Abstract
Heat shock protein 90 (Hsp90) is a molecular chaperone that orchestrates the folding and stability of proteins that regulate cellular signaling, proliferation and inflammation. We have previously shown that Hsp90 controls the production of reactive oxygen species by modulating the activity of Noxes1-3 and 5, but not Nox4. The goal of the current study was to define the regions on Nox5 that bind Hsp90 and determine how Hsp90 regulates enzyme activity. In isolated enzyme activity assays, we found that Hsp90 inhibitors selectively decrease superoxide, but not hydrogen peroxide, production. The addition of Hsp90 alone only modestly increases Nox5 enzyme activity but in combination with the co-chaperones, Hsp70, HOP, Hsp40, and p23 it robustly stimulated superoxide, but not hydrogen peroxide, production. Proximity ligation assays reveal that Nox5 and Hsp90 interact in intact cells. In cell lysates using a co-IP approach, Hsp90 binds to Nox5 but not Nox4, and the degree of binding can be influenced by calcium-dependent stimuli. Inhibition of Hsp90 induced the degradation of full length, catalytically inactive and a C-terminal fragment (aa398–719) of Nox5. In contrast, inhibition of Hsp90 did not affect the expression levels of N-terminal fragments (aa1–550) suggesting that Hsp90 binding maintains the stability of C-terminal regions. In Co-IP assays, Hsp90 was bound only to the C-terminal region of Nox5. Further refinement using deletion analysis revealed that the region between aa490–550 mediates Hsp90 binding. Converse mapping experiments show that the C-terminal region of Nox5 bound to the M domain of Hsp90 (aa310–529). In addition to Hsp90, Nox5 bound other components of the foldosome including co-chaperones Hsp70, HOP, p23 and Hsp40. Silencing of HOP, Hsp40 and p23 reduced Nox5-dependent superoxide. In contrast, increased expression of Hsp70 decreased Nox5 activity whereas a mutant of Hsp70 failed to do so. Inhibition of Hsp90 results in the loss of higher molecular weight complexes of Nox5 and decreased interaction between monomers. Collectively these results show that the C-terminal region of Nox5 binds to the M domain of Hsp90 and that the binding of Hsp90 and select co-chaperones facilitate oligomerization and the efficient production of superoxide.
Keywords: NADPH oxidase, Nox, Hsp90, Hsp70, co-chaperones, vascular biology
Introduction
Reactive oxygen species (ROS) are important bioactive molecules that participate in a wide range of physiological and pathophysiological processes. The aberrant production and persistence of ROS are thought to be key mechanisms underlying the pathogenesis of cardiovascular diseases, including vascular inflammation [1] endothelial dysfunction[2], and atherosclerosis[3]. In blood vessels, the major ROS generators are the NADPH oxidase (Nox) family of enzymes and elevated expression and inappropriate activation of Nox enzymes have been shown to contribute to cancer, stroke, atherosclerosis, pulmonary hypertension, vascular remodeling and endothelial dysfunction [2, 4–8]. Seven closely related Nox enzymes have been identified including Nox1-5 and Duox1 and 2. The Nox isoforms are distinguished by their relative expression in different cell types and the mechanisms governing activation[9]. The importance of ROS and Nox enzymes to numerous disease states has attracted considerable interest in the identification of novel mechanisms regulating Nox enzyme activity with the goal of developing effective tools to selectively target Nox isoforms and regulate ROS production [10].
Nox5 was the last of the Nox enzymes to be identified and is expressed in a number of organs and tissues including the heart, blood vessels, testis, kidney, lymph nodes and spleen,and also various cell types, including endothelial cells, smooth muscle cells, podocytes and primary cardiac fibroblasts[11–14]. The absence of Nox5 from rodent genomes has limited our appreciation of its physiological or pathophysiological significance. In humans, increased expression of Nox5 has been observed in acute myocardial infarction, atherosclerosis, and hypertension, suggesting that dysfunction of Nox5 in cardiovascular system contributes to disease [12, 15–17]. The activation of Nox5 is mediated by changes in intracellular calcium[18] and posttranslational modifications such as phosphorylation [19–23], S-nitrosylation[24], SUMOylation[25] and protein-protein interaction [26] which can modulate Nox5 activity via direct or indirect mechanisms. Recently, we and others have identified the binding of heat shock protein 90 (Hsp90) as a new mechanism that regulates the production of ROS from Nox5 as well as Nox1-3[27–29].
There are 2 major isoforms of Hsp90, designated α (inducible) and β (constitutive). Both are abundant cytosolic proteins that participate in the proper folding of a subset of the proteome primarily involved in cell signaling and survival[30, 31]. The ability of Hsp90 to function as a chaperone and selectively bind client proteins is regulated by numerous co-chaperones [32, 33]. The co-chaperones for Hsp90 include Hsp40, Cdc37, Hop and p23. In the well characterized example of steroid hormone receptors, nascent proteins bind first to Hsp70/Hsp40 and recruit Hop which facilitates the docking of client/chaperone complexes to Hsp90. ATP-driven changes in the conformation of Hsp90 promote constriction of the N-terminus decreasing Hop/Hsp70 binding. P23 then binds to Hsp90 and is essential for stable Hsp90:client protein complexes [31, 34]. Hsp90 proteins can be functionally divided into 3 major domains, an N-terminal nucleotide binding domain, a middle (M) domain and a C-terminal dimerization domain. While the M-domain is considered the primary binding region for most client proteins, the binding of some clients within the N and C domains has been reported [35]. It is not yet known where on Hsp90 the Nox enzymes bind.
We and others have shown that pharmacological and genetic inhibition of Hsp90 using siRNA potently reduces Nox-dependent ROS production in both cells and in isolated blood vessels [27–29]. Co-immunoprecipitation (Co-IP) and bioluminescence resonance energy transfer (BRET) studies revealed that Hsp90 physically associates with Nox5 and together with Hsp70 regulates Nox protein stability [27, 28]. However, we do not yet know how Hsp90 alters Nox5 activity, the respective binding site(s) of Hsp90 and Nox5, the role of Hsp90 co-chaperones, and whether there are factors that influence the binding of Nox proteins to Hsp90. Thus, the major goals of this study were to define the binding sites of Hsp90 and Nox5 on respective proteins, identify the role of Hsp90 co-chaperones in binding to and regulating Nox function including self-assembly and to determine whether stimuli regulating Nox activity alter Hsp90 binding. Collectively this knowledge will advance our understanding of the mechanisms by which molecular chaperones govern Nox5 activity.
Materials and Methods
Cell culture and transfection
COS-7 and HEK293 were grown at 37 °C in 5% CO2 in DMEM as described [27]. The HA-Nox5 HEK293 cell line was generated by Flp Recombinase-Mediated Integration (Invitrogen). Cells were transfected using Lipofectamine 2000 reagent (Invitrogen) as described previously [14]. Cells were exposed to different concentrations of radicicol (RAD) (Tocris Bioscience) or 17-allylamino-17-demethoxygeldanamycin (17-AAG) (Fisher Scientific) for 1–24h.
DNA constructs and purified Hsp90 and co-chaperone proteins
Plasmid DNA coding RFP, Nox5β (AF325189), V5-Nox4, Flag-Hsp90, Hsp70 constructs have been described previously [27, 28, 36–38]. Nox5 truncations, deletion mutants and multiple sites mutations were generated by Q5® Site-Directed Mutagenesis Kit (NEB, Ipswich, MA) and confirmed by DNA sequencing. The Hsp90, Hsp70 and Hop, Hsp40 and p23 recombinant proteins were provided by Dr. Ahmed Chadli.
Transient knockdown of CHIP gene with siRNA
The Silencer Select siRNAs targeting HOP (STIP1, siRNA ID: s21584), Hsp40 (DNAJB1, siRNA ID: s223882), p23 (PTGES3, siRNA ID: s54254) were obtained from Applied Biosystems. Validated control and targeting siRNAs were transfected into HEK293 cells stably expressing Nox5 using Lipofectamine® RNAiMAX (Invitrogen).
Superoxide and hydrogen peroxide measurement
The measurement of superoxide (L-012) and hydrogen peroxide (Amplex Red) have been described [27]. The production of superoxide in cells expressing Nox genes was also assessed using Electron Paramagnetic Resonance (EPR) Spectroscopy. In brief, spin trapping was performed using approximately 106 cells/ml in phenol free medium (Invitrogen) with 50 mM 5,5-dimethyl-1-pyrroline N-oxide (DMPO, Aldrich). EPR spectra were recorded in cells at room temperature with a Bruker e-scan M spectrometer. The amplitude of each EPR spectrum 2nd integral area was determined using ANALYSIS software (version 2.02; Magnettech). Quantitation of the superoxide signals was performed by comparing EPR spectrum amplitudes with that of known concentrations of the 1-Oxyl-2,2,6,6-tetramethyl-4-hydroxypiperidine (TEMPOL, TOCRIS) in phenol free medium (Invitrogen). The specificity of L-012 and EPR Spectroscopy for superoxide was confirmed by transfecting cells with a control plasmid, RFP, or by co-incubation of a superoxide dismutase (SOD, 100 U/ml).
