘Bacteroidetes’) described by Suzuki et al. (2001). To date the genus comprises 21 species derived from a variety of marine environments and marine organisms (Kim et al. 2013; LPSN 2015). Several of the type strains in genus Tenacibaculum have been reported as pathogenic for fish or associated with disease in cultured marine fish (Wakabayashi et al. 1986; Hansen et al. 1992; Piñeiro-Vidal et al. 2008a, b; López et al. 2010; Piñeiro-Vidal et al. 2012). Tenacibaculummaritimum, the causative agent of marine tenacibaculosis, is the best known and most extensively studied fish pathogenic bacterium in the genus (Wakabayashi et al. 1986; Suzuki et al. 2001). The disease has been reported from Europe, Japan, North America and Australia and affects both wild and cultured fish, including Rainbow trout and Atlantic salmon (Toranzo et al. 2005; Avendaño-Herrera et al. 2006; Bruno et al. 2013). T. maritimum has never been isolated in cases of ulcerative disease in Norway (Olsen et al. 2011).
There has been a growing attention regarding the potential role of Norwegian Tenacibaculum spp. in causing ulcerative disease in sea-reared Atlantic salmon, as they are commonly identified from skin lesion in mixed cultures with the bacterium Moritella viscosa or as the apparent sole agent (Olsen et al. 2011; Bornø and Lie 2015). The aim of the present study was to determine the taxonomic position of the fish pathogenic Tenacibaculum strain HFJT using genetic, phenotypic and chemotaxonomic characterisations, a detailed phylogenetic investigation based on 16S rRNA gene sequences and concatenated housekeeping (HK) gene sequences, and DNA–DNA hybridization (DDH).
Materials and methods
A total of 11 isolates from genus Tenacibaculum were included in the present study (Table 1). Strain HFJT was isolated in spring 2013 from a skin lesion of a diseased Atlantic salmon at a seawater site in Finnmark, Norway. Tenacibaculum sp. strains Tsp. 2–7 were collected from skin or gill of Atlantic salmon and cod in Norway. The type strains Tenacibaculum dicentrarchi NCIMB 14598T, Tenacibaculum ovolyticum NCIMB 13127T, Tenacibaculumsoleae NCIMB 14368T and T. maritimum NCIMB 2154T were obtained from TheNational Collection of Industrial, Marine and Food Bacteria (NCIMB). Subcultivation was performed on Marine agar (MA) (Difco 2216) plates at 16 °C for 48 h. The strains were preserved in CryoTube™ vials (Thermo scientific) at −80 °C.
Table 1.
List of Tenacibaculum strains included in the present study
| Bacterial species | Strain | Origin | Host | Tissue | Year |
|---|---|---|---|---|---|
| Tenacibaculum sp. | HFJT | Norway | Atlantic salmon | Skin | 2013 |
| Tenacibaculum sp. | Tsp.2 | Norway | Atlantic salmon | Skin | 2013 |
| Tenacibaculum sp. | Tsp.3 | Norway | Atlantic salmon | Gill | 2014 |
| Tenacibaculum sp. | Tsp.4 | Norway | Atlantic salmon | Skin | 2013 |
| Tenacibaculum sp. | Tsp.5 | Norway | Atlantic salmon | Skin | 2014 |
| Tenacibaculum sp. | Tsp.6 | Norway | Atlantic salmon | Skin | 2009 |
| Tenacibaculum sp. | Tsp.7 | Norway | Farmed Atlantic cod | Skin | 2009 |
| Tenacibaculum maritimum | NCIMB 2154T | Japan | Red sea bream fingerling | Kidney | 1977 |
| Tenacibaculum soleae | NCIMB 14368T | Spain | Senegalese sole | Unknown | 2007 |
| Tenacibaculum ovolyticum | NCIMB 13127T | Norway | Atlantic halibut eggs | Eggs | 1989 |
| Tenacibaculum dicentrarchi | NCIMB 14598T | Spain | European sea bass | Skin | 2009 |
The Tenacibaculum sp. strains were collected from Norwegian field cases, whereas the type strains were obtained from NCIMB
Draft genome sequencing of strain HFJT was carried out by BaseClear B.V (Leiden, The Netherlands.) using Illumina next generation sequencing on a HiSeq 2500™ platform. Extraction of the required concentration (>100 ng/µl) of genomic DNA was performed using an E.Z.N.A. tissue DNA kit™ (Omega Bio-Tek) following the cultured cells protocol. The draft genome sequence obtained for strain HFJT was used for the PCR primer design using primer-BLAST (Ye et al. 2012) and to verify obtained sequences for strain HFJT.
Genomic DNA from all Tenacibaculum sp. strains listed in Table 1 was extracted using an E.Z.N.A. tissue DNA kit™ (Omega Bio-Tek) following the cultured cells protocol. PCR was performed using the 16S rRNA primers 27F and 1518R (Giovannoni et al. 1996) and specific primers for five HK genes (atpD, fusA,pgk, rpoB, and tuf) (Table 2). Amplification was based on a standard reaction mixture containing 2.5 µl Extra buffer, 1.25 mM deoxyribonucleotide triphosphates, 0.75 units (0.15 µL) Taq DNA polymerase (BioLabs, New England), 5 µM (1 µL) of forward and reverse primers; DNase-RNase free water was added to a final volume of 25 µL (16.85 µL H2O). Amplification was performed in a GeneAmp PCR system 2700 (Applied Biosystems) at 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 60-100 s, followed by 72 °C for 10 min. The PCR product was confirmed by gel electrophoresis and enzymatically purified using ExoStar 1-Step ® (GE Healthcare Bio-Sciences Corp) in an Artik Thermal Cycler (Thermo Scientific) at 37 °C for 15 min and at 80 °C for 15 min. The sequencing reaction was performed using a BigDye® version 3.1 reaction in an Arktik Thermal Cycler, at 96 °C for 5 min; 30 cycles of 96 °C for 10 s, 58 °C for 5 s and 60 °C for 4 min. The reaction was composed of a mixture of 5.5 µL deionized water, 1 µL BigDye® Terminator 3.1 version sequencing buffer, 1 µL BigDye Terminator 3.1 version Ready Reaction Premix (2.5X) (Invitrogen), 3.2 pmol (1µL) forward and reverse primers and 1.5 µL purified PCR product. Sequencing was carried out by the Sequencing Facility at Høyteknologisenteret i Bergen (http://www.uib.no/seqlab). Samples were cleaned with Agencourt CleannSeq (Beckman Coulter, Inc.) before being sequenced in a 96-capillary 3730xl DNA Analyzer (Applied Biosystems). The software Vector NTI® v.9.0 (Invitrogen) was used to assemble and align the obtained sequences.
