Abstract
Aneuploidy, the unbalanced segregation of chromosomes during cell division, is recurrent in many tumors and the cause of birth defects and genetic diseases. Centromeric chromatin represents the chromosome attachment site to the mitotic spindle, marked by specialized nucleosomes containing a specific histone variant, CEN-H3/Cse4, in yeast. Mislocalization of Cse4 outside the centromere is deleterious and may cause aberrant chromosome behavior and mitotic loss. For this reason, ubiquitylation by the E3-ubiquitin ligase Psh1 and subsequent proteolysis tightly regulates its restricted localization. Among multiproteic machineries, the SAGA complex is not merely engaged in acetylation but also directly involved in deubiquitylation. In this study, we investigated the role of SAGA-DUB’s Ubp8-driven deubiquitylation of the centromeric histone variant Cse4 in budding yeast. We found that Ubp8 works in concert with the E3-ubiquitin ligase Psh1, and that its loss causes defective deubiquitylation and the accumulation of a short ubiquitin oligomer on Cse4. We also show that lack of Ubp8 and defective deubiquitylation increase mitotic instability, cause faster Cse4 proteolysis and induce mislocalization of the centromeric histone outside the centromere. Our data provide evidence for a fundamental role of DUB-Ubp8 in deubiquitylation and the stability of the centromeric histone in budding yeast.
Keywords: SAGA complex, DUB-Ubp8, deubiquitylation, histone variant Cse4, centromere, mitotic stability
The SAGA (Spt-Ada-Gcn5-acetyltransferase) complex is a key multicomponent regulator of acetylation. Through its deubiquitination (DUB) module it is also involved in deubiquitylation exerted, for example, on histone H2B (Henry et al. 2003; Daniel et al. 2004). The ubiquitin protease Ubp8 (ubiquitin-specific processing protease 8) performs a discrete function in the DUB module (Ingvarsdottir et al. 2005). Its deubiquitylation activity is directly involved in the transcriptional activation of SAGA-responsive genes, in nucleosome eviction and remodelling, and it is often associated with concurrent histone H3 methylation (Shukla et al. 2006). Also, the SAGA DUB module (Sgf73, Sgf11, Sus1, Ubp8) has been shown to couple transcription with mRNA export (Kohler et al. 2006; Klockner et al. 2009). Among the Ubp8 orthologs, Drosophila melanogaster Non-stop is involved in axon guidance in the optic lobe (Weake et al. 2008), and human USP22 represents a signature associated with poor prognosis in various human cancers (Zhang et al. 2008). Also, USP22 silencing promotes apoptosis and cell cycle arrest in human brain gliomas (Li et al. 2013). Taken together, these multifunctional examples demonstrate the biological relevance of the SAGA complex and its DUB in cellular differentiation and human disease.
Chromosomal regions such as centromeres are substrates of epigenetic modifications. Although chromatin modifiers have been mainly linked to the regulation of gene expression, they are also likely to be engaged in the regulation of key processes in mitosis and meiosis. Centromeres are epigenetically marked by specialized nucleosomes and incorporate specific histone variants (CenH3) (Stoler et al. 1995; Keith and Fitzgerald-Hayes 2000; Bernad et al. 2009). CENP-A (histone H3-like centromeric protein A) in humans, CID in flies, and Cse4 (chromosome segregation protein 4) in budding yeast contribute to the formation of specialized nucleosomes making up the centromere, the chromosome attachment site to the mitotic spindle, and set the foundation for the hierarchical assembly of the kinetochore (Meluh and Koshland 1997). Following the first reports that Saccharomyces cerevisiae possesses a single nucleosome structure (Meluh and Koshland 1997; Furuyama and Biggins 2007), more recent findings suggest that there are additional Cse4 molecules and 0.5/3 variant nucleosomes at the centromere (Maresca 2013). Histone variant mislocalization or its overexpression may create putative sites of erroneous attachment of the chromosomes to the mitotic spindle, thus inducing aberrant segregation and aneuploidy (Tomonaga et al. 2003; Heun et al. 2006; Moreno-Moreno et al. 2006). In yeast, the centromeric histone variant Cse4 marks the centromere and regulates the behavior and segregation of chromosomes in mitosis. Mislocalization of centromeric histone causes aberrant attachment sites and mitotic instability. Proteasomal degradation and removal of CenH3 from ectopic localization is therefore a fundamental regulatory step, achieved through the deposition of a poly-Ub chain on Cse4 by Psh1 (Collins et al. 2007; Hewawasam et al. 2010; Ranjitkar et al. 2010). This process ensures the exclusive localization of Cse4 at the centromere (Folco and Desai 2010). Surprisingly, Psh1 deletion does not induce mitotic defects in yeast, implying that additional mechanisms might be involved (Folco and Desai 2010). Since ubiquitylation plays a crucial role in Cse4 proteolysis, we reasoned that a counteracting deubiquitylation activity could also be important. Here we demonstrate, at the genetic level, that Psh1 is epistatic to the Ub-protease Ubp8 and that its absence causes a high rate of mitotic instability and minichromosome loss rescued by further deletion of Psh1. Additionally, we show that Ubp8 directly deubiquitylates Cse4 and that its deletion produces not only the accumulation of a short Ub-oligomer on the centromeric histone but also its faster proteolytic degradation. Finally, our results suggest that the presence of this short ubiquitin mark on Cse4 counteracts the elongation of polyubiquitin chains by Psh1 and represents a poor mark for removal from ectopic localization. In brief, we describe a new function for the SAGA complex and the DUB-Ubp8 component. We propose a model in which Ubp8 is directly involved in the regulation of the ubiquitylation status of the histone variant Cse4, and we suggest that Ubp8 contributes not only to the mechanism ensuring its restricted localization at the centromere but also to the general mitotic stability of the cell.
Materials and Methods
Materials, yeast strains and culture
All media components were either from BD or GIBCO, general reagents from Sigma Aldrich and Roche unless otherwise stated. Yeast strains were from a w303 isogenic background. All strains are listed in Table 1. Gene disruption and protein tagging were performed as previously described (Janke et al. 2004) and controlled by PCR amplification and western blot assays. Growth media were: YPD (1% yeast extract, 2% bactopeptone, 2% glucose), SD (0.67% YNB and 2% glucose) and SC (0.67% YNB, 2% glucose or galactose, selective Drop-Out Mix). Benomyl was used at a final concentration of 10/15μγρ/μl. Antibodies were: anti-Myc (clone 9E10, sc-40, Santa Cruz), anti-ADA2 (sc-6651, Santa Cruz), anti-HA (sc-7392, Santa Cruz), and Anti-6X His tag (ab1187, Abcam).
Table 1. Yeast strains used in Canzonetta et al.