Isolated Nox5 activity assay
The partially purified Nox5 activity assay was performed as described previously[19]. In brief, COS-7 cells transfected with Nox5 were lysed in a MOPS (30 mM, pH 7.2)-based buffer containing Triton X-100 (0.3%) and protease inhibitors (Sigma). Cells were placed on ice, rocked gently in lysis buffer and then washed 3 times with phosphate-buffered saline. The remaining cellular fractions were scraped in MOPS buffer containing 0.3 mM EGTA and centrifuged at 14,000 rpm for 5mins at 4°C. The supernatant was discarded, and the pellet was then resuspended in MOPS buffer with mild sonication. Membrane extracts were then aliquoted into buffers containing 1 mM MgCl2, 100 μM FAD (Sigma), and 1 mM CaCl2. After 1 h incubation with Hsp90 inhibitors or mixtures containing Hsp90 and co-chaperones (Hsp70, Hsp40, HOP, p23), reduced NADPH (Sigma) was injected to a final concentration of 200 μM, and the production of superoxide and hydrogen peroxide was monitored over time using L-012 and the Amplex Red Assay.
Co-immunoprecipitation and Western blotting analysis
Cells were lysed on ice in 20mM Tris-HCl (pH 7.4), 1% Triton X-100, 0.025% SDS, 100mM NaCl, 1mM Na3VO4, 10mM NaF, and 1% protease inhibitor cocktail (Sigma). Soluble extracts were incubated for 2 h at 4C with relevant antibodies: anti-HA (Roche Applied Science), anti-V5 (Invitrogen), anti-Flag (Cell Signaling Technology) and a negative isotype control mouse immunoglobulin (IgG) (Santa Cruz Biotechnology). Immune complexes were precipitated with protein A/G agarose (Santa Cruz Biotechnology). In select experiments, cells expressing Nox5 were crosslinked with formaldehyde. Western blotting was performed as described previously [27] using anti-HA (Roche), anti-V5 (Invitrogen), anti-Hsp90 (BD Biosciences), anti-Hsp70 (Cell Signaling Technology), anti-Flag (Cell Signaling Technology), anti-p22 (Santa Cruz Biotechnology), anti-Hsp40 (Cell Signaling Technology), anti-p23 [39], anti-HOP [39], and GAPDH (Santa Cruz Biotechnology).
Optimization of Formaldehyde Cross-Linking condition for Nox5
To determine the optimal conditions for cross-linking Nox5 protein, approximately 1 × 107 COS-7 cells expressing Nox5 were resuspended in PBS in a 15ml tube and incubated with increasing concentration of formaldehyde (0.30%, 0.60%, 1.20%, 1.8% and 2.4%) for 7mins at room temperature. Cells were then pelleted at 4000rpm at RT for 3 mins, resulting in a total of 10 mins exposure to formaldehyde. Cross-Linking was then stopped with 10 ml ice-cold glycine (1.25M) in PBS and cells washed once with PBS. Cells were then transferred to 1.5ml tubes and lysed for co-immunoprecipitation and Western blotting analysis. Cell lysates were incubated at either 65°C for 5mins or 99°C for 20mins to reverse cross-linking and proteins size fractionated by SDS PAGE.
Proximity ligation assay for co-localization
Proximity Ligation Assays (PLA, Duolink, Sigma) were performed according to the manufacturer’s instructions using primary antibodies (anti-Hsp90, 1:100 and anti-HA, 1:100), followed by a pair of oligonucleotide-labeled secondary antibodies. In this assay, positive signal can only be detected when the epitopes of the target proteins are in close proximity (<40 nm). The signal from each of the detected pair of PLA probes was then counted using fluorescence microscopy (excitation: BP545/25, emission: BP605/70). One primary antibody or both primary antibodies were omitted for negative controls.
Sequence alignment
A structurally-informed multiple sequence alignment of dehydrogenase domains from 204 Nox homologs (including Ferric reductases) and several crystallized dehydrogenase domains was constructed using PromalS3D (PROMALS3D: a tool for multiple sequence and structure alignment with [40] default structural constraints and visualized with Jalview[41] The Nox2 NADPH binding domain crystal (3A1F) and the crystallized dehydrogenase domains were superimposed using the Matchmaker tool in Chimera with default parameters [42]; the structural alignment obtained agreed with the PromalS3D alignment in the NADPH-binding domain, giving high confidence in the accuracy of the overall dehydrogenase alignment.
Statistical Analysis
Data were reported as mean ± SE and statistical analyses were performed using Instat and a two-tailed student’s t-test or ANOVA with a post-hoc test where appropriate. Differences were considered as significant at p < 0.05.
Results
Hsp90 directly regulates Nox5 activity and requires the presence of co-chaperones
The ability of L-012 chemiluminescence to detect Nox5-dependent superoxide was first confirmed using DMPO-mediated EPR. As shown in Figure 1A–C, COS-7 cells expressing Nox5 produced greater amounts of superoxide than cells expressing a control gene (RFP). Superoxide production was significantly decreased in Nox5 expressing COS-7 cells that were pretreated with an Hsp90 inhibitor (radicicol, RAD, 20μM, 1hr) and eliminated in cells pretreated with a scavenger of superoxide, SOD (100U/ml). Results obtained with EPR spectroscopy were consistent with those obtained with L-012 and therefore L-012 was employed to measure Nox5-derived superoxide production in subsequent experiments. Further experiments showed that superoxide signal from Nox5 as measured by L-012 is inhibited by DPI and SOD but not catalase (Supplemental Figure 1A), while hydrogen peroxide production from Nox5 measured by Amplex red is inhibited by DPI and catalase but not SOD (Supplemental Figure 1B). These data confirmed the specificity of L-012 and Amplex red to detect superoxide and hydrogen peroxide production, respectively, and suggest that dismutation of superoxide contributes very little to the production of hydrogen peroxide. Previously we found that in intact cells, the production of superoxide, but not hydrogen peroxide can be reduced by the acute (1h) inhibition of Hsp90 [27, 28]. Therefore we next tested whether Hsp90 can directly influence superoxide and hydrogen peroxide production from Nox5 in an isolated enzyme activity assay. As shown in Figure 1D–F and Supplemental Figure 1C, the acute (1h) inhibition of Hsp90 by RAD inhibited superoxide but not hydrogen peroxide production from purified membranes containing Nox5[19]. In contrast, the incubation of a mixture of Nox5 with purified Hsp90 and purified chaperones and co-chaperones (Hsp70, HOP, Hsp40, and p23) significantly increased superoxide, but not hydrogen peroxide, production (Figure 1G–H). These data suggest that the underlying mechanism by which Hsp90 regulates Nox5 activity is through a direct protein-protein interaction.
Figure 1. Hsp90 directly influences Nox5-dependent ROS production.
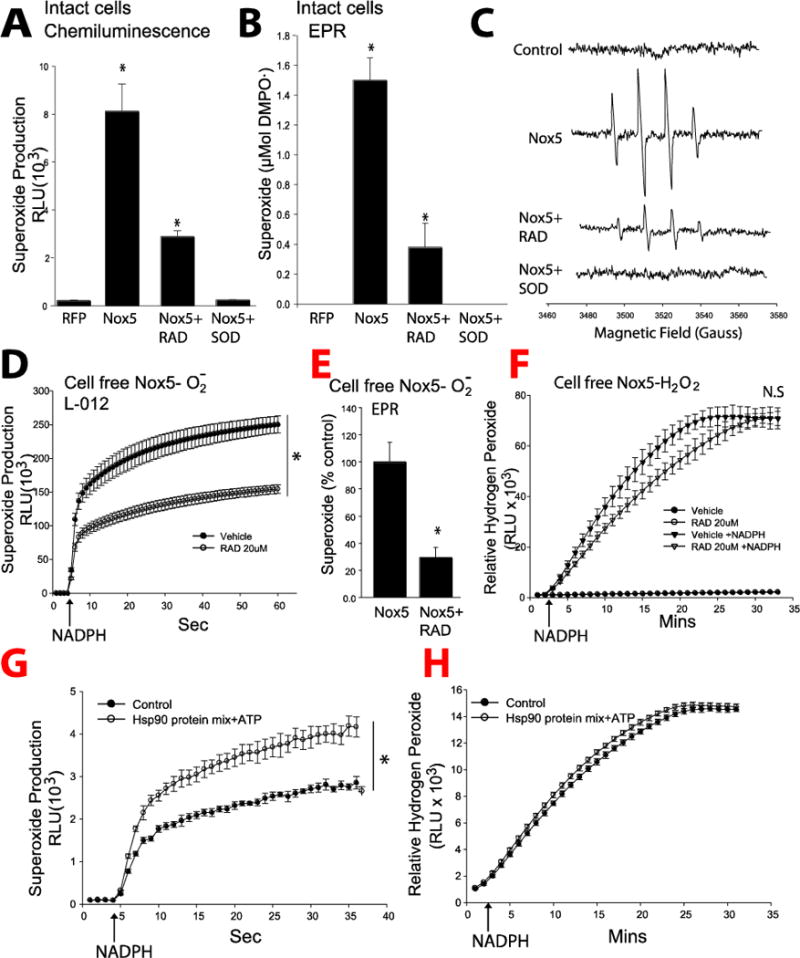
(A–C) COS-7 cells were transfected with cDNAs encoding a non-specific control gene (RFP) or Nox5 were treated with or without RAD (20μM) or SOD (100U/ml) for 1 h, and superoxide production was monitored by L-012-mediated chemiluminescence, and EPR spin trapping. EPR spectra were obtained in the presence of 50 mM DMPO, and representative spectra were shown from triplicate measurements. (D–F) Membrane preparations from COS-7 cells expressing Nox5 were treated with or without RAD (20μM) for 1h, and superoxide and hydrogen peroxide production initiated following the addition of NADPH (200μM). Superoxide was measured using L-012 chemiluminescence and EPR and levels of hydrogen peroxide using the Amplex Red assay. (G–H) Isolated membrane were incubated with 1μmol each of Hsp90β, Hsp70, Hop, Hsp40 (Ydj), and p23 in reaction buffer (20 mm Tris/HCl, pH 7.5, 5 mm MgCl2, 2 mm DTT, 0.01% Nonidet P-40, 50 mm KCl, and 5 mm ATP) and superoxide/hydrogen peroxide measured following NADPH injection. *different from Vehicle or control, p < 0.05 (n = 5–6).
