Abstract
The MAPK-interacting kinases 1 and 2 (MNK1 and MNK2) are activated by extracellular signal-regulated kinases 1 and 2 (ERK1/2) or p38 in response to cellular stress and extracellular stimuli that include growth factors, cytokines, and hormones. Modulation of MNK activity affects translation of mRNAs involved in the cell cycle, cancer progression, and cell survival. However, the mechanism by which MNK selectively affects translation of these mRNAs is not understood. MNK binds eukaryotic translation initiation factor 4G (eIF4G) and phosphorylates the cap-binding protein eIF4E. Using a cell-free translation system from rabbit reticulocytes programmed with mRNAs containing different 5′-ends, we show that an MNK inhibitor, CGP57380, affects translation of only those mRNAs that contain both a cap and a hairpin in the 5′-UTR. Similarly, a C-terminal fragment of human eIF4G-1, eIF4G(1357–1600), which prevents binding of MNK to intact eIF4G, reduces eIF4E phosphorylation and inhibits translation of only capped and hairpin-containing mRNAs. Analysis of proteins bound to m7GTP-Sepharose reveals that both CGP and eIF4G(1357–1600) decrease binding of eIF4E to eIF4G. These data suggest that MNK stimulates translation only of mRNAs containing both a cap and 5′-terminal RNA duplex via eIF4E phosphorylation, thereby enhancing the coupled cap-binding and RNA-unwinding activities of eIF4F.
Keywords: eukaryotic translation initiation, eukaryotic translation initiation factor 4E (eIF4E), eukaryotic translation initiation factor 4G (eIF4G), mRNA, phosphorylation
Introduction
The rate of translational initiation in eukaryotic cells depends on both cis-acting features of individual mRNAs, such as the cap, poly(A) tract, and untranslated regions (UTRs), and trans-acting components, such as initiation factors, protein kinases, and microRNAs (1–3). The recruitment of capped mRNAs to 48S initiation complexes involves several discrete steps that include cap recognition by eukaryotic translation initiation factor 4E (eIF4E),3 mRNA binding by eIF4G, ATP-dependent unwinding of 5′-terminal secondary structure by the helicase eIF4A in concert with eIF4B and eIF4H, recognition of the 3′-terminal poly(A) tract by poly(A)-binding protein (PABP), and binding of eIF4G to the 40S ribosomal subunit through eIF3. Both the primary and secondary structure of the 5′-UTR are capable of modulating translational efficiency (4, 5). mRNAs containing short 5′-UTRs with little secondary structure or terminal oligopyrimidine tracts are more efficiently translated under normal conditions with a limited level of eIF4E. Translational efficiency is diminished in mRNAs containing long, G+C-rich, and highly structured 5′-UTRs because of the necessity for energy-dependent unwinding prior to start codon recognition (6). Translation of these mRNAs requires high levels of eIF4F, the complex of eIF4E, eIF4A, and eIF4G (3, 7). The availability of eIF4E to enter the eIF4F complex is regulated by the PI3K/Akt/mTOR signaling cascade; eIF4E is sequestered by binding to the 4E-BPs, but activation of the mTOR kinase causes phosphorylation of the 4E-BPs and release of eIF4E (8, 9).
The activity of eIF4E can be also regulated by phosphorylation via the mitogen-activated protein kinase-interacting kinases 1 and 2 (MNK1 and -2) (10), which are activated in response to MAPK or stress signaling via ERK1/2 and p38 (10, 11). MNK phosphorylates eIF4E at a single site (Ser-209 in the human protein (12, 13)), and phosphorylation is greatly enhanced when MNK is bound to eIF4G (14, 15). Phosphorylation of eIF4E reduces its affinity for the cap structure (16, 17). There are numerous studies showing positive correlations between eIF4E phosphorylation and increased protein synthesis (18), cell cycle progression (19), cell proliferation (19, 20), tumorigenesis (21–23), cell hypertrophy (24), transformation (25), and metastasis (26) (also reviewed in Ref. 27). The oncogenic effect of eIF4E overexpression in mouse models of lymphoma (28) and prostate cancer (22) is strictly dependent on the phosphorylation status of eIF4E. Activation of MNK contributes to cell proliferation, transformation, and metastasis (25, 26); growth and survival during cancer development (19, 29–33); or serum deprivation (34, 35) and also contributes to cancer chemoresistance (36, 37) (also reviewed in Ref. 38). Despite the fact that deletion of both MNK genes does not affect mouse development (39), MNK1/2 deficiency and lack of phosphorylation of eIF4E delay tumor development in a PTEN-null background (30) and inhibit cell growth, invasion, and migration while inducing apoptosis in breast cancer cells (40).
It has been suggested that MNK affects cellular processes by three different mechanisms: 1) regulation of mRNA translation, 2) nucleo-cytoplasmic transport of mRNA, and 3) stability of mRNA (reviewed in Refs. 41 and 42). Recently, a fourth role of MNK, translocating phosphorylated eIF4E to specific sites of local translation, has been suggested (43, 44). Stimulation of neuronal cells with BDNF activates MNK, leading to phosphorylation of eIF4E; translocation of eIF4E into dendritic mRNA granules; and translational activation of synaptoneurosomal Arc, αCaMKII, PKM-ζ, calmodulin, and BDNF mRNAs.
The question of mRNA-specific translational control by MNK has been examined by overexpression of constitutively active MNK, but the results depend on the cell type used. Overexpression of constitutively active MNK1/2 in human embryonic kidney 293 cells inhibited cap-dependent while stimulating IRES-driven translation (45), but overexpression of constitutively active MNK1 in T-cells stimulated translation of RFLAT-1 mRNA, which contains GC-rich 5′-UTR and whose translational efficiency is dependent on the level of eIF4E, suggesting a cap-dependent mechanism (46). Another study in Aplysia neurons indicated that both cap- and IRES-dependent translation were inhibited by overexpression of either wild-type or constitutively active MNK (47, 48). In adult cardiocytes, overexpression of wild-type MNK1 stimulated translation of mRNA, regardless of the presence of secondary structure in the 5′-UTR (49). In diffuse large B-cell lymphoma cells, modulation of MNK1/2 activity differentially affects mRNAs that utilize eIF4E1- versus eIF4E3-driven translation on the basis of a 5′-UTR motif (50). In breast carcinoma (MDA-MB-435) cells, α6β4-integrin-dependent activation of MNK stimulates translation of VEGF mRNA, which contains a long, highly structured 5′-UTR, but not GAPDH mRNA, which contains a short 5′-UTR with little secondary structure (35).
As the foregoing findings indicate, it is difficult to determine how MNK affects translation per se in whole-cell studies because 1) MNK affects multiple mRNA-related processes (nucleo-cytoplasmic transport, stability, subcellular site of translation, etc.), 2) different cell types give different results, and 3) structural features of mRNA appear to affect susceptibility to MNK. We have therefore utilized a rabbit reticulocyte cell-free system to investigate whether any effect of MNK on translation is dependent on 5′-terminal structures in mRNA. We show that inhibition of eIF4E phosphorylation, whether by use of a kinase inhibitor or by use of an eIF4G fragment that prevents MNK binding to full-length eIF4G, affects translation of only mRNAs that contain both a cap and 5′-terminal hairpin loop. Moreover, the kinase inhibitor did not affect translation if the eIF4G was proteolytically cleaved to separate the eIF4E-binding from the MNK-binding domains, supporting the idea that MNK-regulated translation requires the protein that binds the cap (eIF4E) and the protein that unwinds the hairpin (eIF4A) to be present in the same molecular complex. Finally, the composition of the eIF4F complex changes in the presence of MNK inhibitors, suggesting that MNK regulates cap-dependent translation by altering interactions among eIF4F components.
Experimental Procedures
Materials
The MNK inhibitor CGP57380 (CGP) was provided by Novartis Pharma AG (Basel, Switzerland) and was stored as a 10 mm stock solution in 100% DMSO (MP Biomedicals, LLC); for all experiments in which CGP was added to translation reactions, results were normalized by comparison with control reaction mixtures containing DMSO at the same dilution. Nickel-nitrilotriacetic acid-agarose was purchased from Qiagen. Econo-Pac® 10 DG disposable chromatography columns and the protein assay kit were obtained from Bio-Rad. m7GTP-Sepharose 4B was purchased from Amersham Biosciences. S-protein-agarose was obtained from Novagen. Complete EDTA-free protease inhibitor mixture was from Roche Diagnostics. [35S]Met and [γ-32P]ATP were from ICN-MP Radiochemicals. The “anti-reverse cap analog” (ARCA) m27,3′-OGpppG (51) was a gift from Edward Darzynkiewicz (University of Warsaw). All other reagents were of analytical grade and were purchased from Sigma.
Expression and Purification of Recombinant Proteins
The expression and purification of two fragments of eIF4G-1 (NCBI accession number NP_886553) have been described previously; eIF4G(589–1600) is aa residues 589–1600 with an N-terminal tag consisting of thioredoxin, His6, and S-peptide (52), and eIF4G(1357–1600) is similar except with aa residues 1357–1600 (53). Recombinant human MNK1 and MNK2 were expressed from plasmids pET14-His6-Mnk1 and pET14-His6-Mnk2, respectively (54). Human eIF4E with Ser-209 changed to Ala (eIF4E(S209A)) and with Asp (eIF4E(S209D)) were described previously (12, 55). Human eIF4A dominant negative mutant eIF4A(R362Q) was described previously (56). Proteins were expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen), purified by nickel-nitrilotriacetic acid-agarose chromatography, and passed over an Econo-Pac® 10 DG column to replace the buffer with buffer A (20 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% (v/v) glycerol). Recombinant coxsackievirus 2A protease was expressed and purified as described previously (57). The concentrations of recombinant proteins were determined with the Bio-Rad protein assay kit based on BSA as a standard.
Synthetic mRNAs
The plasmid (CAA)n-GUS encoding β-glucuronidase (GUS) (58, 59) has been described previously. The (CAA)n-GUS plasmid produces mRNA with the 5′-end sequence 5′-GCAAGAA-(CAA)19-CACCAUGG-[GUS], where the start codon is in boldface type. Plasmids (CAA)n-Stem-GUS containing hairpin insertions in the (CAA)n-GUS construct. (CAA)n-Stem-GUS plasmids have the generic structure 5′-G(CAA)14-STEM-(CAA)4CCAUGG … [GUS], in which there is a defined GC-rich hairpin (“STEM”) inserted in the 5′-UTR after 43 unstructured nucleotides (59). Four hairpins with varying stability were inserted to make 5′-(CAA)n-Stem1-GUS (hairpin stability, −5.5 kcal/mol), 5′-(CAA)n-Stem2-GUS (−13.1 kcal/mol), 5′-(CAA)n-Stem3-GUS (−18.9 kcal/mol), and 5′-(CAA)n-Stem4-GUS (−27.6 kcal/mol). Stabilities were predicted using Mfold version 3.0 (60). RNA was synthesized by transcribing plasmids with T7 RNA polymerase (61). Capped mRNAs were produced by lowering the GTP concentration from 0.5 to 0.1 mm and including the ARCA at 1 mm (62), which prevents the cap from being incorporated in the reverse orientation (51). The plasmid pCITE-Luc was used to synthesize firefly luciferase mRNA containing the EMCV IRES upstream of the coding region (IRES-Luc) and no poly(A) tract (63). RNA was purified with an RNeasy® minikit (Qiagen) using the manufacturer's protocol. The concentrations of RNAs were determined spectrophotometrically.