Table 2.
List of PCR primers used in present study
| Target gene | Name | Sequence (5′–3′) | Source |
|---|---|---|---|
| 16S rRNA | B27F | AGAGTTTGATCMTGGCTCAG | Giovannoni et al. (1996) |
| 16S rRNA | A1518R | AAGGAGGTGATCCANCCRCA | Giovannoni et al. (1996) |
| tuf | Tb_tuf F1 | ACCTCCTTCACGGATAGC | Present study |
| tuf | Tb_tuf R1 | TTACGATCGTTCGAAGCCCC | Present study |
| rpoB | Tb_rpoB F1 | ATYTCTCCAAAACGCTGACC | Present study |
| rpoB | Tb_rpoB R1 | AAAACGAATCAAGGWACGAAYA | Present study |
| rpoB | Tb_rpoB F2 | ACCCTTTCCAAGGCATAAAGG | Present study |
| rpoB | Tb_rpoB R2 | GAGCCATYGGTTTTGAAAGAGA | Present study |
| rpoB | Tb_rpoB F3 | CTCTTGCTGTCTCCTCATCTG | Present study |
| rpoB | Tb_rpoB R3 | ATCCACCAAGATATAGCATCCA | Present study |
| pgk | Tb_pgk F1 | GCTCCWCCACCWGTAGAAAC | Present study |
| pgk | Tb_pgk R1 | TYCGTGTAGATTTTAATGTGCCT | Present study |
| atpD | Tb_atpD F1 | TGGYCCAGTWATCGATGTTGA | Present study |
| atpD | Tb_atpD R1 | AATACGYTCTTGCATTGCTC | Present study |
| fusA | Tb_fusA F1 | ATGGTAACTCACCCATTCCAGA | Present study |
| fusA | Tb_fusA R1 | TGGCATGATGCAACACAAGG | Present study |
Three alignments were constructed for phylogenetic analysis. The first, 16S rRNA gene sequence alignment, consisted of 1341 positions and included sequences of HFJT and the 21 published type strains in the genus Tenacibaculum. In this alignment, all sequences were obtained from GenBank with the exception of strain HFJT, T. dicentrarchi NCIMB 14598T, T. ovolyticum NCIMB 13127T, T. soleae NCIMB 14368T and T. maritimum NCIMB 2154T. The second 16S rRNA gene sequence alignment of 1349 positions contained sequences from all strains listed in Table 1. A third alignment, of 6750 positions, was constructed using concatenated sequences of the five HK genes of the strains listed in Table 1; atpD at position 1-807, fusA at position 808–1575, pgk at position 1576–2511, rpoB at position 2512–5778 and tuf at position 5779–6750. All sequences obtained in the present study are available in GenBank with accession numbers presented in Table 3. Alignments were constructed in AlignX in the Vector NTI® v.9.0 (Invitrogen) software package before sequences were adjusted to equal length and correct reading frames in GeneDoc (Nicholas et al. 1997). Concatenation of the HK alignments was performed using KAKUSAN4 (Tanabe 2011). The best fitted evolutionary model for each alignment was calculated using Mega 6 (Tamura et al. 2013). For the Bayesian analysis of the concatenated HK alignment, KAKUSAN4 was used for calculation of substitution rate and the best fit model for the individual loci and codon positions and exported into a Mr. Bayes-block. A phylogenetic analysis of all three alignments were conducted using the Maximum Likelihood (ML) method with the best fitted evolutionary model, 1000 bootstrap replications and default settings in Mega 6. The BEAST package v1.8 (Drummond and Rambaut 2007) was used for Bayesian analysis of the two 16S rRNA gene datasets using the best fitted model, relaxed lognormal molecular clock and a mcmc of 100 000 000 generations. The Bayesian phylogenetic analysis of the HK gene dataset was conducted in Mr.Bayes V.3.2.2 (Ronquist et al. 2012) using the data block with the proportional codon proportional, model from KAKUSAN4 and a mcmc of 67 000 000 generations. Kordia algicida (GenBank accession nr: AB681152) was used as outgroup in the 16S rRNA phylogenetic analysis that included all the type strains, while T. maritimum NCIMB 2154T was used as outgroup in the other phylogenetic analysis. The phylograms for the ML analysis were constructed in Mega 6. The Effective sample size values (ESS) in the Bayesian analysis were inspected using Tracer ver. 1.6 (Rambaut et al. 2014). All ESS values were within the recommended range (above 200) for all parameters. A maximum clade credibility tree was obtained using a 10 % burn-In in Tree-Annotator and viewed using FigTree (Drummond et al. 2012). For 16S rRNA gene sequence similarity analysis, Percent Nucleotide Identity (PNI) was calculated using the distance matrix option in BioEdit (Hall 2011). In the Average Nucleotide Identity (ANI) calculations, the sequences from the concatenated HK gene alignment for all strains listed in Table 1 were uploaded and analysed using the Average Nucleotide Identify option in EzGenome (Kim et al. 2012).
Table 3.
