Abstract
The chloroplast is the site of photosynthesis and many other essential plant metabolic processes, and chloroplast development is an integral part of plant growth and development. Mutants defective in chloroplast development can display various color phenotypes including the intriguing virescence phenotype, which shows yellow/white coloration at the leaf base and greening toward the leaf tip. Through large scale genetic screens, we identified a series of new virescent mutants including virescent3-1 (vir3-1), vir4-1, and vir5-1 in Arabidopsis thaliana. We showed that VIR3 encodes a putative chloroplast metalloprotease by map-based cloning. Through site-directed mutagenesis, we showed that the conserved histidine 235 residue in the zinc binding motif HEAGH of VIR3 is indispensable for VIR3 accumulation in the chloroplast. The chloroplast localization of VIR3 was confirmed by the transient expression of VIR3-GFP in leaf protoplasts. Furthermore, taking advantage of transgenic lines expressing VIR3-FLAG, we demonstrated that VIR3 is an intrinsic thylakoid membrane protein that mainly resides in the stromal lamellae. Moreover, topology analysis using transgenic lines expressing a dual epitope-tagged VIR3 indicated that both the N and C termini of VIR3 are located in the stroma, and the catalytic domain of VIR3 is probably facing the stroma. Blue native gel analysis indicated that VIR3 is likely present as a monomer or part of a small complex in the thylakoid membrane. This work not only implicates VIR3 as a new factor involved in early chloroplast development but also provides more insight into the roles of chloroplast proteases in chloroplast biogenesis.
Keywords: Arabidopsis thaliana, chloroplast, membrane protein, metalloprotease, photosynthesis
Introduction
Chloroplasts not only serve as the site of photosynthesis but also are responsible for the production of many essential metabolites in higher plants. Although a descendant of photosynthetic prokaryotic organisms through endosymbiosis, the chloroplast has evolved many unique features including complex regulatory networks to achieve the coordinated expressions of plastid genome and nuclear genome and the coordination of chloroplast development with leaf development (1, 2). Given the importance of the chloroplast, much research has been directed at dissecting the intricate pathways that regulate chloroplast development. One fruitful way has been the identification of mutants that are defective in nuclear genes for chloroplast proteins. Large scale genetic screens have revealed that mutants of nuclear genes for chloroplast proteins can give rise to myriads of visible leaf color phenotypes ranging from albino, yellow, and pale green to virescence and variegation (3).
Among the many types of mutants that are associated with chloroplast defects are the intriguing virescent mutants. These mutants display young and emerging tissues that are deficient of photosynthetic pigments, a sign that is often associated with underdeveloped plastids (2, 4). As leaves expand, however, these tissues gradually become greener along the leaf proximal-distal axis. At the whole plant level, the center regions of the mutants, mostly emerging tissues, are yellow or white, and the peripheral regions of mutants are greener, rendering a “center yellow” appearance. For many virescent mutants, the phenotype can become difficult to distinguish at late developmental stages because most of the yellow/white tissues turn green. Virescence is quite prevalent in higher plants and has long fascinated plant biologists. Early work has identified virescent mutants from many plant species including maize, cotton, tobacco, peanut, and bean, and genetic analyses have shown that both nuclear and chloroplast mutations can be responsible for the virescent phenotype (5–9).
In the past several years, a growing number of virescent mutants are characterized at the molecular level, especially in the model system Arabidopsis thaliana. At least two major categories of nuclear genes for chloroplast proteins are clearly linked with the virescent phenotype. The first group includes genes encoding subunits of the chloroplast ClpPRS protease complex (10, 11). Mutations in the ClpR1, ClpR2, or ClpR4 subunit can give rise to the virescent phenotype (12–15). The second group includes a number of genes coding for chloroplast pentatricopeptide repeat proteins. At least three such genes, DELAYED GREENING1 (DG1), YELLOW SEEDLINGS 1 (YS1), and ORGANELLE TRANSCRIPT PROCESSING 70 (OTP70) have a virescent mutant phenotype and are involved in regulating plastid transcription activities (16–18). In dg1 mutant, plastid-encoded plastid RNA polymerase transcribed gene expression is reduced, whereas nucleus-encoded plastid RNA polymerase-mediated gene expression is increased (16). ys1 mutation causes an abolished RNA editing site in rpoB transcript, which may lead to defective plastid-encoded plastid RNA polymerase activities (17). Similarly, in otp70, the splicing of rpoC1 is compromised, which also leads to abnormal plastid-encoded plastid RNA polymerase activities (18). In addition to these two categories of genes, virescent mutants have also been observed at rather high frequency in our screens for yellow variegated (var2) suppressors (19). One such suppressor line, ems2505, defines a leaky allele of an essential gene coding for a novel chloroplast protein that is indispensable for chloroplast development and plant survival (20). Identification of ems2505 suggests that a hypomorphic mutation of an essential gene can lead to virescence (20, 21). Virescent mutants were also isolated from a genetic screen for chlorophyll a/b-binding protein (CAB) underexpressed (cue) mutants in Arabidopsis (22). Virescent mutants were also identified in monocot species. For example, mutations in a chloroplast and mitochondrion dual-targeted guanylate kinase conditioned a temperature-sensitive virescent phenotype in rice (23, 24).
Although a growing body of evidence points to the virescent phenotype as a common indicator of chloroplast development defects, little is known regarding the underlying mechanism of this curious phenotype, and only limited effort has been directed at identifying such mutants (25). To gain more insight into the mechanism of virescence, we initiated genetic screens in our activation-tagged and EMS-mutagenized mutant populations to systematically look for Arabidopsis virescent mutants. Here we report the identification of three nonallelic virescent mutants, virescent3-1 (vir3-1), virescent4-1 (vir4-1) and virescent5-1 (vir5-1), following the existing naming system (25). These mutants exhibited yellow/white emerging leaves that gradually turned green as plants developed further. To explore the mechanism of virescence at the molecular level, we cloned the VIRESCENT3 (VIR3) locus and showed that it encodes a putative chloroplast zinc binding metalloprotease. Our results further showed that VIR3 is located in the thylakoid membranes. Our findings shed new light on the mechanisms of plant virescence and also the regulation of chloroplast development.
Experimental Procedures
Plant Materials and Growth Conditions
All Arabidopsis strains used in this study are of the Columbia-0 (Col) background. Arabidopsis seeds were planted on commercial soil mix (Pindstrup) or on half-strength Murashige and Skoog medium (Caisson Laboratories). The mitochondrion marker line has been described previously (26). Plants were maintained under continuous light (∼100 μmol m−2 s−1) at ∼22 °C.
DNA and RNA Techniques
Arabidopsis leaf DNAs were isolated using the CTAB method (27). Total leaf RNAs were purified using the TRIzol RNA reagent (Invitrogen). RNA gel and Northern blot analyses were performed as described (14). For semiquantitative RT-PCR analysis, cDNAs were synthesized from 1 μg of DNase I-treated total cellular RNA using a Transcriptor first strand cDNA synthesis kit following the manufacturer's instructions (Roche). All primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer name | Primer sequence | Notes |
|---|---|---|
| F13N6#1F | 5′-CGA CTC AGT GAA CTC CAA GT-3′ | Indel marker |
| F13N6#1R | 5′-ATT CGC TGC TGA ATT CGA GC-3′ | |
| F13N6#2F | 5′-CTG CAG CAA TTG CAC AGT CT-3′ | Indel marker |
| F13N6#2R | 5′-GCA CAA TCA CAC TAT CAG AG-3′ | |
| F14G9#2F | 5′-GCT ATT AGG TGC ATG ATG AGA ATT-3′ | dCAPs marker with EcoRI |
| F14G9#2R | 5′-TCT CTC TTA CTT CCT TCT CC-3′ | |
| T6H22#7F | 5′-CTG AGC TCA AAA ATA AGG GAG AAT-3′ | dCAPs marker with EcoRI |
| T6H22#7R | 5′-CCA GTT CCC AAA GAA ATT GG-3′ | |
| F14J16#4F | 5′-CTT TCT CTT TAG TCT CAA CTT TGA T-3′ | dCAPs marker with EcoRV |
| F14J16#4R | 5′-GGT CTT TAT CCC AAT CAA TC- 3′ | |
| 56180F | 5′-CAT TAT AGA GCA CCT CTA ATG GCT TTA TCT CC-3′ | Amplifying At1g56180 cDNA |
| 56180R | 5′-CAT TCT AGA ATA CCG AGA GCT TAT CTG CTT CG-3′ | |
| 21960F | 5′-CAT GGATCC acg atg aag cat tta tct gat tc-3′ | Amplifying At2g21960 cDNA |
| 21960R | 5′-CAT GGATCC tca tgt gaa aca att gag tgt tg-3′ | |
| 180CTPF | 5′-CATG GGATCC GCA CCT CTA ATG GCT TTA TCT CC-3′ | Constructing VIR3-cTP-GFP |
| 180CTPR | 5′-CATG CCAT GGA ACC ACC ACC ACC ACC ACC TCT CTT CAC AGC ATC TTC GTA-3′ | |
| VIR3GFPF | 5′-CAT TCT AGA GCA CCT CTA ATG GCT TTA TCT CC-3′ | Constructing VIR3-GFP |
| VIR3GFPR | 5′-CAT TCTAGA ACC ACC ACC ACC ACC ACC TTTGCTTGAAGACATGGCTTCCT-3′ | |
| VIR3-Flag-F | 5′-CCG CTC GAG ATG GCT TTA TCT CCG TCG TCT-3′ | Amplifying VIR3-FLAG |
| VIR3-Flag-R | 5′-CCG CTC GAG CTA CTT ATC ATC ATC ATC TTT ATA ATC TTT GCT TGA AGA CAT GGC TTC-3′ | |
| VIR3-H235L-R | 5′-AAG ATG ACC AGC TTC CAG AAC TAC GAT TCT-3′ | Site-directed mutagenesis |
| VIR3-H235L-F | 5′-AAG ATG ACC AGC TTC CAG AAC TAC GAT TCT-3′ | |
| HA-VIR3-F | 5′-ccgc tcg agT TAC CCA TAC GAT GTT CCA GAT TAC GCT GCA CTC AGG GAA TGG CGG GAG-3′ | Amplifying HA-VIR3-FLAG |
| ACT2F | 5′-TCAAAGACCAGCTCTTCCATCGAGA-3′ | RT-PCR internal control |
| ACT2R | 5′-ACACACAAGTGCATCATAGAAACGA-3′ |
Map-based Cloning of VIR3
Map-based cloning was conducted as described (28). In brief, vir3-1 was crossed with Landsberg erecta (Ler) to generate an F2 mapping population. Bulked segregant analysis first located VIR3 to a region adjacent to SSLP markers NGA280 and CIW1 on chromosome 1 (28, 29). To fine map vir3-1, a mapping population consisting of 1600 F2 mutant plants (3200 chromosomes) were screened by NGA280 and F13N6#1, two markers flanking the region bearing the mutation. New molecular markers were designed based on Indel or SNP polymorphisms between Ler and Col (Table 1) to narrow down the interval containing the mutation to a region of ∼63 kb.