In Vitro Translation
Rabbit reticulocyte lysate (RRL) treated with micrococcal nuclease (Promega) was used for in vitro translation reactions according to the manufacturer's protocol. Reaction mixtures supplemented with [35S]Met contained mRNA at 2 μg/ml and were incubated for 25 min at 30 °C, over which period the rate of protein syntheses was constant. GUS synthesis was measured by incorporation of [35S]Met and detection with a Storm 860 PhosphorImager (GE Healthcare) or by autoradiography. In the experiment with eIF4A(R362Q), 15-μl translation reactions supplemented with [35S]Met were preincubated at 30 °C for 10 min with or without eIF4A(R362Q), followed by the addition of 0.2 μg of mRNA and further incubation for 60 min. Samples were analyzed by SDS-PAGE and subsequent autoradiography. Luciferase synthesis from the IRES-Luc mRNA was measured by detection of enzymatic activity with a Monolight 2010 luminometer. In some experiments, the system was made cap-independent by incubating RRL with recombinant coxsackievirus 2A protease (50 μg/ml for 30 min at 4 °C) before the start of translation (57). To monitor eIF4E phosphorylation, translation reactions were incubated with [γ-32P]ATP at 30 °C for the indicated periods of time and analyzed by either SDS-PAGE followed by PhosphorImager detection or Western blotting with anti-eIF4E antibodies. To determine statistical significance, data were analyzed by Student's t test; p < 0.05 was considered to represent statistical significance.
Protein Binding to m7GTP-Sepharose
Translation reactions (10 μl) containing 1 μm CGP, 10 μm eIF4G(1357–1600), or 0.01% DMSO were incubated at 30 °C for 30 min and then combined with 20 μl of m7GTP-Sepharose (50% slurry in buffer A supplemented with 0.05% Tween 20). After a 2-h incubation at 4 °C with rotation, the resin was washed three times with 200-μl aliquots of buffer B (20 mm Tris-HCl, 150 mm KCl, 0.05% Tween 20, 10% (v/v) glycerol, pH 7.5). Proteins were eluted in 20 μl of SDS loading buffer and analyzed by Western blotting.
Protein Binding to S-protein-Agarose
Reactions containing eIF4G(589–1600) and either eIF4E(S209A) or eIF4E(S209D), all at 50 nm, were mixed with increasing amounts of MNK1 or MNK2 (0, 25, 50, or 100 nm) and then combined with 20 μl of S-protein-agarose (50% slurry in buffer A plus 0.2 mg/ml BSA and 0.05% Tween 20). Incubation, washing, and elution were performed as for m7-GTP-Sepharose.
Immunologic Procedures
Western blotting was performed on translation or binding reactions by SDS-PAGE on 10 or 12% gels and transfer to nitrocellulose membranes (Bio-Rad) using a Mini Trans-Blot cell (Bio-Rad) (64). Rabbit anti-human eIF4G-1 (anti-peptide 7) antibodies were produced as described previously (65). Antibodies against MNK1 and 4E-BP1 were purchased from Cell Signaling Technology, Inc. Antibodies against MNK2 were purchased from Santa Cruz Biotechnology, Inc. Membranes were incubated with a 1:1000 dilution of primary antibodies in buffer C (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% Tween 20) containing 3% BSA overnight at 4 °C, washed three times for 10 min with buffer C, and incubated for 45 min at room temperature with secondary antibodies conjugated with either alkaline phosphatase (to detect eIF4G and eIF4E) or horseradish peroxidase (to detect MNK1, MNK2, and 4E-BP1) (Vector Laboratories, Inc.) at a dilution of 1:1000 in 5% milk proteins in buffer C. The eIF4G and eIF4E blots were developed with the nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate color development reagents (Promega). The MNK1, MNK2, and 4E-BP1 blots were developed with the ECL+ Western blotting development kit (Amersham Biosciences).
Results
CGP Inhibits de Novo Phosphorylation of eIF4E in Translation Reactions
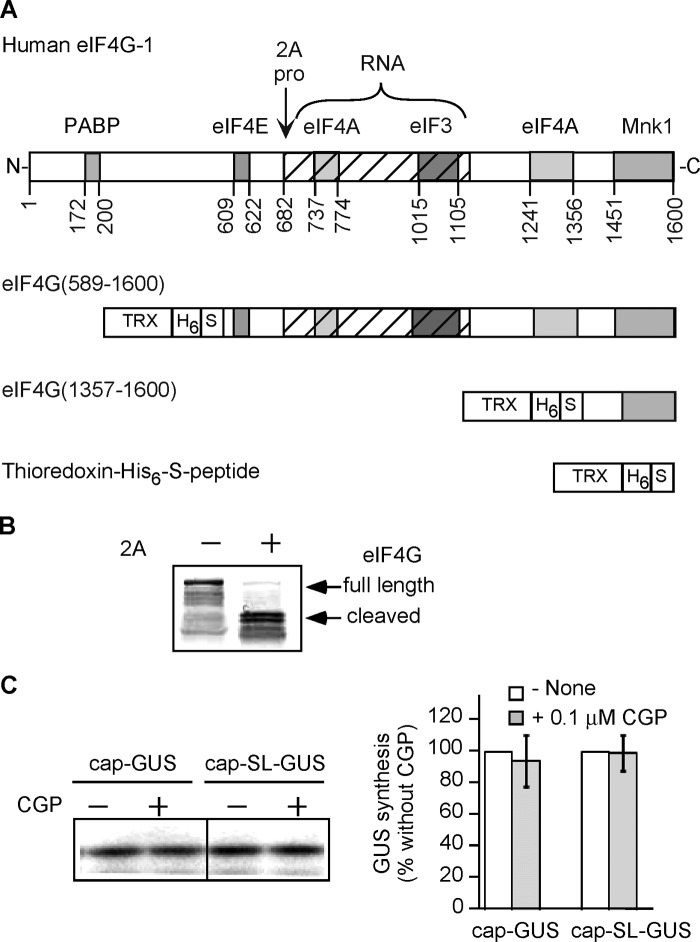
We first monitored the phosphorylation status of eIF4E in translation reactions containing [γ-32P]ATP (Fig. 1A, top). We observed increasing 32P incorporation into eIF4E after 10, 25, 45, and 60 min of incubation. These results indicate that RRL contains active MNK capable of phosphorylating eIF4E. eIF4E phosphorylation increased steadily but began to plateau by 60 min (Fig. 1A, bottom). We chose a 25-min time point for subsequent experiments because the rate of protein syntheses was constant over this period (data not shown). The [32P]eIF4E level was 66 ± 10% of maximum at 25 min.
FIGURE 1.
CGP inhibits phosphorylation of eIF4E during in vitro translation. A, translation reactions were incubated in the presence of [γ-32P]ATP at 30 °C. Aliquots (5 μl) were taken from the reaction mixture at various times and mixed with SDS-loading buffer. Top, autoradiogram of [32P]eIF4E and Western blotting of total eIF4E in the same aliquots. Bottom, quantitation of the [32P]eIF4E data in the top normalized for total eIF4E. Data are expressed as percentage of [32P]eIF4E in the 60 min aliquot (n = 3, mean ± S.E. (error bars)). B, translation reactions were incubated in the presence of [γ-32P]ATP without (lane 1) or with the indicated concentrations of CGP (lanes 2–5) for 25 min at 30 °C. Top, autoradiography of phosphorylated eIF4E (above) and Western blotting with anti-eIF4E antibodies (below). Bottom, quantitation of eIF4E phosphorylation in the presence or absence of CGP from several experiments similar to that in the top. Bars, [32P]eIF4E signal intensity normalized for the total eIF4E as a percentage of the signal in the absence of CGP (n = 3, mean ± S.E.).
Next, we investigated the effect of increasing CGP on in vitro phosphorylation of eIF4E (Fig. 1B). CGP inhibited 32P incorporation into eIF4E in a dose-response manner, the maximum inhibition being observed at 1000 nm CGP (54 ± 5% of DMSO alone). Because MNK is the only known physiological kinase for eIF4E (39), this shows that CGP inhibits MNK in RRL.
Translation of mRNA Containing Hairpin Depends on the eIF4A Activity
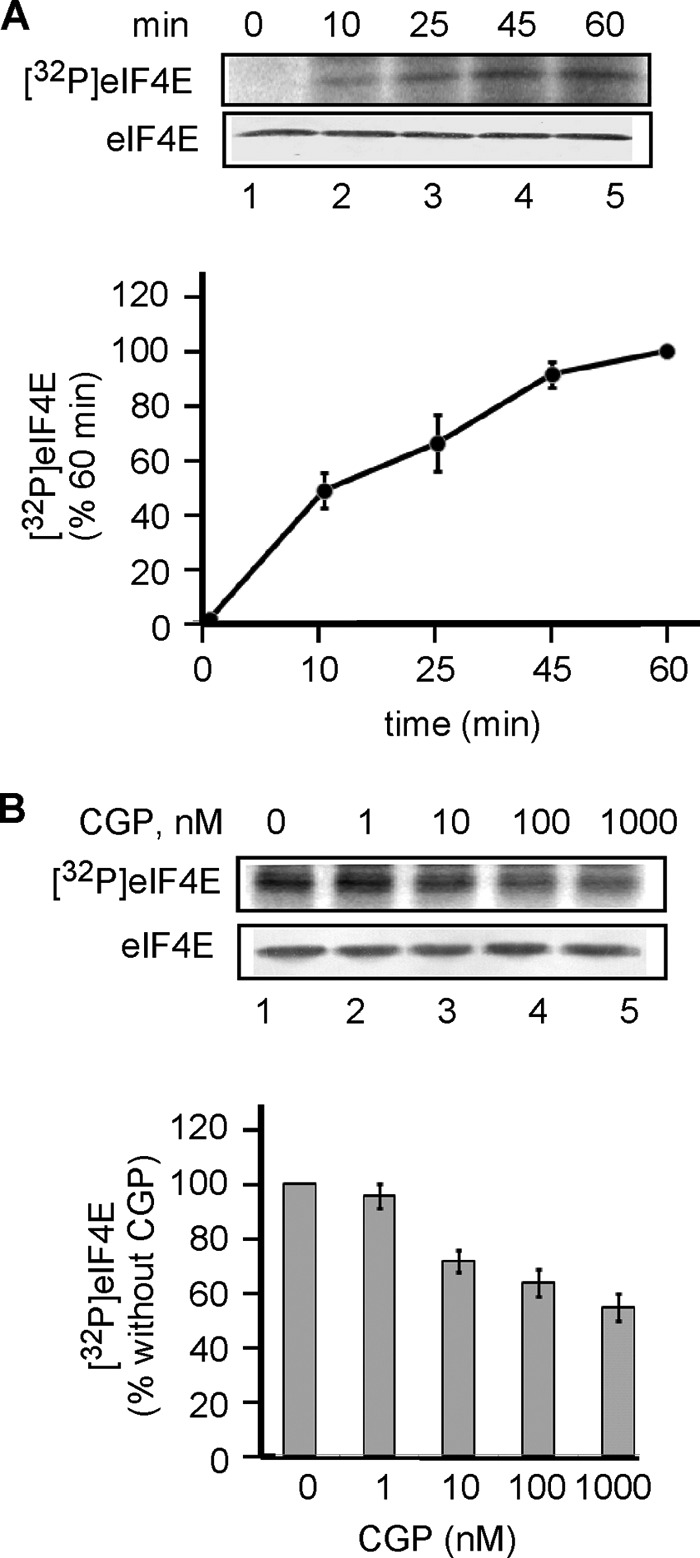
To choose an mRNA construct suitable for our study, we first investigated translation of several mRNAs with or without hairpins in their 5′-UTRs and encoding GUS. One mRNA, (CAA)n-GUS, has a 5′-UTR lacking secondary structure and consists of the sequence 5′-GCAAGAA-(CAA)19-CACCAUGG-[GUS], in which the start codon is in boldface type (Fig. 2A). The single-stranded conformation of the (CAA)n sequence has been established by enzymatic probing (66). Four other mRNAs, (CAA)n-Stem-GUS, contain GC-rich stems with stabilities ranging from −5.5 to −27.6 kcal/mol and flanked by CAA repeats (Fig. 2A). Introduction of GC-rich hairpins of increasing stability progressively inhibited translation of GUS mRNA (Fig. 2B, compare lanes 3, 5, 7, and 9 with lane 1). Previously, it was shown in a reconstituted translation system that 48S initiation complex assembly on these mRNAs requires the presence of eIF4F, eIF4A, and eIF4B (59), indicating the importance of 5′-UTR unwinding for translation of these mRNAs. In agreement with this, preincubation of the RRL with the dominant negative mutant eIF4A(R362Q) inhibited translation of the hairpin-containing mRNAs (Fig. 2B, compare lane 4 with lane 3, lane 6 with lane 5, lane 8 with lane 7, and lane 10 with 9).
