Abstract
CYP1A1 is a phase I xenobiotic-metabolizing enzyme whose expression is mainly driven by AhR. Endosulfan is an organochlorine pesticide used agriculturally for a wide range of crops. In this study, we investigated the effect of endosulfan on CYP1A1 expression and regulation. Endosulfan significantly increased CYP1A1 enzyme activity as well as mRNA and protein levels. In addition, endosulfan markedly induced XRE transcriptional activity. CH-223191, an AhR antagonist, blocked the endosulfan-induced increase in CYP1A1 mRNA and protein expression. Moreover, endosulfan did not induce CYP1A1 gene expression in AhR-deficient mutant cells. Furthermore, endosulfan enhanced the phosphorylation of calcium calmodulin (CaM)-dependent protein kinase (CaMK) and protein kinase C (PKC). In conclusion, endosulfan-induced up-regulation of CYP1A1 is associated with AhR activation, which may be mediated by PKC-dependent pathways.
Keywords: Endosulfan, CYP1A1, Aryl hydrocarbon receptor, Calcium, Protein kinase C
INTRODUCTION
Cytochrome P450s (CYPs) belong to a superfamily of hemoproteins and provide monooxygenase activity in xenobiotic metabolism (1). In response to xenobiotics for the purpose of detoxification, CYP members are induced and are responsible for oxidizing xenobiotics into epoxide derivatives, which constitutes the initial step of xenobiotic conversion into water-soluble derivatives for excretion (2). However, epoxide derivatives may be further metabolized into reactive intermediates that could potentially attack DNA, resulting in DNA mutation and carcinogenesis (3). CYP1A1, the best-known CYP member, is one of the major aryl hydrocarbon receptor (AhR)-sensitive targets, and is highly induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and 3-methylcholanthrene (MC) (3).
AhR is an intracellular mediator of the xenobiotic signaling pathway, and is predominantly located in the cytoplasm and exists as a complex with molecular chaperone Hsp90 (4,5), Hsp90-interacting protein p23 (6), and the immunophilin-like protein XAP2, also known as ARA9 or AIP (7). When environmental contaminants such as TCDD and 3-MC bind to the AhR, the receptor complex accumulates in the nucleus, where AhR dissociates from Hsp90 to bind to the heterodimer partner AhR nuclear translocator (ARNT) (8). In the nucleus, the AhR/ARNT heterodimer directly binds to xenobiotic response elements (XREs) in the promoters of target genes and activates CYP1A1 (9).
Endosulfan (1,4,5,6,7,7-hexachloro-8,9,10-trinorborn-5-en-2,3-ylenebismethylene) has been widely used as a broad-spectrum cyclodiene insecticide (10), and is classified as a moderately hazardous chemical by the World Health Organization (11). Endosulfan is well-absorbed through ingestion, inhalation, and skin contact. It is toxic to fish and aquatic invertebrates (12), and has been implicated in mammalian gonadal toxicity (13), genotoxicity (14), and neurotoxicity (15). Endosulfan also induces oxidative tissue damage resulting from the generation of reactive oxygen species (ROS) (16). Recently, we reported that endosulfan up-regulates the expression of inducible nitric oxide synthase (iNOS) and proinflammatory cytokines in macrophages, which was mediated in part through the NF-κB binding sites of these genes (17). In response to environmental toxins, AhR mediates carcinogenesis, teratogenesis, changes in thymocyte development, T cell-dependent immune reactions, and triggers inflammatory skin lesions (1). However, despite extensive attempts to elucidate the various effects of endosulfan, little is known regarding the physiological function of endosulfan. In this study, we investigated the influence of endosulfan on CYP1A1 expression and its regulatory mechanism.