| Name | Genotype | Source |
|---|---|---|
| W303 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. | (Vernarecci et al. 2008) |
| yFT21 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. ubp8::HIS3MX6 | This work |
| yCB12 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. psh1::HIS3MX6 | This work |
| ySP1090 | MATα, cse4-1 ade2-101 his3D-100 leu2-3 LYS2 trp1D, ura3-52. | (Stoler et al. 1995) |
| yCC11 | MATα, cse4-1 ade2-101 his3D-100 leu2-3 LYS2 trp1D, ura3-52. ubp8::HIS3MX6. | This work |
| yCC12 | MATα, cse4-1 ade2-101 his3D-100 leu2-3 LYS2 trp1D, ura3-52. psh1::HIS3MX6. | This work |
| CBY3 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). | This work |
| CBY8 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). ubp8::HIS3MX6. | This work |
| CBY13 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). psh1::HIS3MX6. | This work |
| CMY13 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). ubp8::HIS3MX6. psh1::KanMX6 | This work |
| CBY4 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). gcn5::KanMX6. | This work |
| CBY9 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). ubp8::HIS3MX6. gcn5::KanMX6. | This work |
| CBY15 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). ada3::HIS3MX6. | This work |
| CBY21 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. pRS415-ADE2 1225 (CEN/ARS LEU2). Sgf73::HIS3MX6. | This work |
| SBY3570 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB816, pGAL-MYC13-CSE4, URA3, 2μ) | (Collins et al. 2004) |
| SBY3571 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB817, pGAL-MYC13-CSE4-16R, URA3, 2μ) | (Collins et al. 2004) |
| FTY22 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB816, pGAL-MYC13-CSE4, URA3, 2μ). ubp8::HIS3MX6. | This work |
| RBY013 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB817, pGAL-MYC13-CSE4-16R, URA3, 2μ). ubp8::HIS3MX6. | This work |
| CBY17 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB816, pGAL-MYC13-CSE4, URA3, 2μ). psh1::HIS3MX6. | This work |
| CMY6 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB817, pGAL-MYC13-CSE4-16R, URA3, 2μ). psh1::HIS3MX6. | This work |
| CBY24 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB816, pGAL-MYC13-CSE4, URA3, 2μ). ubp8::HIS3MX6. psh1::KanMX6 | This work |
| yML48 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB817, pGAL-MYC13-CSE4, URA3, 2μ). ubp8::HIS3MX6. psh1::KanMX6 | This work |
| CMY19 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB816, pGAL-MYC13-CSE4, URA3, 2μ). Ubp10::HIS3MX6. | This work |
| CMY20 | MATa ura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 (pSB817, pGAL-MYC13-CSE4-16R, URA3, 2μ). Ubp10::HIS3MX6. | This work |
| SBY617 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3. | (Buvelot et al. 2003) |
| yFT17 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3 ubp8::HIS3MX6. | This work |
| yCB16 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3 psh1::HIS3MX6. | This work |
| yCC1 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3 ubp8::HIS3MX6, psh1::KanMX6. | This work |
| yCB21 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. (pJD421 His-Ubiquitin::LEU). | This work |
| yCM2 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. CSE4-MYC12::URA3. (pJD421 His-Ubiquitin::LEU). | This work |
| yCM4 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3 ubp8::HIS3MX6. (pJD421 His-Ubiquitin::LEU) | This work |
| YCC9 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3, psh1::HIS3MX6, (pJD421 His-Ubiquitin::LEU) | This work |
| YCC10 | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 can1-100 bar1Δ CSE4-MYC12::URA3, ubp8::HIS3MX6, psh1::KanMX6, (pJD421 His-Ubiquitin::LEU) | This work |
| yCC15 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. CSE4-MYC12::URA3, doa1::KanMX6, (pJD421 His-Ubiquitin::LEU). | This work |
| yCC14 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. CSE4-MYC12::URA3, ubp8::HIS3MX6, doa1::KanMX6, (pJD421 His-Ubiquitin::LEU). | This work |
| yFT33 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. Ubp8-MYC9::HIS3MX6 | This work |
| yCC21 | MATa ade2-1 trp1-1 leu2-3, 112 his3-11,15 ura3 can1-100 ssd1. Ubp8-MYC9::HIS3MX6, CSE4-HA6:: klTrp1 | This work |
Color sectoring
WT (wild-type, CBY3), psh1Δ (CBY13), ubp8Δ (CBY8), ubp8Δpsh1Δ (CMY13), gcn5Δ (CBY4), gcn5Δubp8Δ (CBY9), ada3Δ (CBY24) and sgf73Δ (CBY21) strains containing the centromeric plasmid pRS415-1225 (CEN/ARS LEU2/ADE2) were grown to early log phase in selective ade-leu- minimal medium at 28°. Cells were then diluted to 2 × 103 cells/ml, plated on YPD plates, and grown for 3–4 days. Colonies were then scored as white (pRS415-1225 retention) or red (pRS415-1225 loss) or sectorialized (pRS415-1225 partial loss) (Ma et al. 2010).
Cycloheximide degradation assay
Cells expressing tagged proteins (Cse4-Myc) were grown in 100 ml of YPD to an OD600 of 0.4. A 20 ml aliquot was collected before adding cycloheximide to a final concentration of 10 μg/ml (time zero). Subsequent shares were harvested every 40 min up to 2 hr and processed promptly accordingly to Knop et al. (1999) with minor modifications. Briefly, cells were spun down at 3500 rpm for 5 min, resuspended in 0.3 M NaOH, 140 mM BME, vortexed, and incubated on ice for 15 min with occasional vortexing. TCA (55%) was added to a final concentration of 6.5% followed by further incubation on ice. Samples were centrifuged at 4° at maximum speed for 20 min. Protein pellets were resuspended in HU buffer (8 M urea, 5% SDS, 200 mM Tris pH 6.8, 0.1 mM EDTA, 100 mM DTT, bromophenol blue), and loaded on 10% SDS-PAGE gels. Western blots were hybridized with anti-myc, and anti-Ada2 signal was used as a loading control.
Coimmunoprecipitation
Yeast strains were grown overnight in the appropriate medium, diluted to an OD600 of 0.25 in YPD, and collected to an OD600 of 0.6; 100 ml of the exponentially growing cells were then harvested and incubated in spheroplast buffer [1.2 M sorbitol, 50 mM potassium phosphate (pH 7.4), 1 mM MgCl, 250 μg of zymolyase per ml], with shaking for 1 hr at 30°. Spheroplasts were harvested by centrifugation for 2 min at 5000 rpm, washed with 1.2 M sorbitol, and pelleted at low speed as before. All subsequent steps were performed at 4°. Spheroplasts were lysed for 15 min on ice in IP buffer [50 mM Tris (pH 8), 1 mM EDTA, 50 mM NaCl, 1% NP-40] plus protease inhibitors. Lysates were centrifuged at 13,000 rpm for 5 min and the pellet was discarded. Supernatants were precleared by incubation for 1 hr with protein G magnetic beads; 1/20th of the crude extract was kept as input material. Immunoprecipitation was performed overnight at 4° on a rotating wheel with 10 µl of G-conjugated Dynabeads (Invitrogen) that had been prebound with an anti-myc antibody. Beads were collected in the morning, washed extensively with IP buffer, and resuspended in 30 µl HU-buffer. Proteins were released from the beads after 10 min incubation at 65°. Supernatant was either used immediately or frozen at −80°. Eluates and Inputs were loaded on 7.5% and 10% SDS-PAGE gels, respectively. Western blots were hybridized with anti-myc, Anti-HA, and anti-Ada2 antibodies. Signals were detected by chemiluminescence using LiteAblot Extend (EuroClone) and the ChemiDoc XRS+ System (Biorad).
Purification of ubiquitylated proteins
Cells transformed with a plasmid encoding 6His-ubiquitin under the CUP1 promoter [YEp96-6His-Ub, (Iglesias et al. 2010)] were grown on selective media and stimulated overnight with 0.1 mM CuSO4. Growth was stopped in the morning at an OD600 of 0.8–1. Cells were lysed in guanidinium buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl, pH 8.0, 0.1% Triton X-100, 5 mM imidazole, 10 mM β-mercaptoethanol, 0.1 mM MG132 and protease inhibitors), 1/20th of the lysate was kept aside as input material and TCA (5.5%) precipitated; the rest was incubated on a Ni-NTA agarose resin (QIAGEN) at 4° with rotation. The resin was washed three times with urea buffer (8 M urea, 0.1 M Na2HPO4/Na2HPO4, 0.01 M Tris-HCl, pH 6.4, 3.5 mM β-mercaptoethanol, and 0.1% Triton X-100) before elution in 2X sample buffer (0.125 M Tris-HCl, pH 6.8, 4% SDS, 0.285 M β-mercaptoethanol, 20% SDS, 2% bromophenol blue). Eluates and inputs were loaded on 7.5% and 10% SDS-PAGE gels, respectively. Western blots were hybridized with anti-myc, Anti-6X His tag, and anti-Ada2 antibodies. Signals were detected by chemiluminescence using LiteAblot Extend (EuroClone) and the ChemiDoc XRS+ System (Biorad).
Chromatin immunoprecipitation
ChIP was performed as described (Vernarecci et al. 2008): 100 ml of an exponentially growing culture (OD600 0.3–0.6) were fixed for 60 min with 1% formaldehyde, then harvested and washed twice in 20 ml of Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 150 mM NaCl). Lysis was performed in 500 μl ice cold lysis buffer (1 mM EDTA, 50 mM HEPES, 140 mM NaCl, 1% Triton X-100, 1 mg/ml Na deoxycholate) with glass beads by vortexing 10 times for 30 sec at 4°. Chromatin was collected and shared by sonication (four times, 30 sec ON/30 sec OFF on ice) using a Branson Digital Sonifier (Branson Ultrasonic Corporation, Danbury, CT) on 10% impulse (average fragment size 500 bp) then clarified for 5 min in a microcentrifuge. For immunoprecipitation, solubilized chromatin was incubated overnight at 4° on a rotating wheel with 10 µl of G-conjugated Dynabeads (Invitrogen) that had been prebound with anti-myc or anti-HA antibodies. Immunoprecipitates were collected using a step-wise washing protocol. The immunocomplexes were eluted by adding 0.25 ml Elution buffer (50 mM Tris-Cl, pH 7.5, 10 mM EDTA, 1% SDS), and incubated at 65° for 10 min. Eluates were phenol chloroform and ethanol precipitated. Immunoprecipitated DNA was analyzed by real-time qPCR (Bioline, Sensimix, QT615-02). Real-time qPCR was performed using a BioRad iCycler iQ (BioRad).
