Abstract
Histone proteins and the nucleosomal organization of chromatin are near-universal eukaroytic features, with the exception of dinoflagellates. Previous studies have suggested that histones do not play a major role in the packaging of dinoflagellate genomes, although several genomic and transcriptomic surveys have detected a full set of core histone genes. Here, transcriptomic and genomic sequence data from multiple dinoflagellate lineages are analyzed, and the diversity of histone proteins and their variants characterized, with particular focus on their potential post-translational modifications and the conservation of the histone code. In addition, the set of putative epigenetic mark readers and writers, chromatin remodelers and histone chaperones are examined. Dinoflagellates clearly express the most derived set of histones among all autonomous eukaryote nuclei, consistent with a combination of relaxation of sequence constraints imposed by the histone code and the presence of numerous specialized histone variants. The histone code itself appears to have diverged significantly in some of its components, yet others are conserved, implying conservation of the associated biochemical processes. Specifically, and with major implications for the function of histones in dinoflagellates, the results presented here strongly suggest that transcription through nucleosomal arrays happens in dinoflagellates. Finally, the plausible roles of histones in dinoflagellate nuclei are discussed.
Keywords: chromatin, dinoflagellates, histone code, histones, transcription
A core feature of eukaryotic genome biology is the nucleosomal organization of chromatin. Nucleosomes consist of a histone octamer containing two copies of each of the four core histone proteins H2A, H2B, H3, and H4, wrapped around 147 bp of DNA (Luger et al. 1997). In addition, the linker histone H1 binds to the nucleosome and the linker DNA between individual nucleosomes.
The major exception from this almost universal organization is the dinoflagellate lineage. Dinoflagellates exhibit numerous highly unusual features, such as the organization of their mitochondrial (Waller and Jackson 2009) and plastid (Zhang et al. 1999; Barbrook and Howe 2000) genomes, but their nuclei are particularly striking (Rizzo 2003). Dinoflagellate chromatin does not exhibit a banding pattern upon nuclease digestion, it contains little acid-soluble protein (the ratio of basic proteins to DNA is 10% compared to the typical 1:1; Rizzo and Nooden 1972; Herzog and Soyer 1981), and chromosomes exist in a permanently condensed liquid crystalline state (Rill et al. 1989). Histone proteins are not readily detected in dinoflagellates, and until quite recently they were thought to be completely absent. So unusual is dinoflagellate chromatin that at one time dinoflagellates were suggested to be “mesokaryotes”, i.e., intermediate between prokaryotes and eukaryotes (Dodge 1965). We now know that dinoflagellates firmly belong to the alveolates, together with apicomplexans and ciliates, and that the loss of nucleosomes is a derived feature. But the mystery of dinoflagellate chromatin remains largely unresolved.
Several reports have identified histone-like proteins in dinoflagellates (Chan et al. 2006; Chan and Wong 2007; Wargo and Rizzo 2000; Chudnovsky et al. 2002; Sala-Rovira et al. 1991; Wong et al. 2003; Rizzo and Burghardt 1982). More recently, it was found that dinoflagellates express virus-derived nucleoproteins, completely unrelated to histones (dinoflagellate viral nucleoproteins; DVNPs), which seem to substitute for histones as far as the packaging of DNA is concerned (Gornik et al. 2012). However, multiple reports have also identified histone genes and low levels of histone proteins in several species. These include studies of transcriptomes from Lingulodinium (Roy and Morse 2012), Symbiodinium (Bayer et al. 2012), and Alexandrium catenella (Zhang et al. 2014), the draft genome sequence of S. minutum (Shoguchi et al. 2013), and environmental transcriptomes (Lin et al. 2010).
These observations suggest that histones do play some role in dinoflagellate biology, but its precise nature remains unclear. A somewhat underappreciated fact is that the loss of nucleosomes has far more profound consequences than the mere packaging of DNA, as the post-translational modifications (PTMs) of histone proteins and the “histone code” they constitute (Jenuwein and Allis 2001) play a key role in most aspects of chromatin biology. These modifications happen primarily (but not only) in the N-terminal tails of histones and serve as platforms for the recruitment of specific PTM “reader” domain-containing proteins (Kouzarides 2007). Hundreds of histone modifications have been identified, densely covering histone tails (Huang et al. 2014), which is one explanation for the extreme conservation of their sequence across very deeply diverging lineages of eukaryotes (Waterborg 2012; Postberg et al. 2010; Feng and Jacobsen 2011).
In the light of the deep conservation and fundamental importance of the histone code, it is of significant interest to know the extent to which it is conserved in dinoflagellates given that histones are present but are not the major constituent of chromatin in these organisms. Such insights can shed light on the functional roles of histone proteins in dinoflagellate biology. In this study, these issues are addressed by carrying out a detailed survey of the sequence of histone proteins, as well as the presence or absence of chromatin mark writers, readers, and erasers in available transcriptomic and genomic data from a large number of dinoflagellate species.
Materials and Methods
Genomic and transcriptomic sequence data
Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) transcriptome datasets and assemblies were downloaded on June 19, 2014. Glenodinium foliaceum (Stein 1883) and Kryptoperidinium foliaceum (Lindemann 1924) are listed separately following the submission labels, even though they are considered synonymous (Gómez 2005). Low-quality transcriptome assemblies, featuring very low numbers of assembled transcripts, were removed. A full list of the samples used is provided in Supporting Information, Table S1. In addition, genome assemblies and annotations for Perkinsus marinus (accession number GCF_000006405.1) and S. minutum were used. Additional genome assemblies and annotations were downloaded from the NCBI (Thecamonas trahens: GCA_000142905.1, Acanthamoeba castellanii: GCF_000313135.1, Monosiga brevicollis: GCF_000002865.2, Paramecium caudatum: GCA_000715435.1, Capsaspora owczarzaki: GCF_000151315.1, Trichomonas vaginalis: GCF_000002825.2, Ectocarpus siliculosus: GCA_000310025.1) or from the EnsemblProtists (Plasmodium falciparum, Toxoplasma gondii, Entamoeba histolytica, Chlamydomonas reinhardtii, Micromonas pusilla, Tetrahymena thermophila, Guillardia theta, Emiliania huxleyi, Naegleria gruberi, Dictyostelium discoideum, Phytophthora infestans, Cyanidioschyzon merolae) and EnsemblFungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe) databases.
Sequence analysis
Histone proteins were identified using a combination of BLASTP searches (using histone sequences from Homo sapiens, S. cerevisiae, Drosophila melanogaster, Arabidopsis thaliana, and T. thermophila as queries, and an e-value of as cutoff) and HMMER3.0 (Eddy 2011) scans against the Pfam 27.0 database (Finn et al. 2014) (scanning for histone fold and linker histone domains).
For most of the analyses presented, incomplete hits (i.e., partial sequences without both a start and a stop codon) were removed. For the analysis of H3 histone tails, proteins with complete N-termini but incomplete C-termini were included; in addition, H3 sequences were manually examined and in cases where additional amino acid residues were present in front of an otherwise clearly conventional histone H3 tail, such residues were removed (their most likely origin is the presence of an earlier start codon in the transcript assembly and the most parsimonious explanation is that they are not part of the actual protein). Clear cases of misassembly (such as concatenated copies of histone proteins in the same sequence) were also removed.
Multiple sequence alignments were carried out using MUSCLE (Edgar 2004; version 3.8.31) and visualized with JalView (Waterhouse et al. 2009; version 2.8.2). Phylogenetic analysis was carried out using MEGA 6.0 (Tamura et al. 2013) (selection of best substitution model) and RAxML (Stamatakis 2014) (version 8.0.26; generation of maximum likelihood trees and bootstrap analysis).
Gene expression quantification
Sequencing reads were first mapped (as 2 50 bp sequences) to the transcriptome assemblies using Bowtie (Langmead et al. 2009; version 1.0.1), with the following settings: -v 3 -a -t -X 1000. Transcript-level quantification was then carried out with eXpress (Roberts and Pachter 2013; version 1.5.1). Transcript per million (TPM) values were used for subsequent analysis.
Results
Histone proteins in dinoflagellates
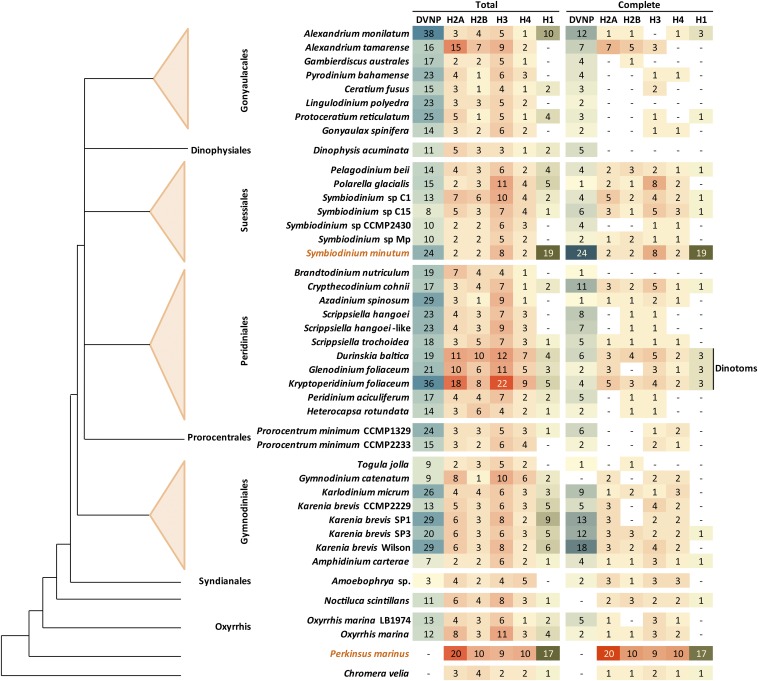
Their often very large size and existing technological limitations have so far prevented the complete sequencing of dinoflagellate genomes, and little is known in detail about their sequence and organization. Fortunately, in recent years, transcriptomic data from a large number of dinoflagellates has become available, in particular through the efforts of the MMETSP (Keeling et al. 2014). MMETSP transcriptome assemblies and translations served as the main dataset for this study. In addition, a draft genomic sequence exists for S. minutum (Shoguchi et al. 2013) even though it is far from complete, and a genome assembly is available from the NCBI database for P. marinus, a member of the early branching, sister to dinoflagellates lineage, the perkinsids; annotated proteins from these genome assemblies were also included. Finally, an MMETSP transcriptome for Chromera velia, a photosynthetic relative of apicomplexans, was used as an outgroup/control. The species considered and their evolutionary relationships are shown in Figure 1, and the full list of datasets can be found in Table S1. In total, the analysis focused on 40 dinoflagellate species/isolates with transcriptome assemblies, two species with genome sequences, and the C. velia transcriptome.
Figure 1.
Detection of histones and DVNP proteins in dinoflagellate transcriptomic and genomic assemblies. “Total” refers to the number of unique proteins detected, while “complete” refers to the subset of full-length proteins (i.e., assembled transcripts in which both start and stop codons are present). Symbiodinium minutum and Perkinsus marinus are colored differently as genome assemblies are available for these species, while only the transcriptomic space has been sampled for all others. The dinotoms are also highlighted separately as they contain an unreduced diatom endosymbiont, meaning that their transcriptomes contain transcripts from two different eukaryotic genomes, only one of which is a dinoflagellate. The cladogram was generated following previously published phylogenies (Orr et al. 2014).
A combination of hidden Markov model (HMM) scans and BLASTP searches against translated transcriptomes (see the Materials and Methods section and figure legends for more details) was used to identify histone proteins. A similar search for DVNPs was also carried out. Part of the reality of working with transcriptome assemblies is that proteins are not always completely assembled, which can confound certain analyses. For these reasons, hits were divided into complete and incomplete sequences, and putative misassemblies were filtered out (details in the Materials and Methods section).
Figure 1 shows the number of fully and incompletely assembled core histone proteins, putative linker histones, and DVNPs in all species studied. All four core histones were identified in all dinoflagellates, usually in multiple distinct variants, with histone H3 exhibiting a particularly great diversity. Putative linker histones were also identified in many species; the breadth of its phylogenetic distribution suggests that histone H1 is present in all dinoflagellates, with the failures to detect it being false negatives.
A unique representative of the complexity of dinoflagellate biology deserves a special mention. The largest number of histones were identified in Durinskia baltica, Kryptoperidinium foliaceum, and Glenodinium foliaceum. These species are known as “dinotoms” (Imanian et al. 2010), as they harbor a tertiary diatom endosymbiont (Dodge 1971; Tomas and Cox 1973; Figueroa et al. 2009), which has not been reduced and retains a large genome. This means that they have both a dinoflagellate nucleus and a diatom one, the latter with conventional nucleosomal organization. Thus, all results that follow should be interpreted with this caveat in mind in the case of dinotoms.
Another potentially confounding factor concerns the purity of the samples studied, many of which were not axenic (Table S1). In some cases, the presence of other eukaryotes is necessary to maintain dinoflagellate cultures. For example, Dinophysis contains kleptoplastids of cryptophyte origin (Schnepf and Elbrächter 1988; Nagai et al. 2008), which apparently do not contain a nucleomorph (Lucas and Vesk 1990), and are extracted from the ciliate Myrionecta rubra (=Mesodinium rubra) (Takishita et al. 2002), which in turn acquires them from cryptophytes (Johnson et al. 2006); Dinophysis is grown together with Myrionecta. The other such example involves Oxyrrhis marina, which is heterotrophic and is often grown with other eukaryotes as prey (Lowe et al. 2011), in this case the diatom Phaeodactylum tricornutum.