Hsp90 binds to Nox5 in intact cells and in a stimulus-dependent manner but does not bind to Nox4
To investigate whether Hsp90 binds directly to Nox5 in cells, we adopted an in situ proximity ligation assay (PLA). As shown in Figure 2A, a strong positive PLA signal can be detected in cells expressing Hsp90 and Nox5 which was absent in cells incubated with either anti- Hsp90 or HA-Nox5 antibodies alone. The ability of Hsp90 to interact with Nox4 and Nox5 was further assessed by co-IP experiments. We have previously shown that the activity of Nox4, which emits only hydrogen peroxide and not superoxide, is not affected by Hsp90 inhibitors [27, 28]. Consistent with these results, we found that Nox5 but not Nox4, robustly bound Hsp90 (Figure 2B). The membrane subunit, p22phox which has been shown to directly interact with Nox4 was used as a positive control and was found within Nox4 immune complexes but not Nox5. Previous studies have shown that Nox5 activity can be stimulated by calcium mobilizing agents such as ionomycin as well as through changes in protein phosphorylation [18–20]. It is not yet known whether interventions that increase Nox5 activity affect the binding of Hsp90. COS-7 cells expressing Nox5 were treated with either vehicle or ionomycin and the degree of Hsp90:Nox5 binding determined by co-IP. Exposure to ionomycin significantly reduced the interaction between Hsp90 and Nox5 (Figure 2C). However, phorbol 12-myristate 13-acetate (PMA) which stimulates Nox5 phosphorylation and activity[19] failed to alter Hsp90 binding (Figure 2C).
Figure 2. Hsp90 binds to Nox5 but not Nox4.
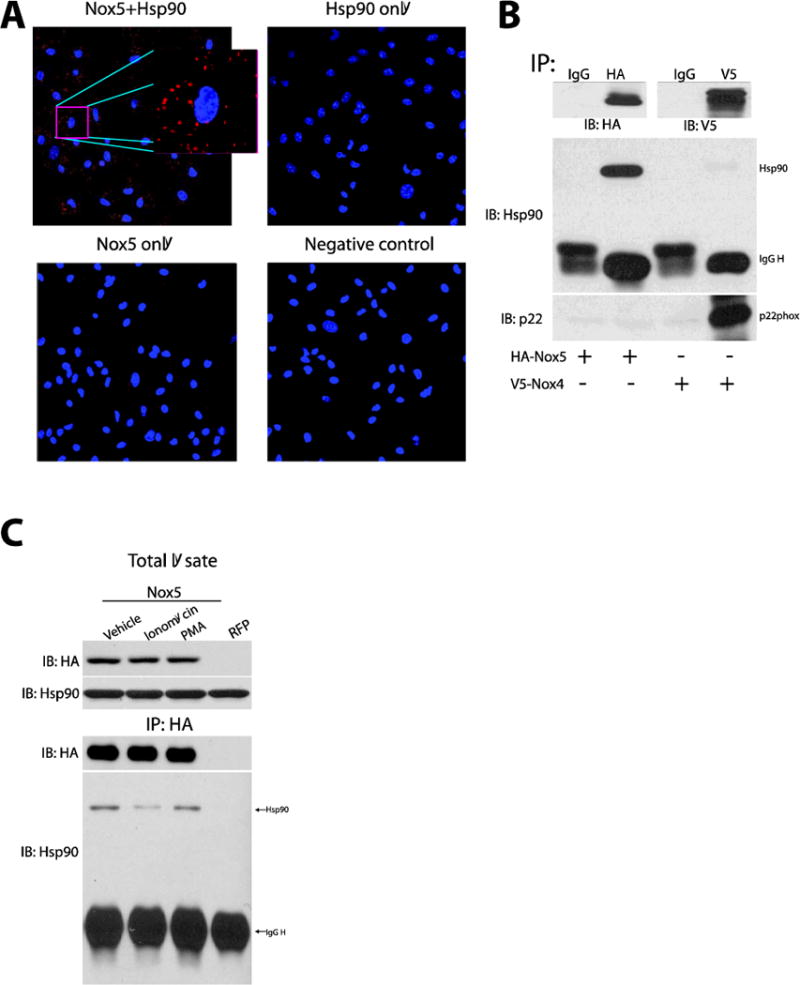
(A) Representative images of proximity ligation assays (PLA) for the co-localization of Nox5 and Hsp90 in Nox5 transfected COS-7 cells. (B) COS-7 cells expressing HA-Nox5 or V5-Nox4 were subject to immunoprecipitation using a negative control IgG, HA or V5 antibody. Immune complexes were immunoblotted for HA, V5, Hsp90 and p22phox. (C) COS-7 cells expressing RFP or HA-Nox5 were treated with vehicle, ionomycin (1μM), or PMA (100nM) for 30mins, Nox5 was immunoprecipitated from cell lysates and immune complexes were immunoblotted for HA-Nox5 and associated Hsp90. Results are representative of at least 3 separate experiments.
Hsp90 binds to Nox5 between amino acids 490-550 and maintains stability of the C-terminus
The binding site of Hsp90 on Nox5 has not yet been established. To assess this we first investigated the loss of protein-stability which requires Hsp90 binding. COS-7 cells expressing full length (WT) or inactive Nox5(H268L), truncated variants of Nox5 encompassing the N-terminus (aa1–397) or the C-terminus (aa398–719), as shown in Figure 3A, were treated with an Hsp90 inhibitor (RAD, 5–20μM, 24h). We found that inhibition of Hsp90 resulted in degradation of both the full length WT, inactive Nox5 constructs and C-terminal Nox5 (Figure 3B, and Supplemental Figure 2). In contrast, expression of the N-terminal truncation mutant was not affected by inhibition of Hsp90, suggesting that Hsp90 binds to and regulates the stability of the C-terminus.
Figure 3. Hsp90 binds to the C-terminal region of Nox5.
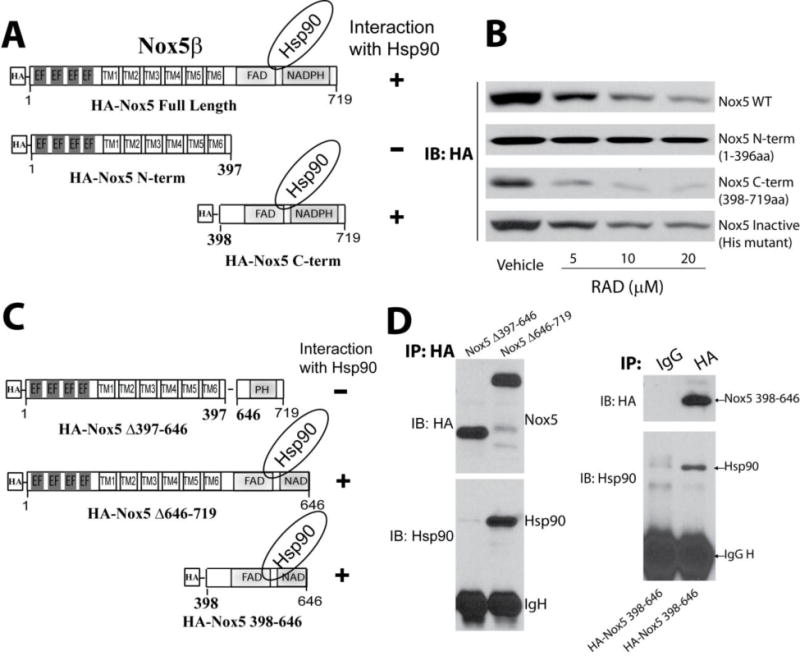
(A) Schematic of various fragments of HA-Nox5β. (B) COS-7 cells expressing Nox5 fragments were exposed to the indicated concentrations of Hsp90 inhibitor, RAD for 24h and expression levels determined by western blot. (C) Schematic of HA-Nox5β deletion constructs. (D) Nox5 deletion constructs were expressed in COS-7 cells, and cell lysates were immunoprecipitated using either control IgG or anti-HA antibody. Immune complexes were immunoblotted for HA or Hsp90 to determine the relative amount of bound Hsp90. Results are representative of at least 3–5 separate experiments.