Primer sequences:
CEN3F 5′-gatcagcgccaaacaatatgg-3′, CEN3R 5′-aacttccaccagtaaacgtttc-3′
TEL6F-5′ccataatgcctcctatatttagccttt-3′, TEL6 R 5′-tgacatatccttcacgaatattgttaga-3′
NTSF-5′atgttcagtaggtgggagtgagag-3′, NTSR-5′catcccggtgccgtaaatgcaaaac-3′
Data analysis and statistical analyses
Each experiment represents the average of at least three different biological replicates unless otherwise stated in the figure legend. Standard errors are shown and asterisks are as follows: ** P < 0.01, * < 0.05. ChIP experiment analysis: equal volumes from input and immunoprecipitated samples were subjected to real-time qPCR. Reactions were carried out in duplicate in 25 μl volumes. The enrichment of a given target sequence was determined as the fold difference between the amount of target sequence in the immunoprecipitated fraction and the amount of target sequence in the input DNA (IP/Input). In each ChIP experiment, fold difference values obtained for the WT strain, with each primer pair, were used for standardization. For this reason, the WT values are set to one and do not present error bars. Average fold enrichment was thus calculated for ubp8Δ and psh1Δ strains and error bars represent the standard errors of the set of values obtained for each PCR amplicon position. Background immunoprecipitation with nonspecific antibody (anti-HA) and no antibody was also determined for each experiment and subtracted to the specific signal for each primer pair. Cycloheximide protein analysis: signals obtained for Cse4-myc and ADA2 were quantified with Optiquant densitometry software. Each value obtained for Cse4-myc at a specific time point was normalized to its equivalent ADA2 signal. For each yeast strain tested, the protein levels were referred to the respective time zero (no cycloheximide).
Data availability
Strains are available upon request.
Results
Mitotic stability is compromised by loss of Ubp8
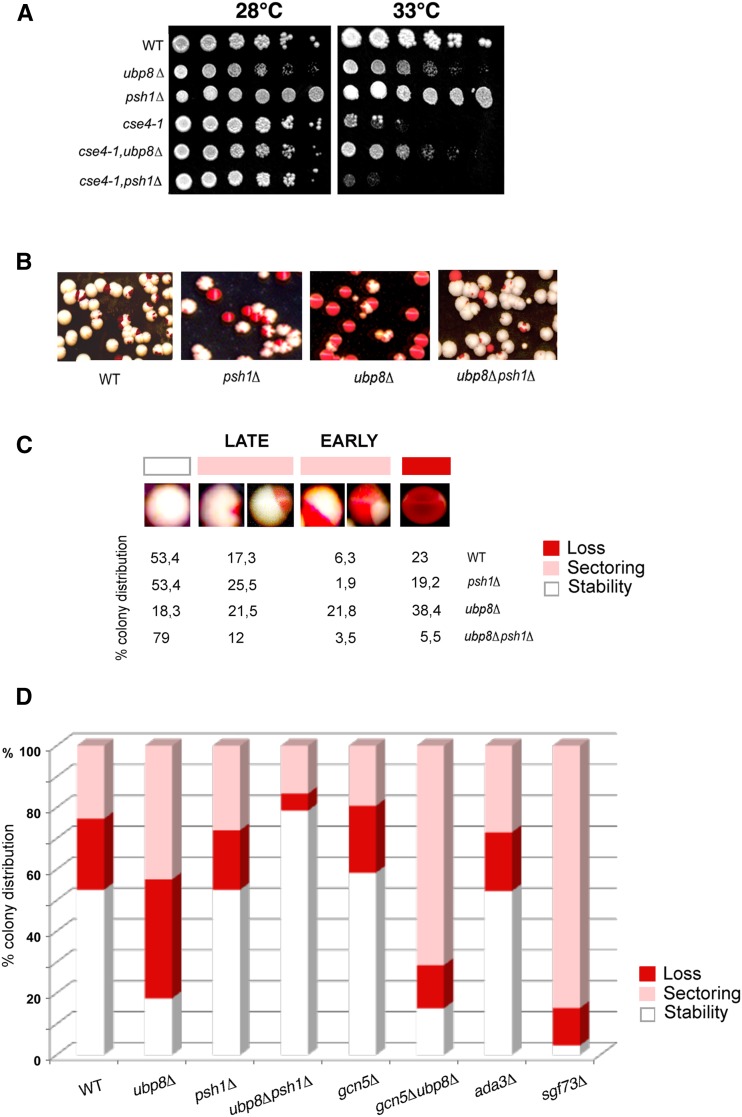
It has been shown that the SAGA DUB subunit Ubp8 deubiquitylates histone H2B (Ingvarsdottir et al. 2005). We wanted to investigate whether Ubp8 might also be functionally active on the centromeric histone variant Cse4. We first assayed, at the genetic level, putative interactions between a mutant carrying the TS-allele cse4-1 and deletion of the DUB Ub-protease Ubp8 and the E3-Ub ligase Psh1, respectively. The growth spot assay in Figure 1A clearly shows that deletion of Ubp8 at a nonpermissive temperature (33°) rescues the growth defect of the TS-allele cse4-1, as shown in the cse4-1,ubp8Δ mutant, suggesting a genetic interaction between Ubp8 and Cse4. Psh1 deletion at a nonpermissive temperature appeared to be synthetically lethal in the cse4-1,psh1Δ mutant, confirming a genetic correlation between Psh1 and Cse4. Lack of Ubp8 rescues the temperature sensitivity of the cse4-1 mutant. Starting from this evidence, we followed several experimental lines in order to demonstrate the genetic interaction found. We examined whether lack of Ubp8 might affect mitotic stability. Using a color-sectoring assay, we were able to visualize the segregation of a stably inherited centromeric plasmid (Zhang et al. 2008). The CEN plasmid was intentionally chosen in order to provide an assay to measure the behavior of the centromere, more than an absolute mitotic stability value. Strains deleted in different SAGA components were grown on YPD complete solid medium, cells with defects in plasmid maintenance and transmission appeared red or partially sectorialized (white/red), while those stably segregating appeared white. WT, psh1Δ, ubp8Δ and ubp8Δpsh1Δ strains are shown in Figure 1B. The percentage of stably segregating white cells was calculated for each strain and compared to the sectorialized and red colonies (Figure 1C). The fraction of white stably segregating colonies was very similar between the WT and psh1Δ strains, confirming the absence of mitotic defects in the Psh1-deleted mutant (Hewawasam et al. 2010). Surprisingly, the ubp8Δ strain showed a drastic increase in plasmid loss, with most of the colonies being sectorialized or red, clearly indicating a significant increase in centromere instability. Interestingly, the double ubp8Δpsh1Δ strain (Figure 1, B–D) completely rescued loss of the CEN plasmid found in the ubp8Δ strain, thus excluding additional indirect effects due to Ubp8 deletion and indicating an epistatic link between Psh1 and Ubp8. Strains deleted in other SAGA components were similarly analyzed. Deletion of the HAT Gcn5 alone did not produce mitotic defects while CEN plasmid loss was drastically increased in the double gcn5Δubp8Δ strain (Figure 1D), underlying once more the effect of Ubp8. We also tested the deletion effects of Sgf73, and of the transcriptional adaptor Ada3 (Figure 1D). The instability observed in the sgf73Δ strain is again linked to Ubp8, since this subunit is required for correct incorporation of Ubp8 in the DUB module (Samara et al. 2010). Lack of Ada3, instead, had no effect, ruling out other putative SAGA-dependent transcriptional defects. This analysis suggests that the centromere instability defect can be specifically attributed to absence of the DUB-Ubp8 deubiquitylase. Collectively, our data show that the histone variant Cse4 interacts at the genetic level with Psh1 and DUB-Ubp8, and that deletion of Ubp8 significantly increases the loss of a stably inherited minichromosome.