MATERIALS AND METHODS
Materials. All chemicals and cell culture materials were obtained from the following sources: endosulfan-alpha (purity, > 99.8%) and 3-MC from Sigma Chemical Co. (St. Louis, MO, USA); 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191), Gö 6983, and CaMK inhibitor W7 (N-(6-Aminohexyl)-5-chloro-1-naphthalenesulfonamide) from Calbiochem (La Jolla, CA, USA); 7-ethoxyresorufin and resorufin from Pierce Chemical Co. (Rockford, IL, USA); MTT assay kit from Roche Co. (London, United Kingdom); Lipofect-AMINE 2000 and Minimum Essential Medium Alpha Medium (α-MEM) from Invitrogen Co. (Carlsbad, CA, USA); fetal bovine serum (FBS), penicillin-streptomycin solution, and trypsin from Life Technologies, Inc. (Carlsbad, CA, USA); pCMV-β-gal from Clontech (Palo Alto, CA, USA); protein assay kit from Bio-Rad Laboratories, Inc. (Hercules, CA, USA); primary antibodies (anti-AhR, CYP1A1, phospho-Thr177-CaMKIα, phospho-PKC and β-actin) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); secondary antibodies (horseradish peroxidase [HRP]-linked anti-rabbit and anti-mouse IgG) from Cell Signaling Technology (Danvers, MA, USA); and enhanced chemiluminescence (ECL) chemiluminescence system and polyvinylidene difluoride (PVDF) membrane from Amersham Pharmacia Biotech (Uppsala, Sweden). Polymerase chain reaction (PCR) oligonucleotide primers were custom synthesized by Bioneer Co. (Daejeon, Korea). All chemicals were of the highest grade commercially available.
Cell culture and treatment. The mouse hepatoma cell line Hepa-1c1c7 and its derivative cell line Tao BpRcl were obtained from the American Type Culture Collection (Rockville, MD, USA). The cells were cultured in α-MEM supplemented with 10% FBS in a humidified 5% CO2 incubator at 37℃. Stock solutions of endosulfan and 3-MC were prepared in dimethylsulfoxide (DMSO) and added directly to the culture media for incubation (3). Control cells were treated with DMSO alone, and the final DMSO concentration was always < 0.2%.
Assay for proliferative activity. Cellular cytotoxicity was examined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction kit according to the manufacturer’s instructions. Briefly, Hepa-1c1c7 and Tao BpRcl cells in α-MEM containing 10% FBS were seeded into 96-well plates. Endosulfan (0.01~100 μM) was added to the wells and plates were incubated at 37℃ for 24 h. The cells were treated with MTT solution for 1 h, the dark blue formazan crystals that formed in intact cells were solubilized with DMSO, and absorbance was measured at 570 nm with a microplate reader (Varioskan; Thermo Electron, Waltham, MA, USA).
Ethoxyresorufin-O-deethylase (EROD) activity assay. Cells were incubated with 3-MC (1 μM) or endosulfan (0.01~1 μM) for 18 h. After incubation, the medium was removed and the wells were washed twice with fresh medium. Ethoxyresorufin-O-deethylase (EROD) activity was measured in intact cells grown in 48-well plates as previously described (18). Fluorescence was measured after 30 min using a FL600 ELISA reader (BIOTEK), with excitation at 530 nm and emission at 590 nm. A standard curve was constructed using resorufin.
mRNA analysis by real-time quantitative PCR. Cells were incubated with 3-MC (1 μM) or endosulfan (0.01~1 μM) for 6 h. Total RNA from the treated cells was prepared with RNAiso Reagent (Takara) according to the manufacturer’s protocol and stored at −80℃ until use. For CYP1A1 detection, total RNA was extracted after stimulation and treatment. PCR was monitored using Sequence Detection System software (ver. 1.7; Applied Biosystems, Foster City, CA). Accumulated PCR products were directly detected by monitoring the increase in reporter dye (SYBR). Expression levels of CYP1A1 in the exposed cells were compared to those in control cells at each collection time point using the comparative cycle threshold (Ct)-method. The quantity of each transcript was calculated as described in the instrument manual and was normalized to the amount of β-actin, a housekeeping gene.
Transfection and luciferase assays. Cells were plated into 24-well plates in α-MEM supplemented with 10% FBS. After 24 h, cells were co-transfected with 0.2 μg pCMV-β-gal and 1 μg pGL3-CYP1A1-Luc or XRE-luc per well using Lipofectamine. The pGL3-CYP1A1-Luc plasmid was created by inserting the mouse CYP1A1 upstream regulatory region (−1395 to +7) PCR product into the pGL3 basic vector. The XRE-driven luciferase reporter plasmid (−1306 to −824 of the murine CYP1A1), which contains four XREs, was used to examine the specific activation of XRE. Four hours after transfection, fresh medium containing 10% FBS was added to the cells along with 3-MC or endosulfan. Following an 18 h exposure, the cells were washed once with 0.5 mL phosphate-buffered saline (PBS) and lysed. The lysed cell preparations were then centri-fuged at 12,000 rpm for 10 min, and the supernatant was assayed for both luciferase and β-galactosidase activity. Luciferase activity was measured using the Luciferase Assay System (Promega) and a luminometer according to the manufacturer’s instructions. Luciferase activity was normalized to β-galactosidase activity and is expressed as the proportion of the activity detected with the vehicle control (3).