Plasmid Constructions and Plant Transformation
Full-length cDNAs of At1g56180 and At2g21960 were amplified with the Pfu Turbo DNA polymerase (Agilent Technologies) using primers 56180F and 56180R, 21960F, and 21960R, respectively. VIR3-FLAG fragment was amplified using primers VIR3-Flag-F and VIR3-Flag-R. Each of the amplified fragments was ligated into pBlueScript KS+ (pBS), sequenced, and subcloned into a binary vector pBI111L (30) to generate P35S::At1g56180, P35S::At2g21960, and P35S::VIR3-FLAG, respectively.
Using pBS-At1g56180 as the template, VIR3 (H235L)-FLAG was generated by site-directed mutagenesis using two pairs of primers: VIR3-Flag-F with VIR3-H235L-R and VIR3-H235L-F with VIR3-Flag-R (31). After sequencing, VIR3 (H235L)-FLAG was subcloned into pBI111L. To generate HA and FLAG dual tagged VIR3, partial coding region of VIR3 excluding its chloroplast transit peptide (cTP, 1–46 amino acids) was amplified with primers HA-VIR3-F and VIR3-Flag-R, cloned into pBS, sequenced, and subcloned downstream of the cTP (1–71 amino acids) of chloroplast translation factor IF3 (At2g24060) in pBI111L.
vir3-1 was transformed with each of the binary vectors using the floral dip method (32). Transgenic lines were screened on 1/2 Murashige and Skoog plates containing 50 mg liter−1 kanamycin.
Bioinformatic Analysis
VIR3-like proteins were identified using the BlastP program at the National Center for Biotechnology Information website. Sequences of VIR3-like genes and their gene products were obtained from the Phytozome (33). The MEGA6 software was used to generate the phylogenetic tree (34). Transmembrane domains of VIR3 were predicted by the TMHMM program.
Whole Plant Chlorophyll Fluorescence Imaging and Starch Staining
The maximum quantum yield of photosystem II (FV/FM) was measured with Open FluorCam FC800-O (Photon Systems Instruments; Czech Republic) following the manufacturer's manual. In brief, 3-week-old wild type and vir3-1 plants were dark-adapted for 15 min before F0 (minimum fluorescence) was measured. 50% light intensity of Super Pulse was used to determine the FM (maximum fluorescence). FV/FM was calculated as (FM − F0)/FM.
Whole plant starch staining was carried out as described (35). In brief, plants were first boiled in 80% (v/v) ethanol to remove pigments and then stained with fresh made iodine solution (10 g liter−1 KI, 1 g liter−1 I2).
Electron Microscopy
Green and yellow leaf materials were collected from 14-day-old wild type and vir3-1 mutant seedlings. Sample preparation for transmission electron microcopy was carried out as described (20). Ultrathin sections (80 nm) were prepared with Ultramicrotome EM UC7 (Leica), stained with uranyl acetate and lead citrate, and examined with Hitachi H-600 transmission electron microscope (Hitachi) operated at 75 kV.
Protoplast Transient Expression Assay
To fuse GFP at the C terminus of VIR3cTP, the coding sequences for the N-terminal region of VIR3 (1–60 amino acids) were amplified using primers VIR3F and VIR3CTPR and cloned into pTF486 vector (14). The resulting construct was designated P35S::VIR3cTP-GFP. Similarly, the full-length VIR3 coding region was amplified using primers VIR3F and VIR3GFPR and cloned into pTF486 to generate P35S::VIR3-GFP. Protoplasts were isolated from rosette leaves of 4-week-old wild type Arabidopsis plants or transgenic Arabidopsis lines expressing a mitochondrion marker ScCOX4-mCherry (26). Transient expressions of GFP fusion proteins were carried out using a PEG-mediated method (36). GFP and chlorophyll autofluorescence signals were captured by an A1 confocal microscope (Nikon).
Chloroplasts Isolation and Fractionation
Three-week-old dark-adapted Arabidopsis plants were used for chloroplast isolation. Intact chloroplast isolation and separation of stroma and membrane fractions were carried out as in Refs. 37 and 38.
To separate thylakoid and envelope membranes, intact chloroplasts were broken by passing through a 24-gauge syringe in hypotonic HM buffer (10 mm Hepes-KOH, pH 7.6, 5 mm MgCl2). Chloroplast lysate was loaded on sucrose gradients (1.2, 1.0, or 0.46 m sucrose in hypotonic buffer) and centrifuged at 58,000 × g for 2 h (SW41 Ti rotor; Beckman). Thylakoids were collected from the bottom of the 1.2 m sucrose phase. The stroma fraction was recovered from the yellowish phase above the 0.46 m sucrose. Env1 (mixed envelope and thylakoids) and Env2 (mainly envelope) fractions were collected at the 1.2/1.0 m interface and the 1.0/0.46 m interface, respectively. Collected membranes were diluted at least three times with hypotonic buffer and centrifuged at 130,000 × g for 1 h (SW41 Ti rotor; Beckman).
To separate the stromal lamellae and the grana thylakoids, thylakoids were solubilized with 0.2% digitonin and subjected to differential centrifugation (39). NaCl, NaSCN, and Na2CO3 treatments were performed as described (40). Trypsin digestion was performed as described (41). All samples were normalized to equal amounts of chlorophyll prior to SDS-PAGE and immunoblot analyses.
Blue Native PAGE Analysis
BN-PAGE3 was performed as described (42). Briefly, thylakoid membranes equivalent to 10 μg of chlorophyll were solubilized in 10 μl of 25BTH20G containing 1.0% (w/v) of n-dodecyl-β-d-maltoside and resolved on 3–14% native PAGE with a constant voltage of 70 V at 4 °C overnight. For two-dimensional PAGE, hand-cut gel lanes were denatured in denaturing buffer (0.125 m Tris-HCl, pH 6.8, 4% SDS, and 1% β-mercaptoethanol) for 30 min and resolved on 12% SDS-PAGE gel.
Antibodies and Immunoblotting
To generate antibodies against VAR2/AtFtsH2, a thylakoid protein with known stroma facing the C terminus (43), the coding sequences for amino acid residues 238–665 of VAR2 was cloned into the bacterial expression vector pET28a. The resulting construct was transformed into Escherichia coli strain BL21 (DE3) to express the His6-VAR2238–665 recombinant protein. After affinity purification, His6-VAR2238–665 was used to raise polyclonal antibodies in rabbits. Monoclonal anti-FLAG was purchased from Sigma-Aldrich. Anti-HA was purchased from Abcam. Antibodies against various chloroplast proteins were purchased from Agrisera.
For immunoblot analysis, proteins separated by SDS-PAGE were transferred onto 0.45-μm nitrocellulose membranes. Blotted membranes were incubated with specific primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies. Signals were detected with Immun-Star chemiluminescence solutions and ChemiDoc Imaging System (Bio-Rad).
Results
The Isolation of Three New Virescent Mutants
We have carried out large scale ethyl methanesulfonate (EMS) and activation tagging mutagenesis looking for genetic suppressors of the Arabidopsis variegation mutant yellow variegated (var2) (44). During the screening process, we isolated a mutant that displayed both a leaf variegation phenotype typical of var2-5 and an additional virescent phenotype, i.e. the bases of young leaves were yellow, and leaf color gradually turned to green toward the leaf tips. Following a previous naming system for virescent mutants, we named this virescent mutation as virescent3–1 (vir3-1) (Fig. 1A and Ref. 25). Subsequently, the var2-5 mutation in the original mutant was removed by backcrossing with wild type, and further characterizations of vir3-1 were carried out in vir3-1 single mutant background. Two additional virescent mutants, vir4-1 and vir5-1, were recovered in an independent EMS mutagenesis (Fig. 1A). Genetic analysis indicated that the three mutants are nonallelic and are results of nuclear recessive mutations (data not shown). vir3-1 was characterized further in this study.
FIGURE 1.
