Abstract
Processive kinesin motors often contain a coiled-coil neck that controls the directionality and processivity. However, the neck coil (NC) of kinesin-3 is too short to form a stable coiled-coil dimer. Here, we found that the coiled-coil (CC1)-forkhead-associated (FHA) tandem (that is connected to NC by Pro-390) of kinesin-3 KIF13A assembles as an extended dimer. With the removal of Pro-390, the NC-CC1 tandem of KIF13A unexpectedly forms a continuous coiled-coil dimer that can be well aligned into the CC1-FHA dimer. The reverse introduction of Pro-390 breaks the NC-CC1 coiled-coil dimer but provides the intrinsic flexibility to couple NC with the CC1-FHA tandem. Mutations of either NC, CC1, or the FHA domain all significantly impaired the motor activity. Thus, the three elements within the NC-CC1-FHA tandem of KIF13A are structurally interrelated to form a stable dimer for activating the motor. This work also provides the first direct structural evidence to support the formation of a coiled-coil neck by the short characteristic neck domain of kinesin-3.
Keywords: intracellular trafficking, kinesin, molecular motor, structural biology, x-ray crystallography, KIF13A, neck coil
Introduction
Intracellular transport is a fundamental biological process that governs the delivery and distribution of cellular components (such as proteins, mRNAs, and membrane vesicles/organelles) and that is often powered by cytoskeleton-dependent molecular motors (1–3). Kinesins are a family of microtubule-based molecular motors that can drive long range intracellular transport and regulate microtubule dynamics (4, 5). To move along microtubules, processive kinesin motors (kinesin-1–3) require a core motor domain (MD,3 referred to as the “head”) and an adjacent neck domain that forms a characteristic “neck” to control the motor domain (5, 6) (Fig. 1A). The motor domain binds to microtubules and undergoes conformational changes upon ATP hydrolysis, whereas the neck domain acts as a mechanical amplifier of the motor domain and controls the directionality and processivity (7, 8). The conventional kinesin-1 contains a neck domain that is composed of a neck linker (NL) and a neck coil (NC) (Fig. 1A). NL is an intriguing structural element that can dock onto or undock from the motor domain (coupled with different nucleotide-bound states) (9–11). NC forms a coiled-coil dimer that brings the two motor domains together to assemble a functional “two-headed” motor (12, 13). The internal strain between the two motor heads (referred to as inter-head strain) together with NL docking and undocking ensures the processive movement of kinesin-1 toward the plus ends of microtubules (14–16).
FIGURE 1.
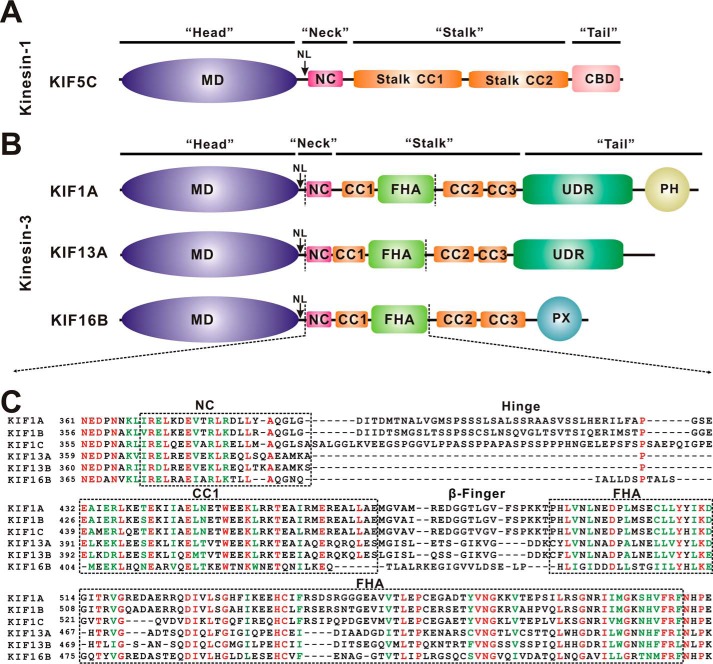
Domain organization of kinesin-1 and kinesin-3. A, domain organization of kinesin-1 KIF5C. KIF5C contains an N-terminal MD, a neck coil (NC), the central stalk CC1 and CC2, and a C-terminal cargo binding domain (CBD). B, domain organization of kinesin-3 KIF1A, KIF13A, and KIF16B. In addition to an N-terminal motor domain and a neck coil, KIF1A, KIF13A, and KIF16B contain non-continuous coiled-coils (CC1-CC3) and an FHA domain in the middle. Both KIF1A and KIF13A contain an undefined region (UDR), and KIF1A and KIF16B contain an extra PH domain and PX domain at the C terminus, respectively. C, sequence alignment of the NC-CC1-FHA tandem from different kinesin-3 motors. The identical residues are colored in red, and the highly conserved residues are colored in green. The regions of NC, CC1, and the FHA domain are marked with dashed boxes.
Kinesin-3 is a subfamily of kinesin motors and plays prominent roles in controlling the transport of synaptic vesicles or endosomes from the cell body to axon tips or cell peripheries (3, 5). In comparison to kinesin-1, the kinesin-3 neck domain contains a similar NL but a shorter NC (∼10 residues shorter than that of kinesin-1) that is followed by a coiled coil (CC1) and the subfamily-specific FHA domain (Fig. 1, B and C). Recent structural studies of the motor domain and NL of KIF1A, the founding member of kinesin-3 subfamily, demonstrated that NL undergoes a similar conformational change upon docking onto the motor domain (17), which suggests that kinesin-3 adopts a processive mechanism similar to that of kinesin-1. In contrast, kinesin-3 NC tends to form a weak coiled-coil dimer (rather than a stable coiled-coil) due to its short length (18, 19). Moreover, this weak dimer can be further regulated by subsequent CC1. Electron microscopy studies of the neck domain of UNC-104 (KIF1A homolog in Caenorhabditis elegans) showed that CC1 can fold back to associate with NC to form a compact bundle-like structure that keeps the motor in a monomeric inactive state (20). This intramolecular interaction between the NC and CC1 of UNC-104 was, therefore, suggested to regulate the monomer-dimer switch of the motor and has been indicated to happen in KIF1A as well (21). More recently, the CC1-mediated regulation of the NC dimer has also been found in other kinesin-3 motors such as KIF13A, KIF13B, and KIF16B (22).
Despite the negative regulation by CC1, the NC-mediated dimerization is required for the processive movement of kinesin-3. Mutations of the hydrophobic residues that are predicted for the NC coiled-coil dimer formation decreased the probability of movement (18, 23). Recent studies of KIF1A demonstrated that the activation of the motor domain needs an additional stretch of residues (∼10 residues) from the subsequent hinge (21), suggesting that the formation of the stable NC coiled-coil dimer for processive movement requires an auxiliary element. Moreover, the FHA domain of KIF1A can also work together with CC1 to form an integrated CC1-FHA dimer that would prevent the CC1-mediated inhibition and further stabilize the motor dimer (24). Thus, the segments immediately after NC are able to facilitate and stabilize the weak NC dimer to assemble a two-headed motor for active transport (21, 24). However, the molecular mechanism underlying the NC coiled-coil dimer formation and how the following CC1-FHA tandem works together with NC for the active motor are both poorly understood.
In this study we characterized the NC-CC1-FHA tandem from the kinesin-3 motor KIF13A. Distinct from other kinesin-3 motors, the CC1-FHA tandem of KIF13A is connected to NC by a proline residue Pro-390 (rather than a flexible hinge in KIF1A) (Fig. 1C). The structure of the CC1-FHA tandem of KIF13A revealed that CC1 and the FHA domain are also integrated to form an extended dimer. By removing Pro-390 between NC and CC1, we obtained the structure of the NC-CC1 tandem of KIF13A. Unexpectedly, it forms a continuous coiled-coil dimer that can be well aligned into the CC1-FHA dimer. The data from molecular dynamics simulations further suggested that the introduction of Pro-390 back into the NC-CC1 tandem breaks the continuous coiled-coil dimer but endows it with the intrinsic flexibility to couple NC with the CC1-FHA tandem to form a stable dimer. Thus, these three elements within the NC-CC1-FHA tandem of KIF13A are structurally correlated for the active conformation of the motor.