Western blotting. Cell lysates were prepared after endosulfan or 3-MC treatment and resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were then electroblotted onto polyvinylidene difluoride membranes (19). The membranes were probed with the appropriate primary antibodies, followed by incubation with HRP-conjugated secondary antibodies. The blots were visualized with an ECL Western blot kit according to the manufacturer’s instructions.
Statistical analysis. All experiments were repeated at least three times. One-way analysis of variance (ANOVA) was used to determine significant differences between treatment groups. Newman-Keuls test was used for multi-group comparisons. Statistical significance was accepted for p values of < 0.01.
RESULTS
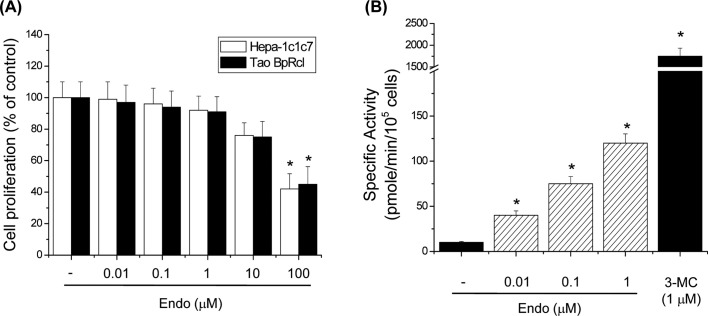
Endosulfan induces CYP1A1 enzyme activity in Hepa-1c1c7 cells. To determine the optimal concentrations for use in our studies, the potential cytotoxicity of endosulfan was tested in Hepa-1c1c7 and Tao BpRc1 cells. Tao BpRcl nonresponsive mutant cells were characterized by relatively low levels of cytosolic and nuclear AhR complex (20). The results showed that 0.01, 0.1, 1, and 10 μM endosulfan did not affect cell proliferation; however, 100 μM caused a decrease in cell viability. Therefore, all subsequent studies were conducted using 0.01~1 μM endosulfan. The CYP1A1 enzyme, encoded by the CYP1A1 gene, can be measured using ethoxyresorufin as a substrate for EROD activity. When cells were treated with endosulfan, there was a significant increase in CYP1A1 enzyme activity (Fig. 1B). 3-MC also significantly increased EROD activity (Fig. 1B). These results demonstrate that endosulfan induced CYP1A1 enzyme activity.
Fig. 1. Effects of endosulfan on CYP1A1 activity. (A) Effects of endosulfan on cytotoxicity in Hepa-1c1c7 and Tao BpRcl cells. Cells were seeded into 96-well plates and treated with various concentrations of endosulfan (Endo) for 24 h. Cell viability was assessed using the MTT assay. (B) Effects of endosulfan on EROD activity in Hepa-1c1c7 cells. EROD activity was measured in cells treated with endosulfan or 3-MC for 18 h. Values represent the means from three independent experiments, each performed in triplicate. * p< 0.01, significantly different from the control, as determined by ANOVA by the Newman-Keuls test.