Figure 1.
Ubp8 interacts with CEN-H3 TS-allele cse4-1 at the genetic level and its loss increases mitotic stability. (A) Comparative growth spot assay of fivefold serial dilutions of WT, ubp8Δ; psh1Δ; cse4-1; cse4-1, ubp8Δ; cse4-1, psh1Δ strains plated on YPD and grown at permissive (28°) and nonpermissive (33°) temperatures for 2 days. Strain with TS-allele cse4-1 was assayed in the absence of DUB-Ubp8 (cse4-1, ubp8Δ) and E3-Ub ligase Psh1 (cse4-1, psh1Δ). (B) WT, ubp8Δ, psh1Δ and ubp8Δpsh1Δ strains carrying the stable ADE/LEU pRS425-1225 CEN/ARS plasmid were grown in selective medium (leu), plated on YPD, grown, and stored at 4° for red color accumulation. (C) Cell percentage (%) showing stable, sectorialized (late and early minichromosome loss), and unstable mitotic phenotype in WT, ubp8Δ, psh1Δ and ubp8Δpsh1Δ. (D) Plasmid loss rates of WT, psh1Δ, ubp8Δ, ubp8Δpsh1Δ, gcn5Δ, gcn5Δubp8Δ, sgf73Δ and ada3Δ strains. Plasmid loss is expressed as the relative percentage of white, red, and sectorialized colonies grown on plates (400-600 colonies counted for each strain). WT, wild-type; YPD, yeast extract peptone dextrose.
Ubp8 interacts with Cse4 at the physical level
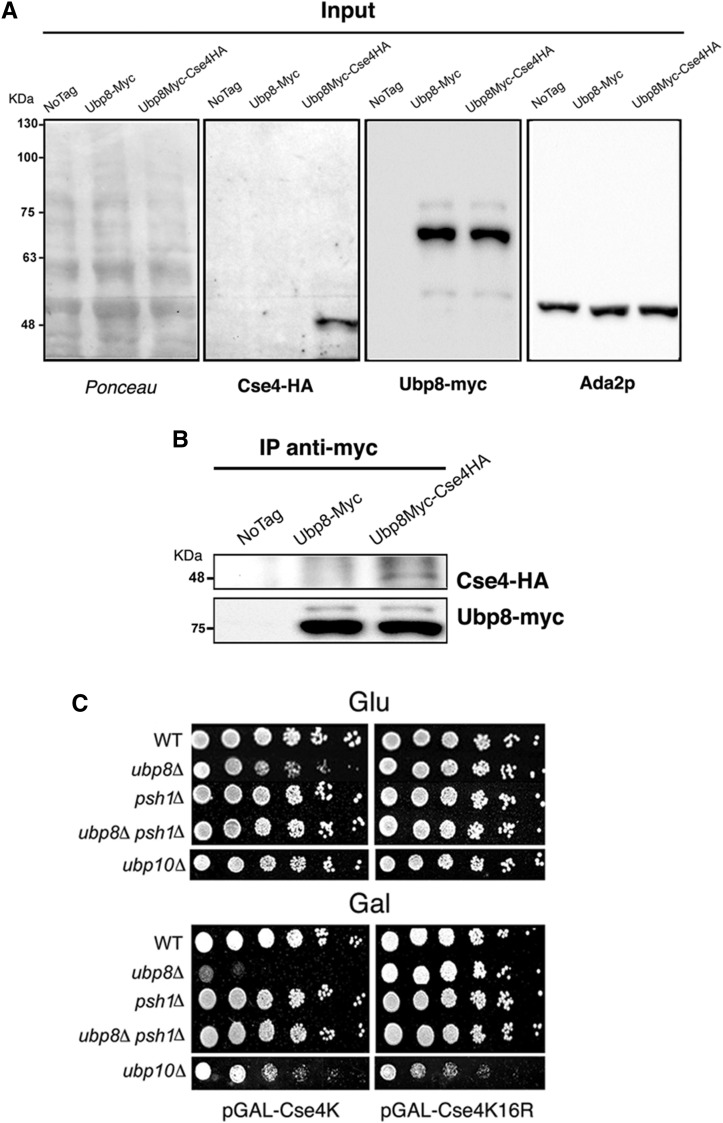
Building on these genetic results, we wanted to exclude off-target effects and determine whether a direct physical interaction existed between Ubp8 and Cse4. The DUB module is composed of the Sgf73 (SaGa associated factor, 73 kDa), Sgf11 (SaGa associated factor 11 kDa), Sus1 (Sl gene upstream of ySa1), and Ubp8 subunits. Sgf73 is required for the incorporation of Ubp8 in the complex and interacts with the structural subunit Spt7 (Han et al. 2014). It has been reported that Spt7 itself interacts with Cse4 (Ranjitkar et al. 2010), suggesting that a direct interaction between the SAGA complex and Cse4 exists. To provide evidence of a physical interaction between Ubp8 and Cse4, coimmunoprecipitation (coIP) was performed on a double epitope-tagged strain carrying both Cse4-6HA and Ubp8-9myc. A single Ubp8-9myc-tagged strain was used as a control. Correct expression of tagged proteins was verified by western blot analysis of total extracts (Figure 2A). Cse4 is not an abundant protein and the tagged Cse4-HA assayed is expressed at physiological levels. Nevertheless, when immunoprecipitation of Ubp8-myc was performed, using an anti-myc antibody, a specific signal corresponding to Cse4-6HA (48 kDa) was observed in the double-tagged strain (Figure 2B) but not in the two negative controls (the untagged and the single Ubp8-9myc strains). Ubp8-myc (75 kDa) was successively detected in both immunoprecipitated strains, confirming that anti-myc immunoprecipitation was successful. Taken together, these results demonstrate that Ubp8 interacts with Cse4 at the genetic and physical levels.
Figure 2.
Ubp8 interacts with Cse4 at a physical level and acts on Cse4 lysines. (A) Inputs from WT (wild-type), Ubp8-myc, and Ubp8myc-Cse4HA strains were run on 10% PAGE, transferred on nitrocellulose membrane, stained with red-ponceau, and consecutively probed with anti-HA (Cse4-HA), anti-myc (Ubp8-myc), and anti-ADA2 (internal protein control) antibodies. (B) Coimmunoprecipitation (coIP) carried out on whole cell extracts shown in (A) indicates that Ubp8-myc is able to coIP Cse4-HA in the double-tagged strain. (C) Growth spot assay. Fivefold serial dilutions of WT, ubp8Δ, psh1Δ, ubp8Δ-psh1Δ, and ubp10Δ strains expressing pGAL-Cse4 or pGAL-Cse4K16R were plated in glucose (GLU) or inducing galactose (GAL) SC-media and grown for 5 d at 28°.
Ubp8 acts on Cse4 lysines, and absence of E3-ubiquitin ligase Psh1 rescues Ubp8 deletion
Our results indicate that Ubp8 genetically interacts with Cse4 and that its absence enhances mitotic instability. Since Cse4 is ubiquitylated and Ub-dependent proteolysis regulates Cse4 localization, we speculated that Ubp8 could be directly engaged in targeting Cse4 lysines. We used a strain overexpressing a WT copy of Cse4 under the control of a Gal promoter (pGAL-Cse4) or a lysine-free version carrying substituted arginines (pGAL-Cse4K16R) that prevents ubiquitylation (Au et al. 2008; Mena et al. 2009). pGAL-Cse4 and pGAL-Cse4K16R were tested in strains lacking the ubiquitin proteases ubp8Δ and ubp10Δ or the E3-Ub ligase psh1Δ. The results are shown in the growth spot assay under noninducing (glucose) or inducing (galactose) conditions (Figure 2C). Overexpression of pGAL-Cse4 was clearly synthetically lethal in ubp8Δ but completely ineffective if the unmodifiable version pGAL-Cse4K16R was expressed. Interestingly, the double-deleted strain ubp8Δpsh1Δ expressing pGAL-Cse4 showed a growth rescue comparable to that obtained by expressing the pGAL-Cse4K16R version in the ubp8Δ strain. Lack of growth defects in the Cse4K16R-substituted mutant or in absence of the Ub-ligase Psh1 clearly indicate a selective role for Ubp8 on Cse4 lysines. It was reported that histone H2B-K123Ub is deubiquitylated by both DUBs, Ubp8 and Ubp10 (Schulze et al. 2011). To assay a putative role of Ubp10 on Cse4, both pGAL-Cse4 and pGAL-Cse4K16R were overexpressed in the ubp10Δ strain. No effect was observed, indicating that Ubp8 and not Ubp10 targets Cse4. Collectively, these results indicate that Ubp8 is acting on Cse4 lysines, since its deletion is ineffective when these are substituted by unmodifiable arginines (Cse4K16R) or when Ub-ligase Psh1 is eliminated and ubiquitin is not deposed. These data reinforce the epistasis and functional interaction between Psh1 and Ubp8, suggesting a role for ubiquitin protease Ubp8 in Cse4 deubiquitylation.
