Abstract
In breast tumors, activation of the nuclear factor κB (NFκB) pathway promotes survival, migration, invasion, angiogenesis, stem cell-like properties, and resistance to therapy—all phenotypes of aggressive disease where therapy options remain limited. Adding an anti-inflammatory/anti-NFκB agent to breast cancer treatment would be beneficial, but no such drug is approved as either a monotherapy or adjuvant therapy. To address this need, we examined whether dimethyl fumarate (DMF), an anti-inflammatory drug already in clinical use for multiple sclerosis, can inhibit the NFκB pathway. We found that DMF effectively blocks NFκB activity in multiple breast cancer cell lines and abrogates NFκB-dependent mammosphere formation, indicating that DMF has anti-cancer stem cell properties. In addition, DMF inhibits cell proliferation and significantly impairs xenograft tumor growth. Mechanistically, DMF prevents p65 nuclear translocation and attenuates its DNA binding activity but has no effect on upstream proteins in the NFκB pathway. Dimethyl succinate, the inactive analog of DMF that lacks the electrophilic double bond of fumarate, is unable to inhibit NFκB activity. Also, the cell-permeable thiol N-acetyl l-cysteine, reverses DMF inhibition of the NFκB pathway, supporting the notion that the electrophile, DMF, acts via covalent modification. To determine whether DMF interacts directly with p65, we synthesized and used a novel chemical probe of DMF by incorporating an alkyne functionality and found that DMF covalently modifies p65, with cysteine 38 being essential for the activity of DMF. These results establish DMF as an NFκB inhibitor with anti-tumor activity that may add therapeutic value in the treatment of aggressive breast cancers.
Keywords: breast cancer, chemical modification, drug action, inflammation, inhibitor, NFκB, covalent modification, cysteine, dimethyl fumarate, mammosphere
Introduction
In the United States, breast cancer is the second most prevalent cancer among women and claims over 40,000 lives each year. Despite major advancements in breast cancer treatment, a successful therapy outcome is limited to early detection of cancer at the primary organ. Therapy options for aggressive breast cancer disease (i.e. advanced stage, therapy-resistant, recurrent, or metastatic) are limited. As a result, the prognosis remains poor, and aggressive disease accounts for more than 90% of breast cancer-related deaths.
Although the underlying mechanisms are not fully understood, inflammation has emerged as a key instigator and driver of aggressive breast cancers (1, 2). More specifically, the nuclear factor κB (NFκB)2 pathway promotes multiple aggressive tumor phenotypes, including cell survival, migration, invasion, angiogenesis, and resistance to therapy (3, 4). The link between the inflammatory NFκB pathway and breast cancer is also supported by the fact that a deregulated, or constitutively active, NFκB pathway is associated with aggressive breast cancer phenotypes and therapy resistance (5–9). More recently, activation of the NFκB pathway has been shown to regulate the survival and propagation of breast cancer stem cells (CSCs) (10–12), which are a small subset of tumor cells that evade all standard therapies and are involved in metastasis and tumor recurrence (13–18). Given that the NFκB pathway is essential for breast cancer progression and aggressiveness, its inhibition can be exploited to eradicate CSCs and other detrimental NFκB-dependent tumor phenotypes. However, to date, there are no such NFκB pathway inhibitors available in the clinic.
Therapeutic targeting of NFκB activity has been directed at inhibiting various players in the pathway (19). The canonical NFκB pathway consists of p65 (RelA) and p50 transcription factors, which are held in the cytoplasm by an inhibitor protein, IκBα. Upon stimulation by inflammatory cytokines, such as TNFα, IL-1β, or other factors, the IκB kinase (IKK) complex, consisting of IKKα, IKKβ, and the scaffolding protein NFκB essential modulator (NEMO), is activated. This leads to phosphorylation and proteasomal degradation of IκBα. As a result, p65/p50 factors are liberated and can translocate to the nucleus, where they bind to DNA and induce gene transcription (20). Therefore, inhibitors targeting the proteasome and upstream kinases have been investigated as a new class of anti-inflammatory drugs, but most have failed because of inhibition of other non-NFκB targets and toxic side effects (21). In addition, given that NFκB is also critical to the innate immune system, most NFκB inhibitors cause long-lasting immune suppression. As a result, the development of safe NFκB inhibitors is even more challenging (22), especially for anti-cancer therapy where continued inhibitor use is required. This raises the issue of how to safely and effectively inhibit the NFκB pathway. One option is to use the anti-inflammatory drug Tecfidera (dimethyl fumarate, DMF). DMF was approved in the United States in March 2013 for multiple sclerosis and is now the number one prescribed oral therapy for relapsing forms of the disease. DMF is neuroprotective and is proposed to act via inhibition of NFκB and activation of Nrf2 pathways (23–26). Most importantly, DMF has a proven safety in humans; it has immune-modulatory properties without significant immune suppression (27). This makes DMF an attractive candidate for NFκB inhibition. Moreover, its therapeutic potential in breast cancer therapy has yet to be explored.
Our studies indicate that DMF inhibits NFκB activity in multiple breast cancer cell lines. Consistent with its anti-NFκB activity, DMF also inhibits mammosphere (MS) formation, cell proliferation, and xenograft tumor growth. Mechanistically, we found that DMF covalently modifies the NFκB transcription factor p65 to block its nuclear translocation and DNA binding activity. These results provide proof-of-principle evidence that DMF can be used to inhibit NFκB activity in breast cancer cells. Understanding the mechanism of action of DMF could provide the needed rationale to advance DMF into the clinic for aggressive breast cancer therapy.
Experimental Procedures
Reagents
TNFα was purchased from R&D Systems. DMF, DMS, NAC and methyl cellulose were purchased from Sigma. IKK7 was purchased from EMD Millipore. Compound 16 was obtained from Dr. Terry Moore (University of Illinois at Chicago). DAPI, ProLong Gold antifade reagent, protein A Dynabeads, and streptavidin M-280 Dynabeads were purchased from Invitrogen. The Click chemistry reagents tris(2-carboxyethyl)phosphine, carboxyrhodamine 110-azide, and biotin-PEG3-azide were purchased from Click Chemistry Tools. Antibodies for p-IKKα/β (catalog no. 2697), IKKα (catalog no. 2682), IKKβ (catalog no. 2370), p-IκBα (catalog no. 2859), IκBα (catalog no. 4814), p-p65 Ser-536 (catalog no. 3033), p-p65 Ser-468 (catalog no. 3039), and TATA-binding protein (TBP) (catalog no. 8515) were purchased from Cell Signaling Technology. The antibody for p65 (catalog no. sc-372) was purchased from Santa Cruz Biotechnology. The antibody for β-actin (catalog no. A5441) was purchased from Sigma. The Alexa Fluor 594-conjugated goat anti-rabbit antibody (catalog no. A11012) was purchased from Invitrogen.
DMF Probe Synthesis
The DMF probe was synthesized as described previously by Götz et al. (28).