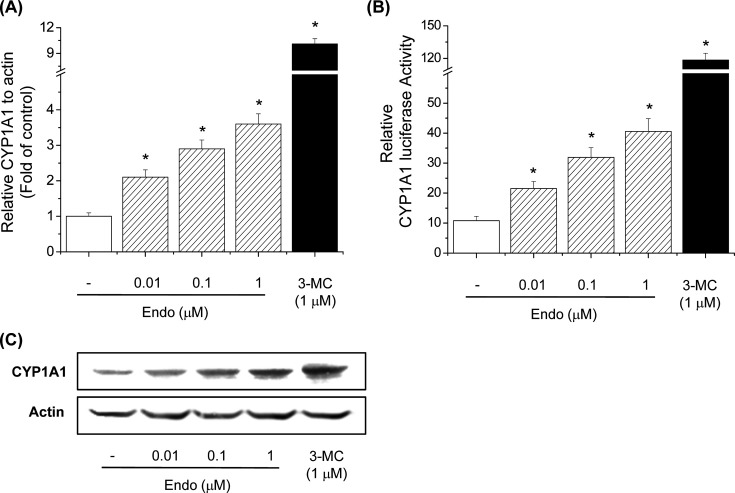
Endosulfan induces CYP1A1 gene expression in Hepa-1c1c7 cells. To further explore whether endosulfan increases CYP1A1 mRNA expression, we performed real-time PCR in Hepa-1c1c7 cells. 3-MC increased CYP1A1 mRNA levels. Endosulfan also significantly increased CYP1A1 mRNA levels, indicating that it may be an AhR agonist like 3-MC (Fig. 2A). To investigate transcriptional activation of the CYP1A1 gene by endosulfan, luciferase assays were performed in which reporter plasmids containing the 5'-flanking region of the CYP1A1 gene were transiently transfected into Hepa-1c1c7 cells. The reporter activity of the −1395/+7 CYP1A1 promoter construct was induced by endosulfan (Fig. 2B). To confirm our hypothesis, the effects of endosulfan on CYP1A1 protein levels were determined by Western blot analysis. Similarly, endosulfan significantly increased CYP1A1 protein levels, indicating that it may function as an AhR agonist (Fig. 2C).
Fig. 2. Effects of endosulfan on CYP1A1 transcriptional activity. (A) Effects of endosulfan on CYP1A1 mRNA expression in Hepa-1c1c7 cells. Cells were treated with endosulfan (Endo) or 3-MC for 6h. The cells were lysed, and total RNA was prepared to analyze CYP1A1 gene expression. PCR amplification of the housekeeping gene, β-actin, was performed for each sample. CYP1A1 mRNA expression was compared between treated and untreated cells at each time point by real-time PCR. Values represent the means from three independent experiments, each performed in triplicate. * p< 0.01, significantly different from the control, as determined by ANOVA by the Newman-Keuls test. (B) Effects of endosulfan on CYP1A1 promoter activity in Hepa-1c1c7 cells. Cells were transfected with pGL3-CYP1A1-Luc and subsequently treated with endosulfan or 3-MC for 18 h. Cells were then harvested and assayed for luciferase activity. Values represent the means from three independent experiments, each performed in triplicate. * p<0.01, significantly different from the control, as determined by ANOVA by the Newman-Keuls test. (C) Effects of endosulfan on CYP1A1 protein levels in Hepa-1c1c7 cells. Cells were treated with endosulfan or 3-MC for 24 h. The membrane was probed with a CYP1A1-specific antibody, and bands were visualized with ECL reagents. Each blot in this figure is representative of three independent experiments with similar results.
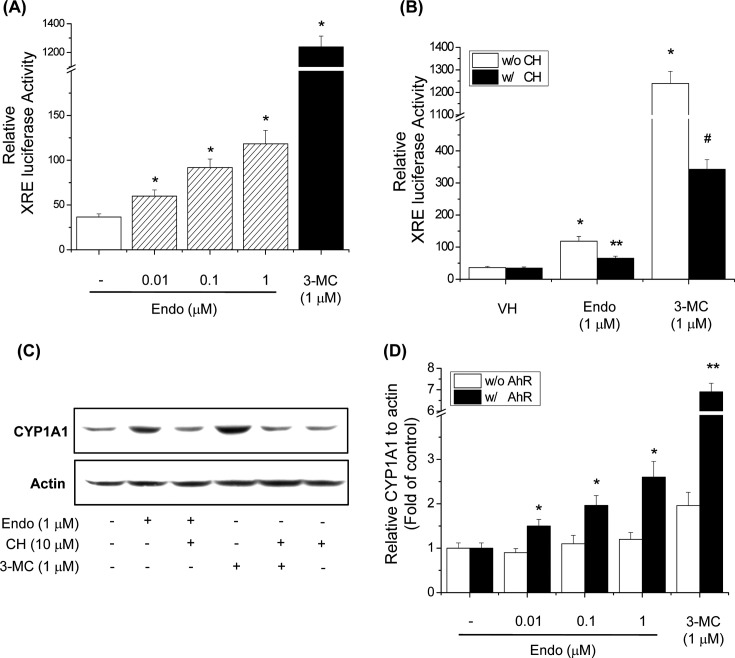
