Abstract
The type I fatty acid synthase (FASN) is responsible for the de novo synthesis of palmitate. Chain length selection and release is performed by the C-terminal thioesterase domain (TE1). FASN expression is up-regulated in cancer, and its activity levels are controlled by gene dosage and transcriptional and post-translational mechanisms. In addition, the chain length of fatty acids produced by FASN is controlled by a type II thioesterase called TE2 (E.C. 3.1.2.14). TE2 has been implicated in breast cancer and generates a broad lipid distribution within milk. The molecular basis for the ability of the TE2 to compete with TE1 for the acyl chain attached to the acyl carrier protein (ACP) domain of FASN is unknown. Herein, we show that human TE1 efficiently hydrolyzes acyl-CoA substrate mimetics. In contrast, TE2 prefers an engineered human acyl-ACP substrate and readily releases short chain fatty acids from full-length FASN during turnover. The 2.8 Å crystal structure of TE2 reveals a novel capping domain insert within the α/β hydrolase core. This domain is reminiscent of capping domains of type II thioesterases involved in polyketide synthesis. The structure also reveals that the capping domain had collapsed onto the active site containing the Ser-101–His-237–Asp-212 catalytic triad. This observation suggests that the capping domain opens to enable the ACP domain to dock and to place the acyl chain and 4′-phosphopantetheinyl-linker arm correctly for catalysis. Thus, the ability of TE2 to prematurely release fatty acids from FASN parallels the role of editing thioesterases involved in polyketide and non-ribosomal peptide synthase synthases.
Keywords: crystal structure, fatty acid synthase (FAS), hydrolase, lipid metabolism, serine esterase, thioesterase
Introduction
The dimeric, type I fatty acid synthase (FASN)2 is the sole enzyme responsible for the biosynthesis of palmitate (16 carbons (C16)) and stearate (C18) in mammals (1, 2). In this process the growing fatty acid chain is attached to the 4′-phosphopantetheinyl (4′-PPT) linker arm of the acyl carrier protein (ACP) domain and ultimately released by the endogenous, C-terminal thioesterase domain (TE1). The expression of FASN is up-regulated in cancer cells by gene amplification and a variety of transcriptional and translational mechanisms (3–7). The enzymatic activity of FASN can also be modulated by phosphorylation (8). In these scenarios either FASN levels are affected or activity level is modulated. In contrast, the monofunctional, type II thioesterase, which is called TE2 for simplicity, is able to interact with FASN to prematurely release fatty acids of shorter chain length.
TE2, also known as oleoyl thioesterase and medium-chain S-acyl FASN thioester hydrolase (E.C. 3.1.2.14; UniProt Q9NV23), was originally identified in rats, and its localization was confirmed to mammary gland epithelial cells (9, 10). FASN and TE2 are up-regulated during lactation to produce lipids as an energy source and metabolic building blocks for offspring (11, 12). Human TE2 has also been used as a marker for cells of breast epithelial origin and a serum marker for mammary cancer (13, 14). The molecular basis for the unique ability of TE2 to modify the chain length of FASN products is not known.
The acyl product distribution of FASN is normally determined by the selectivity of the TE1 domain (15–17). The palmitate-like inhibitor Orlistat binds to a hydrophobic channel of human TE1, which is juxtaposed between the α/β hydrolase core and a unique subdomain that is inserted within a surface loop (18). It is this “specificity channel” that functions as a ruler to preferentially select C16 and C18 acyl substrates for hydrolysis. The interaction of TE2 with FASN, however, shifts the distribution to favor the medium chain fatty acids laurate (C12) and myristate (C14), with the overall lipid composition of breast milk differing between species (19–22).
The ability of TE2 to compete with the endogenous TE1 domain within the FASN megasynthase for acyl substrates is a conundrum, as the intramolecular cleavage reaction by TE1 should be favored based on proximity. Previous studies that used TE2 from rat and duck support that TE2 can directly interact with FASN (23, 24). These studies showed that an acyl chain must be present on the ACP domain and that dissociation of the complex occurs rapidly for the rat enzyme after hydrolysis. At the present time there are no structural insights into this unique interaction, as the crystal structure of human TE2 or any eukaryotic homolog has not been determined. An initial report of crystallization and diffraction studies for rat TE2 was reported many years ago (25). Moreover, the purification and characterization of the substrate specificity of human TE2 and the impact of its interaction with FASN have not been described.
Herein we report a comparison of the activity of human TE2 and the isolated TE1 domain of human FASN against a panel of acyl-CoA substrates and an acyl-ACP substrate, which contains palmitate attached the 4′-PPT moiety. TE1 is significantly more active than TE2 on acyl-CoA substrate mimetics. In contrast, TE2 was shown to be ∼4-fold more active against the C16-ACP substrate using a new quantitative MALDI-TOF MS-based method. Incubation of TE2 with full-length FASN resulted in a fatty acid profile enriched in short chain fatty acids. The 2.8 Å crystal structure of TE2 reveals the canonical α/β hydrolase core and a capping domain that is more like those of thioesterases involved in the processing of polyketides than that of TE1. Taken together, the results support that TE2 utilizes the unique capping domain to interact with the ACP domain of FASN during catalysis in order to prematurely release fatty acids of shorter chain length.
Experimental Procedures
Cloning, Expression, and Purification of Human TE2 and the ACP and TE1 Domains of FASN
Human TE2 was subcloned into the pET151 TOPO/D vector (Life Technologies) using forward 5′-CACCATGGAGAGAGGAGACCAACCTAAGAGAACC-3′ and reverse 5′-TTAAAAATTGGATATCGATGATACTTCTAGACACTTG-3′ primers. The S101A mutant was generated using the QuikChange procedures (Stratagene) and the forward (5′-GCATTTTTTGGCCACGCTATGGGATCCTACATTGC-3′) and reverse (5′-GCAATGTAGGATCCCATAGCGTGGCCAAAAAATGC-3′) primers. The ACP domain (residues 2118–2212) of human FASN was also subcloned into pET151 TOPO/D (Invitrogen) using the forward (5′-CACCTATAGGGACAGGGACAGCCAGCGGG-3′) and reverse (5′-TTAGCTGGGCCAGACCATCCTCCTT-3′) primers.
The N-terminal, His-tagged TE2 (TE2+His) and ACP domain (ACP+His) were expressed in BL21(DE3) GOLD Escherichia coli cells with induction at 16 °C and 0.1–0.2 mm isopropyl 1-thio-β-d-galactopyranoside overnight. The cells were lysed using an Avestin Emulsiflex-C3 homogenizer. The clarified lysate containing 100 μm PMSF and benzamidine, 1 mm MgCl2, 100 μg of DNase powder, 50 mm HEPES, pH 7.9, 500 mm KCl, 5 mm imidazole, 10% glycerol, and 0.1% Triton X-100 was applied to an equilibrated nickel NTA column (Qiagen). The proteins were eluted using a 5–250 mm imidazole gradient in 20 mm HEPES, pH 7.9, and 500 mm KCl. Fractions containing TE2+His and ACP+His were pooled, treated with 5 mm EDTA, 2 mm dithiothreitol (DTT), and 5.0 mg of GST-tagged HRV-3C protease, and dialyzed against 4.0 liters of 20 mm HEPES, pH 7.0, and 1 mm DTT. Removal of the affinity tags was confirmed using a Bruker Autoflex MALDI-TOF MS instrument and saturated sinapinic acid (Fluka Chemika) matrix solution containing 50% acetonitrile and 0.1% formic acid.
The affinity tag-free TE2 and ACP were applied to a Q-Sepharose fast-flow ion-exchange column (GE Healthcare) and eluted with a 0–500 mm NaCl linear gradient in 20 mm HEPES, pH 7.5 (TE2)/7.0 (ACP), and 1 mm DTT. Fractions containing the desired protein were pooled, concentrated to 5 ml, and injected over a Superdex 75 gel filtration column equilibrated in 20 mm HEPES, pH 7.5/7.0, 100 mm (TE2)/250 mm (ACP) NaCl, and 1 mm DTT. The fractions containing TE2 or ACP were pooled, concentrated, and aliquoted for storage at −80 °C. The S101A variant of TE2 was purified using the same protocol. The cloning, expression, and purification strategies for wild-type TE1 have been previously reported (18).