List of GenBank accession numbers of sequences obtained in the present study
| Bacterial species/strain | 16S rRNA | atpD | fusA | pgk | rpoB | tuf |
|---|---|---|---|---|---|---|
| T. finnmarkense sp.nov HFJT | KT270385 | KT270377 | KT270369 | KT270424 | KT270410 | KT270399 |
| T. dicentrarchi NCIMB 14598T | KT270381 | KT270375 | KT270364 | KT270421 | KT270408 | KT270402 |
| T. maritimum NCIMB 2154T | KT270382 | KT270378 | KT270366 | KT270416 | KT270411 | KT270393 |
| T. ovolyticum NCIMB 13127T | KT270383 | KT270379 | KT270367 | KT270423 | KT270412 | KT270395 |
| T. soleae NCIMB 14368T | KT270384 | KT270380 | KT270368 | KT270417 | KT270413 | KT270394 |
| Tenacibaculum sp. Tsp.2 | KT270386 | KT270376 | KT270365 | KT270420 | KT270409 | KT270400 |
| Tenacibaculum sp. Tsp.3 | KT270387 | KT270373 | KT270362 | KT270422 | KT270406 | KT270397 |
| Tenacibaculum sp. Tsp.4 | KT270388 | KT270370 | KT270359 | KT270414 | KT270403 | KT270392 |
| Tenacibaculum sp. Tsp.5 | KT270389 | KT270371 | KT270360 | KT270418 | KT270404 | KT270398 |
| Tenacibaculum sp. Tsp.6 | KT270390 | KT270372 | KT270361 | KT270415 | KT270405 | KT270396 |
| Tenacibaculum sp. Tsp.7 | KT270391 | KT270374 | KT270363 | KT270419 | KT270407 | KT270401 |
Morphological, physiological and biochemical tests were performed as described by Bernardet et al. (2002) for strain HFJT and the phylogenetically closely related type strains T. dicentrarchi NCIMB 14598T and T. ovolyticum NCIMB 13127T as reference strains. All tests were performed on cultures incubated at 16 °C for 48 h unless otherwise stated. Colony shape, margin, elevation, size, texture, appearance, pigmentation and optical property were examined as described by Smibert and Krieg (1994). The ability to stick to agar and viscosity of the colonies was also investigated. Cell morphology was investigated using scanning electron microscopy (SEM), transmission electron microscopy (TEM) and light microscopy. Gliding motility was determined by phase contrast microscopic examination of a Marine broth (MB) (Difco 2216) culture by the hanging drop technique as recommended by Bernardet et al. (2002). Presence of flexirubin type pigments was determined by the bathochromic shift test using a 20 % (w/v) KOH solution (Fautz and Reichenbach 1980). Congo red absorption was tested as described by Bernardet et al. (2002). The Gram reaction were performed with a Fluka 77730 Gram Staining Kit (Fluka® analytical) following the manufacturer`s protocol and the non-staining KOH method (Buck 1982). The Voges-Proskauer reaction was performed as described by Piñeiro-Vidal et al. (2012). Oxidase activity and ability to split indole from tryptophan was tested using BBL™ DrySlide Oxidase and BBL™ DrySlide Indole (BD BBL™, U.S.A), following the manufacturer`s protocol. Catalase activity was examined using the slide (drop) method following the protocol by Reiner (2010). Growth under anaerobic conditions was tested on MA using the GasPak anaerobic system (BBL). Production of H2S was detected by taping a lead acetate impregnated paper strip (Sigma) to the inside of the lid of MA plates, using Parafilm to seal lid and plate. The plates were incubated at 16 °C for 6 days. Growth on blood agar was tested using blood agar containing 2 % NaCl (BAS) (Microbial laboratory, Haukeland University Hospital, Bergen). Degradation of starch (1 % w/v), casein (1 % w/v), and Tween 80 (1 % v/v) was investigated on MA. MB supplemented with gelatin (1 % w/v) was used to investigate degradation of gelatin. Utilisation of carbon sources was tested on basal agar medium [0.2 g NaNO3, 0.2 g NH4Cl, 0.05 g yeast extract, 15 g agar and 36 g red sea salt (Red Sea)] per liter distilled water containing 0.4 % carbon source [d(+)-sucrose, d(−)-ribose, d(+)galactose, d-glucose, l-proline, l-glutamate, l-tyrosine] as described by (Suzuki et al. 2001). Absence of growth after one month of incubation was recorded as negative. Other enzymatic reactions were evaluated in the API ZYM system (bioMerieux) following the manufacturer’s instructions, except that sterile seawater was used as suspension medium. Growth at pH 4–10 (at unit intervals) was assessed in MB; pH was adjusted using 1 M NaOH and 1 M HCl. The temperature range for growth was determined on MA plates incubated at 2, 4, 8, 16, 20, 25, 30 and 37 °C for 7 days. Salinity requirement was determined with saltless MA [per liter distilled water: 5.0 g peptone, 1 g yeast extract and 0.1 g ferric citrate] containing 10, 20, 30, 50, 70 and 100 % strength seawater (100 % seawater = 38.2 g/L red sea salt) or 0.8, 1.0, 3.0, 5.0, 7.0 and 10.0 % (w/v) NaCl (Sigma). Sensitivity to antimicrobials was evaluated by the disc diffusion method following the procedures of The Clinical and Laboratory Standards Institute (CLSI 2005), except that the plates were incubated at 16 °C for 10 days on MA plates due to reduced growth for some strains on the recommended Flexibacter Maritimus Medium (FMM). The tests were performed using commercial discs (Neo-sensitabs™ and Sensi-disc™) containing kanamycin (500 µg), streptomycin (10 µg), gentamicin (30 µg), trimethoprim + sulfamethoxazole (125 + 2375 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), pipemidic acid (30 µg), cefuroxime (30 µg), penicillin G (1U), ampicillin (2 µg), tetracycline (30 µg), erythromycin (15 µg), florfenicol (30 µg), oxolinic acid (10 µg) and oxytetracycline (30 µg). Several of the tests described above were also performed for the other strains included in the present study, except Tsp.7. Strain Tsp.7 was uncultivable after prolonged cryo-storage and was therefore not included in the phenotypic tests.