To further define the region in the C-terminus of Nox5 that interacts with Hsp90, we generated several deletion and truncation mutants of Nox5 (Figure 3C). Using a co-IP approach, Hsp90 was found to bind to the C-terminal region of Nox5 between aa398–646 (Figure 3C–D). Further refinement using stepwise deletion analysis of Nox5 (Figure 4A) showed that aa1–489 do not interact with Hsp90. In contrast, longer fragments of Nox5 (aa 1–550, 1–571, 1–600, 1–646) exhibited increased ability to bind Hsp90 and were susceptible to RAD-mediated degradation (Figure 4A–D). These data reveal that the region between aa 490 and 550 on Nox5 is important for Hsp90 binding.
Figure 4. Refinement of the Hsp90 binding region on Nox5.
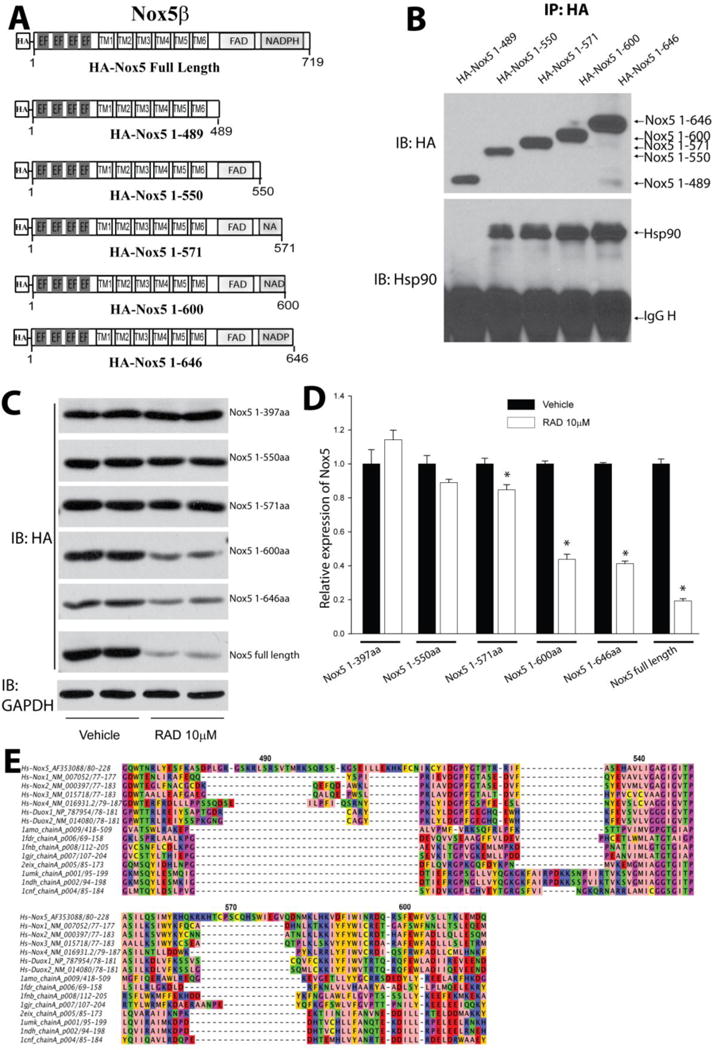
(A) Schematic of various truncation constructs of HA-Nox5β. (B) COS-7 cells were transfected with cDNAs encoding truncated Nox5 enzymes and the degree of Hsp90 binding determined by immunoprecipitation and western blot. (C) COS-7 cells expressing different Nox5 truncations were treated with indicated concentration of RAD for 24h, and expression levels determined by western blot. (D) Relative densitometry of Nox5 truncations vs. GAPDH expression. Results are representative of at least 3 separate experiments, presented as means ± S.E., *p<0.05 versus Vehicle. (E) Sequence analysis of HSP target regions on Nox5. A structurally informed multiple sequence alignment was constructed from dehydrogenase domains of Nox homologs and several crystallized dehydrogenases. A portion of the resulting alignment is shown, with color coding according to amino acid physicochemical properties using the Jalview Zappo scheme, and with Nox5 sequence numbering indicated. Sequence name (protein gi number)from top to bottom: Human Nox5alpha (gi 14211137), Human Nox1 isoform 1 (gi 148536873), Human Nox2 (gi 6996021), Human Nox3 (gi 11136626), Human Nox 4 (gi 8393843), Human Duox1 (gi 28872751), Human Duox2 (gi 132566532), Rat NADPH-cytochrome P450 Reductase (gi 3318958), E. coli Flavodoxin Reductase (gi 157831052), Spinach Ferredoxin Reductase (gi 157831108), Anabaena Ferredoxin NADP+ Reductase (gi 21730170), Physarum polycephalum Cytochrome B5 Reductase (gi 146387239), Human NADH cytochrome B5 Reductase (gi 56554196), Pig Cytochrome b5 Reductase (gi 999817), Corn Nitrate Reductase (gi 1064998).
Based on multiple alignment with other Nox enzymes and molecular modeling (Figure 4E, not shown), region aa 572–600 of Nox5 contains a loop connecting two very well conserved helices in the NADPH-binding domain. This loop has variable length and sequence among the dehydrogenase and Nox homologs, but the Nox5 loop is about twice as long as those of any of the other Noxes and has unique sequence features, again making this loop a good candidate for a HSP target. Nonetheless, mutation of numerous hydrophobic residues and other amino acids in and around this region did not disrupt Hsp90 binding (Supplemental Figure 3). Inhibition of Hsp90 with radicicol promoted the rapid dissociation of Hsp90 from Nox5 (Supplemental Figure 4).
Nox5 binds to the Middle “M” domain of Hsp90
The specific domains on Hsp90 that mediate the binding to Nox5 have not yet been defined. To determine where Nox5 binds to Hsp90 in cells, we used the C-terminal region of Nox5 and deletion mutants of Flag tagged Hsp90 (Figure 5A–B). COS-7 cells were co-transfected with cDNAs encoding either a control gene (GFP) or Flag-tagged Hsp90 constructs and the HA-tagged Nox5 C-terminus, and the degree of interaction assessed using a co-IP approach. As shown in Figure 5C, transfected Flag-Hsp90 constructs are well expressed in COS-7 cells. Co-expression of the HA-tagged Nox5 C-terminus resulted in co-immunoprecipitation of Nox5 with Hsp90 regions aa1–456 and aa1–530, but not aa1–309, aa530–680, or aa540–724 (Figure 5D). This data reveals that the regions of Hsp90 between aa310–529 mediate the interaction with Nox5.
Figure 5. Identification of the Nox5 binding region on Hsp90.
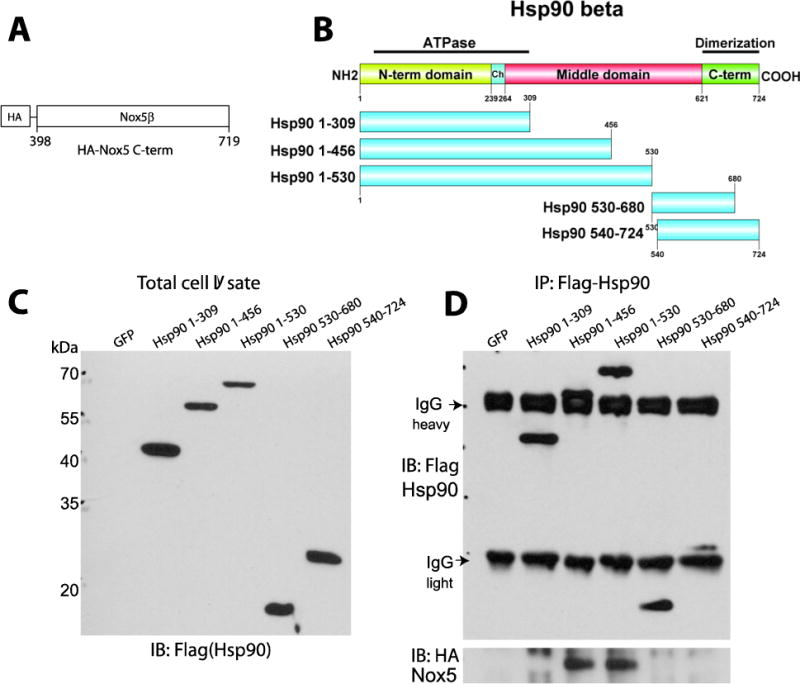
(A–B) Schematic of HA-Nox5β C-terminal construct and Flag-Hsp90 truncation constructs. (C) Relative expression of Hsp90 fragments in transfected COS-7 cells was determined by immunoblotting using Flag antibody. (D) Relative binding of Nox5 to Hsp90 fragments. Different Flag-Hsp90 constructs were immunoprecipitated from COS-7 cells expressing the C-terminal region of Nox5 and the various fragments of Hsp90, and immune complexes were immunoblotted for Flag (upper panel) and Nox5 (lower panel). Results are representative of at least 3 separate experiments.