Thus, there is a possibility that histones and other proteins identified in transcriptome assemblies do not belong to the listed species. Dinoflagellate transcripts are subject to trans-splicing, thus in principle, the presence or absence of splice leaders can be used to identify transcripts of dinoflagellate origin. However, this is best done by using the splice leader to positively select transcripts during library construction (Gavelis et al. 2015), while conventional RNA-seq is highly vulnerable to underrepresentation of the extreme 5 ends of transcripts, meaning that the absence of splice leaders is not a reliable marker for the nondinoflagellate origin of transcripts.
Despite these caveats, two observations argue against contamination being a major issue with the analysis presented here: first, histones are found in both axenic and nonaxenic cultures (Table S1 and Figure 1), and second, as will become clear below, the properties of putative dinoflagellate histones are quite unique, making them unlikely to come from other eukaryotes.
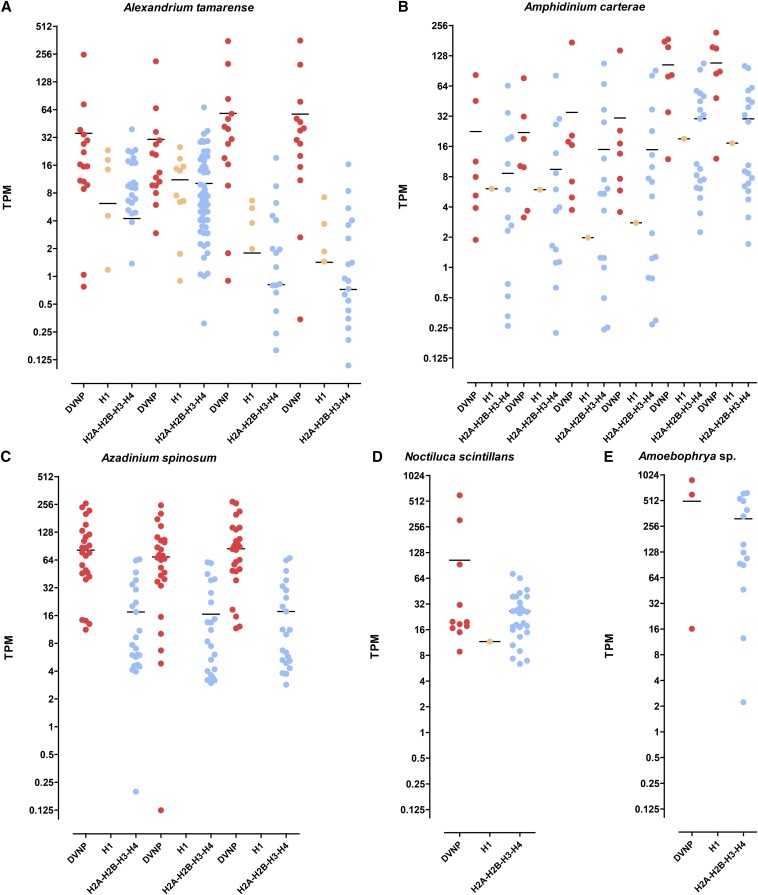
Expression levels of dinoflagellate histones
The expression levels of histones can provide additional information about their functional significance in dinoflagellates. To this end, transcriptomes were quantified by aligning the raw reads back to the assemblies and carrying out transcript-level quantification using eXpress (Roberts and Pachter 2013). Figure 2 and Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, and Figure S7 show the distribution of expression values for core histones, linker histones, and DVNPs. In all nondinotom species, DVNPs are significantly more highly expressed than histones, although considerable variations in the relative levels are observed.
Figure 2.
Expression levels of DVNP, linker histone, and histone genes in dinoflagellates in TPM (transcripts per million). (A) Alexandrium tamarense; from left to right: SRR1296765, SRR1296766, SRR1300221, SRR1300222; (B) Amphidinium carterae; from left to right: SRR1294391, SRR1294392, SRR1294393, SRR1294394, SRR1296757, SRR1296758; (C) Azadinium spinosum; from left to right: SRR1300306, SRR1300307, SRR1300308; (D) Noctiluca scintillans: SRR1296929; (E) Amoebophrya sp.: SRR1296703. See Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, and Figure S7 for data for other species.
While these results should be treated with caution as it is well known that replication-dependent histone mRNAs are not polyadenylated in other eukaryotes (Marzluff 2005), while the datasets studied were generated after polyA-selection, these observations are consistent with histones being present at relatively low levels and DVNPs being the main packaging proteins in dinoflagellates.
Properties of dinoflagellate histones
Previous reports in individual species have noted the increased length of dinoflagellate histones compared to conventional core histones. Figure 3 shows the lengths of all complete H2A, H2B, H3, and H4 proteins identified in this study. Dinoflagellate histones are indeed frequently elongated. However, this is not a universal feature, as many normal-length histones are also observed, and often the same species expresses both long and short histone variants.
Figure 3.
Distribution of histone protein lengths in dinoflagellates. (A) Histone H2A; (B) Histone H2B; (C) Histone H3; (D) Histone H4. Values for Homo sapiens, Drosophila melanogaster, Saccharomyces cerevisiae, and Arabidopsis thaliana core histones are shown at the bottom of each panel for comparison.
The elongation of dinoflagellate histone proteins is not due to the presence of additional protein domains as only core histone domains are readily identifiable (see example in Figure S1), with only two exceptions: a TRAM-LAG1-CLN8 domain in a K. foliaceum H2A, and a NAD(P)-binding domain in an A. tamarense H2A; their functional significance is currently unclear.
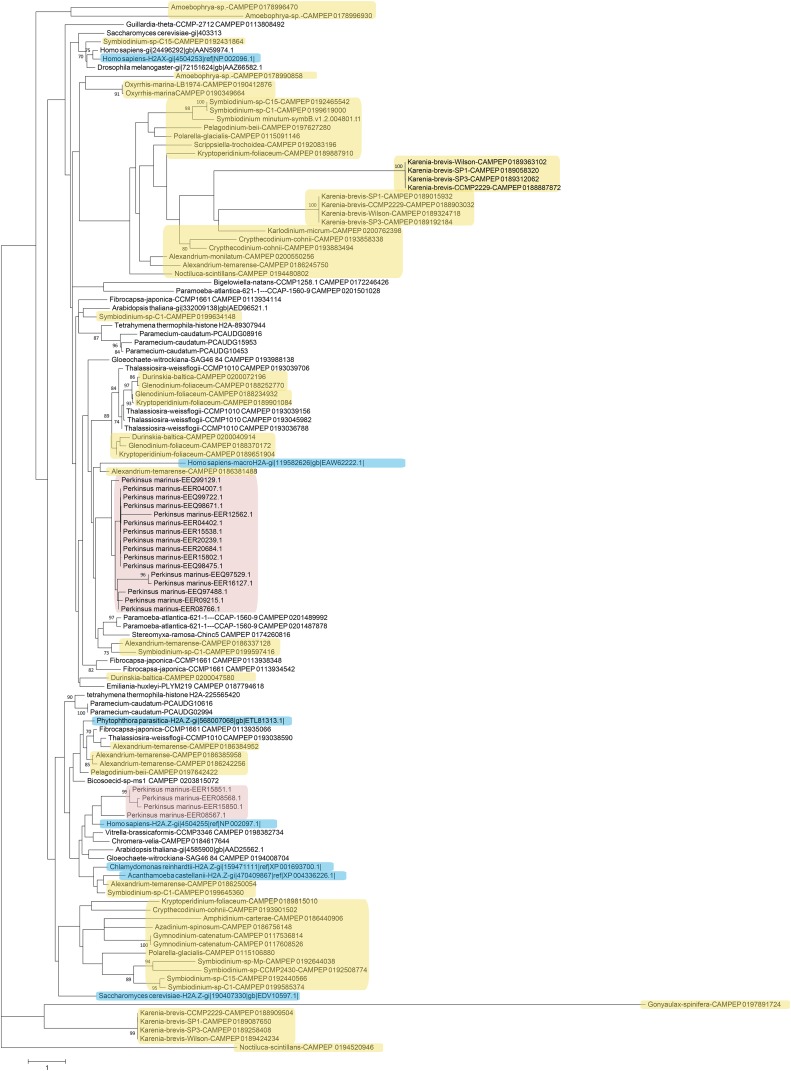
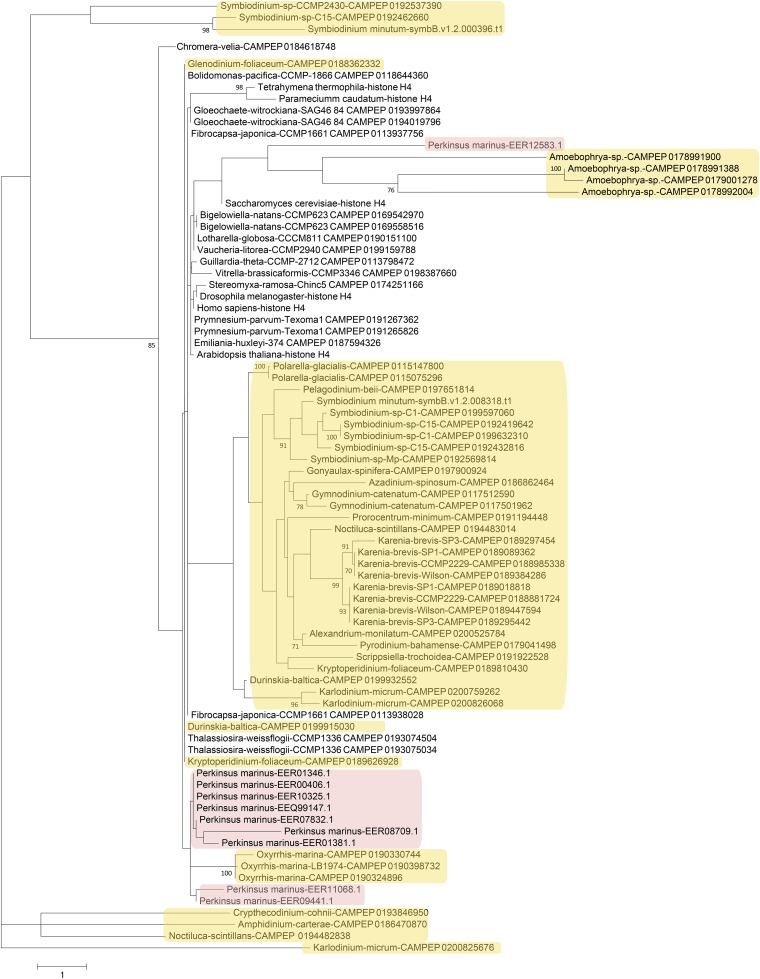
Next, phylogenetic analysis was carried out on histones from dinoflagellates and from a number of other unicellular and multicellular species (Figure 4, Figure 5, Figure 6, and Figure 7). Most eukaryotes express multiple variants of the four core histones (Talbert et al. 2012); some of the additional variants are thought to be ancestral to all eukaryotes. In particular, H2A.Z is often incorporated in nucleosomes surrounding transcription start sites, and specific variants of histone H3 associate with centromeres. H2A.Z and centromeric H3 from several divergent eukaryotes were also included in the analysis in order to identify their putative dinoflagellate homologs (Figure 4 and Figure 6). However, it should be noted that such functional homologs might not be identifiable from sequence alone, as many histone variants are known to be rapidly evolving and/or to be polyphyletic or paraphyletic (Talbert et al. 2012), and numerous cases of loss, gain, and replacement of variants, even between closely related species, have been documented (Baldi and Becker 2013; Drinnenberg et al. 2014; Wang et al. 2011).
Figure 4.
Maximum likelihood phylogenetic tree of H2A protein sequences in dinoflagellates. The tree was generated using RAxML (version 8.0.26) under the LG+G model and with 100 bootstrap replicates. Additional H2A sequences from genome and MMETSP assemblies of other protists were included (the ciliates Paramecium caudatum and Tetrahymena thermophila, the cryptophyte Guillardia theta, the diatom Thalassiosira weissflogii, the discosean amoeba Paramoeba atlantica, another amoebozoan, Stereomyxa ramosa, the bicosoecid Bicosoecid sp., the chlorarachniophyte Bigelowiella natans, the two chromerids Chromera velia and Vitrella brassicaformis, the glaucophyte Gloeochaete witrockiana, the haptophyte Emiliania huxleyi, the raphidophyte Fibrocapsa japonica) as well as sequences from human, yeast, flies, and Arabidopsis. The H2A variants H2A.Z (from multiple eukaryotes), and H2A.X and macroH2A (from Homo sapiens) were also included, and are highlighted in blue. Dinoflagellate sequences are marked in yellow while perkinsid proteins are colored in pink.
Figure 5.
Maximum likelihood phylogenetic tree of H2B protein sequences in dinoflagellates and perkinsids. The tree was generated using RAxML (version 8.0.26) under the LG+G model and with 100 bootstrap replicates. Additional sequences from genome sequences and MMETSP assemblies of other protists were included (the two chromerids Chromera velia and Vitrella brassicaformis, the ciliates Paramecium caudatum and Tetrahymena thermophila, the discosean amoeba Paramoeba atlantica, the bicosoecid Cafeteria roenbergensis, the chlorarachniophyte Bigelowiella natans, the chrysophyte Mallomonas, the cryptophyte Cryptomonas curvata, the diatom Thalassiosira weissflogii, the glaucophyte Gloeochaete witrockiana, the haptophyte Emiliania huxleyi, the raphidophyte Fibrocapsa japonica, the xanthopyte Vaucheria litorea) as well as sequences from human, yeast, flies, and Arabidopsis. Dinoflagellate sequences are highlighted in yellow while perkinsid proteins are marked in pink.
Figure 6.