The following chemotaxonomic and genetic analyses were carried out by the Identification Service of the DSMZ (Braunschweig, Germany): DNA G+C content, DDH, menaquinone and fatty acid methyl ester analysis. All strains were grown in MB at 16 °C for 48 h, except for the DDH test. For DDH, cells were grown in MB at 16 °C for 72 h and the obtained bacterial biomass washed twice in 1× Phosphate Buffered Saline. The cells were disrupted using a Constant Systems TS 0.75 KW (IUL Instruments, Germany) and the DNA in the crude lysate purified by chromatography on hydroxyapatite as described by Cashion et al. (1977). DDH was carried out as described by Ley et al. (1970) under consideration of the modifications described by Huss et al. (1983) using a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostatted 6 × 6 multicell changer and a temperature controller with in situ temperature probe (Varian). Extraction of fatty acid methyl esters, washing of extracts and GC analysis were performed by using the Sherlock MIS (MIDI Inc, Newark, USA) system using the MIDI Sherlock version 6.1 and TSBA40 database.
Results and discussion
A polyphasic approach that integrates phenotypic data with genetic and phylogenetic data was performed in the current study. This approach is recommended by several authors for bacterial taxonomic investigations (Bernardet et al. 2002; Tindall et al. 2010). As it has been regarded as best practice to include more than one representative strain when describing a novel taxon, several Tenacibaculum sp. strains (Tsp.2–7) obtained from Norwegian mariculture were included in the present study. Strains HFJT and Tsp.2 have been shown to be pathogenic to Atlantic salmon reproducing the clinical signs in a challenge study in 2013 (Vold 2014). The bacteria were re-isolated and their identity confirmed by sequencing of the 16S rRNA gene, thus fulfilling Koch’s postulates.
The phylogenetic analysis based on the 16S rRNA gene sequences and the concatenated HK gene sequences (Fig. 1, 2, 3) showed that strains HFJT, Tsp.2, Tsp.5 and Tsp.7 belong to a distinct clade separate from the closely related type strains in the genus Tenacibaculum. Moreover, the analysis showed that strain Tsp.4 forms a clade with T. dicentrarchi NCIMB 14598T and Tsp.6 forms a clade with T. ovolyticum NCIMB 13127T, while the phylogenetic placement of strain Tsp.3 is uncertain. These clades were evident in all phylogenetic trees using both the Bayesian and ML method. All phylogenetic trees showed that strain HFJT is closely related to T. dicentrarchi NCIMB 14598T, both belonging to distinct clades.
Fig. 1.
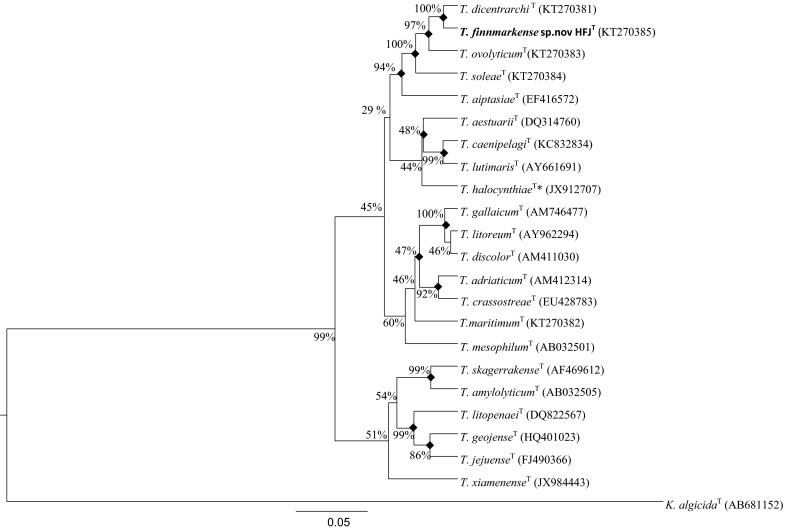
The relationship of the novel species T. finnmarkense sp. nov HFJT and the 21 type strains in genus Tenacibaculum (* = quotation marks denote names that have not been validly published) based on the 16S rRNA gene sequences, using Kordia algicida T as outgroup. The phylogenetic analysis was inferred using the Bayesian method with the best fitted evolutionary model (GTR + G + I). The posterior probability is presented next to each node in percentage. There were a total of 1341 positions in the dataset. Evolutionary analyses were conducted using BEAST package v1.8. Shared nodes identified in corresponding ML analysis are marked with filled squares. Accesion numbers are in parentheses. Scale bar 0.05 substiutions per site
Fig. 2.
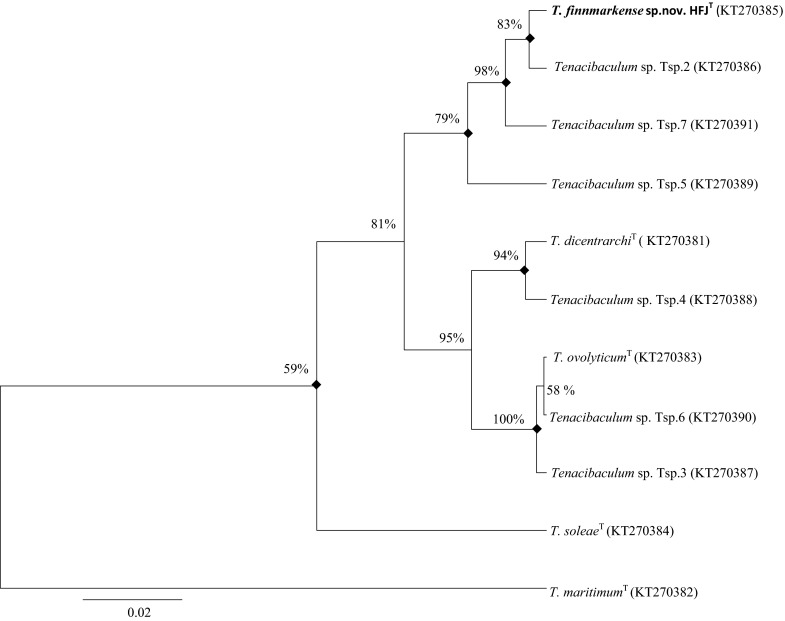
The relationship of the novel species T. finnmarkense sp.nov HFJT, Tenacibaculum sp. strains Tsp. 2–7 and the three closest related type strains based on 16S rRNA gene sequences, using T. maritimum NCIMB 2154T as outgroup. The phylogenetic analysis was inferred using the Bayesian method with the best fitted evolutionary model (HKY + G + I). The posterior probability is presented next to each node in percentage. There were a total of 1349 positions in the dataset. Evolutionary analyses were conducted using BEAST package v1.8. Shared nodes identified in corresponding ML analysis are marked with filled squares. Accesion numbers are in parentheses. Scale bar 0.02 substitutions per site
Fig. 3.