Nox5 binds to and is regulated by Hsp90 co-chaperones
Hsp90 does not act alone in regulating protein folding and is assisted by a number of co-chaperones such as p23, Hsp40, Cdc37 and Hsp70-Hsp90 organizing protein (HOP) which not only contribute to function, but provide client selectivity [43]. For example, p23 is essential for the activation of steroid hormone receptors [44] and Cdc37 regulates kinase activity [45]. Therefore, it is important to understand the role of co-chaperones in regulating Nox5 enzyme activity. We first determined the in vitro binding of co-chaperones to Nox5 using partially purified Nox5 incubated with 1μmol each of Hsp90β, Hsp70, Hop, Hsp40 (Ydj), and p23. The relative binding to Nox5 was determined by co-IP. As we can see in Figure 6A, together with Hsp90 and Hsp70, the co-chaperones including Hop, Hsp40 and p23 are present in Nox5-dependent immune complexes. To verify this relationship in intact cells, Nox5 was immunoprecipitated and immune complex probed for the presence of co-chaperones. We found evidence of strong protein-protein interactions between Nox5 and Hsp70, Hop, Hsp40 and p23 (Figure 6B). The relative binding of Hsp90 and the co-chaperones p23, to Nox5 was reduced in the presence of Hsp90 inhibitor 17-AAG. In contrast, the amount of Hsp70, Hop and Hsp40 bound to Nox5 was increased with 17-AAG (Figure 6C). We next determined the functional significance of co-chaperone binding to ROS production. The selective silencing HOP, Hsp40 or p23 using siRNA reduced Nox5 derived superoxide production (Figure 6D–F). In contrast, the increased expression of Hsp70, but not an inactive ATP binding domain mutant, Hsp70 K71E, inhibited Nox5 activity (Figure 6G).
Figure 6. Hsp90 co-chaperones bind to Nox5 and regulate its activity.
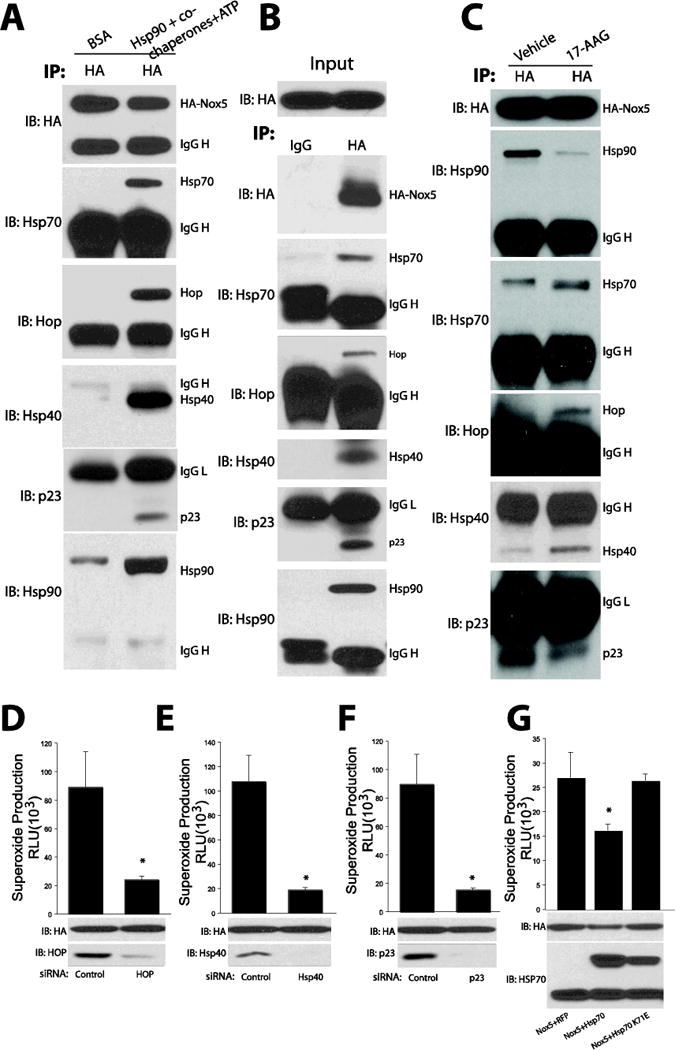
(A) Cell membranes from COS-7 cells expressing HA-Nox5 were incubated with 1μmol each of Hsp90β, Hsp70, Hop, Hsp40 (Ydj), and p23 and the relatively binding was determined by immunoprecipitation of HA and immunoblotting for HA, Hsp90 and co-chaperones with specific antibodies. (B) HA-Nox5 was immunoprecipitated from Intact, transfected COS-7 cells and the degree of chaperone binding determined by immunoprecipitation using either control IgG or anti-HA antibody and immunoblotting for HA, Hsp90 and co-chaperones with specific antibodies. (C) Hsp90 inhibitors alter chaperone binding to Nox5. COS-7 cells expressing HA-Nox5 were incubated with the Hsp90 inhibitor 17-AAG (30μM) and the relative binding of Hsp90, Hsp70 and Hop, Hsp40 and p23 determined by immunoprecipitation of HA-Nox5 and immunoblotting for HA, Hsp90 and co-chaperones with specific antibodies. (D–F) HEK293 cells expressing HA-Nox5 were transfected with control siRNA or chaperone specific siRNA (Hop, Hsp40 and p23) for 48h, Nox5 activity was determined using L-012 chemliluminescence and expression of relevant proteins via WB. (G) COS-7 cells were co-transfected with Nox5 and a nonspecific control gene (RFP), Hsp70 or inactive Hsp70 (K71E) and the relative production of ROS determined by L-012 chemiluminescence. Results are representative of at least 3-5 separate experiments. *Different from control or RFP, p < 0.05 (n = 3–6).
The role of Hsp90 in oligomerization of Nox5
Previous studies have shown that the C-terminal region of Nox5 mediates self-assembly and the formation of multimers including tetramers[46]. To investigate the role of Hsp90 in the multimeric state of Nox5, cells expressing Nox5 were exposed to an Hsp90 inhibitor and proteins cross-linked with increasing amounts of formaldehyde. The ability of Nox5 to form higher molecular weight complexes was significantly reduced in the presence of an Hsp90 inhibitor (Figure 7A). To further understand whether Hsp90 is important for Nox5 oligomerization, we performed co-IP experiments in cells expressing Nox5 variants with different tags and molecular weights, HA-Nox5 and GFP-Nox5 (Figure 7B top panel). The interaction of HA-Nox5 with GFP-Nox5 was decreased in the presence of RAD (Figure 7B lower panel).
Figure 7. Hsp90 inhibition decreases the oligomerization of Nox5.
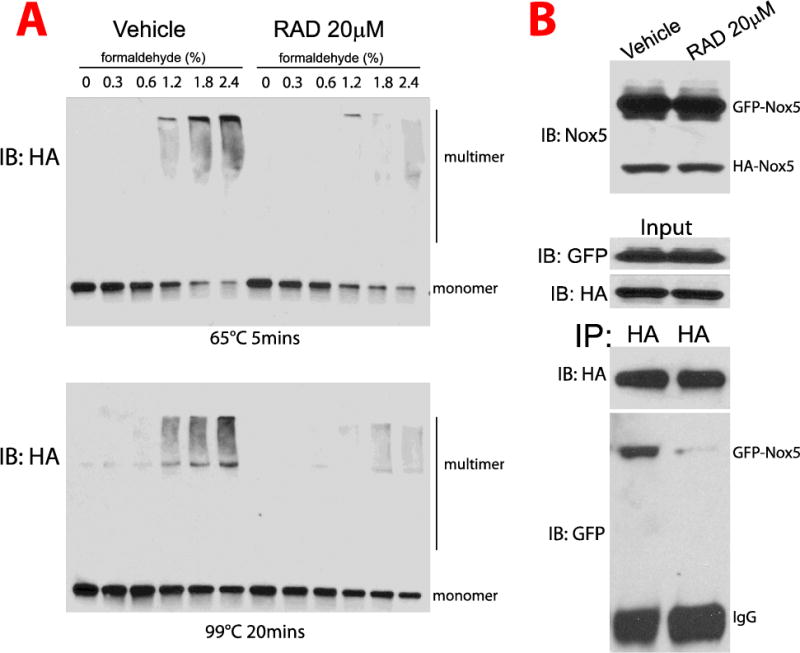
(A) COS-7 cells expressing HA-Nox5 were incubated with the Hsp90 inhibitor RAD for 30mins and proteins cross-linked with increasing concentrations of formaldehyde, 0.30%, 0.60%, 1.20%, 1.8% and 2.4%. Cell lysates were incubated at either 65°C for 5mins (top panel) or 99°C for 20 mins (lower panel) and size fractionated using the SDS-PAGE gel 4–15%. Proteins were then immunoblotted for HA-Nox5. (B) COS-7 cells co-expressing HA-Nox5 and GFP-Nox5 were incubated with vehicle or RAD (20μM) for 30mins and then pelleted and resuspended in PBS. Proteins were then cross-linked with 1.8% formaldehyde, cell lysates were immunoprecipitated using anti-HA antibody. Cell lysates and immune complexes were immunoblotted for HA and GFP. Results are representative of at least 3–5 separate experiments.