Experimental Procedures
Protein Expression and Purification
DNA sequences encoding mouse KIF13A fragments including the NC-CC1 tandem (residues 355–445), the CC1-FHA tandem (residues 386–559), the NC-CC1-FHA tandem (residues 355–559), and various mutants were each cloned into a modified version of the pET32a vector. Point mutations were created using the standard PCR-based mutagenesis method and confirmed by DNA sequencing. Recombinant proteins were expressed in Escherichia coli BL21 Condon Plus (DE3) host cells at 16 °C. The GB1-His6-tagged fusion proteins were purified by Ni2+-Sepharose 6 Fast Flow (GE Healthcare) affinity chromatography followed by size-exclusion chromatography. After the cleavage of the GB1 tag, the resulting proteins were further purified by another step of size-exclusion chromatography.
Crystallization and Data Collection
Native crystals of the CC1-FHA tandem (10 mg/ml in 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 1 mm DTT) and native and selenomethionine crystals of the NC-CC1(ΔPro-390) mutant (15 mg/ml in 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 1 mm DTT) were obtained using the sitting-drop vapor diffusion method at 16 °C. The CC1-FHA tandem was crystallized in 0.2 m calcium acetate, 16% (w/v) PEG3350, whereas the NC-CC1(ΔPro-390) mutant was crystallized in 1.7 m ammonium sulfate, 8% (v/v) isopropyl alcohol. The crystals of the CC1-FHA tandem were cryo-protected by the mother liquor supplemented with 30–40% (w/v) PEG3350, and the crystals of the NC-CC1(ΔPro-390) mutant were cryo-protected by the mother liquor supplemented with 12% (v/v) ethylene glycol, and all the crystals were then flash-frozen by plunging into liquid nitrogen. Diffraction data were collected at the beam-line BL17U of the Shanghai Synchrotron Radiation Facility with a wavelength of 0.979 Å at 100 K and were processed and scaled with HKL2000 (25).
Structural Determination
The structure of the CC1-FHA tandem was determined by the molecular replacement methods using the FHA domain of KIF13B (PDB code 3FM8) as the search model with Phaser (26). Selenium-labeled sites in the crystals of the NC-CC1(ΔPro-390) mutant were determined using the program SHELXD from the CCP4 suite (27). The single-wavelength anomalous dispersion phasing was obtained, and an initial model was built using AutoSol in PHENIX (28). Additional missing residues were manually modeled into the structures according to the 2Fo − Fc and Fo − Fc electron density maps. All the structures were further fitted and rebuilt with COOT (29) and refined with PHENIX (28). The overall quality of the final structural models of the CC1-FHA tandem and the NC-CC1(ΔPro-390) mutant was assessed by PROCHECK (30). The protein structure figures were prepared using the program PyMOL. The statistics for the data collection and structure refinement were summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses refer to the highest resolution shell. –, not available.
| KIF13A CC1-FHA | Se-Met NC-CC1(ΔP390) | Native NC-CC1(ΔP390) | |
|---|---|---|---|
| Diffraction data | |||
| Space group | C 1 2 1 | H 3 2 | H 3 2 |
| Wavelength (Å) | 0.9791 | 0.9791 | 0.9791 |
| Cell dimensions | |||
| a, b, c (Å) | 99.2, 50.0, 66.9 | 160.7, 160.7, 85.0 | 158.6,158.6,82.5 |
| α, β, γ (°) | 90, 92.0, 90 | 90, 90, 120 | 90,90,120 |
| Resolution (Å) | 49.55-1.74 (1.79-1.74) | 50-3.0 (3.16-3.00) | 50–2.82 (2.97–2.82) |
| Unique reflections | 33,409 (2,472) | 8,555 (1229) | 9,716 (1,408) |
| Rmerge (%) | 6.9 (80.5) | 11.5 (62.2) | 10.5 (109.3) |
| Rpim (%)a | 3.4 (38.0) | 3.7 (19.6) | 2.7 (27.3) |
| Mean I/σ(I) | 12.2 (2.6) | 20.7 (5.5) | 18.7 (3.3) |
| Multiplicity | 5.1 (5.3) | 20.7 (21.3) | 16.1 (16.5) |
| Completeness (%) | 99.0 (99.7) | 100.0 (100.0) | 100.0 (100.0) |
| Refinement | |||
| Rwork/Rfreeb(%) | 16.3/22.1 | 18.4/23.3 | |
| Mean B factors (Å2) | |||
| Protein | 37.4 | 86.0 | |
| Ligand | 56.0 | – | |
| Solvent | 47.9 | 74.0 | |
| r.m.s.d.c | |||
| Bond length (Å) | 0.007 | 0.009 | |
| Bond angles (°) | 1.103 | 1.120 | |
| Ramachandran plot (%) | |||
| Favored region | 98.3 | 96.8 | |
| Allowed region | 1.7 | 2.6 | |
| Disallowed region | 0.0 | 0.6 | |
| PDB ID | 5DJO | 5DJN | |
a Rpim is the precision-indicating merging R factor. It measures the quality of the data after averaging the multiple measurements.
b Rwork is the Rfactor for the working dataset. Rfactor = Σ‖Fo − |Fc‖/Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree is the cross-validation Rfactor computed for a randomly chosen subset of reflections, which were not used during refinement.
c Root mean square deviation from ideal values.
Size Exclusion Chromatography Coupled with Multiangle Light Scattering
Protein samples (∼1.0 mg/ml) were analyzed with static light scattering by injection of them into an Agilent HPLC system with a WTC size exclusion chromatography column (Wyatt Technology). The chromatography system was coupled with an 18-angle light-scattering detector (DAWN HELEOS II, Wyatt Technology) and differential refractive index detector (Optilab rEx, Wyatt Technology). Masses (molecular masses) were calculated with ASTRA (Wyatt Technology). Bovine serum albumin (Sigma) was used as the calibration standard.
Cell Culture, Imaging, and Data Analysis
The KIF13A fragments including MD (residues 1–354), MD-NC (residues 1–385), MD-NC-CC1 (residues 1–445), MD-NC-CC1-FHA (residues 1–559), and various mutants were each cloned into a pEGFP-N3 vector. The resulting constructs contained a linker of 14 residues between the KIF13A fragments and the C-terminal GFP tag. N2A (a mouse neuroblastoma cell line) cells were cultured in DMEM containing 10% (v/v) fetal bovine serum. The cells were transfected with the KIF13A fragments by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Fluorescence images were obtained on an Olympus FV1000 laser scanning confocal microscope equipped with a 60× oil-immersion objective lens (NA = 1.42). Confocal settings used for image capture were held constant in comparison experiments. All the fluorescence images were processed and analyzed by ImageJ (National Institutes of Health). The final quantification graphs were generated by Origin (OriginLab). For the cellular distribution data analysis, the specific regions of the cell body (excluding the nucleus) and the tip of each cell were chosen, and the average fluorescence intensities were calculated, respectively.
Molecular Dynamics Simulations
Based on the structures of the CC1-FHA and NC-CC1(ΔPro-390) dimers, the initial structural model of the NC-CC1-FHA dimer of KIF13A was built, and Pro-390 was introduced between NC and CC1. The model structures of the NC-CC1-FHA and NC-CC1(ΔPro-390) dimers were then solvated in a 66 × 85 × 163 Å3 and a 145 × 58 × 58 Å3 water box, respectively, which included 52 Na+ and 34 Cl− ions to neutralize the systems. The NAMD package (31) and CHARMM36 all-atom force field (32–34) were used for energy minimizations and molecular dynamics simulations. Under periodic boundary conditions, a 12 Å cutoff was used for van der Waals interactions, and Particle Mesh Ewald summation was used to calculate the electrostatic interactions. Three independent simulations were performed for each system. For each simulation energy was first carefully minimized in multisteps to avoid any possible clashes. The energy-minimized system was then equilibrated for 5 ns with temperature controlled at 310 K by Langevin dynamics and pressure controlled at 1 atm by the Lagevin piston method (46, 47). With the equilibrated structures, 145- and 130-ns free dynamics simulations were performed for the NC-CC1-FHA and NC-CC1(ΔPro-390) dimers, respectively. The simulation trajectories were analyzed with VMD (35).