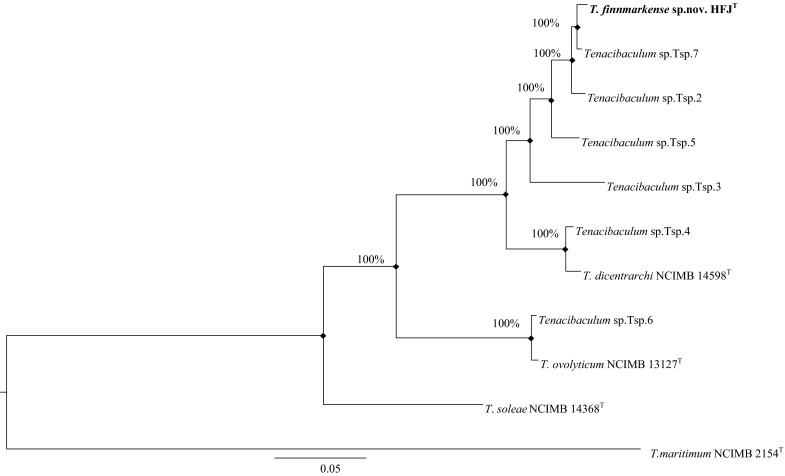
The relationship of the novel species T. finnmarkense sp.nov HFJT, Tenacibaculum sp. strains Tsp. 2–7 and the three closest related type strains based on a concatenated sequences of five HK genes (atpD at position 1–807, fusA at position 808–1575, pgk at position 1576–2511, rpoB at position 2512–5778 and tuf at position 5779–6750), using T. maritimum T as outgroup. The accession numbers for the HK genes used in the concatenated dataset are presented in Table 3. The phylogenetic analysis was inferred using the Bayesian method with the best fitted evolutionary model. The posterior probability is presented next to each node in percentage. There were a total of 6750 positions in the dataset. Evolutionary analyses were conducted using KAKUSAN4 and Mr.Bayes. Shared nodes identified in corresponding ML analysis are marked with filled squares. Scale bar 0.05 substitutions per site
As strain HFJT showed more than 97 % 16S rRNA gene sequence similarity (PNI) to T. dicentrarchi NCIMB 14598T (Table 4), DDH was performed as recommended (Stackebrandt and Goebel 1994; Tindall et al. 2010). The DDH tests revealed that the DNA relatedness of strain HFJT was 54.8 (52.0) % to T. dicentrarchi NCIMB 14598T and 36.6 (39.7) % to T. ovolyticum NCIMB 13127T. Results from repeated tests are shown in parentheses. When considering the threshold value of 70 % DNA–DNA similarity for delineation of bacterial species proposed by the ad hoc committee (Wayne et al. 1987), strain HFJT does not belong to the species T. dicentrarchi NCIMB 14598T or T. ovolyticum NCIMB 13127T. It is generally accepted that an ANI value of 95–96 % corresponds to a DDH threshold value of 70 % and can be used as a boundary for species delineation (Goris et al. 2007; Richter and Rosselló-Móra 2009). Furthermore, a study by Kim et al. (2014) revealed that a PNI of 98.65 % corresponded to an ANI value of 95–96 %. The calculated ANI and PNI values between strain HFJT and T. dicentrarchi NCIMB 14598T were 94.6 and 97.2 % respectively. By applying both the ANI and PNI threshold on all 11 strains included in this study (Table 4) we found that strains HFJT, Tsp.2, Tsp.5 and Tsp.7 belong to the same species; Tsp.4 belongs to the species T. dicentrarchi, while Tsp.6 belongs to the species T. ovolyticum. These findings correspond to results from the phylogenetic analysis and underpin that strain HFJT represents a novel species in genus Tenacibaculum.
Table 4.