Discussion
The goals of the current study were to identify the region(s) of Nox5 that bind to Hsp90, the Nox5 binding region on Hsp90, whether the association of Hsp90 with Nox5 enzymes is affected by stimuli, whether chaperones regulate oligomerization and the role of co-chaperones in the regulation of Nox5 activity. We found that the ability of Hsp90 to regulate Nox5 activity depends on the presence of the co-chaperones HOP, Hsp40 and p23 which also bind to Nox5. Calcium-mobilizing stimuli decrease the relative association of Hsp90 and Nox5. The binding region of Hsp90 was mapped to the C-terminal region of Nox5, between aa 490–550 and Hsp90 binding and activity is important for the assembly of oligomers of Nox5. Nox5 was found to bind to the middle domain of Hsp90. Collectively, these results suggest that Nox5 is regulated by Hsp90 and co-chaperones in a manner analogous to that described for the steroid hormone receptors.
How proteins adopt complex and dynamic tertiary structures to enable specific functions remains an important but incompletely understood question. Molecular chaperones and their co-chaperones help fold nascent proteins and prevent protein aggregation[47]. Hsp90 binds to a more selective group of substrates that includes signaling molecules such as receptors and kinases, transcription factors, nitric oxide synthases etc [48]. The binding of Hsp90 enables the proper folding of critical domains of client proteins to enable specific functions. In the example of steroid receptors, Hsp90 stabilizes the ligand binding pocket which enables steroids to bind and activate the receptor. In the examples of iNOS, nNOS and sGC, Hsp90 has been shown to modulate the substrate binding cleft, facilitate the insertion of heme and promote catalytic activity [49–51]. Hsp90 is also an important regulator of eNOS activity but this occurs through changes in phosphorylation and protein:protein interactions [37, 52]. The mechanism by which Hsp90 impacts the activity of the Nox enzymes (Nox1-3 and 5) is not yet known. The activity of Noxes1-3 and 5 are stimulus dependent and result in both superoxide and hydrogen peroxide production. In contrast, Nox4 is constitutively active and rapidly converts superoxide to emit only hydrogen peroxide [53–55]. Hsp90 binds to and regulates the activity of Nox 2 and 5 (and presumably 1 and 3 based on their degradation in the presence of Hsp90 inhibitors) [27, 28] but as shown in this study, it does not bind to or regulate the activity of Nox4. Therefore it remains possible that Hsp90 binding helps to coordinate the stimulus-dependent activation of the Nox enzymes and or regulate the relative production of superoxide versus hydrogen peroxide.
To identify how Hsp90 impacts the function of Nox5, we first assessed the enzymatic regions on Nox5 where Hsp90 binds. The binding of Hsp90 to WT-Nox5 has been demonstrated previously [27, 28] and confirmed in this study in intact cells using the proximity ligation assay and also in vitro using a co-IP approach with purified proteins. To determine where Hsp90 binds to Nox5, we generated novel Nox5 constructs using stepwise deletion and truncation analysis based on functional domains. We found that the region aa 490–550 of Nox5 is necessary for Hsp90 binding. Given the ability of Hsp90 inhibitors to trigger the degradation of Hsp90-client proteins including Nox enzymes [27, 28] we utilized the loss of Nox protein expression in the presence of Hsp90 inhibitors to probe the dependency on Hsp90 binding. We found that Hsp90 inhibitors promoted the degradation of both WT and an inactive mutant of Nox5, suggesting that Nox activity is not a requirement for Hsp90 binding. Previous studies have shown that the C-terminus of Nox5 physically associates with Hsp90 [27] and consistent with this, we found that Hsp90 inhibitors robustly decrease the expression levels of a C-terminal Nox5 fragment (aa 398–719). In contrast, expression of an N-terminal truncation mutant of Nox5 (aa 1–397) was not affected by Hsp90 inhibition. Further mapping revealed that aa 572–600 are necessary for the loss of Nox5 protein expression in the presence of Hsp90 inhibitors. While the Co-IP experiments suggest that the region aa490–550 is critical for binding Hsp90, we cannot rule out an additional binding region between aa 551–646. This is based on results with the Nox5Δ646–719 and C-terminal fragment, aa 398–646 which also bind to Hsp90. The mechanisms underlying the difference between the identified Hsp90 binding site using Co-IP (aa 490–550) and the regions of Nox5 necessary for protein degradation in the presence of Hsp90 inhibitors (aa 572–600) are not yet clear but may relate to the aforementioned presence of multiple binding regions for Hsp90, the binding of Hsp70 and CHIP, which are induced by Hsp90 inhibitors and necessary for Nox protein degradation [28] or critical sites of protein ubiquitination which mediate the proteasome-dependent degradation of Nox proteins. Collectively these results suggest the C-terminal region of Nox5 (aa490–600) requires Hsp90 to maintain protein stability.
Sequence analysis of Nox isoforms and homologous crystallized dehydrogenase domains reveals that the N-terminal portion of the aa 490–550 region of Nox5 (and the corresponding regions of other Noxes) includes an insertion in the FAD binding domain of Noxes compared to the crystallized dehydrogenase domains (Figure 4E). Nox5 has a significantly longer insertion here than Nox1–4, making this region an excellent candidate for an HSP binding target. The N-terminal portion of this region is adjacent to the FAD binding site. Interestingly, peptides derived from the homologous region of Nox2 bind p67phox [56] suggesting an evolutionary conservation of regulatory protein:protein interactions in this area. The region between aa 572–600 contains a loop connecting helices in the NADPH-binding domain. However, mutation of hydrophobic residues in and around this region did not affect Hsp90 binding. This lack of a highly defined binding site for Hsp90 is consistent with other studies. Binding sites for Hsp90 been found to be ambiguous with regard to amino acid sequence and complex in that Hsp90 recognizes exposed hydrophophic amino acids that may be distributed across 3 dimensional space[48, 57]. While the C-terminal portion of the 490–550 region also spans the linker connecting the FAD-binding domain and the NADPH domain, molecular modeling suggests that much of that linker is likely to be buried by interaction with the transmembrane domain and thus unavailable for binding to HSP90.
The identification of the specific domains within Hsp90 that bind to client proteins is challenging due to the dynamics of client protein binding and the multiple regions of Hsp90 involved in folding and the absence of a clearly identifiable domain. Substrates have been found to bind to both the N-terminal, middle domains and C-terminal domains [48]. The specific domains of Hsp90 that bind to Nox5 have not previously been defined. We found that the middle domain of Hsp90 and more specifically, aa 310–529 of Hsp90 mediate the binding to Nox5. This is consistent with studies showing that other heme containing proteins bind to the middle domain of Hsp90 [37, 58] as well as kinases such as Akt [59]. The middle domain of Hsp90 can be regulated by phosphorylation, acetylation and the binding of co-chaperones. It remains possible that post-translational modification of this region could affect substrate binding and folding and therefore affect Nox activity and ROS levels. In contrast it is unlikely that the phosphorylation of Nox5 influences Hsp90 binding as PMA, which induces the phosphorylation of Nox5β at Thr494 and Ser498 [19] within the region identified to bind Hsp90, did not modify Hsp90 binding. However, the addition of a calcium mobilizing agent, ionomycin which stimulates Nox5 activity did promote the dissociation of Hsp90 from Nox5. Other studies have shown that calcium-mobilizing stimuli can promote Hsp90 association [52] to facilitate enzyme activation. Thus the significance of decreased Hsp90 binding to Nox5 is not yet known and may represent a negative feedback mechanism.