Cloning, Expression, and Purification of Accessory Proteins Used to Generate the Acyl-ACP Substrate
Generation of the C16-ACP substrate required two enzymes to sequentially add in vitro the 4′-PPT moiety and the fatty acid palmitate. The gene for the phosphopantetheinyl transferase from Streptomyces coelicolor (UniProt # O86785, Loader+His) was subcloned into pET151 TOPO/D vector using forward (5′-CACCATGAGCATCATCGGGGTCGGG-3′) and reverse (5′-CTATCCCTCCGCGATCACCACCG-3′) primers (26). The protein was purified using the same procedure as TE2, except that the His tag was not removed to facilitate its removal from the subsequent reactions. The E. coli holo acyl-ACP synthase (HAAS+His) expression construct (pET28b) was kindly provided by Dr. John Shanklin, Brookhaven National Laboratories. HAAS+His purification strategy was slightly modified from what was previously reported (27). First, cell cultures were grown at 37 °C to mid-log phase and then cooled to room temperature before induction. Second, HAAS+His was eluted from a NTA column with 250 mm imidazole in 50 mm Tris, pH 8.0, 20 mm MgCl2, 2% Triton X-100. The HAAS+His was dialyzed overnight into 50 mm Tris, pH 8.0, 20 mm MgCl2, and 2% Triton X-100 and then aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C until use.
Synthesis and Purification of the C16-ACP Substrate
The 4′-phosphopantetheinyl-ACP (ACP-SH) was produced using a modified procedure (26). Fifty milligrams of ACP was resuspended in 5 ml of reaction buffer containing 50 mm Tris, pH 8.0, 1 mm MgCl2, 10 mm coenzyme A (CoA), 1.6 mg/ml Loader+His, and 5 mm β-mercaptoethanol. The reaction was incubated at room temperature with intermittent mixing for 1.5 h. ACP loading was monitored via MALDI-TOF MS until only the fully loaded, ACP-SH peak was observed at 11,524 m/z. The completed reaction was diluted with 5 ml of 20 mm HEPES, pH 7.9, 500 mm KCl and then applied to a NTA column equilibrated with the same buffer. Fractions containing ACP-SH were dialyzed overnight against 2.0 liters 50 mm Tris, pH 8.0, 1 mm DTT, concentrated using a 5000 molecular weight cutoff spin concentrator, aliquoted, and stored at −80 °C.
Loading of palmitate onto ACP-SH was optimized by first performing small scale (100 μl) reactions composed of 1 mg/ml ACP-SH, 0.3 mg/ml HAAS+His, 100 mm Tris, pH 8.0, 10 mm MgCl2, 1% Triton X-100, and 3 μl of saturated palmitate in 100% methanol. ACP-SH loading was monitored over 2 h via MALDI-TOF MS until only the acyl-ACP peak was observed at 11,763 m/z. Samples were desalted using C18 ZipTip (Millipore) procedures before MALDI-TOF MS analyses. The reactions were scaled proportionally to generate larger batches of C16-ACP in 10 ml of reaction buffer. A 2-ml NTA column, equilibrated in 50 mm Tris, pH 8.0, and 0.5% Triton X-100, was used to remove the accessory proteins from the 0.45-μm-filtered reaction solution. The flow-through was collected and dialyzed overnight into 2 liters of stabilization buffer, 20 mm sodium acetate, pH 5.5, and 0.5% Tween 20 at 4 °C. The C16-ACP was concentrated using a 5000 molecular weight cutoff spin concentrator, aliquoted, and stored at −80 °C.
Coupled Assay for Monitoring Acyl-CoA Hydrolysis
The release of CoA from acyl-CoA substrates was performed using a modified procedure (28). Thiamine pyrophosphate, α-ketoglutarate dehydrogenase, and α-ketoglutarate were purchased from Sigma. Acyl-CoA substrates were purchased from Avanti Polar Lipids, Inc. Fresh stocks of NAD+ and NADH were prepared, and the concentrations were measured at 260 nm (ϵ = 18 mm−1 cm−1) and 340 nm (ϵ = 6.22 mm−1 cm−1), respectively. All kinetic measurements were performed on a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies, Santa Clara, CA) at 30 °C. The voltage gain was set to achieve the maximum relative fluorescence units (RFUs) when 10 μm NADH was measured. The different thioesterases (25 nm TE1 and 125 nm TE2) were assayed for 5 min in triplicate on multiple days; these enzyme concentrations ensured linear rates. Reaction rates were measured using 10–320 μm acyl-CoA substrates in 250 mm Na2HPO4/KH2PO4, pH 7.5. The rates were converted from RFU/min to μmol of acyl-CoA hydrolyzed/min using the slope of the RFU versus NADH standard curve (0–10 μm). Specific activities (nmol/min/mg) were calculated by dividing the enzyme activities by the amount of enzyme in the reaction.
Quantitative MALDI-TOF MS Assay for Monitoring Acyl-ACP Hydrolysis
To rapidly monitor the release of palmitate from C16-ACP by TE1 and TE2, a MALDI-TOF MS method was developed and validated (see Fig. 3). The same MS instrument and matrix described above were used. Reactions were initiated by the addition of C16-ACP and monitored for 10 min. The final reaction contained 25 mm Hepes, pH 7.5, 100 mm NaCl, 25 nm TE1, or 10 nm TE2, which had been pre-reduced with 1 mm DTT and desalted, and 20 μm C16-ACP. The reactions were quenched by the direct addition of matrix. The positive ion mode was used to collect and sum 1500 spectra. The ratio of ACP-SH peak intensities to the sum of the observed ACP-SH and C16-ACP peak intensities was determined by the equation IACP-SH/(IACP-SH + IC16-ACP)). The amount ACP-SH formed per min was determined by applying the equation above to the observed MALDI-TOF MS spectra. Specific activities (nmol/min/mg) were calculated by the amount of enzyme in the reaction. The experiments were completed in triplicate on multiple days.
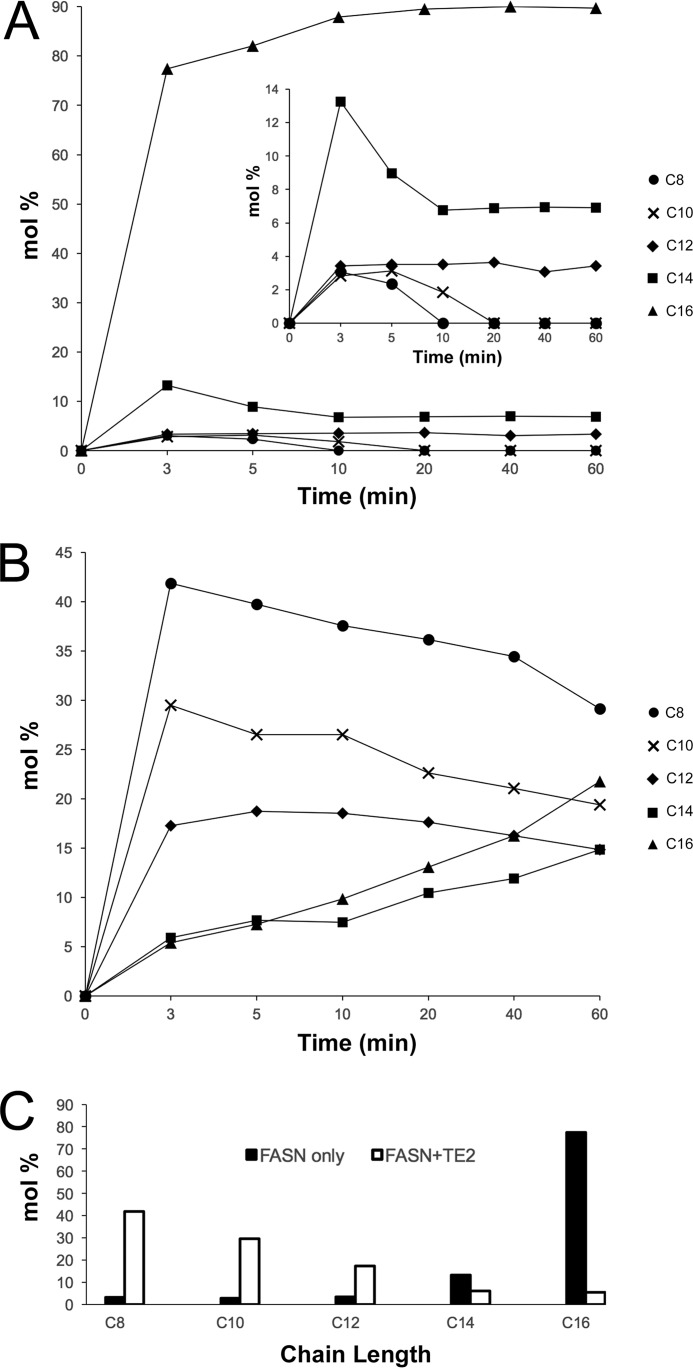
FIGURE 3.