Results from the PNI and ANI analysis performed for all strains listed in Table 1
| Tsp.2 | Tsp.3 | Tsp.4 | Tsp.5 | Tsp.6 | Tsp.7 | Strain HFJT | T.dicentrarchi T | T.ovolyticum T | T.soleae T | T.maritimum T | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain HFJT | 99.3 (98.7) | 95.7 (95.4) | 97.3 (94.8) | 98.6 (97.4) | 96.0 (91.2) | 98.8 (99.3) | 97.2 (94.6) | 96.0 (91.1) | 96.4 (89.5) | 93.6 (83.7) | |
| T. dicentrarchi T | 97.1 (94.6) | 96.8 (94.0) | 99.6 (99.0) | 98.2 (95.2) | 97.0 (90.7) | 97.0 (94.6) | 97.2 (94.6) | 97.0 (90.6) | 96.5 (89.3) | 93.1 (84.4) | |
| T. ovolyticum T | 96.6 (91.1) | 99.7 (91.0) | 97.0 (90.6) | 96.3 (90.9) | 100 (99.4) | 96.8 (91.0) | 96.0 (91.1) | 97.0 (90.6) | 96.7 (89.8) | 93.2 (85.2) | |
| T. soleae T | 96.6 (89.6) | 96.5 (89.4) | 96.3 (89.3) | 96.9 (89.5) | 96.7 (89.8) | 97.1 (89.5) | 96.4 (89.5) | 96.5 (89.3) | 96.7 (89.8) | 94.0 (84.3) | |
| T. maritimum T | 93.7 (83.9) | 92.9 (83.9) | 93.1 (84.2) | 93.3 (84.0) | 93.2 (85.3) | 93.8 (84.0) | 93.6 (83.7) | 93.1 (84.4) | 93.2 (85.2) | 94.0 (84.3) |
The pairwise 16S rRNA sequence identities and ANI values are presented as percent (%) similarity. ANI values are shown in parentheses
Cells of strain HFJT were observed to be rod-shaped, 0.5 µm wide and 5–25 µm in length and Gram-stain negative. Considerably longer filamentous cells and spherical degenerative cells were frequently observed in older cultures. A rapid decrease in viability was found to occur with prolonged incubation (>96 h). Differential phenotypic characteristics between all strains listed in Table 1, except strain Tsp.7, are summarised in Table 5 and are included in the species description. The G+C content of strain HFJT was determined to be 34.1 mol% which is within the range reported for other type strains in the genus Tenacibaculum (29.8–35.2 mol%). The major fatty acids (>5 % of the total fatty acids) for strain HFJT were identified as summed feature 3 (comprising C16:1 ω7c and/or iso-C15:0 2-OH), iso-C15:0, anteiso-C15:0, Iso-C15:1 and iso-C15:0 3-OH. Results from the fatty acid analysis for strain HFJT and T. dicentrarchi NCIMB 14598T are listed in Table 6. The respiratory quinone was identified as menaquinone 6 (100 %) while flexirubin-type pigments were found to be absent. This is in accordance with the chemotaxonomic characteristics of the members of the genus Tenacibaculum (Suzuki et al. 2001). In the API ZYM system, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystein arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase were found to be present. Lipase (C14), trypsin, α-chymotrypsin and all enzymes related to the metabolism of carbohydrates were found to be absent. Strain HFTT was found to be susceptible to trimethoprim-sulfamethoxazole, ceftazidime, ciprofloxacin, pipemidic acid, cefuroxime, penicillin G, ampicillin, tetracycline, erythromycin, florfenicol, oxytetracycline and oxolinic acid, but resistant to kanamycin, gentamicin and streptomycin.
Table 5.
Differential characteristics of all strains listed in Table 1, except strain Tsp.7
| Characteristic | Tsp.2 | Tsp.3 | Tsp.4 | Tsp.5 | Tsp.6 | HFJT | T.dicentrarchi T | T.ovolyticum T | T.soleae T | T.maritimum T |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell size | 3–30 µm | 2–20 µm | 2–40 µm | 2–25 µm | 2–15 µm | 5–25 × 0.5 µm | 2–40 µm | 2–10 µm | 2–25 µm | 2–30 µm |
| Gram stain | − | − | − | − | − | − | − | − | − | − |
| Gliding motility | + | + | + | + | + | + | + | + | + | + |
| Colony morphology | Circular | Circular | Circular | Circular | Circular | Circular | Circular | Circular | Circular | Circular |
| Color | Pale yellow | Bright Yellow | Pale yellow | Yellow | Pale yellow | Yellow | Brownish yellow | Pale yellow | Yellow | Pale yellow |
| Growth temp °C | 8–16b | 8–16b | 8–16b | 8–16b | 8–16b | 2–20 | 2–25 | 8–25 | 8–16b | 8–16b |
| Salinity range | ||||||||||
| NaCl | Nt | Nt | Nt | Nt | Nt | Ng | Ng | Ng | Nt | Nt |
| Seawater %a | Nt | Nt | Nt | Nt | Nt | 50–100 | 50–100 | 70–100 | Nt | Nt |
| pH range | Nt | Nt | Nt | Nt | Nt | 4–9 | 5–10 | Nt | Nt | Nt |
| Catalase | w | w | w | w | w | w | w | w | w | + |
| H2S | Nt | Nt | Nt | Nt | Nt | − | + | + | Nt | Nt |
| Antimicrobial drugs | ||||||||||
| Ceftazidime | s | r | s | s | r | s | s | r | r | s |
| Pipemidic acid | s | s | s | s | s | s | s | s | r | s |
| Penicillin G | s | r | s | s | r | s | s | r | r | s |
| Ampicillin | s | s | s | s | r | s | s | r | r | s |
| Growth on | ||||||||||
| l-tyrosin | Nt | Nt | Nt | Nt | Nt | − | − | + | Nt | Nt |
| d-glucose | Nt | Nt | Nt | Nt | Nt | − | − | + | Nt | Nt |
| Blood agar (2 % NaCl) | − | + | − | − | − | − | +c | − | +d | + |
| Hydrolization of | ||||||||||
| Tween 80 | Nt | Nt | Nt | Nt | Nt | − | + | + | Nt | Nt |
| API-ZYM | ||||||||||
| Esterase (C4) | + | + | + | + | + | + | + | − | + | − |
| Cystein arylamidase | + | + | + | + | + | + | + | − | − | − |
| Trypsin | − | + | − | − | + | − | − | + | − | − |
| N-acetyl-β-glucosaminidase | − | + | − | − | + | − | − | + | − | − |
| G+C (mol%) | Nt | Nt | Nt | Nt | Nt | 34.1 | 31.3 | 30.3 | Nt | Nt |
All data is from this study, except the DNA G+C contents of the two reference strains taken from Piñeiro-Vidal et al. (2012) and Suzuki et al. (2001)
+ positive, − negative, w weakly positive, Nt not tested, Ng no growth, r resistant, s susceptible. All strains are oxidase positive and indole negative
aPercent calculated using a relation of 100 % seawater = 38.2 g red sea salt L−1
bOnly tested at 8 and 16 °C
cInduces β-hemolysis or hemedigestion (CDC 2013) on blood agar containing 2 % NaCl
dInduces α-hemolysis on blood agar containing 2 % NaCl
Table 6.