Nox1-3 and 5 produce a combination of superoxide and hydrogen peroxide, while Nox4 emits primarily only hydrogen peroxide. We have previously shown that Hsp90 inhibition specifically reduces the production of superoxide but not hydrogen peroxide from Noxes 1-3 and 5 in cells [27, 28] but the mechanisms by which this occurs are not known. As the detection of specific reactive oxygen species is heavily dependent on methods with varying degrees of sensitivity and specificity, we employed multiple approaches including L-012 chemiluminescence, EPR to measure superoxide and amplex red for hydrogen peroxide along with appropriate controls. Using both L-012 and EPR we found that acute inhibition of Hsp90 reduces Nox5 derived superoxide in both cells and in isolated enzyme activity assays. In contrast, the addition of a mixture of Hsp90 and the co-chaperones Hsp70, HOP, Hsp40, and p23 with Nox5 significantly increased superoxide production. This data suggest that the underlying mechanism by which Hsp90 regulates superoxide production from Nox5 occurs through a direct protein-protein interaction rather than a change in intracellular location. Consistent with previous findings in intact cells, we found that Hsp90 inhibitors did not alter the ability of Nox5 to produce hydrogen peroxide in isolated membranes. However, the mechanisms by which Hsp90 specifically influences superoxide but not hydrogen peroxide production is still unclear. Others have shown that hydrogen peroxide production from Nox enzymes results from the actions of extracellular loops of Nox proteins which promote the conversion of superoxide to hydrogen peroxide [60–62]. At this time it is unclear how Hsp90, which binds Nox5 on the intracellular C-terminal region would influence the function of extracellular loops. However, evidence exists to suggest that intracellular domains may also influence the production of hydrogen peroxide. For example fusion of the N-terminus of superoxide-producing Nox1 or Nox2 (which contains the extracellular loops) to the C-terminus of the hydrogen peroxide-producing Nox4 results in hydrogen peroxide production [27, 63]. The region where Hsp90 binds is close to the identified phosphorylation sites regulating calcium-dependent activation of Nox5. This region may influence binding of the N-terminal region of Nox5 to the C-terminal Regulatory EF-hand Binding Domain (REFBD, aa 638–661 Nox5β, Nox5V2) [64], and it is possible that Hsp90 binding facilitates a structural change to facilitate enzyme activation via this mechanism. However, as both calcium and phosphorylation-dependent activation of Nox5 increase both superoxide and hydrogen peroxide without an obvious bias [20], it is unlikely to be the underlying mechanism. Alternatively, Nox5 has been shown to assemble into oligomers, presumably a tetramer via interactions within the C-terminal region[46]. The ability of inactive Nox5 constructs to function as dominant negatives [11, 46] suggests that oligomerization of Nox5 may be important for ROS formation. We found that short-term inhibition of Hsp90 reduced the formation of higher molecular weight Nox5 complexes, but it remains to be shown whether the degree of oligomerization impacts the amount of ROS produced and/or the balance of superoxide versus hydrogen peroxide.
Hsp90 works in concert with a number of chaperones and co-chaperones that modulate the activity of Hsp90 and the binding and conformation of its client proteins [65, 66]. We have previously found that Hsp70 is a negative regulator of Nox5[28], however, the roles of other important co-chaperones like HOP, Hsp40 and p23 have not been studied. We found that the co-chaperones including Hop, Hsp40 and p23 can directly interact with Nox5. In the presence of an Hsp90 inhibitor, the relative association of Hsp90 and p23 was reduced, while Hsp70, Hsp40 and Hop were increased. These data suggest that nhibition of Hsp90 promotes the transition of a stable enzyme complex to a proteasome-targeted form that results in enhanced degradation. These results are consistent with those observed with steroid hormone receptors. The Hsp90 co-chaperones are important for the function of Nox5 as genetic silencing robustly decreases the ability of Nox5 to generate superoxide. In contrast Hsp70, negatively regulates Nox function as shown previously [28]. Thus, Hsp90 should not be considered a solitary regulator of Nox5 function and instead operates in concert and/or opposition with multiple chaperones.
In summary, our study adds significantly to our understanding of how molecular chaperones influence Nox enzyme stability and ROS production. We show that Nox5, but not Nox4, directly binds to Hsp90 and that the binding of an active folding complex containing Hsp90 and other chaperones to Nox5 positively regulates the production of superoxide, but not hydrogen peroxide. Binding studies reveal a region of Nox5, between aa 490–550 that mediates binding to Hsp90 and a more distal region, aa 572–600 that may also contribute to Hsp90 binding and is required to facilitate Hsp90-dependent protein degradation. The C-terminal region of Nox5 bound to the M domain of Hsp90 (aa 310–529) and post-translational modifications of Hsp90 within this region have the potential to alter the ability Nox activity. In addition to Hsp90, Nox5 was found to bind the co-chaperones Hsp70, Hop, p23 and Hsp40 which are required to support enzyme activity. While this study has focused on the role of Nox5 and Nox4, Hsp90 also regulates the activity and stability of Nox1-3 and it remains to be determined if Hsp90 binds to the C-terminal region of other Hsp90-regulated Nox enzymes. Greater appreciation of the nature of the interaction between Hsp90 and Nox enzymes may provide critical new insights into the development of modalities to better target Nox-derived superoxide production and more selectively regulate the aberrant production of reactive oxygen species.
Supplementary Material
Supplemental Figure 1. Measurement of superoxide and hydrogen peroxide levels with L-012 chemiluminescence and amplex red fluorescence in cells and cell membranes. Cells were transfected with a control gene (β-gal) or Nox5, and superoxide (A) and hydrogen peroxide (B) were measured using L-012 or amplex red, respectively, in the presence and absence of SOD (200 U/ml), DPI (3μM), or catalase (200 U/ml). (C) Membrane preparations from COS-7 cells expressing Nox5 were treated with or without RAD (20μM), SOD (200 U/ml) or catalase (200 U/ml) for 30 mins, and superoxide production initiated following the addition of NADPH (200μM). Superoxide was measured using L-012 chemiluminescence. *different from Vehicle, p < 0.05 (n = 5–6).
Supplemental Figure 2. Densitometry analysis of Nox5 expression in COS-7 cells in the presence and absence of vehicle or the Hsp90 inhibitor RAD (5–20μM) for 24h.
Supplemental Figure 3. Mutation of amino acids in and around the distal loop in the Hsp90 binding region (aa490–600) does not alter Hsp90 binding. COS-7 cells expressing RFP, HA-Nox5 WT, or the indicated HA-Nox5 mutants were subject to immunoprecipitation using an anti-HA antibody. Immune complexes were immunoblotted for HA and associated Hsp90. Results are representative of at least 2 separate experiments.
Supplemental Figure 4. The Hsp90 inhibitor RAD potently decreases the interaction between Nox5 and Hsp90. COS-7 cells expressing RFP or Nox5 were treated with or without RAD for 30min. Cell lysates were immunoprecipitated using anti-HA antibody, and immune complexes were immunoblotted for HA-Nox5 and Hsp90. Results are representative of at least 3–5 separate experiments.
Highlights.
In isolated Nox5 enzyme activity assays, Hsp90 inhibitors selectively decrease superoxide, but not hydrogen peroxide.
Hsp90 alone modestly increases Nox5 activity but in combination with co-chaperones, it robustly stimulated superoxide, but not hydrogen peroxide
Proximity ligation assays reveal that Nox5 and Hsp90 interact in intact cells. Co-IP reveals that Hsp90 binds to Nox5 but not Nox4. Calcium-disrupts Hsp90 binding.
Inhibition of Hsp90 promotes the degradation of a C-terminal fragment (aa398–719) of Nox5, but not N-terminal fragments (aa1–550)
The region between aa490–550 of Nox5 binds to the M domain of Hsp90
Nox5 binds co-chaperones Hsp70, HOP, p23 and Hsp40 which are important for superoxide production.
Inhibition of Hsp90 results in the loss of higher molecular weight complexes of Nox5 and decreased interaction between monomers.
