Abstract
Small nonprotein-coding microRNAs (miRNAs) are present in most eukaryotes and are central effectors of RNA silencing-mediated mechanisms for gene expression regulation. In plants, DICER-LIKE1 (DCL1) is the founding member of a highly conserved family of RNase III-like endonucleases that function as core machinery proteins to process hairpin-like precursor transcripts into mature miRNAs, small regulatory RNAs, 21–22 nucleotides in length. Zinc finger nucleases (ZFNs) were used to generate single and double-mutants of putative soybean DCL1 homologs, DCL1a and DCL1b, to confirm their functional role(s) in the soybean miRNA pathway. Neither DCL1 single mutant, dcl1a or dcl1b plants, exhibited a pronounced morphological or molecular phenotype. However, the dcl1a/dcl1b double mutant expressed a strong morphological phenotype, characterized by reduced seed size and aborted seedling development, in addition to defective miRNA precursor transcript processing efficiency and deregulated miRNA target gene expression. Together, these findings indicate that the two soybean DCL1 paralogs, DCL1a and DCL1b, largely play functionally redundant roles in the miRNA pathway and are essential for normal plant development.
Keywords: dicer-like, miRNA, genome engineering, ZFN, soybean
Plants produce two main small RNA (sRNA) populations, microRNAs (miRNAs) and small-interfering RNAs (siRNAs) (Chen 2010). miRNAs are generated from hairpin-like precursor transcripts that are partially self-complementary; the 21–22 nucleotide (nt) mature miRNAs are crucial for directing the targeting of endogenous messenger RNA (mRNA) transcripts for RNA-induced silencing complex (RISC)-mediated cleavage or, in some cases, translational repression (Millar and Waterhouse 2005; Bartel 2009; Voinnet 2009). siRNAs, which represent approximately 90–95% of the global sRNA population in plants, are generated from perfectly double-stranded RNA (dsRNA) derived from replicating viruses, transposons and other classes of repetitive DNA, or introduced transgenes (Waterhouse et al. 1998; Baulcombe 2004; Voinnet 2009). In addition, secondary siRNAs (21–22 nt) are derived from either mRNAs of protein-coding genes, or from long nonprotein-coding RNAs. The enzyme family responsible for processing a sRNA from its precursor transcript is the Dicer (Dcr) family of RNase III-like endonucleases (Bernstein et al. 2001). Many eukaryotes encode multiple Dcr family members, and each Dcr can mediate specialized, yet often redundant roles. In Caenorhabditis elegans, however, a single Dcr is responsible for both miRNA and siRNA production (Duchaine et al. 2006). Drosophila melanogaster encodes two Dcrs: Dcr1 is required for miRNA biogenesis and Dcr2 is responsible for siRNA production (Tomari and Zamore 2005). The first plant Dcr identified was in Arabidopsis thaliana (Arabidopsis) via the characterization of a mutant plant originally termed carpel factory (caf), due to the floral meristem defects expressed by caf mutants (Jacobsen et al. 1999). Subsequently, the CAF gene was determined to encode a protein with similarities to animal Dcrs, and was therefore renamed DICER-LIKE1 (DCL1). Furthermore, the severe developmental phenotype expressed by caf plants has been shown to result from the ectopic expression of genes that are normally under miRNA regulation. In brief, the dcl1 mutation led to reduced miRNA precursor transcript processing (cleavage) efficiency, reduced mature miRNA accumulation, and deregulated miRNA target transcript expression.
To date, several Arabidopsis dcl1 alleles have been reported, including the weak dcl1-7 (short integument1-1) hypomorphic mutant and the embryo lethal dcl1-5 (suspensor1-5) mutant (Schauer et al. 2002). In rice, hairpin directed DCL1 knock-down transformants express severe developmental phenotypes in strongly silenced lines and lesser pleiotropic developmental defects in weakly silenced lines (Liu et al. 2005). In common bean, Phaseolus vulgaris, the authors demonstrated disruption of miR399 expression in composite plants transformed with a PvDCL1 hairpin reagent (Valdes-Lopez et al. 2008). A maize dcl1 mutant plant named fuzzy tassel (fzt) was recently identified by screening an ethyl methane sulfonate-mutagenized population and fzt plants displayed severe developmental defects (Thompson et al. 2014). The model moss species, Physcomitrella patens encodes four DCL proteins, including two DCL1 homologs, DCL1a and DCL1b. Curiously, PpDCL1a and PpDCL1b have been demonstrated to play distinct functional roles in the P. patens miRNA pathway (Khraiwesh et al. 2010). The PpDCL1a protein functions identically to its Arabidopsis ortholog AtDCL1, processing miRNA precursor transcripts as well as being involved in the production of the phased class of siRNAs, termed pasiRNAs. However, the PpDCL1a paralog, PpDCL1b, is not required for miRNA production. Instead, PpDCL1b is involved in the miRNA-directed target transcript cleavage stage of the P. patens miRNA pathway (Khraiwesh et al. 2010).
The genome of the paleopolyploid legume Glycine max (soybean) has retained a high level of gene duplication, with approximately 75% of the genes present in more than a single copy (Schmutz et al. 2010; Roulin et al. 2013). This identifies soybean as an interesting system for investigating the divergence of ancient homologous genes, with the potential to exhibit patterns of subfunctionalization or neofunctionalization (Force et al. 1999; Curtin et al. 2012). Soybean has retained two putatively functional DCL1 gene copies that encode proteins which are highly conserved orthologs of the well-characterized AtDCL1. The GmDCL1a and GmDCL1b loci are located in a large syntenous block between chromosomes 3 and 19 (Schmutz et al. 2010; Curtin et al. 2012). GmDCL1a was designated with the gene identifier Glyma03g42290 in the version 1 genome assembly (Schmutz et al. 2010) and Glyma.03g262100 in the version 2 assembly. GmDCL1b was designated the identifiers Glyma19g45060 and Glyma.19g261200 in the first and second genome assemblies, respectively (Schmutz et al. 2010). While some transcriptional differences have been noted for the two GmDCL1 gene copies (Curtin et al. 2012), the duplicated nature of the two soybean DCL1 genes makes it difficult to define the functional role each plays in miRNA-directed RNA silencing in soybean.
While a number of soybean mutant lines have been generated via random mutagenesis (Cooper et al. 2008; Hancock et al. 2011; Cui et al. 2013; Bolon et al. 2014), mutants for these two GmDCL1 loci are not readily available. Therefore, targeted mutagenesis using site-specific nucleases (SSNs) offers a promising approach for creating site-specific mutations in these duplicate genes, as has been previously demonstrated in soybean (Curtin et al. 2011; Haun et al. 2014; Jacobs et al. 2015; Sun et al. 2015). SSNs are synthetic enzymes with programmable DNA target specificities that are used to introduce double-strand breaks at specific loci. The double-stranded break is rapidly repaired by the host’s error-prone nonhomologous end joining (NHEJ) repair pathway, a repair mechanism that often results in the introduction of small insertion-deletions (indels) that range in size from one to many hundreds of base pairs (Voytas 2013). In a previous study, we engineered a zinc finger nuclease (ZFN) to target both GmDCL1 copies and demonstrated its capacity to generate mutations in somatic hairy-root tissue. Here, the DCL1-targeting ZFN was used to generate heritable dcl1a and dcl1b mutations in whole soybean plants. Furthermore, the mutations were combined via a standard genetic crossing approach to generate the dcl1a/dcl1b double mutant. The miRNA pathway was assessed in single and double mutant plants to gain new insights into the functional redundancies exhibited by the GmDCL1 paralogs in this gene expression regulatory pathway that is central to plant development.