Ubiquitin mediated proteolysis of Cse4 is enhanced in the absence of Ubp8
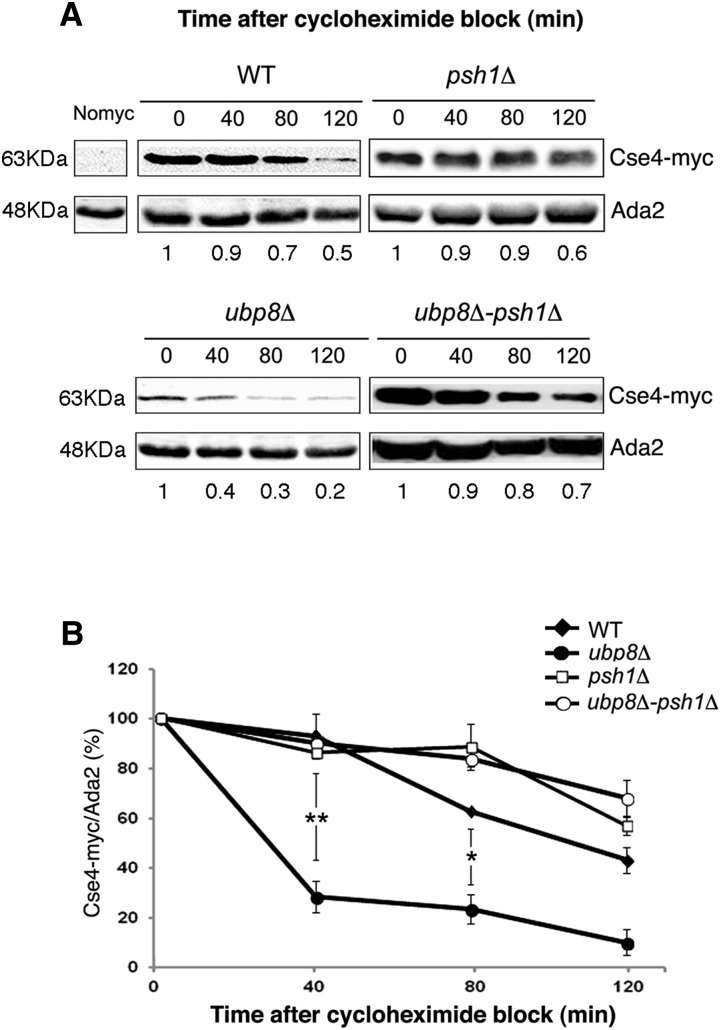
CenH3 Cse4 is an unstable protein whose ubiquitin-mediated proteolysis contributes to its exclusive localization at the centromere (Collins et al. 2004). Since our data provided evidence for both genetic and physical interactions between Ubp8 and Cse4, we reasoned that DUB Ubp8 might play a direct role in Cse4 deubiquitylation and thus interfere with the protein degradation rate. Cse4-myc steady-state levels were assessed upon de novo protein translation block with cycloheximide in WT, ubp8Δ, psh1Δ, and ubp8Δpsh1Δ strains (Figure 3A). Interestingly, in the absence of Ubp8, Cse4-myc levels were drastically reduced after 40 min of treatment (Figure 3A). Previous reports have shown that proteolytic degradation depends on lysine ubiquitylation (Ranjitkar et al. 2010); similarly, we noted that Cse4p is stabilized in psh1Δ cells (Figure 3A). Based on these observations, we expected that a subsequent deletion of the E3-Ub ligase Psh1 in the ubp8Δ strain would reverse Cse4p faster proteolysis. Consistent with this assumption, Cse4p was highly stabilized and its degradation significantly reduced in the ubp8Δpsh1Δ strains. Band intensities were quantified and the results summarized in the histograms in Figure 3B. The y axis reports the relative amount of Cse4-myc/ADA2 at each point referred to the 100% value at time 0 for each strain. This observation, our genetic data and coimmunoprecipitation experiments all suggested a direct link between DUB-Ubp8 and the centromeric histone Cse4. Moreover, the strong recovery in protein stability observed in the double-deleted ubp8Δpsh1Δ strain clearly indicated a role for Ubp8 in removing ubiquitin from Cse4 lysines downstream of Psh1. These results prompted us to investigate the status of Cse4 ubiquitylation at the biochemical level in order to directly demonstrate this novel activity of the Ub-protease Ubp8.
Figure 3.
Lack of Ubp8 causes faster proteolysis of Cse4-myc rescued by concomitant deletion of the E3-Ub ligase Psh1. (A) Proteins were extracted after adding cycloheximide (CHX) (10 μg/ml) from WT (wild-type), ubp8Δ, psh1Δ, and ubp8Δpsh1Δ strains containing an integrated copy of Cse4-myc. Lysates prepared at indicated time points (min) were probed with anti-myc and anti-ADA2 antibodies. The relative amount of Cse4-myc at each time point was quantified and normalized to Ada2 levels as indicated under each lane. ADA2-normalized Cse4 protein levels at each time point of CHX treatment are presented as a fraction of the remaining amount relative to the sample t = 0. (B) The graph shows the percentage of Cse4-myc signal normalized to Ada2 in each strain at the indicated time points. WT closed diamonds, ubp8Δ closed circles, psh1Δ open squares, ubp8Δ-psh1Δ open circles. Standard errors of the means are shown. Asterisks are as follows: ** P < 0.01, * < 0.05.