FIGURE 2.
The presence of a hairpin in the 5′-UTR affects translation of 5′-(CAA)n-STEM-GUS mRNAs. A, sequences of stem-loops inserted into (CAA)n-Stem-GUS mRNAs showing the site of insertion, structure, and stability (59). The hairpins, called stems 1–4, have increasing stability (predicted by using Mfold version 3.0 (60)). Reproduced with permission from Pisareva, V. P., Pisarev, A. V., Komar, A. A., Hellen, C. U., and Pestova, T. V. (2008) Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135, 1237–1250. B, phosphor image of [35S]GUS synthesized from mRNA without hairpin [(CAA)nGUS] (lanes 1 and 2) or mRNAs containing stems 1–4 (lanes 3–10) in the absence (lanes 1, 3, 5, 7, and 9) or presence of eIF4A(R362Q) (lanes 2, 4, 6, 8, and 10).
For subsequent experiments, we chose (CAA)n-Stem2-GUS mRNA (ΔG = −13.1 kcal/mol). This choice was based on two observations: (i) mRNA with stem 2 was more efficiently translated than those with stronger hairpins (stems 3 and 4 with ΔG = −18.9 and −27.6 kcal/mol, respectively) (Fig. 2B, compare lane 7 with lanes 3 and 9), and (ii) mRNA with stem 2 was more dependent on the eIF4F helicase activity than those without a hairpin or a weaker hairpin (stem 1; ΔG = −5.5 kcal/mol) (Fig. 2B, compare lane 8 with lanes 2 and 6).
CGP Inhibits Translation of Only Capped mRNA with 5′-Hairpin
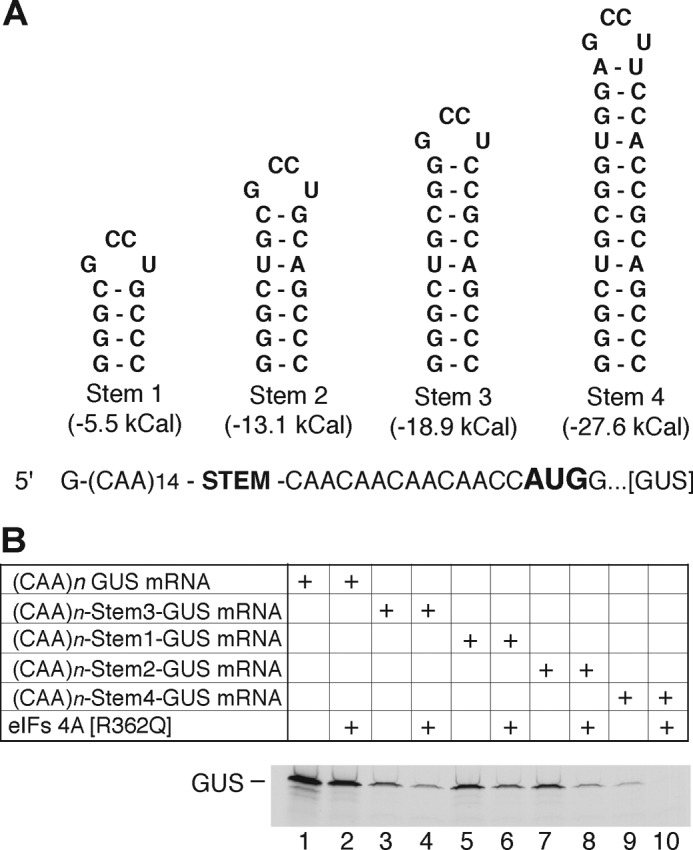
To investigate effect of cap and hairpin structure on the regulation of translation by MNK, we measured the translation of mRNAs that differ in the presence or absence of a cap and hairpin structure in the 5′-UTR (Fig. 3A). The unc-GUS mRNA is uncapped (CAA)n-GUS mRNA encoding GUS (the in vitro transcription reaction in the absence of an ARCA produces mRNA with a 5′-triphosphate). The unc-SL-GUS mRNA is also uncapped but contains a stem 2 structure in the 5′-UTR with stability of −13.1 kcal/mol ((CAA)n-Stem2-GUS mRNA, Fig. 2A). The cap-GUS mRNA has a cap structure and an unstructured 5′-UTR. Finally, cap-SL-GUS mRNA contains both a cap structure and the stem-loop stability of −13.1 kcal/mol. The fifth mRNA was used for IRES-driven translation and consisted of the EMCV IRES followed by the firefly luciferase (Luc) coding region.
FIGURE 3.
CGP inhibits translation of cap-SL-GUS but not cap-GUS mRNA. A, schematic representation of mRNAs used in translation reactions. Unc-GUS mRNA representing the (CAA)n-GUS mRNA has no cap and an unstructured 5′-UTR. Unc-SL-GUS mRNA representing the (CAA)n-Stem2-GUS mRNA (Fig. 2A) is also uncapped but contains a hairpin structure inserted in its 5′-UTR. Cap-GUS mRNA representing the (CAA)n-GUS mRNA is capped and has an unstructured 5′-UTR. Cap-SL-GUS representing the (CAA)n-Stem2-GUS mRNA contains both a cap and a hairpin. IRES-Luc mRNA contains the EMCV IRES but encodes firefly luciferase. B, phosphor image of [35S]GUS synthesized from cap-GUS and cap-SL-GUS mRNAs in the presence of DMSO only (lanes 1 and 6) or 1, 10, 100, or 1000 nm CGP (lanes 2–5 and 7–10, respectively). C, quantitation of the data in B, expressed as a percentage of the GUS signal in the presence of DMSO alone (n = 4, mean ± S.E. (error bars)). Open bars, translation of cap-GUS; gray bars, cap-SL-GUS. *, p < 0.01.
The effect of a stem-loop structure on the regulation of translation by MNK was investigated by comparing translation of cap-SL-GUS with cap-GUS mRNA in the presence or absence of increasing amounts of CGP (Fig. 3, B and C). CGP inhibited translation of cap-SL-GUS in a dose-dependent manner (Fig. 3, B (lanes 6–10) and C (gray bars)) but not translation of cap-GUS mRNA (Fig. 3, B (lanes 1–5) and C (open bars)). Because eIF4F/4A/4B are required for formation of a functional 48S complex at the initiation codon for GUS mRNA that contains a stem 2 in the 5′-UTR (59) and also translation of this mRNA is inhibited by the dominant negative mutant eIF4A(R362Q) (Fig. 2B, lanes 7 and 8), we interpret these findings to mean that MNK stimulates the helicase activity of eIF4F. It follows that translation of mRNA requiring this helicase activity is diminished when MNK is inhibited by CGP.
The effect of a cap on the regulation of translation by MNK was investigated by comparing unc-GUS and unc-SL-GUS mRNAs in the RRL system (Fig. 4, A and B). CGP did not affect translation of either uncapped mRNA (Fig. 4, A (lanes 1–5 and 6–10) and B (open and black bars)) over the same concentration range where cap-SL-GUS translation was inhibited (Fig. 4, A (lanes 11–15) and B (gray bars)).
FIGURE 4.
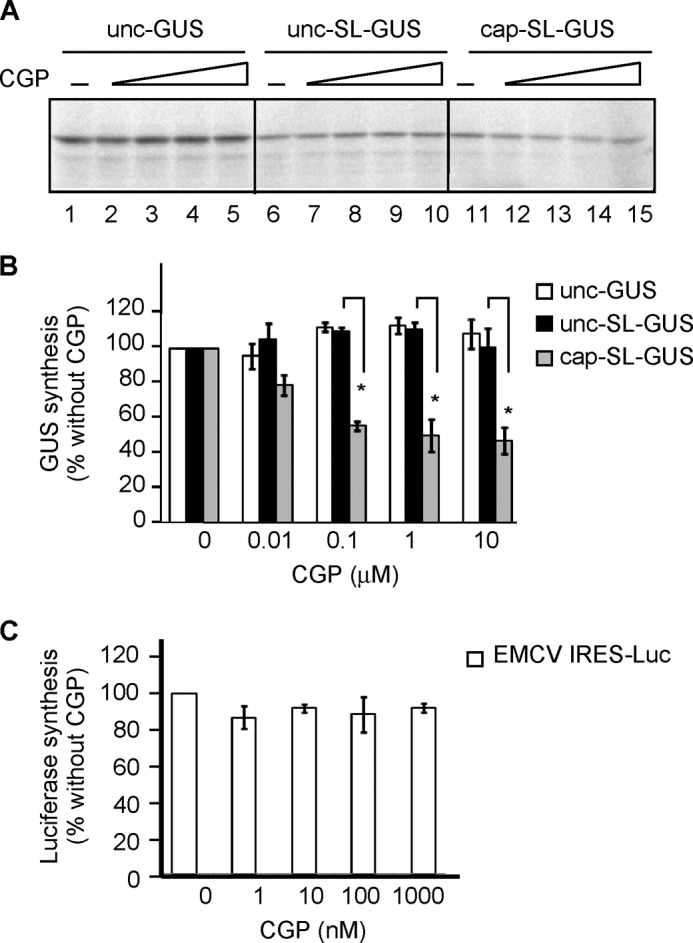
CGP does not affect translation of uncapped mRNA regardless of the 5′-UTR structure. A, phosphor image of [35S]GUS synthesized from the indicated mRNAs as in Fig. 3B. The concentrations of CGP were 0, 0.01, 0.1, 1, and 10 μm (lanes 1–5 for unc-GUS mRNA, lanes 6–10 for unc-SL-GUS mRNA, and lanes 11–15 for cap-SL-GUS mRNA, respectively). B, quantitation as in Fig. 3C of the GUS synthesized from unc-GUS, unc-SL-GUS, and cap-SL-GUS mRNAs in experiments similar to that in Fig. 4A (n = 3, mean ± S.E. (error bars)). Open bars, translation of unc-GUS; black bars, unc-SL-GUS; gray bars, cap-SL-GUS mRNAs. C, quantitation of IRES-Luc mRNA translation as in Fig. 3C except that luciferase activity rather than [35S]GUS was measured (n = 3, mean ± S.E.).