Material and Methods
The transformation of zinc finger nucleases targeting DCL1
The identification of the DCL1 target site, construction of the DCL1 zinc finger nuclease, and functional confirmation in somatic soybean tissue for targeting both copies of the DCL1a and DCL1b nuclease has been previously reported (Curtin et al. 2011). The ZFN transgene was transformed into Agrobacterium rhizogenes strain 18r12, and the DCL1-ZFN harboring A. rhizogenes culture was used to transform whole plant soybean cultivar ‘Bert’ as described previously (Paz et al. 2006; Curtin et al. 2011).
Detection of ZFN-induced mutations
DNA was extracted from soybean leaf chads of ZFN-transformed plants using the previously described CTAB extraction method (Curtin et al. 2008). To detect ZFN-induced mutations, both the DCL1a and DCL1b targeted loci were amplified using primers; 5′-ATCTGTACTAATGCAGAGGATCTGG-3′ and 5′-CAAGTGATCCAGG CAGTGGGTGTACG-3′ for DCL1a, and 5′-ATCCAGCCAAGCCTTCCGTATCCAC-3′ and 5′-AAGTTTTGTCATGTGTCTCTTCG-3′ for DCL1b. The resulting amplicons were digested for 6 hr at 37° with PsiI and visualized via standard gel electrophoresis. Cleavage-resistant amplicons were gel purified, cloned into the pGEM-T Easy vector (Promega), and sequenced. Subsequent genotyping of the DCL1 loci in nontransgenic progeny was performed using the PCR amplicon digestion strategy previously described in Curtin et al. (2011).
Development of single and double mutants and phenotypic analyses
T0 plants harboring mutations were allowed to self-pollinate to homozygous specific mutated alleles and remove the ZFN transgenes through genetic segregation in subsequent generations. Different combinations of alleles at the DCL1a and DCL1b loci were stacked together through a series of cross-fertilization and selfing generations. Plants were genotyped for the introduced transgene using PCR and genotyped for the mutant alleles using PCR followed by restriction digestion assays. The single and double-mutants were monitored for gross morphological phenotypes compared to the fully wild-type segregants. For the single mutant phenotypic analyses, seeds were germinated in sterile Metro Mix, inoculated with Bradyrhizobium USDA110 strain, and cultivated in a temperature controlled growth chamber for 14 d.
Small RNA and RNA-Seq library sequencing and informatics analyses
Total RNA from plant leaves or whole-plants was isolated using the Purelink Plant RNA Reagent (Invitrogen; Cat#12322-012). Small RNA libraries were constructed using the Illumina TruSeq Small RNA sample preparation kit (RS-200-0012) and RNA-Seq libraries were prepared using the Illumina TruSeq RNA sample preparation kit (RS-122-2001). All libraries were sequenced on an Illumina HiSeq2000 instrument at the Delaware Biotechnology Institute of the University of Delaware. The raw sequences were processed by bioinformatic removal of linker adaptor sequences and the trimmed reads matched to the soybean genome, Wm82.a2.v1 (Schmutz et al. 2010). To facilitate library comparison between each assessed mutant background, the count/abundance of each mutant population was normalized to its corresponding control based on a sequencing read depth and visualized using the gplots package in R statistical environment (R Development Core Team 2013). Raw RNA-Seq libraries were processed using Tophat (Trapnell et al. 2009) to map the reads to the soybean genome, Wm82.a2.v1 (Schmutz et al. 2010). Genome mapped reads were assembled and a ‘merged’ transcriptome assembly was generated using Cufflinks. Quantification of gene expression levels was done using featureCounts (Liao et al. 2014) to create count tables. These count tables were imported to R statistical environment (R Development Core Team 2013) where lowly expressed reads were filtered out and the data were normalized using DESeq to identify differentially expressed genes. Due to the difficulty of generating replicates, we needed to calculate dispersion under the assumption that the mean of the libraries being compared would be a good predictor for the dispersion. Under this assumption, it is possible to calculate differentially expressed transcripts from the samples (Love et al. 2014). While several miRNA targets were previously identified (Arikit et al. 2014), their analysis was done on a previous version of the soybean genome. In order to identify differentially expressed miRNA target genes with the current genome annotation, it was necessary to predict and validate individual miRNA target genes. The identification of miRNA targets was performed using the sPARTA package (Kakrana et al. 2014) built-in target prediction module mirFerno. Nine PARE libraries were utilized to validate those targets and targets were determined ‘real’ if the target had an adjusted P-value <0.05. The validated targets were then mapped back to the previously identified differentially expressed transcripts to confirm differentially expressed miRNA target genes.
Quantitative real time PCR (RT-qPCR)
RT-qPCR was used to quantify the relative expression of protein-encoding transcripts determined to be miRNA target genes. Reactions were performed using FastStart Universal SYBR Green Mix kit (Roche) on a LightCycler 480 Instrument (Roche). Primers were designed for a small subset of miRNA target transcripts based on previously published reports (Kulcheski et al. 2011; Song et al. 2011). ACTIN (gene model Glyma.15G050200) and ELONGATION FACTOR1 ALPHAGENE (Glyma.17G186600) were used as reference genes to normalize miRNA target gene expression. RT-qPCR reactions consisted of 0.3 μM of each primer and 1× FastStart Universal SYBR Green Mix in a final reaction volume of 10 μl with the cDNA template obtained from 2000 ng of total RNA. Primer efficiencies and Cq values were determined using the LingRegPCR v2013.0 software (Ruijter et al. 2009). Normalized copy number and expression were calculated using previously-described equations (Hellemans et al. 2007) for multiple reference genes, and the results were standardized using an adapted version of the Microsoft Excel Qgene template (Muller et al. 2002). Three technical replicates were performed for each of the two biological replicates. Error bars represent standard error of the mean between biological replicates.
Data availability
RNA-Seq raw data are available on Sequence Read Archive under the accession GSE76036 or can be accessed through the soybean gene analysis link (https://mpss.udel.edu/dbs/index.php?SITE=soy_RNAseq).