Impact of TE2 activity on the fatty acid product profile of intact, human FASN. A, distribution of free fatty acids produced by FASN alone. The molar percentage (mol%) or proportion of free fatty acids produced over time, ranging from C8 to C16 carbons in length, was determined by GC-MS as described under “Experimental Procedures.” Each time point was completed in triplicate and averaged; S.D. <15%. B, distribution of free fatty acids produced by FASN in the presence of TE2. A ratio of 1:2 (FASN:TE2) was used. C, comparison of the lipid profiles at the 3-min time point. TE2 shifts the product distribution significantly toward short-chain fatty acids.
GC-MS Analysis of Human FASN Product Profile
Human, full-length FASN was a generous gift from GlaxoSmithKline (29). FASN was incubated at 30 °C in the absence and presence of TE2 under the following reaction conditions: 0.5 μm FASN, 1 μm TE2, 25 mm Hepes, pH 7.5, 100 mm NaCl, 1 mm NADPH, 200 μm acetyl-CoA, 400 μm malonyl-CoA, 0.5 mm EDTA, and 5 mm DTT. FASN was preincubated with DTT for 5 min before initiating catalysis with the addition of malonyl-CoA and acetyl-CoA. Triplicate reactions were quenched at different time points by the addition of 50% fresh KOH (60 μl of sample: 300 μl of KOH) and stored at −80 °C. The amount of C8, C10, C12, C14, and C16 free fatty acids was determined by GC-MS using a modified protocol by Zheng et al. (30). The protocol was modified to use C9 and C15 fatty acids (Nuchek Prep, Inc., Elysian, MN) as internal standards, which were doped into each sample before derivatization with propyl chloroformate, such that the final concentration was 800 pg/μl at injection. Samples were adjusted to ∼pH 8 before derivatization. The samples were analyzed using a Thermo-Scientific TSQ Quantum XLS GC-MS, automated by a TriPlus auto-sampler. Fatty acid derivatives were separated on an HP-5ms capillary column (5% phenylmethylpolysiloxane, 30-m × 250-μm inner diameter, 0.25-μm film thickness, Agilent Technologies). The initial oven temperature was held at 50 °C for 5 min, ramped to 100 °C at a rate of 10 °C/min, held at 100 °C for 2 min, ramped to 200 °C at a rate of 7 °C/min, held at 200 °C for 2 min, ramped to 260 °C at a rate of 15 °C/min, and finally held at 260 °C for 5 min. Helium was used as a carrier gas at a constant flow rate of 1.5 ml/min through the column. The temperatures of the front inlet, transfer line, and electron impact ion source were set at 260, 260, and 230 °C, respectively. The electron energy was −70 eV, and data were collected in a full scan mode over the range m/z 50–500.
Crystallization and Structure Determination
Rod-shaped crystals of TE2 (10 mg/ml with 10 mm fresh DTT) were grown using the vapor diffusion method at 22 °C using a 1:1 drop to well ratio. The well solutions contained 1.64 m NaH2PO4, 0.42 m K2HPO4, and 50 mm HEPES, pH 7.6. X-ray diffraction data were collected as 0.5° oscillation images on an in-house Rigaku RA-Micro 007 generator and an RaxisIV detector. Crystals were cryoprotected by transfer to paratone. The d*Trek software was used to process the dataset, and PHASER within the CCP4i software suite was used to identify the initial non-isomorphous molecular replacement solution (31, 32). The closest homologue of TE2, RifR (30%, PDB code 3FLB), was successfully used as an initial search model when only the α/β hydrolase core was used (residues 5–123, 191–248). All interconnecting loops and the active site region were rebuilt using iterative cycles of model building in COOT and refinement in PHENIX (simulated annealing of torsion angles at early steps, xyz coordinates, and group B values) (33, 34). At this stage the capping domain was built and refined. The final model (Table 1), composed of residues 17–58, 75–157, and 168–265, 1 chloride ion, and no water molecules, was refined with 6 TLS groups and individual B values. The structure was validated using MolProbity within the PHENIX suite (35).
TABLE 1.
Data collection and refinement statistics
| Data Processing | |
| Space group | I422 |
| Cell parameters (Å) | 96.1, 96.1, 160.0 (α = β = γ = 90°) |
| Resolution (Å) | 67.9-2.8 (2.9-2.8) |
| Total reflections | 112,317 |
| Unique reflections | 9,601 |
| Completeness (%) | 99.9 (100)a |
| Multiplicity | 11.7 (11.8) |
| Rmeasured (%) | 9.0 (48.7) |
| CC1/2 (%) | 99.9 (97.4) |
| 〈I/σI〉 | 12.8 (2.9) |
| Wilson B (Å2) | 66.5 |
| Refinement | |
| Resolution (Å) | 24.8-2.8 |
| Completeness (%) | 97.6 |
| Rwork/Rfree (%) | 29.7/34.5 |
| Root mean square | |
| Bonds (Å) | 0.003 |
| Angles (°) | 0.725 |
| Average B (Å2) | 93.3 |
| Molprobity validation | |
| Ramachandran plot | |
| Most favored (%) | 93.3 |
| Additionally allowed (%) | 6.7 |
| Disallowed (%) | 0 |
| Rotamer outliers (%) | 0 |
| Clash score | 1.2 |
| PDB code | 4XJV |
a Values given in parentheses correspond to the outer shell of data.
Results
Hydrolysis of Acyl-CoA Substrate Mimetics by TE1 and TE2
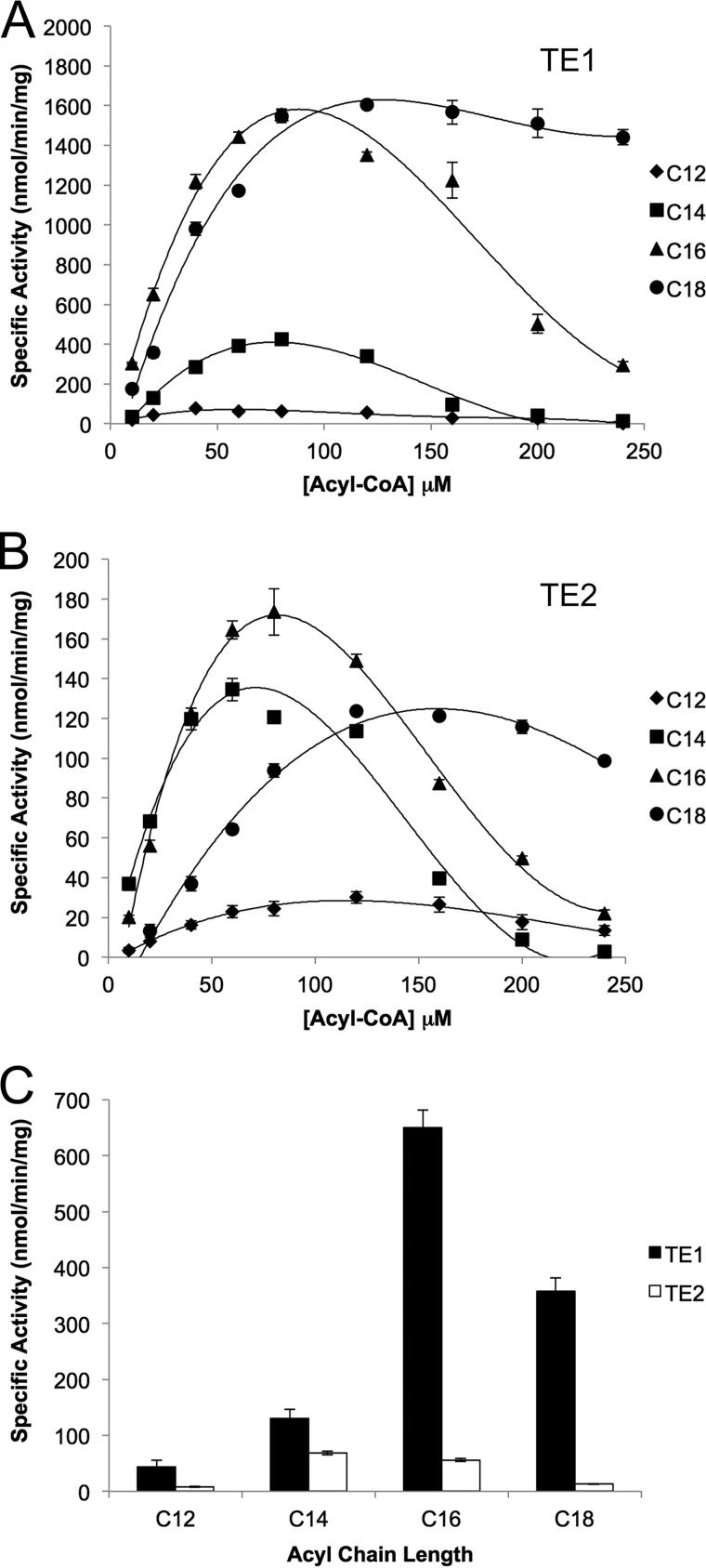
As a first step toward understanding the unique properties of human TE2, a panel of acyl-CoA substrates, with chain lengths ranging from 12 to 18 carbons, was analyzed. A coupled assay that included α-ketoglutarate dehydrogenase was used to monitor the release of CoA. For both enzymes and all substrates tested, the activity increased to a maximal value and then decreased (Fig. 1, A and B). These observations are consistent with previous studies on rat TE1 and TE2 (9, 36). For example, the maximal specific activities determined by the coupled assay for TE2 against C14-CoA are similar to those determined by a radiochemical method (∼600 versus ∼140 nmol min−1 mg−1) despite the differences in method, buffer conditions, and enzyme source (9). The loss in activity has long been attributed to the detergent-like properties of the acyl-CoA substrates, which precludes the determination of Michaelis-Menten kinetic parameters (37). In an effort to minimize the influence of these complex inhibitory effects, the activity of TE1 and TE2 was compared at 20 μm substrate (Fig. 1C), a concentration far from any inhibitory affects, and TE1 exhibited the highest activity for C16-containing substrate followed by C18. TE2 had the highest activity for C14 followed by C16, similar to the fatty acid distribution in milk (19–22). However, the specific activity of TE2 against acyl-CoA substrates was 5–10-fold lower than that of TE1.