Results
The Structure of the CC1-FHA Tandem of KIF13A
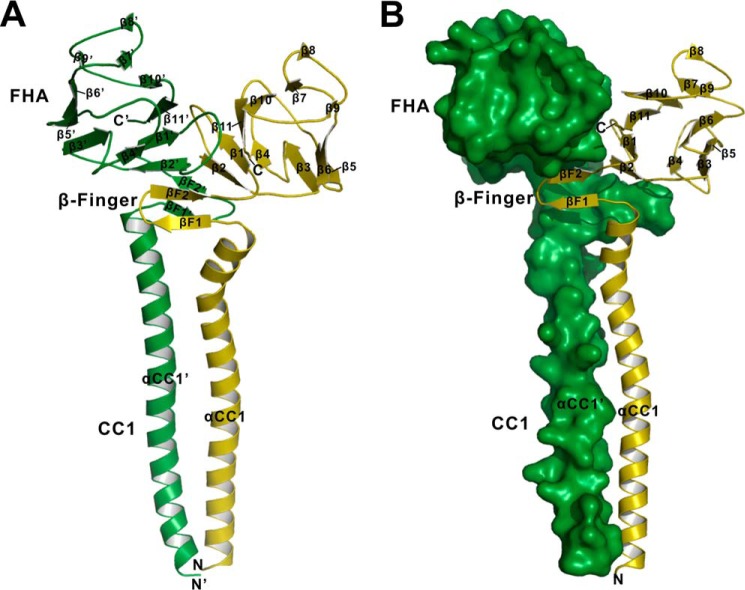
To investigate how the CC1-FHA tandem affects NC within kinesin-3 motors, we initiated this study by structural characterization of the CC1-FHA tandem from KIF13A (Fig. 1B). The structure of the CC1-FHA tandem of KIF13A was determined by the molecular replacement method and was refined to 1.74 Å (Table 1). From the overall structure, CC1 and the FHA domain within the CC1-FHA tandem are integrated by a covalent linker to form an elongated parallel dimer (Fig. 2A), which is consistent with the previous structural studies of the CC1-FHA tandem of KIF1A (24). In this dimer structure, the FHA domain adopts a classical FHA-fold with 11 strands (β1-β11) to form a β-sandwich, whereas CC1 adopts an α-helical structure (αCC1) to form an extended coiled coil (Fig. 2A). The covalent linker between CC1 and the FHA domain forms two antiparallel strands (βF1 and βF2, referred to as the β-finger) that pair with the β-sandwich of the FHA domain and also contact the C-terminal end of CC1 (Fig. 2). Thus, the β-finger linker likely acts as “glue” to stick CC1 and the FHA domain together to form an integrated dimer.
FIGURE 2.
The overall structure of the CC1-FHA dimer of KIF13A. A, a ribbon diagram of the structure of the CC1-FHA dimer. The two subunits of the CC1-FHA dimer are colored in green and olive, respectively. The secondary structures of the FHA domain, the β-finger, and the CC1 helix are labeled, and both the N and C termini are also marked. B, a combined surface and ribbon representation of the dimer structure. One of the two subunits is in the surface representation (colored in green), and the other is in the ribbon representation (colored in olive).
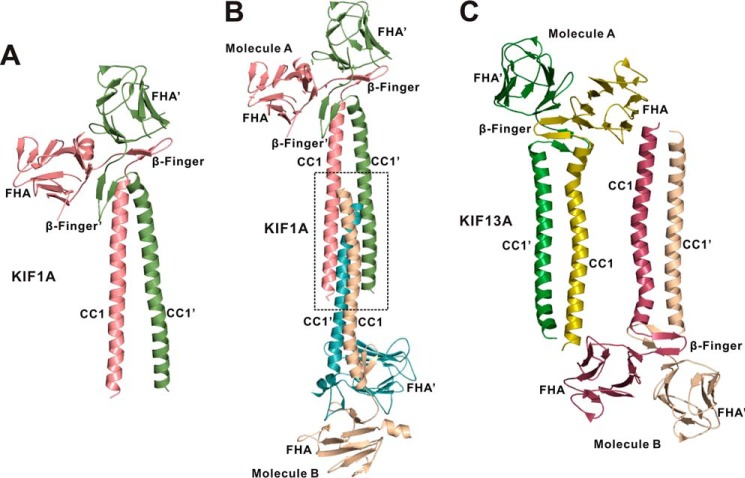
The overall structure of the CC1-FHA dimer of KIF13A is similar to that of KIF1A. However, the β-finger regions of the two dimer structures are different. In the CC1-FHA dimer of KIF1A, the two β-fingers are largely separated, and one of them intimately covers the C-terminal end of CC1, whereas in that of KIF13A the two β-fingers pack with each other to form the central core without extensive contacts with CC1 (Figs. 2A and 3A). This structural difference may be caused by the different lengths of the β-finger regions of KIF1A and KIF13A (Fig. 1C). Additionally, the conformations of CC1 in the two structures are also different, i.e. the two CC1 helices contact each other in the CC1-FHA dimer of KIF13A but are separated in that of KIF1A (Figs. 2A and 3A). To reconcile this difference, we further analyzed the crystal packing of the CC1-FHA tandem of KIF1A and found that the two CC1 helices from one dimer pack with the ones from the other dimer to form a four-helix bundle (Fig. 3B), which would somewhat distort the CC1 coiled-coil dimer. Thus, the separation of the two CC1 helices in the CC1-FHA dimer of KIF1A is most likely to be crystal-packing artifacts. In contrast, the CC1 coiled-coil dimer remains intact in the crystal packing of the CC1-FHA tandem of KIF13A (Fig. 3C).
FIGURE 3.
Analysis of the crystal packing of the CC1-FHA dimers of KIF1A and KIF13A. A, a ribbon diagram of the structure of the CC1-FHA dimer of KIF1A (PDB code 4EGX). The two subunits of the dimer are colored in green and red, respectively. B, analysis of the crystal stacking of the CC1-FHA dimer of KIF1A. In the crystal stacking the two CC1 helices from one dimer (Molecule A) pack with the ones from the other dimer (Molecule B) to form a four-helix bundle, which induces the separation of the N-terminal halves of the two CC1 helices. C, analysis of the crystal stacking of the CC1-FHA dimer of KIF13A. In the crystal stacking, the two CC1 helices from one dimer (molecule A) do not interfere with the ones from the other dimer (molecule B).
The Interaction Interface between the CC1-FHA Dimer of KIF13A
In the CC1-FHA dimer of KIF3A, the dimer interface (buried with ∼5200 Å2) is contributed by the FHA domain, the β-finger, and CC1. Among them, the β-finger is the core component that intimately pairs with the first two β-strands (β1 and β2) of the FHA domain to form a central β-sandwich in the interface (Fig. 2A). Specifically, βF2 of the β-finger augments β2 of the FHA domain in an antiparallel manner, and the hydrophobic packing core of the central β-sandwich is constructed by Ile-433 and Leu-435 from βF1, Ile-440 and Val-442 from βF2, Tyr-448 and Val-450 from β1, and Leu-462, Val-463, and Tyr-465 from β2 (Fig. 4, A–C). Moreover, one open site of the central β-sandwich is capped by the C-terminal ends of the two CC1 helices, i.e. Leu-428 and Met-431 from CC1 make the hydrophobic contacts with Ile-433 and Leu-435 from βF1 of the β-finger (Fig. 4, A–C), which would further stabilize the central dimer interface.
FIGURE 4.
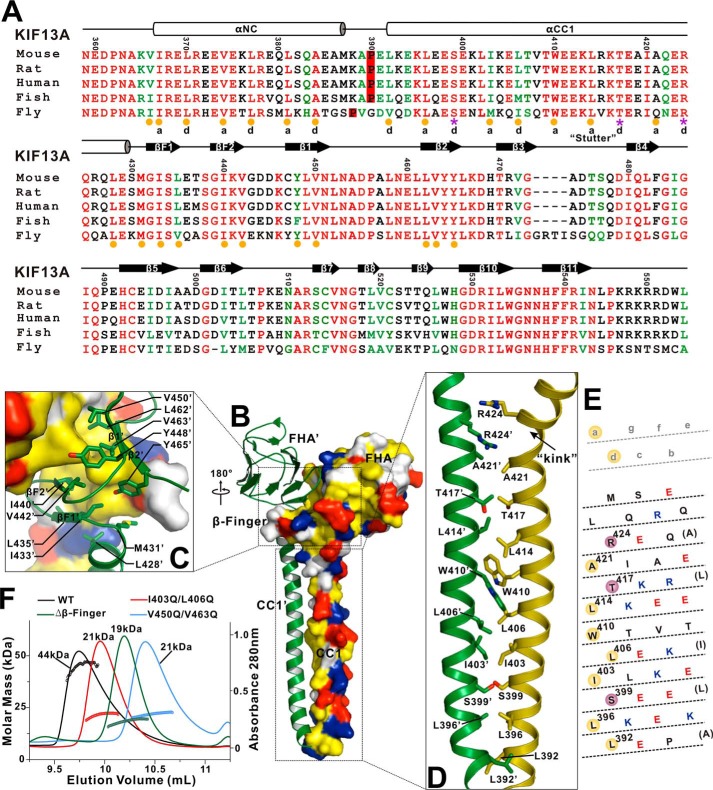
The interaction interface between the CC1-FHA dimer. A, structure-based sequence alignment of the NC-CC1-FHA tandem of KIF13A from different species. The identical residues are colored in red, and the highly conserved residues are in green. The secondary structures and residue numbers are marked on the top. The hydrophobic residues responsible for the formation of the NC-CC1(ΔPro-390) and CC1-FHA dimers are highlighted by yellow circles at the bottom. The hydrophilic residues that are located in the interhelical packing between the CC1 dimer are marked by purple asterisks. The a and d sites of the NC and CC1 helices for interhelical packing are also marked. B, a combined surface and ribbon representation of the CC1-FHA dimer. In this surface drawing the hydrophobic, positively charged, negatively charged residues and remaining residues are colored in yellow, blue, red, and white, respectively. The interaction interface between the dimer can be divided into two parts (highlighted by boxes). C, a combined surface, ribbon and stick model illustrates the dimer interface mediated by the FHA domain and β-finger. The side chains of the residues involved in the dimer packing are shown as sticks. D, a combined ribbon and stick model illustrates the dimer interface mediated by CC1. The side chains of the residues involved in the dimer packing are shown as sticks. E, heptad repeat register of the residues for the CC1 dimer packing. The CC1 helix has been cut and opened flat to give a two-dimensional representation. The hydrophobic residues and hydrophilic residues in the packing core are highlighted by yellow and red circles, respectively. F, size exclusion chromatography-multiangle light scattering analysis of the CC1-FHA tandem and its mutants. The calculated molecular mass of each fragment is marked.
