Abstract
The intracellular infections of Mycobacterium tuberculosis, which is the causative agent of tuberculosis, are regulated by many cyclic dinucleotide signaling. Rv2837c from M. tuberculosis is a soluble, stand-alone DHH-DHHA1 domain phosphodiesterase that down-regulates c-di-AMP through catalytic degradation and plays an important role in M. tuberculosis infections. Here, we report the crystal structure of Rv2837c (2.0 Å), and its complex with hydrolysis intermediate 5′-pApA (2.35 Å). Our structures indicate that both DHH and DHHA1 domains are essential for c-di-AMP degradation. Further structural analysis shows that Rv2837c does not distinguish adenine from guanine, which explains why Rv2837c hydrolyzes all linear dinucleotides with almost the same efficiency. We observed that Rv2837c degraded other c-di-NMPs at a lower rate than it did on c-di-AMP. Nevertheless, our data also showed that Rv2837c significantly decreases concentrations of both c-di-AMP and c-di-GMP in vivo. Our results suggest that beside its major role in c-di-AMP degradation Rv2837c could also regulate c-di-GMP signaling pathways in bacterial cell.
Keywords: cyclic diadenosine monophosphate (c-di-AMP), enzyme degradation, Mycobacterium tuberculosis, phosphodiesterases, structure-function
Introduction
Mycobacterium tuberculosis is an intracellular pathogen that causes tuberculosis, which remains one of the world's deadliest communicable diseases. There were an estimated 9.0 million people developed tuberculosis in 2013, and 1.5 million people died from the infection (1). The infections of M. tuberculosis are regulated by many signaling molecules, in which c-di-NMPs3 act as triggers for innate immune during tuberculosis infection (2).
c-di-NMPs (c-di-AMP, c-di-GMP, and c-GAMP) have been known to get involved in the regulation and coordination of many cellular processes (3–5). c-di-GMP regulates processes such as biofilm formation, motility, virulence, cell cycle, and type I interferon stimulation (6–9). c-di-AMP regulates many cellular processes like sporulation, fatty acid synthesis, cell wall homeostasis, DNA damage response, potassium transport, and virulence in a wide variety of organisms (10–15). Cellular c-di-AMP is generated by diadenylate cyclases containing DisA_N domain (Pfam accession PF02457) that produce c-di-AMP from two molecules of ATP or ADP. Specific phosphodiesterases (PDE) associated with DHH-DHHA1 or HD domain hydrolyze c-di-AMP into pApA or AMP (16–18). Recently, many proteins with PDE activity have been identified. One group is GdpP-like protein family, which usually contains a tandem PAS domain, a degenerate GGDEF domain, and a catalytic DHH-DHHA1 domain (19). The other group is recently discovered PgpH protein family, which contains an extracellular 7TM receptor-like domain and a cytoplasmic HD domain (18). They all hydrolyze c-di-AMP into linear dinucleotide 5′-pApA. There is also a PDE family that only contains a stand-alone DHH-DHHA1 domain, they hydrolyze c-di-AMP in a two-step process: first to linear 5′-pApA, which is subsequently hydrolyzed into two 5′-AMP molecules (16, 17, 20). All types of PDE enzymes require ions for catalytic activity. The degradation mechanism of HD domain has been well studied, but how DHH-DHHA1 domain degrades c-di-AMP is not clarified.
M. tuberculosis only harbors a single cytosolic c-di-AMP phosphodiesterase Rv2837c, which contains a stand-alone DHH-DHHA1 domain. It does not encode GdpP homologous proteins or HD domain proteins. Rv2837c exhibits bifunctional 3′-5′ exonuclease, as well as CysQ-like phosphatase (pAp) activity (21). It hydrolyzes different RNA substrates or single-stranded DNA in the 3′-5′ direction. The 2-mer oligonucleotides are preferred substrates of Rv2837c (21, 22). It has been reported that deletion of Rv2837c led to reduced virulence in M. tuberculosis infections (23). Rv2837c hydrolyzes c-di-AMP efficiently in vitro, but it has low activity on c-di-GMP (15). It is clear that Rv2837c plays an important role in maintaining c-di-AMP homeostasis. Because M. tuberculosis has very few enzymes for c-di-GMP metabolism (24), there is the possibility that Rv2837c also plays a role in c-di-GMP homeostasis.
In this study, we first present the structure of Rv2837c at 2.0 Å resolution. We then obtained the structure of Rv2837c in complex with the c-di-AMP hydrolysis intermediates 5′-pApA by soaking Rv2837c crystal with c-di-AMP. The complex structure presents a precise enzymatic model that the DHH domain has the PDE activity catalytic core and the DHHA1 domain contributes to substrate recognition and substrate stabilization. Structure analyses combined with biochemistry studies allow us to propose a two-metal ion catalytic mechanism for c-di-NMP hydrolysis. Moreover, with the support of in vivo data, we speculate that Rv2837c may also regulate c-di-GMP signaling pathways beside its major in down-regulating intracellular c-di-AMP concentration.
Experimental Procedures
Cloning and Site-directed Mutagenesis
The genes encoding full-length Rv2837c and the Rv2837c DHH domain which contains amino acids 1–206 were amplified by PCR from the M. tuberculosis H37Rv genomic DNA. To get the crystal, the Rv2837c fragment which contains amino acids 10–336 with an additional amino acids sequence (LTRAPPPPPLRSGC) in the C-terminal (designated “Rv2837c-C”) was also PCR-amplified. These PCR products were cloned into the BamHI and XhoI sites of pET-28a-SUMO vector. With Rv2837c-pET-28a-SUMO as the template, all mutants were produced using the QuikChange site-directed mutagenesis and were confirmed by DNA sequencing. All constructed vectors were overexpressed in Escherichia coli strain BL21 (DE3).
Protein Expression and Purification
For protein purification, the transformed cells were incubated in LB medium at 37 °C to an A600 of 0.8. Following overnight induction with 0.12 mm isopropyl-d-1-thiogalactopyranoside at 16 °C, the cells were harvested at 6000 × g for 15 min at 4 °C. The pellet was resuspended in 25 mm Tris-HCl buffer at pH 8.0 containing 200 mm NaCl and was lysed by sonication. After ultracentrifugation at 28,370 × g for 45 min at 4 °C, the supernatant was loaded onto nickel-nitrilotriacetic acid affinity resin (Qiagen) for affinity chromatography of the N-terminal His-SUMO-tagged protein and was eluted with 25 mm Tris-HCl buffer at pH 8.0 containing 100 mm NaCl and 250 mm imidazole. The eluted protein was incubated with the protease UlpI at a 100:1 ratio (w/w) and dialyzed against the reaction buffer (10 mm Tris-HCl, pH 8.0, 100 mm NaCl) at 4 °C overnight. The cleavage mixture was loaded onto nickel-nitrilotriacetic acid affinity resin again to remove the cleaved His6-SUMO tag and the His6-UlpI tag. Subsequently, untagged protein was purified by size exclusion chromatography (Superdex 200; GE Healthcare) with 10 mm Tris-HCl buffer at pH 8.0 containing 100 mm NaCl. Finally, 2 mm MnCl2 was added to the protein for crystal screen. Se-Met-Rv2837c-C was overexpressed as was the native Rv2837c-C but using M9 medium based on a methionine-biosynthesis inhibition method. Se-Met-Rv2837c-C protein was then purified by following the same protocol as used for native protein.