Results
Targeted mutagenesis of DCL1a and DCL1b with zinc finger nucleases
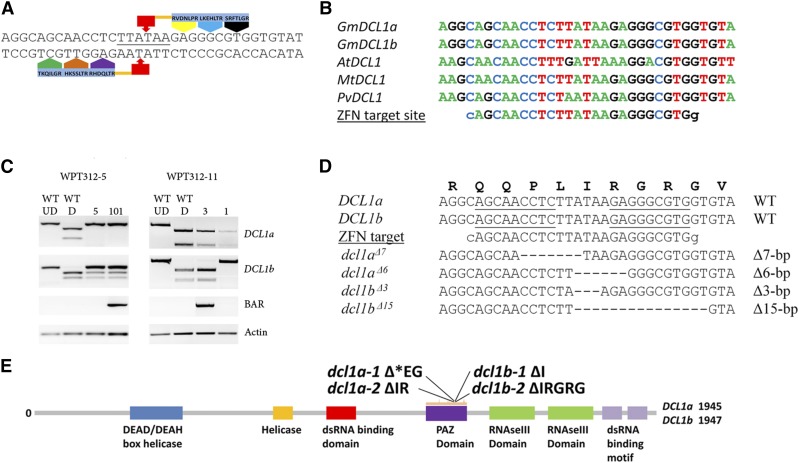
Previous work investigating the relative transcription of DCL1a and DCL1b across a panel of stress treatments indicated only minor expression differences for these two genes (Curtin et al. 2012). Publicly available RNA-Seq data from various soybean tissues (Nakano et al. 2006; Severin et al. 2010) further confirmed that DCL1a and DCL1b transcript levels are relatively similar to one another with no obvious signs of transcriptional specialization. Mutations for these genes were therefore desirable to investigate potential divergences between the DCL1 gene copies. Having previously reported a highly active CoDA-engineered ZFN (Sander et al. 2011) that can introduce somatic mutations in DCL1a and DCL1b (Curtin et al. 2011), we sought to introduce these reagents into whole plants with the purpose of creating knock-out mutations in both DCL1 loci (Figure 1A). Since the targeted region is located in the seventeenth and eighteenth exon of DCL1a and DCL1b respectively, coding sequences that encode for the functionally crucial PAZ domain, it was presumed that frameshift mutations at the targeted site would result in the generation of the desired functional knock-outs. Furthermore, the ZFN target site is highly conserved across multiple plant species, including Arabidopsis, Medicago and common bean (Figure 1B), further suggesting that the introduction of in-frame mutations in this region would perturb normal gene function.
Figure 1.
Generation of whole plant dcl1 mutants in soybean using zinc finger nuclease. (A) Schematic of zinc finger nuclease (ZFN) monomers bound to the target DNA. The target sequence of both left and right zinc finger arrays (ZFAs) recognize a 9 bp sequence, with underlined sequences representing the PsiI restriction site used in conjunction with a PCR-based assay to identify putative transformants harboring the desired mutations. (B) Sequence alignment of the homologous DCL1a and DCL1b ZFN target sites between legume species and Arabidopsis, highlighting the high level of sequence conservation of the targeted region. (C) The PCR digestion assays of T1 progeny from putative transformant lines, WPT312-5 and WPT312-11. Undigested bands indicate mutated DNA sequences. The presence of the ZFN transgene was detected by an additional PCR screen specific for the BAR transgene. ‘D’ and ‘UD’ denotes digested and undigested amplicons, respectively. (D) Sequence confirmation of the dcl1a and dcl1b mutant alleles identified in ZFN-transformed whole-plants. (E) A schematic of the DCL1 protein, highlighting the location of the four mutations within the exon encoding the functionally essential PAZ domain.
Two soybean T0 plants, WPT312-5 and WPT312-11, determined to putatively harbor site-specific mutations, were recovered from a pool of 12 transformant lines (Supporting Information, File S1). Respective 6 bp and 7 bp deletions were identified at the two DCL1a homologs in the T0 plant WPT312-5. This plant was also found to have a mono-allelic 3 bp deletion of one DCL1b locus. Therefore, the genotype of the T0 plant WPT312-5 was denoted as dcl1aΔ6/dcl1aΔ7/DCL1b/dcl1bΔ3 (Figure 2). Mutations were also confirmed in the second T0 plant, WPT312-11, which harbored a mono-allelic 15 bp deletion at the DCL1b locus (genotype was denoted as DCL1a/DCL1a/DCL1b/dcl1bΔ15) (File S1). Both plants were self-fertilized and the resulting T1 progeny screened to confirm heritable transmission of the mutated alleles, along with the removal of one or more transgenes by genetic segregation (Figure 1C). Six progeny were identified that carried the introduced mutations, but did not carry the transgenes. Since these mutant segregants were nontransgenic, this was considered the M1 generation (Figure 2). Seed from these M1 plants were bulk harvested for further analyses. The names and mutant allele composition of these plants are displayed in Table 1. The mutated alleles in the M1 plants were sequenced and heritable transmission of each mutation was confirmed (Figure 1, D and E, and File S2).
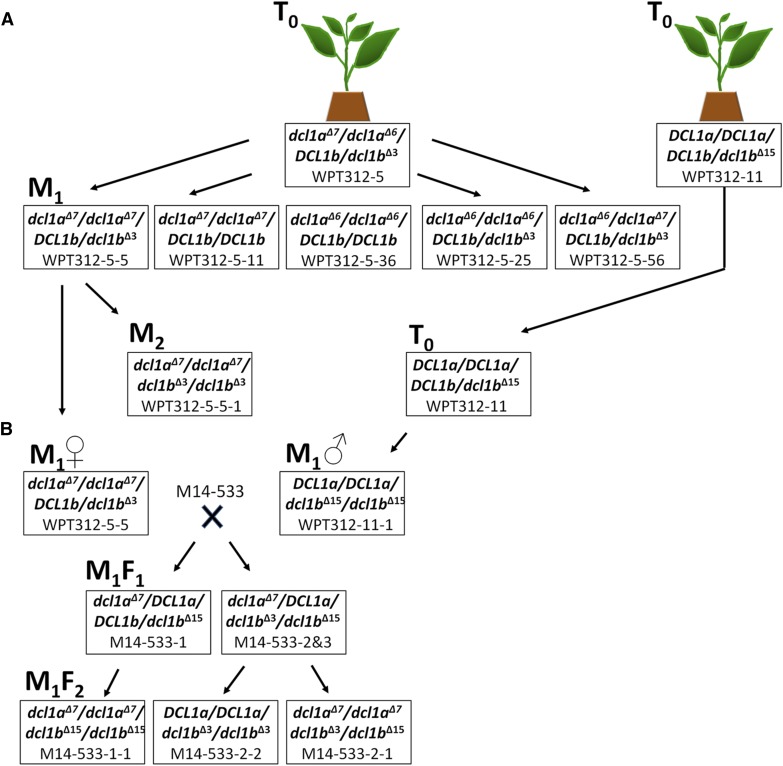
Figure 2.
Schematic outlining the experimental approach used to generate single and double dcl1 mutants. (A) Single and double dcl1 mutations were identified from whole plant transformants, WPT312-5 and WPT312-11. The two T0 plants were self-fertilized, and progeny screened for both single and double mutations (among the dcl1aΔ7, dcl1aΔ6, dcl1bΔ15 alleles) as well as removal of the ZFN (zinc finger nuclease) transgenes by genetic segregation. (B) The WPT312-5-5 (dcl1aΔ7/dcl1aΔ7/DCL1b/dcl1bΔ3) and WPT312-11-1 (DCL1a/DCL1a/dcl1bΔ15/dcl1bΔ15) plants were used as parents for a cross-fertilization experiment to introduce the dcl1bΔ15 mutant allele into the dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b background, and to also isolate the dcl1bΔ3 mutant allele in a wild-type DCL1a background.