Besides the central β-sandwich formed by the FHA domain and the β-finger, CC1 forms an extended coiled-coil dimer that facilitates the dimer formation (Fig. 2A). Based on the heptad repeat pattern (a–g) analysis, the interhelical packing between the coiled-coil dimer is largely mediated by the hydrophobic residues (Leu-392, Leu-396, Ile-I403, Leu-406 and Leu-414) in the a and d sites of the CC1 helix (Fig. 4, A, D, and E). However, a number of hydrophilic residues (Ser-399, Thr-417, and Arg-424) were found in the interhelical packing interface (Fig. 4, A, D, and E), which would somewhat destabilize the CC1 coiled coil. The repulsion between the positively charged side chains of the two Arg-424 residues is also likely to induce an obvious kink at the C terminus of one of the two CC1 helices (Fig. 4D), which results in an asymmetric conformation of the CC1-FHA dimer (Fig. 2A). Moreover, the steric hindrance between the bulky aromatic side chains of the two Trp-410 residues in the middle would produce another twist in the CC1 helix (Fig. 4D). In addition to the above potential defects in the interhelical packing, the coiled-coil stacking in the middle of the CC1 helix is also irregular, i.e. there is a “stutter” break around Trp-410 with the 3-4-4-3 pattern (versus the classical 3-4-3-4 pattern, and the number indicates the interval of packing residues in the interhelical interface) (36) (Fig. 4, A and E). Thus, CC1 is not a perfect coiled coil for dimerization but possesses intrinsic unusual features to distort the coiled-coil formation.
To evaluate the role of the hydrophobic packing in the interaction interface for the dimer formation, we next made point mutations or deletions in CC1 (I403Q/L406Q), the β-finger (Δβ-finger), or the FHA domain (V450Q/V463Q). We characterized the CC1-FHA tandem and its mutants by using size exclusion chromatography coupled with multiangle light scattering assay. Consistent with the above structural analysis, the CC1-FHA tandem of KIF13A formed a dimer in solution (with the calculated molecular mass of ∼44 kDa), whereas the mutants existed in a monomeric state (with the calculated molecular mass of ∼21 kDa (the I403Q/L406Q mutant), ∼19 kDa (the Δβ-finger mutant), and ∼21 kDa (the V450Q/V463Q mutant) (Fig. 4F), indicating that all these mutations significantly impaired and destabilized the dimeric conformation of the CC1-FHA tandem. Thus, the hydrophobic interaction interface formed by CC1, the β-finger, and the FHA domain is essential for the CC1-FHA dimer formation.
The Structure of the NC-CC1(ΔPro-390) Mutant of KIF13A
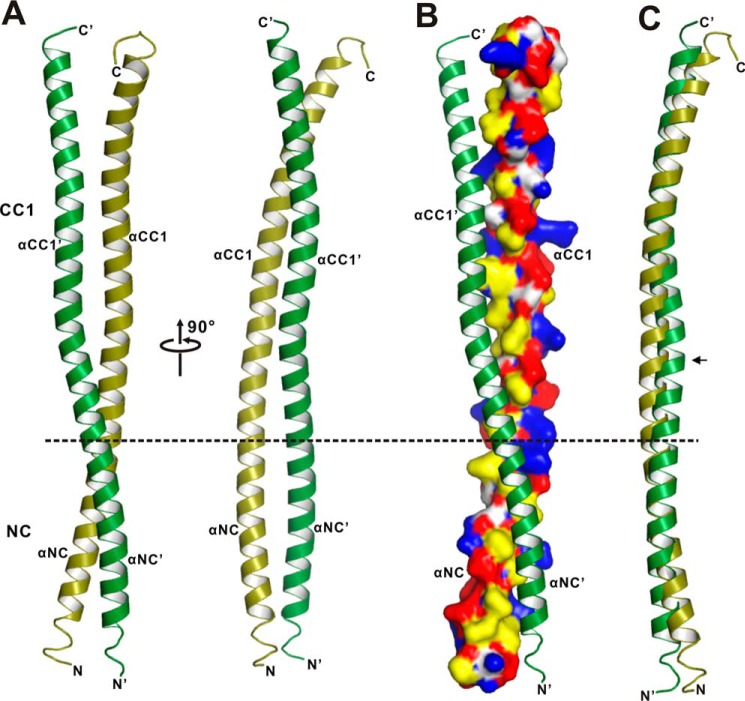
Based on the structure of the CC1-FHA dimer (Fig. 2), NC, immediately preceding CC1, would also form a coiled-coil dimer to tether the two motor domains together. To investigate the mechanism underlying the NC coiled-coil dimer formation, we next performed crystal screening of NC and the NC-CC1-FHA tandem. However, neither of the two fragments could be crystallized, and we then resorted to the NC-CC1 tandem. Recent studies of KIF13A demonstrated that although CC1 could fold back to inhibit the NC dimer, the deletion of Pro-390 between NC and CC1 (the NC-CC1(ΔPro-390) mutant) could produce a coiled-coil dimer that can activate the motor (22), indicating that the NC-CC1(ΔPro-390) mutant might be a stable dimer suitable for further structural studies. We then focused on the NC-CC1(ΔPro-390) mutant and successfully obtained the high quality crystals of this mutant. The crystal structure of the NC-CC1(ΔPro-390) mutant was solved by the single-wavelength anomalous dispersion method and was refined to 2.82 Å (Table 1). The NC-CC1(ΔPro-390) mutant unexpectedly formed a continuous extended coiled-coil dimer integrated by NC and CC1 (i.e. without breaks between NC and CC1 due to the removal of Pro-390) (Fig. 5, A and B). In this coiled-coil dimer structure, the two subunits are similar except for the two flexible terminal ends (Fig. 5C).
FIGURE 5.
The structure of the NC-CC1(ΔPro-390) dimer. A, a ribbon diagram of the structure of the NC-CC1(ΔPro-390) dimer. The two subunits of the dimer are colored in green and olive, respectively. The division site between NC and CC1 is highlighted by a dashed line. The secondary structures of NC and CC1 are labeled, and the N and C termini are also marked. B, a combined surface and ribbon representation of the NC-CC1(ΔPro-390) dimer. In this surface drawing the color schemes follow that of Fig. 4B. C, superposition of the two subunits of the NC-CC1(ΔPro-390) dimer. Notably, one helix is slightly twisted in the middle (marked with an arrow).