Crystallization, Data Collection, and Structure Determination
The recombinant proteins of Rv2837c-C and Se-Met-labeled Rv2837c-C were concentrated to 10 mg ml−1. All crystals were obtained by using the hanging drop vapor diffusion method at 20 °C. The native crystals were grown in solution containing equal volume of protein solution and 10% (w/v) PEG 10,000, 5% MPD, 0.1 m HEPES, pH 7.5. The Se-Met-labeled crystals were grown in solution containing equal volumes of protein solution and 2.0 m sodium formate, 0.1 m Bis-tris propane, pH 7.0. To prevent radiation damage, all crystals were cryoprotected with reservoir buffer containing 15% (v/v) glycerol by flash-cooling in liquid nitrogen. Nucleotide-Rv2837c-C complexes were obtained by soaking Rv2837c-Ccrystals in cryoprotected buffer with 5 mm c-di-AMP for 30 s.
X-ray diffraction data were collected on Beamline BL17U1 at the Shanghai Synchrotron Radiation Facility. All data were processed using HKL-2000 (25). The phase problem for the Se-Met-labeled crystal was solved by the single-wavelength anomalous dispersion phasing. Phases were calculated using AutoSol implemented in PHENIX (26). The initial model was built automatically using AutoBuild in PHENIX. The initial model was then used as search model for molecular replacement against the native data set using MOLREP in CCP4 (27). Structural refinement and manual model rebuilding were carried out alternately by using REFMAC (28), Phenix.Refine, and Coot (29). All molecular structures in the figures were rendered by PyMOL.
Phosphodiesterase Activity Assay
The reaction buffer contains 50 mm Tris-HCl at pH 7.5, 1 mm MnCl2, and 0.2 mm indicated nucleotide substrate. The reaction was initiated by adding 1 μm enzyme (wild type Rv2837c or other mutants) to 10 μl of reaction buffer and incubating for 1 h at 37 °C. Subsequently, reactions were terminated by adding 1 μl of 0.5 m EDTA and diluted 5-fold with water. Finally, 20 μl of sample was injected and separated by reverse phase HPLC with a C-18 column (4.6 × 250 mm Sapphire) equilibrated with 20 mm potassium phosphate, pH 6.0, and 6% ethanol. Same buffer was used in the eluting program at a flow rate of 0.7 ml min−1. Nucleotides were monitored at 254 nm. The assay buffer used for cation screening contained 1 mm CaCl2, CoCl2, FeCl2, FeCl3, MgCl2, MnCl2, NiCl2, or ZnCl2, respectively. For pH dependence determination, reaction buffer containing 1 mm MnCl2 at different pH conditions of 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5 were used. c-di-NMPs and pApA and pGpG standards were purchased from BioLog. ATP, AMP, and GMP were purchased from Sigma.
Determination of Intracellular c-di-AMP and c-di-GMP Concentrations of Mycobacterium smegmatis and E. coli MG1655 by LC-MS/MS
M. smegmatis strains were grown to logarithmic phase (20 h) at 42 °C in 7H9 broth. E. coli MG1655 strains were grown to logarithmic phase (6 h) at 42 °C in LB medium. The cells were immediately harvested at 4 °C and washed twice by distilled water, respectively. Nucleotide was extracted from the cell pellets using the method previously reported (30). Detection of c-di-AMP and c-di-GMP were performed on a Thermo UltiMate3000 liquid chromatography system followed by a Bruker HD Impact tandem mass spectrum system. Purified c-di-AMP and c-di-GMP (BIOLOG, Bremen, Germany) were used to generate the standard curve. Intracellular c-di-AMP and c-di-GMP levels were normalized by the corresponding wet cell weight.
Circular Dichroism
CD spectra were acquired using a Chirascan Plus spectrometer (Applied Photophysics, Leatherhead, UK). The instrument was flushed with pure nitrogen gas during the measurements (all at 25 °C). Far UV spectra were recorded from 250 to 190 nm (1-nm bandwidth, 1-nm step, and 0.5-s interval) Rv2837c and its mutants were diluted in Superdex buffer (10 mm Tris-HCl, pH 8.0, with 100 mm NaCl) to a final concentration of ∼1.0 mg ml−1. During data processing, the spectrum of the “protein-free solution” was subtracted from other spectra as buffer background.
Synthesis of c-di-AMP
c-di-AMP was synthesized by 2 mm ATP with 10 μm of the protein DncV in reaction buffer (50 mm Tris-HCl, pH 8.0, 120 mm NaCl, and 10 mm MgCl2) at room temperature overnight (31). The reaction was incubated for 10 min at boiling water and centrifuged at 28,370 × g for 30 min to remove protein. The supernatant was separated and purified by HPLC with a C18 column (21.2 × 250 mm; Sapphire). The following buffers were used in the gradient program: buffer A (10 mm NH4Ac, pH 5.0) and buffer B (100% methyl cyanide). The following protocol was used for separation (the values are times in minutes and percentage of buffer B used): 0.0, 3%; 20.0, 30%; 25.0, 30%; 26.0, 3%; 38.0, 3% at a flow rate of 10 ml min−1; with a diode array detector at 254 nm, the samples containing c-di-AMP were gathered. The organic solvent was removed by distillation, and then pure sample powder was acquired by freeze-thaw cycles.
Crystallization of c-di-AMP
The purified c-di-AMP was diluted in 5 mm, the crystal of c-di-AMP was grown in solution containing equal volume of c-di-AMP solution and 3.5 m ammonium chloride, 0.1 m sodium acetate trihydrate, pH 4.6, using the sitting drop method. The details of data collection and structure determination are not shown.
Motility Assays
E. coli MG1655 overnight cultures grown in LB medium at 42 °C without shaking were diluted to the same concentration. 0.5 μl of each were inoculated directly into LB agar (0.5% agar) and incubated for 10h at 42 °C, and the diameter of the diffuse growth zone was measured.
Results
Rv2837c Is a c-di-AMP Phosphodiesterase
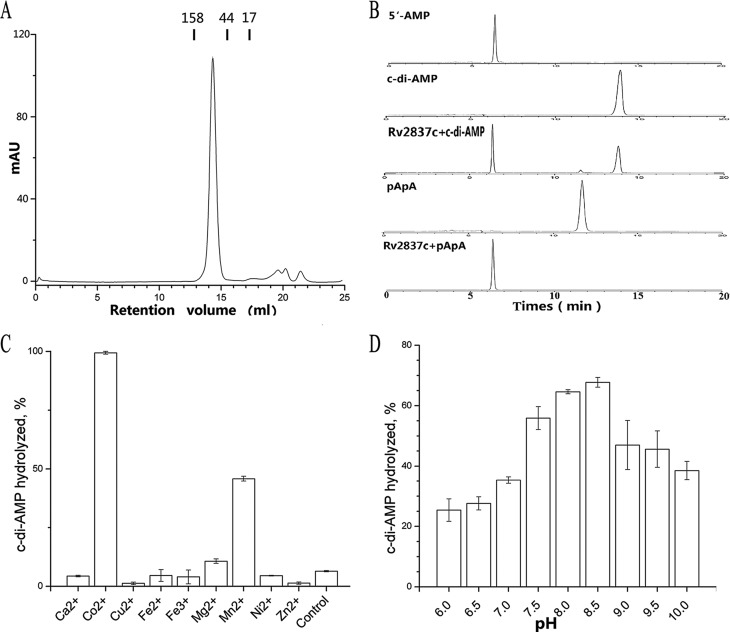
Size exclusion chromatography analysis indicated that Rv2837c (molecular mass = 34 kDa) forms a stable dimer in solution (Fig. 1A). To directly examine Rv2837c phosphodiesterase activity toward c-di-AMP, we incubated the protein with c-di-AMP under different conditions and analyzed the reactions using HPLC. Our results showed that Rv2837c hydrolyzed both c-di-AMP and 5′-pApA into 5′-AMP quickly (Fig. 1B). Co2+ or Mn2+ is necessary for Rv2837c PDE activity toward c-di-AMP (Fig. 1C), with Co2+-associated catalytic activity being as twice high as that of Mn2+. In contrast, Streptococcus pneumoniae Pde2 binding Mn2+ has the highest efficiency (16). The enzymatic activity of Rv2837c adapts to a broad pH range (Fig. 1D) and peaks at pH 8.5.