Maximum likelihood phylogenetic tree of H3 protein sequences in dinoflagellates and perkinsids. The tree was generated using RAxML (version 8.0.26) under the LG+G model and with 100 bootstrap replicates. Additional sequences from genome sequences and MMETSP assemblies of other protists were included (the eight H3 variants from the ciliate Stylonychia lemnae, H3 sequences from two other ciliates, Paramecium caudatum and Tetrahymena thermophila, the bicosoecid Bicosoecid sp., the chlorarachniophyte Bigelowiella natans, the two chromerids Chromera velia and Vitrella brassicaformis, the diatoms Thalassiosira weissflogii and Fragilariopsis kerguelensis, the glaucophyte Gloeochaete witrockiana, the haptophytes Emiliania huxleyi and Prymnesium parvum, the amoebozoans Stereomyxa ramosa and Vexillifera sp., the Bolidomonas heterokont, the cercozoan Minchinia chitonis) as well as sequences from human, yeast, flies, and Arabidopsis. Centromeric histone H3 variants from Homo sapiens, Saccharomyces cerevisiae, Arabidopsis, and Tetrahymena thermophila were also included and are highlighted in gray. Dinoflagellate sequences are highlighted in yellow while perkinsid proteins are marked in pink.
Figure 7.
Maximum likelihood phylogenetic tree of H4 protein sequences in dinoflagellates and perkinsids. The tree was generated using RAxML (version 8.0.26) under the LG+G model and 100 bootstrap replicates. Additional sequences from genome sequences and MMETSP assemblies of other protists (the two ciliates, Paramecium caudatum and Tetrahymena thermophila, the amoebozoan Stereomyxa ramosa, the Bolidomonas heterokont, the chlorarachniophytes Bigelowiella natans and Lotharella globosa, the two chromerids Chromera velia and Vitrella brassicaformis, the cryptophyte Guillardia theta, the diatom Thalassiosira weissflogii, the glaucophyte Gloeochaete witrockiana, the haptophytes Emiliania huxleyi and Prymnesium parvum, the raphidophyte Fibrocapsa japonica, the xanthopyte Vaucheria litorea) were included as well as sequences from human, yeast, flies, and Arabidopsis. Dinoflagellate sequences are highlighted in yellow while perkinsid proteins are marked in pink.
The patterns emerging from these comparisons are complex, with the deep branches of all trees being poorly resolved. This is particularly true for H2A and H2B, which is not surprising as these are in general the most variable core histones within eukaryotes and exhibit the highest degree of innovation in terms of novel histone variants in different lineages.
Within H2A histones, a group of putative H2A.Z variants is observed (Figure 4), but in addition to it, there are several other very deep branches consisting of proteins that might serve as novel variants with currently unknown functions. One other histone variant can be identified from sequence – H2A.X, which is best known for its role during DNA damage response (Ismail and Hendzel 2008). H2A.X is characterized by the presence of a SQ(E/D) phosphorylation motif at the C-terminus of the protein (Talbert et al. 2012), the exact sequence of which varies but is generally constant within the broad divisions of eukaryotes. A number of dinoflagellate histones do contain similar motifs at their C-terminus, but a great deal of diversity is observed among them (Table S2): SQEF, SQEY, SQQY, and SQDF motifs are all observed. Of note, most of the H2A variants in the P. genome contain SQ(E/D) motifs, one being SQEI, and eight SQEM; the functional significance of having so many putative H2A.X variants is currently unclear.
Excluding putative H2A.Z variants, a few dinoflagellate H2A proteins cluster closely with conventional H2A, but most of these are from dinotoms and very similar to H2A sequences from the diatom Thalassiosira, suggesting that they are of endosymbiont origin. The majority of dinoflagellate H2A sequences are highly divergent. Similar patterns are observed for histone H2B (Figure 5), with a dinotom-specific cluster, many highly derived sequences from both dinotoms and other dinoflagellates, and a small number of proteins clustering with conventional core histone H2B.
Deeply diverging sequences also constitute the majority of dinoflagellate histone H3 proteins (Figure 6). No group of dinoflagellate histones that robustly clusters with centromeric H3 sequences from other eukaryotes could be identified, but given the very rapid evolution of centromeric H3 proteins, this is not surprising. Thus, the presence of centromeric histones in dinoflagellates cannot be excluded. Notably, several deeply diverging H3 variants are also observed in P. marinus.
Histones H3.1 and H3.3 form a pair of sequence variants shared between many eukaryotes. H3.1 is deposited during the S-phase of the cell cycle while H3.3 replaces it as a result of transcriptional activity outside of it (Hake and Allis 2006; Otero et al. 2014). H3.3 and H3.1 are distinguished by a very small number of residues, in particular and classically, the presence at position 31 of an S (metazoans) or T (plants) in H3.3 vs. A in H3.1; some other variations on that theme are also observed in other eukaryotes (Talbert et al. 2012). In a few dinoflagellates, including A. tamarense, some Symbiodinium isolates, and Heterocapsa rotundata, pairs of very similar histones distinguished by the presence or absence of a phosphorylatable amino acid at position 31, are observed (Figure S8). Although there are only a few such species and their phylogenetic distribution is patchy, these observations do suggest that such pairs of H3 variants might be present in dinoflagellates, especially if one of the other distinctions between H3.3 and H3.1 also holds, namely that the former is replication-independent and with polyadenylated mRNAs while the other is replication-dependent and with mRNAs lacking polyA tails (Marzluff et al. 2008), and is thus systematically under-represented in polyA-selected RNA-seq libraries.
Histone H4 displays the most striking divergence among dinoflagellate histones (Figure 7). Aside from the dinotom-specific group, most dinoflagellate H4 proteins form a well-defined group of highly derived sequences. In addition, a Symbiodinium-specific deeply divergent variant is observed, and Amoebophrya expresses four even more divergent variants. Curiously, such variants are also found in the P. marinus genome, in addition to the more conventional H4 variants it contains. Many of these are elongated variants. As H4 is the most conserved of all histones and also the one with the fewest known variants (Talbert et al. 2012), these are especially intriguing observations in terms of their functional implications.
Conservation and divergence of the histone code in dinoflagellates
As discussed above, the question of the functional importance of dinoflagellate histones is closely related to the extent of conservation and divergence of the dinoflagellate histone code. To address this question, the conservation of key post-translationally modified histone residues relative to other eukaryotes was assessed. Before the results from these comparisons are described, a brief overview of the significance of these residues is presented.
The histone code:
Hundreds of histone modifications have been identified (Huang et al. 2014), including monoubiquitination, acetylation, mono-, di-, and trimethylation of lysines, mono- and symmetric and asymmetric dimethylation of arginines, phosphorylation of serines, threonines, and tyrosines, isomerization of prolines, and others (Figure S9). While the great majority have not been characterized in any detail, the role of a significant number is fairly well understood, which has revealed both the functional importance of the histone code and the strong constraints that it imposes on the corresponding residues across eukaryotes. The presence or absence of these residues in dinoflagellate histones can be highly informative about the conservation of the relevant aspects of the histone code and chromatin biology. This approach has some limitations, as conservation of amino acid residues need not be solely due to their PTMs, and does not on its own mean that the relevant modifications are in fact deposited in vivo and have a conserved function. However, the absence of an important modified residue does mean that the corresponding histone marks are not conserved, and that their functionality has been lost (or, possibly, adopted by other residues elsewhere on nucleosomes).
The key histone marks in eukaryotes and their functions, as revealed by studies in yeast, mammals, and other model systems will be briefly described before the state of the histone code in dinoflagellates is discussed. The list is by no means exhaustive and the discussion is by necessity simplified as the complexity of the histone code is immense. The focus will be on a subset of marks associated with the following processes: transcriptional activation, initiation, and elongation, the formation of heterochromatin and other repressive chromatin environments, and the regulation of chromatin dynamics during mitosis.
Repression and heterochromatin:
Heterochromatinization is a major mechanism for the permanent repression of transposable elements and some host genes in eukaryotes; heterochromatin is also often found in telomeric and centromeric regions. The main mechanisms for establishing and maintaining heterochromatin are deeply conserved (including in other alveolates; Liu et al. 2007; Schwope and Chalker 2014) and likely ancestral to all eukaryotes, with histone marks playing a key role. The classic heterochromatin histone mark is the trimethylation of lysine 9 of histone H3 (H3K9me3). H3K9me3 recruits the heterochromatin protein HP1 (Lachner et al. 2001; Bannister et al. 2001), which leads to compaction of chromatin. It is also bound by H3K9me3 methyltransferases (Tachibana et al. 2001), which can further methylate neighboring H3K9 residues, and has a positive feedback loop relationship with cytosine DNA methylation (Stancheva 2005).
Another mechanism for gene repression is mediated by the H3K27me3 mark and involves the action of Polycomb proteins, which both deposit and are recruited by it. The process has been extensively characterized in both plants and metazoans (Zheng and Chen 2011; Simon and Kingston 2013), and also involves the monoubiquitination of histone H2A (H2AK119ub1; Shilatifard 2006).
Additional histone marks associated with repressive chromatin include H4K20me3 (Balakrishnan and Milavetz 2010) and H3K64me3 (Daujat et al. 2009). It should be noted that heterochromatin is not a homogeneous entity, and that different marks or combinations of marks may characterize distinct types of repressive chromatin.
Transcription activation and initiation:
The first histone modification to be assigned potential functional significance was histone acetylation, which usually exercises a general stimulating effect on transcription by reducing the positive charge of histones and loosening histone–DNA interactions (Allfrey et al. 1964). Many lysines on histones are acetylated (Figure S9; the ones discussed in some detail here include: H3K9ac, H3K14ac H3K18ac, H3K23ac, H3K27ac, H3K56ac, H3K64ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac, H4K59ac, H4K79ac, H4K91ac, H2AK4ac, H2AK8ac, H2AK12ac, H2BK5ac, H2BK12ac, H2BK16ac, and H2BK20ac), but this general activating effect is not their sole function. For example, H3K27ac has been identified as a marker of active enhancer elements in metazoans (Rada-Iglesias et al. 2010; Creyghton et al. 2010), H3K56ac plays a role in the activation of transcription but is also important for histone exchange and nucleosome assembly (Rufiange et al. 2007; Xu et al. 2005; Xie et al. 2009), etc.
In all eukaryotes studied so far, active promoters are specifically marked by H3K4me3. The exact biochemical mechanisms in which H3K4me3 is involved have not yet been completely elucidated (Shilatifard 2012), but its deep conservation suggests that it plays a key role in the process of initiation.
In addition, multiple histone marks have been correlated with transcription activation, but the mechanistic details of their role are not entirely clear. These include the phosphorylation of threonine 6 on histone H3 (H3T6ph), which prevents the demethylation of H3K4me3 (Metzger et al. 2010), H3S28ph, which marks active promoters and transcribed gene bodies (Drobic et al. 2010; Sawicka et al. 2014) and cross-talks negatively with H3K27me3 (Fischle et al. 2003), and several arginine methylations. H3R2 is known to be both symmetrically and asymmetrically methylated, with asymmetric methylation (H3R2me2a) being mutually exclusive with H3K4me3 (Guccione et al. 2007; Kirmizis et al. 2007) while H3R2me2s correlates positively with it (Yuan et al. 2012). Similarly, H4R3me2s is associated with repression (Tsutsui et al. 2013; Dhar et al. 2012), and H4R3me2a with activation (Sun et al. 2011).
Transcription elongation:
The eukaryote transcription cycle involves a complexly choreographed sequence of nucleosome remodeling events and PTMs on histones and the polymerase itself. Because nucleosomes are inhibitory to it, the nucleosome barrier has to be overcome for transcription to proceed. This is achieved through a combination of histone acetylation and the action of the FACT complex (Reinberg and Sims 2006; Pavri et al. 2006; Zhu et al. 2005), which partially disassembles nucleosomes, allowing for the polymerase to proceed, and then reassembles them. The action of FACT is associated with the transient monoubiquitination of histone H2B (H2BK120ub1 in mammals).
The role of H3K36me3 in transcription elongation is particularly well understood mechanistically (Wagner and Carpenter 2012). The acetylation marks deposited during elongation need to be erased if cryptic transcription from within the now more open chromatin is to be prevented (Kaplan et al. 2003). H3K36me3 is deposited by methyltransferases (KMTs) associated with the elongating polymerase, and then serves as a recruitment mark for histone deacetylases (HDACs), which remove the acetylation marks and close the cycle. However, this is not the only function of H3K36me3, as it has also been shown to play a significant role in the process of splicing in metazoans (Kolasinska-Zwierz et al. 2009; Schwartz et al. 2009).
Another site of modification associated with elongation is H3K79 (H3K79me2/3; Nguyen and Zhang 2011). H3K79me is also notable for being catalyzed by the Dot1 KMT (Feng et al. 2002) rather than a SET-domain methyltransferase as all other lysine methylations on histones.
An additional mark involved in elongation is H2AY57ph, which appears to be required for the H2BK120 ubiquitination/deubiquitination cycle (Basnet et al. 2014).
Finally, the isomerization of prolines 30 and 38 in H3 (H3P30iso, H3P38iso) can regulate the activity of H3K36 methyltransferases (Nelson et al. 2006).
Mitosis:
Chromatin undergoes dramatic transformations during mitosis, as it is duplicated in the S-phase and then compacted in the M phase. Histone marks play a key role in these processes (Desvoyes et al. 2014; Doenecke 2014), in particular several phosphorylatable residues on histones H3, H4, and H2A (Sawicka and Seiser 2012).