Table 1. Combinations of dcl1a and dcl1b mutant alleles.
| Plant Identifier | Genotype | Generation | Transgene |
|---|---|---|---|
| WPT312-5 | dcl1aΔ7/dcl1aΔ6/DCL1b/dcl1bΔ3 | T0 | Yes |
| WPT312-11 | DCL1a/DCL1a/DCL1b/dcl1bΔ15 | T0 | Yes |
| WPT312-5-11 | dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b | M1 | None |
| WPT312-5-36 | dcl1aΔ6/dcl1aΔ6/DCL1b/DCL1b | M1 | None |
| WPT312-11-1 | DCL1a/DCL1a/dcl1bΔ15/dcl1bΔ15 | M1 | None |
| WPT312-5-5 | dcl1aΔ7/dcl1aΔ7/DCL1b/dcl1bΔ3 | M1 | None |
| WPT312-5-25 | dcl1aΔ6/dcl1aΔ6/DCL1b/dcl1bΔ3 | M1 | None |
| WPT312-5-56 | dcl1aΔ6/dcl1aΔ7/DCL1b/dcl1bΔ3 | M1 | None |
| WPT312-5-5-1 | dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 | M2 | None |
| M14-533-1 | DCL1a/dcl1aΔ7/DCL1b/dcl1bΔ15 | M1F1 | None |
| M14-533-2 & 3 | DCL1a/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 | M1F2 | None |
| M14-534-1 & 2 | DCL1a/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 | M1F2 | None |
| M14-533-1-1 | dcl1aΔ7/dcl1aΔ7/ dcl1bΔ15/dcl1bΔ15 | M1F2 | None |
| M14-533-2-1 | dcl1aΔ7/dcl1aΔ7/ dcl1bΔ3/dcl1bΔ15 | M1F2 | None |
A series of self-pollination and cross-fertilization experiments were conducted to isolate single homozygous mutant individuals and combine dcl1a and dcl1b alleles in a dcl1a/dcl1b double mutant (Figure 2, A and B). This approach resulted in the identification of several homozygous single mutants including plants with the allele combinations of dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b, dcl1aΔ6/dcl1aΔ6/DCL1b/DCL1b, DCL1a/DCL1a/dcl1bΔ3/dcl1bΔ3, and DCL1a/DCL1a/dcl1bΔ15/dcl1bΔ15. Furthermore, several double mutant combinations were also identified, including plants with the allele combinations dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 and the homozygous genotypes dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3, and dcl1aΔ7/dcl1aΔ7/dcl1bΔ15/dcl1bΔ15 (File S3 and Table 1).
Phenotypic analysis of single and double dcl1 mutants
Phenotypic analyses were performed on the single homozygous mutant plants dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b, dcl1aΔ6/dcl1aΔ6/DCL1b/DCL1b, and DCL1a/ DCL1a/dcl1bΔ15/dcl1bΔ15. These plants were evaluated for developmental abnormalities, including height differences, and/or altered nodulation characteristics. No noticeable phenotype was observed in any of the analyzed single mutants (Figure 3A), revealing no evidence for subfunctionalization or neofunctionalization of the soybean DCL1 homologs.
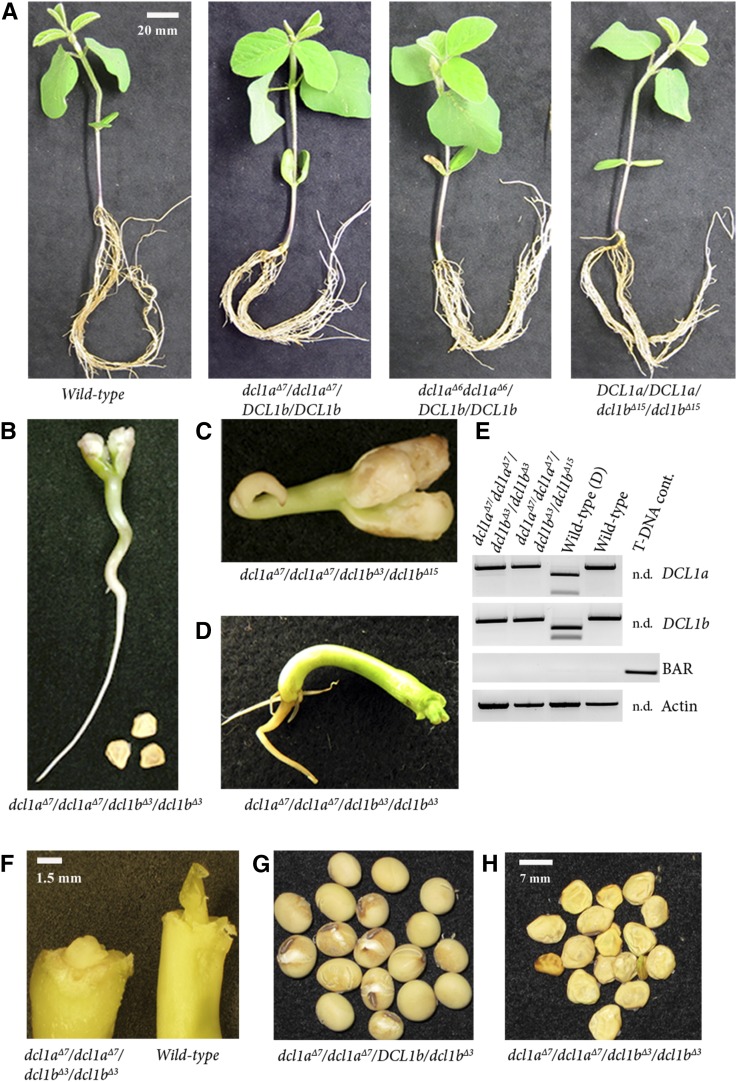
Figure 3.
Phenotypic analysis of single and double dcl1 mutants. (A) Fourteen-day-old single homozygous dcl1a or dcl1b mutant plants exhibiting no obvious developmental abnormalities. (B–D) Developmental abnormalities were observed in 14-day-old dcl1a/dcl1b double mutant plants: dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/ dcl1bΔ3 and dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15. A dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 plant with removed cotyledons revealed an apical meristem that could not differentiate. Heterozygote plants with a wild-type allele of DCL1b had a wild-type appearance. (E) PCR restriction-digestion assays showed that both assessed plants (B–D) are double-mutants (PCR bands are resistant to digestion) and that they are nontransgenic (no band was amplified in the BAR PCR reaction, while a positive control transgenic plant produced the BAR band). (F) Four-day-old dcl1aΔ7/dcl1aΔ7 /dcl1bΔ3/dcl1bΔ3 mutant and wild-type seedlings after removal of the cotyledons to expose the shoot apical meristem (SAM). The mutant SAM could not initiate leaves, presumably due to defective miRNA processing. (G–H) Seed phenotypes displayed by dcl1aΔ7/dcl1aΔ7/DCL1b/dcl1bΔ3 and dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 plants. Seed from the plant with one wild-type DCL1b allele, dcl1aΔ7/dcl1aΔ7/DCL1b/dcl1bΔ3 genotype, exhibit a wild-type phenotype while seed of the homozygous double-mutant plant, dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3, are shrunken and shriveled and can only germinate under specific induction conditions.
The previous segregation analysis of the progeny of T0 plant WPT312-5 (dcl1aΔ6/dcl1aΔ7/DCL1b/dcl1bΔ3) did not identify a viable homozygous double mutant dcl1 plant (Figure 2A). Approximately one-sixteenth of segregating T1 seed failed to germinate with the planted seeds consequently rotting in soil. This strongly indicated that plants harboring the double dcl1 mutation had an embryonic lethal phenotype, or that the mutations exacerbated embryonic dormancy. To rule out seed dormancy, we introduced a seed stratification step, namely 5 d incubation at 4° on sterile filter paper, followed by 3 d room temperature incubation in the dark. Two d poststratification, three seeds germinated, as evidenced by the emergence of the embryonic axis. The seed coats were gently removed to facilitate complete germination.