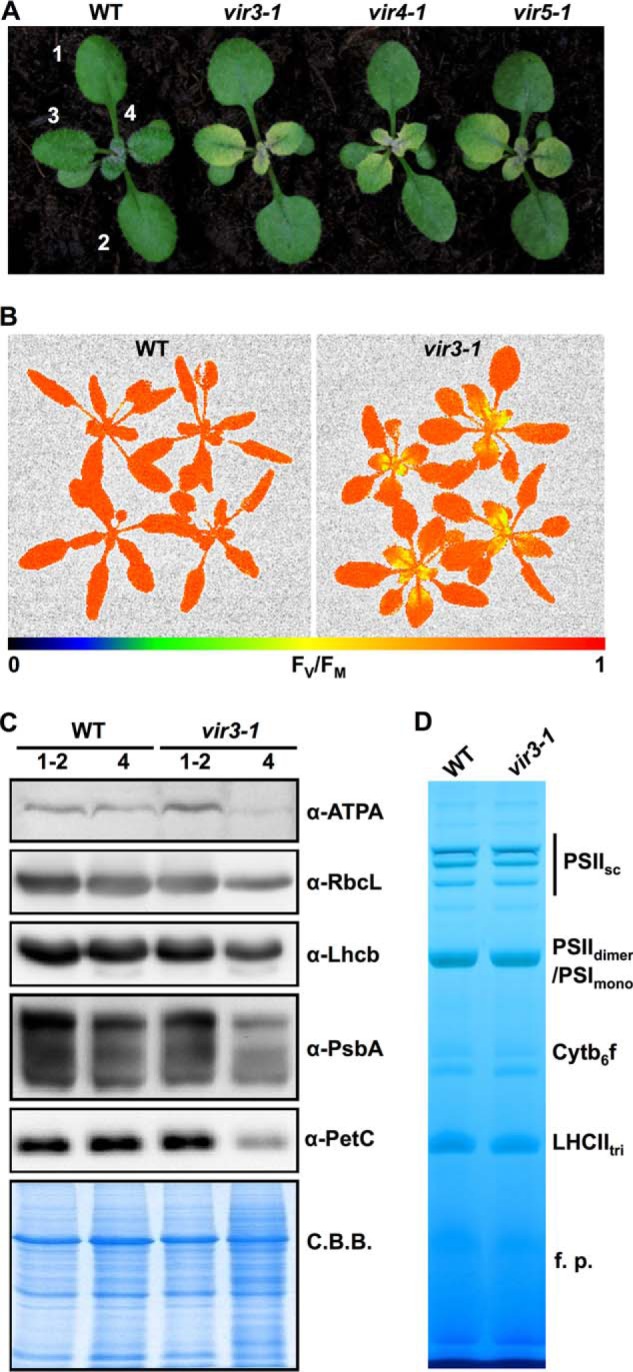
Phenotypes of three Arabidopsis virescent mutants. A, representative 2-week-old seedlings of wild type and three virescent mutants, vir3-1, vir4-1, and vir5-1. Plants were maintained under continuous light (∼100 μmol m−2 s−1) at 22 °C. Numbers indicate the order of true leaves. B, whole plant chlorophyll fluorescence imaging of 3-week-old wild type and vir3-1. Images were pseudocolored based on the FV/FM values. C, accumulations of chloroplast proteins in 2-week-old wild type and vir3-1. Total cellular proteins were purified from indicated leaf materials. Lanes 1-2, first two true leaves; lanes 4, the fourth true leaf. Protein samples were loaded on a fresh leaf weight basis and probed with indicated antibodies. Coomassie Brilliant Blue (C.B.B.)-stained gel was used for loading control. D, photosynthetic complexes accumulation in wild type and vir3-1. Thylakoids were isolated from 2-week-old whole seedlings of wild type and vir3-1. Thylakoid samples equivalent to 5 μg of chlorophyll were resolved on 3–14% BN-PAGE. PSIIsc, PSII supercomplexes; PSIIdi, PSII dimer; PSImono, PSI monomer; Cytb6f, cytochrome b6f complex; LHCtri, Light-harvesting complex II trimer; f.p., free proteins.
To assess the photosynthesis activities in vir3-1, we measured the maximum quantum yield of PSII (FV/FM) of 3-week-old wild type and vir3-1 using a whole plant chlorophyll fluorescence imaging system (Fig. 1B). The average FV/FM value of wild type plants was ∼0.84, whereas the FV/FM value of vir3-1 young yellow leaves was only ∼0.66 (Fig. 1B). On the other hand, the average FV/FM value of vir3-1 mature green tissues was 0.82. These results suggest that the quantum yield of PSII photochemistry was impaired in young vir3-1 leaves but was gradually recovered as vir3-1 leaves turned green (Fig. 1B).
Next, we compared the accumulations of individual photosynthetic proteins in wild type and vir3-1. Immunoblotting analyses were carried out using specific antibodies against subunits of key photosynthetic complexes in the chloroplast. We found that ATPA (the α-subunit of the chloroplast ATP synthase), RbcL (the large subunit of the ribulose-bisphosphate carboxylase/oxygenase), Lhcb (light-harvesting chlorophyll a/b binding protein), PsbA (D1 protein of PSII), and PetC (Rieske iron-sulfur subunit of the cytochrome b6f complex) accumulated to wild type levels in the green tissues of vir3-1 but were reduced in the yellow tissues of vir3-1 leaves (Fig. 1C). In addition, BN-PAGE analysis showed that major photosynthetic complexes, such as the PSII dimer, the photosystem I (PSI) monomer, and the light-harvesting complexes in vir3-1 accumulated to levels that are comparable with those of wild type (Fig. 1D). These observations suggest that chloroplast defects caused by vir3-1 closely correlated with mutant leaf developmental stages.
Starch Accumulation Is Impaired in vir3-1
To check the status of chloroplast development in vir3-1, the ultrastructures of plastids in wild type, vir3-1 green tissues and vir3-1 yellow tissues were examined with transmission electron microcopy (Fig. 2A). Compared with wild type, thylakoid structures including grana and stromal lamellae were not grossly affected in the green or the yellow tissues of vir3-1 mutant (Fig. 2A). Interestingly, starch granules were reduced in both the green and yellow tissues of vir3-1 compared with those of the wild type (Fig. 2A). To validate the observations of electron microcopy, we analyzed the starch accumulation in 3-week-old wild type and vir3-1 using the iodine staining method (Fig. 2B). The characteristic purple color of starch stained by iodine solution was observed in wild type but not in vir3-1 (Fig. 2B), confirming the starch accumulation defects in vir3-1. Taken together, these data indicate that although the chloroplasts in vir3-1 display seemingly normal ultrastructures, their photosynthetic capacities are probably dampened.
FIGURE 2.
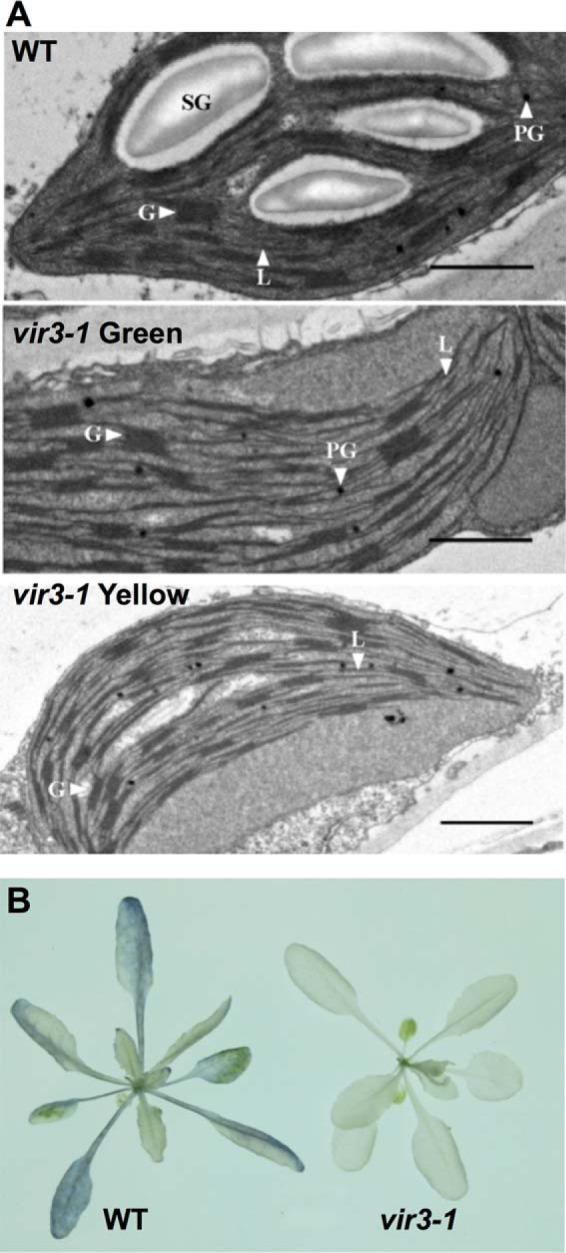
Starch accumulation defects in vir3-1. A, chloroplast ultrastructures of wild type and vir3-1. Chloroplast ultrastructure was obtained via transmission electron microcopy. All leaf materials were collected from 2-week-old plants. Leaf samples of wild type and vir3-1 green were collected from the first two true leaves, whereas vir3-1 yellow was collected from the yellow parts of young vir3-1 leaves. SG, starch granule; PG, plastoglobule; G, granum; L, lamellae. Bars, 1 μm. B, comparison of starch distribution in 3-week-old wild type and vir3-1. Pigments of 3-week-old plants were removed by ethanol, and starch was stained with iodine solution.
The Molecular Cloning of VIR3
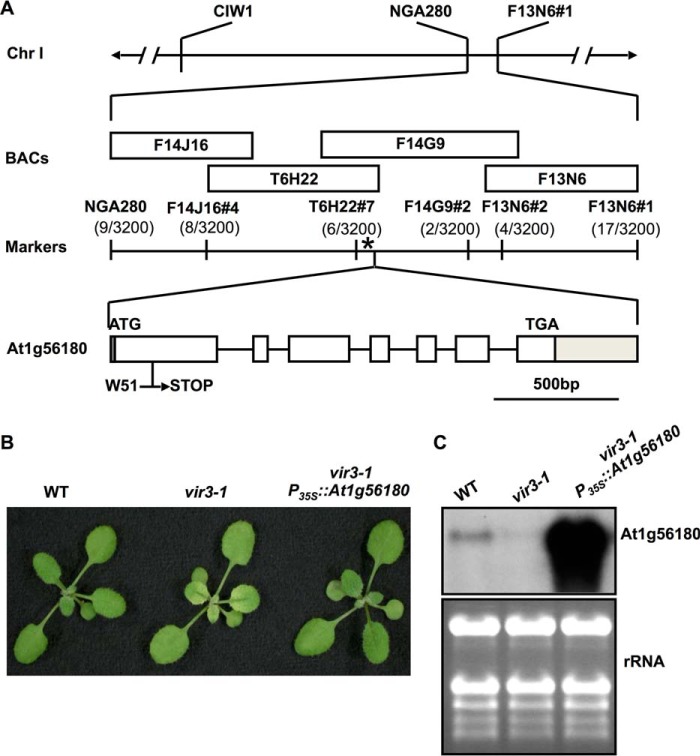
To identify the mutation in vir3-1, a mapping population was generated by crossing vir3-1 (in Col-0 background) with Ler. Bulked segregant analysis first linked the vir3-1 mutation to two SSLP markers, CIW1 and NGA280, on chromosome 1 (29) (Fig. 3A). Further fine mapping with additional molecular markers narrowed down the chromosomal region containing the vir3-1 mutation to an interval of ∼63 kb (Fig. 3A). Based on the virescent phenotype, we reasoned that VIR3 likely encodes a chloroplast protein. Genomic regions of three genes encoding putative chloroplast proteins located in the interval were sequenced. A single G to A transition was identified at the +153 position from ATG in At1g56180 in vir3-1. This mutation converts a tryptophan codon (TGG, Trp51) to a stop codon (TGA) (Fig. 3A). Furthermore, the overexpression of a wild type version of At1g56180 cDNA in vir3-1 background (vir3-1 P35S::At1g56180) fully complemented the virescent phenotype of vir3-1 (Fig. 3B). Northern blot analysis confirmed that At1g56180 transcripts were greatly reduced in vir3-1 and dramatically increased in one complementation line (Fig. 3C). Taken together, these data indicate that At1g56180 is VIR3.