FIGURE 1.
Characterization of Rv2837c. A, size exclusion chromatograph of Rv2837c using a Superdex 200 column. The molecular mass (in kDa) and the retention volumes are indicated. B, Rv2837c was incubated with c-di-AMP or pApA, as indicated. Each sample was characterized by HPLC and monitored at 254 nm. Purified c-di-AMP, pApA, and AMP were used as standards. C and D, the c-di-AMP hydrolysis by purified Rv2837c in the presence of indicated metal cations (C) at different pH levels (D). The reaction in control of C was not supplemented with any cation. The data shown in C and D are mean values of three independent experiments. The error bars indicate the S.E.
Overall Structure of Rv2837c
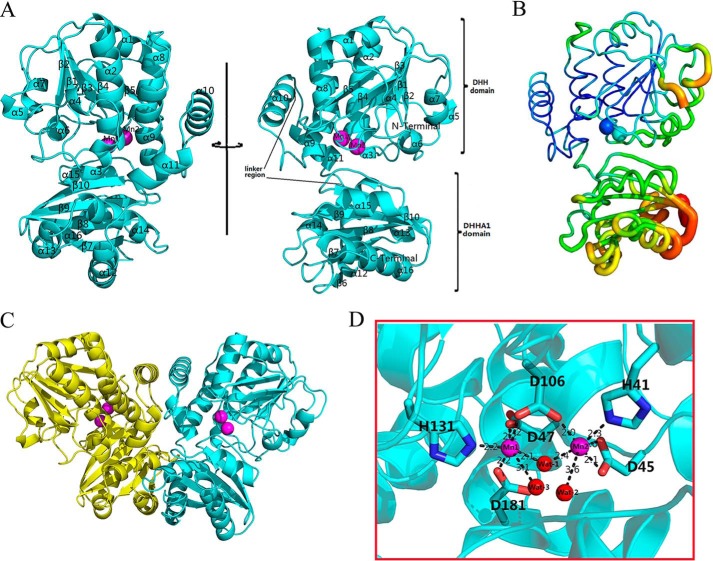
The final model of Rv2837c is refined to 2.0 Å resolution and contains one Rv2837c molecule in an asymmetric unit (Fig. 2A). Each monomer misses 15 residues at the N terminus and had 6 extra residues of the 14-amino acids peptide at the C terminus. Two crystallographic symmetry related monomers make up the biologically active dimer (Fig. 2C). The details of the data collection and refinement statistics are given in Table 1. The dimeric assembly of Rv2837c resembles its homologue, MSMEG-2630, which has the unique subunit packing and large domain interface (22).
FIGURE 2.
Overall structure of Rv2837c. A, cartoon diagram depicting the binary Rv2837c, in which Rv2837c is shown in cyan. Secondary structure elements of Rv2837c mentioned in the text are labeled. Rv2837c consists of the N-terminal DHH domain and the C-terminal DHHA1 domain. A linker region connects two distinct domains. B, distribution of the B-factor of the Rv2837c-C structure. The B-factor is colored from high (red) to low (blue). The DHH and DHHA1 domains are indicated. C, the predicated structure of the Rv2837c homodimer by PISA. Each subunit is shown as a cartoon and in different colors. The metal ions are shown as magenta spheres. D, stereo view of Mn2+ cations at the active site of Rv2837c. The residues in contact with the metal ions are shown as sticks along the Rv2837c ribbon (cyan). In addition, Wat1 is a bridging ligand to both Mn2+ ions. Wat2 and Wat3 are present to complete the coordination state of two Mn2+ ions. Mn12+ and Mn22+ are denoted by magenta spheres, whereas water molecules are shown as red spheres.
TABLE 1.
X-ray diffraction data collection and refinement statistics
| Rv2837c-C native | Rv2837c-C/5′-pApA | Rv2837c-C SAD | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9791 | 0.9791 | 0.9791 |
| Space group | P21212 | P212121 | P21212 |
| Unit cell parameters | |||
| a, b, c (Å) | 58.9, 98.6, 55.1 | 58.4, 99.3, 108.7 | 58.6, 98.2, 54.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å)a | 50–2.0 (2.07–2.00) | 50–2.35 (2.43–2.35) | 50–2.10 (2.18–2.10) |
| Total no. of reflections | 160,863 | 148,741 | 213,723 |
| No. of unique reflections | 22,675 | 25,317 | 15,582 |
| Completeness (%)a | 99.9 (100.0) | 96.1 (100.0) | 99.5 (100.0) |
| Redundancya | 7.1 (7.2) | 5.9 (6.8) | 7.4 (7.5) |
| I/σ(I)a | 23.94 (5.19) | 21.03 (16.31) | 27.76 (18.20) |
| Rmerge (%)b | 8.4 (45.3) | 8.3 (16.9) | 12.5 (37.6) |
| Refinement | |||
| Rwork/Rfree (%)c | 19.60/22.90 | 23.00/28.50 | |
| No. atoms | |||
| Protein | 2,425 | 4,806 | |
| 5′-pApA | 0 | 90 | |
| Mn2+ | 2 | 4 | |
| Water | 175 | 114 | |
| Average B-factors (Å2) | |||
| Protein | 34.78 | 32.84 | |
| 5′-pApA | 44.40 | ||
| Mn2+ | 28.01 | 24.39 | |
| Water | 38.00 | 34.25 | |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.007 | 0.008 | |
| Bond angles (°) | 1.070 | 1.153 | |
| Ramachandran plot (%) | |||
| Favored | 96.32 | 94.75 | |
| Allowed | 3.68 | 4.78 | |
| Generally allowed | 0 | 0.46 | |
a The values in parentheses refer to the high resolution shell.
b Rmerge = Σ Σ|I(k) − <I>|/Σ I(k), where I(k) and <I> represent the diffraction intensity values of the individual measurements and the corresponding mean values, respectively. The summation is over all unique measurements.
c Rwork = Σ||Fobs| − k|Fcalc||/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
A monomer of Rv2837c consists of two distinct domains (Fig. 2A). Its N-terminal DHH and C-terminal DHHA1 domains consist of residues 15–202 and 220–336, respectively. Two domains are linked by a longer flexible loop. The DHH domain has a five-parallel strand β-sheet (β1–β5), which is sandwiched by 10 α-helices (with α1, α2, α3, α4, α8, α9, and α10 on one side and α5, α6, and α7 on the other side). Structurally similar to DHH, the DHHA1 domain has an anti-parallel β-sheet (β6-β10) that is sandwiched by five α-helices (with α12–α14 and α15–α16 on both sides, respectively). The high average B-factor of the DHHA1 domain suggests that it is more flexible than DHH domain (Fig. 2B). The two active site-binding Mn2+ ions of the enzyme were observed in DHH domain.