FIGURE 1.
Activity of human TE1 and TE2 against acyl-CoA substrates. A, TE1 thioesterase domain of human FASN. B, human TE2. C, comparison of specific activity at 20 μm for each acyl-CoA substrate. Acyl chains 12–18 carbons in length were tested. The concentrations of TE1 and TE2 used were 25 and 125 nm, respectively.
Hydrolysis of C16-ACP by TE1 and TE2
To determine whether or not there was a difference in the activity of TE1 and TE2 against an acyl-ACP substrate, human C16-ACP was made in a two-step process. First, the 4′-PPT moiety was transferred from CoA using the phosphopantetheinyl transferase from S. coelicolor. Second, palmitate was added to the linker arm via the action of E. coli holo acyl-ACP synthase. Each step of this process was readily monitored by MALDI-TOF MS. Importantly, the choice of preparing and evaluating the C16-ACP for both enzymes was necessary, as TE1 shows limited activity against C14-containing substrates.
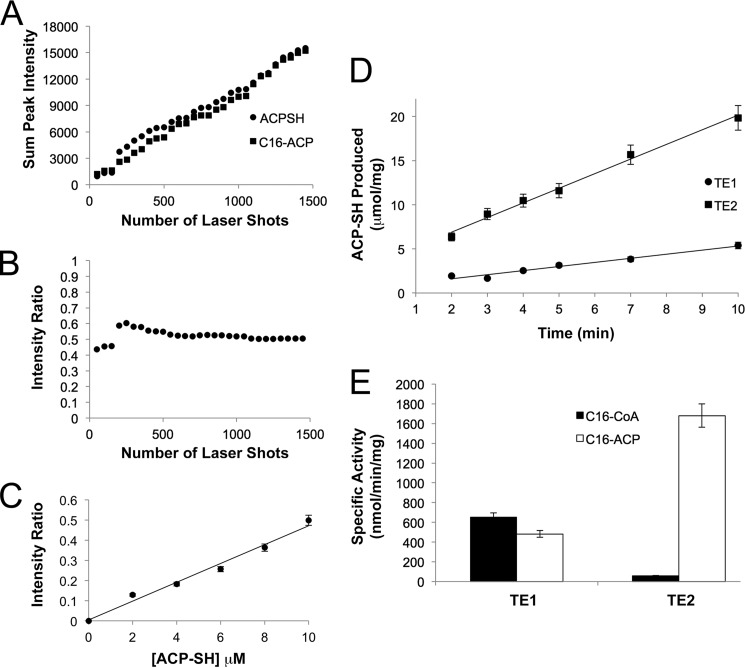
Instead of using available radiochemical and HPLC-based methods for monitoring the release of fatty acids from either intact FASN or acyl-CoA substrate (9, 38), we developed an efficient MALDI-TOF assay. In this assay the acidic MALDI matrix was used to directly quench the reaction. This discontinuous assay was readily able to quantitate the amount of ACP-SH produced over time. The chief concerns of any MALDI-TOF MS-based assay, which had to be addressed during validation, were the potential for ion suppression of either ACP-SH or C16-ACP, unequal sampling of substrate and product during the recrystallization in matrix, limited sample detection, and the potential to saturate the mass detector. Fig. 2A demonstrates the linearity of the sum of the peak intensity for an equimolar mixture (10 μm) of ACP-SH and C16-ACP from the acquisition of spectra in 50 laser pulse increments. The ratio of ACP-SH peak intensities to the sum of the observed ACP-SH and C16-ACP peak intensities was determined by the equation IACP-SH/(IACP-SH + IC16-ACP)). A plot of this intensity ratio (Fig. 2B) versus the number of laser shots demonstrated that each species was detected equally and that the ratio stabilized above the compilation of 500 spectra. A sum of 1500 spectra was used for all subsequent experiments. The linearity of the ACP-SH response was also confirmed from 2–10 μm ACP-SH, within a constant background of 10 μm C16-ACP (Fig. 2C).
FIGURE 2.
Quantitative MALDI-TOF MS analysis of TE1 and TE2 activity against the C16-ACP substrate. A, sum of peak intensity for lipid-free ACP-SH and C16-ACP. The peak intensities were summed at 50 shot intervals for an equimolar mixture (10 μm). B, intensity ratio versus number of laser shots. The intensity ratio is defined as IACP-SH/(IACP-SH + IC16-ACP), the peak intensity of ACP-SH divided by the total intensity of both ACP-SH and C16-ACP species. C, standard curve for ACP-SH present in a mixture containing 10 μm C16-ACP. D, determination of the rate of C16-ACP hydrolysis by TE1 and TE2. TE1, TE2, and C16-ACP were used at a concentration of 25 nm, 10 nm, and 20 μm, respectively. E, comparison of specific activities for the hydrolysis of C16-CoA and C16-ACP substrates by TE1 and TE2. Both substrates were used at a concentration of 20 μm.
Once the assay was validated, the rate of ACP-SH formation from 20 μm C16-ACP was determined for TE1 and TE2 (Fig. 2D). In contrast to the analysis of the acyl-CoA substrates, the concentration of TE2 used had to be lowered from 125 to 10 nm. A comparison of the specific activities of TE1 and TE2 against acyl-CoA and the C16-ACP substrates at 20 μm (Fig. 2E) revealed that TE2 is ∼30-fold more active against C16-ACP than it is against C16-CoA (1681 versus 56 nmol/min/mg). The activity of TE2 was also ∼4-fold higher than TE1 on C16-ACP (1681 versus 482 nmol/min/mg), whereas TE1 exhibited comparable activity (482 versus 650 nmol/min/mg) on both substrates. Altogether, these data support that TE2 preferentially hydrolyzes the thioester bond of acyl-ACP substrates and that this MS method is able to monitor specific activity values comparable to previous methods (9).
Release of Short Chain Fatty Acids from Intact FASN by TE2
Although the previous results clearly demonstrate the specificity differences between TE1 and TE2 on substrate mimetics and an acyl-ACP fragment of FASN, we next sought to determine the ability of TE2 to modify the products of intact human FASN during catalysis. Previous work by Libertini and Smith (9) determined the distribution of lipids, mole percentage (mol %), using different ratios of rat FASN:TE2 and monitoring the incorporation of [14C]acetyl-CoA and [14C]malonyl-CoA into products. We, however, increased the sensitivity of the assay by performing GC-MS analysis after derivatization with propyl chloroformate as described under “Experimental Procedures.” We also performed a time-course study that contrasts with the previous work where only the reaction end point was analyzed. Moreover, this alternative approach enabled us to use significantly lower protein concentrations (0.5–1 μm) than those used in previous studies, which ranged from 2 to 16 μm rat FASN and 3 to13 μm rat TE2.