The foregoing experiments show that eIF4E phosphorylation is important for translation of an mRNA containing both a cap and a 5′-terminal hairpin that was previously demonstrated to require the helicase activity of eIF4F (Fig. 2B). Another way to test the requirement for a cap in MNK action is with the EMCV IRES, which confers efficient initiation of translation without a cap (67) yet requires the unwinding activity of eIF4F (7, 68). We found that CGP fails to inhibit translation of IRES-Luc mRNA (Fig. 4C). This provides further evidence that an mRNA must be capped for MNK to exert an effect on its translation.
CGP Does Not Interfere with Cap Binding by eIF4E per se
One hypothesis to explain the observation that CGP inhibits translation of cap-SL-GUS but not cap-GUS mRNA (Fig. 3, B and C) is that (i) cap-GUS mRNA translation does not require cap recognition by eIF4E, (ii) cap-SL-GUS mRNA does require cap recognition by eIF4E, and (iii) CGP inhibits the cap-eIF4E interaction. We ruled this out by adding m7GTP, which competes with capped mRNA for binding to eIF4E (69) and eIF4F (70). Translation of both cap-GUS and cap-SL-GUS mRNA was inhibited ∼50% by 5 μm m7GTP (Fig. 5A). However, in the same translation experiment, cap-SL-GUS but not cap-GUS mRNA was inhibited by 0.1 μm CGP (Fig. 5B). Thus, cap-GUS mRNA translation requires cap binding by eIF4E, so the failure of CGP to inhibit cap-GUS mRNA translation is not caused by it blocking the eIF4E-cap interaction.
FIGURE 5.
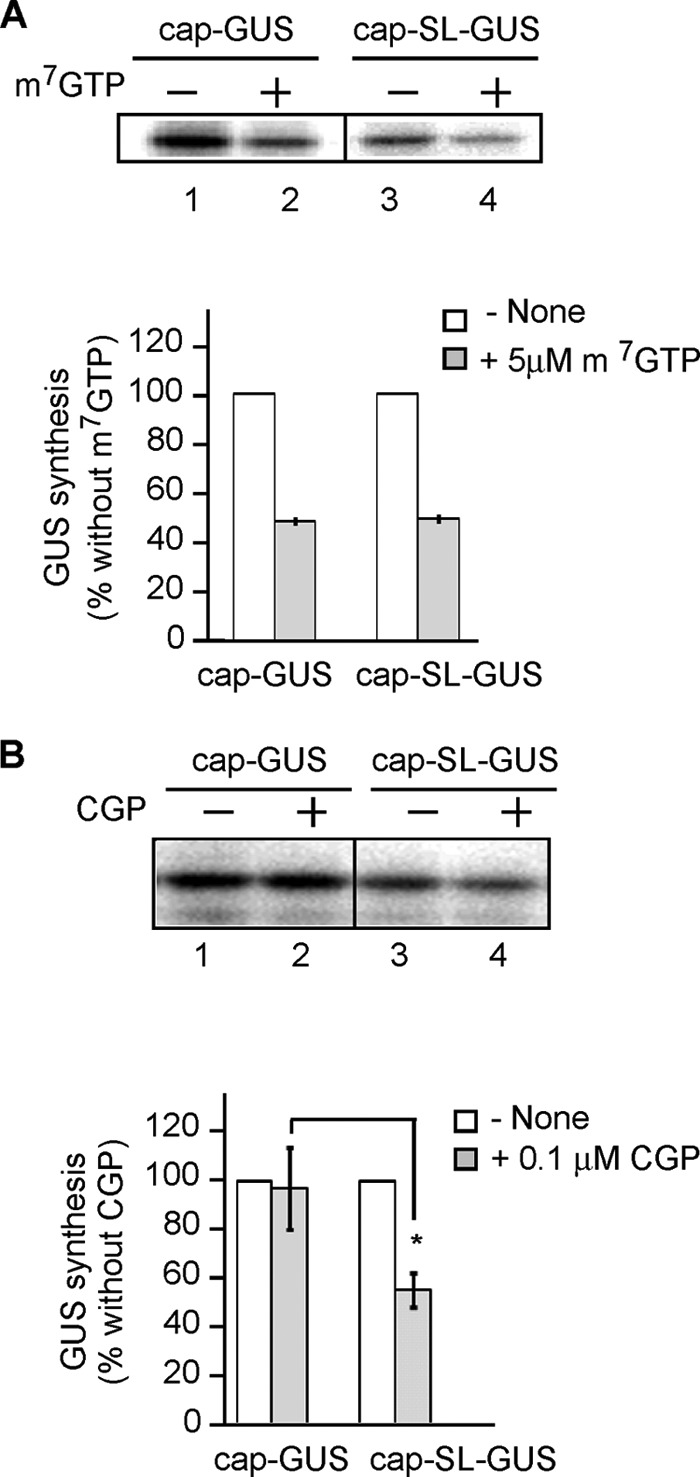
Effect of CGP on translation is different from the effect of a cap analog. A, translation reactions containing cap-GUS or cap-SL-GUS mRNA were incubated in the presence or absence of 5 μm m7GTP. Top, phosphor image of [35S]GUS in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of cap analog. Bottom, the GUS signal in the absence (open bars) or presence (filled bars) from similar experiments is expressed as a percentage of reactions containing no m7GTP (n = 3, mean ± S.E. (error bars)). B, same as A except 0.1 μm CGP was used instead of m7GTP. Top, phosphor image of [35S]GUS. Bottom, quantitation of the data from similar experiments (n = 3, mean ± S.E.). *, p < 0.02.
eIF4F Integrity Is Required for the Effect of CGP on Translation
The foregoing results indicate that both a cap and a 5′-terminal stem-loop are required for inhibition of translation by CGP. The cap is recognized by eIF4E, whereas secondary structure is unwound in an ATP-dependent manner by eIF4A, both of which have specific and high affinity binding sites on eIF4G (71–74) (Fig. 6A). Coxsackievirus 2A protease cleaves eIF4G at a single peptide bond that causes separation of the N-terminal one-third of eIF4G containing the eIF4E-binding site from the C-terminal two-thirds of eIF4G containing the two eIF4A-binding sites (57) (Fig. 6A, arrow). The C-terminal fragment also binds eIF3, mRNA, and MNK, serving as cap-independent “ribosome recruitment core” (14, 57, 75) (Fig. 6A). To test whether inhibition of cap-SL-GUS mRNA translation by CGP requires eIF4E and eIF4A to be functionally linked through eIF4G, we pretreated RRL with recombinant coxsackievirus 2A protease. Under these conditions, most of the eIF4G is cleaved (Fig. 6B). The multiple bands of uncleaved eIF4G are due to alternative translation initiation sites on eIF4G-1 mRNA (76, 77). After eIF4G cleavage, CGP did not inhibit the translation of either cap-SL-GUS or cap-GUS mRNAs (Fig. 6C). This result indicates that CGP does not affect helicase-dependent translation per se. We know this because RNA unwinding is needed for translation of mRNA containing stem 2 (Fig. 2B); after eIF4G cleavage, this is carried out by the C-terminal two-thirds of eIF4G in complex with eIF4A, yet cap-SL-GUS mRNA translation is not affected by CGP (Fig. 6C). This means that the CGP-inhibitory effect requires intact eIF4G. Based on these results, we propose that CGP targets the coordinated action of eIF4E and eIF4A, in complex with eIF4G, to catalyze initiation of an mRNA containing a cap (requiring eIF4E) and a 5′-terminal stem-loop (requiring eIF4A-eIF4G) but does not target the actions of eIF4E or eIF4A-eIF4G individually.
FIGURE 6.
Effect of CGP on translation requires intact eIF4G. A, schematic representation of human eIF4G-1 and recombinant eIF4G(1357–1600), eIF4G(589–1600), and T-H-S used in this study. Binding sites for initiation factors, MNK, and RNA, and the cleavage site for coxsackievirus 2A protease (arrow) are shown. eIF4G(1357–1600) and eIF4G(1357–1600) contain the T-H-S at their N termini. B, Western blotting analysis of eIF4G in RRL pretreated (+) or not (−) with 2A protease. Positions of full-length and cleaved eIF4G are shown. The multiple bands of uncleaved eIF4G are due to alternative translation initiation sites on eIF4G-1 mRNA (76, 77). C, left, phosphor image of [35S]GUS synthesized from either cap-GUS or cap-SL-GUS mRNA in RRL treated with coxsackievirus 2A protease prior to the addition of mRNA and either 0.1 μm CGP (+) or 0.001% DMSO (−). Right, quantitation of these data from similar experiments (n = 3, mean ± S.E. (error bars)).
Preventing MNK Binding to eIF4G Inhibits eIF4E Phosphorylation and Translation Only of mRNA Containing Both a Cap and a 5′-Terminal Hairpin
Although CGP inhibits MNK activity with high specificity, other kinases are also inhibited by CGP, albeit with lower selectivity (78–80). We therefore took a second approach to inhibit eIF4E phosphorylation that did not involve CGP. We generated a recombinant fragment of human eIF4G-1 containing the MNK-binding site, eIF4G(1357–1600) (Fig. 6A). This fragment would be expected to compete with full-length eIF4G for binding to MNK and thus specifically inhibit eIF4E phosphorylation by MNK, because eIF4E phosphorylation is more efficient when MNK is bound to eIF4G (14, 15). As a control, we used the N-terminal tag present on all eIF4G fragments, thioredoxin-His6-S-peptide (T-H-S) (Fig. 6A). We first determined the effect of eIF4G(1357–1600) and the T-H-S on the phosphorylation of eIF4E. eIF4G(1357–1600) but not the T-H-S reduces incorporation of [32P] into eIF4E by ∼50% (Fig. 7A). This indicates that eIF4G(1357–1600) inhibits MNK activity during translation, similar to CGP.
FIGURE 7.