A DALI search (32) for the homologues of Rv2837c was performed against the Protein Data Bank (PDB). The output includes a number of DHH superfamily proteins, with the closest homologue being MSMEG-2630 (Z score = 39.6, root mean square deviation = 4.9 Å for 319 Cα atoms, PDB entry 4LS9). Structural comparison of Rv2837c with MSMEG2630 indicates that the high root mean square deviation comes from domain shift (Fig. 3A). Structure analysis shows that the space between the DHH and DHHA1 domain in MSMEG-2630 is much larger than that in Rv2837c. Previous studies have shown that MSMEG-2630 has unique subunit packing that leads to the ability to bind many different substrates (22). Rv2837c is also a versatile enzyme that can hydrolyze various substrates, such as nanoRNA, single-stranded DNA, pAp, and c-di-AMP (15, 21, 22, 33). The long linker region between DHH and DHHA1 domains may give Rv2837 flexibility to bind various substrates.
FIGURE 3.
Structural comparison of Rv2837c. A, structural comparison of Rv2837c and M. smegmatis MSMEG_2630. The cartoon views of Rv2837c and MSMEG_2630 are colored cyan and wheat. B, effects of mutants in metal ligands and the DHH motif on c-di-AMP hydrolysis. C, CD spectra of Rv2837c and its mutants. In general, alanine replacement mutagenesis was constructed. The glycine was replaced by arginine. D, structural comparison of Rv2837c and Rv2837c/pApA complex. The cartoon views of Rv2837c and Rv2837c/pApA complex are colored cyan and orange, respectively. The residues in contact with the metal ions and pApA are shown as sticks. All metal ions are shown as spheres and colored by the structure.
Two-metal Ion Active Site of Rv2837c
The active site of Rv2837c is characterized by two Mn2+ ions (Mn1 and Mn2). A water molecule (Wat1) serves as a bridge to connect these two Mn2+ ions. Both Mn1 and Mn2 are present in an octahedral geometry with six coordination states. Mn1 are coordinated by Asp47, Asp106, His131, Asp181, Wat1, and Wat3. Mn2 are coordinated by His41, Asp45, Asp106, Wat1, and Wat2 (Fig. 2D). The six chelating residues of Rv2837c are all conserved in the DHH-DHHA1 domain proteins (Fig. 4). We have mutated these residues into alanine and found the mutations had greatly reduced the enzymatic activity of Rv2837. Mutations on Mn12+ ligands, D47A, D106A, H131A, and D181A completely abolished the catalytic activity, indicating their importance in maintaining the enzymatic activity. In contrast, mutations on Mn22+ ligands, such as H41A and D45A, do not eliminate Rv2837c activity completely. The above-mentioned results indicate that Mn1 could be more important than Mn2 in performing Rv2837 enzymatic activity (Fig. 3B).
FIGURE 4.
Multiple sequences alignment of Rv2837c. The alignment of M. tuberculosis Rv2837c, M. smegmatis MSMEG_2630 (PDB code 4LS9), Bacteroides fragilis WP_005791024.1 (PDB code 3W5W), Mycoplasma pneumoniae Mpn140, S. hemeolyticus WP_011275520.1 (PDB code 3DEV), and S. pneumoniae SPD_1153 is performed using ESPript 3.0. The secondary structure for Rv2837c is shown above the alignment. Also, the highly conserved residues are indicated with the black triangles. The residues involved in 5′-pApA binding are shown as aqua circles.
The 130DHH132 motif of Rv2837c has been found highly conserved in all DHH superfamily members, the right conformation of the 130DHH132 loop, which enables His131 to coordinate Mn12+. Accordingly, both D130A and H132A completely lose their catalytic activity (Fig. 3B). CD spectra of these mutants indicate that the losses of catalytic activity are not due to misfolding of Rv2837c (Fig. 3C). The structure and our biochemical data also suggest that Rv2837c follows a two-metal ion catalytic mechanism.
Structure of Rv2837c in Complex with 5′-pApA
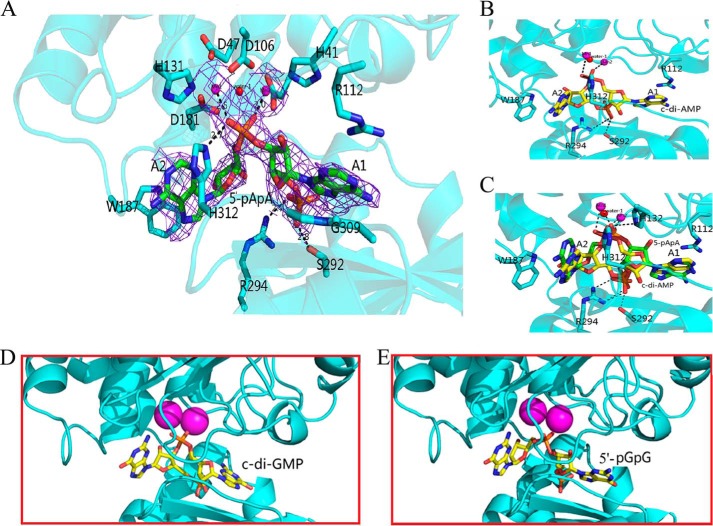
The structure of Rv2837 in complex with 5′-pApA (Table 1) reveals a dimer similar to the previous structure (Fig. 2C), with an exception that the side chain of Arg-112 shifts dramatically to stabilize 5′-pApA. In addition, two water molecules, Wat2 and Wat3 in coordination with the two Mn2+ ions in the substrate-free structure are replaced by two oxygens of phosphate-2 from a 5′-pApA molecule (Fig. 3D). The 5′-pApA molecule, an intermediate of the hydrolysis of c-di-AMP to AMP, was unambiguously identified by the difference electron density. It sits in a large tunnel between the DHH and DHHA1 domains (Fig. 5A) and is stabilized by several kinds of interactions. The nonbridging oxygen of the esterase phosphate group of 5′-pApA directly chelates two Mn2+ ions. The two adenine bases of 5′-pApA molecule are perpendicular to each other, with nucleotide A1 being sandwiched by the 309GGGH312 motif and Arg112. The adenine base of A1 is readily oriented by the cation–π interaction with Arg112. The phosphate group of A1 forms H-bonds with Ser292 and the conserved Arg294. The adenine base of nucleotide A2 is positioned at one end of substrate binding tunnel and forms a π-π interaction with the benzene group of His312, the signature residue of the conserved 309GGGH312 motif (Fig. 5A). Notably, His312 forms an H-bond with the phosphate group of the phosphodiester bond of 5′-pApA. The two ribose groups of 5′-pApA have no interaction with protein.
FIGURE 5.
The pApA binding structure of Rv2837c. A, stereo view of 5′-pApA bound to the active site of Rv2837c. Bonding interactions are represented by dashed lines with distances in Å. Electron density for 5′-pApA and two Mn2+ ions from the 2Fo − Fc map is contoured at 1 σ. B, the docking result of c-di-AMP in the active site of Rv2837c. C, superposition of the structures of c-di-AMP and 5′-pApA in the active site. Two Mn2+ ions are denoted by magenta spheres, the catalytic water is denoted by red spheres, and substrates are shown as sticks along with the protein ribbon (cyan). D and E, the docking results for c-di-GMP (D) and 5′-pGpG (E). In the cartoon representation of Rv2837c monomer, the ligands are represented in stick mode and colored by atom type.