Cell Lines and Culture Conditions
The human estrogen receptor (ER)-positive breast cancer cell lines MCF-7 and T47D and the ER+/Her2+ cell line BT474 were obtained from Dr. Debra Tonetti (University of Illinois at Chicago). Constitutively active IKKβ (CA-IKKβ) cells are stably transfected MCF-7 cells engineered to overexpress a doxycycline (Dox)/tetracycline-inducible, constitutively active form of IKKβ (S177E/S181E) (29). Briefly, cells were derived using the Retro-X Tet-On advanced inducible expression system from Clontech. The IKKβ expression vector was purchased from InvivoGen. Mutations were introduced using site-directed mutagenesis (Stratagene), and the CA-IKKβ plasmid was then subcloned into the puromycin-resistant Tet-On vector pRetroX-Tight-Pur. Retrovirus generation and infection of MCF-7-rtTA cells, which were stably transduced with a geneticin-resistant vector encoding the Tet activator rtTA, were performed according to a protocol published previously (30). Single cell clones were selected using geneticin and puromycin and fully characterized for NFκB activity (data not shown). MCF-7, T47D, BT474, and CA-IKKβ cells were maintained routinely in Roswell Park Memorial Institute 1640 medium (Invitrogen Life Technologies) with phenol red supplemented with 10% FBS, 1% non-essential amino acids, 2 mmol/liter l-glutamine, 1% penicillin-streptomycin, and 6 ng/ml insulin. The ER breast cancer cell line MDA-MB-231 was obtained from Dr. Clodia Osipo (Loyola University Chicago) and maintained routinely in Improved Minimum Essential Medium (Corning) supplemented with 5% FBS, 1% non-essential amino acids, 2 mm l-glutamine, and 1% penicillin-streptomycin.
Luciferase Reporter Assay
MCF-7 cells were transiently co-transfected with an NFκB-RE luciferase construct (Clontech) along with the Renilla luciferase construct pGL4.70 (Promega), and Dual-Luciferase assays were carried out as described previously (31). The mammalian expression vectors containing cDNAs for wild-type p65 and mutant C38S-p65 were a gift from Dr. Thomas Gilmore (Boston University) and have been described previously in detail (32).
RT-Quantitative PCR (QPCR)
RNA isolation was carried out using TRIzol according to the instructions of the manufacturer (Invitrogen). Total RNA (0.5 μg) was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Invitrogen). The resulting product was diluted to 100 μl with double-distilled water, and 2 μl was used for each subsequent quantitative PCR reaction. Quantitative PCR was carried out and analyzed as described previously (33). All QPCR primers used have been validated and reported previously (33). The -fold change was calculated using the ΔΔCt method, with ribosomal protein 36B4 mRNA serving as the internal control.
MS Assay
Breast cancer cells were seeded at single-cell density on low attachment plates in medium described by Dontu et al. (37) supplemented with 1% methyl cellulose to prevent cellular aggregation. After 7 days, the diameter of MS was measured, and MS ≥ 75 μm in diameter were counted. For MS formation studies, inhibitors were added the day after seeding. For RNA, p65 DNA binding activity, and protein studies, MS were grown for 7 days, and inhibitors were added for the last 3–6 h.
Crystal Violet Proliferation Assay
Briefly, cells were seeded in 24-well plates and treated for 7 days with varying concentrations of DMF. Cells were then stained with crystal violet (0.5% in 20% methanol) for 15 min and solubilized using 1% sodium dodecyl sulfate solution, and an absorbance reading was taken at 570 nm.
Xenograft Study
All mouse experiments were carried out at the University of Illinois at Chicago animal facility. All mouse experiments were conducted in accordance with institutional procedures and guidelines and prior approval from the Institutional Animal Care and Use Committee. Female athymic nude mice (nu/nu), 4–5 weeks old, were purchased from Harlan. Five million MDA-MB-231 cells were injected orthotopically into the thoracic mammary glands (n = 14–16 injections/group). Tumor formation was monitored by palpation, and when tumors were detected, mice were randomized into either vehicle control or DMF groups. Mice were gavaged daily with vehicle (0.8% methyl cellulose) or DMF (30 mg/kg suspended in 0.8% methyl cellulose). Tumor sizes were measured daily with an electronic caliper, and tumor volume was calculated as (length × width2) × π/2. Tumor growth was monitored until the total tumor burden reached humane end point criteria.
p65 DNA Binding Assay
The p65 DNA binding activity of nuclear proteins extracted from MCF-7 cells or of recombinant p65 protein (catalog no. 31102, Active Motif) was measured via an ELISA (Active Motif) according to the guidelines of the manufacturer.
ChIP Assay
The ChIP assay was performed as described previously with some modifications (34). Briefly, MCF-7 cells were cross-linked with 2 mm disuccinimidyl glutarate followed by 1% formaldehyde. For the precipitations, beads were coated with antibody prior to pulldown, and pulldowns occurred while rotating for 16 h at 4 °C. Beads were then washed with TSE I (20 mm Tris/HCl, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, and 2 mm EDTA), twice with TSE III (10 mm Tris/HCl, 250 mm LiCl, 1% IGEPAL CA-630, 0.7% deoxycholate, and 1 mm EDTA), and twice with TE followed by elution from the beads using elution buffer (0.1 m NaHCO3 and 1% SDS). Elutions were subsequently de-cross-linked overnight at 65 °C, and DNA was purified and used for QPCR. QPCR primer sequences are available upon request.
Western Blotting
Whole-cell extracts were prepared using M-PER reagent (Thermo Scientific). Proteins were separated by SDS-PAGE (Bio-Rad), transferred to nitrocellulose membranes (Thermo Scientific), blocked for 1 h in buffer containing 5% nonfat dry milk (Lab Scientific) or 5% bovine serum albumin, and incubated with the appropriate primary antibody overnight. The next day, secondary antibody was applied, and the signal was visualized on a Molecular Imager ChemidocXRS (Bio-Rad) using Pierce Supersignal West Pico chemiluminescent substrate (Thermo Scientific). Images were obtained using Quantity One software (Bio-Rad).
Immunohistochemistry (IHC)
Cells were seeded on 0.1% gelatin-coated coverslips. After treatment, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized using 0.1% Triton X-100 for 1 min, and blocked with 10% serum in PBS for 1 h. Cells were then stained overnight at 4 °C with p65 antibody (dilution 1:200), followed by 1-h incubation with Alexa Fluor 594-conjugated secondary antibody (dilution 1:1000). The coverslips were mounted with ProLong Gold antifade reagent with DAPI. Images were then acquired at ×63 magnification using an LSM710 confocal microscope.
In-gel Fluorescence
In situ labeled recombinant p65 protein and its in-gel fluorescence activity were measured as described previously with some modifications (35). Briefly, p65 protein, with or without preincubation with 50 μm DMF for 30 min, was reacted with the DMF probe (50 μm) at 37 °C for 30 min. After click chemistry reaction (36) using CuSO4, tris(2-carboxyethyl)phosphine, and carboxyrhodamine 110-azide, the rhodamine-labeled p65 protein was separated by SDS-PAGE. The gel was then visualized by in-gel fluorescence scanning using the Typhoon system and is shown in a gray scale.