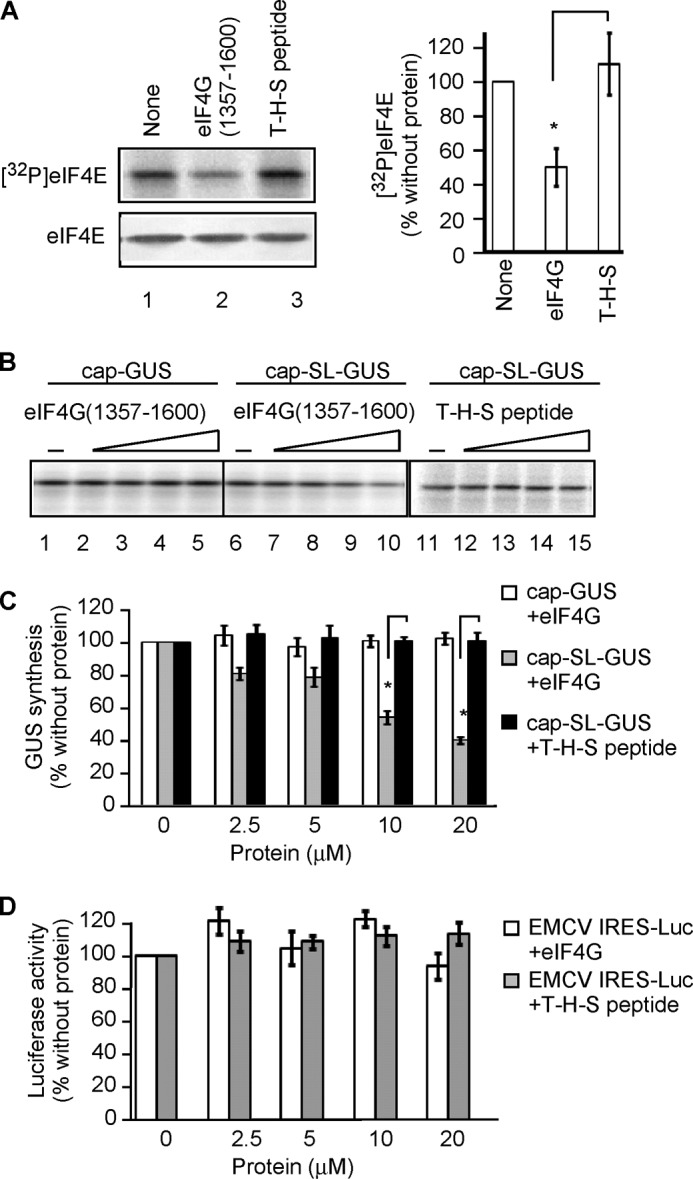
eIF4G(1357–1600) inhibits translation only of an mRNA containing both a cap and 5′-terminal hairpin. A, left, phosphor image of [32P]eIF4E and Western blotting of eIF4E in RRL containing [γ-32P]ATP and incubated in the presence or absence of 10 μm eIF4G(1357–1600) or 20 μm T-H-S for 25 min. Right, quantitation of the [32P]eIF4E signal intensity normalized to total eIF4E in similar experiments expressed as a percentage of the signal in translation reactions without additional proteins (n = 3, mean ± S.E. (error bars)). *, p < 0.01. B, eIF4G(1357–1600) inhibits translation of cap-SL-GUS but not cap-GUS mRNA. Shown is a phosphor image of [35S]GUS synthesized from either cap-GUS or cap-SL-GUS mRNAs in RRL in the presence of 0, 2.5, 5, 10, or 20 μm eIF4G(1357–1600) (lanes 1–5 and lanes 6–10, respectively). Cap-SL-GUS mRNA was translated in the presence of the same concentrations of the T-H-S (lanes 11–15). C, quantitation of data from experiments similar to B (three with eIF4G(1357–1600) and two with the T-H-S) expressed as a percentage of the GUS signal in reactions without additional proteins. *, p < 0.01. Open bars, cap-GUS mRNA with eIF4G(1357–1600); gray bars, cap-SL-GUS mRNA with eIF4G(1357–1600); black bars, cap-SL-GUS mRNA with T-H-S. D, luciferase synthesis from IRES-Luc mRNA in the presence or absence of either eIF4G(1357–1600) (open bars) or the T-H-S (filled bars). Data are expressed as percentage of the luciferase signal in reactions without additional proteins (n = 3, mean ± S.E.).
Translation of cap-SL-GUS mRNA was inhibited in a dose-dependent manner by eIF4G(1357–1600) (Fig. 7, B (lanes 6–10) and C (gray bars)), but translation of cap-GUS mRNA was unaffected (Fig. 7, B (lanes 1–5) and C (open bars)). Equimolar concentrations of the T-H-S failed to inhibit translation of cap-SL-GUS mRNA (Fig. 7, B (lanes 11–15) and C (solid black bars)). Thus, preventing eIF4E phosphorylation with either CGP (Fig. 3, B and C) or eIF4G(1357–1600) (Fig. 7, B and C) has the same effect: inhibition of cap-SL-GUS but not cap-GUS mRNA translation.
We next investigated the effect of eIF4G(1357–1600) on translation of IRES-Luc mRNA. Neither eIF4G(1357–1600) nor T-H-S inhibited translation of EMCV IRES-Luc (Fig. 7D). This was the same result obtained with CGP (Fig. 4C). Thus, both reagents that prevent eIF4E phosphorylation, the kinase inhibitor CGP and the interfering eIF4G fragment eIF4G(1357–1600), affect translation of only mRNA containing both cap and 5′-terminal hairpin.
eIF4E Phosphorylation Enhances Its Binding to eIF4G
One possible mechanism by which eIF4E phosphorylation may enhance translation of cap-SL-GUS mRNA is by more efficient assembly of the eIF4F complex, which carries out cap-dependent unwinding of mRNA (81). Knockdown of protein phosphatase PP2A in human lung cancer cells increases both phosphorylation of eIF4E and retention of eIF4G on m7GTP-Sepharose (82). Furthermore, treatment of these cells with CGP diminishes eIF4G binding to m7GTP-Sepharose (82). Both observations suggest that eIF4E phosphorylation promotes eIF4F assembly. We tested this in the simpler cell-free translation system. Both CGP (Fig. 8A, lane 2) and eIF4G(1357–1600) (lane 3) decreased the amount of eIF4G retained on m7GTP-Sepharose, supporting this model. Interestingly, there was no decrease in 4E-BP1 retention on m7GTP-Sepharose, although eIF4G and 4E-BP1 bind to nearly the same site on the dorsal side of eIF4E (83).
FIGURE 8.
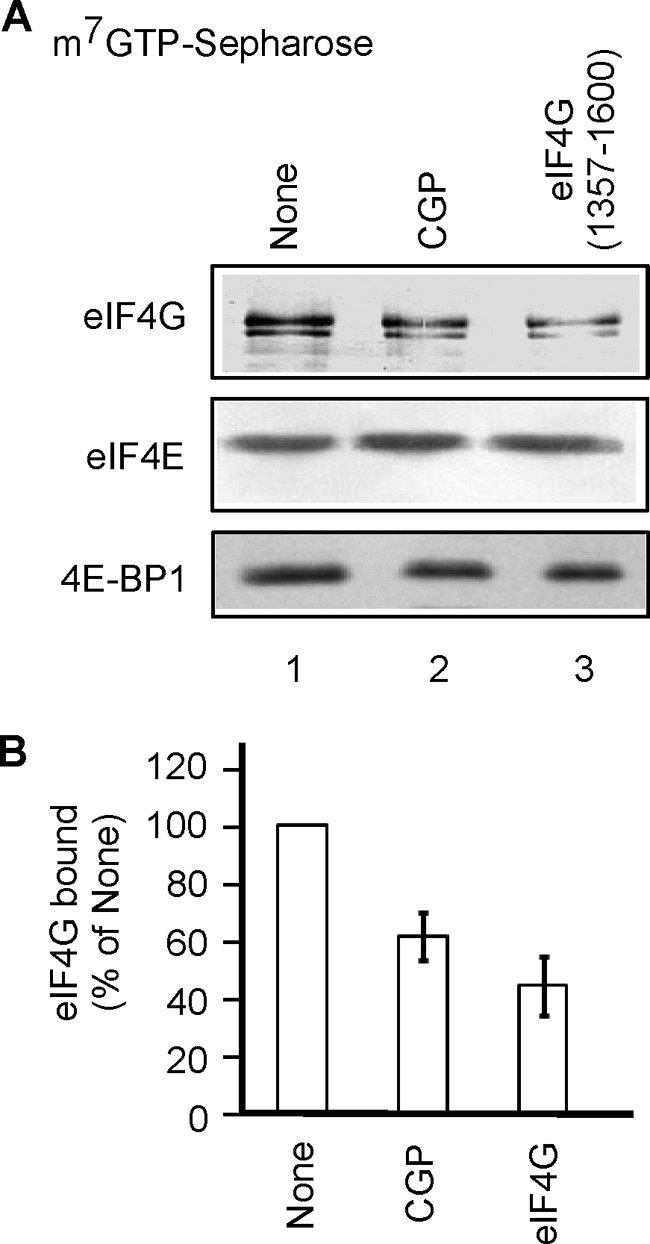
CGP and eIF4G(1357–1600) diminish eIF4G retention on m7GTP-Sepharose. A, RRL was incubated in the presence or absence of 0.01% DMSO, 1 μm CGP, or 10 μm eIF4G(1357–1600) for 30 min and then mixed with m7GTP-Sepharose. Bound proteins were eluted and analyzed by Western blotting as described under “Experimental Procedures.” B, quantification of eIF4G data from experiments similar to A (four for CGP and two for eIF4G(1357–1600)) normalized for eIF4E and expressed as a percentage of eIF4G retained on m7GTP-Sepharose in the absence of inhibitors (mean ± S.E. (error bars)).
MNK Binding to eIF4G per se Does Not Affect eIF4G Binding to eIF4E
Because both eIF4E and MNK bind to eIF4G (Fig. 6A), it is conceivable that MNK increases eIF4E binding to eIF4G through a mechanism not involving eIF4E phosphorylation (e.g. by altering eIF4G conformation). To investigate the relationship between phosphorylation of eIF4E and its binding to eIF4G, we incubated the S-tagged eIF4G(589–1600) fragment, which contains the eIF4E-binding site (see Fig. 6A), with an equimolar concentration of either eIF4E(S209A) or eIF4E(S209D) in the presence of increasing amounts of recombinant MNK1 (Fig. 9A) or MNK2 (Fig. 9B) and then captured the eIF4G-bound complexes on S-protein-agarose. In the absence of MNK, there was no difference in eIF4G binding to either eIF4E phosphorylation mutant (lanes 1 and 5). The amount of eIF4E(S209A) (lanes 1–4) and eIF4E(S209D) lanes 5–8) bound to S-tagged eIF4G(589–1600) did not significantly change as either MNK1 or MNK2 was increased up to a 2-fold molar excess over eIF4E and eIF4G. These results indicate that MNK binding to eIF4G in the absence of eIF4E phosphorylation does not increase eIF4E binding to eIF4G.
FIGURE 9.
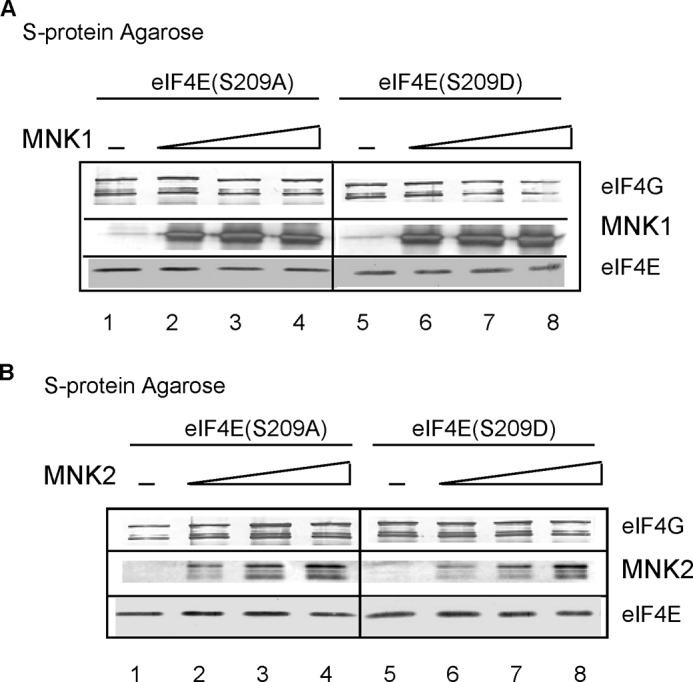
MNK binding to eIF4G per se does not affect eIF4E binding to eIF4G. Binding mixtures containing constant amounts of eIF4G(589–1600) and eIF4E(S209A) (lanes 1–4) or eIF4E(S209D) (lanes 5–8) were incubated with S-protein-agarose in the absence or presence of increasing amounts of MNK1 (A) or MNK2 (B). Bound proteins were purified and analyzed by Western blotting as described under “Experimental Procedures.” Results are representative of two independent experiments.