The Binding of c-di-AMP to Rv2837c Based on Molecular Docking
The structure of 5′-pApA binding Rv2837c provides very important information for us to understand the second step of the catalytic reaction. However, to clarify the whole process of the reaction, we need to know how c-di-AMP reacts with the enzyme. Because it is difficult to get the structure of Rv2837 in complex with c-di-AMP because of its fast transition into 5′-pApA, we modeled the interaction of c-di-AMP with Rv2837 by the AUTODOCK program (version 4.2) (34), based on the structure of Rv2837c in complex with 5′-pApA (Fig. 5B).
In the calculated model of c-di-AMP-Rv2837 complex, the central ring of c-di-AMP compared with that of 5′-pApA, turns 180° horizontally around the center of the molecule. At this moment, the 5′-phosphate group of nucleotide A1 in the c-di-AMP has not been cut off from the nucleotide A2 and is bound to two Mn2+ ions. Similar to the 5′-phosphate group in the 5′-pApA, the other phosphate group is stabilized by Ser292 and Arg294. Two adenine groups of c-di-AMP are sandwiched by the same residues as those in the Rv2837c/5′-pApA complex and the two ribose groups of c-di-AMP do not show any major interaction with their surrounding residues.
Because c-di-AMP has two symmetrical phosphodiester bonds, it has to take two steps to hydrolyze both phosphodiester bonds to produce AMP finally. Our molecular docking result, in combination with the structure of Rv2837c/5′-pApA, suggests that a flip should happen to the substrate between the two reaction steps: a c-di-AMP first binds to the large tunnel of Rv2837c. One phosphodiester bond in contact with two Mn2+ ions is then hydrolyzed and results in the linear 5′-pA1pA2. This intermediate product will flip its ribose and phosphate groups to make the second phosphodiester bond contact the catalytic center of Rv2837c, which was captured in the Rv2837-pApA complex structure. Finally, the second phosphodiester bond breaks and releases two AMP molecules (Fig. 5C). At this step, the freedom of ribose groups may facilitate an easy turnover of the molecules.
Molecular Mechanism of PDE Activity of Rv2837c
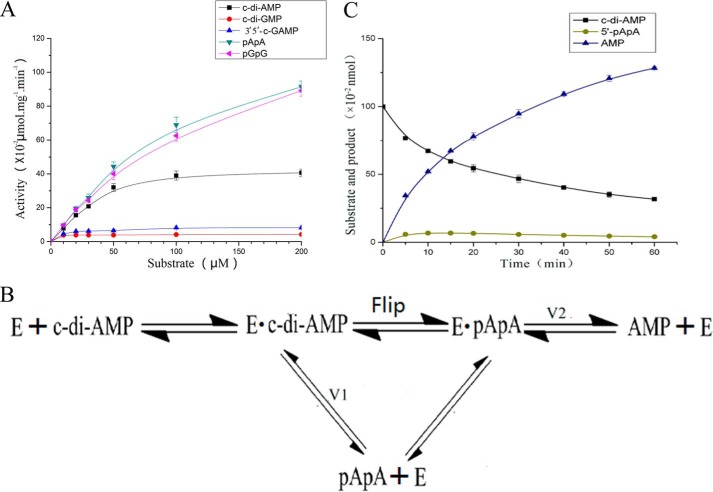
To evaluate the proposed reaction catalyzed by Rv2837c from c-di-AMP into AMP, we have conducted a series of mutagenesis study on residues involved in the catalytic process. Our data showed the mutants that affect substrates/intermediates binding usually impaired enzymatic activity. Mutations of H292A, R294A, G309R, and H312A decrease catalytic capability of Rv2837c dramatically. Mutations of R112A, W187A/H312A, G309R/G310R/G311R, and deletion of the DHHA1 domain revoked PDE activity (Fig. 6A). Intriguingly, mutation of W187A resulted in a doubled catalytic activity compared with WT protein. Because Trp187 interacts with the A2 adenine group, W187A may produce a spacious room to expedite the turnover of 5′-pApA.
FIGURE 6.
Hydrolysis activities of Rv2837c and mutants for different substrates analysis. A, effects of mutants in the DHH and GGGH motif and other residues involved in 5′-pApA binding on c-di-AMP hydrolysis. B, effects of Rv2837c mutants involved in 5′-pApA binding on 5′-pGpG hydrolysis. Different mutagenesis containing the amino acid substitutions as indicated above each lane. C–E, the same amount of Rv2837c was incubated with c-di-GMP (C), 3′3′-cGAMP (D), and 2′3′-cGAMP (E) 1, 1, and 2 h, respectively. The bottom panels in C–E are the diagrams showing the cleavage bonds of each c-di-NMP by Rv2837c.
Because 5′-pApA is a perfect nanoRNA, the complex structure of Rv2837c/5′-pApA also provides an excellent model of Rv2837c hydrolyzing nanoRNA. To test whether this model also applies to other nonoRNA, we use 5′-pGpG as a nanoRNA substrate to test the catalytic activity of Rv2837c (Fig. 6B). It is indicated that the hydrolysis of 5′-pGpG by Rv2837c is quite similar to the hydrolysis of c-di-AMP by Rv2837c in vitro. This suggests that Rv2837c degrades c-di-AMP and other nanoRNA with the same catalytic mechanism. The W187A mutant did not show any difference in hydrolyzing linear 5′-pGpG compared with Rv2837c WT, because 5′-pGpG, like 5′-pApA need not flip during the hydrolysis process.
The structure of Rv2387c-5′-pApA complex has several important structural features: First, Arg112 and the 309GGGH312 motif stabilize the binding of 5′-pApA through cation-π, H-bonds and π-π interactions. Second, Mn12+ and Mn22+ are directly chelated by the side chains of residues His41, Asp45, Asp47, Asp106, Hia131, and Asp181, as well as the two oxygens of 5′-pApA phosphate-2. Third, the catalytic water molecule WAT1, bonding with both Mn2+ ions, is only 3.0 Å from the phosphate group. Finally, the carboxyl group of Asp181 interacts with both Mn12+ and WAT1. The important role of Asp181 in activating the WAT1 has been confirmed by the destructive effect of its mutation D181A on the catalytic activity of Rv2837c (Fig. 3B).
Based on the structure of Rv2837c-5′-pApA complex and biochemical data, we propose a simplified catalytic mechanism for phosphodiester bond hydrolyzation by Rv2837c: the Wat1 molecule is activated by Asp181, together with two Mn2+ ions, generating a nucleophile to attack the phosphorus atom of 5′-pA1pA2. Then it attacks the 3′-phosphate-ester bond. After the reaction, the 3′-hydroxyl of nucleotide A2 is the leaving group and is likely to be protonated by a solvent water molecule. Finally, two AMP molecules are released (Fig. 7).
FIGURE 7.

Diagram showing the coordination of 5′-pApA and Mn2+ in the active site of Rv2837c and the proposed reaction mechanism of 5′-pApA hydrolysis. Mn1 and Mn2 are proposed to be responsible for the activation of the catalytic water molecule Wat1 that is used to produce the nucleophilic hydroxyl. The arrowheads show the directions of nucleophilic attack. A soluble water molecule Wat2 is proposed to function as a proton donor for the leaving O3′ group of A2. The numbers represent interatomic distances (in Å).