When 0.5 μm human FASN was incubated alone with acetyl-CoA, malonyl-CoA, NADPH, and DTT, the primary product was C16 (Fig. 3A). The abundance of the remaining lipids analyzed, C8-C14, was consistent with the sequential addition of two carbon units (Fig. 3, inset). To mimic the increased expression of TE2 in breast cells during lactation, we added TE2 to FASN in a ratio of 2:1 (1 μm TE2: 0.5 μm FASN). The addition of TE2 to FASN resulted in the production of C8, C10, and C12 fatty acids (Fig. 3B). A comparison of the lipid profiles at the earliest time point (Fig. 3C) clearly demonstrates the dramatic reversal in fatty acid distribution, with TE2 successfully outcompeting the endogenous TE1 domain to release fatty acids of shorter chain length. These data are completely consistent with those involving rat TE2 and FASN.
Crystal Structure of Human TE2
To visualize the TE2 active site and to identify structural determinants that may influence the interaction of TE2 with the ACP domain of FASN, we determined the crystal structure of human TE2 to 2.8 Å resolution. Based on our previous structural work on human TE1, we hypothesized that a loop insertion within the α/β-fold would generate a surface subdomain or capping domain near the active site. After screening several different crystal forms, the structure of TE2 was solved from crystals exhibiting space group I422 and one molecule in the asymmetric unit (Table 1). The best crystal exhibited a high Matthews coefficient and solvent content (3.1 and 61%, respectively). A further, initial indication of poor packing and possible motion within the structure, despite cryocooling, was the Wilson B-value of 67 Å2.
A BLAST search indicated that the RifR (29%, rifamycin synthesis pathway) and RedJ (26%, prodiginine pathway) thioesterases from Amycolatopsis mediterranei and S. coelicolor, respectively, had the highest sequence identity. We tested the α/β hydrolase domains of RifR (PDB code 3FLB), RedJ (PDB code 3QMV), and human TE1 (PDB code 2PX6; 18% sequence identity) as molecular replacement search models with PHASER (18, 31, 38, 39). Importantly, in an effort to reduce model bias, only the α/β core was used, and portions of the active site containing the catalytic residues were removed. The TE1 search model failed. The solution with RifR trial was used as the starting point for the careful rebuilding of the active site, all interconnecting loops, and the capping domain with COOT and iterative refinement cycles with PHENIX (33, 34).
The final model only contained residues 25–58, 77–158, and 166–265. The model exhibits excellent geometric and Molprobity validation statistics (Table 1; no Ramachandran or rotamer outliers; low clash score). However, no electron density was observed for ∼21% of the protein (56/271 residues) and the remnants of the affinity tag. Based on these observations, the final Rwork/Rfree values of 29.7/34.5% and an average B-factor value of 93 Å2 for the final model are consistent with a moderately diffracting, poorly packed crystal that contains significant motion within the molecule, as described in more detail below.
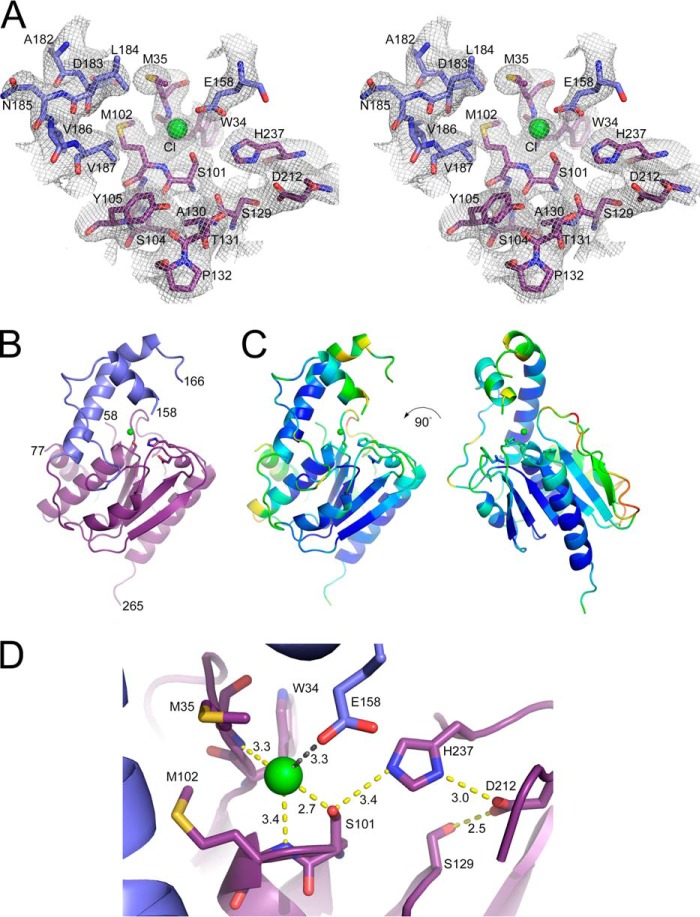
In contrast to the missing parts of the structure, the electron density for the α/β hydrolase core and the active site were unambiguous (Fig. 4A). As expected, a surface loop insertion was observed, residues 135–192 (Fig. 4B). This insert or capping domain contains one long helix (residues 178–192), with B-values directly comparable with the α/β hydrolase core (Fig. 4C). The remaining coil and short helical sections of the capping domain and the backside of the molecule exhibit 2–3-fold higher B-values or are missing in the electron density (residues 59–76 and 159–165).
FIGURE 4.
Crystal structure of human TE2. A, stereoview of the 2.8 Å resolution, 2Fo − Fc electron density map near the Ser-101–His-237–Asp-212 catalytic triad. The map is contoured at 1.2 σ. A chloride ion (green) was bound to the oxyanion hole, the backbone nitrogen atoms of Met-35 and Met-102. The carbon atoms are colored based on the subdomains shown in panel B. B, a schematic diagram illustrating the α/β hydrolase core (purple) and capping (blue, residues 135–192) domains. C, B-factor analysis. The schematic diagram is rainbow-colored based on a low-to-high, blue-to-red scheme, as implemented by PyMOL. D, active site details. Ser-101 makes imperfect interactions with the other components of the catalytic triad, His-237 and Asp-212. Putative hydrogen bonds are indicated in yellow. The tip of the capping domain has collapsed onto the active site, placing Glu-158 near the Cl− ion (distance indicated in gray).
A closer inspection of the active site (Fig. 4D) reveals the canonical catalytic triad, composed of residues Ser-101, His-237, and Asp-212. A 6-σ-positive electron density peak was observed within the oxyanion hole that is generated by the amide nitrogen atoms of Met-35 and Met-102. Given the longer distances between atoms and comparable B-values, the placement of a larger chloride ion is supported at this location. The capping domain blocked access to the catalytic triad. Clear electron density for a short helix (residues 149–158) positioned Glu-158 within 3.3 Å of the chloride ion. The geometry of the catalytic triad is also suboptimal for catalysis, with the nitrogen atoms of His-237 being 3.4 and 3.0 Å away from Ser-101 and Asp-212, respectively. The S101A mutant of TE2 showed no activity with C14-CoA (data not shown), consistent with previous studies on the mutagenesis of the Ser-101 and His-237 triad in the rat enzyme (40, 41). This study confirms Asp-212 as part of the catalytic triad for the first time. For TE2, the hydrogen-bonding network of the catalytic triad is also extended to include the Ser-129-Asp-212 interaction.