Discussion
Activation of MNK occurs in response to growth factors, cellular stress, and cytokines. One mechanism by which MNK alters cell fate is by altering the translation of specific mRNAs. To date, there have been numerous mRNAs identified as translationally regulated targets of MNK, including those encoding cytokines IL-17 (32) and pro-inflammatory TNFα (79), the anti-apoptotic protein Mcl-1 (28), ribosomal proteins S19 and L32 (19), proteins involved in cell proliferation (CDK2, CDK8, CDK9, HIF-1α, KAP1, proliferating cell nuclear antigen, PIAS1, and RASSF1 (19)), proteins involved in cell cycle progression (cyclins A, B, D1, and D3 (19, 78)), cancer-related proteins (HER2 (21), VEGF (35), and IRF-1 (84)), transcription factors (RFLAT-1 (46), CHOP (85), and β-catenin (25)), and neuronal proteins (Arc, αCaMKII, PKM-ζ, calmodulin, BDNF (86), and PEM1 (20)). Translation of many of these mRNAs is also responsive to eIF4E levels, indicating a cap- and eIF4E-dependent mechanism of translation: HIF-1α (19), HER2 (21), Mcl-1 (28), cyclin D1 (87, 78), VEGF (88), RFLAT-1 (46), β-catenin (25), and Arc (86). Recently, ribosomal profiling of IL-6-stimulated multiple myeloma cells revealed more than 160 mRNA targets that were translationally inhibited by expression of the phosphodefective eIF4E variant eIF4E(S209A) (89). The fact that their inhibition was abolished by overexpression of wild-type eIF4E indicates a cap-dependent mechanism of translation (89).
The purpose of our study was to understand the mechanism underlying these MNK-dependent effects on translation by investigating different mRNA structural features. Our use of a cell-free translation system allowed us to study effects of MNK on translation per se without complications of mRNA export from the nucleus, localized translation, mRNA stability, changes in MNK1/2 levels due to nucleo-cytoplasmic transport, phosphorylation by MNK of nuclear targets (hnRNPA1 and PSF), and down-regulation of eIF4E levels by MNK, all of which are MNK-dependent events other than translation (see Introduction). Our use of reticulocyte lysate pretreated with micrococcal nuclease eliminated competition of exogenous mRNA with cellular mRNAs. We found that CGP efficiently inhibits the activity of MNK in RRL, and inhibition of MNK by CGP or eIF4G(1357–1600) affects translation of only mRNAs that contain both a cap and a stem-loop in their 5′-UTR. Our data contrast with another in vitro study that showed no difference between wild-type eIF4E and two phosphorylation site variants, eIF4E(S209D) and eIF4E(S209A), for translation of capped luciferase mRNA containing a stem-loop in the 5′-UTR (55). To understand this disparity, it is important to note two differences between the work of McKendrick et al. (55) and the current study. First, the activity of MNK was not altered in the earlier study. Second, McKendrick et al. (55) used a longer stem-loop (ΔG = −21.3 kcal/mol) that inhibited translation more than 20-fold, whereas ours (ΔG = −13.1 kcal/mol) inhibited translation <1.5-fold. The increase in RNA unwinding activity by eIF4F caused by MNK phosphorylation of eIF4E is apparently insufficient to overcome such severe inhibition of translation.
We did not observe an inhibitory effect of either CGP or eIF4G(1357–1600) on IRES-driven translation. This contrasts with several previous studies in whole cells in which either MNK was overexpressed (45, 47) or the signaling cascade leading to its activation was inhibited with rapamycin (90). Interpreting these studies with respect to an effect on translation per se is complicated by the ability of MNK to shuttle between the nucleus and cytoplasm and also to phosphorylate hnRNPA1 and PSF (91, 92), which are RNA-binding proteins that could potentially affect IRES-driven translation. An additional, recently identified MNK target, the Ser/Arg-rich protein kinase, is involved in mRNA splicing, export, stability, and translational initiation of type I picornovirus IRESs (93, 94); Ser/Arg-rich protein kinase phosphorylation could potentially account for MNK-related differences in IRES-driven translation between whole-cell and cell-free systems.
We found that translational sensitivity of mRNA containing a cap and 5′ stem-loop to MNK activity requires intact eIF4F, because cleavage of eIF4G with coxsackievirus 2A protease abrogates the inhibitory effect of CGP. It has been suggested that phosphorylation of eIF4E by MNK increases translational initiation by stimulating the release of eIF4F from capped mRNA (16) or by inhibiting the rebinding of eIF4F to the cap (17), the idea being that once mRNA is recruited to eIF4F, release of the cap is needed before scanning to the initiation codon can occur. This model is based on the fact that phosphorylation of eIF4E reduces its affinity for the cap, although the mechanism is controversial, one group claiming an increase of koff (16) and another claiming a decrease in kon (17). This model predicts that inhibiting MNK would inhibit eIF4E- and cap-dependent translation. However, we did not observe translational inhibition by either CGP or eIF4G(1357–1600) of mRNA containing a cap but no 5′-UTR secondary structure, although initiation of this mRNA was shown to be cap-dependent based on its inhibition by m7GTP. Rather, these MNK inhibitors affected translation of only mRNA containing a cap and 5′ stem-loop, which requires both the cap recognition and unwinding activities of eIF4F. We conclude that inhibition of MNK affects the coupled cap recognition and RNA-unwinding activities of eIF4F.
We hypothesize that MNK affects eIF4F assembly. In support of this idea, the current study and those of others have shown that modulation of MNK activity affects the amount of eIF4G bound to eIF4E (Fig. 8 and Refs. 44, 82, and 86). It also affects co-immunoprecipitation of phospho-eIF4E with MNK, which presumably reflects the presence of both entities in a complex with eIF4G (89). However, there are several studies showing no effect of inhibition of MNK activity on the level of eIF4G bound to eIF4E in cells (19, 45, 78, 95). These discrepancies could be due to different degrees of MNK inactivation and/or higher cellular levels of eIF4G compared with eIF4E. It was also shown that MNK activity may contribute to the phosphorylation of 4E-BP1 (96) and 4E-BP2 (44) and thereby affect eIF4F assembly. Moreover, inhibition of MNK activity with CGP increased binding of eIF4E to 4E-BP1 on m7GTP-Sepharose in glioma cells (96). However, we failed to observe any effect of CGP on retention of 4E-BP1 on m7GTP-Sepharose (Fig. 8A) or the level of 4E-BP1 phosphorylation in RRL (data not shown), suggesting that MNK effects on eIF4F assembly are not due to 4E-BP1 in our system.
It was recently suggested that phosphorylation of eIF4E itself stimulates association of eIF4E with eIF4G, based on the positive correlation between eIF4E phosphorylation and increased eIF4F formation (97) and the observation that phosphorylated eIF4E has lower affinity for 4E-BP1 (98). In the present study, we did not observe a difference in binding of recombinant eIF4G to either eIF4E(S209A) or eIF4E(S209D), suggesting that phosphorylation of eIF4E per se does not modulate its binding to eIF4G. It is conceivable that binding of MNK to eIF4G induces a conformational change in eIF4G that promotes its binding to eIF4E. However, we failed to observe any changes in eIF4G and eIF4E binding in the presence of a 2-fold molar excess of inactive MNK over eIF4G and eIF4E (Fig. 9). It was shown that MNK binding to eIF4G is regulated not only by MNK activation status but also by phosphorylation of substrates, binding to ATP analogs, the presence of RNA, and phosphorylation of eIF4G (15, 53, 99, 100–104). Any of these events could conceivably induce conformational changes in eIF4G that promote the formation of eIF4F.
Two of the best studied signal transduction pathways regulating translational initiation are the MNK/eIF4E and mTOR/4E-BP1 pathways, which act through different mechanisms. Considering that activation of both pathways increases assembly of eIF4F, one might expect the consequences of these two mechanisms to be the same. However, in prostate cancer cells, rapamycin and CGP inhibit different spectra of cyclin mRNAs (19). Also, the combination of inhibitors suppressing the mTOR/4E-BP1 and MNK/eIF4E pathways triggers apoptosis in cutaneous lymphomas (105), reduces glioblastoma growth (96), and suppresses proliferation and induces apoptosis in leukemia cells (106), but this does not occur with either inhibitor separately. These results suggest that the MNK/eIF4E and mTOR/4E-BP1 pathways affect the translation of different spectra of mRNAs, despite the fact that they both increase eIF4F assembly.
Author Contributions
N. L. K. designed and monitored all experiments, contributed to the data presented in Figs. 1 and 5–9, and wrote the manuscript. A. S. performed the experiments presented in Figs. 3 and 4. R. E. R. contributed to the experiment design, data interpretation, and also manuscript writing. H. G. and M. A. E. provided valuable advice on experiment planning and data discussion. All authors read and approved the final manuscript.
Acknowledgments
We thank Tatyana Pestova (SUNY, Downstate Medical Center, Brooklyn, NY) for plasmids to synthetize (CAA)n-GUS and (CAA)n-Stem1–4-GUS mRNAs and also for providing data for Fig. 2B; Edward Darzynkiewicz (Department of Biophysics, Warsaw University, Poland) for providing the ARCA cap analog; and Maria Quimis for technical assistance.
This work was supported by NIGMS, National Institutes of Health, Grant 2R01GM20818 (to R. E. R.). Hermann Gram is an employee and shareholder of Novartis AG. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- eIF
- eukaryotic translation initiation factor
- 4E-BP
- eIF4E-binding protein
- mTOR
- mammalian target of rapamycin
- MNK
- mitogen-activated protein kinase-interacting kinase
- CGP
- MNK inhibitor CGP57380
- EMCV
- encephalomyocarditis virus
- GUS
- β-glucuronidase
- IRES
- internal ribosomal entry site
- Luc
- firefly luciferase
- T-H-S
- thioredoxin-His6-S-peptide
- RRL
- rabbit reticulocyte lysate
- ARCA
- anti-reverse cap analog
- aa
- amino acid(s).