FIGURE 3.
The molecular cloning of VIR3. A, schematic representation of the map-based cloning of VIR3. vir3-1 mutation was linked to markers CIW1 and NGA280 on chromosome 1. Numbers of recombinants are listed under each marker. The asterisk represents the position of VIR3 gene, At1g56180. Boxes represent exons, and solid lines represent introns in the gene model. The 5′- and 3′-UTRs are shaded. B, representative 2-week-old wild type, vir3-1, and a representative vir3-1 complementation line, designated vir3-1 P35S::At1g56180. C, RNA gel blot of At1g56180 mRNA accumulations in wild type, vir3-1, and the complementation line vir3-1 P35S::At1g56180. Total leaf RNAs were extracted from 14-day-old plants. Equal amounts of RNAs (3 μg) were loaded onto each lane. After electrophoresis and transfer, nylon membranes were probed with 32P-labeled At1g56180 cDNA sequences. Ethidium bromide-stained gel was shown as a loading control.
VIR3 Encodes a Putative Chloroplast Protease
VIR3 is annotated to encode a protein of 389 amino acids with unknown function in the Arabidopsis Information Resource. However, conserved domains/motifs searches revealed that VIR3 may contain two transmembrane domains and is likely a member of the metalloprotease M41 superfamily (45, 46). The signature zinc binding motif HEXXH for this type of metalloprotease is found near the C terminus of VIR3 (amino acids 235–239) (46). A prominent example of the chloroplast membrane localized M41 type of metalloprotease in higher plants is VAR2/AtFtsH2 (43). In contrast to VAR2/AtFtsH2, VIR3 lacks the ATP-binding domain.
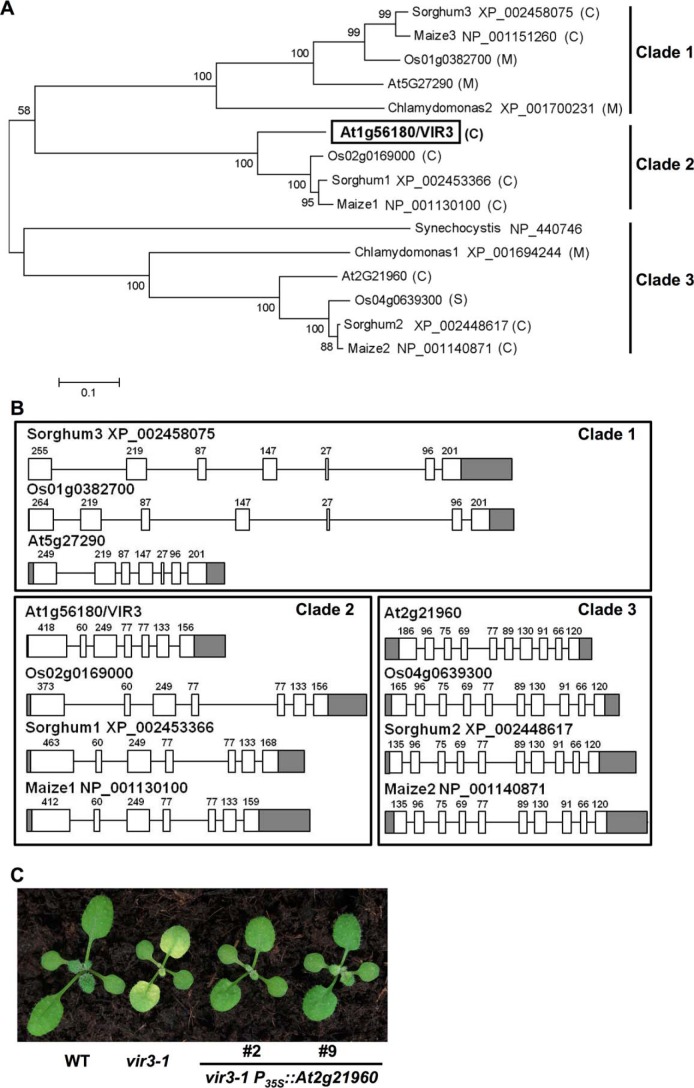
Next, we obtained sequences of VIR3 and VIR3-like proteins from the National Center for Biotechnology Information and analyzed their evolutionary relationship and putative subcellular localizations. Database searches showed that VIR3-like proteins are present in photosynthetic prokaryotic and eukaryotic organisms. Most VIR3-like proteins in higher plant species were predicted to be targeted to the chloroplast or the mitochondrion (47) (Fig. 4A). Phylogenetic analysis of VIR3-like proteins from representative photosynthetic species showed that VIR3-like proteins evolved into three distinct clades (Fig. 4A). Each clade has one VIR3 or VIR3-like homolog belonging to representative dicot (Arabidopsis) and monocot (rice, maize, and sorghum) plant species. Comparison of the gene structures of VIR3 and VIR3-like genes revealed that the number of exons and the length of each exon are highly conserved within each clade but are diversified between different clades (Fig. 4B). These analyses suggest that the three lineages of VIR3-like proteins were diverged before the separation of monocots and dicots.
FIGURE 4.
Conservation of VIR3 and VIR3-like genes. A, phylogenetic analysis of VIR3 and VIR3-like proteins. Full-length protein sequences from A. thaliana, Oryza sativa, Zea mays, Sorghum bicolor, Chlamydomonas reinhardtii, and Synechocystis sp. PCC6803 were obtained from the National Center for Biotechnology Information. National Center for Biotechnology Information GenBankTM accession numbers or gene codes are listed. The phylogenetic tree was constructed using the boot strap method with 1000 trials by MEGA6 (32). Putative subcellular localizations of VIR3 and VIR3-likes were predicted by TargetP (46). M, mitochondrion; C, chloroplast. B, structures of VIR3 and VIR3-like genes in different clades. Gene structures were drawn based on annotations in Phytozome. Exons and introns were represented by boxes and lines, respectively. Gray boxes represent the UTRs. Lengths of exons excluding the UTRs were marked on top of each box. C, phenotypes of 14-day-old wild type, vir3-1, and two independent lines that overexpressing At2g21960 in vir3-1 background.
At least two other VIR3-like proteins, At5g27290 and At2g21960, are present in Arabidopsis (Fig. 4A). Like VIR3, At2g21960 is a putative chloroplast localized protein, whereas At5g27290 is predicted to be targeted to the mitochondrion. To test whether there is functional redundancy between VIR3 and At2g21960, we overexpressed At2g21960 in the vir3-1 mutant. As shown in Fig. 4C, multiple overexpression lines showed greatly reduced virescence and greener leaves compared with that of vir3-1. These observations suggest that the overexpression of the close homolog of VIR3, At2g21960, could compensate for the loss of VIR3 gene, at least partially.
Histidine 235 Is Essential for VIR3 Accumulation
To test whether the putative zinc binding motif 235HEXXH239 is necessary for VIR3 functions, His235 was replaced by a leucine residue via site-directed mutagenesis, and a FLAG tag was added to the C terminus of this mutated form of VIR3 to generate P35S::VIR3 (H235L)-FLAG. As a control, a FLAG tag was also added to a wild type form of VIR3 (P35S::VIR3-FLAG). These two constructs were transformed into the vir3-1 mutant, respectively. Independent vir3-1 P35S::VIR3-FLAG lines showed green true leaves and accumulated VIR3-FLAG protein, suggesting that the C-terminal FLAG tag does not interfere with VIR3 functions and FLAG-tagged VIR3 is functional in plants (Fig. 5, A and B). In contrast, vir3-1 P35S::VIR3 (H235L)-FLAG lines showed leaf virescence similar to that of vir3-1 (Fig. 5C). In addition, the accumulation of VIR3 (H235L)-FLAG protein was nondetectable despite the dramatically increased VIR3 (H235L)-FLAG transcripts in these lines (Fig. 5, D and E). These results indicate that the putative zinc binding site of VIR3 is indispensable for its function and/or stability.
FIGURE 5.