Immunoprecipitation
MDA-MB-231 cells were lysed using radioimmune precipitation assay buffer, and the lysate was purified using a 10-kDa cutoff Millipore Amicon column. Samples containing about 500 μg of total protein were reacted with the DMF probe or vehicle control (dimethyl sulfoxide) as described above. Click chemistry was performed on the samples using CuSO4, tris(2-carboxyethyl)phosphine, and biotin-PEG3-azide. Biotinylated proteins were incubated with streptavidin beads, and pulldown occurred while rotating for 30 min at room temperature. The beads were then washed twice with PBS, and proteins were eluted with 0.1% SDS at 95 °C for 5 min.
Statistical Analysis
Data are presented as mean ± S.E. from at least three independent determinations. Statistical analysis consisted of one- or two-way analysis of variance followed by Tukey post test or t test, as appropriate.
Results
Anti-NFκB Activity of DMF in Breast Cancer Cells
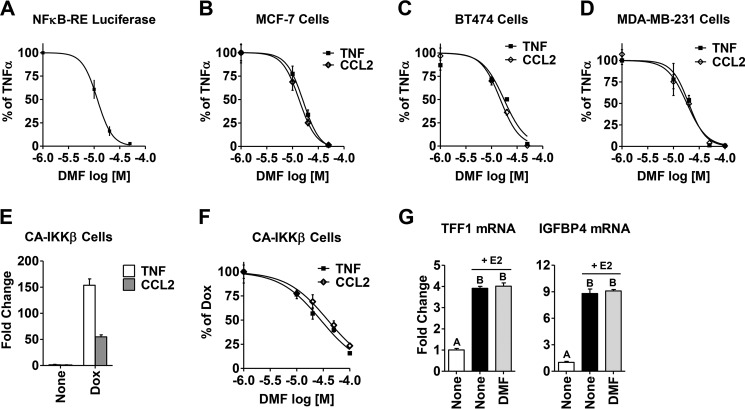
To determine whether DMF inhibits the NFκB pathway in breast cancer cells, we measured DMF activity on several NFκB end points (Fig. 1, A–D). Following TNFα-induced activation of the NFκB pathway, we found that DMF inhibits both NFκB-RE activity (Fig. 1A) and expression of NFκB target genes, such as CCL2 and TNF (Fig. 1, B–D), in a dose-dependent manner, with a calculated IC50 value of ∼20 μm. Moreover, the inhibitory effect of DMF is shown in three different breast cancer cell lines (MCF-7, BT474, and MDA-MB-231), representing different breast cancer subtypes: estrogen receptor-positive, Her2-positive, and triple-negative subtype, respectively. In an alternative, non-cytokine-induced model, we tested DMF activity in stably transfected MCF-7 cells with CA-IKKβ, a key kinase in the NFκB pathway. Upon adding Dox, the NFκB pathway was activated, as shown by elevated target gene expression (Fig. 1E). Similar to cytokine-induced activation of NFκB, we found that adding DMF blocks Dox-induced gene expression in CA-IKKβ cells in a dose-dependent manner (Fig. 1F). Therefore, DMF inhibits NFκB activity across multiple breast cancer cell lines under various stimuli that activate the NFκB pathway. To test the effect of DMF on other transcription factors, we chose the estrogen receptor, given its prominent role in breast cancer. We found that DMF (20 μm) has no effect on classical estrogen receptor-target genes such as TFF1 and IGFBP4, as shown in Fig. 1G. This suggests that DMF does not exert a general nonspecific effect on transcription factors in breast cancer cells.
FIGURE 1.
DMF inhibits TNFα-induced and constitutively active NFκB activity in breast cancer cells. A, NFκB-RE activity was measured in MCF-7 cells following TNFα (10 ng/ml) treatment for 4 h. B–D, expression of the NFκB target genes TNF and CCL2 following TNFα treatment for 2 h was measured by quantitative RT-PCR in MCF-7 cells (B), BT474 cells (C), and MDA-MB-231 cells (D). Increasing concentrations of DMF were added 2 h prior to treatment with TNFα, and the inhibitory activity of DMF is plotted as percent of TNFα alone. E, Dox-induced expression of the NFκB target genes TNF and CCL2, measured by quantitative RT-PCR, is shown in CA-IKKβ cells. F, DMF inhibits expression of the TNF and CCL2 genes in CA-IKKβ cells in a dose-dependent manner. The inhibitory activity of DMF is plotted as percent of Dox alone. IC50 values were calculated with GraphPad software using normalized data. G, DMF has no effect on estrogen receptor target gene TFF1 and IGFBP4 mRNA, measured in MCF-7 cells pretreated with DMF (20 μm) for 2 h, followed by estrogen treatment (E2, 10 nm) for another 2 h. The different letters above bars indicate significant difference between treatments, p < 0.001.
DMF Inhibits Cell Proliferation, MS Formation, and Xenograft Tumor Growth
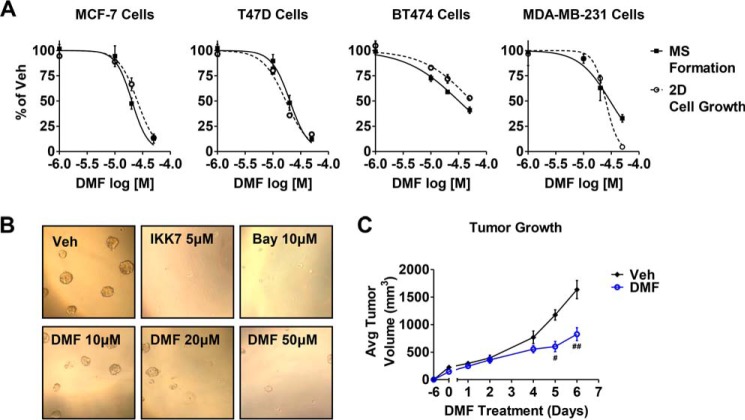
Given that breast CSC survival and propagation has been shown to be dependent on NFκB activity (10–12), we next explored whether DMF could affect the formation of MS, which are enriched for cells with the stem cell-like properties of self-renewal and anchorage-independent growth (37, 38). Two NFκB pathway inhibitors, IKK7 and Bay117082, were used as controls (Fig. 2B) on MS formation. Similar to the known NFκB inhibitors, we found that DMF abrogates MS formation in a dose-dependent manner in all breast cancer cell lines examined (Fig. 2, A, solid line, and B). IC50 values for MS inhibition across the different cell lines is ∼20 μm, the same IC50 value observed for inhibition of the cytokine-induced NFκB pathway in adherent monolayer cultures.
FIGURE 2.
DMF has anti-breast cancer activity both in vitro and in vivo. A, MS formation (solid line) and 2D cell growth (dashed line) in the indicated cell lines were measured after treatment with varying concentrations of DMF. The effect of DMF is plotted as percent of dimethyl sulfoxide vehicle (veh) control. B, representative pictures at ×10 magnification of MCF-7 MS formation upon treatment with DMF or the NFκB inhibitors IKK7 (5 μm) and Bay117082 (Bay, 10 μm). C, effect of DMF (30 mg/kg daily) on MDA-MB-231 xenograft tumor growth is indicated. #, p = 0.0002; ##, p = 0.003. Avg, average.