Rv2837c Specifically Targets 3′-5′ Phosphodieste Bond
Our structure shows that Rv2837c interacts with the adenine base of 5′-pApA mainly through cation-π and π-π interactions. Because guanine and adenine are structurally similar, we speculate that Rv2837c may not distinguish between 5′-pApA and 5′-pGpG or between c-di-AMP and c-di-GMP (or cGAMP) during hydrolysis. This hypothesis was first supported by molecular docking results. Both 5′-pGpG and c-di-GMP fit well into the active site Rv2837c just like 5′-pApA and c-di-AMP, respectively (Fig. 5, D and E). We then tested the PDE activities of Rv2837c toward 5′-pApA and 5′-pGpG and found that Rv2837c hydrolyzed them with all most the equivalent efficiencies (Fig. 8A).
FIGURE 8.
Two possible mechanisms of the hydrolysis by Rv2837c. A, kinetics of Rv2837c in hydrolysis of c-di-NMPs, 5′-pApA, and 5′-pGpG. All substrates at indicated concentrations were incubated with 0.1 μm Rv2837c for 30 min. The enzymatic activities were determined using HPLC. From the curves, Rv2837c yielded a kcat of 0.23 ± 0.02 s−1 and a Michaelis-Menten constant (km) of 30.89 ± 6.71 μm for c-di-AMP and kcat of 0.87 ± 0.05 s−1 and km of 129.88 ± 12.95 μm for 5′-pApA. The error bars represent the S.E. from at least three independent replicates. B, schematic diagram of two possible mechanisms for c-di-AMP hydrolysis. The top line shows that the intermediate 5′-pApA flips before the second reaction, and the bottom line shows that 5′-pApA is released first and then binds back to Rv2837c. C, 0.1 μm Rv2837c was incubated at 37 °C with 100 μm of c-di-AMP, and then the reaction products were subjected to HPLC analysis at different time points. The 5′-pApA concentration is at highest at 10 min. We get the observed values for V1 = 1.64 × 10−2 nmol·min−1 and V2 = 1.56 × 10−2 nmol·min−1 according to the slope of the curve of c-di-AMP reduction and AMP production at this moment.
Base on the activities of Rv2837c toward 5′-pApA and 5′-pGpG, we infer that Rv2837c may hydrolyze other c-di-NMPs as it does c-di-AMP. The relevant experiments show that Rv2837c breaks down c-di-AMP very fast and produces AMP as the final product. It can also degrade other c-di-NMPs but with much lower activity (Fig. 8A). Importantly, Rv2837c hydrolyzes c-di-GMP and 3′3′-cGAMP, producing single nucleotides as the final products, whereas it can only degrade 2′3′-cGAMP into pGpA (2′-5′), a linear dinucleotides (Fig. 6, C–E). Because c-di-AMP, c-di-GMP, and 3′3′-cGAMP all have two 3′-5′ phosphodiester bonds and 2′3′-cGAMP only has one, it is obvious that Rv2837c specifically targets 3′-5′ phosphodiester bond.
Rv2837c Hydrolyzes Both c-di-AMP and c-di-GMP in Vivo
Rv2837c has long been known as a c-di-AMP regulator. However, our data show that it can also degrade c-di-GMP although with a lower activity. This raises the question of whether Rv2837c also plays a role in c-di-GMP signaling in vivo. To explore the c-di-NMP phosphodiesterase activity of Rv2837c in vivo, we overexpressed Rv2837c in M. smegmatis of the MSMEG_2630 deletion mutant strain. Intracellular c-di-AMP and c-di-GMP levels of ΔMSMEG_2630 and overexpression Rv2837c (OERv2837c) strains were determined by LC-MS/MS.
According to our results, intracellular c-di-AMP concentration declined very significantly (∼10-fold) that it is hard to detect it in OERv2837c strain. The c-di-GMP concentration also declined significantly (∼4-fold) compared with ΔMSMEG_2630 strain (Fig. 9A). This indicates that Rv2837c can hydrolyze both c-di-AMP and c-di-GMP considerably in M. smegmatis. Although the decline in c-di-GMP concentration is smaller than the decline in c-di-AMP concentration, it still could be large enough to produce a physiological effect. However, the potential physiological effect of c-di-GMP cannot be easily distinguished from that of c-di-AMP.
FIGURE 9.
Rv2837c decreases both c-di-AMP and c-di-GMP levels in vivo. A, determination of intracellular c-di-AMP and c-di-GMP concentrations of ΔMSMEG_2630/pMV261 and OERv2837c strains by LC-MS/MS. 50 μg ml−1 of kanamycin was added to stabilize the pMV261 plasmids. B, determination of intracellular c-di-GMP concentrations of E. coli MG1655/pMV261 and OERv2837c strains by LC-MS/MS. C, swimming phenotypic effects of OERv2837c. Swimming motility in LB after 10 h at 42 °C was measured; the diameter of the diffuse growth zone is indicated by the histogram.
Finally we overexpressed Rv2837c in E. coli K-12 MG1655 strain to see whether Rv2837c can trigger a physiological effect through degrading c-di-GMP. The results showed that the intracellular c-di-GMP level of OERv2837c declined by 45% compared with wild type E. coli MG1655 (Fig. 9B). Because c-di-GMP is known to modulate swimming motility in various bacterial species (35), we further examined the impact of Rv2837c on this phenotype of E. coli MG1655. Indeed, overexpression of Rv2837c resulted in a significant increase in the swimming zone of E. coli MG1655 (Fig. 9C). Taken together, these observations suggest that Rv2837c may play a role in both c-di-AMP and c-di-GMP signaling in vivo.
Discussion
Rv2827 Hydrolyze Its Substrate with a Two Metal-based Mechanism
Rv2837c is a soluble, stand-alone DHH-DHHA1 phosphodiesterase from M. tuberculosis that hydrolyzes c-di-AMP to AMP efficiently (15). In this study, we have solved the crystal structure of Rv2837c and its complex with hydrolysis intermediate 5′-pApA. We have conducted structural and biochemical characterization of Rv2837c from M. tuberculosis. Our data lead to a two metal-based mechanism for the DHH-DHHA1 to hydrolyze c-di-NMP to pNpN and finally to NMP. The two-metal ion mechanism could be conserved in the DHH-DHHA1 superfamily (20). Although the crystal structure of MSMEG-2630 contained only one Mn2+ ion, the sequence alignment of Rv2837c and MSMEG-2630 shows that all residues involved in metal ions coordination and substrate binding are highly conserved (Fig. 4). In Rv2837c, the mutations on these conserved sites abolished the catalytic activity for the enzyme (Fig. 3B). Although few enzymatic analysis are available for the MSMEG-2630 mutants, the residue conservation of the active site makes us believe that MSMEG-2630 would also need two Mn2+ ions in its active site. The Mn22+ in MSMEG-2630 could be lost during the process of purification and crystallization.
Rv2837c Primarily Adopts the Flipping Mechanism
c-di-AMP has two phosphodiester bonds. According our structures, there is no way for Rv2837c to cleave the two phosphodiester bonds simultaneously. Therefore Rv2837c needs to sequentially catalyze the hydrolysis of phosphodiester bond twice before reaching the final product AMP. This raises the question of whether Rv2837c would complete the two-step reaction and produce AMP once c-di-AMP binds to its active site or release the intermediate product 5′-pApA first and finish the reaction the next time when 5′-pApA binds back to the active site (Fig. 8B). The first mechanism requires intermediate 5′-pApA to flip before it can get the remaining phosphodiester bond close to the two Mn2+, whereas the second does not. The small amount of intermediate product (5′-pApA) observed during the reaction seems favor the second mechanism (Fig. 1B). However, the low concentration of 5′-pApA cannot simply exclude the first mechanism without quantitative study.