Comparisons of Human TE2 to Structural Homologues
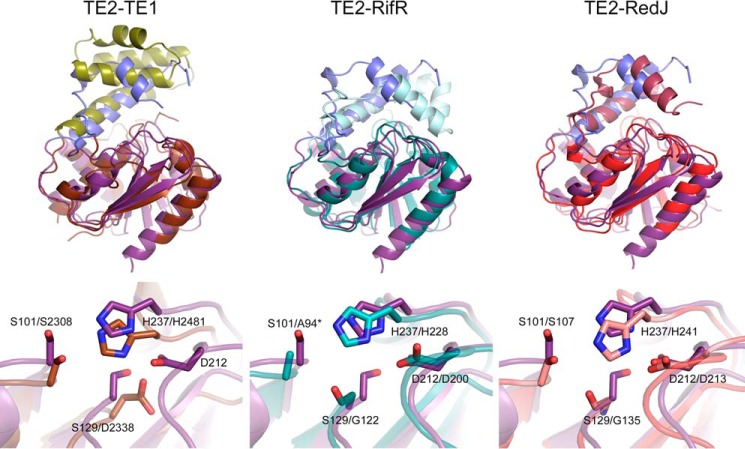
Superposition of the TE2 structure onto those of FASN TE1, RifR, and RedJ (Fig. 5) demonstrates the conservation of the α/β serine hydrolase core and the presence of a capping or lid domain that covers that active site (18, 38, 39). The capping domains of RifR and RedJ are also more mobile, with B-values 2–3-fold higher than the α/β hydrolase core. Although the relative position of the long helix within the latter subdomain is conserved, the location, number, and structure of additional helices and coil regions are quite different. TE1 is the most divergent with the presence a 4-helical bundle. The locations of the Ser and His residues within the catalytic triad are also conserved, but the position of the Asp residue is variable. TE2 places Asp-212 in the canonical position, similar to RifR and RedJ. In contrast, TE1 uses Asp-2338 as part of its catalytic triad. The position of Asp-2338 in TE1 is the same as Ser-129 of TE2, which extends the hydrogen-bonding network of the catalytic triad in TE2.
FIGURE 5.
Comparison of TE2 to structural homologs. The top and bottom panels depict the overlay of the structures and the active site regions, respectively. The root mean square deviation values for the superpositions are: TE2-TE1, 2.6 Å; TE2-RifR, 1.9 Å; TE2-RedJ, 2.2 Å. Protein Data Bank files used: human TE1 (2PX6), A. mediterranei RifR (3FLB), and Streptomyces coelicolor RedJ (3QMV). The α/β hydrolase core and capping domains of TE1, RifR, and RedJ are colored brown/olive, teal/cyan, and red/light purple, respectively. *, the active Ser94 residue of RifR has been mutated to Ala.
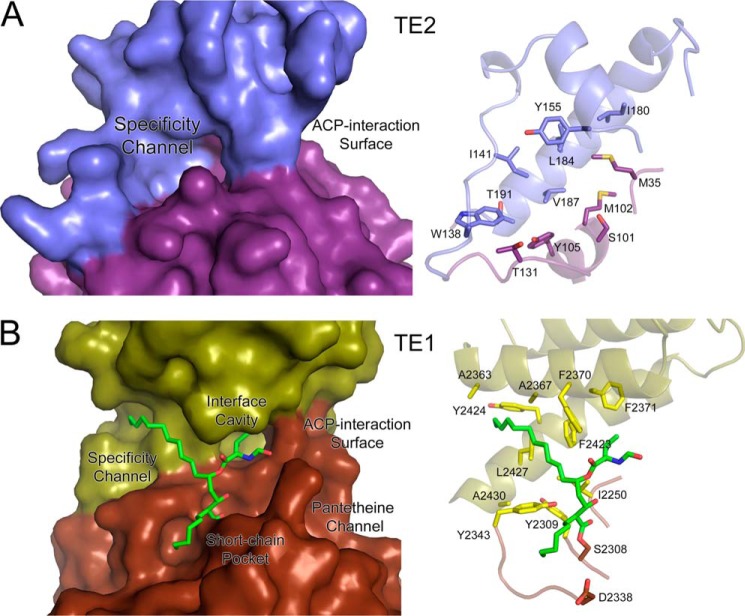
A comparison of the surface representation of TE2 to TE1 provides insight into the putative binding sites of the acyl chain, the 4′-PPT moiety, and the ACP domain of FASN. For TE2 (Fig. 6A), the capping domain collapsed onto the active site (Fig. 4), blocking access to the catalytic triad. In contrast, the Orlistat complex of TE1 possesses an open active site (Fig. 6B) with a pocket, termed the specificity channel, that binds the C16, palmitate-like moiety of Orlistat (18). The interface cavity and short-chain pocket bind additional substituents of the drug. An extended hydrophilic channel and a concave surface, adjacent to the catalytic triad, are the most likely binding sites for the 4′-PPT linker arm and the ACP domain, respectively. The C16 moiety of Orlistat interacts with several hydrophobic residues, including Phe-2370, Phe-2423, Tyr-2424, and Leu-2427. A constellation of hydrophobic residues is also present within the putative specificity channel of TE2: Trp-138, Ile-141, Tyr-155, Leu-184, Val-187 of the capping domain, and Met-102 and Tyr105 of the hydrolase core. However, if the C16 moiety of Orlistat represents where the acyl chain binds to TE2, considerable structural changes in the TE2 active site and the capping domain would be required. This is because a coil region containing Trp-138 and Ile-141 blocks the end of the cavity. It is entirely possible that when the conformation of the capping domain of TE2 switches from the closed to the open conformation to allow the ACP domain and its associated 4′-PPT group to bind, this coil region also rearranges to accept the acyl chain.
FIGURE 6.
Comparison of the putative specificity channel of TE2 to that of TE1. A, surface representation of TE2 and the underlying residues that contribute to the putative specificity channel. The capping domain occludes access to the catalytic triad and surface channels. B, the Orlistat-TE1 complex and the residues that contribute to the specificity channel. In both cases the majority of the residues that interact with the acyl chain are located in the capping domain, a unique surface-loop insert for both enzymes. Orlistat also interacts with the TE1 surface through the interface cavity and short chain pocket, as described by Pemble et al. (PDB code 2PX6) (18). The putative pantetheine channel and ACP-interaction surface are also indicated.
Discussion
The impetus for this study was to understand how TE2 modulates the chain length of fatty acids produced by FASN. TE2 was 5–10-fold less active against acyl-CoA substrates (Fig. 1) and demonstrated a preference for shorter acyl chains than TE1. The specific activities observed and the data trends are directly comparable with the literature studying the homologous domains from rat and duck (9, 36). TE2 showed a significant increase in activity over TE1 for the C16-ACP substrate (Fig. 2), supporting the need for an interaction between TE2 and the ACP domain. The addition of TE2 to intact human FASN, the biological substrate, completely shifted the fatty acid product profile to favor the short chain fatty acids C8-C12 (Fig. 3). Importantly, this switch in product distribution is consistent with analogous studies with the rat enzymes and with the lipid profile found in the breast milk of humans and other mammals (9, 11, 19, 42). For example, human milk fat contains ∼50% saturated fatty acids of all chain lengths from C4 to C18. The mol % of the C4, C6, C8, C10, C12, C14, C16, and C18 chain lengths is 0.2, 0.3, 1.0, 2.4, 7.8, 11.5, 21.6, and 6.4%, respectively. If one considers the proportion of just the saturated fatty acids, the C8–14 fatty acids represent ∼45% of the lipids. Thus, TE2 plays a key role in producing short chain fatty acids, but the structure of the enzyme from humans or other animals was not determined before this work.
The crystal structure of TE2 (Fig. 4) revealed the α/β hydrolase-fold, residues involved in the catalytic triad, and the presence of a mobile capping or lid domain that occluded the active site. This latter domain exhibited greater similarity to the domains observed in the type II, monomeric thioesterases involved in polyketide synthesis (Fig. 5). Moreover, comparisons of the closed conformation of the TE2 structure to the Orlistat complex of TE1 (Fig. 6) suggest that the capping domain must move and an adjacent coil region rearrange for the acyl-ACP to dock and catalysis to occur.
All of these observations are consistent with the polyketide synthase and non-ribosomal peptide synthase systems. The FASN, polyketide synthase, and non-ribosomal peptide synthase systems function as modular assembly lines to build lipids, macrolides, and peptides from different acyl and amino acid precursors (43–46). One of the common attributes between these macromolecular machines is the tethering of the appropriate building blocks to the 4′-PPT linker arm of an ACP or related peptidyl carrier protein domain. Like FASN, a C-terminal thioesterase domain typically releases the final product. The type II thioesterases function in the polyketide synthase/non-ribosomal peptide synthase systems to edit or release aberrant reaction and misprimed intermediates. The action of TE2 within the specialized compartment of the mammary gland does not release damaged fatty acids per se but does allow for the production of a broader distribution of lipid in milk fat. One study also suggests that the levels of TE2 in breast adenocarcinoma cells correlates with changes in the lipid profiles of these cells, although future studies are clearly needed (47).