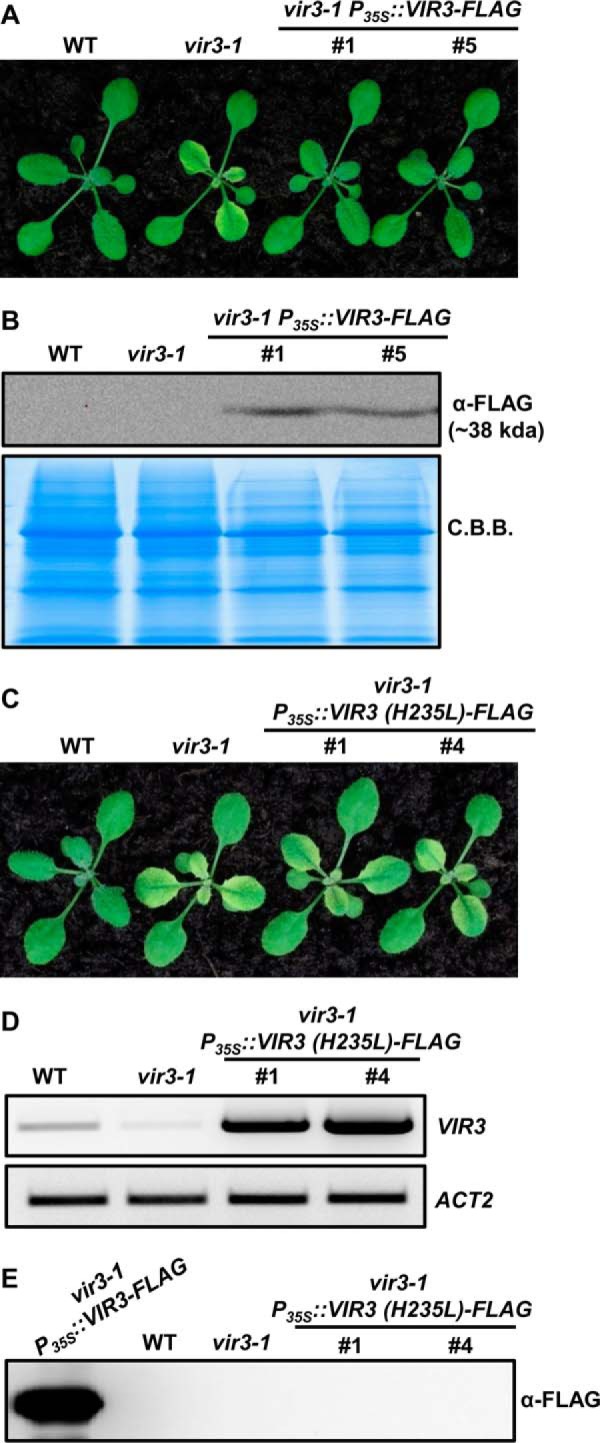
His235 is essential for VIR3 accumulation. A, 14-day-old seedlings of wild type, vir3-1, and two independent vir3-1 P35S::VIR3-FLAG lines. B, immunoblot analysis of VIR3-FLAG levels in plants shown in A. Total protein was extracted from 14-day-old seedlings and resolved on 12% SDS-PAGE gel. Coomassie Brilliant Blue (C.B.B.)-stained gel served as a loading control. C, 14-day-old seedlings of wild type, vir3-1, and two independent vir3-1 P35S::VIR3 (H235L)-FLAG lines. D, semiquantitative RT-PCR analysis of VIR3 transcripts levels in plants shown in C. Expression of ACT2 was used as a control. E, immunoblot analysis of the accumulation of VIR3 (H235L)-FLAG in plants shown in C. A protein sample of vir3-1 P35S::VIR3-FLAG plant was included as a positive control.
VIR3 Is a Chloroplast Protein
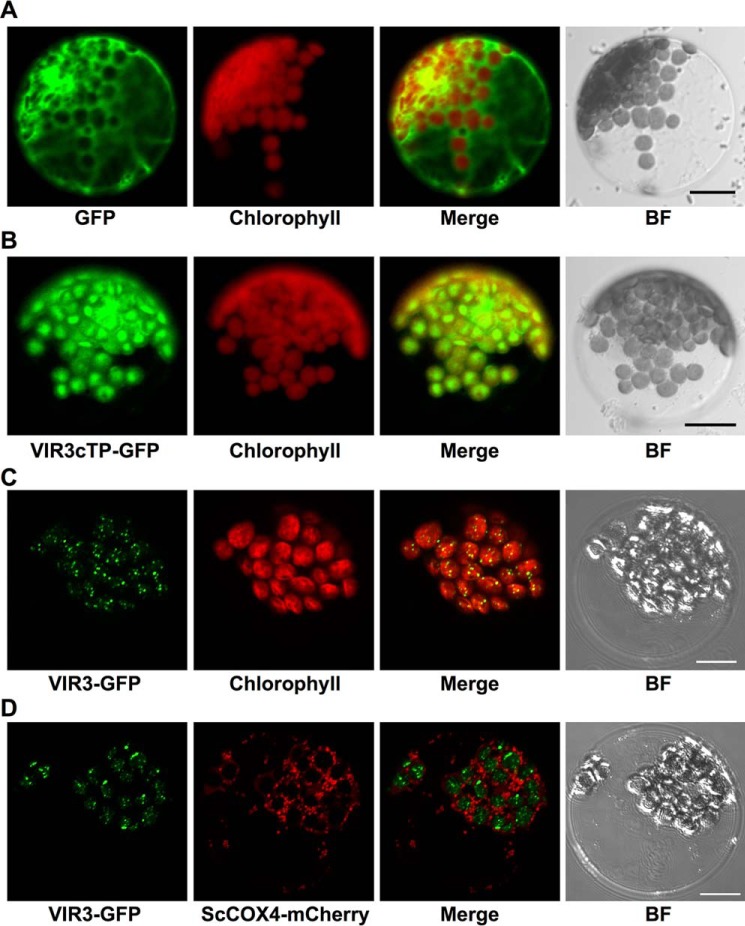
To confirm the chloroplast localization of VIR3, VIR3 N-terminal region (1–60 amino acids, harboring the putative cTP) or the full-length VIR3 was fused with GFP and transiently expressed in Arabidopsis leaf protoplasts. Although the expression of the control vector (P35S::GFP) leads to a distribution of GFP signals in the cytoplasm and the nucleus (Fig. 6A), VIR3cTP-GFP signals overlapped nicely with the chlorophyll autofluorescence, suggesting that VIR3cTP can direct GFP into chloroplasts (Fig. 6B). Next, we fused GFP at the C terminus of the full-length VIR3 protein and expressed the VIR3-GFP fusion protein in leaf protoplasts. VIR3-GFP appeared as discrete foci (Fig. 6C). Merged images of VIR3-GFP and chlorophyll autofluorescence showed that all VIR3-GFP signals located inside chloroplasts (Fig. 6C). To test whether VIR3 could also be targeted to the mitochondrion, we expressed VIR3-GFP in protoplasts isolated from transgenic plants expressing ScCOX4-mCherry, a red fluorescent protein-tagged mitochondrion marker (Fig. 6D) (26). As shown in Fig. 6D, no overlapped signals of VIR3-GFP and ScCOX4-mCherry were observed, indicating that VIR3-GFP is likely not targeted to the mitochondrion. Taken together, our results indicate that VIR3 is a chloroplast protein.
FIGURE 6.
Subcellular localization of VIR3. A, transient expression of GFP in wild type leaf protoplasts. B, transient expression of VIR3cTP-GFP in wild type leaf protoplasts. C, transient expression of VIR3-GFP fusion protein in wild type leaf protoplasts. D, transient expression of VIR3-GFP fusion protein in protoplasts isolated from plants stably expressing a mitochondrion marker ScCOX4-mCherry. Fluorescent signals were monitored with confocal microscopy using the following emission filter settings: GFP, 500–550 nm; ScCOX4-mCherry, 570–620 nm; and chlorophyll autofluorescence, 663–738 nm. A single representative protoplast is shown for each transformation. BF, bright field. Bars, 10 μm.
VIR3 Is a Thylakoid Membrane Protein
We next investigated the subchloroplast localization of VIR3. Intact chloroplasts from vir3-1 P35S::VIR3-FLAG complementation lines were isolated and separated into the soluble stroma fraction and the membrane fraction (Fig. 7A). As expected, RbcL was detected largely in the soluble stroma fraction, whereas PSII light-harvesting complex Lhcb was present in the membrane fraction (Fig. 7A). Immunoblotting with FLAG antibody showed the presence of VIR3-FLAG signals in the intact chloroplasts and the membrane fraction, but not in the stroma fraction, similar to Lhcb signals. These results suggest that VIR3 is associated with chloroplast membranes.
FIGURE 7.