To clarify the catalytic mechanism, we perform a kinetic study of the c-di-AMP hydrolysis by Rv2837c. The idea is that using 5′-pApA as substrate we can measure the time evolution of both 5′-pApA and AMP and calculate the km and the catalytic rate constant. Then with the km and the catalytic rate constant value for 5′-pApA and the 5′-pApA concentration at a given instant in the reaction of c-di-AMP, we can calculate the theoretical reaction rate for AMP production. Suppose the second mechanism is right and the first wrong; then the theoretical reaction rate for AMP production should be comparable to the observed value. If not, that means the first mechanism also happens. The difference between the theoretical value and the observed value reflects how much c-di-AMP takes the first mechanism or the second mechanism. The detailed kinetic analysis for two mechanisms as follows. The second mechanism shows that c-di-AMP and 5′-pApA would compete for enzyme to degrade. In the reaction system, we have
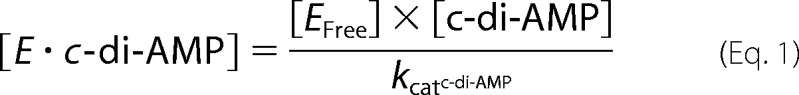 |
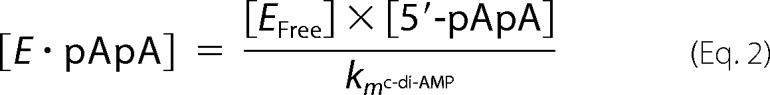 |
Therefore the ratio of enzyme binding to c-di-AMP to enzyme binding to 5′-pApA is
 |
The ratio of the calculated rate for c-di-AMP to that of 5′-pApA is
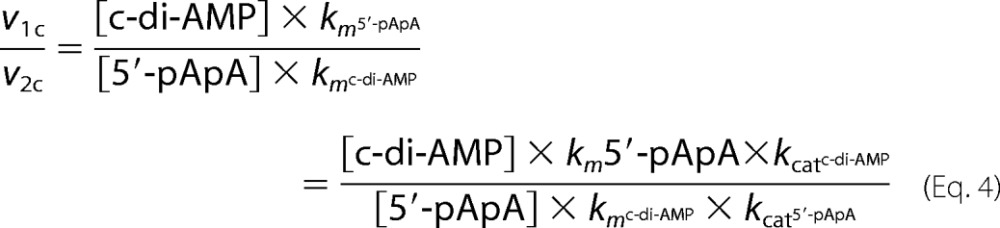 |
Because the molarity of the enzyme is much smaller than c-di-AMP and 5′-pApA at the instant we pick for calculation, the molarities for c-di-AMP and 5′-pApA can be measured directly without much error as in Fig. 8C.
Substituting the kcat and km values for c-di-AMP and 5′-pApA from Fig. 8A, we have
 |
According to c-di-AMP time evolution during the degradation (Fig. 8C), we can get the observed values for V1 and V2 according to c-di-AMP reduction and AMP production.
 |
The great difference between the observed value and the calculated value strongly suggests that Rv2837c primarily adopts the flipping mechanism.
The Oligomeric State of c-di-NMPs Affects Rv2837c Activity
Rv2837c is a 3′-5′ PDEase for many c-di-NMPs. Notably, its activity on c-di-AMP is much higher than on c-di-GMP and other c-di-NMPs, because Rv2837c hydrolyzes 5′-pApA and 5′-pGpG at almost the same rate (Fig. 8A). The difference in the activities on different substrates in vitro may come from their first step of the reactions. Our structure shows that Rv2837c is unlikely to distinguish between c-di-AMP and c-di-GMP (or cGAMP) rigorously during hydrolysis. Thus we speculate that the difference may derive from the particular properties of different substrates.
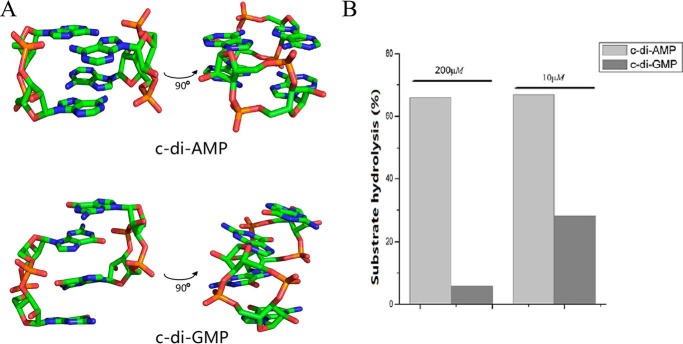
It has been reported that c-di-GMP forms heterogeneous oligomers (including monomer, dimer, tetramer, and octamer) in solution (36). Because only the monomer of c-di-NMP can enter the active site, we further tested whether the difference in reaction rate for c-di-AMP and c-di-GMP is related to the effective diffusion of their monomers into the active site of Rv2837c. We determined the crystal structure of c-di-AMP and compared it with that of c-di-GMP (37, 38). Crystal structure shows that two monomers of c-di-GMP form an interlocked dimer in crystal structure, whereas two monomers of c-di-AMP have parallel matching with each other (Fig. 10A). The structure clearly shows that c-di-GMP tends to form dimer at high concentration, whereas c-di-AMP forms monomer. These data suggest that c-di-AMP is more prone to the isolated monomers than c-di-GMP in solution. In vivo all c-di-NMPs tend to exist as monomers because of their low physiological concentrations (<10 μm). The difference between hydrolysis rates for c-di-AMP and c-di-GMP would then be smaller. Indeed, the ratio of degradation of c-di-AMP to that of c-di-GMP, catalyzed by Rv2837c, significantly dropped at their diluted concentrations (Fig. 10B). Consequently, we guess Rv2837c could also play a role in c-di-GMP signaling regulation in addition to its function in c-di-AMP signaling because intracellular c-di-NMPs generally have a low concentration.
FIGURE 10.
The oligomeric state of c-di-AMP and c-di-GMP affect Rv2837c's activity. A, overview of the crystal structures of dimeric c-di-AMP and c-di-GMP. B, the different ratios of reaction rate of c-di-AMP to that of c-di-GMP by Rv2837c in high (200 μm) and low (10 μm) concentrations.
Rv2837c May Regulate Both c-di-AMP and c-di-GMP Signaling Pathways in M. tuberculosis
Both c-di-AMP and c-di-GMP are important intracellular signaling molecules, and their homeostasis processes are regulated by many proteins in bacteria cells (3–5). c-di-GMP is down-regulated by EAL domain and HD-GYP domain, resulting in 5′-pGpG and GMP, respectively (6, 39), whereas c-di-AMP is broken down by stand-alone DHH-DHHA1 domain and membrane-bound DHH-DHHA1, as well as HD domain to AMP and 5′-pApA, respectively (16, 18, 40). M. tuberculosis does not have HD-GYP domain protein and seems lack an essential link in c-di-GMP metabolism (41). On this issue, the ability of Rv2837c to hydrolyze 5′-pGpG efficiently may fill this important link. What is more, Rv2837c can also use c-di-GMP as substrate with a lower activity, and our in vivo data have indicated that it decreases the intracellular c-di-GMP significantly enough to produce obvious physiological phenotype. Because there is no membrane-bound DHH-DHHA1 or HD domain protein for c-di-AMP regulation discovered so far in M. tuberculosis. Rv2837c, with its activity to down-regulate both c-di-AMP and c-di-GMP, is of central importance in regulating both signaling pathways in M. tuberculosis. Moreover, the ability of Rv2837c to degrade 2′3′-cGAMP may raise a possibility that Rv2837c works as an innate immune system suppressor during the infection of M. tuberculosis.