At similar potency as inhibition of the NFκB pathway and MS formation, DMF acted to inhibit cancer cell proliferation, as measured by the crystal violet assay (Fig. 2A, dashed line). These in vitro effects of DMF prompted us to examine DMF activity on xenograft tumor growth. We found that DMF (30 mg/kg daily) significantly impaired MDA-MB-231 tumor growth in athymic nude mice (Fig. 3C) without affecting animal weight (data not shown). Together, these data indicate, for the first time, the efficacy of DMF on breast cancer phenotypes both in vitro and in vivo.
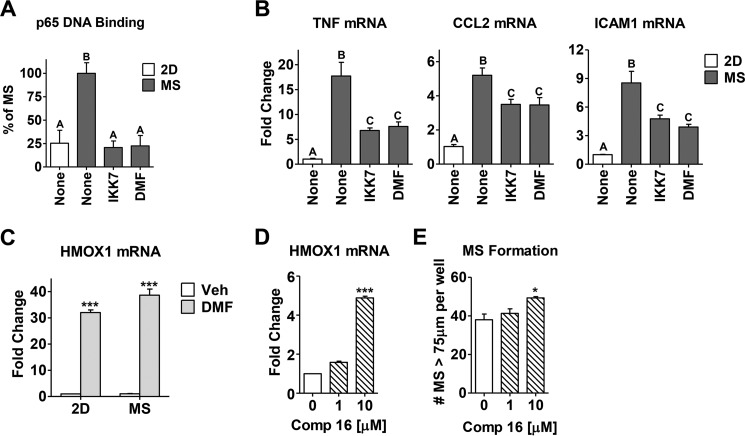
FIGURE 3.
DMF inhibits the intrinsic NFκB activity in MS culture of breast cancer cells. A, p65 DNA binding activity was measured via ELISA in a conventional adherent 2D culture of MCF-7 cells or MS culture with or without inhibitors (IKK7 1 μm or DMF 50 μm) added for the last 3 h. B, expression of the TNF, CCL2, and ICAM1 genes after 6 h of drug treatment was measured in the same groups described in A. The different letters above the columns indicate significant difference between treatments (p < 0.001). C, DMF (20 μm) up-regulates the expression of HMOX1 mRNA in both 2D and MS of MCF-7 cells. Data are shown as -fold change compared with vehicle (Veh) control. D, compound 16 (Comp 16, 1 and 10 μm) up-regulates the expression of HMOX1 mRNA in MCF-7 cells. E, MS formation is measured in MCF-7 cells treated with compound 16. *, p < 0.01; ***, p < 0.001.
DMF Inhibits the High Intrinsic NFκB Activity in MS
To determine whether DMF inhibits NFκB activity in MS culture, MS were allowed to form over 7 days, and inhibitors were added for the last 3 or 6 h of culture. IKK7, a known IKKα/β inhibitor, was used as a positive control. MS displayed elevated levels of p65 DNA binding activity (Fig. 3A) and high NFκB target gene expression (Fig. 3B) compared with untreated breast cancer cells cultured in standard monolayer two-dimensional (2D) conditions. All of these end points were attenuated by DMF or IKK7 to the same extent (Fig. 3, A and B), suggesting that DMF can abrogate MS formation by inhibiting the NFκB pathway.
Besides NFκB inhibition, DMF has also been proposed to activate Nrf2 (23–26). Indeed, we found that DMF significantly up-regulates the Nrf2 target gene, heme oxygenase 1 (HMOX1) mRNA, both in 2D and in MS (Fig. 3C). However, an alternative Nrf2 activator, compound 16 (39), does not inhibit MS formation (Fig. 3E), indicating that Nrf2 activation is not likely to contribute to DMF inhibitory activity in MS.
DMF Blocks p65 DNA Binding, Its Transcriptional Activity, and Its Nuclear Translocation
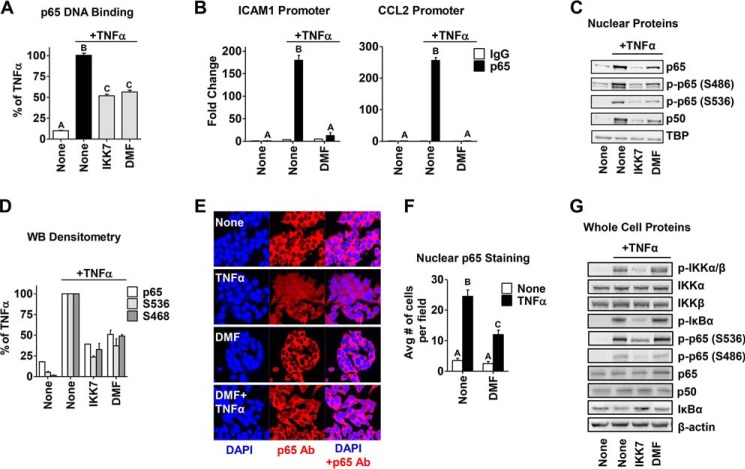
To determine where in the NFκB pathway DMF may be acting, we first examined the DNA binding activity of the main NFκB family member, p65 (RelA), upon TNFα-induced activation in MCF-7 cells. DMF attenuates p65 DNA binding by ∼50%, which is comparable, in this assay, with the known IKK α/β inhibitor IKK7 (Fig. 4A). We next examined the effect of DMF on TNFα-induced p65 DNA recruitment and occupancy on the promoters of the NFκB target genes, ICAM1 and CCL2, via ChIP assay. We found that DMF significantly reduced p65 occupancy on both gene promoters (Fig. 4B), indicating that DMF inhibits p65 transcriptional activity. Interestingly, we also found that total and phosphorylated p65 nuclear protein levels are reduced to the same extent upon treatment, suggesting that DMF exerts its effect on p65 independently of phosphorylation status (Fig. 4, C and D). This observation was corroborated by IHC studies where DMF significantly prevented p65 nuclear localization upon TNFα activation (Fig. 4, E and F). However, the cellular content of upstream components in the NFκB signaling pathway, such as IKKα/β phosphorylation, IκBα phosphorylation and degradation, and p65 phosphorylation, are not affected by DMF. In contrast, IKK7 reduced the nuclear levels of phosphorylated and total p65 similarly to DMF (Fig. 4C) and also significantly attenuated upstream NFκB signaling, as indicated in Fig. 4G, consistent with its known inhibitory effect on IKKα/β. This suggests that the inhibitory action of DMF on p65 nuclear localization and its transcriptional activity are mediated by a target downstream of IKK/IκBα.
FIGURE 4.