The stratification, incubation, and seed coat removal steps allowed for the observation of early developmental phenotypes in dcl1 double mutants. Two different double mutant genotypes were evaluated, including the dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 and dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 backgrounds. Seedlings grew to approximately 5–7 cm in height and exhibited striking developmental phenotypes. These included a lopsided differentiation of the vascular tissue, a hallmark of miRNA deregulation (Lobbes et al. 2006; Allen et al. 2007; Eamens et al. 2012), resulting in a wavy stem phenotype. There was also no sign of radial root growth or leaf initiation at the shoot apical meristem. Removal of the atrophied cotyledons revealed a shoot apical meristem that was either delayed or not capable of shoot initiation (Figure 3, B–D). A PCR digestion assay revealed digestion-resistant amplicons from the DCL1a and DCL1b loci, confirming the biallelic mutations harbored by the plant (Figure 3E). Sequencing of these amplicons confirmed the genotype status of the double mutation (File S3). In addition, apical meristem disruption and a shrunken seed phenotype were readily observed to correlate with double-mutant genotypes (Figure 3, F–H).
Assessment of miRNA processing and function in single and double dcl1 mutants
miRNAs require DCL1 for their maturation from their hairpin structured precursor transcripts. Upon maturation, miRNAs are key regulators of gene expression in development and in responses to the environment, such as abiotic stress or pathogen attack (Liu et al. 2008). Furthermore, disrupted miRNA biogenesis is a hallmark of previously characterized dcl1 mutants in multiple plant species (Schauer et al. 2002; Liu et al. 2005; Valdes-Lopez et al. 2008; Khraiwesh et al. 2010; Thompson et al. 2014). We therefore expected to detect a global reduction of total miRNA accumulation, combined with globally elevated miRNA target gene expression in our soybean dcl1 mutants. To test this, sRNA populations were profiled from two biological replicates of 14-day-old leaf material for wild-type plants and the homozygous single mutants; dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b, dcl1aΔ6/dcl1aΔ6/DCL1b/DCL1b, and DCL1a/ DCL1a/dcl1bΔ15/dcl1bΔ15. The mutants exhibited minimal differences in miRNA levels compared with the wild-type control, indicating probable functional redundancy between the homologous DCL1a and DCL1b genes (Figure 4 and Table S1).
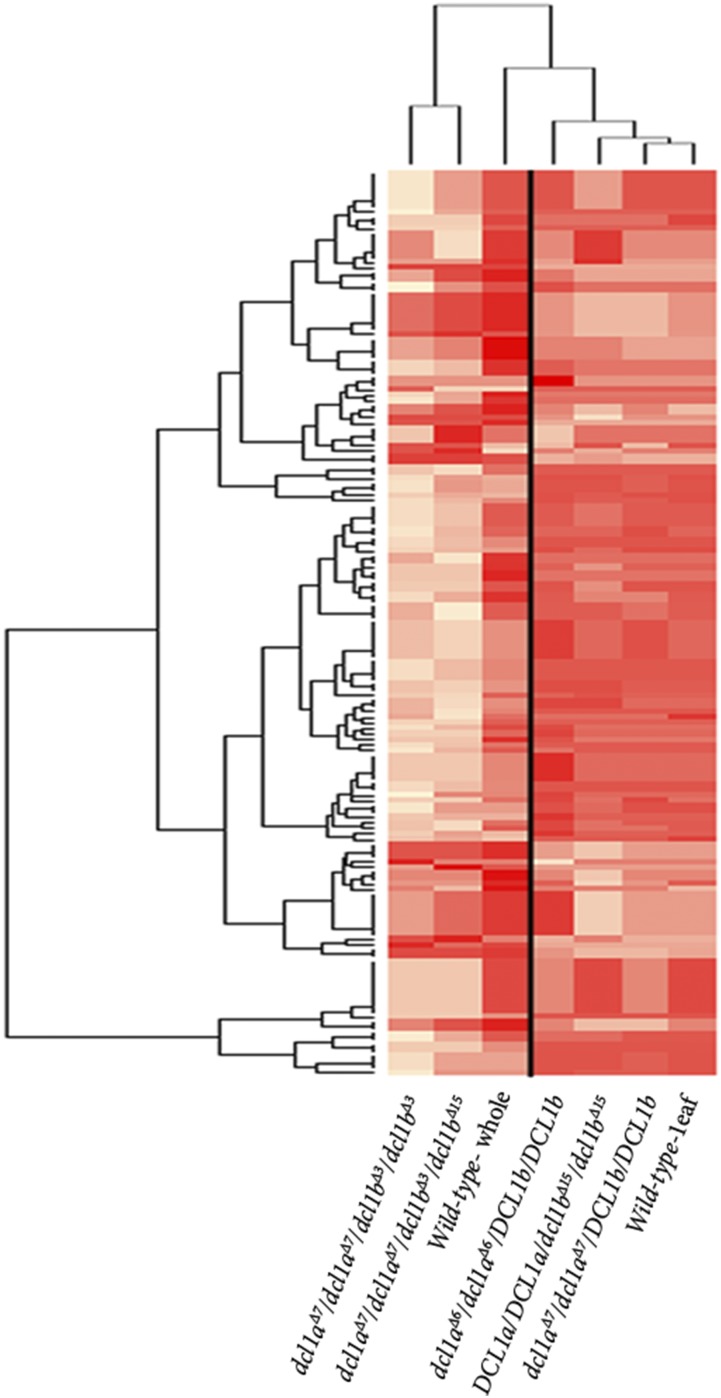
Figure 4.
Small RNA sequence profiling of miRNA accumulation in single and double dcl1 mutants. Small RNA sequence profiling of soybean miRNAs in 14-day-old single and double dcl1 mutant plants. The rows indicate miRNA accumulation in each mutant plant (Table S1 and Table S2).
A similar comparison was performed on the double mutant backgrounds dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 and dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15. However, since the double mutants only produced cotyledons, stems, and roots, the sRNA fraction was instead extracted from available 14-day-old ‘whole plant’ materials, for both the wild-type and mutants. Analysis of the double mutants revealed an almost global reduction in miRNA accumulation, ranging from negligible to a maximum 7.5-fold difference, indicating a vast reduction in mature miRNA levels in the double mutants (Figure 4 and Table S2). Among the most highly reduced miRNAs were the closely related miRNAs miR319f and miR159a, miRNAs that target a subset of genes encoding the developmentally important TCP and GAMYB-like transcription factors, respectively.