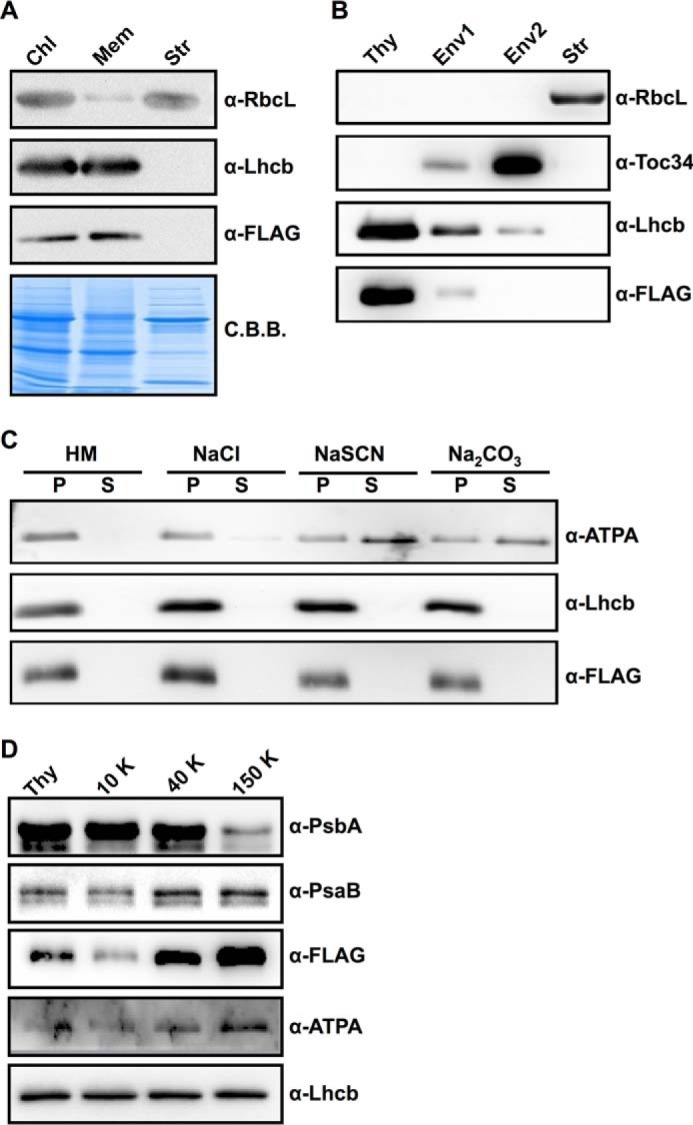
Subchloroplast localization of VIR3. A, protein samples of intact chloroplasts (Chl), stroma (Str), and chloroplast membranes (Mem) from vir3-1 P35S::VIR3-FLAG lines were normalized to equal amounts of chlorophyll and probed with indicated antibodies. C.B.B., Coomassie Brilliant Blue. B, protein samples of thylakoids (Thy), mixed envelope and thylakoids (Env1), mainly envelope (Env2), and stroma (Str) fractions from vir3-1 P35S::VIR3-FLAG lines were normalized to equal amounts of chlorophyll and probed with indicated antibodies. RbcL and Lhcb were used as stroma and thylakoid controls, respectively. Toc34 was used as a control for chloroplast envelope protein. C, thylakoids isolated from vir3-1 P35S::VIR3-FLAG lines were treated with 2 m NaCl, 2 m NaSCN, or 0.1 m Na2CO3. HM, thylakoids in Hepes-KOH MgCl2 buffer. After incubation and centrifugation, supernatants (S) and membrane fractions (P) were resolved on SDS-PAGE for immunoblotting with indicated antibodies. Lhcb and ATPA were used as the integral membrane protein and the peripheral membrane protein controls, respectively. D, thylakoid membranes from vir3-1 P35S::VIR3-FLAG lines were fractionated to grana thylakoids (10,000 × g, 10 K), intermediate membranes (40,000 × g, 40 K), and stromal lamellae (150,000 × g, 150 K) with differential centrifugation after digitonin treatment. Each fraction was normalized to an equal amount of chlorophyll and probed with indicated antibodies. PsaB and ATPA were used as controls for proteins enriched in the stromal lamellae, whereas PsbA was used as a control for proteins enriched in the grana thylakoids.
To distinguish whether VIR3 resides in the thylakoid membrane or in the chloroplast envelope, intact chloroplasts were further fractionated into thylakoids, Env1 (mixed thylakoid and envelope membranes), Env2 (mostly envelope membranes), and stroma fractions via sucrose gradient centrifugation. As expected, RbcL was only detected in the stroma fraction (Fig. 7B). Lhcb was detected largely in the thylakoid fraction (Fig. 7B). Toc34, a subunit of the chloroplast translocon at the chloroplast envelope membrane, were enriched in the Env2 fraction (Fig. 7B). VIR3-FLAG was mainly detected in the thylakoid fraction, and trace amounts of VIR3-FLAG could also be detected in the Env1 fraction, a pattern of signals similar to that of Lhcb (Fig. 7B). In contrast, neither in the stroma fraction nor in the Env2 fraction could VIR3-FLAG be detected (Fig. 7B). These results suggest that VIR3 is likely a thylakoid membrane protein.
To further confirm whether VIR3 is an integral membrane protein, thylakoids of P35S::VIR3-FLAG lines were incubated with different salt solutions. Thylakoid-associated proteins, such as ATPA, can be effectively removed from the thylakoids by chaotropic salts or the alkaline pH (Fig. 7C). In contrast, intrinsic membrane proteins, such as Lhcb, remained in the thylakoids after the treatments (Fig. 7C). VIR3-FLAG was retained in the thylakoid membrane fractions after the washes, similar to Lhcb (Fig. 7C). These data indicate that VIR3 is an intrinsic thylakoid membrane protein (Fig. 7C).
To check whether VIR3 is localized to specific regions of thylakoids, digitonin-treated thylakoids were separated by ultracentrifugation into different fractions enriched in grana thylakoids (10,000 × g) or stromal lamellae (150,000 × g). Fig. 7D showed that the PSII reaction center protein PsbA was enriched in grana thylakoids as reported (48). On the other hand, the PSI reaction center protein PsaB and the chloroplast ATP synthase subunit ATPA were enriched in the stromal lamellae (Fig. 7D). VIR3-FLAG was enriched in the stromal lamellae, resembling the patterns of PsaB and ATPA (Fig. 7D), suggesting that the majority of VIR3 localizes to the stromal lamellae.
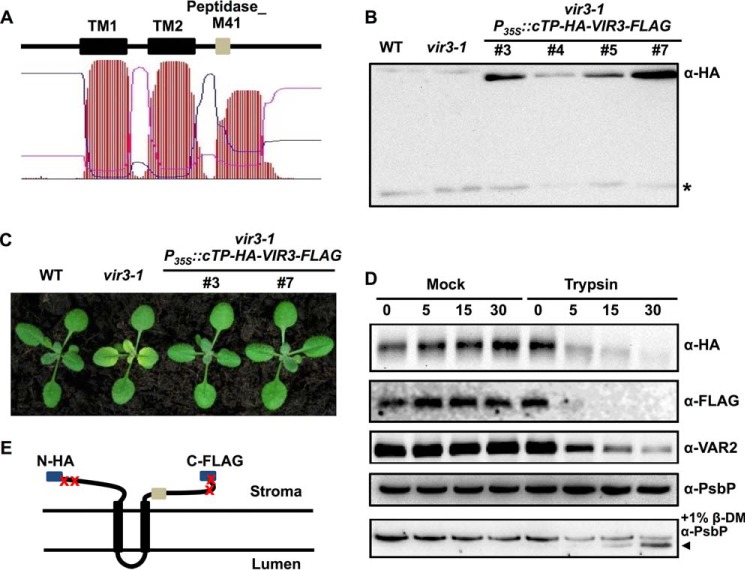
The Catalytic Domain of VIR3 Is Likely Facing the Stroma
VIR3 contains two predicted transmembrane domains (TM1 and TM2) and an additional hydrophobic region harboring the peptidase M41 domain (Fig. 8A). To determine the membrane topology of VIR3, a construct expressing a modified version of VIR3 with a N-terminal HA tag (after a cTP) and a C-terminal FLAG tag was generated (P35S::cTP-HA-VIR3-FLAG) and introduced into the vir3-1 mutant. Accumulation of HA-VIR3-FLAG proteins was detected in multiple transgenic lines (Fig. 8B). Moreover, the virescent phenotype of vir3-1 was fully complemented by the transgene, indicating that the dual epitope-tagged HA-VIR3-FLAG is functional (Fig. 8C). Thylakoids of vir3-1 P35S::cTP-HA-VIR3-FLAG lines were treated with trypsin-treated or mock treated with buffer. Both the N-terminal HA and the C-terminal FLAG signals became undetectable after trypsin treatment, indicating that trypsin is able to access and degrade both epitopes (Fig. 8D). To verify the trypsin treatment results, we also monitored the degradation of VAR2/AtFtsH2, a known stroma-exposed thylakoid protein, using an antibody raised against the C-terminal stromal region of VAR2/AtFtsH2 (anti-VAR2) (43). As shown in Fig. 8D, the changes in VAR2 signals were similar to those of HA and FLAG, thus confirming that the HA and FLAG tags were exposed to the stroma. On the contrary, PsbP (OEC23 subunit of the oxygen-evolving complexes), a thylakoid lumen protein, was protected by the thylakoid membranes and was not degraded (Fig. 8D). When the detergent n-dodecyl-β-d-maltoside was added to solubilize thylakoids, PsbP also became accessible to trypsin (Fig. 8D). Meanwhile, in samples containing mock treated thylakoids, levels of HA, FLAG, VAR2, and PsbP all remained stable over the time points we examined, confirming that there are no endogenous proteolytic activities in our assays (Fig. 8D). Together, these results suggest that both the N and C termini of VIR3 are facing the stroma. Based on these findings, a possible model of VIR3 topology in the thylakoids was proposed (Fig. 8E). In this model, VIR3 is an intrinsic membrane protein with two transmembrane domains: TM1 and TM2, and the proteolytic domain bearing the putative zinc binding motif is located in the stroma.
FIGURE 8.
Membrane topology of VIR3. A, prediction of the hydrophobic regions in VIR3 using TMHMM program. TM1 and TM2 indicated two putative transmembrane domains. The Peptidase_M41 domain was in the third hydrophobic region. B, immunoblot analysis of the accumulation of HA-VIR3-FLAG in four independent P35S::cTP-HA-VIR3-FLAG lines using the anti-HA antibody. Wild type and vir3-1 served as negative controls. The asterisk marks a band that is due to a nonspecific cross reaction with the HA antibody in Arabidopsis. C, phenotypes of 14-day-old wild type, vir3-1, and two independent vir3-1 P35S::cTP-HA-VIR3-FLAG lines. D, thylakoids isolated from P35S::cTP-HA-VIR3-FLAG lines were treated with trypsin or mock treated with buffer for 5, 15, and 30 min. Samples were analyzed by immunoblots probed with indicated antibodies. To expose PsbP, thylakoids were solubilized with 1% n-dodecyl-β-d-maltoside. The degradation product of PsbP was labeled with the black arrowhead. E, model of the possible topology of VIR3 on thylakoid membranes. Both N-terminal HA and C-terminal FLAG were accessible to trypsin as shown in D. The Peptidase_M41 domain (gray box) was on the stromal side of the thylakoid membrane.
Detection of VIR3 in the Thylakoid Membrane with BN-PAGE Analysis
To determine whether VIR3 forms protein complexes, two-dimensional BN-PAGE was performed with thylakoid fractions isolated from vir3-1 P35S::VIR3-FLAG lines. BSA (∼66 kDa) and β-casein (∼26 kDa) were included as the internal molecular weight controls (Fig. 9). Migrations of photosynthetic complexes, such as PSI monomer, light-harvesting complex II, and cytochrome b6f in the BN-gel were monitored with antibodies against PsaB, Lhcb, and PetC, respectively. VIR3-FLAG (∼38 kDa) was detected at a position between BSA and β-casein in the second dimension BN-PAGE gel (Fig. 9), suggesting that VIR3 functions as a monomer or forms a small complex in the thylakoid membrane.