Ubp8 deubiquitylates Cse4 and removes a short Ub-oligomer
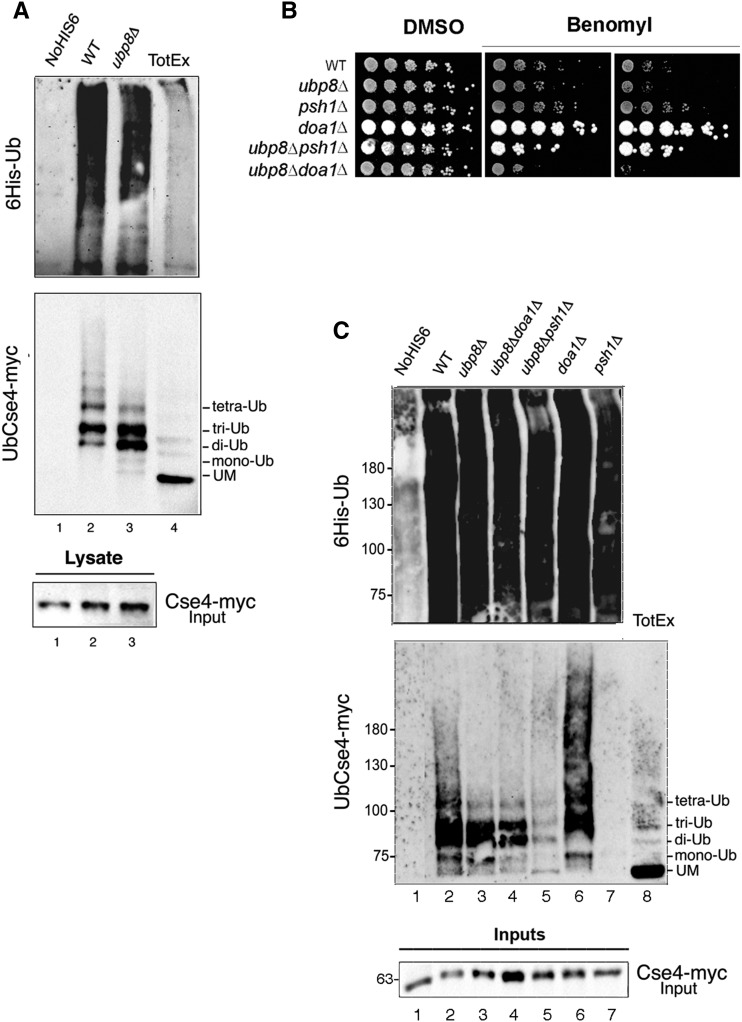
Polyubiquitination is a complex and versatile posttranslational modification, in which ubiquitin is linked to a specific lysine to form distinct polymers characterized by multivalent and polybranched ubiquitin chains. It has been proposed that many DUBs, such as yeast Ubp12 and Ubp15 or human USP7, preferentially remove monoubiquitin residues from their target substrates (Schaefer and Morgan 2011), thus opposing chain elongation by the E3 ligases. In yeast, Cse4 is ubiquitylated by Psh1 at its C-terminus in vitro (Hewawasam et al. 2010; Ranjitkar et al. 2010). Moreover, it has been reported that Doa1, a regulator of ubiquitin homeostasis, may also contribute to its ubiquitylation (Au et al. 2013). Live microscopy has then revealed a de novo replacement of Cse4 at the centromere in S-phase, underlying the dynamic behavior of the unstable Cse4p during the cell cycle (Wisniewski et al. 2014). These observations and the ubiquitin-mediated proteolysis of Cse4 advocate a precise role for deubiquitylation in Cse4 regulation. Our findings suggest that Ubp8 interacts with Cse4 by acting on its lysines. Remarkably, lack of Ubp8 causes faster proteolytic degradation of the centromeric histone, which is reverted by deletion of the E3-Ub ligase Psh1 (Figure 3). Since our results point to a novel deubiquitylating activity of Ubp8 on Cse4, we wanted to provide an unbiased, biochemical demonstration of the variations in the Cse4 polyubiquitylation pattern. Yeast strains expressing histidine-tagged ubiquitin (PCUP1:6HIS-Ubiquitin plasmid) and carrying Cse4-myc were created and analyzed (Iglesias et al. 2010). 6His-Ub-proteins were purified on nickel columns and analyzed by immunoblotting. Upon induction of the PCUP1 promoter (CuSO4+), tagged ubiquitin was efficiently incorporated and retained on a Ni+column. A regular ladder of 6His-Ub-Cse4-myc conjugate, corresponding to 9.5 kDa multimers, was obtained in the WT strain, confirming an efficient purification step (Figure 4A lane 2; Supporting Information, Figure S1, lanes 3 and 7). Protein extracts from a control WT strain lacking 6His-Ub, and WT and ubp8Δ strains expressing the tagged ubiquitin, were eluted and run in parallel with a WT total extract (Figure 4, A and C). Absence of Ubp8 induced a clear change in the Cse4-myc ubiquitylation pattern. The high molecular weight ladder corresponding to longer polyubiquitin chains found in the WT strain (Figure 4, A and C, lane 2) was absent in the ubp8Δ strain (Figure 4, A and C, lane 3) which, instead, presented a strong increase in di and tri Ub-Cse4-myc oligomers (Figure 4, A and C, lane 3). This finding suggests that Ubp8 removes a Ub oligomer that may correspond to a K48-linked short Ub-chain, which has a recognized role in proteasome degradation (Kulathu and Komander 2012), in full agreement with the proteolysis pattern obtained (Figure 3). Published results suggest that Doa1 might have a role in Cse4 ubiquitylation (Au et al. 2013). We therefore reasoned that it may also affect Ubp8 activity. Microtubule shock under benomyl growth is a useful assay to assess centromeric or mitotic defects. Growth spot assays in the presence of the microtubule interfering agent were therefore performed in ubp8Δ, psh1Δ, doa1Δ, and the respective double mutants ubp8Δpsh1Δ and ubp8Δdoa1Δ strains (Figure 4B). Deletion of Ubp8, alone or in combination with Doa1, inhibited growth suggesting that Ubp8 and Doa1 may act on Cse4 through two independent pathways. In contrast, deletion of Psh1 and Ubp8 showed amelioration of the growth phenotype, thus emphasizing the epistatic link between Psh1 and Ubp8 previously shown (Figure 2A). To further investigate the effects of these deletions, we analyzed Cse4 polyubiquitylation patterns in the entire set of mutant strains (Figure 4C). In the ubp8Δpsh1Δ strain, the high molecular weight poly-Ub ladder was almost completely cleared, leaving a minor lasting signal corresponding to the short Ub oligomer (Figure 4C, lane 5). This persistent short ubiquitin ladder may represent a Psh1-independent polyubiquitylation mechanism, whose effector remains to be discovered and that is not fully degraded in the absence of Ubp8. The polyubiquitylation pattern was also analyzed in the ubp8Δdoa1Δ double mutant strain (Figure 4C, lane 6). Persistency of the short ubiquitin chain indicated that Doa1 is not directly implicated in deposing the Ub-trimer on Cse4 and excluded any putative link between Ubp8 and Doa1. We cannot rule out, however, that Doa1 might work in parallel with Psh1, ultimately contributing to ubiquitin homeostasis. As expected, deletion of Psh1 completely eliminated the Cse4 poly-Ub ladder (Figure 4C, lane 7), suggesting that, when the E3 ubiquitin ligase is not present and the long poly-ubiquitin mark is not deposed, Ubp8 processes the residual short Ub-chain. The increase in an ubiquitin oligomer obtained in the absence of Ubp8 directly indicates that SAGA DUB Ubp8 is responsible for its processing and removal from Cse4. The absence of a high molecular weight poly-Ub ladder in the ubp8Δ-deleted strain further implies that the presence of a Ub oligomer may interfere with and impair the deposition of additional ubiquitin molecules and prevent the formation of longer poly-Ub chains by Psh1 (Figure 4D).
Figure 4.
SAGA-DUB Ubp8 removes a short ubiquitin oligomer from the centromeric histone Cse4. (A) Cse4-myc tagged strains containing no vector, lane 1, or a PCUP1:6HIS-Ubiquitin plasmid (WT, 2; ubp8Δ, 3). 6HIS-Ub conjugates purified on a Ni+ column were run in parallel with a wild-type (WT) total extract, lane 4. Western blot was sequentially hybridized with anti-myc and anti-6His antibodies; 1/20th of total extracts were run separately and hybridized with anti-myc for Input quantitation (lanes 1–3). (B) Growth spot assay. Fivefold serial dilutions of WT, ubp8Δ, psh1Δ, doa1Δ, and ubp8Δdoa1Δ strains were plated on YPD-DMSO and YPD-benomyl (12 and 18 μg/μl, respectively) and grown for 2 d at 28°. (C) The same procedure as in (A) was carried out with, respectively, WT (2); ubp8Δ (3); ubp8Δdoa1Δ (4); ubp8Δpsh1Δ (5); doa1Δ (6); and psh1Δ (7). Western blot was sequentially hybridized with anti-myc and anti-6His antibodies; 1/20th of total extracts were run separately and hybridized with anti-myc for Input quantitation. YPD, yeast extract peptone dextrose; DMSO, dimethyl sulfoxide.