H3S10ph, although it can also occur in other contexts, is the best characterized such mark, involved in chromosomal condensation during mitosis (Hendzel et al. 1997). A key event during this process is the establishment of an interaction between the N-terminal tail of histone H4 and an acidic patch on H2A-H2B dimers on neighboring nucleosomes, which, however, is prevented by the presence of H4K16ac; H3S10ph recruits HDACs, which remove acetylation marks from H4 tails, allowing compaction to happen (Wilkins et al. 2014). H3S10ph also has the effect of excluding HP1, which is ejected from chromatin during the M phase, as HP1 cannot bind to H3 tails containing both H3K9me3 and H3S10ph (Fischle et al. 2005).
Additional marks implicated in the regulation of mitotic chromatin dynamics include H3T3ph (Polioudaki et al. 2004; Wang et al. 2010), H3T11ph (Preuss et al. 2003), which is also involved in transcriptional activation (Metzger et al. 2008), H3S28ph, H4S1ph and H2AS1ph (Barber et al. 2004), H2AS122ph (Yamagishi et al. 2010), H2AT120ph (van der Horst et al. 2015), and others. Most of these marks are conserved between plants and humans although the precise patterns of their chromosomal distribution may differ somewhat (Houben et al. 2007; Manzanero et al. 2000).
Histone marks also play a role in nucleosome assembly during and independently of replication. H3K56ac, which occurs in the core histone domain rather than the tail, facilitates histone exchange (Rufiange et al. 2007); other marks suggested to play such a role include H4K79ac and H4K91ac (Yang et al. 2011).
Conservation and divergence of dinoflagellate histone tails:
In the light of the histone code, many dinoflagellate histones present a curious mixture of conserved and divergent characters (Figure 8 shows H3 tails in Gymnodinium catenatum). The most well-known H3 variants are centromeric histones, and those usually have highly divergent histone tails (Figure S10). There are many examples of such tails in dinoflagellates, but there are also histones with tails fairly similar to the conventional state, and all sorts of variations between these two extremes. Thus G. catenatum expresses a variant that is a close match to the typical H3 sequence (CAMPEP_0117531604), and two very divergent ones (CAMPEP_0117479984 and CAMPEP_0117494448), one or both of which exhibit substitutions at many key residues of the histone code, including H3K9, H3K27, H3K36, and H3K79, as well as several insertions within the N-terminal tails and the core histone domain. The other variants in Gymnodinium show an intermediate level of divergence, with the region between residues 12 and 31 being particularly variable.
Figure 8.
Multiple sequence alignments of the N-terminal regions of histone H3 sequences from Gymnodinium catenatum and of core histone H3 proteins from several other eukaryotes. Sequences were aligned using MUSCLE (Edgar 2004; version 3.8.31) and visualized using JalView (Waterhouse et al. 2009; version 2.8.2).
Similar observations can be made in many other species, although because of the certainly incomplete sampling of variants in transcriptome assemblies, such a comprehensive set of variations is not always seen.
The histone code in dinoflagellates:
To assess the conservation of the histone code, multiple sequence alignments of all variants of each core histone in each species and a reference (taken to be the human core histones, with histone H3.1 used in the case of H3) were carried out. All histone sequences with complete N-terminal tails were used for this analysis (i.e., a protein was allowed to be incomplete at its C-terminus and not just the “complete” sequences shown in Figure 1 were included). This was done in order to capture all the histone tail diversity for which there is evidence in the data.
Conservation was then scored against the reference sequence as follows: for a given position i in the reference protein and a radius r, a precise match of the sequence with coordinates was required to score that residue as conserved, i.e., when , only a conservation of the residue itself is required, but when and , the 3-amino acid and 5-amino acid contexts were considered. This approach was adopted as histone marks are often deposited and read according to their sequence context. Thus, conservation of the context makes it more plausible that the modification is also conserved. Of course, there are limitations to such interpretations: residues and their context need not be conserved because of conservation of a particular histone mark, and the absence of strict conservation of context does not necessarily mean that the mark is not deposited (its reader and writer proteins might have evolved and adapted accordingly). Nevertheless, it remains true that the complete absence of a residue is almost certain evidence that the corresponding histone marks are absent too, and that strong conservation of its context boosts confidence in the preservation of at least the capacity to deposit them.
The results from these comparisons for histones H3, H4, H2A, and H2B are shown in Figure 9, Figure 10, Figure 11, and Figure 12, respectively.
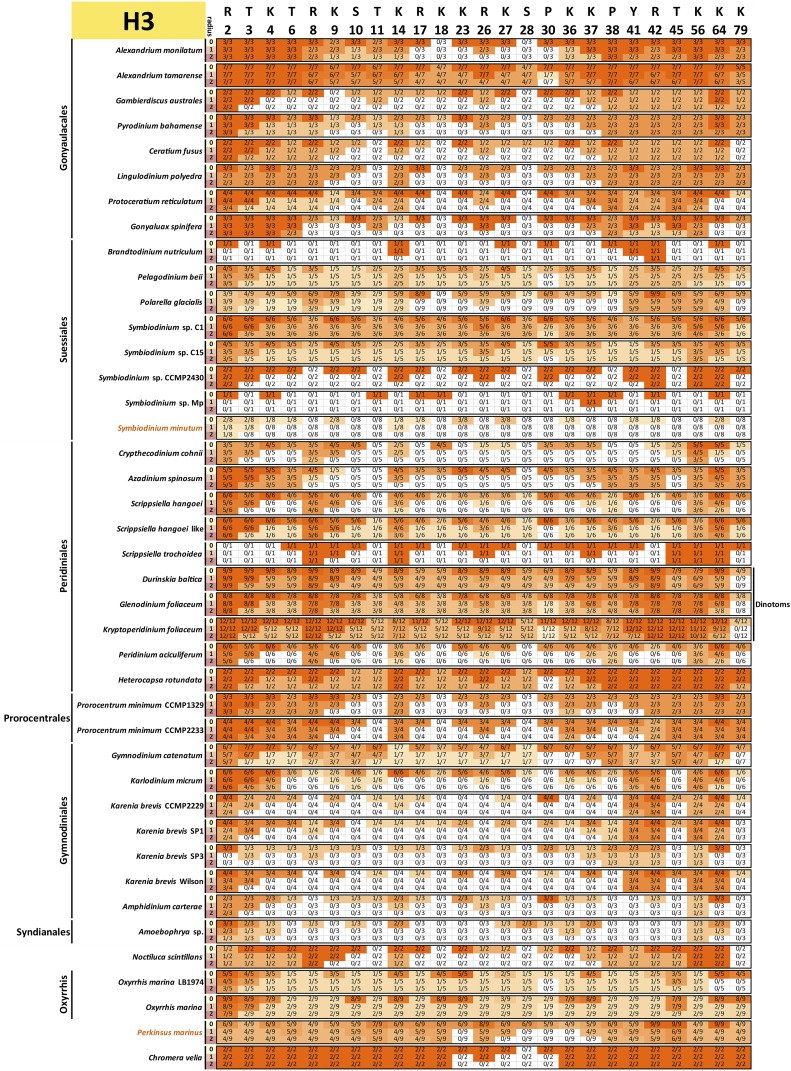
Figure 9.
Conservation of key post-transcriptionally modified residues in dinoflagellate histone H3 proteins. The radius r refers to the size of the context considered when scoring conservation. When , only the residue itself is considered; when , a perfect match to the three-amino acid peptide also including the flanking residues on each side is required; when , the five-amino acid peptide also including the two flanking residues on each side is considered. The fractions indicate the number of histone H3 proteins C with conserved residues according to the criteria specified by the radius relative to the total number of histone H3 proteins N for each species. All histone H3 sequences with complete N-terminal tails identified in the transcriptomic data are included (the C-terminal portions of the protein were allowed to be incomplete for the purposes of this analysis, but the N factor in the fractions was adjusted accordingly if necessary in cases when gaps in the alignments were due to an incomplete sequence).
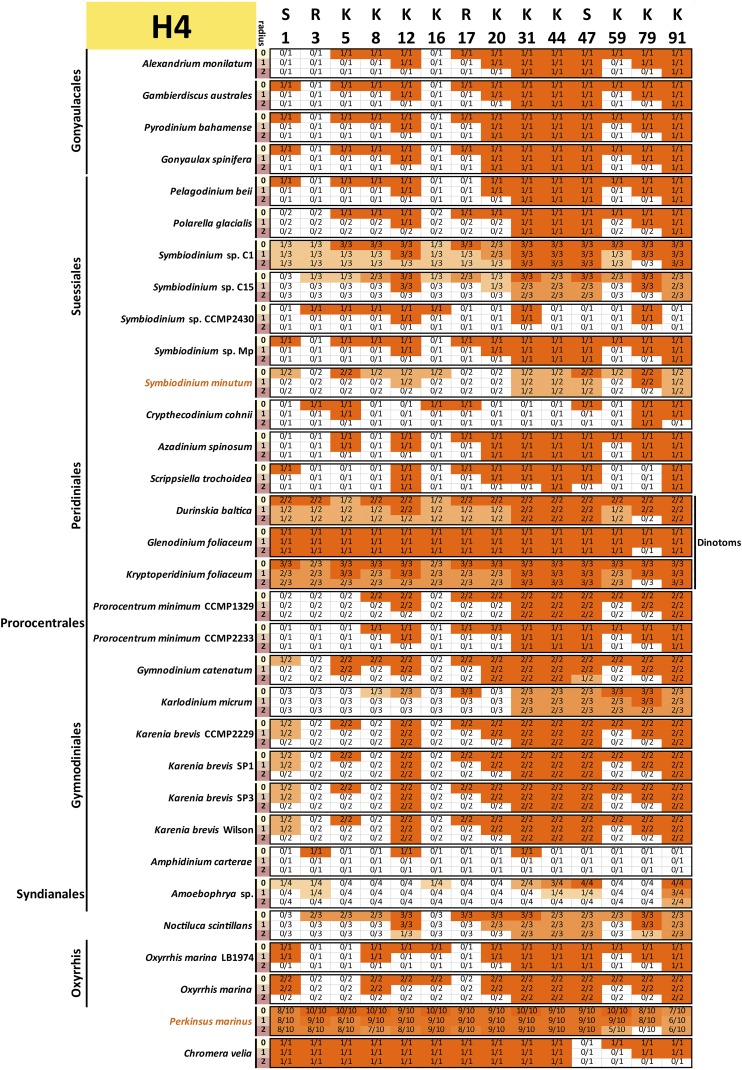
Figure 10.
Conservation of key post-transcriptionally modified residues in dinoflagellate histone H4 proteins. The radius r refers to the size of the context considered when scoring conservation. When , only the residue itself is considered; when , a perfect match to the three-amino acid peptide also including the flanking residues on each side is required; when , the five-amino acid peptide also including the two flanking residues on each side is considered. The fractions indicate the number of histone H4 proteins C with conserved residues according to the criteria specified by the radius relative to the total number of histone H4 proteins N for each species. All histone H4 sequences with complete N-terminal tails identified in the transcriptomic data are included (the C-terminal portions of the protein were allowed to be incomplete for the purposes of this analysis, but the N factor in the fractions was adjusted accordingly if necessary in cases when gaps in the alignments were due to an incomplete sequence).
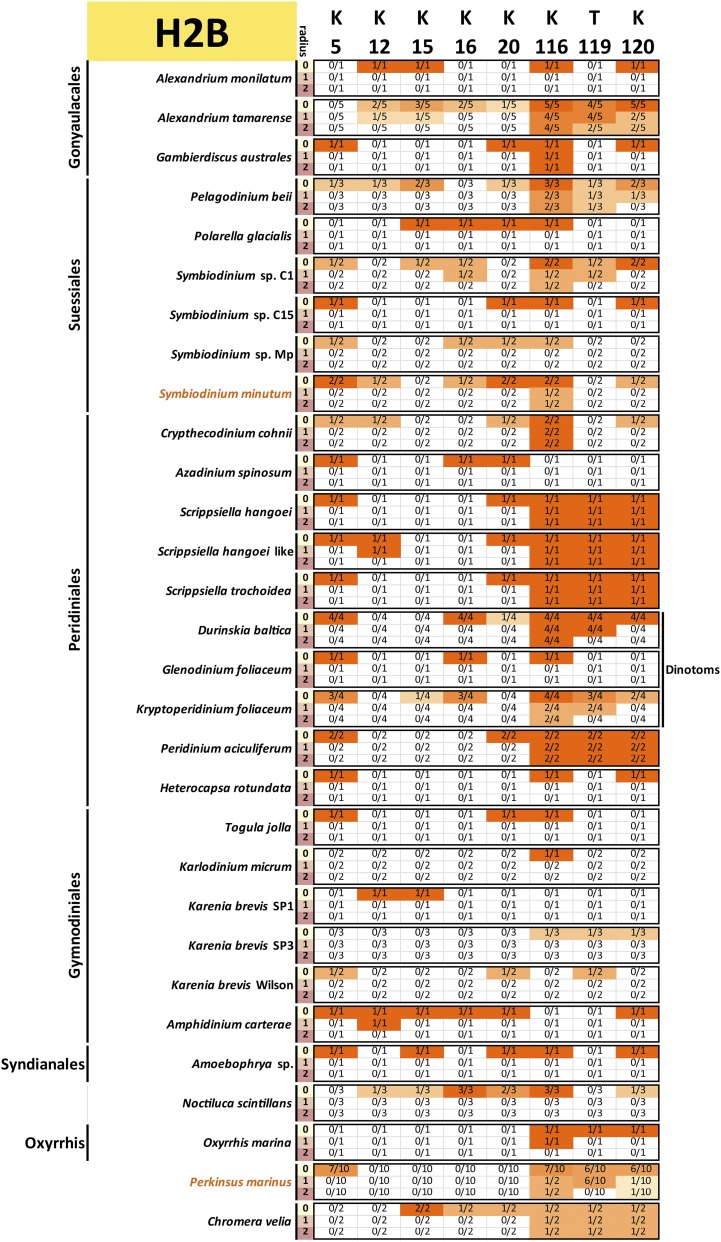
Figure 11.