DMF inhibits p65 DNA binding and transcriptional activity and its nuclear translocation in an IKK/IκBα-independent manner. A, p65 DNA binding activity was measured in MCF-7 cells treated with IKK7 (1 μm) or DMF (50 μm) for 2 h, followed by TNFα treatment for 15 min. B, ChIP assays were carried out for p65 and the IgG control following treatment of MCF-7 cells with TNFα for 45 min with or without DMF (50 μm) added 2 h prior to TNFα. The -fold-increase in IgG or p65 occupancy at the ICAM1 (left panel) and CCL2 (right panel) promoters were calculated from the percent input of each pulldown and comparison of each treatment to vehicle controls. C, nuclear extracts of cells treated as in A were prepared, and NFκB transcription factors were examined by Western blotting. Representative Western blotting analyses from three independent experiments are shown. TBP served as a loading control. D, densitometry of nuclear proteins relative to TBP. Data are plotted as percent of TNFα alone. WB, Western blotting. E, representative pictures of IHC staining for nuclear p65 in MCF-7 cells treated with DMF (50 μm) followed by TNFα for 15 min. IHC was performed using an anti-p65 antibody (Ab, red) and DAPI (blue) for nuclear staining and visualized using a Zeiss laser-scanning microscope. F, IHC quantitation of the nuclear p65 content in E. Avg, average. G, whole-cell extracts of cells treated as in A were prepared, and NFκB signaling proteins were examined by Western blotting. Representative Western blotting analyses from three independent experiments are shown. β-Actin served as a loading control. The different letters above the columns in A, B, and E indicate a significant difference between treatments (p < 0.001).
DMF Inhibits NFκB by Direct Covalent Modification of p65
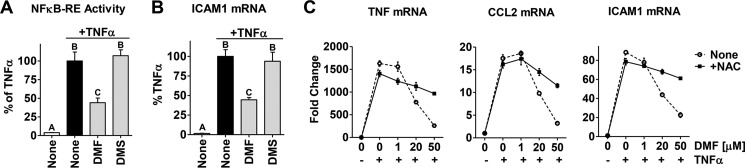
DMF is a cell-permeable α,β-unsaturated electrophilic Michael acceptor that can covalently react with reactive cellular nucleophiles, notably protein cysteine residues (40, 41). First, to determine whether the fumarate Michael acceptor is responsible for activity, we tested DMS, the saturated analog of DMF devoid of the double bond of the fumarate (therefore, it is unable to form covalent protein adducts). We found that DMS at 20 μm (corresponding to the IC50 value of DMF) was unable to inhibit the NFκB pathway in MCF-7 cells (Fig. 5, A and B). This indicates that the electrophilic reactivity associated with the double bond of fumarate is required for the anti-NFκB activity of DMF. Second, to trap DMF before reaction with protein cysteine residues, we utilized the cell-permeable small-molecule thiol NAC, which reacts directly with DMF. Pretreatment of cells with NAC reverses DMF inhibition of NFκB target genes (Fig. 5C, solid line), further supporting the notion that the activity of DMF is caused by covalent modification of a cellular target.
FIGURE 5.
The double bond reactivity of DMF is required to inhibit the NFκB pathway in breast cancer cells. A and B, NFκB-RE activity (A) and ICAM1 (B) gene expression were measure in MCF-7 cells upon treatment with DMF or DMS, 20 μm each, as described in Fig. 1. Data are plotted as percent of TNFα alone. The different letters above the columns indicate a significant difference between treatments (p < 0.001). C, NAC (dashed line) reverses the inhibitory effect of DMF on the TNFα-induced expression of the NFκB target genes TNF, CCL2, and ICAM1 in MCF-7 cells. NAC (500 μm) was added 30 min prior to DMF and TNFα treatment.
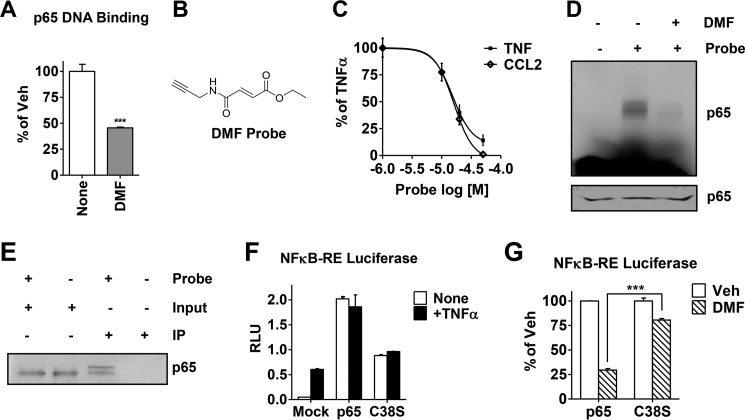
So far, the data obtained from using DMS and NAC indicate that the electrophilic reactivity of fumarate, and, therefore, its ability to form covalent protein adducts, drives the anti-NFκB activity of DMF. As presented above, our data indicate that the inhibition of p65 activity by DMF is mediated by a protein target downstream of IKK/IκBα. The hypothesis that p65 may be targeted directly by DMF is supported by the fact that p65 contains numerous reactive cysteines susceptible to covalent modification by electrophiles (32, 42–45). Therefore, we asked whether the effect of DMF on p65 activity is the result of direct inactivation as opposed to an indirect effect. To test this, we preincubated recombinant p65 protein with DMF and observed a significant attenuation of p65 DNA binding activity, indicating a direct interaction between p65 and DMF (Fig. 6A). On the basis of our data and the nature of DMF, we hypothesized that DMF inhibits the NFκB pathway via direct covalent modification of p65 cysteine residues. To test this hypothesis, we synthesized a novel chemical probe of DMF, shown in Fig. 6B, designed to replicate the biological activity of DMF. The small alkynyl modification to DMF is designed to allow visualization and immunoprecipitation (IP) of covalently modified proteins without loss or deviation of the specific bioactivity of DMF (35). We find that the alkynyl-DMF probe recapitulates the inhibitory activity of DMF on classical NFκB target genes such as TNF and CCL2 (Fig. 6C). The probe was then incubated with recombinant p65 protein, followed by click chemistry cross-linking to the azido-rhodamine reporter tag using copper-catalyzed cycloaddition (36). The probe-labeled p65 protein was then visualized by SDS-PAGE using in-gel fluorescence scanning (35), as shown in Fig. 6D, indicating a significant labeling of p65 (second lane). Preincubation of p65 with DMF before addition of the alkynyl-DMF probe reduces p65 labeling by the probe, indicating that DMF and the probe compete for and covalently modify the same site on p65. These data show that DMF covalently modifies recombinant p65. To determine whether this finding also applies to breast cancer cells, MDA-MB-231 cell lysates were incubated with the alkynyl-DMF probe and, in this experiment, cross-linked by click chemistry to an azido-biotin reporter tag, allowing IP using streptavidin beads to capture biotinylated protein. Cell lysates prior to IP showed significant levels of p65 using an antibody for p65 (Fig. 6E, first and second lanes). The eluate from streptavidin bead IP also showed significant alkynyl-DMF-modified p65 (Fig. 6E, third lane). The control experiment, in which the alkynyl-DMF probe was omitted from the experiment, showed no p65 protein from the eluate after IP (Fig. 6E, fourth lane). Together, the data demonstrate that DMF covalently modifies p65 both recombinant protein and in cell lysates.
FIGURE 6.