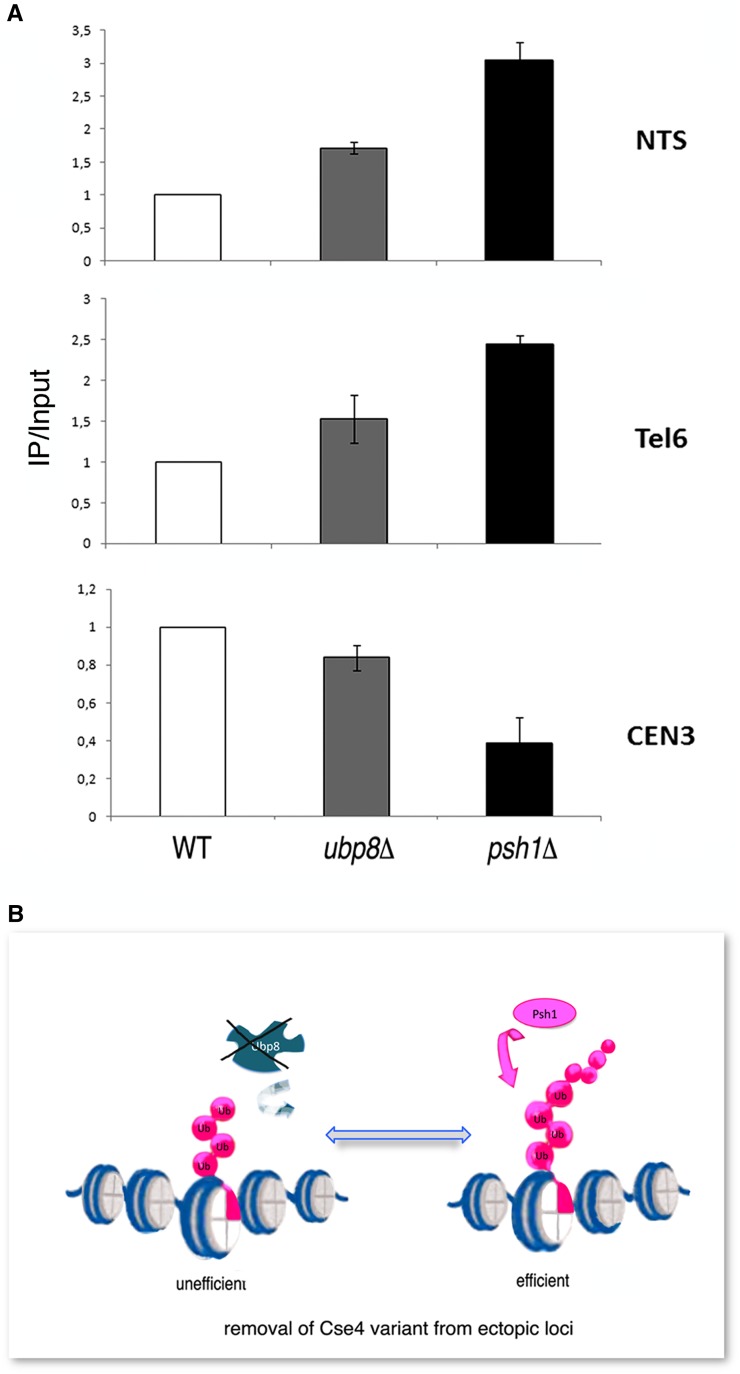
Ectopic localization of CenH3-Cse4 is enhanced in ubp8Δ strain
Cse4 is temporarily dislodged from the centromere in early S-phase and its loading is dynamically regulated (Zhou et al. 2011). The ubiquitin chain represents the recognition signal for Cse4 removal, which is fundamental in avoiding dangerous misincorporations outside of the centromere (Hewawasam and Gerton 2011). Consequently, in the absence of E3-Ub ligase Psh1, overexpressed Cse4 is increasingly found at ectopic chromosomal loci, underlining the role of ubiquitin in CenH3 restricted localization (Hewawasam et al. 2010). Based on our collected results, we decided to assay whether the presence of the short Ub-oligomer accumulating in the absence of Ubp8 might be able to affect CenH3-Cse4 localization on the chromosome. Chromatin immunoprecipitation (ChIP) was performed in WT, ubp8Δ, and psh1Δ strains carrying an integrated copy of myc-tagged Cse4 expressed at physiological levels (Figure 5A). Chromatin was extracted, purified, and immunoprecipitated with an anti-myc antibody. Quantitative real-time PCR was employed to investigate Cse4 distribution at the centromeric locus CEN3, and at the two noncentromeric sites rDNA-NTS (ribosomal region) (Camahort et al. 2009) and Tel6 (telomere). ChIP analyses were carried out in at least three biological replicates. In each experiment, amplification values obtained for each primer pair in the WT strain were used to normalize the values obtained for the ubp8Δ and psh1Δ strains. WT values were therefore set equal to one. Our results showed an increase in Cse4 localization at the ectopic sites rDNA-NTS and Tel6, both in the ubp8Δ and psh1Δ strains compared to WT (Figure 5A, NTS and Tel6 panels). The increase of ectopic localization in the absence of Cse4 ubiquitylation in a psh1Δ asynchronous cell population had been previously reported (Hewawasam et al. 2010), while it had never been described in association with the deletion of the ubiquitin protease Ubp8. We have shown that Ubp8 is required for the removal of a short Ub-oligomer on Cse4 (Figure 4A), which counteracts further polyubiquitylation by Psh1 (Figure 4C). Collectively these findings suggest that the short Ub oligomer, accumulating in the absence of Ubp8, represents a poor signal for the efficient removal of Cse4 from ectopic sites, enlightening a novel step in the control and regulation of CenH3 localization at the centromere in budding yeast (Figure 5B).
Figure 5.
Loss of Ubp8 increases mislocalization of Cse4 at rDNA-NTS and Tel6 regions. (A) ChIP, followed by qPCR, was performed to analyze Cse4-myc localization in wild-type (WT), ubp8Δ, and psh1Δ strains in order to determine Cse4 localization at the centromere (Cen3) and at the ectopic sites rDNA-NTS and telomere Tel6. The values on the y-axis are arbitrary units and represent the fold difference between the amount of target sequence in the immunoprecipitated fraction and the amount of target sequence in the input DNA (IP/Input). Standard error of the mean is shown. (B) Model depicting inefficient removal of Ub-oligomer-Cse4 from ectopic loci in the absence of DUB-Ubp8.
Discussion
Centromere identity is epigenetically defined by targeted and exclusive recruitment of a specialized histone variant, Cse4, in budding yeast. Polyubiquitylation of Cse4 by the E3 RING finger ligase Psh1 has been widely studied, and has been recognized as a central mechanism ensuring the localization of Cse4 at centromeric regions and its removal from ectopic sites (Hewawasam et al. 2010; Hewawasam and Gerton 2011). The peculiar lack of any mitotic defect induced by deletion of Psh1 led us to hypothesize the existence of additional processes involved in Cse4 regulation that were yet to be discovered (Folco and Desai, 2010). Therefore, the aim of our study was to elucidate the effects of deubiquitylation, its underlying mechanism, and its putative role in centromeric histone deposition and turnover. In S. cerevisiae, the SAGA complex DUB subunit Ubp8 is implicated in the deubiquitylation of histone H2B (Henry et al. 2003). The human ortholog Usp22 has been shown to be able to act on other substrates, including SIRT1 (Lin et al. 2012). In this investigation, we report the novel finding that Ubp8 is implicated in the deubiquitylation of the centromeric histone variant Cse4. We found that deletion of Ubp8 and Psh1 genetically interacts with a cse4-1 mutation, that Ubp8 interacts with Cse4 at the physical level, and that its loss induces high mitotic instability, as shown by an ARS/CEN minichromosome assay. We demonstrated that Ubp8 selectively acts on Cse4 lysines by showing that its deletion is lethal if wild-type Cse4 (pGAL-Cse4) is overexpressed, but that it becomes ineffective on an arginine substituted version pGAL-Cse4K16R or when Cse4 ubiquitylation is abrogated by deletion of the Ub-ligase Psh1. The results obtained from the proteolytic profile of Cse4-myc (Figure 3) support these findings and show faster Cse4p proteolysis in the ubp8Δ strain, rescued by the subsequent deletion of Psh1. The lethal overexpression of pGAL-Cse4 in the Upb8-deleted strain can be retrospectively attributed to an increased amount of an altered Cse4 version marked by a short Ub-chain (3/4mer) as shown in Figure 4. We can hypothesize that the persistence in the cell of an anomalous version of Ub-Cse4 may also affect its function. Polymerization of polyubiquitin chains is affected by the choice of the lysine linkage that ultimately determines its structure. The variety of ubiquitin chain versions represent not only a signal for the proteasome but also a versatile three-dimensional code, controlling many different cellular functions (Heride et al. 2014). There is still a great debate regarding the precise role of Ub-chain species, how their different chain length and linkage type can affect the way DUBs process them, and the meaning and translation of this ubiquitin code into biological processes (Shekhawat et al. 2014). The investigation of the mechanisms and the players involved represent a great challenge for the future. Between the vast array of poly-Ub chains that can be assembled on the seven ubiquitin’ lysines, for example, K63 poly-Ub is not a substrate for proteolysis, as reported for NF-κB (Chen and Chen 2013), while a minimal chain length at K48 signals for degradation (Thrower et al. 2000; Braun and Madhani 2012). In this study we aimed to demonstrate a novel role for the Ub-protease Ubp8 in the deubiquitylation of Cse4, an epistatic link between E3Ub-ligase Psh1 and Ubp8, and the involvement of this process in centromere function. We have provided an unbiased demonstration at the biochemical level that, in the absence of Ubp8, Cse4 is marked by a short Ub-oligomer. Our analysis reveals a variation in Cse4 ubiquitylation and in the length of ubiquitin chain upon Ubp8 deletion. We also observe that, in the ubp8Δpsh1Δ strain Cse4, polyubiquitylation is almost completely cleared, strengthening the epistasis between Psh1 and Ubp8. Based on our proteolysis data, we suggest that the short Ub-oligomer accumulating on Cse4 in the ubp8Δ strain may likely correspond to a Ub-K48 chain that does not preclude proteolytic degradation (Sadowski and Sarcevic 2010). We propose that DUB Ubp8 is implicated in removing this short Ub-oligomer and we speculate that, in the presence of the short Ub chain, Psh1 activity or subsequent rounds of ubiquitin deposition may be impaired. Indeed, it has been reported that Ub-K48 is able to block further chain elongation on Met-4 (Flick et al. 2004). The accuracy of variant nucleosome incorporation at the centromere is of fundamental importance in the control of balanced chromosome segregation in mitosis. Therefore, CenH3 mislocalization is both dangerous and has to be avoided. We asked whether the anomalous short Ub-oligomer accumulating on Cse4 in the absence of Ubp8 might affect its deposition across the chromosome. Our ChIP data clearly show that the short Ub-oligomer is not very effective in signaling Cse4 removal and, accordingly, it increases localization at ectopic positions. Collectively, we demonstrate, for the first time, a novel function of the SAGA DUB subunit Ubp8 as a Ub-protease involved in the removal of a short ubiquitin chain on Cse4 affecting its proteolysis and deposition. A future challenge will be to investigate whether the short Cse4 Ub-oligomer may also be engaged in other epigenetically regulated pathways.