Conservation of key post-transcriptionally modified residues in dinoflagellate histone H2A proteins. The radius r refers to the size of the context considered when scoring conservation. When , only the residue itself is considered; when , a perfect match to the three-amino acid peptide also including the flanking residues on each side is required; when , the five-amino acid peptide also including the two flanking residues on each side is considered. The fractions indicate the number of histone H2A proteins C with conserved residues according to the criteria specified by the radius relative to the total number of histone H2A proteins N for each species. All histone H2A sequences with complete N-terminal tails identified in the transcriptomic data are included (the C-terminal portions of the protein were allowed to be incomplete for the purposes of this analysis, but the N factor in the fractions was adjusted accordingly if necessary in cases when gaps in the alignments were due to an incomplete sequence).
Figure 12.
Conservation of key post-transcriptionally modified residues in dinoflagellate histone H2B proteins. The radius r refers to the size of the context considered when scoring conservation. When , only the residue itself is considered; when , a perfect match to the three-amino acid peptide also including the flanking residues on each side is required; when , the five-amino acid peptide also including the two flanking residues on each side is considered. The fractions indicate the number of histone H2B proteins C with conserved residues according to the criteria specified by the radius relative to the total number of histone H2B proteins N for each species. All histone H2B sequences with complete N-terminal tails identified in the transcriptomic data are included (the C-terminal portions of the protein were allowed to be incomplete for the purposes of this analysis, but the N factor in the fractions was adjusted accordingly if necessary in cases when gaps in the alignments were due to an incomplete sequence).
Before these results are discussed, it should be noted that only very divergent H3 histones are found in the assembled S. minutum contigs (even the residues that are conserved in one of the annotated variants according to the criteria used above are found in the context of an otherwise very divergent N-terminal tail). However, the transcriptomes of other Symbiodinium isolates contain more conventional histone H3 sequences, a discrepancy that, if contamination is to be excluded, is best explained as being due to the incompleteness of the S. minutum assembly, which is known to capture only a fraction of the total genomic sequence (Shoguchi et al. 2013).
Repression and heterochromatin:
H3K9 displays an intriguing pattern of presence and absence in dinoflagellates. It is present in at least one variant in the majority of species (with the exception of a few transcriptomes, in which only a single variant was assembled, and Amoebophrya and Gambierdiscus australes, in which all three and two H3 variants, respectively, do not have it), although its sequence context is often not well conserved. However, most species also have multiple variants without H3K9. Many of these belong to the group of variants with extended protein sequence and very divergent tails, which are more likely to have specialized functions such as those of centromeric H3 histones in other eukaryotes. Yet quite remarkably, H3K9 is not absent solely from H3 tails that are highly divergent; for example, A. monilatum expresses a 167-amino acid histone H3 (the increased length is due to a C-terminal extension), the N-terminal portion of which is overall a close match to human histone H3 with the notable exception of H3K9, which is substituted by a methionine (Figure S11). Functional homologs of HP1 have been identified in the two other major alveolate lineages, ciliates (Schwope and Chalker 2014) and apicomplexans (Flueck et al. 2009), but no clear homologs (defined as proteins with a pair of chromodomains and a chromo shadow domain) were detected in dinoflagellate transcriptomes. None were found in C. velia or P. marinus either, but at least in the case of C. velia this is most likely a false negative case. As H3K9 is also subject to numerous other post-transcriptional modifications, the possibility that its conservation is for other reasons and that the overall H3K9me3/HP1 heterochromatin formation mechanism is not conserved cannot be dismissed.
Almost all dinoflagellates possess at least one histone H3 variant with H3K27 (more than half of the variants possessing H3K27 is the typical condition), although as in the case of H3K9, its sequence context is not well conserved (because the neighboring H3S28 is usually missing). The number of H2A variants assembled is often low, thus it is not certain that all are present in the assemblies, but in most species a variant with H2AK119 (often ubiquitinated in concert with H3K27me3 deposition) is present.
H4K20 is observed in most dinoflagellate H4 histones with the exception of the four variants in Amoebophrya and the two variants in the S. minutum genome assembly.
H3K64 is a remarkably well conserved residue (although not always in its sequence context), being present in at least one (usually most) H3 variant in all dinoflagellates. As H3K64 resides in the core histone domain, this might be due to constraints other than its role in heterochromatin formation, but its strong conservation suggests that if nucleosomal heterochromatin does form in dinoflagellates, it might be playing a role in the process.
Overall, the analysis of heterochromatin-associated histone mark-bearing residues suggests that while the capacity to deposit these marks has been retained in most dinoflagellates, the constraints on their sequence context seem to have been relaxed. This is possibly due to a significant reduction of the amount and functional importance of nucleosomal heterochromatin, or even its complete loss.
Transcription activation and initiation:
With the exception of H4K12, the majority of acetylated lysines in the N-terminal histone tails display poor conservation across dinoflagellates. This is not entirely surprising as these lysines are known to be redundant with each other (Dion et al. 2005; Martin et al. 2004), and could be lost if constraints on histone sequence are relaxed.
The status of conservation of H3K4 is of particular interest given its strong association with active transcription start sites (TSSs) in other eukaryotes. It is found in all dinoflagellates (with the exception of the single divergent H3 variant assembled in Brandtodinium nutriculum), usually in most variants, and often in a well-conserved sequence context. Variants without it are also observed, and as with H3K9, these are not always of the elongated highly divergent varieties, but overall these observations suggest retention of the functional importance of H3K4me3 in dinoflagellates.
Other, less well characterized residues such as H3R2 and H3T6 are also fairly well conserved, but it could be that this is due to constraints on the H3K4 sequence context. Another methylated arginine, H4R3, displays a remarkable absence of conservation, being found in few dinoflagellates outside of dinotoms (but well conserved in Perkinsus and Chromera).
The conservation status of these marks suggests preservation of the pivotal role of H3K4me3 and a derived state for some other components of the histone code associated with the activation and repression of transcription.
Transcription elongation:
H3K36 is observed in the majority of dinoflagellates (with the possible exception of Karenia brevis among the species with a large number of assembled H3 variants), suggesting the possible conservation of its role in transcription elongation. H3K79 is less well conserved although still found in most species. The presence/absence pattern of H2BK120 is patchy, but this might well be due to incomplete assemblies as the number of observed H2B variants is often low.
The two prolines subject to isomerization, H3P30 and H3P38, are also found in most dinoflagellates but not in all tails and not always together.
Somewhat surprisingly, the residue implicated in transcriptional elongation that displays the strongest conservation is H2AY57.
Overall, these observations suggest that the capacity to deposit the key histone marks involved in transcriptional elongation is present, with some divergence, and the processes they are involved in might be conserved in dinoflagellates.
Mitosis:
The phosphorylatable residues involved in mitosis (H3S10ph, H3T3ph, H3T11ph, H3S28ph, H4S1ph, H2AS1ph, H2AS122ph, and H2AT120ph) display quite poor conservation in dinoflagellates. H3T3ph is an exception but this could be because of constraints on the neighboring H3K4. H3S10 is also present in at least one variant in many species, but is also completely absent in numerous others. Also remarkable is the absence of conservation of H4K16, the residue involved in the control of nucleosome compaction. Given these observations, it is quite likely that the H3K9me3S10ph switch does not operate in dinoflagellates and that histone phosphorylation plays a reduced (if any) role in mitosis.
Of note, all these residues are best conserved in O. marina, the earliest branching dinoflagellate included in this study. It is therefore possible that the corresponding processes are most preserved in Oxyrrhis while other dinoflagellates are in an even more derived state.
H3K56, H4K79, and H4K91, the acetylation of which has been implicated in the process of nucleosome assembly, are very well conserved across dinoflagellates, including at the level of their sequence context.
Histone mark writers, erasers, and readers, and chromatin remodelers
Histone marks are deposited and erased by specific enzymes and read by proteins containing “reader” domains, recognizing particular modifications. Histone acetylation is carried out by several families of lysine acetyltransferases: KATs, or, in the context of histones, HATs (Marmorstein and Zhou 2014; Wang et al. 2008; Thomas and Voss 2007; Doyon and Côté 2004). Acetylation marks are removed by HDACs, of which there are also several classes (Mariadason 2008; Vaquero 2009), one of which includes members of the sirtuin protein family. Lysine methylation is carried out by SET-domain-containing proteins (Dillon et al. 2005), with the exception of H3K79 methylation and Dot1. Lysine methylation is primarily erased through the action of demethylases of the Jumonji (Jmj) family (Takeuchi et al. 2006). Arginine methyltransferases include PRMT5 and CARM1-type proteins (Karkhanis et al. 2011; Wysocka et al. 2006).
A variety of reader domains are known (Musselman et al. 2012). These include the bromodomain and the chromodomain, which bind to acetylated and methylated lysines, respectively (Sanchez et al. 2014; Blus et al. 2011), the PHD finger, which binds to a variety of unmodified and modified histone tail substrates, WD40 domains, which bind to trimethylated lysines among other targets, Tudor domains, which bind to both methylated lysines and arginines, PWWP domains, which bind to methylated lysines, and others.
ATP-dependent chromatin remodeling complexes, which move nucleosomes along DNA, are another important component of the eukaryote chromatin toolkit. Four major families of remodelers – SWI/SNF, ISWI, CHD, and INO80 – are known, distinguished by characteristic combinations of protein domains (Clapier and Cairns 2009).
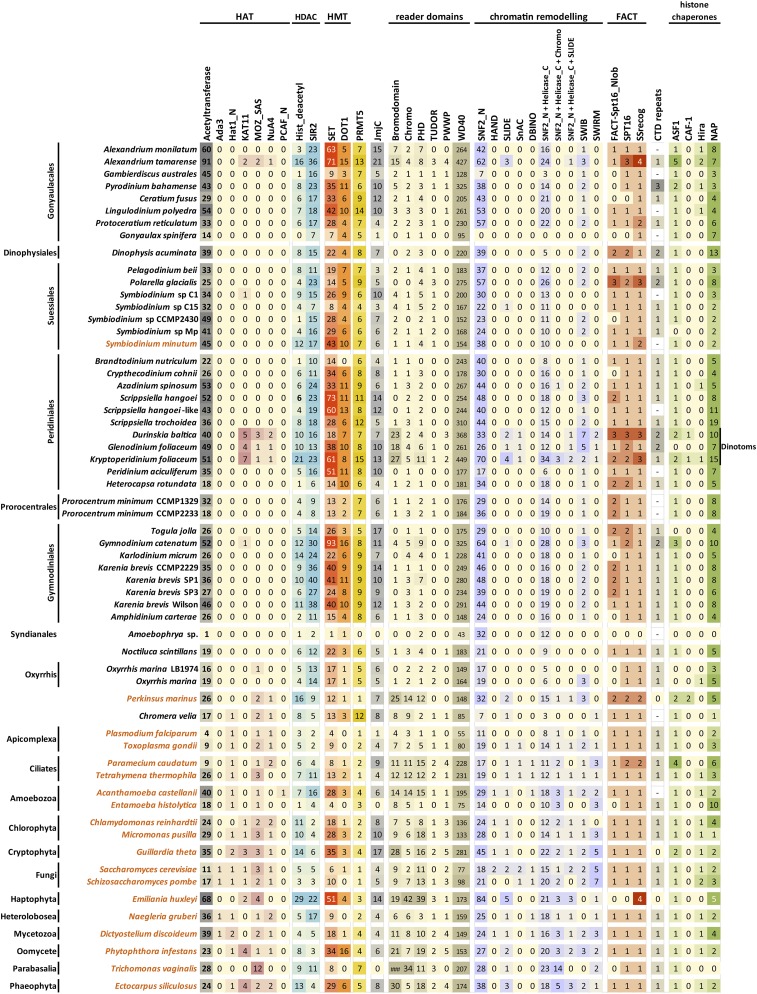
To evaluate the composition of the set of writer, eraser, and reader proteins and chromatin remodelers in dinoflagellates, putative homologs were identified by scanning datasets for the presence of their characteristic domains, or combinations of domains (Figure 13). In addition to Perkinsus and Chromera, the same analysis was also carried out on the sequenced and annotated genomes of a diverse set of unicellular eukaryotes, in order to compare the number of proteins in each group to what is observed in other protozoans.
Figure 13.