In each of the systems presented, the type II thioesterase must compete with a type I thioesterase domain. The structural analysis of other type II thioesterases, including RifR, RedJ, and the surfactin thioesterase II, has demonstrated that the movement of the capping or lid domain is a hallmark of these enzymes (38, 39, 48). Moreover, a change from a closed to an open structure most likely enables the type II thioesterase to select the appropriate ACP within the context of the respective megasynthase during catalysis. Based on our observations of increased activity for the C16-ACP substrate (Fig. 2), the ability of TE2 to readily outcompete the TE1 domain of intact FASN during catalysis (Fig. 3), and a closed conformation in TE2 (Figs. 4 and 6), a similar scenario most likely occurs for TE2 to interact with the ACP domain of FASN. This property of TE2 is even more remarkable given the open conformation and rigid capping domain observed for TE1 (Fig. 6). The closed conformation of TE2 may also explain its low activity against acyl-CoA substrates. Perhaps the inability of the smaller acyl-CoA substrates to transition TE2 to the open conformation prevents TE2 for altering the acyl-CoA pool, an essential component of peroxisomal lipid, triglyceride, and glycerolphospholipid metabolism (49, 50).
The issue of ACP selectivity and recognition is universal, as this domain is responsible for shuttling building blocks from one catalytic domain to another in type I megasynthases and the homologous type II synthetic systems (46). The ACP domain of the type II fatty acid synthase system in bacteria contains a 4-helix bundle that can encapsulate the acyl chain, thus protecting it from solution. In contrast, studies of the isolated rat ACP domain of FASN suggest that the acyl chain is not buried, which likely facilitates turnover in the FASN megasynthase context (51). Studies on human ACP with regard to this issue are still needed. Nonetheless, the 4′-PPT moiety, attached to a Ser-2156 residue on the apex of the α2 helix and the surface of ACP, most likely makes specific interactions with the TE2. These interactions would include presenting the acyl chain and the thioester bond in the correct orientation near the catalytic triad for hydrolysis to occur. Support for this notion comes from studies of the E. coli ACP-LpxD, E. coli ACP-FabA, and human phosphopantetheinyl transferase in complex with human apo-ACP (52–54). The former two studies collectively demonstrate a mechanism that involves 4′-PPT cofactor interactions to prime either the recipient hydrolase or β-carbon processing enzyme for substrate loading followed by conformational changes of the ACP. The loading of the 4′-PPT moiety onto ACP by the phosphopantetheinyl transferase also involves specific recognition of the ACP domain.
In conclusion, human TE2 is a unique type II thioesterase that can selectively interact with the ACP domain of FASN during catalysis. This interaction requires a transition from a closed to open conformation for the 4′-PPT and the fatty acid to be loaded into the active site. These features are similar to the roles of type II thioesterases involved in the rescue of polyketide synthase and non-ribosomal peptide synthase stalled and aberrant intermediates. In contrast, the TE2-FASN interaction results in the preferential release of shorter chain fatty acids than FASN would otherwise produce. The resulting broad distribution of lipids in breast milk fulfills a key biological need for infants and may represent a potential focus for future studies in breast cancer cells.
Author Contributions
M. K. R., S. J. K., and W. T. L. designed the study and wrote the paper. M. K. R. purified and crystallized the TE2 protein, performed all kinetic and MS analyses, and determined the x-ray structure. L. C. J. and C. W. P. contributed to expression constructs and kinetic assay design, protein expression, and purification. J. E. C. performed all experiments with intact FASN and TE2 and helped with protein purifications. C. M. F. and B. E. F. developed and performed all GC-MS assays. All authors analyzed the data and approved the final version of the manuscript.
Acknowledgments
We thank Dr. John Shanklin of Brookhaven National Laboratories for providing the E. coli holo acyl-ACP synthase expression construct and Alan Rendina of GlaxoSmithKline for providing full-length, human fatty acid synthase. We also acknowledge the support of the Crystallography and Computational Biophysics and Proteomics and Metabolomics Shared Resources of the Comprehensive Cancer Center of Wake Forest Baptist Medical Center.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA411401 (NCI), Comprehensive Cancer Center of Wake Forest Baptist Medical Center (NCI Cancer Center Support Grant P30CA012197), and the Department of Defense (PC08165). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 4XJV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FASN
- fatty acid synthase
- 4′-PPT
- 4′-phosphopantetheinyl
- ACP
- acyl carrier protein
- TE
- thioesterase
- NTA
- nitrilotriacetic acid
- HAAS
- holo acyl-ACP synthase
- ACP-SH
- 4′-phosphopantetheinyl-ACP
- RFU
- relative fluorescence units.
References
- 1. Maier T., Leibundgut M., and Ban N. (2008) The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 2. Asturias F. J., Chadick J. Z., Cheung I. K., Stark H., Witkowski A., Joshi A. K., and Smith S. (2005) Structure and molecular organization of mammalian fatty acid synthase. Nat. Struct. Mol. Biol. 12, 225–232 [DOI] [PubMed] [Google Scholar]
- 3. Little J. L., and Kridel S. J. (2008) Fatty acid synthase activity in tumor cells. Subcell. Biochem. 49, 169–194 [DOI] [PubMed] [Google Scholar]
- 4. Wu X., Qin L., Fako V., and Zhang J. T. (2014) Molecular mechanisms of fatty acid synthase (FASN)-mediated resistance to anti-cancer treatments. Adv. Biol. Regul. 54, 214–221 [DOI] [PubMed] [Google Scholar]
- 5. Jensen-Urstad A. P., and Semenkovich C. F. (2012) Fatty acid synthase and liver triglyceride metabolism: housekeeper or messenger? Biochim. Biophys. Acta 1821, 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zadra G., Photopoulos C., and Loda M. (2013) The fat side of prostate cancer. Biochim. Biophys. Acta 1831, 1518–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shah U. S., Dhir R., Gollin S. M., Chandran U. R., Lewis D., Acquafondata M., and Pflug B. R. (2006) Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum. Pathol. 37, 401–409 [DOI] [PubMed] [Google Scholar]
- 8. Jensen-Urstad A. P., Song H., Lodhi I. J., Funai K., Yin L., Coleman T., and Semenkovich C. F. (2013) Nutrient-dependent phosphorylation channels lipid synthesis to regulate PPARα. J. Lipid Res. 54, 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Libertini L. J., and Smith S. (1978) Purification and properties of a thioesterase from lactating rat mammary gland which modifies the product specificity of fatty acid synthetase. J. Biol. Chem. 253, 1393–1401 [PubMed] [Google Scholar]
- 10. Nolin J. M., Thompson B. J., and Smith S. (1982) Localization of thioesterase II, the chain-length regulatory enzyme of milk fatty acid synthesis, in rat mammary gland epithelial cells. J. Endocrinol. 94, 251–256 [DOI] [PubMed] [Google Scholar]
- 11. German J. B. (2011) Dietary lipids from an evolutionary perspective: sources, structures and functions. Matern. Child Nutr. 7, 2–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson S. M., Rudolph M. C., McManaman J. L., and Neville M. C. (2007) Key stages in mammary gland development: secretory activation in the mammary gland; it's not just about milk protein synthesis! Breast Cancer Res. 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pawlak J., and Smith S. (1986) Evaluation of thioesterase II as a serum marker for rat mammary cancer. Cancer Res. 46, 4712–4719 [PubMed] [Google Scholar]
- 14. Smith S., Pasco D., Pawlak J., Thompson B. J., Stampfer M., and Nandi S. (1984) Thioesterase II, a new marker enzyme for human cells of breast epithelial origin. J. Natl. Cancer Inst. 73, 323–329 [DOI] [PubMed] [Google Scholar]
- 15. Pazirandeh M., Chirala S. S., Huang W. Y., and Wakil S. J. (1989) Characterization of recombinant thioesterase and acyl carrier protein domains of chicken fatty acid synthase expressed in Escherichia coli. J. Biol. Chem. 264, 18195–18201 [PubMed] [Google Scholar]