Author Contributions
Q. H., F. W., and L. G. designed the study. Q. H., F. W., S. L., H. C., D. Z., F. G., H. W., Z. L., J. L., and L. G. performed experiments. Q. H., F. W., J. L., and L. G. analyzed the data. Q. H. and F. W. wrote the original draft. Q. H., F. W., B. L., Z. L., J. L., and L. G. edited the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Prof. Xiuhua Pang from Shandong University for providing us the M. tuberculosis genomic DNA. We thank Prof. Jin He at Huazhong Agricultural University for providing us the MSMEG-2630 knock-out strain and the pMV261 vector. We also gratefully acknowledge the staff at the Beamline BL17U1 at the Shanghai Synchrotron Radiation Facility for supporting the data collection.
This work was supported by National Natural Science Foundation of China Grant 31470732 (to L. G.) and Shandong Provincial Funds for Distinguished Young Scientists Grant JQ201307 (to L. G.), This work was also supported by Zhejiang Natural Science Foundation Grant LR12C05001 (to Z. L.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5CET and 5CEU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- c-di-NMP
- cyclic dinucleotide
- PDE
- phosphodiesterase
- PDB
- Protein Data Bank.
References
- 1. Zumla A., George A., Sharma V., Herbert R. H., Baroness Masham of Ilton, Oxley A., and Oliver M. (2015) The WHO 2014 global tuberculosis report: further to go [J]. Lancet Global Health 3, e10–e12 [DOI] [PubMed] [Google Scholar]
- 2. Dey B., Dey R. J., Cheung L. S., Pokkali S., Guo H., Lee J. H., and Bishai W. R. (2015) A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med. 21, 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gomelsky M. (2011) cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all! Mol. Microbiol. 79, 562–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., Guo M., Roembke B. T., and Sintim H. O. (2013) Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341 [DOI] [PubMed] [Google Scholar]
- 5. Gao J., Tao J., Liang W., Zhao M., Du X., Cui S., Duan H., Kan B., Su X., and Jiang Z. (2015) Identification and characterization of phosphodiesterases that specifically degrade 3′ 3′-cyclic GMP-AMP. Cell Res. 25, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Römling U., Gomelsky M., and Galperin M. Y. (2005) c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57, 629–639 [DOI] [PubMed] [Google Scholar]
- 7. Hengge R. (2009) Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 8. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., and Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Römling U., Galperin M. Y., and Gomelsky M. (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bejerano-Sagie M., Oppenheimer-Shaanan Y., Berlatzky I., Rouvinski A., Meyerovich M., and Ben-Yehuda S. (2006) A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125, 679–690 [DOI] [PubMed] [Google Scholar]
- 11. Zhang L., Li W., and He Z.-G. (2013) DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J. Biol. Chem. 288, 3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witte C. E., Whiteley A. T., Burke T. P., Sauer J.-D., Portnoy D. A., and Woodward J. J. (2013) Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 4, e00282–00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oppenheimer-Shaanan Y., Wexselblatt E., Katzhendler J., Yavin E., and Ben-Yehuda S. (2011) c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12, 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corrigan R. M., Campeotto I., Jeganathan T., Roelofs K. G., Lee V. T., and Gründling A. (2013) Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 9084–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang J., Bai Y., Zhang Y., Gabrielle V. D., Jin L., and Bai G. (2014) Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 93, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bai Y., Yang J., Eisele L. E., Underwood A. J., Koestler B. J., Waters C. M., Metzger D. W., and Bai G. (2013) Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J. Bacteriol. 195, 5123–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manikandan K., Sabareesh V., Singh N., Saigal K., Mechold U., and Sinha K. M. (2014) Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS One 9, e86096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huynh T. N., Luo S., Pensinger D., Sauer J. D., Tong L., and Woodward J. J. (2015) An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U.S.A. 112, E747–E756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rao F., See R. Y., Zhang D., Toh D. C., Ji Q., and Liang Z.-X. (2010) YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye M., Zhang J. J., Fang X., Lawlis G. B., Troxell B., Zhou Y., Gomelsky M., Lou Y., and Yang X. F. (2014) DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect. Immun. 82, 1840–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Postic G., Danchin A., and Mechold U. (2012) Characterization of NrnA homologs from Mycobacterium tuberculosis and Mycoplasma pneumoniae. RNA 18, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Srivastav R., Kumar D., Grover A., Singh A., Manjasetty B. A., Sharma R., and Taneja B. (2014) Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases. Nucleic Acids Res. 42, 7894–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dey B., and Bishai W. R. (2014) Crosstalk between Mycobacterium tuberculosis and the host cell. Semin. Immunol. 26, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar M., and Chatterji D. (2008) Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology 154, 2942–2955 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burhenne H., and Kaever V. (2013) Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. Methods Mol. Biol. 1016, 27–37 [DOI] [PubMed] [Google Scholar]
- 31. Zhu D., Wang L., Shang G., Liu X., Zhu J., Lu D., Wang L., Kan B., Zhang J. R., and Xiang Y. (2014) Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Mol. Cell 55, 931–937 [DOI] [PubMed] [Google Scholar]
- 32. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang Q., Luo Y., Zheng C., Yin K., Ali M. K., Li X., and He J. (2015) Functional analysis of a c-di-AMP-specific phosphodiesterase MsPDE from Mycobacterium smegmatis. Int. J. Biol. Sci. 11, 813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Römling U., and Amikam D. (2006) Cyclic di-GMP as a second messenger. Curr. Opin Microbiol. 9, 218–228 [DOI] [PubMed] [Google Scholar]
- 36. Gentner M., Allan M. G., Zaehringer F., Schirmer T., and Grzesiek S. (2012) Oligomer formation of the bacterial second messenger c-di-GMP: reaction rates and equilibrium constants indicate a monomeric state at physiological concentrations. J. Am. Chem. Soc. 134, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 37. Egli M., Gessner R. V., Williams L. D., Quigley G. J., van der Marel G. A., van Boom J. H., Rich A., and Frederick C. A. (1990) Atomic-resolution structure of the cellulose synthase regulator cyclic diguanylic acid. Proc. Natl. Acad. Sci. U.S.A. 87, 3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guan Y., Gao Y. G., Liaw Y. C., Robinson H., and Wang A. H. (1993) Molecular structure of cyclic diguanylic acid at 1 A resolution of two crystal forms: self-association, interactions with metal ion/planar dyes and modeling studies. J. Biomol. Struct. Dyn. 11, 253–276 [DOI] [PubMed] [Google Scholar]
- 39. Sultan S. Z., Pitzer J. E., Boquoi T., Hobbs G., Miller M. R., and Motaleb M. A. (2011) Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect. Immun. 79, 3273–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Corrigan R. M., Abbott J. C., Burhenne H., Kaever V., and Gründling A. (2011). c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7, e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bharati B. K., Sharma I. M., Kasetty S., Kumar M., Mukherjee R., and Chatterji D. (2012) A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158, 1415–1427 [DOI] [PubMed] [Google Scholar]