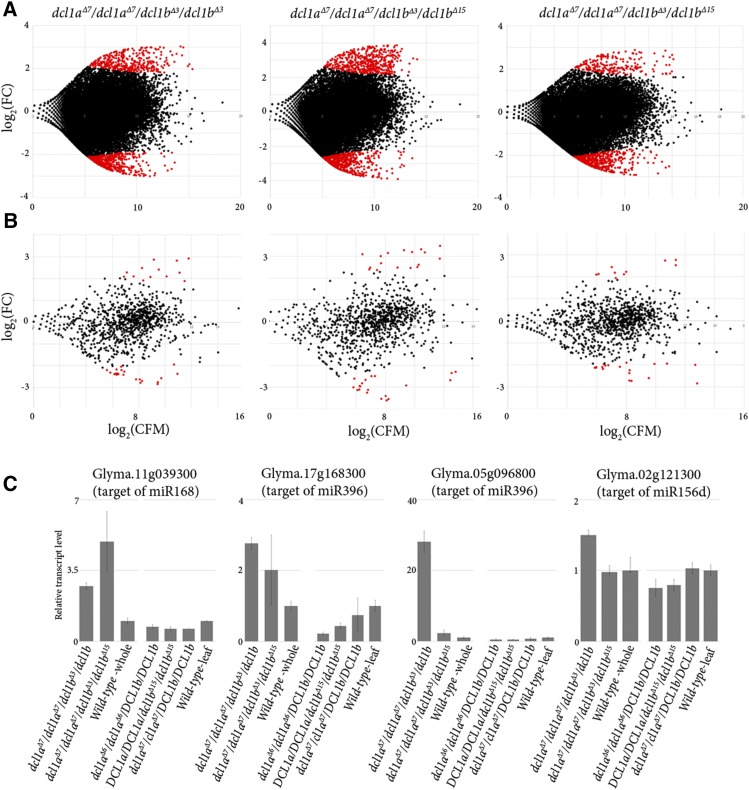
We next performed standard RNA-Seq (mRNA profiling) on the same dcl1 double mutant samples in order to identify and quantify differential expression of target transcripts in the double mutants. This analysis identified approximately 844 transcripts with significantly altered expression (P <0.05) from 34,959 transcripts detected in the dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 mutant, 1034 transcripts from the 34,830 transcripts detected for the dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 mutant, and 735 significantly (P <0.05) altered transcripts from the 34,965 transcripts detected from the second dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15 mutant (Figure 5A and Table S3). To improve the resolution of these plots, we carried out an analysis that identified predicted miRNA target transcripts using the sPARTA package (Kakrana et al. 2014) and combined publicly available PARE (Parallel Analysis of RNA Ends) libraries (Arikit et al. 2014) to map these miRNA target transcripts to the differentially expressed transcripts depicted in Figure 5A. This approach identified a total of 985, 977, and 981 distinct miRNA target transcripts in the RNA-Seq datasets analyzed from the three assessed mutant backgrounds and approximately 35, 38, and 30 of these were determined significantly differentially expressed (P <0.05), respectively. Considerable overlap was observed for the differentially expressed miRNA target genes between the different mutant backgrounds assessed (Figure 5B, Table S4, and Table S5). Finally, quantitative reverse transcription PCR (RT-qPCR) was used to confirm the expression of a small subset of previously published miRNA target transcripts (Kulcheski et al. 2011) and this quantitative analysis aligned closely with the RNA-Seq generated data. Intriguingly, some miRNA target transcripts in the dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 mutant background, notably the miR396 target gene Glyma05g20930 (Glyma.05g096800 in the version 2 genome assembly), remained at wild-type levels. This curious finding suggests that the dcl1bΔ15 mutant allele might function differently to the dcl1bΔ3 allele on specific miRNA-associated substrates.
Figure 5.
The analysis of predicted miRNA targets in the double dcl1 mutant. (A) Three MA plots of the dcl1 double mutants with one replicate of mutant dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ3 and two replicates of mutant dcl1aΔ7/dcl1aΔ7/dcl1bΔ3/dcl1bΔ15. The plots show the differentially expressed transcripts in the dcl double mutant compared to wild-type, with the red dots indicating significantly perturbed transcript expression (P <0.05). (B) Three MA plots indicating differential expression for a subset of transcripts in (A). This subset includes only genes validated as miRNA target transcripts. A total of 985, 977 and 981 distinct miRNA targets were found to be expressed in each of the three assessed mutants, respectively. The red dots indicate the 35, 38 and 30 differentially expressed miRNA target transcripts (P <0.05) in each mutant when compared to wild-type controls. (C) RT-qPCR was used to determine the expression of a subset of miRNA target transcripts in single and double dcl1 mutant backgrounds. CPM, counts per million; FC, fold change.
Discussion
Functional genomic studies in soybean can be challenging due to the duplicated paleopolyploid nature of the soybean genome. Gene redundancy among homologous copies is often sufficient for full complementation of the loss of one homolog. The soybean DCL1 homologs are almost identical to one another at the nucleotide level and, further, have almost identical expression patterns in most soybean tissues. Our data shows that miRNA biogenesis is largely unaffected in plant lines harboring a mutation in one of the two soybean DCL1 loci. This strongly suggests that the DCL1a and DCL1b homologs perform redundant functions in the soybean miRNA pathway. This is in direct contrast to a recent report in P. patens (Khraiwesh et al. 2010) where the two highly similar DCL1 enzymes were shown to perform highly specialized roles at functionally distinct stages of the P. patens miRNA pathway. This strongly suggests that neofunctionalization of an ancient P. patens DCL1 occurred prior to the duplication of this ancestral gene (Khraiwesh et al. 2010).
The isolation of a dcl1 double mutant was required to overcome the hurdle of DCL1a/DCL1b redundancy, and to confirm a role for each DCL1 homolog in the soybean miRNA pathway. To achieve this goal, ZFN-based mutagenesis allowed for the production and recovery of stable, nontransgenic dcl1a and dcl1b mutants, and the subsequent generation of multiple allelic combinations of the dcl1a/dcl1b double mutant. The first double mutant identified in this study was isolated from a single T0 plant that harbored mutations targeted to three of the four DCL1 loci. This included distinct mutations for each of the DCL1a homologs (i.e., biallelic) and a monoallelic mutation for DCL1b. The second double mutant allele was generated by traditional cross-fertilization of the DCL1a/DCL1a/dcl1bΔ15/dcl1bΔ15 mutant with the dcl1aΔ7/dcl1aΔ7/DCL1b/dcl1bΔ3 mutant.
While frameshift dcl1a mutant alleles were identified in this work, there have been no out-of-frame dcl1b mutant alleles identified to date. Both dcl1b alleles from this study were in-frame mutations that result from a one and a five amino acid residue deletion, respectively. Therefore, at this stage, we cannot confirm whether an out-of-frame single dcl1b mutant alone would be sufficient to disrupt the soybean miRNA pathway. However, the in-frame dcl1b mutations clearly disrupt protein function, as the phenotypically normal dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b single mutant exhibits severe seedling phenotypes when combined with the dcl1b in-frame mutations. Furthermore, initial screening of the dcl1aΔ6/dcl1aΔ7/DCL1b/dcl1bΔ3 T0 plant failed to recover viable double mutants with combinations of the in-frame mutant alleles (dcl1aΔ6 and dcl1bΔ3). This would suggest that in-frame mutations also appear to disrupt the function of DCL1, otherwise at least one or more viable dcl1aΔ6/dcl1aΔ6/dcl1bΔ3/dcl1bΔ3 or heterozygote variations of the double mutant would have been recovered. An out-of-frame dcl1b mutation combined with the dcl1aΔ7 allele would likely be embryo lethal and not viable. Such a plant would be consistent with the previously reported embryo defective Arabidopsis dcl1-4 (sus1-4) mutant resulting from a T-DNA insertion disrupting the PAZ domain (McElver et al. 2001).