Endosulfan induces CYP1A1 expression via the XRE binding site. To demonstrate that endosulfan-induced expression of CYP1A1 resulted in XRE activation, an XRE reporter assay was performed. Endosulfan significantly increased XRE promoter activity (Fig. 3A) using 3-MC as a positive control. To confirm that the induction of XRE luciferase activity by endosulfan was dependent on AhR, the effect of the AhR antagonist CH-223191 on the XRE reporter assay was examined. CH-223191 blocked the binding of TCDD to AhR and inhibited TCDD-mediated nuclear translocation and DNA binding of AhR (21). CH-223191 inhibited the endosulfan-induced increase in XRE luciferase activity (Fig. 3B). To further investigate whether endosulfan increases CYP1A1 mediated through AhR activation, we investigated the effect of the AhR antagonist CH-223191 on CYP1A1 protein level. CH-223191 treatment decreased endosulfan-induced CYP1A1 protein expression in Hepa-1c1c7 cells (Fig. 3C). These results demonstrate that endosulfan increased CYP1A1 expression through AhR activation. To further assess the role of AhR in endosulfan-induced CYP1A1 expression, we tested endosulfan inducibility in the AhR-deficient mouse hepatoma cell line, Tao BpRc1. The results showed that, in the wild-type mouse hepatoma cell line Hepa1c1c7, CYP1A1 mRNA expression was induced by endosulfan, and the effects were dose-dependent. However, endosulfan did not induce CYP1A1 mRNA expression in Tao BpRc1 cells (Fig. 3D). These results imply that endosulfan induces CYP1A1 gene expression through an AhR-dependent pathway.
Fig. 3. Effects of endosulfan on the CYP1A1 regulatory mechanism. (A) Effects of endosulfan on XRE promoter activity in Hepa-1c1c7 cells. Cells were transfected with the XRE-Luc construct and subsequently treated with endosulfan (Endo) or 3-MC for 18 h. Cells were then harvested and assayed for luciferase activity. Values represent the means from three independent experiments, each performed in triplicate. * p<0.01, significantly different from the control, as determined by ANOVA by the Newman-Keuls test. (B) Effects of an AhR antagonist on XRE transcriptional activity by endosulfan in Hepa-1c1c7 cells. Cells were transfected with XRE-Luc and treated with CH-223191 (CH), endosulfan or 3-MC for 18 h. Cells were then harvested and assayed for luciferase activity. Values represent the means from three independent experiments, each performed in triplicate. * p< 0.01, ** p < 0.01, and # p < 0.01, significantly different from the control, 3-MC, and endosulfan, as determined by ANOVA by the Newman-Keuls test. (C) Effects of an AhR antagonist on CYP1A1 protein levels by endosulfan in Hepa-1c1c7 cells. Cells were pretreated with CH-223191 for 30 min and then treated with endosulfan or 3-MC for 24 h. The membrane was probed with a CYP1A1-specific antibody, and bands were visualized with ECL reagents. (D) Effects of endosulfan on CYP1A1 mRNA levels in Tao BpRcl cells. Hepa-1c1c7 and Tao BpRcl cells were treated with endosulfan or 3-MC for 6 h. Values represent the means from three independent experiments, each performed in triplicate. * p< 0.01, significantly different from the control, as determined by ANOVA by the Newman-Keuls test.