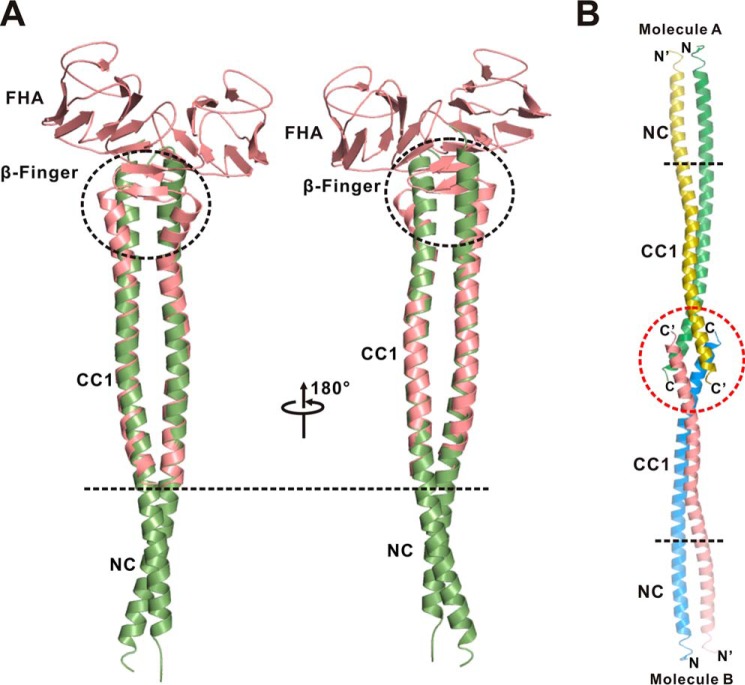
Interestingly, the dimeric CC1 in the structure of the NC-CC1(ΔPro-390) mutant could be well superimposed with that in the CC1-FHA tandem (Fig. 6A), indicating that CC1 adopts a similar coiled-coil conformation in the two dimers. The structural analysis of the NC-CC1(ΔPro-390) dimer showed that CC1 indeed utilized the same set of residues for the interhelical packing (Fig. 4A) and that a similar twist also occurs in the middle of the CC1 helix (Figs. 2A and 5A). Despite these similarities, the C-terminal ends of the two CC1 helices from the two dimer structures are slightly different, i.e. CC1 is not twisted, and a part of the β-finger in the CC1-FHA dimer forms a coiled coil in the NC-CC1(ΔPro-390) dimer (Fig. 6A). Further analysis of the NC-CC1(ΔPro-390) crystal packing revealed that the two C-terminal ends of CC1 from the two dimers form a short four-helix bundle (Fig. 6B), which suggests that the difference of CC1 might also be caused by crystal packing artifacts due to the truncation of the FHA domain.
FIGURE 6.
The extended conformation of the NC-CC1-FHA(ΔPro-390) dimer of KIF13A. A, a structural model of the NC-CC1-FHA(ΔPro-390) dimer built by superimposing the structures of the NC-CC1(ΔPro-390) (green) and CC1-FHA (red) dimers based on CC1. The two CC1 dimers from the two structures can be well aligned with each other except for the extreme C-terminal ends (highlighted by a black circle). B, analysis of the crystal stacking of the NC-CC1(ΔPro-390) dimer of KIF13A. In the crystal stacking the two C-terminal ends of CC1 from one dimer (molecule A) pack with the ones from the other dimer (molecule B), which leads to the difference of the C-terminal end of CC1 between the two dimers.
The Interhelical Packing between the NC Coiled-coil Dimer
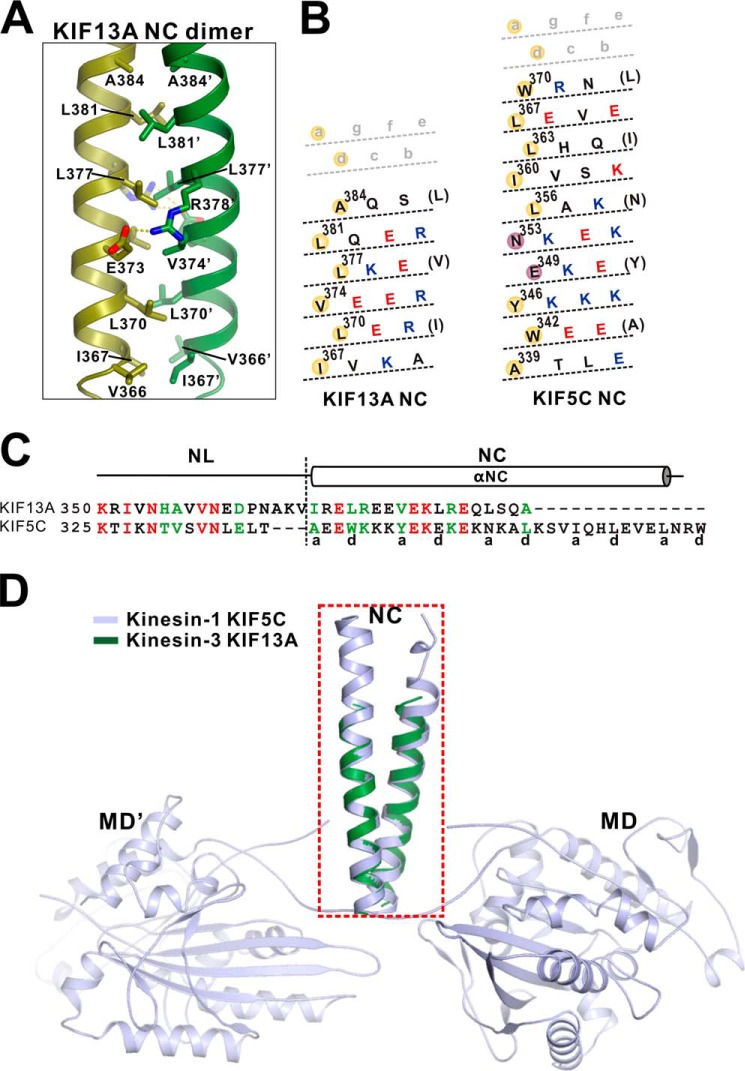
In the NC-CC1(ΔPro-390) structure, NC adopts a classical dimeric coiled coil (Fig. 7A). The interhelical packing between the NC coiled-coil dimer is mediated by the hydrophobic residues from the a and d sites, i.e. Ile-367, Leu-370, Val-374, Leu-377, Leu-381, and Ala-384 form the hydrophobic packing core of the coiled-coil dimer (Figs. 4A and 7, A and B). In addition to the hydrophobic packing, the charge-charge interactions between the two NC helices further stabilize the coiled-coil formation (Fig. 7A). Thus, despite short length, the interhelical packing between the NC coiled-coil dimer matches the classical coiled-coil stacking perfectly.
FIGURE 7.
The interhelical packing between the NC coiled-coil dimer. A, a combined ribbon and stick model illustrates the interaction interface between the NC coiled-coil dimer. The side chains of the residues involved in the interhelical packing are shown as sticks. B, heptad repeat register of the residues for the coiled-coil packing of the NC dimer of KIF13A and KIF5C. The representation mode and color scheme follow that of Fig. 4E. C, structure-based sequence alignment of the neck domain (NL and NC) of kinesin-3 KIF13A and kinesin-1 KIF5C. The identical residues are colored in red, and the highly conserved residues are colored in green. The a and d sites of the NC coiled-coil are marked at the bottom. D, superposition of the NC coiled-coil structure of KIF13A with that of KIF5C (PDB code 3KIN). The NC coiled-coil dimer of KIF13A (colored in green) can be well aligned with that of KIF5C (colored in light blue) but is much shorter.
To the best of our knowledge this is the first time the structure of the dimeric NC coiled-coil of kinesin-3 was obtained. Because NC is a well known key component of kinesin family motors for controlling the motor processive movement, we next compared the structure of the NC coiled-coil dimer of KIF13A with that of the conventional kinesin-1 KIF5C. Interestingly, the two NC coiled-coil dimers can be well aligned with each other (Fig. 7, C and D), supporting that kinesin-3 adopts a similar processive mechanism to that of kinesin-1. However, based on the sequence alignment of NC, kinesin-3 NL is three residues longer than that of kinesin-1 (Fig. 7C), which would decrease the interhead strain and interfere with the processive movement. Consistent with this feature, recent studies demonstrated that the shortening of kinesin-3 NL would restore its processivity similar to that of kinesin-1 (16). In addition to the variation of the NL length, the interhelical packing of the two NC coiled-coil dimers is also different, i.e. some a and d sites of KIF5C NC are occupied by hydrophilic residues, whereas the a and d sites of KIF13A NC are all hydrophobic residues (Fig. 7B). More significantly, the length of the NC helix of KIF13A is much shorter than that of KIF5C (Fig. 7, B–D), indicating the intrinsic instability of the NC coiled-coil dimer of kinesin-3. Taken together, although kinesin-3 NC adopt a similar coiled-coil structure to that of kinesin-1 NC, the NC coiled-coil dimer of kinesin-3 possesses distinct characteristics that may be essential for controlling processive movement.