Presence and absence of histone mark writer, eraser and reader proteins, chromatin remodeling complexes, histone chaperones, the FACT complex, and RNA polymerase II CTD repeats in dinoflagellates and representative unicellular eukaryotes. There are multiple different HAT families and homologs from these were searched for separately. Except for the yeast Hat1 and the human PCAF protein, the domains found in GNAT-type acetyltransferases are also found in many acetyltransferase enzymes whose activity has little to do with chromatin, but the catalytic core of GNAT-type HAT complexes is formed by several proteins (Ada2, Ada3, Sgf29, Gcn5 in the case of the SAGA complex), and the adapter proteins (Muratoglu et al. 2003) were searched for in addition to acetyltransferase domains. The KAT11 domain is found in numerous HATs such as the metazoan p300 and CBP and the yeast Rtt109 (Wang et al. 2008). MYST-family HATs (Thomas and Voss 2007) were identified using searches for the MOZ_SAS domain, and; in addition, a search for the NuA4 domain, part of the EAF6 component of the NuA4 complex of which MYST HATs are often part of (Doyon and Côté 2004), was carried out. There are four classes of HDACs (Mariadason 2008), one of which, the sirtuins, is characterized by the presence of a SIR2 domain (although sirtuin specificity is not restricted to histones). Two classes of lysine methyltransferases are known too – the majority are of the SET-domain type, Dot1 is the exception. Known arginine methylation enzymes include PRMT5- and CARM1-type proteins (Karkhanis et al. 2011; Wysocka et al. 2006); CARM1 is not shown as no CARM1 proteins are found in any of the unicellular species shown here. Aside from the monoamine oxidase LSD1 (Rudolph et al. 2013), lysine demethylases contain a Jumonji domain (JmjC) (Johansson et al. 2014). Bromodomains (Sanchez et al. 2014) and chromodomains (Blus et al. 2011) are readers of lysine acetylation and methylation, respectively. WD40 domains bind to trimethylated lysines among other substrates, Tudor domains can bind to both methylated lysines and arginines, PWWP domains bind to methylated lysines, and PHD fingers can bind to a variety of unmodified and modified histone tail substrates (Musselman et al. 2012). The chromatin remodeling complexes of the SWI/SNF, ISWI, CHD, and INO80 families are characterized by the presence in their ATPase subunits of a combination of a SWI2_N and a Helicase_C domain (although the domains themselves are individually not necessarily restricted to proteins with such functions), plus additional domains in each of the families. These domains include HAND, SLIDE, SnAC, and DBINO as shown in the figure. In addition, the number of detected proteins containing both a SWI2_N and a Helicase_C domain is shown, as well as those containing all three of Chromo, SWI2_N, and Helicase_C domains (i.e., putative CHD-family ATPases) and proteins with all three of the SLIDE, SWI2_N, and Helicase_C domains (putative ATPases in the ISWI family). SWIB and SWIRM are domains found in some of the noncatalytic components of SWI/SNF chromatin remodeling complexes. The FACT complex consists of two proteins, Spt16 and SSRP1; the latter contains the SSrecog domain. In addition to nucleosome chaperoning during transcription elongation, multiple histone chaperones are involved in replication-coupled and replication-independent nucleosome assembly and histone exchange (Burgess and Zhang 2013); among them, NAP1 loads H2A-H2B dimers (Mosammaparast et al. 2002), while CAF-1 and HIRA transfer H3-H4 dimers (Verreault et al. 1996; Ray-Gallet et al. 2002) during and independent of replication, respectively, which are in turn loaded onto them by ASF1 (English et al. 2006). The figure shows the number of proteins detected containing each of the domains (a threshold of was applied using HMMER3.0), with the exception of the CTD repeats of the Rpb1 subunit of RNA polymerase II, which were manually annotated in putative Rpb1 homologs (note that in that case “0” means that CTD repeats are not apparent in the Rpb1 homolog, while “–” refers to absence of detection of an Rpb1 homolog). The results for the organisms labeled in orange are based on available genome assemblies in contrast to those lettered in black, which are derived from transcriptome assemblies.
Remarkably, most dinoflagellates contain a larger number of sirtuins and HDACs than most other eukaryotes. While these deacetylases need not be acting on histones (especially sirtuins), these observations support the occurrence of histone acetylation and deacetylation in dinoflagellates.
The corresponding HAT enzymes are more difficult to identify. There are multiple families of HATs, and not all of them act specifically on histones. In particular, the GCN5-related N-acetyltransferases type (GNAT-type) HATs also acetylate a wide variety of other substrates and are not even restricted to eukaryotes (Xie et al. 2014); these are abundant in dinoflagellates but it is not certain that they acetylate histones. For these reasons, the adapter components of the chromatin modifying complexes, in which they are usually found, were searched for. However, none were detected in any of the dinoflagellate species examined. Both KAT11 and MYST HATs were found in dinotoms, in P. marinus, and in A. tamarense among the dinoflagellates; a KAT11 HAT is found in one of the Symbiodinium isolates and in Gymnodinium, and a MYST HAT is found in Oxyrrhis. No other obvious HATs were found, thus the most likely explanation is that dinoflagellate HATs primarily belong to the GNAT family, and might participate in chromatin modifying complexes of derived composition.
The number of SET-domain proteins in dinoflagellates is strikingly large and exceeds anything observed in other eukaryotes. Not only that, but numerous Dot1 proteins are also observed, as is an expansion of the PRMT5 family. Accordingly, the diversity of Jmj proteins is also large. These are certainly underestimates given that they are based on transcriptomes and not complete genomes, although the extent of actual functional diversification is not yet clear. Of note, these observations are confirmed in the S. minutum genome assembly; therefore it is unlikely that they are due to diversity of alternative transcript products in the transcriptome.
In contrast to the expansion of lysine and arginine methyltransferases, the number of proteins with classical “reader” domains is reduced in dinoflagellates, with the exception of the WD40 domain. Proteins with bromodomains, chromodomains, PHD fingers, Tudor or PWWP domains are found throughout the dinoflagellate phylogeny, but they are fewer in number compared to other protozoans, with the exception of dinotoms. Dinoflagellates do express a large number of proteins with WD40 domains; however, the WD40 domain is by no means restricted to the context of reading chromatin marks, and is found in many other proteins in the cell (Xu and Min 2011).
Finally, putative chromatin remodelers in dinoflagellates were identified. The number of candidate SWI/SNF ATPases is comparable to that in other protozoans, although clear CHD and ISWI homologs are detected in few species other than dinotoms (Figure 13).
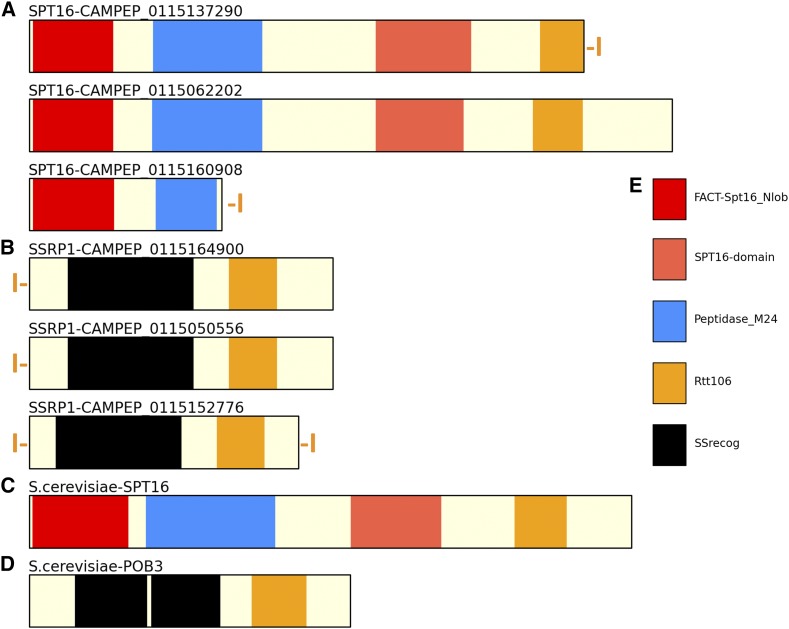
The FACT complex
The FACT complex consists of two subunits, Spt16 and SSRP1, and plays a key role in transcription through chromatinized DNA templates (Orphanides et al. 1998; Reinberg and Sims 2006). Its presence or absence is therefore highly informative of the role that histones might play in dinoflagellates. Strikingly, homologs of the components of FACT are detected in almost all dinoflagellates (Figure 13), with an identical domain structure to that of FACT subunits in yeast and human (Figure 14), strongly implying that they indeed constitute a bona fide FACT complex. Furthermore, while the existence of multiple variants of each subunit in dinotoms is expected (Figure S12), a diversity of subunits is also observed in other species, as well as in Perkinsus. Intriguingly, in some cases variant SSRP1 subunits also contain HMG boxes, a DNA binding domain (Figure S13), which is not observed in FACT subunits of other eukaryotes; its functional significance is currently not clear.
Figure 14.
FACT complex subunits and their domain organization in Polarella glacialis. (A) Polarella glacialis SPT16 proteins; (B) Polarella glacialis SSRP1 proteins; (C) Saccharomyces cerevisiae SPT16; (D) Saccharomyces cerevisiae SSRP1 (POB3); (E) Domain color code. An orange “I” in front and/or after the protein indicates that the protein sequence is known to be not represented completely in the transcriptome assembly.
The ubiquitous presence of the FACT complex throughout the dinoflagellate phylogeny suggests that transcription through nucleosomal arrays does occur in dinoflagellate nuclei.
Histone chaperones
While the FACT complex disassembles and reassembles nucleosomes during transcription, other histone chaperones act during replication-dependent and replication-independent nucleosome assembly and histone exchange (Burgess and Zhang 2013). NAP1 loads H2A-H2B dimers (Mosammaparast et al. 2002), while CAF-1 is the replication-dependent and Hira the replication-independent chaperone for H3-H4 dimers (Verreault et al. 1996; Ray-Gallet et al. 2002); both receive the dimers from another loading factor, ASF1 (English et al. 2006).
Figure 13 shows the number of detected homologs for each factor. CAF-1 is only found in dinotoms and in Perkinsus, while Hira is detected in few but phylogenetically widely distributed dinoflagellate species. Given that ASF1 is found in almost all dinoflagellates, the absence of detection of Hira can be interpreted as a probable case of widespread false negatives. If, however, CAF-1 is genuinely absent in dinoflagellates (while Hira is present), that would have very intriguing implications for the role of nucleosomes in dinoflagellate chromatin (see Discussion section).
Remarkably, the majority of dinoflagellates express a large number of NAP1 proteins (Figure 13); this is also observed in a few other unicellular eukaryotes, but rarely to the same extent. One explanation for the expansion of the NAP1 family is that it might be due to an underlying diversification of NAP1 substrates, in the form of different H2A/H2B variants and dimers.
RNA polymerase II CTD tails
The C-terminal domain of the largest subunit of RNA polymerase II (Rpb1) and its PTMS play a crucial role in coordinating numerous processes during the transcriptional cycle. In almost all eukaryotes (Yang and Stiller 2014) the tail consists of multiple copies of a consensus heptad sequence, YSPTSPS, which contains five phosphorylation sites and two proline isomerization sites. Specific modifications are deposited on and removed from these repeats during each phase of the transcriptional cycle, and they serve to recruit various effector and regulatory proteins (Buratowski 2009), constituting the so-called CTD code (Eick and Geyer 2013), operating in concert with the histone code.
Rpb1 homologs with CTD tails were identified in most dinoflagellates; curiously, in a few cases more than one homolog was found, even in nondinotom species. The heptad repeats are recognizable in most species (Figure 14); however, it is only in dinotom Rpb1 proteins that they closely match the consensus sequence (Figure S14B). In all other dinoflagellates, they are highly divergent (example shown in Figure S14A). This can be interpreted as a sign that some of the classical functionality of the CTD code has been lost in dinoflagellates, although divergent CTD repeats are seen in other eukaryotes too. The PTMs of the tails have not been directly studied in any such group, which will be necessary to clarify the roles they play in transcription.
Discussion
This study presents the most comprehensive characterization of dinoflagellate histone proteins carried out so far. The presence of histones, despite their low abundance in dinoflagellate chromatin, is confirmed and generalized to all species for which genomic or transcriptomic sequence data are available. The properties of their sequences are integrated with the phylogenetic distribution of key components of the histone modification, chromatin remodeling, and transcriptional machineries. These analyses reveal that:
Core histones are present in all dinoflagellates, typically in multiple variants. The linker histone H1 is also detected in most species. Dinoflagellate histone variants include putative homologs, or at least functional analogs, of well-known variants in other eukaryotes, including H2A.Z, H2A.X, and possibly even H3.1/H3.3.
Overall, dinoflagellates express the most divergent histone proteins among all autonomous eukaryote nuclei: their histones exhibit frequent loss of key residues highly conserved among all other eukaryotes and many variants are elongated. The latter, however, is not a universal feature, as a fairly conventional set of histones (in terms of protein length, but not necessarily sequence conservation) is found in many species.
Dinoflagellate histones are expressed at lower levels than DVNPs, consistent with the limited role of histones in chromatin packaging.
The histone code exhibits significant divergence from the conventional eukaryote state but some of its key elements are conserved. These include H3K4 and some of the residues involved in transcription elongation. Histone marks associated with heterochromatin might also be conserved. The components of the histone code involved in chromatin dynamics during mitosis are among the least conserved.
Dinoflagellates express members of most protein families comprising the chromatin mark reader, writer, and eraser toolkit. Remarkably, lysine and arginine methyltransferases and demethylases have even expanded in the group. In contrast, the reader proteins display reduced diversity.
There is evidence for the presence of ATPases involved in chromatin remodeling in dinoflagellates.
Histone chaperones are definitely present and the NAP1 family has even expanded. It is possible that no replication-dependent H3-H4 chaperone is present, although that remains to be confirmed.
Remarkably, a complete FACT complex is also present, occasionally in multiple variants, meaning that transcription through nucleosome arrays occurs in dinoflagellates.
The RNA polymerase II CTD tails exhibit a high degree of divergence, making it unclear to what extent the CTD code is conserved.
Finally, Perkinsus, the sister lineage of dinoflagellates, has a mostly conventional set of histones, yet some of its histone variants exhibit divergence features that resemble what is observed in core dinoflagellates. This includes the otherwise most highly conserved and with very few variants histone H4. It is plausible that although perkinsid chromosomes are organized into typical nucleosomal chromatin, the first steps in the process of evolution toward the derived state seen in dinoflagellates might have taken place prior to the separation of the two groups.