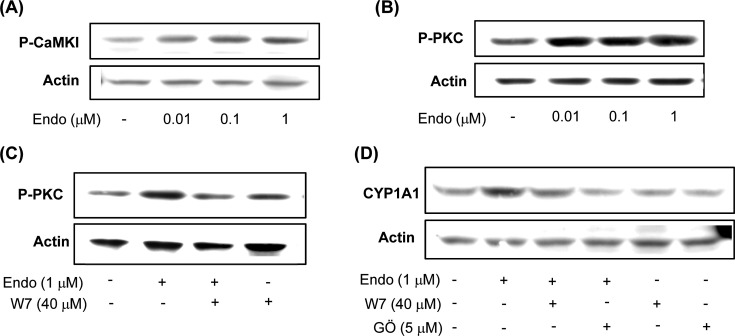
Endosulfan activates the CaMKI and PKC signaling pathways. Recent studies reported that AhR activation is dependent on AhR phosphorylation by PKC (22). In addition, increasing the intracellular concentrations of Ca2+ and calmodulin kinase (CaMK) participates in the signal transduction elicited by AhR agonists (23). To further assess the effect of endosulfan on AhR upstream kinases, activation of CaMKI and PKC was tested in Hepa-1c1c7 cells. Treatment with endosulfan increased CaMKI (Thr177) phosphorylation in a dose-dependent manner (Fig. 4A). Moreover, endosulfan enhanced PKC phosphorylation in Hepa-1c1c7 cells (Fig. 4B). In addition, the CaM antagonist W7 reduced endosulfan-induced PKC activation (Fig. 4C). To clarify the endosulfan-induced AhR activation signaling cascade underlying CYP1A1 expression, the effects of specific inhibitors of CaM (W7) and PKC (Gö 6983) on endosulfan-induced CYP1A1 protein expression in Hepa-1c1c7 cells were examined. W7 and Gö 6983 inhibited endosulfan-induced CYP1A1 protein level increases (Fig. 4C), indicating that the induction of CYP1A1 protein expression by endosulfan is regulated by CaMK and PKC kinases.
Fig. 4. Effects of endosulfan on the AhR upstream signaling pathway. (A) Effects of endosulfan on CaMK protein activation in Hepa-1c1c7 cells. Cells were treated with endosulfan (Endo) for 5min. Phosphorylated CaMKIa at threonine 177 (P-CaMKIα) was then analyzed by Western blotting, and β-actin expression was measured as a loading control. (B-C) Effects of endosulfan on PKC phosphorylation in Hepa-1c1c7 cells. Cells were treated with endosulfan for 10 min (B). Cells were pretreated with W7 and subsequently treated with endosulfan for 10 min (C). P-PKC was then analyzed by Western blotting, and β-actin expression was measured as a loading control. (D) Cells were pretreated with W7 or Gö 6983 (Gö) and treated with endosulfan for 24 h. CYP1A1 protein levels were then analyzed by Western blotting, and β-actin expression was measured as a loading control. Each blot in this figure is representative of three independent experiments with similar results.
DISCUSSION
Liver biotransformations mediate most of the biological effects of xenobiotics such as organochlorine insecticides (OCs). The cytochrome P450-dependent monooxygenase (CYP) enzymes are the most important of the phase I systems in these biotransformation reactions. Biotransformation of OCs including endosulfan is a complex process that involves meditation by xenobiotic-metabolizing phase I CYP enzymes. They represent a super gene family of monooxygenases that carry out both the activation and detoxification of many hormones, drugs and environmental agents (24). Endosulfan has been widely used as a broad-spectrum cyclodiene insecticide (10), and its widespread use has caused concern over the effects of persistent exposure of human and animal systems. Endosulfan is well-absorbed through the skin as well as respiratory and oral routes. In addition, endosulfan persists in the environment and accumulates in animals and plants, leading to instances of food contamination and dietary exposure in humans. Therefore, this study was performed to determine whether endosulfan affects the expression of CYP1A1, and to identify the regulatory pathway preceding CYP1A1 gene expression. We demonstrated that endosulfan can induce CYP1A1 gene expression as a ligand for AhR and increase its transformation by inducing the phosphorylation of AhR, likely via PKC and CaMK.
Endosulfan significantly increased EROD activity and the levels of CYP1A1 mRNA, transcriptional activity, and protein in Hepa-1c1c7 cells, indicating that it may function as an AhR agonist. EROD activity is used to monitor CYP1A1 enzyme induction (17). Ligands bind to AhR, resulting in its translocation to the nucleus. AhR then forms a heterodimer complex with AhR nuclear translocator protein (ARNT), which functions as a transcriptional activator by binding to consensus sequences (XRE) in the 5'-flanking region of numerous genes, including CYP1A1 (3,25). To gain insight into the mechanisms of endosulfan-induced CYP1A1 expression, we performed an XRE reporter assay for the mouse CYP1A1 promoter. Endosulfan enhanced XRE luciferase activity in Hepa-1c1c7 cells. To confirm that the effects of endosulfan treatment on CYP1A1 were AhR-dependent, we utilized the potent and selective AhR antagonist CH-223191. CH-223191 significantly inhibited endosulfan-induced XRE transcriptional activity and CYP1A1 protein levels. To further confirm that the effects of endosulfan on CYP1A1 were AhR-mediated, we investigated CYP1A1 mRNA expression in the mutant cell line Tao BpRc1, which expresses low levels of AhR. Endosulfan increased CYP1A1 mRNA levels in Hepa-1c1c7 cells, but not in Tao BpRc1 cells. Therefore, endosulfan is able to induce CYP1A1 and its effect is dependent on the AhR signaling pathway.