The Dynamic Conformations of the NC-CC1-FHA Dimer of KIF13A
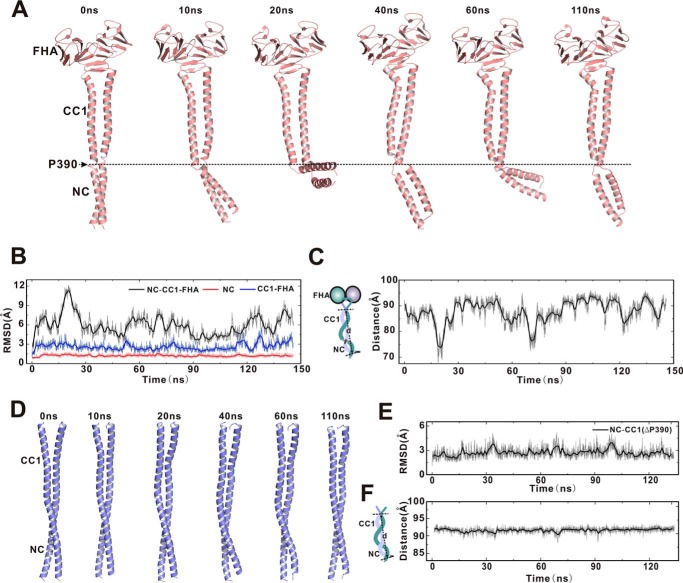
The superimposition of the structures of the CC1-FHA tandem and NC-CC1(ΔPro-390) mutant allowed us to build an extended NC-CC1-FHA(ΔPro-390) dimeric model, which may tether the two motor domains together for activation (Fig. 6A). Unfortunately, the continuous NC-CC1(ΔPro-390) coiled-coil dimer is caused by the removal of Pro-390 between NC and CC1. The reverse introduction of Pro-390 into the NC-CC1(ΔPro-390) coiled-coil dimer would somewhat distort the coiled-coil formation. Consistent with this assumption, the previous studies of KIF13A demonstrated that the NC-CC1 tandem (without the FHA domain) tends to adopt a folded-back conformation to prevent the NC dimer formation, and Pro-390 is located exactly in the essential turn between NC and CC1 (22). However, upon attachment to the FHA domain, the formation of the CC1-FHA dimer would drive CC1 to form a coiled-coil dimer (based on the structure of the CC1-FHA tandem), which would release the NC inhibition and promote the formation of the NC coiled-coil dimer. Thus, the NC-CC1-FHA tandem would still be capable of forming an extended stable dimer, although Pro-390 could break the NC-CC1 coiled-coil dimer.
To evaluate the conformation of the NC-CC1-FHA dimer, we next introduced Pro-390 back into the NC-CC1-FHA(ΔPro-390) dimer and performed the molecular dynamics simulations in solution. As expected, the addition of Pro-390 between NC and CC1 breaks the extended coiled-coil and separates the NC-CC1-FHA dimer into the two linked dimers, i.e. the short NC coiled-coil dimer and the CC1-FHA dimer (Fig. 8A). During the simulations, the two separated dimers undergo little conformational changes (Fig. 8, A and B), indicating that each of them within the NC-CC1-FHA dimer is relatively stable and maintains the dimeric conformation. In contrast, the overall conformation of the NC-CC1-FHA dimer is highly dynamic with the various orientations of the NC coiled-coil dimer relative to the CC1-FHA dimer (Fig. 8C), which is likely caused by the intrinsic flexibility of the Pro-390 linker. On the other hand, we also performed the molecular dynamics simulations of the NC-CC1(ΔPro-390) dimer. As expected, the overall structure of the NC-CC1(ΔPro-390) dimer exhibits no significant conformational changes during the simulations (Fig. 8, D–F), further supporting that the dynamic conformations of the NC-CC1-FHA dimer are most likely caused by Pro-390 between NC and CC1. Thus, the NC-CC1-FHA dimer of KIF13A exhibits dynamic conformations that may be important for active transport (see “Discussion”).
FIGURE 8.
Molecular dynamics simulations of the NC-CC1-FHA and NC-CC1(ΔPro-390) dimers. A, snapshots of a representative simulation of the NC-CC1-FHA dimer with simulation time indicated. The extended NC-CC1-FHA dimer is broken into two linked dimers (the NC dimer and the CC1-FHA dimer) by Pro-390 that is highlighted by a dashed line. B, time course of the root mean square deviation of the NC dimer (red), the CC1-FHA dimer (blue), and the NC-CC1-FHA dimer (black). Both the NC dimer and the CC1-FHA dimer undergo little conformational changes, but the overall conformation of the NC-CC1-FHA dimer is dynamic during simulations. RMSD, root mean square deviation. C, time course of the distance between the N-terminal end of NC and the C-terminal end of CC1 (as indicated in the left panel), indicating the significant fluctuations of the overall conformation of the NC-CC1-FHA dimer. D, snapshots of a representative simulation of the NC-CC1(ΔPro-390) dimer with simulation time indicated. E, time course of the root mean square deviation of the NC-CC1(ΔPro-390) dimer. F, time course of the distance between the N-terminal end of NC and the C-terminal end of CC1, indicating no significant conformational changes of the NC-CC1(ΔPro-390) dimer.
The Dimeric NC-CC1-FHA Tandem of KIF13A Is Essential for Motor Activation
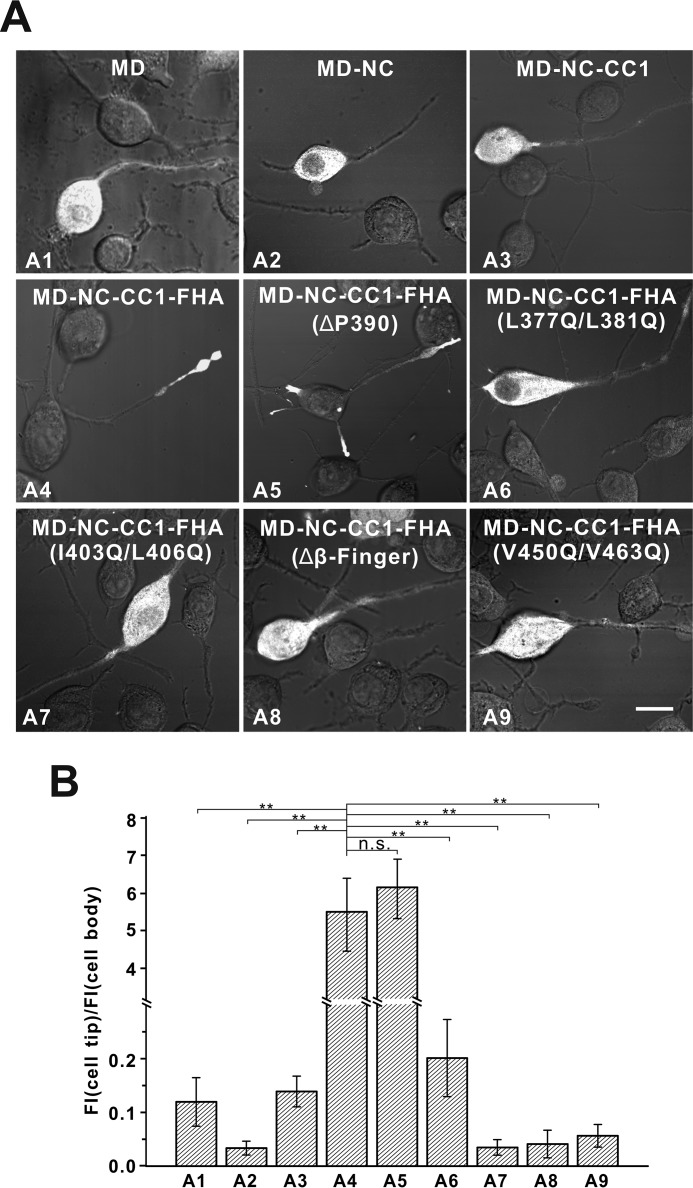
All the above data demonstrated that NC, CC1, and the FHA domain would work together to form a dynamic NC-CC1-FHA dimer for the active motor (Fig. 8). To investigate the role of the NC-CC1-FHA tandem for motor activation, we next evaluated the cellular localization of the different fragments of KIF13A including the motor domain and the NC-CC1-FHA tandem (Fig. 9). In this cell-based assay, the active fragments can be largely localized to the cell periphery, whereas the inactive ones cannot (21, 24). As expected, the motor domain alone was inactive and enriched in the cell body (Fig. 9). The extension of the motor domain to NC or CC1 resulted in the MD-NC or MD-NC-CC1 fragment. Consistent with the previous studies (22), these two fragments of KIF13A were both largely localized in the cell body (Fig. 9), indicating that neither NC nor the NC-CC1 tandem can activate the motor domain. In contrast, the further inclusion of the FHA domain led to the MD-NC-CC1-FHA fragment that was highly enriched at the cell periphery (Fig. 9), suggesting that the NC-CC1-FHA tandem of KIF13A is the minimal essential element for motor activation. To further validate the essential role of the NC-CC1-FHA dimer for motor activation, we made the mutations in NC (L377Q/L381Q), CC1 (I403Q/L406Q), β-finger (Δβ-finger), or the FHA domain (V450Q/V463Q) to destabilize the dimer (Figs. 4 and 7). We also made the deletion of Pro-390 between NC and CC1 to enhance the dimerization. As expected, the MD-NC-CC1-FHA(ΔPro-390) mutant was active and accumulated at the cell periphery, whereas the MD-NC-CC1-FHA mutants with the mutations in either NC, CC1, β-finger, or the FHA domain were all highly enriched in the cell body (Fig. 9), indicating that each component within the NC-CC1-FHA tandem is essential for motor activation. Taken together, all the data demonstrated that the activation of the motor domain of KIF13A requires the correlation of NC with the CC1-FHA tandem to form the minimal essential NC-CC1-FHA dimer.