FIGURE 9.
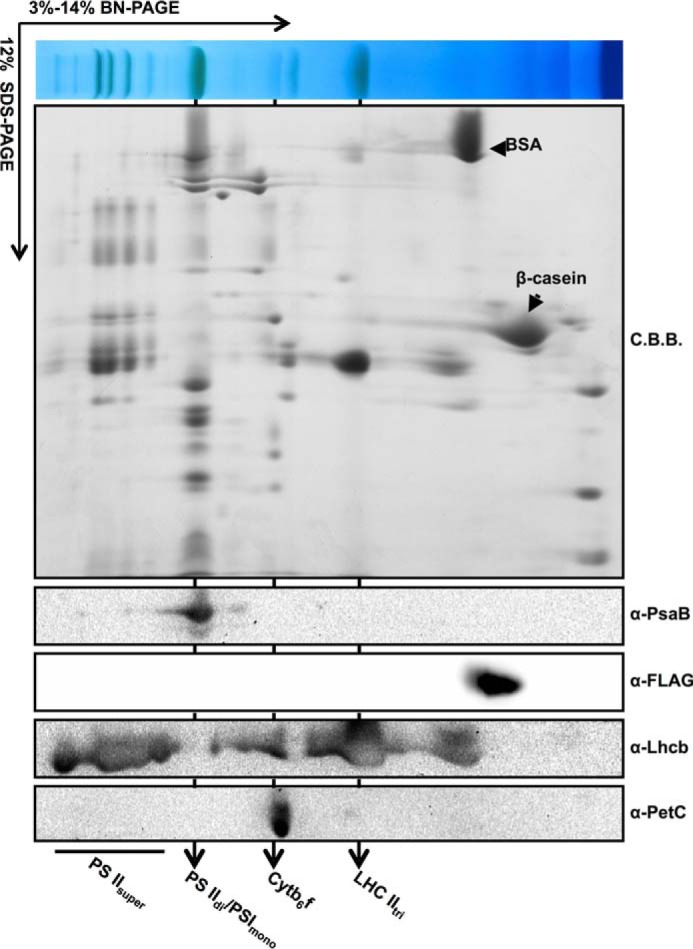
Detection of VIR3 in thylakoid membranes with BN-PAGE analysis. Solubilized thylakoids of P35S::VIR3-FLAG lines were resolved with 3–14% BN-PAGE. β-Casein and BSA were added as internal molecular weight controls. Individual first dimension lanes were subjected to 12% SDS-PAGE gel and probed with indicated antibodies. Major photosynthetic complexes were labeled as in Fig. 1D. C.B.B., Coomassie Brilliant Blue.
Discussion
In the plant cell, chloroplasts are important metabolic hubs where many essential reactions such as photosynthesis, lipid, amino acid, and hormone biosynthesis take place. Given its importance, it came as no surprise that the development of a functional chloroplast is regulated by a large number of genetic factors, as well as environmental cues (1). When functions of these regulatory factors are compromised through mutations, mutant plants may display visual defects that can be identified as one of a spectrum of leaf color phenotypes ranging from albino and pale green to virescent, reticulated, or variegated (22, 49). We are interested in dissecting the genetic regulation of chloroplast development and have carried out screens looking for components that are involved in chloroplast biogenesis (14). Here we report the isolation of three Arabidopsis mutants with unique chloroplast developmental defects, virescent3-1 (vir3-1), vir4-1, and vir5-1 (Fig. 1). In vir3-1 mutants, young leaves and leaf bases show chlorosis and abnormal chloroplast protein accumulation, whereas green tissues do eventually form in leaf tips and mature leaves (Fig. 1). We demonstrate that VIR3 codes for a chloroplast thylakoid membrane localized putative zinc-binding metalloprotease. The virescent phenotype is intriguing in that the leaf color phenotype is not uniformly distributed, i.e. different leaf tissues can have distinct chloroplast developmental stages despite a uniform mutant genetic background. In higher plants, chloroplasts are developed from proplastids in the shoot apical meristem, and this process coincides with the formation of leaves from leaf primordia. It is generally believed that leaf bases are the areas where cells are still undergoing rapid cell divisions, whereas toward the tip region, cell division ceases, and cell expansion accounts for the enlargement of the leaf blade (50). Considering that the color pattern of the virescent phenotype matches well with the pattern of leaf cell division, it is possible that leaf development and chloroplast development may be coupled to a certain extent in higher plants. First, it is conceivable that at early stages of leaf development, rapid cell divisions pose a greater demand for certain metabolic processes in chloroplasts and in turn for certain gene products, such as VIR3. As cell division slows down, the need for these gene products decreases. This scenario would be reminiscent of the threshold model for the var2-mediated leaf variegation (30). Alternatively, there might be compensating activities inside the leaves that enable chloroplasts to form in the absence of VIR3 activities, especially in tissues with slower cell divisions. Functional complementation of vir3-1 by the homolog of VIR3, At2g21960, suggests that At2g21960 might be a possible source of the compensating activities for the lack of VIR3 (Fig. 4C). The functional redundancy of At2g21960 and VIR3 could also contribute to the relatively mild phenotype of vir3-1.
Chloroplasts are believed to be evolved from ancient photosynthetic cyanobacteria through endosymbiosis (2). One consequence of endosymbiosis is the need to coordinate the nuclear genome and the chloroplast genome at many levels. At the protein level, modern-day chloroplasts can only translate a small portion of its proteome, whereas the majority of the chloroplast proteome has to be translated in the cytosol, transported into the chloroplast, and often assembled with protein subunits translated inside the chloroplast. This complex interplay necessitates the employment of an array of proteases to ensure protein quality control and homeostasis (51). Counterparts of almost all prokaryotic protease systems, such as the ATP-dependent Clp protease, the ATP-dependent FtsH protease, and the Deg serine peptidase, have been identified in higher plant chloroplasts (10, 51, 52, 54). A growing list of additional chloroplast localized proteases, including SPP, PreP, CND41, TPP, EGY1/2, and Ctp, has been identified (53, 54–62). In this work, we identified a putative new chloroplast protease VIR3. Although VIR3 is not found in searches against the chloroplast proteomic databases such PPDB and AtChloro, the chloroplast localization of VIR3 was verified by transient expression of VIR3-GFP fusion protein in leaf protoplasts and by biochemical identification of VIR3-FLAG in isolated chloroplasts (Figs. 6 and 7). The lack of proteomic identification of VIR3 may indicate a low level of endogenous VIR3.
VIR3 possesses a conserved zinc-binding motif 235HEAGH239, indicating that VIR3 may be a zinc metalloprotease (45). We were unsuccessful in trying to establish in vitro protease activities of either the refolded His6-VIR3 recombinant protein or the soluble GST-VIR3 using BSA or β-casein as substrates (data not shown). However, substitution of His235 with a leucine residue abolishes the accumulation of VIR3 (Fig. 5), suggesting that His235 and the integrity of the zinc binding site are critical for VIR3 function and/or stability.
Chloroplast proteases have been identified at all subchloroplast compartments, including the envelope, the stroma, the thylakoid membrane, and the lumen. For instance, the Clp protease complex, which shares a similar structure with the eukaryotic 26S proteasome, is localized to the stroma, whereas EGY1 and FtsH are integral thylakoid membrane proteins (10, 61). Our biochemical evidence supports the notion that VIR3 exerts its functions in the thylakoid membranes. First, epitope-tagged VIR3 was detected in the thylakoid membrane fraction and is resistant to salt washes, consistent with the presence of transmembrane domains in VIR3 (Fig. 7, A–C). In addition, further thylakoid fractionation assay indicated that VIR3 is more abundantly localized in the stromal lamellae (Fig. 7D). Second, trypsin protection assays showed that both the N and C termini of VIR3 are likely located in the stroma (Fig. 8). Based on the biochemical evidence and the bioinformatics analysis, we propose a model for the topology of VIR3 in the thylakoid membrane (Fig. 8). In our model, VIR3 has two transmembrane domains, and the potential zinc-binding catalytic site HEAGH is likely situated in the stroma (Fig. 8). This is in contrast to EGY1, a member of the M50 metalloprotease family, in which the catalytic site HEXXH is trapped in the thylakoids as a part of the transmembrane domain (61). Lastly, unlike FtsH protease, which forms complexes in the thylakoid membrane, we showed that VIR3 is probably a monomer or part of small complexes in the thylakoid membrane (Fig. 9). At present, one of the biggest challenges regarding VIR3, as well as other proteases, is the determination of its natural proteolytic substrates. As more progress is made toward this end, it will become possible in the future to deduce the exact roles these proteases play in chloroplast protein homeostasis and development.
Author Contributions
F. Y. conceived and coordinated the study. X. L. and Y. L. designed, performed, and analyzed the experiments shown in Figs. 1–3. Y. Q. and J. Z. designed, performed, and analyzed the experiments shown in Figs. 5, 7, 8, and 9. S. L. designed, performed, and analyzed the experiments shown in Fig. 4. R. W. and S. L. designed, performed, and analyzed the experiments shown in Fig. 6. J. S. and L. A. provided technical assistance and contributed to the preparation of the figures. F. Y., Y. Q., and X. L. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by National Natural Science Foundation of China Grants 31170219 (to F. Y.) and 31300988 (to Y. Q.), by Natural Science Foundation of Shaanxi Province Grants 2014JQ3093 (to J. S.), and by Fundamental Research Funds for the Central Universities Grants QN2012022 (to Y. Q.), QN2013034 (to J. S.), and 2014YB036 (to L. A.). The authors declare that they have no conflicts of interest with the contents of this article.
- BN
- blue native
- PSII
- photosystem II
- PSI
- photosystem I
- EMS
- ethyl methanesulfonate.