According to a previous study, phorbol 12-myristate 13-acetate (PMA), a potent PKC activator, dramatically enhances transcriptional activation of the CYP1A1 gene induced by various AhR ligands, indicating that PKC activity is required for nuclear events in the CYP1A1 transcriptional pathway (26). PKC-mediated signal transduction takes place through a family of PKC isoforms and requires intracellular calcium for activation (27). Treatment with endosulfan activates CaMK and PKC kinases. These kinases can increase AhR activation and activate AhR/ARNT, causing its translocation to the nucleus. Specific inhibitors of CaM (W7) and PKC (GÖ 6983; GÖ) reduced endosulfan-mediated CYP1A1 protein induction. Taken together, these results suggest that activation of PKC and CaKM may be responsible for endosulfan-induced CYP1A1 induction.
In summary, endosulfan-induced translocation of AhR/ARNT to the nucleus causes the complex to react with the XRE site in the CYP1A1 promoter, resulting in elevated CYP1A1 protein expression in mouse hepatoma Hepa-1c1c7 cells. Endosulfan was also found to activate CaMK and PKC kinases. PKC in turn enhances AhR transactivation, which ultimately increases CYP1A1 protein expression.
Acknowledgments
This work was supported by research fund of Chungnam National University.
References
- 1.Nelson D.R., Koymans L., Kamataki T., Stegeman J.J., Feyereisen R., Waxman D.J., Waterman M.R., Gotoh O., Coon M.J., Estabrook R.W., Gunsalus I.C., Nebert D.W. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. (1996);6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Dipple A. Reactions of polycyclic aromatic hydrocarbons with DNA. IARC Sci. Publ. (1994);125:107–129. [PubMed] [Google Scholar]
- 3.Han E.H., Kim H.G., Im J.H., Jeong T.C., Jeong H.G. Up-regulation of CYP1A1 by rutaecarpine is dependent on aryl hydrocarbon receptor and calcium. Toxicology. (2009);266:38–47. doi: 10.1016/j.tox.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Denis M., Cuthill S., Wikström A.C., Poellinger L., Gustafsson J.A. Association of the dioxin receptor with the Mr 90,000 heat shock protein: a structural kinship with the glucocorticoid receptor. Biochem. Biophys. Res. Commun. (1988);155:801–807. doi: 10.1016/S0006-291X(88)80566-7. [DOI] [PubMed] [Google Scholar]
- 5.Perdew G.H. Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. (1988);263:13802–13805. [PubMed] [Google Scholar]
- 6.Kazlauskas A., Poellinger L., Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J. Biol. Chem. (1999);274:13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 7.Meyer B.K., Pray-Grant M.G., Vanden Heuvel J.P., Perdew G.H. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol. Cell. Biol. (1998);18:978–988. doi: 10.1128/MCB.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eguchi H., Ikuta T., Tachibana T., Yoneda Y., Kawajiri K. A nuclear localization signal of human aryl hydrocarbon receptor nuclear translocator/hypoxia-inducible factor 1beta is a novel bipartite type recognized by the two components of nuclear pore-targeting complex. J. Biol. Chem. (1997);272:17640–17647. doi: 10.1074/jbc.272.28.17640. [DOI] [PubMed] [Google Scholar]
- 9.Kolluri S.K., Weiss C., Koff A., Göttlicher M. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. (1999);13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omurtag G.Z., Tozan A., Sehirli A.O., Sener G. Melatonin protects against endosulfan-induced oxidative tissue damage in rats. J. Pineal Res. (2008);44:432–438. doi: 10.1111/j.1600-079X.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- 11.Sethunathan N., Megharaj M., Chen Z., Singh N., Kookana R.S., Naidu R. Persistence of endosulfan and sulfate in soil as affected by moisture regime and organic matter addition. Bull. Environ. Contam. Toxicol. (2002);68:725–731. doi: 10.1007/s001280314. [DOI] [PubMed] [Google Scholar]