Supplementary Material
Acknowledgments
We thank Giuseppe Pisaneschi of IBPM-CNR (Institute of Molecular Biology and Pathology-National Research Council), Rome, for his technical assistance. We also thank S. Biggins, C. Bonnet, and C. Dargemont for providing the yeast strains, 6His-Ub expression plasmids and protocols. This work was supported by The Institute Pasteur Cenci Bolognetti Foundation, C.C. was supported by a FIRB (Basic Research Investment Fund)-Cenci Bolognetti international grant. The authors have no conflict of interest to declare.
Author contributions: P.F., P.B. and C.C. conceived the study and designed the experiments. C.C., S.V., M.I., C.B., and C.M. performed the experiments. C.C., P.F., and S.V. analyzed the results. P.F. and C.C., with input from S.V. and P.B., wrote the manuscript.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.024877/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Au W. C., Crisp M. J., DeLuca S. Z., Rando O. J., Basrai M. A., 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179(1): 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Dawson A. R., Rawson D. W., Taylor S. B., Baker R. E., et al. , 2013. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 194(2): 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., Sánchez P., Losada A., 2009. Epigenetic specification of centromeres by CENP-A. Exp. Cell Res. 315(19): 3233–3241. [DOI] [PubMed] [Google Scholar]
- Braun S., Madhani H. D., 2012. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 13(7): 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S., 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. The Journal of cell biology 160(3): 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., et al. , 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35(6): 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chen Z. J., 2013. Regulation of NF-kappaB by ubiquitination. Curr. Opin. Immunol. 25(1): 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Furuyama S., Biggins S., 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14(21): 1968–1972. [DOI] [PubMed] [Google Scholar]
- Collins K. A., Camahort R., Seidel C., Gerton J. L., Biggins S., 2007. The overexpression of a Saccharomyces cerevisiae centromeric histone H3 variant mutant protein leads to a defect in kinetochore biorientation. Genetics 179(1): 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. A., Torok M. S., Sun Z. W., Schieltz D., Allis C. D., et al. , 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279(3): 1867–1871. [DOI] [PubMed] [Google Scholar]
- Flick K., Ouni I., Wohlschlegel J. A., Capati C., McDonald W. H., et al. , 2004. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 6(7): 634–641. [DOI] [PubMed] [Google Scholar]
- Folco H. D., Desai A., 2010. A PSHaver for centromeric histones. Mol. Cell 40(3): 351–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S., Biggins S., 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104(37): 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Luo J., Ranish J., Hahn S., 2014. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33(21): 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., et al. , 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17(21): 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heride C., Urbe S., Clague M. J., 2014. Ubiquitin code assembly and disassembly. Curr. Biol. 24(6): R215–R220. [DOI] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M. D., Weiss S., Skora A. D., et al. , 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10(3): 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G. S., Gerton J. L., 2011. Cse4 gets a kiss-of-death from Psh1. Cell Cycle 10(4): 566–567. [DOI] [PubMed] [Google Scholar]
- Hewawasam G., Shivaraju M., Mattingly M., Venkatesh S., Martin-Brown S., et al. , 2010. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol. Cell 40(3): 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N., Tutucci E., Gwizdek C., Vinciguerra P., Von Dach E., et al. , 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24(17): 1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K., Krogan N. J., Emre N. C., Wyce A., Thompson N. J., et al. , 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25(3): 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21(11): 947–962. [DOI] [PubMed] [Google Scholar]
- Keith K.C., Fitzgerald-Hayes M., 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics 156 (3):973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockner C., Schneider M., Lutz S., Jani D., Kressler D., et al. , 2009. Mutational uncoupling of the role of Sus1 in nuclear pore complex targeting of an mRNA export complex and histone H2B deubiquitination. J. Biol. Chem. 284(18): 12049–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., et al. , 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15(10B): 963–972. [DOI] [PubMed] [Google Scholar]
- Kohler A., Pascual-Garcia P., Llopis A., Zapater M., Posas F., et al. , 2006. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell 17(10): 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y., Komander D., 2012. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13(8): 508–523. [DOI] [PubMed] [Google Scholar]
- Li Z. H., Yu Y., Du C., Fu H., Wang J., et al. , 2013. RNA interference-mediated USP22 gene silencing promotes human brain glioma apoptosis and induces cell cycle arrest. Oncol. Lett. 5(4): 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lin Z., Yang H., Kong Q., Li J., Lee S. M., et al. , 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 46(4): 484–494. [DOI] [PubMed] [Google Scholar]
- Ma L., Zhai Y., Feng D., Chan T. C., Lu Y., et al. , 2010. Identification of novel factors involved in or regulating initiation of DNA replication by a genome-wide phenotypic screen in Saccharomyces cerevisiae. Cell Cycle 9(21): 4399–4410. [DOI] [PubMed] [Google Scholar]
- Maresca T. J., 2013. Chromosome segregation: not to put too fine a point (centromere) on it. Curr. Biol. 23(19): R875–R878. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D., 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11(24): 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M. A., Rodriguez-Navarro J. A., de Yebenes J. G., 2009. The multiple mechanisms of amyloid deposition: the role of parkin. Prion 3(1): 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., Azorin F., 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34(21): 6247–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M. O., Yi X., Baker R., MacCoss M. J., et al. , 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 40(3): 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M., Sarcevic B., 2010. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., et al. , 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328(5981): 1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J. B., Morgan D. O., 2011. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J. Biol. Chem. 286(52): 45186–45196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J. M., Hentrich T., Nakanishi S., Gupta A., Emberly E., et al. , 2011. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 25(21): 2242–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhawat S. S., Pham G. H., Prabakaran J., Strieter E. R., 2014. Simultaneous detection of distinct ubiquitin chain topologies by 19F NMR. ACS Chem. Biol. 9(10): 2229–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S. R., 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26(9): 3339–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9(5): 573–586. [DOI] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M., 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19(1): 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., et al. , 2003. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63(13): 3511–3516. [PubMed] [Google Scholar]
- Vernarecci S., Ornaghi P., Bagu A., Cundari E., Ballario P., et al. , 2008. Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol. Cell. Biol. 28(3): 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake V. M., Lee K. K., Guelman S., Lin C. H., Seidel C., et al. , 2008. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 27(2): 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J., Hajj B., Chen J., Mizuguchi G., Xiao H., et al. , 2014. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife 3: e02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., et al. , 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell 29(1): 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Feng H., Zhou B. R., Ghirlando R., Hu K., et al. , 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472(7342): 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request.