DMF covalently modifies p65 both in vitro and in cell lysates, and cysteine 38 is the key residue responsible for DMF activity on the NFκB pathway. A, recombinant p65 DNA binding activity was measured after incubation with DMF (50 μm) for 30 min. Data are plotted as percent vehicle (Veh) control. ***, p < 0.001. B, the chemical structure of the novel alkyne-based DMF probe. C, expression of the NFκB target genes TNF and CCL2 in MCF-7 cells was measured by quantitative RT-PCR upon treatment with varying concentrations of the DMF probe, followed by TNFα. Data are plotted as percent of TNFα alone. D, gel image for labeling of recombinant p65 with the DMF probe (50 μm) measured by in-gel fluorescence. Third lane, DMF (50 μm) was added for 30 min prior to incubation with the DMF probe. Bottom panel, Coomassie staining of the gel indicating equal protein loading. E, biotin IP was carried out in MDA-MB-231 cell lysates cross-linked in the presence or absence of the DMF probe (50 μm). Total p65 protein was then immunoblotted and compared between IPs and inputs (10% of the protein lysate load). F, NFκB-RE activity (shown in relative luciferase units, RLU) was measured in MCF-7 cells transfected with wild-type p65 or C38S-p65 or mock-transfected and then treated with or without TNFα for 4 h. G, NFκB-RE activity was measured in MCF-7 cells transfected with wild-type p65 or C38S-p65 and treated with DMF (50 μm). Data are presented as percent inhibition relative to the vehicle (Veh) control. ***, p < 0.001.
The transcription factor p65 has multiple reactive cysteines (46, 47), but, in particular, cysteine 38 has been shown to be alkylated by electrophiles similar in nature to DMF (32, 42–45). Because Cys-38 participates in DNA binding of p65 (42, 48) and because its covalent modification has been shown to inhibit p65 nuclear localization (43), we examined whether the action of DMF was dependent on Cys-38 covalent modification by expressing wild-type p65 or C38S-p65. In the mutant, the thiol functional group is replaced by an alcohol, which can no longer react with DMF. We found that the transfected cells overexpressing the wild-type or the C38S mutant exhibited elevated p65 activity, as measured by NFκB-RE luciferase, and no stimulation by TNFα was needed (Fig. 6F). DMF treatment significantly reduced wild-type p65 activity. However, C38S activity was significantly less inhibited by DMF (Fig. 6G). These data demonstrate that cysteine 38 is a key residue mediating the effect of DMF on p65 activity.
Discussion
In this study, we demonstrated that DMF can be used effectively to inhibit NFκB activity in breast cancer cells. Importantly, we showed that DMF attenuates MS formation by inhibiting their intrinsically high NFκB activity. This indicates that the anti-NFκB activity of DMF can be exploited to eradicate breast CSCs, given their reliance on the NFκB pathway for survival and propagation, making DMF a candidate for anti-CSC therapy in breast cancer. This is in agreement with our prior findings showing that fumarate-based drugs are effective anti-breast CSC agents (49).
Similarly, one can envision that DMF may have additional anti-tumor activities by inhibiting other NFκB-dependent phenotypes, such as tumor cell proliferation and survival. Indeed, we demonstrated that DMF significantly impaired the growth of MDA-MB-231 xenograft tumors. Because MDA-MB-231 cells represent the aggressive triple-negative breast cancer subtype, this is highly significant. Triple-negative breast cancers have aggressive clinical manifestations and lack targeted therapy, and, as a result, patient outcome remains poor. Furthermore, the triple-negative subtype is enriched with CSC markers more than other subtypes (50) and displays higher NFκB activity (5), suggesting that application of DMF therapy may be highly beneficial.
Although DMF is an approved immune-modulatory drug that has been shown to inhibit NFκB signaling in a variety of cell lines and tissues (23–26, 52), the activity in breast cancer cells was unknown, and the specific mechanism of action with respect to the NFκB pathway was unclear. Previous reports have suggested that, upon DMF treatment, phosphorylation of NFκB transcription factors and subsequent nuclear translocation are attenuated (26, 52). This occurs in an IKK/IκBα-independent manner and via other kinases, such as MSK-1, which phosphorylates p65 at Ser-468 (24, 26, 52). In agreement with these reports, we found that, in breast cancer cells, DMF reduces the nuclear content of phosphorylated and total p65 in an IKK/IκBα-independent manner. However, when examining the cellular content of phosphorylated p65, including the Ser-468 site, no change is observed, suggesting that a kinase-mediated effect on p65 is unlikely in breast cancer cells, at least at the concentration used in our study (50 μm DMF versus 100 μm in the study by Peng et al. (26)). Instead, our data demonstrate that DMF inhibits NFκB activity in breast cancer cells by covalently modifying the main transcription factor of the NFκB family, p65, on cysteine 38, which, in turn, blocks p65 nuclear translocation and DNA binding activity.
Reactive protein cysteine residues are expected to be modified by DMF because they are the most intrinsically nucleophilic amino acids in proteins. The p65 transcription factor has multiple such cysteines. Nine cysteine residues are clustered in the Rel domain, and six of them are highly conserved among all other known Rel-related proteins (46, 47). In particular, cysteines 38 and 120 have been shown to be alkylated by electrophiles similar in nature to DMF (32, 42–45). Our results show more than one modification site, as indicated in Fig. 6E, consistent with multiple reactive cysteine residues in p65. Intriguingly, Cys-38 participates in DNA binding of p65 by forming a hydrogen bond with the sugar/phosphate DNA backbone (42, 48). Moreover, covalent modification of Cys-38 has been shown to inhibit the nuclear localization of p65 (43). Therefore, p65 Cys-38 alkylation may contribute to attenuate NFκB activity in breast cancer cells, suggesting that this may be the main mode of action for DMF. In this work, we use the point mutant to prove that Cys-38 of p65 is a key amino acid required for DMF-mediated NFκB inhibition in breast cancer cells.
Drugs that are covalent inhibitors, like DMF, exhibit multiple advantages over conventional non-covalent drugs, such as improved biochemical efficiency because competition with endogenous substrates is reduced; lower, less frequent dosing, resulting in a lower overall patient burden; and potential prevention of drug resistance because of continuous target suppression (51). Determining the mechanism of action of DMF is important because it may enable one to dose better through monitoring of the effects of the drug on the target pathway in patients, to predict potential side effects, and to stratify clinical trials to focus on patients most likely to respond. Together, we conclude that DMF represents an effective way to inhibit NFκB in breast cancer cells. Furthermore, we demonstrate that DMF has anti-tumor activity in a breast cancer xenograft model of the triple-negative subtype. Our findings have a tremendous clinical impact by establishing DMF as a viable NFκB inhibitor and both an anti-CSC and anti-tumor agent. Understanding the mechanism of action of DMF sets the stage for advancing DMF into clinical testing to treat aggressive breast cancers.
Author Contributions
I. K. conceived and coordinated the study, performed and analyzed most of the experiments, and wrote the paper. M. A. S. synthesized the alkynyl-DMF probe shown in Fig. 6B. E. L. C. G. performed daily gavage and assisted with the animal study data collection. L. E. S. performed the IHC staining shown in Fig. 4E. G. G. performed the Dual-Luciferase assay shown in Fig. 6, F and G, and assisted with the crystal violet assay shown in Fig. 3B. E. N. T. optimized the click chemistry procedure shown in Fig. 6D. G. R. J. T. and J. F. contributed to the preparation of the figures and drafting of the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Thomas Gilmore for the wild-type and mutant p65 plasmids and Shuangping Zhao, Bryant Marure, Jeanne Danes, Ye Li, and Daniel D. Lantvit for technical assistance.