Cellular fatty acid composition (%) of strain HFJT and T. dicentrarchi NCIMB 14598T
| Fatty acid | 1 | 2 |
|---|---|---|
| Straight chain | ||
| C14:0 | 1.3 | 1.0 |
| C15:0 | 1.8 | 3.8 |
| Branched chain | ||
| iso-C13:0 | 1.3 | 1.3 |
| iso-C14:0 | 1.6 | 2.5 |
| iso-C15:0 | 17.1 | 15.2 |
| anteiso-C15:1 | 17.7 | 13.3 |
| iso-C15:1 | 9.5 | 9.0 |
| anteiso-C15:1 | 1.9 | 1.9 |
| iso-C16:0 | Tr | 1.0 |
| iso-C16:1 | Tr | 2.8 |
| Unsaturated | ||
| C15:1ω6c | 3.3 | 3.1 |
| C16:1ω5c | 1.8 | 1.6 |
| C17:1ω6c | Tr | 2.0 |
| Hydroxylated | ||
| iso-C15:0 3-OH | 12.4 | 11.6 |
| C15:0 2-OH | 1.0 | 1.3 |
| C15:0 3-OH | 1.5 | 2.2 |
| iso-C16:0 3-OH | 2.9 | 5.3 |
| C16:0 3-OH | 4.0 | 3.8 |
| iso-C17:0 3-OH | 2.4 | 2.7 |
| Summed feature 3a | 9.5 | 10.3 |
Strains 1 HFJT, 2 T. dicentrarchi NCIMB 14598T
All data are from this study. Fatty acids amounting to <1 % of the total fatty acids in all strains are not shown. Tr, Trace (<1 %)
aSummed feature are groups of two or three fatty acids that cannot be separated by GLC using the MIDI system. Summed feature 3 comprises C16:1 ω7c and/or iso-C15:0 2-OH
Results from the phenotypic and chemotaxonomic tests show that strain HFJT differs significantly from T. dicentrarchi NCIMB 14598T and T. ovolyticum NCIMB 13127T (Table 5). The fatty acid composition analysis (Table 6) shows that strain HFJT has a very similar profile compared to that of T. dicentrarchi NCIMB 14598T. Moreover, the G+C content of strain HFJT is higher than those reported for T. dicentrarchi NCIMB 14598T and T. ovolyticum NCIMB 13127T. Strain HFJT and T. ovolyticum NCIMB 13127T do not grow on BAS, in contrast to T. dicentrarchi NCIMB 14598T. T. ovolyticum NCIMB 13127T is positive for the enzymes trypsin and N-acetyl-glucosaminidase, while strain HFJT and T. dicentrarchi NCIMB 14598T are negative. T. ovolyticum NCIMB 13127T was unique in being resistant to the antimicrobial drugs ceftazidime, penicillin G and ampicillin. The above mentioned differences further support strain HFJT as representative of a novel species in the genus Tenacibaculum. Cell length was the only characteristic shown to correspond to the three clades inferred in the phylogenetic analysis. Results showed a length of 2–40 µm for strain Tsp.4 and T. dicentrarchi NCIMB 14598T, 2–30 µm for strains HFJT, Tsp.2, and Tsp.5, and 2–15 µm for strain Tsp.6 and T. ovolyticum NCIMB 13127T.
In conclusion, the differential genetic, phylogenetic, phenotypic and chemotaxonomic data presented shows that strain HFJT should be classified as a novel species in genus Tenacibaculum, for which the name Tenacibaculum finnmarkense sp.nov. is proposed. This novel species also includes strains Tsp.2 and Tsp.5.
Description of Tenacibaculum finnmarkense sp. nov.
Tenacibaculum finnmarkense (finn.mark.en’se. N.L. neut.adj. finnmarkense of Finnmark, Norway, referring to the place of isolation).
Cells are strictly aerobic, Gram-stain negative, straight rods, 0.5 µm in diameter and 2–30 µm in length (filamentous cells >100 µm long may occur in older cultures) and motile by gliding. Degenerative spherical cells are observed in ageing cultures. Colonies on MA are circular, convex, pale yellow or yellow pigmented with translucent edges, have entire and/or undulating margins and a smooth texture with a shiny and sometimes nacreous appearance. The colonies are slightly viscous and do not stick to agar. Congo red absorption is negative. Growth occurs in media containing 50–100 % strength seawater but not in media supplemented with NaCl only. No growth occurs on BAS. Growth occurs at 2, 4, 8, 16 and 20 °C, but not at 25, 30 and 37 °C. Growth occurs at pH 4.0–9.0 (optimum pH 6–8). Catalase and cytochrome oxidase activities are present. Gelatin and casein are hydrolysed, but Tween 80 and starch are not. The Voges–Proskauer and flexirubin tests are negative. No anaerobic growth is observed. H2S and indole are not produced. l-Proline and l-glutamate are utilised but d(+)-sucrose, d(−)-ribose, d(+)-galactose, d(+)-glucose and l-tyrosine are not. The major fatty acids (>5 % of the total fatty acids) are summed feature 3 (comprising C16:1 ω7c and/or iso-C15:0 2-OH), iso-C15:0, anteiso-C15:0, Iso-C15:1 and iso-C15:0 3-OH. The respiratory quinone is menaquinone 6. The DNA G+C content of the type strain is 34.1 mol%.
The type strain is HFJT (=DSM 28541T = NCIMB 42386T), isolated from diseased Atlantic salmon (Salmo salar L.) in Norway. The GenBank accession number for the 16S rRNA gene sequence of strain HFJT is KT270385.