FIGURE 9.
The NC-CC1-FHA tandem is essential for motor activation. A, cellular localizations of the MD, MD-NC, MD-NC-CC1, and MD-NC-CC1-FHA fragments and various MD-NC-CC1-FHA mutants. The MD, MD-NC, and MD-NC-CC1 fragments were all enriched in the cell body (A1–A3), whereas the MD-NC-CC1-FHA fragment was predominantly localized to the cell periphery (A4). The MD-NC-CC1-FHA mutants with the mutations to disrupt the dimer were localized in the cell body (A6–A9), but the NC-CC1-FHA(ΔPro-390) mutant was localized to the cell periphery (A5). Scale bar: 20 μm. B, quantification of the cellular distribution data shown in panel A. The ratio of the tip to cell body average fluorescence intensity (FI) was quantified for each construct for more than 15 cells (n > 15). Each bar represents the mean value ± S.D. **, p < 0.05. n.s., not significant.
Discussion
The CC1-FHA Dimer of Kinesin-3
We have demonstrated that the CC1-FHA tandem of KIF1A can form a stable dimer for the dimerization and activation of the motor (24) (Fig. 3A). In this study the structure of the CC1-FHA tandem from the kinesin-3 motor KIF13A revealed a similar extended CC1-FHA dimer integrated by the β-finger linker between CC1 and the FHA domain (Figs. 2 and 3). Thus, as suggested in our previous studies (24), the CC1-FHA-mediated dimerization is likely to be a general feature for kinesin-3 motors.
In the structure of the CC1-FHA dimer of KIF13A, the interhelical packing interface between the CC1 coiled-coil dimer contains some significant defects (Fig. 4). The intrinsic imperfectness of CC1 may endow versatile roles of this segment in regulating motor activity. Consistent with this assumption, CC1 has been demonstrated to fold back to interact with NC and inhibit its coiled-coil formation (22). However, with the help of the FHA domain and the β-finger linker, the two CC1 helices in the CC1-FHA dimer of KIF13A are forced to form a coiled-coil dimer (Fig. 2), which would prevent it from folding back to interact with NC. Thus, as in previous suggestions (24), the formation of the CC1-FHA dimer would somewhat release the CC1-mediated inhibition (Figs. 8 and 9).
Because the CC1-FHA dimer of kinesin-3 motors is essential for the motor activation, the modulations of this dimer would be a potential mechanism to regulate the motor activity. Interestingly, recent studies of KIF1A demonstrated that it can be phosphorylated by the kinase Cdk5 and the phosphorylation site (-487SPKK490-) is located between the β-finger and the FHA domain of KIF1A and is in the interface of the CC1-FHA dimer (37) (Fig. 1C). The phosphorylation-mimic mutation was found to disrupt the CC1-FHA dimer and deactivate the motor (37), suggesting that the Cdk5-mediated phosphorylation of the CC1-FHA tandem could be essential for the motor regulation. Moreover, given that the FHA domain is a well known phosphopeptide binding module (38, 39) and is directly involved in the dimer formation (Fig. 2), the binding of target proteins to this domain would be another potential regulation of the CC1-FHA dimer formation, although the FHA domain of KIF13B (the KIF13A homolog in mammals) was reported to bind to its target proteins rather in a phospho-independent manner (40). Nevertheless, the FHA domain binding partner-mediated potential regulation of the CC1-FHA dimer assembly is of great interest and still needs further investigations.
The Coiled-coil Dimer Formed by Kinesin-3 NC
Similar to kinesin-1 NC, kinesin-3 NC is also an essential component for the motor dimerization and activation (18, 21). However, due to its short length, it is difficult to obtain the structure of the NC coiled-coil dimer of kinesin-3. In this study we determined the structure of the NC-CC1(ΔPro-390) mutant of KIF13A and obtained the structural information of NC, which provides the first structural evidence to support the formation of the coiled-coil dimer by kinesin-3 NC (Figs. 5 and 7). As predicted, the interhelical packing between the NC coiled-coil dimer is predominantly mediated by the hydrophobic residues (Fig. 7B). However, in comparison to kinesin-1, the length of the NC coiled-coil dimer of kinesin-3 is relatively short (Fig. 7, C and D), which may be the major cause that leads to the instability of this dimer. Consistent with this feature, UNC-104 was found to often pause during the processive movement likely due to the temporally unwinding of the NC dimer (18).
It is interesting to note that the NC coiled-coil dimer of kinesin-1 also contains some defects (e.g. some hydrophilic residues are located in interhelical interface (Ref. 41; Fig. 7, B and C)) that would somewhat destabilize the dimer. This intrinsic instability/flexibility has been demonstrated to be essential for the processive movement of kinesin-1 under load (42), and one possible explanation is that it may facilitate the motor to skip the obstacles on the microtubule tracks. Thus, the intrinsic instability of the NC coiled-coil dimer of kinesin-3 may be also essential for facilitating its processive movement. Consistently, the previous studies of kinesin-3 motors demonstrated that they are marathon runners in cells with much longer run length (22).
The Active Conformation of Kinesin-3 Mediated by the NC-CC1-FHA Tandem
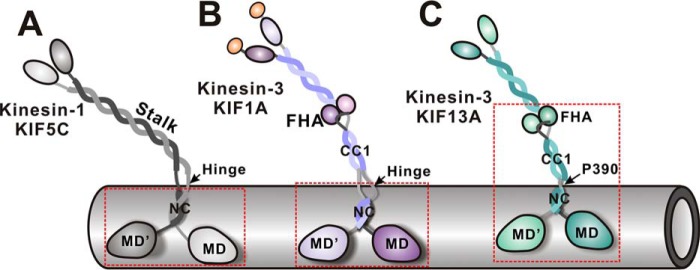
Given that the coiled-coil dimer formed by kinesin-3 NC is unstable, the formation of the active motor often required the subsequent segments to stabilize the NC coiled-coil dimer (Fig. 1B). In KIF1A/UNC-104, a large flexible hinge immediately followed NC, and the residues from this hinge are essential for motor activation (21) (Fig. 10). A similar scenario happens in KIF16B (22). However, this essential flexible hinge is missing from KIF13A, and the CC1-FHA tandem is linked to NC by Pro-390 (Fig. 1C). Moreover, the NC-CC1-FHA tandem (but not NC or the NC-CC1 tandem) of KIF13A was able to activate the motor domain (Fig. 9). Thus, distinct from other kinesin-3 motors, the full activation of KIF13A needed the correlation of NC with the CC1-FHA tandem (rather than a few residues from the flexible hinge) to form an extended dimer (Fig. 10).
FIGURE 10.
Schematic models illustrating the active states of kinesin-1 and kinesin-3. In the active state of kinesin-1 KIF5C, the NC dimer alone is sufficient to activate the motor domain and the flexible linker between the neck and stalk is essential for processive movement (A). In contrast, the NC dimer of kinesin-3 KIF1A is unable to activate the motor domain and needs the subsequent segments (i.e. a part of the NC/CC1-hinge) to stabilize the dimer for motor activation (B). Due to the lack of the NC/CC1 hinge, the whole CC1-FHA tandem of KIF13A is required for correlating with the NC dimer to activate the motor domain (C). The minimum active fragments of KIF5C, KIF1A, and KIF13A are marked with dashed boxes. The intrinsic flexibility between the neck and stalk of kinesin-3 (provided by the NC/CC1-hinge of KIF1A and the Pro-390 linker of KIF13A) is also likely to be essential for active transport.