How are we to interpret these observations? The presence of the FACT complex is key to the piecing together of a working model as it means that transcription through nucleosomes occurs in dinoflagellates. Less clear is the context in which it is happening. The permanent condensation and liquid crystalline state of dinoflagellate chromosomes is expected to not be particularly permissive to transcription. Thus, models of transcription in dinoflagellates have proposed that most of it happens on decondensed extrachromosomal chromatin loops that stick out of the permanently condensed general chromatin mass (Wisecaver and Hackett 2011). If that model is correct, it could be that these temporary loops associate with nucleosomes, and transcription proceeds in a more or less conventional manner, with promoters defined by the presence of H3K4me3 and maybe even H2A.Z, and a transcription elongation cycle involving the action of FACT and the deposition of H3K36me3 and some of the other marks usually associated with active transcription. The possible absence of replication-dependent histone chaperones also fits such a picture, as they would not be necessary if the primary mode of nucleosome assembly is related to active transcription outside of the S-phase (however, it is not directly compatible with the possible presence of replication-dependent and replication-independent H3.1/H3.3 variants). The observation that histone marks involved in chromatin condensation during mitosis are poorly conserved in dinoflagellates is also consistent with it.
Of course, many unknowns remain, for example, how the formation and identity of chromatin loops is regulated, which histones exactly associate with them, among the numerous variants observed, and what the role of the ones (if any) associating with other areas of the genome is. It is tempting and natural to think that the least derived histones are the ones forming nucleosomal arrays on loops.
This is the most plausible explanation, but it need not be true. Examples of significant innovation in the composition of the nucleosome are known; for example, in bdelloid rotifers, a metazoan lineage with otherwise normal chromatin, conventional histone H2A has apparently been completely replaced by an elongated H2A variant (Van Doninck et al. 2009). Given the uniqueness and extreme divergence of dinoflagellate nuclear organization, the possibility of analogous innovations cannot be rejected, including some that are restricted to individual lineages and not common to all dinoflagellates.
Additional clues might be provided by sperm cells, the other notable example of nuclei in which histones do not play a primary packaging role, a system that has been studied much more extensively. Chromatin undergoes a dramatic transformation during spermatogenesis as histones are largely replaced by protamines and it becomes condensed (Rathke et al. 2014), a condition reminiscent of that of dinoflagellate nuclei. However, not all histones are removed – recent genome-wide mapping studies have revealed that they remain associated with a small fraction of the genome (Hammoud et al. 2009; Erkek et al. 2013; Carone et al. 2014), in particular the promoters of developmental regulators. Of note, it seems that nucleosomal patterns upon nuclease digestion of sperm chromatin only become apparent under preparation conditions that stabilize protein–DNA interactions (Carone et al. 2014). Sperm is also the site of expression of many unique histone variants (Wiedemann et al. 2010; Soboleva et al. 2011; Schenk et al. 2011), thought to play a role in both the process of histone replacement and in the final condensed chromatin. Parallels can be drawn between that system and dinoflagellates: variant histones might be associated with specific regions of dinoflagellate genomes even within an otherwise permanently condensed chromatin mass.
Specialized histone variants from other organisms might also have analogs in dinoflagellates, and these are not limited to centromeric histones. The group that might be most informative is the kinetoplastids. Dinoflagellates and kinetoplastids have evolved convergently in many aspects of their biology (Lukes et al. 2009), including the polycistronic organization of genes and the ubiquity of trans-splicing. Histone marks and variants have been studied in some kinetoplastids and intriguing discoveries have been made. For example, in Trypanosoma brucei novel variants of all four core histones mark the boundaries of polycistronic transcription units (Siegel et al. 2009), and another histone H3 variant is associated with telomeres (Lowell and Cross 2004). Analogous variants might be present in dinoflagellates. Chromosome dynamics during mitosis in kinetoplastids is also unique and apparently either highly derived or very deeply diverging (Akiyoshi and Gull 2013); trypanosomatid H3 histones lack H3S10 (Sullivan et al. 2006), which might be related to this fact, and has similarities to the poor conservation of histone phosphorylation sites in dinoflagellates.
The restriction of histones to certain sections of the genome possibly combined with their diversification and subfunctionalization can explain the relaxation of the constraints on their sequence imposed by the histone code. The functional role of DVNPs is highly relevant for clarifying to what extent that hypothesis is true. The focus of this study has been on histones, but DVNPs and any other histone-like proteins in dinoflagellates are no less interesting. They could well function as much more than mere packaging proteins and be involved in a completely new “DVNP code”, analogous to the histone code. Not only that but a large number of distinct DVNPs are expressed in all dinoflagellates; the divisions of functions between them are completely unknown at present. A separate DVNP code might provide an explanation for the expansion of some portions of the epigenetic writer and eraser toolkit, as DVNPs are most likely methylated by novel, specialized KMT enzymes. Less clear in such context is why proteins with classical reader domains are reduced in number. Some possible ways out of this conundrum include the takeover of such functionalities by proteins with other domains (such as WD40) and the evolution of entirely novel ones.
DVNPs and permanent condensation of chromatin could also explain the poor conservation of histone phosphorylation during the mitotic cycle – dinoflagellate histones are not involved in a nucleosomal compaction and decompaction cycle during mitosis. Similarly, DVNPs are likely to be the main component of what is the equivalent of heterochromatin, which, if true, makes it unclear what functional roles might be left for histone-based heterochromatinization; this is the reason why the putative conservation of heterochromatin marks is the most uncertain among the modifications discussed here.
An enormous amount remains to be learned about dinoflagellate chromatin, transcription, and transcriptional and post-transcriptional regulation. The answers to these questions will derive from the application of the genomics and proteomics tools that have successfully been applied to the study of chromatin structure and histone modifications in model eukaryote systems. These include the use of targeted mass-spectrometry analysis to reveal the exact PTMs deposited in vivo onto histones and DVNPs (Tan et al. 2011), and of functional genomic assays such as ChIP-seq (Johnson et al. 2007), ATAC-seq (Buenrostro et al. 2013) and DNAse-seq (Hesselberth et al. 2009), and Hi-C/ChIA-PET (Lieberman-Aiden et al. 2009; Fullwood et al. 2009) to characterize chromatin structure, the distribution of histones, histone marks, and other chromatin-associated proteins along dinoflagellate genomes, and their three-dimensional organization. A prerequisite for such studies is the availability of high-quality and reasonably complete genome sequences from several species, which should be facilitated by the ongoing advances in genome sequencing technologies.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation grant NSF-MCB-1518060 to M.L. It is also based upon work supported by the National Science Foundation under Grant No. CNS-0521433.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.023275/-/DC1
Communicating editor: C. Nislow
Literature Cited
- Akiyoshi B., Gull K., 2013. Evolutionary cell biology of chromosome segregation: insights from trypanosomes. Open Biol. 3(5): 130023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey V. G., Faulkner R., Mirsky A. E., 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan L., Milavetz B., 2010. Decoding the histone H4 lysine 20 methylation mark. Crit. Rev. Biochem. Mol. Biol. 45(5): 440–452. [DOI] [PubMed] [Google Scholar]
- Baldi S., Becker P. B., 2013. The variant histone H2A.V of Drosophila – three roles, two guises. Chromosoma 122(4): 245–258. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., et al. , 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410(6824): 120–124. [DOI] [PubMed] [Google Scholar]
- Barber C. M., Turner F. B., Wang Y., Hagstrom K., Taverna S. D., et al. , 2004. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 112(7): 360–371. [DOI] [PubMed] [Google Scholar]
- Barbrook A. C., Howe C. J., 2000. Minicircular plastid DNA in the dinoflagellate Amphidinium operculatum. Mol. Gen. Genet. 263: 152–158. [DOI] [PubMed] [Google Scholar]
- Basnet H., Su X. B., Tan Y., Meisenhelder J., Merkurjev D., et al. , 2014. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 516(7530): 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T., Aranda M., Sunagawa S., Yum L. K., Desalvo M. K., et al. , 2012. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7(4): e35269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blus B. J., Wiggins K., Khorasanizadeh S., 2011. Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 46(6): 507–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10(12): 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36(4): 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. J., Zhang Z., 2013. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20(1): 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone B. R., Hung J. H., Hainer S. J., Chou M. T., Carone D. M., et al. , 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell 30(1): 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. H., Wong J. T., 2007. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acids Res. 35(8): 2573–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. H., Kwok A. C., Tsang J. S., Wong J. T., 2006. Alveolata histone-like proteins have different evolutionary origins. J. Evol. Biol. 19(5): 1717–1721. [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y., Li J. F., Rizzo P. J., Hastings J. W., Fagan T. F., 2002. Cloning, expression, and charactertization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum (Dinophyceae). J. Phycol. 38: 1–9. [Google Scholar]
- Clapier C. R., Cairns B. R., 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., et al. , 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107(50): 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S., Weiss T., Mohn F., Lange U. C., Ziegler-Birling C., et al. , 2009. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat. Struct. Mol. Biol. 16(7): 777–781. [DOI] [PubMed] [Google Scholar]
- Desvoyes B., Fernández-Marcos M., Sequeira-Mendes J., Otero S., Vergara Z., et al. , 2014. Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 5: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S. S., Lee S. H., Kan P. Y., Voigt P., Ma L., et al. , 2012. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 26(24): 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S. C., Zhang X., Trievel R. C., Cheng X., 2005. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6(8): 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M. F., Altschuler S. J., Wu L. F., Rando O. J., 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102(15): 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge J. D., 1965. Chromosome structure in the dinoflagellates and the problem of the mesocaryotic cell. Prog. Protozool 2: 264. [Google Scholar]
- Dodge J. D., 1971. A dinoflagellate with both a mesokaryotic and a eukaryotic nucleus: Part 1 fine structure of the nuclei. Protoplasma 73: 145–157. [DOI] [PubMed] [Google Scholar]
- Doenecke D., 2014. Chromatin dynamics from S-phase to mitosis: contributions of histone modifications. Cell Tissue Res. 356(3): 467–475. [DOI] [PubMed] [Google Scholar]
- Doyon Y., Côté J., 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14(2): 147–154. [DOI] [PubMed] [Google Scholar]
- Drinnenberg I. A., deYoung D., Henikoff S., Malik H. S., 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3: . 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobic B., Pérez-Cadahía B., Yu J., Kung S. K., Davie J. R., 2010. Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res. 38(10): 3196–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R., 2011. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7(10): e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5): 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Geyer M., 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113(11): 8456–8490. [DOI] [PubMed] [Google Scholar]
- English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K., 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127(3): 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S., Hisano M., Liang C. Y., Gill M., Murr R., et al. , 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 20(7): 868–875. [DOI] [PubMed] [Google Scholar]
- Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., et al. , 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12(12): 1052–1058. [DOI] [PubMed] [Google Scholar]
- Feng S., Jacobsen S. E., 2011. Epigenetic modifications in plants: an evolutionary perspective. Curr. Opin. Plant Biol. 14(2): 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa R. I., Bravo I., Fraga S., Garcés E., Llaveria G., 2009. The life history and cell cycle of Kryptoperidinium foliaceum, a dinoflagellate with two eukaryotic nuclei. Protist 160(2): 285–300. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42(Database issue): D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Allis C. D., 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425(6957): 475–479. [DOI] [PubMed] [Google Scholar]
- Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., et al. , 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438(7071): 1116–1122. [DOI] [PubMed] [Google Scholar]
- Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A. M., et al. , 2009. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 5(9): e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., et al. , 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462(7269): 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavelis G. S., White R. A., Suttle C. A., Keeling P. J., Leander B. S., 2015. Single-cell transcriptomics using spliced leader PCR: evidence for multiple losses of photosynthesis in polykrikoid dinoflagellates. BMC Genomics 16: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez F., 2005. A list of free-living dinoflagellate species in the world’s oceans. Acta Bot Croat 64(1): 129–212. [Google Scholar]
- Gornik S. G., Ford K. L., Mulhern T. D., Bacic A., McFadden G. I., et al. , 2012. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr. Biol. 22(24): 2303–2312. [DOI] [PubMed] [Google Scholar]
- Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., et al. , 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449(7164): 933–937. [DOI] [PubMed] [Google Scholar]
- Hake S. B., Allis C. D., 2006. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 103(17): 6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., et al. , 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460(7254): 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., et al. , 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106(6): 348–360. [DOI] [PubMed] [Google Scholar]
- Herzog M., Soyer M. O., 1981. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species Prorocentrum micans E. Eur. J. Cell Biol. 23(2): 295–302. [PubMed] [Google Scholar]
- Hesselberth J. R., Chen X., Zhang Z., Sabo P. J., Sandstrom R., et al. , 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6(4): 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A., Demidov D., Caperta A. D., Karimi R., Agueci F., et al. , 2007 Phosphorylation of histone H3 in plants: a dynamic affair. Biochim. Biophys. Acta 1769(5–6): 308–315. [DOI] [PubMed] [Google Scholar]
- Huang H., Sabari B. R., Garcia B. A., Allis C. D., Zhao Y., 2014. SnapShot: histone modifications. Cell 159(2): 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B., Pombert J. F., Keeling P. J., 2010. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One 5(5): e10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail I. H., Hendzel M. J., 2008. The γ-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ. Mol. Mutagen. 49(1): 73–82. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D., 2001. Translating the histone code. Science 293(5532):1074–1080. [DOI] [PubMed] [Google Scholar]