Acknowledgments
This study was partially funded by The Research Council of Norway (Project nr: 241364/O30) and Cermaq Norway.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Avendaño-Herrera R, Toranzo AE, Magariños B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis Aquat Org. 2006;71:255–266. doi: 10.3354/dao071255. [DOI] [PubMed] [Google Scholar]
- Bernardet J-F, Nakagawa Y, Holmes B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evolut Microbiol. 2002;52:1049–1070. doi: 10.1099/00207713-52-3-1049. [DOI] [PubMed] [Google Scholar]
- Bornø G, Lie LM. Fish health report 2014. Harstad: The Norwegain Veterinary Institute; 2015. [Google Scholar]
- Bruno DW, Noguera PA, Poppe TT. A color Atlas of salmonid diseases. 2. Dordrecht: Springer; 2013. [Google Scholar]
- Buck JD. Nonstaining (KOH) method for determination of gram reactions of marine bacteria. Appl Environ Microbiol. 1982;44:992–993. doi: 10.1128/aem.44.4.992-993.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashion P, Holder-Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- CLSI (2005) Methods for antimicrobial disc suseptibility testing of bacteria isolated from aquatic animals; proposed guidelines. Clinical and Laboratory standards institute M42-P Vol.25 No. 21
- Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fautz E, Reichenbach H. A simple test for flexirubin-type pigments. FEMS Microbiol Lett. 1980;8:87–91. doi: 10.1111/j.1574-6968.1980.tb05056.x. [DOI] [Google Scholar]
- Giovannoni SJ, Rappé MS, Vergin KL, Adair NL. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the Green Non-Sulfur bacteria. Proc Natl Acad Sci. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evolut Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2:60–61. [Google Scholar]
- Hansen GH, Bergh O, Michaelsen J, Knappskog D. Flexibacter ovolyticus sp. nov., a pathogen of eggs and larvae of Atlantic halibut, Hippoglossus hippoglossus L. Int J Syst Bacteriol. 1992;42:451–458. doi: 10.1099/00207713-42-3-451. [DOI] [PubMed] [Google Scholar]
- Huss VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Kim O-S, Cho Y-J, Lee K, Yoon S-H, Kim M, Na H, Park S-C, Jeon YS, Lee J-H, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kim Y-O, Park S, Nam B-H, Jung Y-T, Kim D-G, Jee Y-J, Yoon J-H. Tenacibaculum halocynthiae sp. nov., a member of the family Flavobacteriaceae isolated from sea squirt Halocynthia roretzi. Antonie Van Leeuwenhoek. 2013;103:1321–1327. doi: 10.1007/s10482-013-9913-5. [DOI] [PubMed] [Google Scholar]
- Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:1825. doi: 10.1099/ijs.0.064931-0. [DOI] [PubMed] [Google Scholar]
- Ley JD, Cattoir H, Reynaerts A. The Quantitative Measurement of DNA Hybridization from Renaturation Rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- López JR, Piñeiro-Vidal M, García-Lamas N, De La Herran R, Navas JI, Hachero-Cruzado I, Santos Y. First isolation of Tenacibaculum soleae from diseased cultured wedge sole, Dicologoglossa cuneata (Moreau), and brill, Scophthalmus rhombus (L.) J Fish Dis. 2010;33:273–278. doi: 10.1111/j.1365-2761.2009.01105.x. [DOI] [PubMed] [Google Scholar]
- LPSN (2015) List of prokaryotic names with standing in nomenclature. http://www.bacterio.net/tenacibaculum.html [DOI] [PMC free article] [PubMed]
- Nicholas KB, Nicholas HBJ, Deerfield DWI. GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- Olsen AB, Nilsen H, Sandlund N, Mikkelsen H, Rum H, Colquhoun DJ. Tenacibaculum sp. associated with winter ulcers in sea-reared Atlantic salmon Salmo salar. Dis Aquat Org. 2011;94:189–199. doi: 10.3354/dao02324. [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Carballas CG, Gómez-Barreiro O, Riaza A, Santos Y. Tenacibaculum soleae sp. nov., isolated from diseased sole (Solea senegalensis Kaup) Int J Syst Evol Microbiol. 2008;58:881–885. doi: 10.1099/ijs.0.65539-0. [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Riaza A, Santos Y. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int J Syst Evol Microbiol. 2008;58:21–25. doi: 10.1099/ijs.0.65397-0. [DOI] [PubMed] [Google Scholar]
- Piñeiro-Vidal M, Gijón D, Zarza C, Santos Y. Tenacibaculum dicentrarchi sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from European sea bass. Int J Syst Evol Microbiol. 2012;62:425–429. doi: 10.1099/ijs.0.025122-0. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer v1. 6. Available from http://beast.bio.ed.ac.uk/Tracer
- Reiner K (2010) Catalse Test Protocol. American Society For Microbiology. http://www.microbelibrary.org/library/laboratory-test/3226-catalase-test-protocol. 2014
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist FT, van der Mark M, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert MR, Krieg RN. Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood AW, Krieg RN, editors. Methods for general and molecular bacteriology. Washington D.C: American Society for Microbiology; 1994. [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- Suzuki M, Nakagawa Y, Harayama S, Yamamoto S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int J Syst Evol Microbiol. 2001;51:1639–1652. doi: 10.1099/00207713-51-5-1639. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution doi:10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed]
- Tanabe AS. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 2011;11:914–921. doi: 10.1111/j.1755-0998.2011.03021.x. [DOI] [PubMed] [Google Scholar]
- Tindall BJ, Rosselló-Móra R, Busse H-J, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- Toranzo AE, Magariños B, Romalde JL. A review of the main bacterial fish diseases in mariculture systems. Aquaculture. 2005;246:37–61. doi: 10.1016/j.aquaculture.2005.01.002. [DOI] [Google Scholar]
- Vold V (2014) Challenge experiment with field isolates of Tenacibaculum spp. isolated from moribound Atlantic salmon (Salmo salar L). Master thesis. University of Bergen, 2014
- Wakabayashi H, Hikida M, Masumura K. Flexibacter maritimus sp. nov., a Pathogen of Marine fishes. Int J Syst Bacteriol. 1986;36:396–398. doi: 10.1099/00207713-36-3-396. [DOI] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]