Due to the Pro-390 linker, the overall conformation of the NC-CC1-FHA dimer of KIF13A is dynamic with the various orientations between NC and the CC1-FHA tandem (Fig. 8). The NC-CC1-FHA tandem of KIF1A would possess this feature, and its conformation would be much more dynamic because of a larger flexible hinge between NC and the CC1-FHA tandem (Fig. 10). It is interesting to note that a flexible hinge also exists between the NC dimer and the stalk coiled-coil of kinesin-1, which would provide certain flexibility between the neck and stalk (Figs. 1 and 10) and has been demonstrated to be essential for processive movement (43, 44). In addition, the flexibility between the neck and stalk of kinesin-1 would endow the different orientations between them. The previous studies of kinesin-1 demonstrated that, during processive movement, the neck is parallel to the microtubule tracks, whereas the stalk seems somewhat more perpendicular to the tracks for holding cargoes (45). As for kinesin-3, NC forms the neck, whereas the CC1-FHA tandem may resemble the stalk of kinesin-1 (Fig. 10). Thus, the dynamic conformations of the NC-CC1-FHA dimer of kinesin-3 may also be essential for processive movement, and the active conformation of kinesin-3 for cargo transport would be much similar to that of kinesin-1 upon walking along microtubules.
Author Contributions
J. R., L. H., and W. F. conceived and designed the experiments. J. R., L. H., W. W., Y. Z., and W. L. performed the research. J. L. and T. X. gave technical assistance on the MD simulation and cell imaging, respectively. W. F. coordinated the entire research project and wrote the paper.
Acknowledgment
We thank the beam-line BL17U of the Shanghai Synchrotron Radiation Facility for the beam time.
This work was supported by National Major Basic Research Program of China Grants 2011CB910503 and 2014CB910202 and National Natural Science Foundation of China Grants 31300611, 31190062, 31470746, and 31200577. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5DJO and 5DJN) have been deposited in the Protein Data Bank (http://wwpdb.org/), respectively.
- MD
- motor domain
- NL
- neck linker
- NC
- neck coil
- CC1
- coiled-coil
- FHA
- forkhead-associated.
References
- 1. Vale R. D. (2003) The molecular motor toolbox for intracellular transport. Cell 112, 467–480 [DOI] [PubMed] [Google Scholar]
- 2. Soldati T., and Schliwa M. (2006) Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 7, 897–908 [DOI] [PubMed] [Google Scholar]
- 3. Hirokawa N., Niwa S., and Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 4. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 5. Verhey K. J., Kaul N., and Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 [DOI] [PubMed] [Google Scholar]
- 6. Vale R. D., and Fletterick R. J. (1997) The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13, 745–777 [DOI] [PubMed] [Google Scholar]
- 7. Vale R. D., Case R., Sablin E., Hart C., and Fletterick R. (2000) Searching for kinesin's mechanical amplifier. Philos. Trans. R Soc. Lond. B Biol. Sci. 355, 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Endow S. A. (1999) Determinants of molecular motor directionality. Nat. Cell Biol. 1, E163–E167 [DOI] [PubMed] [Google Scholar]
- 9. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., and Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 10. Sindelar C. V., Budny M. J., Rice S., Naber N., Fletterick R., and Cooke R. (2002) Two conformations in the human kinesin power stroke defined by x-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 9, 844–848 [DOI] [PubMed] [Google Scholar]
- 11. Asenjo A. B., Weinberg Y., and Sosa H. (2006) Nucleotide binding and hydrolysis induces a disorder-order transition in the kinesin neck-linker region. Nat. Struct. Mol. Biol. 13, 648–654 [DOI] [PubMed] [Google Scholar]
- 12. Kozielski F., Sack S., Marx A., Thormählen M., Schönbrunn E., Biou V., Thompson A., Mandelkow E. M., and Mandelkow E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91, 985–994 [DOI] [PubMed] [Google Scholar]
- 13. Woehlke G., and Schliwa M. (2000) Walking on two heads: the many talents of kinesin. Nat. Rev. Mol. Cell Biol. 1, 50–58 [DOI] [PubMed] [Google Scholar]
- 14. Yildiz A., Tomishige M., Gennerich A., and Vale R. D. (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyazono Y., Hayashi M., Karagiannis P., Harada Y., and Tadakuma H. (2010) Strain through the neck linker ensures processive runs: a DNA-kinesin hybrid nanomachine study. EMBO J. 29, 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shastry S., and Hancock W. O. (2011) Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253–16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Atherton J., Farabella I., Yu I. M., Rosenfeld S. S., Houdusse A., Topf M., and Moores C. A. (2014) Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife 3, e03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomishige M., Klopfenstein D. R., and Vale R. D. (2002) Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263–2267 [DOI] [PubMed] [Google Scholar]
- 19. Rashid D. J., Bononi J., Tripet B. P., Hodges R. S., and Pierce D. W. (2005) Monomeric and dimeric states exhibited by the kinesin-related motor protein KIF1A. J. Pept. Res. 65, 538–549 [DOI] [PubMed] [Google Scholar]
- 20. Al-Bassam J., Cui Y., Klopfenstein D., Carragher B. O., Vale R. D., and Milligan R. A. (2003) Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J. Cell Biol. 163, 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hammond J. W., Cai D., Blasius T. L., Li Z., Jiang Y., Jih G. T., Meyhofer E., and Verhey K. J. (2009) Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLos Biol. 7, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soppina V., Norris S. R., Dizaji A. S., Kortus M., Veatch S., Peckham M., and Verhey K. J. (2014) Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. U.S.A. 111, 5562–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huckaba T. M., Gennerich A., Wilhelm J. E., Chishti A. H., and Vale R. D. (2011) Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J. Biol. Chem. 286, 7457–7467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huo L., Yue Y., Ren J., Yu J., Liu J., Yu Y., Ye F., Xu T., Zhang M., and Feng W. (2012) The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure 20, 1550–1561 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., Macarthur M. W., Moss D. S., and Thornton J. M. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., and Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 33. MacKerell A. D. Jr., Feig M., and Brooks C. L. 3rd (2004) Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 126, 698–699 [DOI] [PubMed] [Google Scholar]
- 34. Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr. (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ, and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Humphrey W., Dalke A., and Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 36. Brown J. H., Cohen C., and Parry D. A. (1996) Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins 26, 134–145 [DOI] [PubMed] [Google Scholar]
- 37. Liu B., Yue Y., Yu Y., Ren J. Q., Feng W., Huo L., and Xu T. (2014) Potential phosphorylation site modulates the dimerization and activity of KIF1A. Prog. Biochem. Biophys. 41, 870–876 [Google Scholar]
- 38. Mahajan A., Yuan C., Lee H., Chen E. S., Wu P. Y., and Tsai M. D. (2008) Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal 1, re12. [DOI] [PubMed] [Google Scholar]
- 39. Liang X., and Van Doren S. R. (2008) Mechanistic insights into phosphoprotein-binding FHA domains. Acc. Chem. Res. 41, 991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong Y., Tempel W., Wang H., Yamada K., Shen L., Senisterra G. A., MacKenzie F., Chishti A. H., and Park H. W. (2010) Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1. Proc. Natl. Acad. Sci. U.S.A. 107, 20346–20351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thormählen M., Marx A., Sack S., and Mandelkow E. (1998) The coiled-coil helix in the neck of kinesin. J. Struct. Biol. 122, 30–41 [DOI] [PubMed] [Google Scholar]
- 42. Jaud J., Bathe F., Schliwa M., Rief M., and Woehlke G. (2006) Flexibility of the neck domain enhances Kinesin-1 motility under load. Biophys. J. 91, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grummt M., Woehlke G., Henningsen U., Fuchs S., Schleicher M., and Schliwa M. (1998) Importance of a flexible hinge near the motor domain in kinesin-driven motility. EMBO J. 17, 5536–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Crevenna A. H., Madathil S., Cohen D. N., Wagenbach M., Fahmy K., and Howard J. (2008) Secondary structure and compliance of a predicted flexible domain in kinesin-1 necessary for cooperation of motors. Biophys. J. 95, 5216–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin D. S., Fathi R., Mitchison T. J., and Gelles J. (2010) FRET measurements of kinesin neck orientation reveal a structural basis for processivity and asymmetry. Proc. Natl. Acad. Sci. U.S.A. 107, 5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martyna G. J., Tobias D. J., and Klein M. L. (1994) Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 101, 4177–4189 [Google Scholar]
- 47. Feller S. E., Zhang Y. H., Pastor R. W., and Brooks B. R. (1995) Constant-pressure molecular-dynamics simulation-the Langevin piston method. J. Chem. Phys. 103, 4613–4621 [Google Scholar]