- Johansson, C., A. Tumber, K. Che, P. Cain, R. Nowaket al. , 2014 The roles of Jumonji-type oxygenases in human disease. Epigenomics 6(1): 89–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B., 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316(5830): 1497–1502. [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Tengs T., Oldach D., Stoecker D. K., 2006. Sequestration, performance, and functional control of cryptophyte plastids in the ciliate Myrionecta rubra (Ciliophora). J. Phycol. 42: 1235–1246. [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301(5636): 1096–1099. [DOI] [PubMed] [Google Scholar]
- Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., Sif S., 2011. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36(12): 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J., Burki F., Wilcox H. M., Allam B., Allen E. E., et al. , 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12(6): e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Vermeulen M., et al. , 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449(7164): 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., et al. , 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 41(3): 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., 2007. Chromatin modifications and their function. Cell 128(4): 693–705. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410(6824): 116–120. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3): R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950): 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Zhang H., Zhuang Y., Tran B., Gill J., 2010. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc. Natl. Acad. Sci. USA 107(46): 20033–20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann E., 1924. Der Bau der Hülle bei Heterocapsa und Kryptoperidinium foliaceum (Stein) n. nom. (Zugleich eine vorläufige Mitteilund). Botanisches Archiv 5: 114–120. [Google Scholar]
- Liu Y., Taverna S. D., Muratore T. L., Shabanowitz J., Hunt D. F., et al. , 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 21(12): 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C. D., Martin L. E., Roberts E. C., Watts P. C., Wootton E. C., et al. , 2011. Collection, isolation and culturing strategies for Oxyrrhis marina. J. Plankton Res. 33(4): 569–578. [Google Scholar]
- Lowell J. E., Cross G. A., 2004. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J. Cell Sci. 117(Pt 24): 5937–5947. [DOI] [PubMed] [Google Scholar]
- Lucas I. A. N., Vesk M., 1990. The fine structure of two photosynthetic species of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol. 26: 345–357. [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389(6648): 251–260. [DOI] [PubMed] [Google Scholar]
- Lukes J., Leander B. S., Keeling P. J., 2009. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc. Natl. Acad. Sci. USA 106(Suppl 1): 9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanero S., Arana P., Puertas M. J., Houben A., 2000. The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109(5): 308–317. [DOI] [PubMed] [Google Scholar]
- Mariadason J. M., 2008. HDACs and HDAC inhibitors in colon cancer. Epigenetics 3(1): 28–37. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Zhou M. M., 2014. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6(7): a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. M., Pouchnik D. J., Walker J. L., Wyrick J. J., 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167(3): 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., 2005. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 17(3): 274–280. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Wagner E. J., Duronio R. J., 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9(11): 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E., Yin N., Wissmann M., Kunowska N., Fischer K., et al. , 2008. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat. Cell Biol. 10(1): 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E., Imhof A., Patel D., Kahl P., Hoffmeyer K., et al. , 2010. Phosphorylation of histone H3T6 by PKC controls demethylation at histone H3K4. Nature 464(7289): 792–796. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Ewart C. S., Pemberton L. F., 2002. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21(23): 6527–6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratoglu S., Georgieva S., Pápai G., Scheer E., Enünlü I., et al. , 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23(1): 306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman C. A., Lalonde M. E., Côté J., Kutateladze T. G., 2012. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19(12): 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Nitshitani G., Tomaru Y., Sakiyama S., Kamiyama T., 2008. Predation by the toxic dinoflagellate Dinophysis fortii on the ciliate Myrionecta rubra and observation of sequestration of ciliate chloroplasts. J. Phycol. 44: 909–922. [DOI] [PubMed] [Google Scholar]
- Nelson C. J., Santos-Rosa H., Kouzarides T., 2006. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126(5): 905–916. [DOI] [PubMed] [Google Scholar]
- Nguyen A. T., Zhang Y., 2011. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25(13): 1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D., 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92(1): 105–116. [DOI] [PubMed] [Google Scholar]
- Orr R. J., Murray S. A., Stüken A., Rhodes L., Jakobsen K. S., 2014. When naked became armored: an eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS One 7(11): e50004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero S., Desvoyes B., Gutierrez C., 2014. Histone H3 dynamics in plant cell cycle and development. Cytogenet. Genome Res. 143(1–3): 114–124. [DOI] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., et al. , 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125(4): 703–717. [DOI] [PubMed] [Google Scholar]
- Polioudaki H., Markaki Y., Kourmouli N., Dialynas G., Theodoropoulos P. A., et al. , 2004. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 560(1–3): 39–44. [DOI] [PubMed] [Google Scholar]
- Postberg J., Forcob S., Chang W. J., Lipps H. J., 2010. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 10: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss U., Landsberg G., Scheidtmann K. H., 2003. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31(3): 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., et al. , 2010. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470(7333): 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C., Baarends W. M., Awe S., Renkawitz-Pohl R., 2014. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 1839(3): 155–168. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., et al. , 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9(5): 1091–1100. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Sims R. J., 3rd, 2006. de FACTo nucleosome dynamics. J. Biol. Chem. 281(33): 23297–23301. [DOI] [PubMed] [Google Scholar]
- Rill R. L., Livolant F., Aldrich H. C., Davidson M. W., 1989. Electron microscopy of liquid crystalline DNA: direct evidence for cholesteric-like organization of DNA in dinoflagellate chromosomes. Chromosoma 98(4): 280–286. [DOI] [PubMed] [Google Scholar]
- Rizzo P. J., 2003. Those amazing dinoflagellate chromosomes. Cell Res. 13: 215–217. [DOI] [PubMed] [Google Scholar]
- Rizzo P. J., Burghardt R. C., 1982. Histone-like protein and chromatin structure in the wall-less dinoflagellate Gymnodinium nelsoni. Biosystems 15(1): 27–34. [DOI] [PubMed] [Google Scholar]
- Rizzo P. J., Nooden L. D., 1972. Chromosomal proteins in the dinoflagellate alga Gyrodinium cohnii. Science 176: 796–797. [DOI] [PubMed] [Google Scholar]
- Roberts A., Pachter L., 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10(1): 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Morse D., 2012. A full suite of histone and histone modifying genes are transcribed in the dinoflagellate Lingulodinium. PLoS One 7(4): e34340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T., Beuch S., Reuter G., 2013. Lysine-specific histone demethylase LSD1 and the dynamic control of chromatin. Biol. Chem. 394(8): 1019–1028. [DOI] [PubMed] [Google Scholar]
- Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A., 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27(3): 393–405. [DOI] [PubMed] [Google Scholar]
- Sala-Rovira M., Geraud M. L., Caput D., Jacques F., Soyer-Gobillard M.-O., et al. , 1991. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCc) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta). Chromosoma 100: 510–518. [DOI] [PubMed] [Google Scholar]
- Sanchez R., Meslamani J., Zhou M. M., 2014 The bromodomain: from epigenome reader to druggable target. Biochim. Biophys. Acta 1839(8): 676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka A., Seiser C., 2012. Histone H3 phosphorylation: a versatile chromatin modification for different occasions. Biochimie 94: 2193–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka A., Hartl D., Goiser M., Pusch O., Stocsits R. R., et al. , 2014. H3S28 phosphorylation is a hallmark of the transcriptional response to cellular stress. Genome Res. 24(11): 1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk R., Jenke A., Zilbauer M., Wirth S., Postberg J., 2011. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120(3): 275–285. [DOI] [PubMed] [Google Scholar]
- Schnepf E., Elbrächter M., 1988. Cryptophycean-like double membrane-bound chloroplast in the dinoflagellate, Dinophysis Ehrenb.: evolutionary, phylogenetic and toxicological implications. Bot. Acta 101: 196–203. [Google Scholar]
- Schwartz S., Meshorer E., Ast G., 2009. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16(9): 990–995. [DOI] [PubMed] [Google Scholar]
- Schwope R. M., Chalker D. L., 2014. Mutations in Pdd1 reveal distinct requirements for its chromodomain and chromoshadow domain in directing histone methylation and heterochromatin elimination. Eukaryot. Cell 13(2): 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A., 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75: 243–269. [DOI] [PubMed] [Google Scholar]
- Shilatifard A., 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., et al. , 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23(15): 1399–1408. [DOI] [PubMed] [Google Scholar]
- Siegel T. N., Hekstra D. R., Kemp L. E., Figueiredo L. M., Lowell J. E., et al. , 2009. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 23(9): 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E., 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49(5): 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva T. A., Nekrasov M., Pahwa A., Williams R., Huttley G. A., et al. , 2011. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol. 19(1): 25–30. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I., 2005. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem. Cell Biol. 83(3): 385–395. [DOI] [PubMed] [Google Scholar]
- Stein, F., 1883 Die Naturgeschichte der Arthrodelen Flagellaten, Vol. III Pt. 2: Der Organismus der Infusionstiere, pp. 1–30. W. Engelmann, Leipzig. [Google Scholar]
- Sullivan W. J., Jr, Naguleswaran A., Angel S. O., 2006. Histones and histone modifications in protozoan parasites. Cell. Microbiol. 8(12): 1850–1861. [DOI] [PubMed] [Google Scholar]
- Sun L., Wang M., Lv Z., Yang N., Liu Y., et al. , 2011. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc. Natl. Acad. Sci. USA 108(51): 20538–20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Fukushima T., Shinkai Y., 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276(27): 25309–25317. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Watanabe Y., Takano-Shimizu T., Kondo S., 2006. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 235(9): 2449–2459. [DOI] [PubMed] [Google Scholar]
- Takishita K., Koike K., Maruyama T., Ogata T., 2002. Molecular evidence for plastid robbery (Kleptoplastidy) in Dinophysis, a dinoflagellate causing diarrhetic shellfish poisoning. Protist 153: 293–302. [DOI] [PubMed] [Google Scholar]
- Talbert P. B., Ahmad K., Almouzni G., Ausió J., Berger F., et al. , 2012. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30(12): 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Luo H., Lee S., Jin F., Yang J. S., et al. , 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146(6): 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Voss A. K., 2007. The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle 6(6): 696–704. [DOI] [PubMed] [Google Scholar]
- Tomas R. N., Cox E. R., 1973. Observations on symbiosis of Peridinium Balticum and its intracellular alga. 1. Ultrastructure. J. Phycol. 9: 304–323. [Google Scholar]
- Tsutsui T., Fukasawa R., Shinmyouzu K., Nakagawa R., Tobe K., et al. , 2013. Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J. Biol. Chem. 288(29): 20955–20965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., Vromans M. J., Bouwman K., van der Waal M. S., Hadders M. A., et al. , 2015. Inter-domain cooperation in INCENP promotes Aurora B relocation from centromeres to microtubules. Cell Reports 12(3): 380–387. [DOI] [PubMed] [Google Scholar]
- Van Doninck K., Mandigo M. L., Hur J. H., Wang P., Guglielmini J., et al. , 2009. Phylogenomics of unusual histone H2A variants in Bdelloid rotifers. PLoS Genet. 5(3): e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., 2009. The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 53(2–3): 303–322. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P. D., Kobayashi R., Stillman B., 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87(1): 95–104. [DOI] [PubMed] [Google Scholar]
- Wagner E. J., Carpenter P. B., 2012. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13(2): 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller R. F., Jackson C. J., 2009. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. BioEssays 31: 237–245. [DOI] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J. R., Niedzialkowska E., Banerjee B., et al. , 2010. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330(6001): 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., He Q., Liu F., Cheng Z., Talbert P. B., et al. , 2011. Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid Brassica species. Chromosoma 120(4): 353–365. [DOI] [PubMed] [Google Scholar]
- Wang L., Tang Y., Cole P. A., Marmorstein R., 2008. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18(6): 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo M. J., Rizzo P. J., 2000. Characterization of Gymnodinium mikimotoi (Dinophyceae) nuclei and identification of the major histone-like protein, HGm. J. Phycol. 36: 584–589. [DOI] [PubMed] [Google Scholar]
- Waterborg J. H., 2012. Evolution of histone H3: emergence of variants and conservation of post-translational modification sites. Biochem. Cell Biol. 90(1): 79–95. [DOI] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., 2009. Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9): 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann S. M., Mildner S. N., Bánisch C., Israel L., Maiser A., et al. , 2010. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 190(5): 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins B. J., Rall N. A., Ostwal Y., Kruitwagen T., Hiragami-Hamada K., et al. , 2014. A cascade of histone modifications induces chromatin condensation in mitosis. Science 343(6166): 77–80. [DOI] [PubMed] [Google Scholar]
- Wisecaver J. H., Hackett J. D., 2011. Dinoflagellate genome evolution. Annu. Rev. Microbiol. 65: 369–387. [DOI] [PubMed] [Google Scholar]
- Wong J. T., New D. C., Wong J. C., Hung V. K., 2003. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell 2(3): 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Allis C. D., Coonrod S., 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11: 344–355. [DOI] [PubMed] [Google Scholar]
- Xie L., Zeng J., Luo H., Pan W., Xie J., 2014. The roles of bacterial GCN5-related N-acetyltransferases. Crit. Rev. Eukaryot. Gene Expr. 24(1): 77–87. [DOI] [PubMed] [Google Scholar]
- Xie W., Song C., Young N. L., Sperling A. S., Xu F., et al. , 2009. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell 33(4): 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Min J., 2011. Structure and function of WD40 domain proteins. Protein Cell 2(3): 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M., 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121(3): 375–385. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y., 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330(6001): 239–243. [DOI] [PubMed] [Google Scholar]
- Yang C., Stiller J. W., 2014. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc. Natl. Acad. Sci. USA 111(16): 5920–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu W., Shi L., Sun L., Liang J., et al. , 2011. HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44(1): 39–50. [DOI] [PubMed] [Google Scholar]
- Yuan C. C., Matthews A. G., Jin Y., Chen C. F., Chapman B. A., et al. , 2012. Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Reports 1(2): 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Sui Z., Chang L., Kang K., Ma J., et al. , 2014. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 537(2): 285–293. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Green B. R., Cavalier-Smith T., 1999. Single gene circles in dinoflagellate chloroplast genomes. Nature 400: 155–159. [DOI] [PubMed] [Google Scholar]
- Zheng B., Chen X., 2011. Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14(2): 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., et al. , 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20(4): 601–611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.