- 16. Lin C. Y., and Smith S. (1978) Properties of the thioesterase component obtained by limited trypsinization of the fatty acid synthetase multienzyme complex. J. Biol. Chem. 253, 1954–1962 [PubMed] [Google Scholar]
- 17. Joshi A. K., Witkowski A., Berman H. A., Zhang L., and Smith S. (2005) Effect of modification of the length and flexibility of the acyl carrier protein-thioesterase interdomain linker on functionality of the animal fatty acid synthase. Biochemistry 44, 4100–4107 [DOI] [PubMed] [Google Scholar]
- 18. Pemble C. W. 4th, Johnson L. C., Kridel S. J., and Lowther W. T. (2007) Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat. Struct. Mol. Biol. 14, 704–709 [DOI] [PubMed] [Google Scholar]
- 19. Thompson B. J., and Smith S. (1985) Biosynthesis of fatty acids by lactating human breast epithelial cells: an evaluation of the contribution to the overall composition of human milk fat. Pediatr. Res. 19, 139–143 [DOI] [PubMed] [Google Scholar]
- 20. Rogers L., Kolattukudy P. E., and deRenobales M. (1982) Purification and characterization of S-acyl fatty acid synthase thioester hydrolase which modifies the product specificity of fatty acid synthase in the uropygial gland of mallard. J. Biol. Chem. 257, 880–886 [PubMed] [Google Scholar]
- 21. Knudsen J., Grunnet I., and Dils R. (1981) Medium-chain fatty acyl-S-4′-phosphopantetheine-fatty acid synthase thioester hydrolase from lactating rabbit and goat mammary glands. Methods Enzymol. 71, 200–229 [DOI] [PubMed] [Google Scholar]
- 22. Dils R. R., and Knudsen J. (1980) Milk fat composition and biosynthesis in non-ruminants and ruminants. Biochem. Soc. Trans. 8, 292–294 [DOI] [PubMed] [Google Scholar]
- 23. Mikkelsen J., Witkowski A., and Smith S. (1987) Interaction of rat mammary gland thioesterase II with fatty acid synthetase is dependent on the presence of acyl chains on the synthetase. J. Biol. Chem. 262, 1570–1574 [PubMed] [Google Scholar]
- 24. Foster R. J., Bonsall R. F., Poulose A. J., and Kolattukudy P. E. (1985) Interaction of S-acyl fatty acid synthase thioester hydrolase with fatty acid synthase: direct measurement of binding by fluorescence anisotropy. J. Biol. Chem. 260, 1386–1389 [PubMed] [Google Scholar]
- 25. Buchbinder J. L., Witkowski A., Smith S., and Fletterick R. J. (1995) Crystallization and preliminary diffraction studies of thioesterase II from rat mammary gland. Proteins 22, 73–75 [DOI] [PubMed] [Google Scholar]
- 26. Cox R. J., Crosby J., Daltrop O., Glod F., Jarzabek M. E., Nicholson T. P., Reed M., Simpson T. J., Smith L. H., Soulas F., Szafranska A. E., and Westcott J. (2002) Streptomyces coelicolor phosphopantetheinyl transferase: a promiscuous activator of polyketide and fatty acid synthase acyl carrier proteins. J. Chem. Soc. Perkin Trans. 1 2002, 1644–1649 [Google Scholar]
- 27. Shanklin J. (2000) Overexpression and purification of the Escherichia coli inner membrane enzyme acyl-acyl carrier protein synthase in an active form. Protein Expr. Purif. 18, 355–360 [DOI] [PubMed] [Google Scholar]
- 28. Molnos J., Gardiner R., Dale G. E., and Lange R. (2003) A continuous coupled enzyme assay for bacterial malonyl-CoA:acyl carrier protein transacylase (FabD). Anal. Biochem. 319, 171–176 [DOI] [PubMed] [Google Scholar]
- 29. Hardwicke M. A., Rendina A. R., Williams S. P., Moore M. L., Wang L., Krueger J. A., Plant R. N., Totoritis R. D., Zhang G., Briand J., Burkhart W. A., Brown K. K., and Parrish C. A. (2014) A human fatty acid synthase inhibitor binds β-ketoacyl reductase in the keto-substrate site. Nat. Chem. Biol. 10, 774–779 [DOI] [PubMed] [Google Scholar]
- 30. Zheng X., Qiu Y., Zhong W., Baxter S., Su M., Li Q., Xie G., Ore B. M., Qiao S., Spencer M. D., Zeisel S. H., Zhou Z., Zhao A., and Jia W. (2013) A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naggert J., Witkowski A., Wessa B., and Smith S. (1991) Expression in Escherichia coli, purification and characterization of two mammalian thioesterases involved in fatty acid synthesis. Biochem. J. 273, 787–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Powell G. L., Grothusen J. R., Zimmerman J. K., Evans C. A., and Fish W. W. (1981) A re-examination of some properties of fatty acyl-CoA micelles. J. Biol. Chem. 256, 12740–12747 [PubMed] [Google Scholar]
- 38. Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., and Smith J. L. (2009) Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 284, 5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whicher J. R., Florova G., Sydor P. K., Singh R., Alhamadsheh M., Challis G. L., Reynolds K. A., and Smith J. L. (2011) Structure and function of the RedJ protein, a thioesterase from the prodiginine biosynthetic pathway in Streptomyces coelicolor. J. Biol. Chem. 286, 22558–22569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Witkowski A., Naggert J., Wessa B., and Smith S. (1991) A catalytic role for histidine 237 in rat mammary gland thioesterase II. J. Biol. Chem. 266, 18514–18519 [PubMed] [Google Scholar]
- 41. Witkowski A., Naggert J., Witkowska H. E., Randhawa Z. I., and Smith S. (1992) Utilization of an active serine 101-cysteine mutant to demonstrate the proximity of the catalytic serine 101 and histidine 237 residues in thioesterase II. J. Biol. Chem. 267, 18488–18492 [PubMed] [Google Scholar]
- 42. Martin M. A., Lassek W. D., Gaulin S. J., Evans R. W., Woo J. G., Geraghty S. R., Davidson B. S., Morrow A. L., Kaplan H. S., and Gurven M. D. (2012) Fatty acid composition in the mature milk of Bolivian forager-horticulturalists: controlled comparisons with a US sample. Matern. Child Nutr. 8, 404–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hur G. H., Vickery C. R., and Burkart M. D. (2012) Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 29, 1074–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Condurso H. L., and Bruner S. D. (2012) Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery. Nat. Prod. Rep. 29, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Keatinge-Clay A. T. (2012) The structures of type I polyketide synthases. Nat. Prod. Rep. 29, 1050–1073 [DOI] [PubMed] [Google Scholar]
- 46. Crosby J., and Crump M. P. (2012) The structural role of the carrier protein–active controller or passive carrier. Nat. Prod. Rep. 29, 1111–1137 [DOI] [PubMed] [Google Scholar]
- 47. Libertini L. J., Lin C. Y., Abraham S., Hilf R., and Smith S. (1980) Medium chain fatty acid synthesis in rodent mammary adenocarcinomas in vitro. Biochim. Biophys. Acta 618, 185–191 [DOI] [PubMed] [Google Scholar]
- 48. Koglin A., Löhr F., Bernhard F., Rogov V. V., Frueh D. P., Strieter E. R., Mofid M. R., Güntert P., Wagner G., Walsh C. T., Marahiel M. A., and Dötsch V. (2008) Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature 454, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hunt M. C., Tillander V., and Alexson S. E. (2014) Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie 98, 45–55 [DOI] [PubMed] [Google Scholar]
- 50. Yamashita A., Hayashi Y., Matsumoto N., Nemoto-Sasaki Y., Oka S., Tanikawa T., and Sugiura T. (2014) Glycerophosphate/acylglycerophosphate acyltransferases. Biology 3, 801–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Płoskoń E., Arthur C. J., Evans S. E., Williams C., Crosby J., Simpson T. J., and Crump M. P. (2008) A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J. Biol. Chem. 283, 518–528 [DOI] [PubMed] [Google Scholar]
- 52. Bunkoczi G., Pasta S., Joshi A., Wu X., Kavanagh K. L., Smith S., and Oppermann U. (2007) Mechanism and substrate recognition of human holo ACP synthase. Chem. Biol. 14, 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Masoudi A., Raetz C. R., Zhou P., and Pemble C. W. 4th (2014) Chasing acyl carrier protein through a catalytic cycle of lipid A production. Nature 505, 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nguyen C., Haushalter R. W., Lee D. J., Markwick P. R., Bruegger J., Caldara-Festin G., Finzel K., Jackson D. R., Ishikawa F., O'Dowd B., McCammon J. A., Opella S. J., Tsai S. C., and Burkart M. D. (2014) Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 505, 427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]