This work was supported by National Institutes of Health Grants R01 CA130932 and R01 CA200669 (to J. F.) and R01 CA121107 (to G. R. J. T.) and by Postdoctoral Fellowship Grant PDF12229484 from Susan G. Komen for the Cure (to I. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NFκB
- nuclear factor κB
- CSC
- cancer stem cell
- IKK
- IκB kinase
- DMF
- dimethyl fumarate
- MS
- mammosphere(s)
- DMS
- dimethyl succinate
- NAC
- N-acetyl l-cysteine
- ER
- estrogen receptor
- CA
- constitutionally active
- Dox
- doxycycline
- QPCR
- quantitative PCR
- IHC
- immunohistochemistry
- IP
- immunoprecipitation
- 2D
- monolayer two-dimensional.
References
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Baumgarten S. C., and Frasor J. (2012) Inflammation: an instigator of more aggressive estrogen receptor (ER) positive breast cancers. Mol. Endocrinol. 26, 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karin M., Cao Y., Greten F. R., and Li Z. W. (2002) NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2, 301–310 [DOI] [PubMed] [Google Scholar]
- 4. Kim H. J., Hawke N., and Baldwin A. S. (2006) NF-κB and IKK as therapeutic targets in cancer. Cell Death Differ. 13, 738–747 [DOI] [PubMed] [Google Scholar]
- 5. Nakshatri H., Bhat-Nakshatri P., Martin D. A., Goulet R. J. Jr., and Sledge G. W. Jr. (1997) Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell. Biol. 17, 3629–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sovak M. A., Bellas R. E., Kim D. W., Zanieski G. J., Rogers A. E., Traish A. M., and Sonenshein G. E. (1997) Aberrant nuclear factor-κB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 100, 2952–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakshatri H., and Goulet R. J. Jr. (2002) NF-κB and breast cancer. Curr. Probl. Cancer 26, 282–309 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Y., Eppenberger-Castori S., Marx C., Yau C., Scott G. K., Eppenberger U., and Benz C. C. (2005) Activation of nuclear factor-κB (NFκB) identifies a high-risk subset of hormone-dependent breast cancers. Int. J. Biochem. Cell Biol. 37, 1130–1144 [DOI] [PubMed] [Google Scholar]
- 9. Jones R. L., Rojo F., A'Hern R., Villena N., Salter J., Corominas J. M., Servitja S., Smith I. E., Rovira A., Reis-Filho J. S., Dowsett M., and Albanell J. (2011) Nuclear NF-κB/p65 expression and response to neoadjuvant chemotherapy in breast cancer. J. Clin. Pathol. 64, 130–135 [DOI] [PubMed] [Google Scholar]
- 10. Cao Y., Luo J. L., and Karin M. (2007) IκB kinase α kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15852–15857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iliopoulos D., Hirsch H. A., and Struhl K. (2009) An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinohara K., Kobayashi S., Kanauchi H., Shimizu S., Nishioka K., Tsuji E., Tada K., Umezawa K., Mori M., Ogawa T., Inoue J., Tojo A., and Gotoh N. (2012) ErbB receptor tyrosine kinase/NF-κB signaling controls mammosphere formation in human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 6584–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., and Clarke M. F. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X., Lewis M. T., Huang J., Gutierrez C., Osborne C. K., Wu M. F., Hilsenbeck S. G., Pavlick A., Zhang X., Chamness G. C., Wong H., Rosen J., and Chang J. C. (2008) Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 100, 672–679 [DOI] [PubMed] [Google Scholar]
- 15. Croker A. K., and Allan A. L. (2008) Cancer stem cells: implications for the progression and treatment of metastatic disease. J. Cell. Mol. Med. 12, 374–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hollier B. G., Evans K., and Mani S. A. (2009) The epithelial-to-mesenchymal transition and cancer stem cells: a coalition against cancer therapies. J. Mammary Gland Biol. Neoplasia 14, 29–43 [DOI] [PubMed] [Google Scholar]
- 17. Diehn M., Cho R. W., Lobo N. A., Kalisky T., Dorie M. J., Kulp A. N., Qian D., Lam J. S., Ailles L. E., Wong M., Joshua B., Kaplan M. J., Wapnir I., Dirbas F. M., Somlo G., Garberoglio C., Paz B., Shen J., Lau S. K., Quake S. R., Brown J. M., Weissman I. L., and Clarke M. F. (2009) Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Velasco-Velázquez M. A., Popov V. M., Lisanti M. P., and Pestell R. G. (2011) The role of breast cancer stem cells in metastasis and therapeutic implications. Am. J. Pathol. 179, 2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilmore T. D., and Herscovitch M. (2006) Inhibitors of NF-κB signaling: 785 and counting. Oncogene 25, 6887–6899 [DOI] [PubMed] [Google Scholar]
- 20. Hayden M. S., and Ghosh S. (2008) Shared principles in NF-κB signaling. Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 21. Garber K. (2006) The second wave in kinase cancer drugs. Nat. Biotechnol. 24, 127–130 [DOI] [PubMed] [Google Scholar]
- 22. Greten F. R., Arkan M. C., Bollrath J., Hsu L. C., Goode J., Miething C., Göktuna S. I., Neuenhahn M., Fierer J., Paxian S., Van Rooijen N., Xu Y., O'Cain T., Jaffee B. B., Busch D. H., Duyster J., Schmid R. M., Eckmann L., and Karin M. (2007) NF-κB is a negative regulator of IL-1β secretion as revealed by genetic and pharmacological inhibition of IKKβ. Cell 130, 918–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vandermeeren M., Janssens S., Wouters H., Borghmans I., Borgers M., Beyaert R., and Geysen J. (2001) Dimethylfumarate is an inhibitor of cytokine-induced nuclear translocation of NF-κ B1, but not RelA in normal human dermal fibroblast cells. J. Invest. Dermatol. 116, 124–130 [DOI] [PubMed] [Google Scholar]
- 24. Seidel P., Merfort I., Hughes J. M., Oliver B. G., Tamm M., and Roth M. (2009) Dimethylfumarate inhibits NF-κB function at multiple levels to limit airway smooth muscle cell cytokine secretion. Am. J. Physiol. Lung Cell Mol. Physiol. 297, L326–339 [DOI] [PubMed] [Google Scholar]
- 25. Wilms H., Sievers J., Rickert U., Rostami-Yazdi M., Mrowietz U., and Lucius R. (2010) Dimethylfumarate inhibits microglial and astrocytic inflammation by suppressing the synthesis of nitric oxide, IL-1β, TNF-α and IL-6 in an in-vitro model of brain inflammation. J. Neuroinflammation 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng H., Guerau-de-Arellano M., Mehta V. B., Yang Y., Huss D. J., Papenfuss T. L., Lovett-Racke A. E., and Racke M. K. (2012) Dimethyl fumarate inhibits dendritic cell maturation via nuclear factor κB (NF-κB) and extracellular signal-regulated kinase 1 and 2 (ERK1/2) and mitogen stress-activated kinase 1 (MSK1) signaling. J. Biol. Chem. 287, 28017–28026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoefnagel J. J., Thio H. B., Willemze R., and Bouwes Bavinck J. N. (2003) Long-term safety aspects of systemic therapy with fumaric acid esters in severe psoriasis. Br. J. Dermatol. 149, 363–369 [DOI] [PubMed] [Google Scholar]