References
- 1. Sonenberg N., and Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Topisirovic I., Svitkin Y. V., Sonenberg N., and Shatkin A. J. (2011) Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 [DOI] [PubMed] [Google Scholar]
- 3. Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 [DOI] [PubMed] [Google Scholar]
- 4. Pelletier J., and Sonenberg N. (1987) The involvement of mRNA secondary structure in protein synthesis. Biochem. Cell Biol. 65, 576–581 [DOI] [PubMed] [Google Scholar]
- 5. Patursky-Polischuk I., Stolovich-Rain M., Hausner-Hanochi M., Kasir J., Cybulski N., Avruch J., Rüegg M. A., Hall M. N., and Meyuhas O. (2009) The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 29, 640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson R. J. (1991) The ATP requirement for initiation of eukaryotic translation varies according to the mRNA species. Eur. J. Biochem. 200, 285–294 [DOI] [PubMed] [Google Scholar]
- 7. Svitkin Y. V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G. J., and Sonenberg N. (2001) The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7, 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gingras A. C., Raught B., and Sonenberg N. (2004) mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279, 169–197 [DOI] [PubMed] [Google Scholar]
- 9. Ma X. M., and Blenis J. (2009) Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 10. Waskiewicz A. J., Flynn A., Proud C. G., and Cooper J. A. (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukunaga R., and Hunter T. (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joshi B., Cai A.-L., Keiper B. D., Minich W. B., Mendez R., Beach C. M., Stepinski J., Stolarski R., Darzynkiewicz E., and Rhoads R. E. (1995) Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J. Biol. Chem. 270, 14597–14603 [DOI] [PubMed] [Google Scholar]
- 13. Flynn A., and Proud C. G. (1995) Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J. Biol. Chem. 270, 21684–21688 [DOI] [PubMed] [Google Scholar]
- 14. Pyronnet S., Imataka H., Gingras A.-C., Fukunaga R., Hunter T., and Sonenberg N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 18, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shveygert M., Kaiser C., Bradrick S. S., and Gromeier M. (2010) Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol. Cell. Biol. 30, 5160–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scheper G. C., van Kollenburg B., Hu J., Luo Y., Goss D. J., and Proud C. G. (2002) Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277, 3303–3309 [DOI] [PubMed] [Google Scholar]
- 17. Slepenkov S. V., Darzynkiewicz E., and Rhoads R. E. (2006) Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J. Biol. Chem. 281, 14927–14938 [DOI] [PubMed] [Google Scholar]
- 18. Flynn A., and Proud C. G. (1996) Insulin and phorbol ester stimulate initiation factor eIF-4E phosphorylation by distinct pathways in Chinese hamster ovary cells overexpressing the insulin receptor. Eur. J. Biochem. 236, 40–47 [DOI] [PubMed] [Google Scholar]
- 19. Bianchini A., Loiarro M., Bielli P., Busà R., Paronetto M. P., Loreni F., Geremia R., and Sette C. (2008) Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis 29, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 20. Paix A., Le Nguyen P. N., and Sardet C. (2011) Bi-polarized translation of ascidian maternal mRNA determinant pem-1 associated with regulators of the translation machinery on cortical endoplasmic reticulum (cER). Dev. Biol. 357, 211–226 [DOI] [PubMed] [Google Scholar]
- 21. Yoon S.-O., Shin S., and Lipscomb E. A. (2006) A novel mechanism for integrin-mediated Ras activation in breast carcinoma cells: the α6β4 integrin regulates ErbB2 translation and transactivates epidermal growth factor receptor/ErbB2 signaling. Cancer Res. 66, 2732–2739 [DOI] [PubMed] [Google Scholar]
- 22. Furic L., Rong L., Larsson O., Koumakpayi I. H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L. A., Pandolfi P. P., Saad F., and Sonenberg N. (2010) eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferrandiz-Pulido C., Masferrer E., Toll A., Hernandez-Losa J., Mojal S., Pujol R. M., Ramon y Cajal S., de Torres I., and Garcia-Patos V. (2013) mTOR signaling pathway in penile squamous cell carcinoma: pmTOR and peIF4E over expression correlate with aggressive tumor behavior. J. Urol. 190, 2288–2295 [DOI] [PubMed] [Google Scholar]
- 24. Das F., Ghosh-Choudhury N., Bera A., Kasinath B. S., and Choudhury G. G. (2013) TGFbeta-induced PI 3 kinase-dependent Mnk-1 activation is necessary for Ser-209 phosphorylation of eIF4E and mesangial cell hypertrophy. J. Cell. Physiol. 228, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim S., Saw T. Y., Zhang M., Janes M. R., Nacro K., Hill J., Lim A. Q., Chang C. T., Fruman D. A., Rizzieri D. A., Tan S. Y., Fan H., Chuah C. T., and Ong S. T. (2013) Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc. Natl. Acad. Sci. U.S.A. 110, E2298–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng J., Li J., Xu L., Xie G., Wen Q., Luo J., Li D., Huang D., and Fan S. (2014) Phosphorylated Mnk1 and eIF4E are associated with lymph node metastasis and poor prognosis of nasopharyngeal carcinoma. PLoS One 9, e89220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carroll M., and Borden K. L. (2013) The oncogene eIF4E: using biochemical insights to target cancer. J. Interferon Cytokine Res. 33, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wendel H.-G., Silva R. L. A., Malina A., Mills J. R., Zhu H., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Teruya-Feldstein J., Pelletier J., and Lowe S. W. (2007) Dissecting eIF4E action in tumorigenesis. Genes Dev. 21, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chrestensen C. A., Shuman J. K., Eschenroeder A., Worthington M., Gram H., and Sturgill T. W. (2007) MNK1 and MNK2 regulation in HER2-overexpressing breast cancer lines. J. Biol. Chem. 282, 4243–4252 [DOI] [PubMed] [Google Scholar]
- 30. Ueda T., Sasaki M., Elia A. J., Chio I. I., Hamada K., Fukunaga R., and Mak T. W. (2010) Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc. Natl. Acad. Sci. U.S.A. 107, 13984–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konicek B. W., Stephens J. R., McNulty A. M., Robichaud N., Peery R. B., Dumstorf C. A., Dowless M. S., Iversen P. W., Parsons S., Ellis K. E., McCann D. J., Pelletier J., Furic L., Yingling J. M., Stancato L. F., Sonenberg N., and Graff J. R. (2011) Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 71, 1849–1857 [DOI] [PubMed] [Google Scholar]
- 32. Noubade R., Krementsov D. N., Del Rio R., Thornton T., Nagaleekar V., Saligrama N., Spitzack A., Spach K., Sabio G., Davis R. J., Rincon M., and Teuscher C. (2011) Activation of p38 MAPK in CD4 T cells controls IL-17 production and autoimmune encephalomyelitis. Blood 118, 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altman J. K., Szilard A., Konicek B. W., Iversen P. W., Kroczynska B., Glaser H., Sassano A., Vakana E., Graff J. R., and Platanias L. C. (2013) Inhibition of Mnk kinase activity by cercosporamide and suppressive effects on acute myeloid leukemia precursors. Blood 121, 3675–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chrestensen C. A., Eschenroeder A., Ross W. G., Ueda T., Watanabe-Fukunaga R., Fukunaga R., and Sturgill T. W. (2007) Loss of MNK function sensitizes fibroblasts to serum-withdrawal induced apoptosis. Genes Cells 12, 1133–1140 [DOI] [PubMed] [Google Scholar]
- 35. Korneeva N. L., Soung Y. H., Kim H. I., Giordano A., Rhoads R. E., Gram H., and Chung J. (2010) Mnk mediates integrin (α6β4) dependent eIF4E phosphorylation and translation of VEGF mRNA. Mol. Cancer Res. 8, 1571–1578 [DOI] [PubMed] [Google Scholar]
- 36. Astanehe A., Finkbeiner M. R., Krzywinski M., Fotovati A., Dhillon J., Berquin I. M., Mills G. B., Marra M. A., and Dunn S. E. (2012) MKNK1 is a YB-1 target gene responsible for imparting trastuzumab resistance and can be blocked by RSK inhibition. Oncogene 31, 4434–4446 [DOI] [PubMed] [Google Scholar]
- 37. Adesso L., Calabretta S., Barbagallo F., Capurso G., Pilozzi E., Geremia R., Delle Fave G., and Sette C. (2013) Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene 32, 2848–2857 [DOI] [PubMed] [Google Scholar]
- 38. Proud C. G. (2015) Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta 1849, 766–773 [DOI] [PubMed] [Google Scholar]
- 39. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., and Fukunaga R. (2004) Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramalingam S., Gediya L., Kwegyir-Afful A. K., Ramamurthy V. P., Purushottamachar P., Mbatia H., and Njar V. C. (2014) First MNKs degrading agents block phosphorylation of eIF4E, induce apoptosis, inhibit cell growth, migration and invasion in triple negative and Her2-overexpressing breast cancer cell lines. Oncotarget 5, 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buxade M., Parra-Palau J. L., and Proud C. G. (2008) The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front. Biosci. 13, 5359–5373 [DOI] [PubMed] [Google Scholar]
- 42. Joshi S., and Platanias L. C. (2014) Mnk kinase pathway: cellular functions and biological outcomes. World J. Biol. Chem. 5, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smart F. M., Edelman G. M., and Vanderklish P. W. (2003) BDNF induces translocation of initiation factor 4E to mRNA granules: evidence for a role of synaptic microfilaments and integrins. Proc. Natl. Acad. Sci. U.S.A. 100, 14403–14408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Panja D., Kenney J. W., D'Andrea L., Zalfa F., Vedeler A., Wibrand K., Fukunaga R., Bagni C., Proud C. G., and Bramham C. R. (2014) Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep. 9, 1430–1445 [DOI] [PubMed] [Google Scholar]
- 45. Knauf U., Tschopp C., and Gram H. (2001) Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol. Cell. Biol. 21, 5500–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikolcheva T., Pyronnet S., Chou S. Y., Sonenberg N., Song A., Clayberger C., and Krensky A. M. (2002) A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J. Clin. Invest. 110, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ross G., Dyer J. R., Castellucci V. F., and Sossin W. S. (2006) Mnk is a negative regulator of cap-dependent translation in Aplysia neurons. J. Neurochem. 97, 79–91 [DOI] [PubMed] [Google Scholar]
- 48. Dyer J., and Sossin W. S. (2013) Characterization of the role of eIF4G in stimulating cap- and IRES-dependent translation in aplysia neurons. PLoS One 8, e74085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tuxworth W. J. Jr., Saghir A. N., Spruill L. S., Menick D. R., and McDermott P. J. (2004) Regulation of protein synthesis by eIF4E phosphorylation in adult cardiocytes: the consequence of secondary structure in the 5′-untranslated region of mRNA. Biochem. J. 378, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Landon A. L., Muniandy P. A., Shetty A. C., Lehrmann E., Volpon L., Houng S., Zhang Y., Dai B., Peroutka R., Mazan-Mamczarz K., Steinhardt J., Mahurkar A., Becker K. G., Borden K. L., and Gartenhaus R. B. (2014) MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nat. Commun. 5, 5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., and Rhoads R. E. (2001) Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogues 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA 7, 1486–1495 [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao X., Lamphear B. J., Xiong D., Knowlton K., and Rhoads R. E. (2003) Protection of cap-dependent protein synthesis in vivo and in vitro with an eIF4G-1 variant highly resistant to cleavage by coxsackievirus 2A protease. J. Biol. Chem. 278, 4449–4457 [DOI] [PubMed] [Google Scholar]
- 53. Orton K. C., Ling J., Waskiewicz A. J., Cooper J. A., Merrick W. C., Korneeva N. L., Rhoads R. E., Sonenberg N., and Traugh J. A. (2004) Phosphorylation of Mnk1 by caspase-activated Pak2/g-PAK inhibits phosphorylation and interaction of eIF4G with Mnk. J. Biol. Chem. 279, 38649–38657 [DOI] [PubMed] [Google Scholar]