We also performed small RNA sequencing to establish miRNA accumulation profiles for the three single dcl1 mutant alleles, including mutant lines dcl1aΔ7/dcl1aΔ7/DCL1b/DCL1b (out-of-frame mutation), dcl1aΔ6/dcl1aΔ6/DCL1b/ DCL1b (in-frame mutation), and DCL1a/ DCL1a/dcl1bΔ15/dcl1bΔ15 (in-frame mutation). Compared to wild-type plants, no major alterations to miRNA accumulation were observed in any of the analyzed single mutants. However, in the three assessed double mutants, we observed an almost global reduction in miRNA accumulation. Intriguingly, miRNA accumulation was not uniformly reduced, or completely abolished in any assessed double mutant, including several miRNAs that are largely unaffected in these backgrounds. Possible explanations for this curious finding include: 1) disruption of the so-called Dicer ’molecular ruler’; 2) disassociation of various interacting proteins that either bind to single and double-stranded transcripts or to DCL1 itself; 3) gene redundancies between the five other soybean DCL paralogs (Curtin et al. 2012); or 4) a defective DCL1b mutant protein that is still functionally competent to process a small subset of miRNAs (MacRae et al. 2006; Rogers and Chen 2013). We discuss each of these possibilities in more detail below.
Previous crystal structure analysis of the Dcr enzyme revealed a region between the PAZ and the first RNase III domain that essentially functions as a ‘molecular ruler’ by governing the size of the processed sRNAs (MacRae et al. 2006). The dcl1b mutation could have potentially disrupted this ruler resulting in shorter, misprocessed miRNAs. However, the small RNA sequencing analysis was able to distinguish sRNA sizes ranging from 18 –35 nt and there was no indication of short miRNA sequences in either the single or double mutant datasets. This suggests that disruption to the molecular ruler of the two soybean DCL1s is unlikely to have been the cause of incomplete loss of miRNA accumulation. The precise molecular mechanisms of how DCL1 recognizes, interacts with, and processes precursor molecules, namely primary-miRNAs (pri-miRNAs) and precursor-miRNAs (pre-miRNAs) to miRNA/miRNA* duplexes is not completely understood. Two protein cofactors, including the double-stranded (ds)RNA-binding protein DRB1 (HYL1) and the C2H2-type zinc finger domain-containing protein SERRATE (SE), have been shown to interact with both DCL1 and pri-miRNAs for accurate and efficient processing of miRNAs from their precursor transcripts (Rogers and Chen 2013). It is plausible that interactions between DCL1 and these protein cofactors are disrupted by the dcl1b mutations. In turn, this could affect the loading of the duplex miRNA into the RISC by SE or DRB1, or the accuracy of DCL1-mediated pri-miRNA and pre-miRNA processing (Eamens et al. 2009; Rogers and Chen 2013).
Another possible explanation for incomplete loss of miRNA accumulation in the double mutant is gene redundancy by other DCL enzymes such as DCL2 or DCL4. There are seven functional DCL enzymes in soybean including two DCL2 (DCL2a and DCL2b) and two DCL4 (DCL4a and DCL4b) proteins (Curtin et al. 2012). Reports from Arabidopsis have shown that the DCL4/DRB4 functional partnership can process certain miRNAs (Rajagopalan et al. 2006). This possibility is further supported by other demonstrations in Arabidopsis; that is, DRB2 appears capable of forming functional partnerships with either DCL1 or DCL4 for miRNA and siRNA production, respectively (Eamens et al. 2012). Therefore, the higher than expected levels observed for some miRNAs in the double mutant backgrounds may result from DCL4 activity, with the functional assistance of either DRB2 or DRB4, partnerships that would have increased access to miRNA precursor transcripts in the absence of functionally competent DCL1a and DCL1b.
An additional explanation involves irregularities with the plant pri-miRNA processing step. Typically, pri-miRNAs are cleaved by the DCL1 enzyme in the canonical ‘base-to-loop’ manner; the first cleavage event is at the base of the hairpin to produce the pre-miRNA, and the second cleavage event liberates the miRNA/miRNA* duplex from the loop structure of the pre-miRNA hairpin. In plants, there is also a highly conserved, yet ‘noncanonical’ pathway that is initiated and proceeds in the opposite direction to the two-step canonical pri-miRNA processing pathway. In the noncanonical ‘loop-to-base’ pri-miRNA processing pathway, four DCL1-mediated cleavage steps are required for pri-miRNA to pre-miRNA processing, and subsequent miRNA/miRNA* duplex liberation of the pre-miRNA. Currently, only two Arabidopsis miRNAs have been demonstrated to be processed by this more complex pathway, including miR159 and mR319 (Bologna et al. 2009). Coincidentally, the two most differentially expressed miRNAs in our data set are miR159 and miR319, a finding that suggests that the noncanonical pri-miRNA processing pathway may be more severely affected in the dcl1a/dcl1b double mutant background. If this is the case, then other miRNAs in our data set with highly differentiated expression states, such as miR403a, miR403b, miR1535b, may also be processed by the noncanonical loop-to-base pathway.
Lastly, RNA-Seq of the double mutant and wild-type plants provides insight into the extent of miRNA accumulation and target gene disruption in these plants. Altered expression of protein-encoding genes that would normally be targets of miRNA regulation was observed in each of the three assessed double mutants, including 35, 38, and 30 putative miRNA target genes, respectively. However, there were approximately 980 putative miRNA protein-encoding target genes identified in total in these analyses. Furthermore, considerable overlap of these differentially expressed miRNA target genes was observed across the assessed mutant backgrounds. Therefore, these results indicate that ∼3%–5% of the total predicted gene targets for each of the currently known soybean miRNAs were differentially expressed. Several possible explanations might account for this low number of affected target transcripts. These include insufficient sample replication resulting in diminished statistical power for the differential expression analysis, thereby contributing to an increase occurrence of false negatives. In addition, it is possible (though unlikely given the severity of the mutant phenotype) that a hypomorphic DCL1 enzyme remains active on some substrates. However, the severe developmental phenotype expressed by the double mutants (i.e., no leaves were available for sampling, with only stem, root, and shriveled cotyledons available for double mutant analyses) makes the differential expression analysis difficult to fully interpret. A future experiment focused on a well-replicated and microdissected SAM, or on germinating seed tissue, would likely yield an improved and more biologically relevant indication of the differentially expressed miRNA target transcript landscape resulting from the introduced mutations.
In conclusion, this study demonstrates targeted mutagenesis using site-specific nucleases as a reverse genetics tool for functional genomics studies in soybean. We generated dcl1a/dcl1b double mutants and demonstrated that the introduction of mutations into both DCL1 loci severely disrupted the soybean miRNA pathway, including altered miRNA accumulation and deregulated miRNA target gene expression. The work presented here provides the soybean community with an important genetic resource to aid in the further study of miRNA-directed RNA silencing-mediated processes, including mounting defense responses against abiotic stress and plant viral, fungal, oomycete, insect, and bacterial pathogens. For example, soybean plants grown in the Mid-West of the United States are highly susceptible to the stem and root rot pathogen Phytopthora sojae, causing millions of dollars in lost revenue annually. It was recently demonstrated that P. sojae secretes proteins that suppress RNA silencing pathways of the host plant, namely the miRNA pathway, to aid in the pathogen’s infection and colonization of soybean (Qiao et al. 2013). The current mechanism for how P. sojae subverts the RNA silencing pathways in soybean is unknown, but genetic resources such as dcl1 and dcl4 mutants may be important for the elucidation of this plant-pathogen interaction. Furthermore, mutant alleles for the remaining soybean DCL genes may be generated using more efficient SSN platforms in the future. This will be critical for generating double mutants in what would otherwise be a difficult and time-consuming undertaking using the more traditional mutagenesis, mutation identification, and stacking methodologies. The ZFN and TALEN SSN platforms are fast becoming outdated due to the ease, efficiency and multiplex capabilities of the CRISPR/Cas9 platform, which will likely be the ‘go-to’ platform in the foreseeable future (Baltes and Voytas 2015).