- 28. Götz M. G., James K. E., Hansell E., Dvorák J., Seshaadri A., Sojka D., Kopácek P., McKerrow J. H., Caffrey C. R., and Powers J. C. (2008) Aza-peptidyl Michael acceptors: a new class of potent and selective inhibitors of asparaginyl endopeptidases (legumains) from evolutionarily diverse pathogens. J. Med. Chem. 51, 2816–2832 [DOI] [PubMed] [Google Scholar]
- 29. Mercurio F., Zhu H., Murray B. W., Shevchenko A., Bennett B. L., Li J., Young D. B., Barbosa M., Mann M., Manning A., and Rao A. (1997) IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science 278, 860–866 [DOI] [PubMed] [Google Scholar]
- 30. Kastrati I., Canestrari E., and Frasor J. (2015) PHLDA1 expression is controlled by an estrogen receptor-NFκB-miR-181 regulatory loop and is essential for formation of ER+ mammospheres. Oncogene 34, 2309–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pradhan M., Baumgarten S. C., Bembinster L. A., and Frasor J. (2012) CBP mediates NF-κB-dependent histone acetylation and estrogen receptor recruitment to an estrogen response element in the BIRC3 promoter. Mol. Cell. Biol. 32, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang M. C., Bardhan S., Pace E. A., Rosman D., Beutler J. A., Porco J. A. Jr., and Gilmore T. D. (2006) Inhibition of transcription factor NF-κB signaling proteins IKKβ and p65 through specific cysteine residues by epoxyquinone A monomer: correlation with its anti-cancer cell growth activity. Biochem. Pharmacol. 71, 634–645 [DOI] [PubMed] [Google Scholar]
- 33. Frasor J., Danes J. M., Komm B., Chang K. C., Lyttle C. R., and Katzenellenbogen B. S. (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144, 4562–4574 [DOI] [PubMed] [Google Scholar]
- 34. Stender J. D., Kim K., Charn T. H., Komm B., Chang K. C., Kraus W. L., Benner C., Glass C. K., and Katzenellenbogen B. S. (2010) Genome-wide analysis of estrogen receptor α DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol. Cell. Biol. 30, 3943–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Speers A. E., Adam G. C., and Cravatt B. F. (2003) Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am. Chem. Soc. 125, 4686–4687 [DOI] [PubMed] [Google Scholar]
- 36. Rostovtsev V. V., Green L. G., Fokin V. V., and Sharpless K. B. (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. 41, 2596–2599 [DOI] [PubMed] [Google Scholar]
- 37. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., and Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charafe-Jauffret E., Monville F., Ginestier C., Dontu G., Birnbaum D., and Wicha M. S. (2008) Cancer stem cells in breast: current opinion and future challenges. Pathobiology 75, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marcotte D., Zeng W., Hus J. C., McKenzie A., Hession C., Jin P., Bergeron C., Lugovskoy A., Enyedy I., Cuervo H., Wang D., Atmanene C., Roecklin D., Vecchi M., Vivat V., Kraemer J., Winkler D., Hong V., Chao J., Lukashev M., and Silvian L. (2013) Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg. Med. Chem 21, 4011–4019 [DOI] [PubMed] [Google Scholar]
- 40. Linker R. A., Lee D. H., Ryan S., van Dam A. M., Conrad R., Bista P., Zeng W., Hronowsky X., Buko A., Chollate S., Ellrichmann G., Brück W., Dawson K., Goelz S., Wiese S., Scannevin R. H., Lukashev M., and Gold R. (2011) Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain 134, 678–692 [DOI] [PubMed] [Google Scholar]
- 41. Sullivan L. B., Martinez-Garcia E., Nguyen H., Mullen A. R., Dufour E., Sudarshan S., Licht J. D., Deberardinis R. J., and Chandel N. S. (2013) The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol. Cell 51, 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. García-Piñeres A. J., Castro V., Mora G., Schmidt T. J., Strunck E., Pahl H. L., and Merfort I. (2001) Cysteine 38 in p65/NF-κB plays a crucial role in DNA binding inhibition by sesquiterpene lactones. J. Biol. Chem. 276, 39713–39720 [DOI] [PubMed] [Google Scholar]
- 43. Tamura R., Morimoto K., Hirano S., Wang L., Zhao M., Ando M., and Kataoka T. (2012) Santonin-related compound 2 inhibits the nuclear translocation of NF-κB subunit p65 by targeting cysteine 38 in TNF-α-induced NF-κB signaling pathway. Biosci. Biotechnol. Biochem 76, 2360–2363 [DOI] [PubMed] [Google Scholar]
- 44. Yamamoto M., Horie R., Takeiri M., Kozawa I., and Umezawa K. (2008) Inactivation of NF-κB components by covalent binding of (-)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J. Med. Chem. 51, 5780–5788 [DOI] [PubMed] [Google Scholar]
- 45. Anand P., Kunnumakkara A. B., Harikumar K. B., Ahn K. S., Badmaev V., and Aggarwal B. B. (2008) Modification of cysteine residue in p65 subunit of nuclear factor-κB (NF-κB) by picroliv suppresses NF-κB-regulated gene products and potentiates apoptosis. Cancer Res. 68, 8861–8870 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Gilmore T. D. (1990) NF-κB, KBF1, dorsal, and related matters. Cell 62, 841–843 [DOI] [PubMed] [Google Scholar]
- 47. Ruben S. M., Dillon P. J., Schreck R., Henkel T., Chen C. H., Maher M., Baeuerle P. A., and Rosen C. A. (1991) Isolation of a rel-related human cDNA that potentially encodes the 65-kD subunit of NF-κ B. Science 251, 1490–1493 [DOI] [PubMed] [Google Scholar]
- 48. Chen F. E., Huang D. B., Chen Y. Q., and Ghosh G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391, 410–413 [DOI] [PubMed] [Google Scholar]
- 49. Kastrati I., Litosh V. A., Zhao S., Alvarez M., Thatcher G. R., and Frasor J. (2015) A novel aspirin prodrug inhibits NFκB activity and breast cancer stem cell properties. BMC Cancer 15, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herschkowitz J. I., Zhao W., Zhang M., Usary J., Murrow G., Edwards D., Knezevic J., Greene S. B., Darr D., Troester M. A., Hilsenbeck S. G., Medina D., Perou C. M., and Rosen J. M. (2012) Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2778–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mah R., Thomas J. R., and Shafer C. M. (2014) Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 24, 33–39 [DOI] [PubMed] [Google Scholar]
- 52. Loewe R., Holnthoner W., Gröger M., Pillinger M., Gruber F., Mechtcheriakova D., Hofer E., Wolff K., and Petzelbauer P. (2002) Dimethylfumarate inhibits TNF-induced nuclear entry of NF-κ B/p65 in human endothelial cells. J. Immunol. 168, 4781–4787 [DOI] [PubMed] [Google Scholar]