References
- 1. Pogson B. J., and Albrecht V. (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarvis P., and López-Juez E. (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 [DOI] [PubMed] [Google Scholar]
- 3. Bryant N., Lloyd J., Sweeney C., Myouga F., and Meinke D. (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155, 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopez-Juez E., and Pyke K. A. (2005) Plastids unleashed: their development and their integration in plant development. Int. J. Dev. Biol. 49, 557–577 [DOI] [PubMed] [Google Scholar]
- 5. Archer E. K., and Bonnett H. T. (1987) Characterization of a virescent chloroplast mutant of Tobacco. Plant Physiol. 83, 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kay R. E., and Phinney B. O. (1956) The control of plastid pigment formation by a virescent gene, Pale-Yellow-1, of maize. Plant Physiol. 31, 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benedict C. R., and Ketring D. L. (1972) Nuclear gene affecting greening in virescent peanut leaves. Plant Physiol. 49, 972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benedict C. R., and Kohel R. J. (1968) Characteristics of a virescent cotton mutant. Plant Physiol. 43, 1611–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Polacco M. L., Chang M. T., and Neuffer M. G. (1985) Nuclear, virescent mutants of Zea mays L. with high levels of chlorophyll (a/b) light-harvesting complex during thylakoid assembly. Plant Physiol. 77, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adam Z., Rudella A., and van Wijk K. (2006) Recent advances in the study of Clp, FtsH, other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9, 234–240 [DOI] [PubMed] [Google Scholar]
- 11. Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P. D., and van Wijk K. J. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21, 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koussevitzky S., Stanne T. M., Peto C. A., Giap T., Sjögren L. L., Zhao Y., Clarke A. K., and Chory J. (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63, 85–96 [DOI] [PubMed] [Google Scholar]
- 13. Rudella A., Friso G., Alonso J. M., Ecker J. R., and van Wijk K. J. (2006) Down regulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18, 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu F., Liu X., Alsheikh M., Park S., and Rodermel S. (2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20, 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu W., Zhu Y., Ma Z., Sun Y., Quan Q., Li P., Hu P., Shi T., Lo C., Chu I. K., and Huang J. (2013) Proteomic evidence for genetic epistasis: ClpR4 mutations switch leaf variegation to virescence in Arabidopsis. Plant J. 76, 943–956 [DOI] [PubMed] [Google Scholar]
- 16. Chi W., Ma J., Zhang D., Guo J., Chen F., Lu C., and Zhang L. (2008) The pentatricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou W., Cheng Y., Yap A., Chateigner-Boutin A. L., Delannoy E., Hammani K., Small I., and Huang J. (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96 [DOI] [PubMed] [Google Scholar]
- 18. Chateigner-Boutin A. L., des Francs-Small C. C., Delannoy E., Kahlau S., Tanz S. K., de Longevialle A. F., Fujii S., and Small I. (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 65, 532–542 [DOI] [PubMed] [Google Scholar]
- 19. Park S., and Rodermel S. (2004) Mutations in ClpC2/Hsp100 suppress the requirement for fish in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu F., Park S. S., Liu X., Foudree A., Fu A., Powikrowska M., Khrouchtchova A., Jensen P. E., Kriger J. N., Gray G. R., Rodermel S. R. (2011) SUPPRESSOR OF VARIEGATION4, a new var2 suppressor locus, encodes a pioneer protein that is required for chloroplast biogenesis. Mol. Plant 4, 229–240 [DOI] [PubMed] [Google Scholar]
- 21. Qiao J., Ma C., Wimmelbacher M., Börnke F., and Luo M. (2011) Two novel proteins, MRL7 and its paralog MRL7-L, have essential but functionally distinct roles in chloroplast development and are involved in plastid gene expression regulation in Arabidopsis. Plant Cell Physiol. 52, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 22. López-Juez E., Jarvis R. P., Takeuchi A., Page A. M., and Chory J. (1998) New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 118, 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sugimoto H., Kusumi K., Noguchi K., Yano M., Yoshimura A., and Iba K. (2007) The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 52, 512–527 [DOI] [PubMed] [Google Scholar]
- 24. Sugimoto H., Kusumi K., Tozawa Y., Yazaki J., Kishimoto N., Kikuchi S., and Iba K. (2004) The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol. 45, 985–996 [DOI] [PubMed] [Google Scholar]
- 25. Brusslan J. A., and Tobin E. M. (1995) Isolation and initial characterization of virescent mutants of Arabidopsis thaliana. Photosyn. Res. 44, 75–79 [DOI] [PubMed] [Google Scholar]
- 26. Nelson B. K., Cai X., and Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 27. Wetzel C. M., Jiang C. Z., Meehan L. J., Voytas D. F., and Rodermel S. R. (1994) Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 6, 161–175 [DOI] [PubMed] [Google Scholar]
- 28. Lukowitz W., Gillmor C. S., and Scheible W. R. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bell C. J., and Ecker J. R. (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144 [DOI] [PubMed] [Google Scholar]
- 30. Yu F., Park S., and Rodermel S. (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37, 864–876 [DOI] [PubMed] [Google Scholar]
- 31. Atanassov I. I., Etchells J. P., and Turner S. R. (2009) A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clough S. J., and Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 33. Goodstein D. M., Shu S., Howson R., Neupane R., Hayes R. D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., and Rokhsar D. S. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X., Myers A. M., and James M. G. (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138, 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoo S. D., Cho Y. H., and Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 37. Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 82, 43–48 [DOI] [PubMed] [Google Scholar]
- 38. Qi Y., Armbruster U., Schmitz-Linneweber C., Delannoy E., de Longevialle A. F., Rühle T., Small I., Jahns P., and Leister D. (2012) Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J. Exp. Bot. 63, 1251–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ossenbühl F., Hartmann K., and Nickelsen J. (2002) A chloroplast RNA binding protein from stromal thylakoid membranes specifically binds to the 5′ untranslated region of the psbA mRNA. Eur. J. Biochem. 269, 3912–3919 [DOI] [PubMed] [Google Scholar]
- 40. Karnauchov I., Herrmann R. G., and Klösgen R. B. (1997) Transmembrane topology of the Rieske Fe/S protein of the cytochrome b6/f complex from spinach chloroplasts. FEBS Lett. 408, 206–210 [DOI] [PubMed] [Google Scholar]
- 41. Armbruster U., Rühle T., Kreller R., Strotbek C., Zühlke J., Tadini L., Blunder T., Hertle A. P., Qi Y., Rengstl B., Nickelsen J., Frank W., and Leister D. (2013) The photosynthesis affected mutant68-like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 10, 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Järvi S., Suorsa M., Paakkarinen V., and Aro E. M. (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439, 207–214 [DOI] [PubMed] [Google Scholar]
- 43. Chen M., Choi Y., Voytas D. F., and Rodermel S. (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22, 303–313 [DOI] [PubMed] [Google Scholar]
- 44. Putarjunan A., Liu X., Nolan T., Yu F., and Rodermel S. (2013) Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosynth. Res. 116, 437–453 [DOI] [PubMed] [Google Scholar]
- 45. Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Tasneem A., Thanki N., Yamashita R. A., Zhang D., Zhang N., and Bryant S. H. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rawlings N. D., Waller M., Barrett A. J., and Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 48. Allen J. F., and Forsberg J. (2001) Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6, 317–326 [DOI] [PubMed] [Google Scholar]
- 49. Yu F., Fu A., Aluru M., Park S., Xu Y., Liu H., Liu X., Foudree A., Nambogga M., and Rodermel S. (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30, 350–365 [DOI] [PubMed] [Google Scholar]
- 50. Poethig R. (1997) Leaf morphogenesis in flowering plants. Plant Cell 9, 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sakamoto W. (2006) Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57, 599–621 [DOI] [PubMed] [Google Scholar]
- 52. Sokolenko A., Pojidaeva E., Zinchenko V., Panichkin V., Glaser V. M., Herrmann R. G., and Shestakov S. V. (2002) The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41, 291–310 [DOI] [PubMed] [Google Scholar]
- 53. Che Y., Fu A., Hou X., McDonald K., Buchanan B. B., Huang W., and Luan S. (2013) C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 16247–16252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kato Y., and Sakamoto W. (2010) New insights into the types and function of proteases in plastids. Int. Rev. Cell Mol. Biol. 280, 185–218 [DOI] [PubMed] [Google Scholar]
- 55. Richter S., and Lamppa G. (1998) A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. U.S.A. 95, 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Richter S., and Lamppa G. (2003) Structural properties of the chloroplast stromal processing peptidase required for its function in transit peptide removal. J. Biol. Chem. 278, 39497–39502 [DOI] [PubMed] [Google Scholar]
- 57. Moberg P., Ståhl A., Bhushan S., Wright S. J., Eriksson A., Bruce B. D., and Glaser E. (2003) Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J. 36, 616–628 [DOI] [PubMed] [Google Scholar]
- 58. Bhushan S., Ståhl A., Nilsson S., Lefebvre B., Seki M., Roth C., McWilliam D., Wright S. J., Liberles D. A., Shinozaki K., Bruce B. D., Boutry M., and Glaser E. (2005) Catalysis, subcellular localization, expression and evolution of the targeting peptides degrading protease, AtPreP2. Plant Cell Physiol. 46, 985–996 [DOI] [PubMed] [Google Scholar]
- 59. Murakami S., Kondo Y., Nakano T., and Sato F. (2000) Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett. 468, 15–18 [DOI] [PubMed] [Google Scholar]
- 60. Chaal B. K., Mould R. M., Barbrook A. C., Gray J. C., and Howe C. J. (1998) Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana: implications for the origin and catalytic mechanism of the enzyme. J. Biol. Chem. 273, 689–692 [DOI] [PubMed] [Google Scholar]
- 61. Chen G., Bi Y. R., and Li N. (2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 41, 364–375 [DOI] [PubMed] [Google Scholar]
- 62. Chen G., Law K., Ho P., Zhang X., and Li N. (2012) EGY2, a chloroplast membrane metalloprotease, plays a role in hypocotyl elongation in Arabidopsis. Mol. Biol. Rep. 39, 2147–2155 [DOI] [PubMed] [Google Scholar]