Supplementary Material
Acknowledgments
We thank Kit Leffler for photography, Grant Barthel and Gail Celio for microscopy assistance, Sue Miller for the Bradyrhizobium strain and inoculation protocol, and Kevin Silverstein, Jeremy Yoder, and Rachel Hillmer for bioinformatics assistance. J.G.H. thanks the Fundación Alfonso Martin Escudero for his postdoctoral fellowship. Work on genome engineering in the Stupar lab is supported by funding from the United Soybean Board (project #1520-532-5601) and the Minnesota Research and Promotion Council (project #18-15C). Work on soybean small RNAs in the Meyers lab is supported by funding from the United Soybean Board and the North Central Soybean Research Program. The authors declare that they have no competing interests.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.022137/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Allen R. S., Li J., Stahle M. I., Dubroué A., Gubler F., et al. , 2007. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 104: 16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S., Xia R., Kakrana A., Huang K., Zhai J., et al. , 2014. An Atlas of Soybean Small RNAs Identifies Phased siRNAs from Hundreds of Coding Genes. Plant Cell 26: 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N. J., Voytas D. F., 2015. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33: 120–131. [DOI] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: Target Recognition and Regulatory Functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D., 2004. RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bologna N. G., Mateos J. L., Bresso E. G., Palatnik J. F., 2009. A loop‐to‐base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28: 3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon Y.-T., Stec A. O., Michno J.-M., Roessler J., Bhaskar P. B., et al. , 2014. Genome Resilience and Prevalence of Segmental Duplications Following Fast Neutron Irradiation of Soybean. Genetics 198: 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., 2010. Small RNAs – secrets and surprises of the genome. Plant J. 61: 941–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. L., Till B. J., Laport R. G., Darlow M. C., Kleffner J. M., et al. , 2008. TILLING to detect induced mutations in soybean. BMC Plant Biol. 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Barampuram S., Stacey M. G., Hancock C. N., Findley S., et al. , 2013. Tnt1 retrotransposon mutagenesis: a tool for soybean functional genomics. Plant Physiol. 161: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Watson J. M., Smith N. A., Eamens A. L., Blanchard C. L., et al. , 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 582: 2753–2760. [DOI] [PubMed] [Google Scholar]
- Curtin S. J., Zhang F., Sander J. D., Haun W. J., Starker C., et al. , 2011. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156: 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin S. J., Kantar M. B., Yoon H. W., Whaley A. M., Schlueter J. A., et al. , 2012. Co-expression of soybean Dicer-like genes in response to stress and development. Funct. Integr. Genomics 12: 671–682. [DOI] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Eamens A. L., Smith N. A., Curtin S. J., Wang M.-B., Waterhouse P. M., 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens A. L., Kim K. W., Curtin S. J., Waterhouse P. M., 2012. DRB2 is required for microRNA biogenesis in Arabidopsis thaliana. PLoS One 7: e35933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y., et al. , 1999. Preservation of Duplicate Genes by Complementary. Degenerative Mutations. Genet. 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock C. N., Zhang F., Floyd K., Richardson A. O., LaFayette P., et al. , 2011. The Rice Miniature Inverted Repeat Transposable Element mPing Is an Effective Insertional Mutagen in Soybean. Plant Physiol. 157: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W., Coffman A., Clasen B. M., Demorest Z. L., Lowy A., et al. , 2014. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12: 934–940. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J., 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T., LaFayette P., Schmitz R., Parrott W., 2015. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S. E., Running M. P., Meyerowitz E. M., 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243. [DOI] [PubMed] [Google Scholar]
- Kakrana A., Hammond R., Patel P., Nakano M., Meyers B. C., 2014. sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res. 42: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M. A., Seumel G. I., Ossowski S., Weigel D., et al. , 2010. Transcriptional control of gene expression by microRNAs. Cell 140: 111–122. [DOI] [PubMed] [Google Scholar]
- Kulcheski F. R., de Oliveira L. F., Molina L. G., Almerão M. P., Rodrigues F. A., et al. , 2011. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2014. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Liu B., Li P., Li X., Liu C., Cao S., et al. , 2005. Loss of function of OsDCL1 affects MicroRNA accumulation and causes developmental defects in rice. Plant Physiol. 139: 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-H., Tian X., Li Y.-J., Wu C.-A., Zheng C.-C., 2008. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D., Rallapalli G., Schmidt D. D., Martin C., Clarke J., 2006. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 7: 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae I. J., Zhou K., Li F., Repic A., Brooks A. N., et al. , 2006. Structural Basis for Double-Stranded RNA Processing by Dicer. Sci. 311: 195–198. [DOI] [PubMed] [Google Scholar]
- McElver J., Tzafrir I., Aux G., Rogers R., Ashby C., et al. , 2001. Insertional Mutagenesis of Genes Required for Seed Development in Arabidopsis thaliana. Genet. 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A., Waterhouse P., 2005. Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics 5: 129–135. [DOI] [PubMed] [Google Scholar]
- Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z., 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1374, 1376, 1378–1379. [PubMed] [Google Scholar]
- Nakano M., Nobuta K., Vemaraju K., Tej S. S., Skogen J. W., et al. , 2006. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 34: D731–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz M. M., Martinez J. C., Kalvig A. B., Fonger T. M., Wang K., 2006. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 25: 206–213. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Liu L., Xiong Q., Flores C., Wong J., et al. , 2013. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45: 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2013 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P., 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X., 2013. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A., Auer P. L., Libault M., Schlueter J., Farmer A., et al. , 2013. The fate of duplicated genes in a polyploid plant genome. Plant J. 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M. H., Karlen Y., Bakker O., et al. , 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., et al. , 2011. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8: 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer S. E., Jacobsen S. E., Meinke D. W., Ray A., 2002. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491. [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., et al. , 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Severin A. J., Woody J. L., Bolon Y.-T., Joseph B., Diers B. W., et al. , 2010. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q.-X., Liu Y.-F., Hu X.-Y., Zhang W.-K., Ma B., et al. , 2011. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Hu Z., Chen R., Jiang Q., Song G., et al. , 2015. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5: 10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. E., Basham C., Hammond R., Ding Q., Kakrana A., et al. , 2014. The dicer-like1 homolog fuzzy tassel is required for the regulation of meristem determinacy in the inflorescence and vegetative growth in maize. Plant Cell 26: 4702–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y., Zamore P. D., 2005. MicroRNA biogenesis: drosha can’t cut it without a partner. Curr. Biol. 15: R61–R64. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Lopez O., Arenas-Huertero C., Ramirez M., Girard L., Sanchez F., et al. , 2008. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 31: 1834–1843. [DOI] [PubMed] [Google Scholar]
- Voinnet O., 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- Voytas D. F., 2013. Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64: 327–350. [DOI] [PubMed] [Google Scholar]
- Waterhouse P. M., Graham M. W., Wang M. B., 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95: 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq raw data are available on Sequence Read Archive under the accession GSE76036 or can be accessed through the soybean gene analysis link (https://mpss.udel.edu/dbs/index.php?SITE=soy_RNAseq).