- 12.Sunderam R.I.M., Cheng D.M.H., Thompson G.B. Toxicity of endosulfan to native and introduced fish in Australia. Environ. Toxicol. Chem. (1992);11:1469–1476. doi: 10.1002/etc.5620111012. [DOI] [Google Scholar]
- 13.Sinha N., Narayan R., Saxena D.K. Effect of endosulfan on testis of growing rats. Bull. Environ. Contam. Toxicol. (1997);58:79–86. doi: 10.1007/s001289900303. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhuri K., Selvaraj S., Pal A.K. Studies on the genotoxicology of endosulfan in bacterial system. Mutat. Res. (1999);439:63–67. doi: 10.1016/S1383-5718(98)00174-0. [DOI] [PubMed] [Google Scholar]
- 15.Paul V., Balasubramaniam E. Effect of single and repeated administration of endosulfan on behaviour and its interaction with centrally acting drugs in experimental animals. Environ. Toxicol. Pharmacol. (1997);3:151–157. doi: 10.1016/S1382-6689(97)00009-4. [DOI] [PubMed] [Google Scholar]
- 16.Hincal F., Gurbay A., Giray B. Induction of lipid peroxidation and alteration of glutathione redox status by endosulfan. Biol. Trace Elem. Res. (1995);47:321–326. doi: 10.1007/BF02790133. [DOI] [PubMed] [Google Scholar]
- 17.Han E.H., Hwang Y.P., Kim H.G., Jeong H.G. Inflammatory effect of endosulfan via NF-kappaB activation in macrophages. Biochem. Biophys. Res. Commun. (2007);355:860–865. doi: 10.1016/j.bbrc.2007.01.062. [DOI] [PubMed] [Google Scholar]
- 18.Kim K.W., Chung Y.H. Hepatotoxicity in rats treated with dimethylformamide or toluene or both. Toxicol. Res. (2013);29:187–193. doi: 10.5487/TR.2013.29.3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C.M., Yoon M.S., Kim Y.C. Effects of pueraria lobata root ethanol extract on adipogenesis and lipogenesis during 3T3-L1 differentiation into adipocytes. Toxicol. Res. (2015);31:191–201. doi: 10.5487/TR.2015.31.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.G., Han E.H., Jeong H.G. Effect of troglitazone on CYP1A1 induction. Toxicology. (2008);246:166–171. doi: 10.1016/j.tox.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Kim S.H., Henry E.C., Kim D.K., Kim Y.H., Shin K.J., Han M.S., Lee T.G., Kang J.K., Gasiewicz T.A., Ryu S.H., Suh P.G. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol. Pharmacol. (2006);69:1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- 22.Iida M., Bak S.M., Murakami Y., Kim E.Y., Iwata H. Transient suppression of AHR activity in early red seabream embryos does not prevent the disruption of peripheral nerve projection by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Aquat. Toxicol. (2014);154:39–47. doi: 10.1016/j.aquatox.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Tan Z., Huang M., Puga A., Xia Y. A critical role for MAP kinases in the control of Ah receptor complex activity. Toxicol. Sci. (2004);82:80–87. doi: 10.1093/toxsci/kfh228. [DOI] [PubMed] [Google Scholar]
- 24.Rendie S., Di Carlo F.J. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab. Rev. (1997);29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 25.Denison M.S., Whitlock J.P. Xenobiotic-inducible transcription of cytochrome P450 genes. J. Biol. Chem. (1995);270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y.H., Tukey R.H. Protein kinase C modulates regulation of the CYP1A1 gene by the aryl hydrocarbon receptor. J. Biol. Chem. (1996);271:26261–26266. doi: 10.1074/jbc.271.42.26261. [DOI] [PubMed] [Google Scholar]
- 27.Machemer D.E., Tukey R.H. The role of protein kinase C in regulation of TCDD-mediated CYP1A1 gene expression. Toxicol. Sci. (2005);87:27–37. doi: 10.1093/toxsci/kfi220. [DOI] [PubMed] [Google Scholar]