- 54. Tschopp C., Knauf U., Brauchle M., Zurini M., Ramage P., Glueck D., New L., Han J., and Gram H. (2000) Phosphorylation of eIF-4E on Ser 209 in response to mitogenic and inflammatory stimuli is faithfully detected by specific antibodies. Mol. Cell. Biol. Res. Commun. 3, 205–211 [DOI] [PubMed] [Google Scholar]
- 55. McKendrick L., Morley S. J., Pain V. M., Jagus R., and Joshi B. (2001) Phosphorylation of eukaryotic initiation factor 4E (eIF4E) at Ser209 is not required for protein synthesis in vitro and in vivo. Eur. J. Biochem. 268, 5375–5385 [DOI] [PubMed] [Google Scholar]
- 56. Pause A., Méthot N., Svitkin Y., Merrick W. C., and Sonenberg N. (1994) Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13, 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lamphear B. J., Yan R., Yang F., Waters D., Liebig H.-D., Klump H., Kuechler E., Skern T., and Rhoads R. E. (1993) Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268, 19200–19203 [PubMed] [Google Scholar]
- 58. Pestova T. V., and Kolupaeva V. G. (2002) The roles of individual eukariotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pisareva V. P., Pisarev A. V., Komar A. A., Hellen C. U., and Pestova T. V. (2008) Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mathews D. H., Sabina J., Zuker M., and Turner D. H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288, 911–940 [DOI] [PubMed] [Google Scholar]
- 61. Grudzien E., Kalek M., Jemielity J., Darzynkiewicz E., and Rhoads R. E. (2006) Differential inhibition of mRNA degradation pathways by novel cap analogs. J. Biol. Chem. 281, 1857–1867 [DOI] [PubMed] [Google Scholar]
- 62. Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., and Rhoads R. E. (2003) Novel “anti-reverse” cap analogues with superior translational properties. RNA 9, 1108–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ryman K. D., Meier K. C., Nangle E. M., Ragsdale S. L., Korneeva N. L., Rhoads R. E., MacDonald M. R., and Klimstra W. B. (2005) Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by α/β interferon priming of dendritic cells. J. Virol. 79, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Korneeva N. L., First E. A., Benoit C. A., and Rhoads R. E. (2005) Interaction between NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 280, 1872–1881 [DOI] [PubMed] [Google Scholar]
- 65. Yan R., Rychlik W., Etchison D., and Rhoads R. E. (1992) Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J. Biol. Chem. 267, 23226–23231 [PubMed] [Google Scholar]
- 66. Tzareva N. V., Makhno V. I., and Boni I. V. (1994) Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337, 189–194 [DOI] [PubMed] [Google Scholar]
- 67. Elroy-Stein O., Fuerst T. R., and Moss B. (1989) Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. U.S.A. 86, 6126–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lomakin I. B., Hellen C. U. T., and Pestova T. V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20, 6019–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sonenberg N., Morgan M. A., Merrick W. C., and Shatkin A. J. (1978) A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. U.S.A. 75, 4843–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., and Merrick W. C. (1987) The ATP-dependent interaction of eukaryotic initition factors with mRNA. J. Biol. Chem. 262, 3826–3832 [PubMed] [Google Scholar]
- 71. Lamphear B. J., Kirchweger R., Skern T., and Rhoads R. E. (1995) Mapping of functional domains in eIF4G with picornaviral proteases: implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270, 21975–21983 [DOI] [PubMed] [Google Scholar]
- 72. Imataka H., and Sonenberg N. (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17, 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Korneeva N. L., Lamphear B. J., Hennigan F. L. C., Merrick W. C., and Rhoads R. E. (2001) Characterization of the two eIF4A binding sites on human eIF4G-1. J. Biol. Chem. 276, 2872–2879 [DOI] [PubMed] [Google Scholar]
- 74. Niedzwiecka A., Marcotrigiano J., Stepinski J., Jankowska-Anyszka M., Wyslouch-Cieszynska A., Dadlez M., Gingras A.-C., Mak P., Darzynkiewicz E., Sonenberg N., Burley S. K., and Stolarski R. (2002) Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319, 615–635 [DOI] [PubMed] [Google Scholar]
- 75. De Gregorio E., Preiss T., and Hentze M. W. (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′-end-dependent. RNA 4, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bradley C. A., Padovan J. C., Thompson T. L., Benoit C. A., Chait B. T., and Rhoads R. E. (2002) Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 277, 12559–12571 [DOI] [PubMed] [Google Scholar]
- 77. Byrd M. P., Zamora M., and Lloyd R. E. (2002) Generation of multiple isoforms of eukaryotic tranlsation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22, 4499–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang M., Fu W., Prabhu S., Moore J. C., Ko J., Kim J. W., Druker B. J., Trapp V., Fruehauf J., Gram H., Fan H. Y., and Ong S. T. (2008) Inhibition of polysome assembly enhances imatinib activity against chronic myelogenous leukemia and overcomes imatinib resistance. Mol. Cell. Biol. 28, 6496–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Buxadé M., Parra J. L., Rousseau S., Shpiro N., Marquez R., Morrice N., Bain J., Espel E., and Proud C. G. (2005) The Mnks are novel components in the control of TNF α biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23, 177–189 [DOI] [PubMed] [Google Scholar]
- 80. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., and Thach R. E. (1985) ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260, 7651–7658 [PubMed] [Google Scholar]
- 82. Li Y., Yue P., Deng X., Ueda T., Fukunaga R., Khuri F. R., and Sun S. Y. (2010) Protein phosphatase 2A negatively regulates eukaryotic initiation factor 4E phosphorylation and eIF4F assembly through direct dephosphorylation of Mnk and eIF4E. Neoplasia 12, 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Marcotrigiano J., Gingras A.-C., Sonenberg N., and Burley S. K. (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716 [DOI] [PubMed] [Google Scholar]
- 84. Joshi S., Sharma B., Kaur S., Majchrzak B., Ueda T., Fukunaga R., Verma A. K., Fish E. N., and Platanias L. C. (2011) Essential role for Mnk kinases in type II interferon (IFNγ) signaling and its suppressive effects on normal hematopoiesis. J. Biol. Chem. 286, 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen Y. J., Tan B. C., Cheng Y. Y., Chen J. S., and Lee S. C. (2010) Differential regulation of CHOP translation by phosphorylated eIF4E under stress conditions. Nucleic Acids Res. 38, 764–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Panja D., Dagyte G., Bidinosti M., Wibrand K., Kristiansen A. M., Sonenberg N., and Bramham C. R. (2009) Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J. Biol. Chem. 284, 31498–31511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rousseau D., Kaspar R., Rosenwald I., Gehrke L., and Sonenberg N. (1996) Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. U.S.A. 93, 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kevil C. G., De Benedetti A., Payne D. K., Coe L. L., Laroux F. S., and Alexander J. S. (1996) Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: implication for tumor angiogenesis. Int. J. Cancer 65, 785–790 [DOI] [PubMed] [Google Scholar]
- 89. Shi Y., Frost P., Hoang B., Yang Y., Bardeleben C., Gera J., and Lichtenstein A. (2014) MNK1-induced eIF-4E phosphorylation in myeloma cells: a pathway mediating IL-6-induced expansion and expression of genes involved in metabolic and proteotoxic responses. PLoS One 9, e94011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90. Shi Y., Frost P., Hoang B., Yang Y., Fukunaga R., Gera J., and Lichtenstein A. (2013) MNK kinases facilitate c-myc IRES activity in rapamycin-treated multiple myeloma cells. Oncogene 32, 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jo O. D., Martin J., Bernath A., Masri J., Lichtenstein A., and Gera J. (2008) Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J. Biol. Chem. 283, 23274–23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cobbold L. C., Spriggs K. A., Haines S. J., Dobbyn H. C., Hayes C., de Moor C. H., Lilley K. S., Bushell M., and Willis A. E. (2008) Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 28, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Brown M. C., Dobrikov M. I., and Gromeier M. (2014) Mitogen-activated protein kinase-interacting kinase regulates mTOR/AKT signaling and controls the serine/arginine-rich protein kinase-responsive type 1 internal ribosome entry site-mediated translation and viral oncolysis. J. Virol. 88, 13149–13160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Brown M. C., Bryant J. D., Dobrikova E. Y., Shveygert M., Bradrick S. S., Chandramohan V., Bigner D. D., and Gromeier M. (2014) Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J. Virol. 88, 13135–13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Waskiewicz A. J., Johnson J. C., Penn B., Mahalingam M., Kimball S. R., and Cooper J. A. (1999) Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Grzmil M., Huber R. M., Hess D., Frank S., Hynx D., Moncayo G., Klein D., Merlo A., and Hemmings B. A. (2014) MNK1 pathway activity maintains protein synthesis in rapalog-treated gliomas. J. Clin. Invest. 124, 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Y., Li Y., and Yang D. Q. (2008) Phosphorylation of eIF-4E positively regulates formation of the eIF-4F translation initiation complex following DNA damage. Biochem. Biophys. Res. Commun. 367, 54–59 [DOI] [PubMed] [Google Scholar]
- 98. Pons B., Peg V., Vázquez-Sánchez M. A., López-Vicente L., Argelaguet E., Coch L., Martínez A., Hernández-Losa J., Armengol G., and Ramon Y Cajal S. (2011) The effect of p-4E-BP1 and p-eIF4E on cell proliferation in a breast cancer model. Int. J. Oncol. 39, 1337–1345 [DOI] [PubMed] [Google Scholar]
- 99. Lawson S. K., Dobrikova E. Y., Shveygert M., and Gromeier M. (2013) p38α mitogen-activated protein kinase depletion and repression of signal transduction to translation machinery by miR-124 and -128 in neurons. Mol. Cell. Biol. 33, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Scheper G. C., Morrice N. A., Kleijn M., and Proud C. G. (2001) The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21, 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Parra J. L., Buxadé M., and Proud C. G. (2005) Features of the catalytic domains and C termini of the MAPK signal-integrating kinases Mnk1 and Mnk2 determine their differing activities and regulatory properties. J. Biol. Chem. 280, 37623–37633 [DOI] [PubMed] [Google Scholar]
- 102. Cuesta R., Xi Q., and Schneider R. J. (2004) Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 78, 7707–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dobrikov M., Dobrikova E., Shveygert M., and Gromeier M. (2011) Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase Cα regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 31, 2947–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dobrikov M. I., Shveygert M., Brown M. C., and Gromeier M. (2014) Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:cyclin B inhibits eIF4A helicase complex binding with RNA. Mol. Cell. Biol. 34, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marzec M., Liu X., Wysocka M., Rook A. H., Odum N., and Wasik M. A. (2011) Simultaneous inhibition of mTOR-containing complex 1 (mTORC1) and MNK induces apoptosis of cutaneous T-cell lymphoma (CTCL) cells. PLoS One 6, e24849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106. Teo T., Yu M., Yang Y., Gillam T., Lam F., Sykes M. J., and Wang S. (2015) Pharmacologic co-inhibition of Mnks and mTORC1 synergistically suppresses proliferation and perturbs cell cycle progression in blast crisis-chronic myeloid leukemia cells. Cancer Lett. 357, 612–623 [DOI] [PubMed] [Google Scholar]