Acknowledgments
This work was supported by NIH grants R01HL092446, HL124773 (DF, DWS) and P01 HL101902-01A1 (DF, AV) and awards from the AHA (FC and YW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith JA. Neutrophils, host defense, and inflammation: a double-edged sword. Journal of leukocyte biology. 1994;56:672–686. doi: 10.1002/jlb.56.6.672. [DOI] [PubMed] [Google Scholar]
- 2.Ali MI, Ketsawatsomkron P, Belin de Chantemele EJ, Mintz JD, Muta K, Salet C, Black SM, Tremblay ML, Fulton DJ, Marrero MB, Stepp DW. Deletion of protein tyrosine phosphatase 1b improves peripheral insulin resistance and vascular function in obese, leptin-resistant mice via reduced oxidant tone. Circ Res. 2009;105:1013–1022. doi: 10.1161/CIRCRESAHA.109.206318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 4.Liu JQ, Zelko IN, Erbynn EM, Sham JS, Folz RJ. Hypoxic pulmonary hypertension: role of superoxide and NADPH oxidase (gp91phox) American journal of physiology Lung cellular and molecular physiology. 2006;290:L2–10. doi: 10.1152/ajplung.00135.2005. [DOI] [PubMed] [Google Scholar]
- 5.Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q, Malik P, Pandey D, Gupta S, Jagnandan D, Belin de Chantemele E, Banfi B, Marrero MB, Rudic RD, Stepp DW, Fulton DJ. Paradoxical activation of endothelial nitric oxide synthase by NADPH oxidase. Arterioscler Thromb Vasc Biol. 2008;28:1627–1633. doi: 10.1161/ATVBAHA.108.168278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szocs K, Lassegue B, Sorescu D, Hilenski LL, Valppu L, Couse TL, Wilcox JN, Quinn MT, Lambeth JD, Griendling KK. Upregulation of Nox-based NAD(P)H oxidases in restenosis after carotid injury. Arterioscler Thromb Vasc Biol. 2002;22:21–27. doi: 10.1161/hq0102.102189. [DOI] [PubMed] [Google Scholar]
- 8.Jackman KA, Miller AA, De Silva TM, Crack PJ, Drummond GR, Sobey CG. Reduction of cerebral infarct volume by apocynin requires pretreatment and is absent in Nox2-deficient mice. Br J Pharmacol. 2009;156:680–688. doi: 10.1111/j.1476-5381.2008.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 10.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10:453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey D, Patel A, Patel V, Chen F, Qian J, Wang Y, Barman SA, Venema RC, Stepp DW, Rudic RD, Fulton DJ. Expression and functional significance of NADPH oxidase 5 (Nox5) and its splice variants in human blood vessels. Am J Physiol Heart Circ Physiol. 2012;302:H1919–1928. doi: 10.1152/ajpheart.00910.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedard K, Jaquet V, Krause KH. NOX5: from basic biology to signaling and disease. Free Radic Biol Med. 2012;52:725–734. doi: 10.1016/j.freeradbiomed.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Holterman CE, Thibodeau JF, Towaij C, Gutsol A, Montezano AC, Parks RJ, Cooper ME, Touyz RM, Kennedy CR. Nephropathy and elevated BP in mice with podocyte-specific NADPH oxidase 5 expression. J Am Soc Nephrol. 2014;25:784–797. doi: 10.1681/ASN.2013040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Barman S, Yu Y, Haigh S, Wang Y, Dou H, Bagi Z, Han W, Su Y, Fulton DJ. Caveolin-1 is a negative regulator of NADPH oxidase-derived reactive oxygen species. Free Radic Biol Med. 2014;73:201–213. doi: 10.1016/j.freeradbiomed.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzik TJ, Chen W, Gongora MC, Guzik B, Lob HE, Mangalat D, Hoch N, Dikalov S, Rudzinski P, Kapelak B, Sadowski J, Harrison DG. Calcium-dependent NOX5 nicotinamide adenine dinucleotide phosphate oxidase contributes to vascular oxidative stress in human coronary artery disease. J Am Coll Cardiol. 2008;52:1803–1809. doi: 10.1016/j.jacc.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn NE, Meischl C, Kawahara T, Musters RJ, Verhoef VM, van der Velden J, Vonk AB, Paulus WJ, van Rossum AC, Niessen HW, Krijnen PA. NOX5 expression is increased in intramyocardial blood vessels and cardiomyocytes after acute myocardial infarction in humans. Am J Pathol. 2012;180:2222–2229. doi: 10.1016/j.ajpath.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Holterman CE, Thibodeau JF, Kennedy CR. NADPH oxidase 5 and renal disease. Curr Opin Nephrol Hypertens. 2015;24:81–87. doi: 10.1097/MNH.0000000000000081. [DOI] [PubMed] [Google Scholar]
- 18.Banfi B, Molnar G, Maturana A, Steger K, Hegedus B, Demaurex N, Krause KH. A Ca(2+)-activated NADPH oxidase in testis, spleen, and lymph nodes. J Biol Chem. 2001;276:37594–37601. doi: 10.1074/jbc.M103034200. [DOI] [PubMed] [Google Scholar]
- 19.Jagnandan D, Church JE, Banfi B, Stuehr DJ, Marrero MB, Fulton DJ. Novel mechanism of activation of NADPH oxidase 5. calcium sensitization via phosphorylation. J Biol Chem. 2007;282:6494–6507. doi: 10.1074/jbc.M608966200. [DOI] [PubMed] [Google Scholar]
- 20.Serrander L, Jaquet V, Bedard K, Plastre O, Hartley O, Arnaudeau S, Demaurex N, Schlegel W, Krause KH. NOX5 is expressed at the plasma membrane and generates superoxide in response to protein kinase C activation. Biochimie. 2007;89:1159–1167. doi: 10.1016/j.biochi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.El Jamali A, Valente AJ, Lechleiter JD, Gamez MJ, Pearson DW, Nauseef WM, Clark RA. Novel redox-dependent regulation of NOX5 by the tyrosine kinase c-Abl. Free Radic Biol Med. 2008;44:868–881. doi: 10.1016/j.freeradbiomed.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey D, Gratton JP, Rafikov R, Black SM, Fulton DJ. Calcium/calmodulin-dependent kinase II mediates the phosphorylation and activation of NADPH oxidase 5. Mol Pharmacol. 2011;80:407–415. doi: 10.1124/mol.110.070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey D, Fulton DJ. Molecular regulation of NADPH oxidase 5 via the MAPK pathway. Am J Physiol Heart Circ Physiol. 2011;300:H1336–1344. doi: 10.1152/ajpheart.01163.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J, Chen F, Kovalenkov Y, Pandey D, Moseley MA, Foster MW, Black SM, Venema RC, Stepp DW, Fulton DJ. Nitric oxide reduces NADPH oxidase 5 (Nox5) activity by reversible S-nitrosylation. Free Radic Biol Med. 2012;52:1806–1819. doi: 10.1016/j.freeradbiomed.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey D, Chen F, Patel A, Wang CY, Dimitropoulou C, Patel VS, Rudic RD, Stepp DW, Fulton DJ. SUMO1 negatively regulates reactive oxygen species production from NADPH oxidases. Arterioscler Thromb Vasc Biol. 2011;31:1634–1642. doi: 10.1161/ATVBAHA.111.226621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tirone F, Cox JA. NADPH oxidase 5 (NOX5) interacts with and is regulated by calmodulin. FEBS Lett. 2007;581:1202–1208. doi: 10.1016/j.febslet.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 27.Chen F, Pandey D, Chadli A, Catravas JD, Chen T, Fulton DJ. Hsp90 regulates NADPH oxidase activity and is necessary for superoxide but not hydrogen peroxide production. Antioxid Redox Signal. 2011;14:2107–2119. doi: 10.1089/ars.2010.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F, Yu Y, Qian J, Wang Y, Cheng B, Dimitropoulou C, Patel V, Chadli A, Rudic RD, Stepp DW, Catravas JD, Fulton DJ. Opposing actions of heat shock protein 90 and 70 regulate nicotinamide adenine dinucleotide phosphate oxidase stability and reactive oxygen species production. Arterioscler Thromb Vasc Biol. 2012;32:2989–2999. doi: 10.1161/ATVBAHA.112.300361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madrigal-Matute J, Fernandez-Garcia CE, Gomez-Guerrero C, Lopez-Franco O, Munoz-Garcia B, Egido J, Blanco-Colio LM, Martin-Ventura JL. HSP90 inhibition by 17-DMAG attenuates oxidative stress in experimental atherosclerosis. Cardiovascular research. 2012;95:116–123. doi: 10.1093/cvr/cvs158. [DOI] [PubMed] [Google Scholar]
- 30.Sharma K, Vabulas RM, Macek B, Pinkert S, Cox J, Mann M, Hartl FU. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Molecular & cellular proteomics: MCP. 2012;11:M111-014654. doi: 10.1074/mcp.M111.014654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grad I, McKee TA, Ludwig SM, Hoyle GW, Ruiz P, Wurst W, Floss T, Miller CA, 3rd, Picard D. The Hsp90 cochaperone p23 is essential for perinatal survival. Mol Cell Biol. 2006;26:8976–8983. doi: 10.1128/MCB.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez MP, Chadli A, Toft DO. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J Biol Chem. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- 35.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 36.Chen F, Fulton DJ. An inhibitor of protein arginine methyltransferases, 7,7’-carbonylbis(azanediyl)bis(4-hydroxynaphthalene-2-sulfonic acid (AMI-1), is a potent scavenger of NADPH-oxidase-derived superoxide. Mol Pharmacol. 2010;77:280–287. doi: 10.1124/mol.109.061077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontana J, Fulton D, Chen Y, Fairchild TA, McCabe TJ, Fujita N, Tsuruo T, Sessa WC. Domain mapping studies reveal that the M domain of hsp90 serves as a molecular scaffold to regulate Akt-dependent phosphorylation of endothelial nitric oxide synthase and NO release. Circ Res. 2002;90:866–873. doi: 10.1161/01.res.0000016837.26733.be. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, Yu Y, Haigh S, Johnson J, Lucas R, Stepp DW, Fulton DJ. Regulation of NADPH oxidase 5 by protein kinase C isoforms. PLoS One. 2014;9:e88405. doi: 10.1371/journal.pone.0088405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patwardhan CA, Fauq A, Peterson LB, Miller C, Blagg BS, Chadli A. Gedunin inactivates the co-chaperone p23 protein causing cancer cell death by apoptosis. J Biol Chem. 2013;288:7313–7325. doi: 10.1074/jbc.M112.427328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim Biophys Acta. 2012;1823:607–613. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Felts SJ, Karnitz LM, Toft DO. Functioning of the Hsp90 machine in chaperoning checkpoint kinase I (Chk1) and the progesterone receptor (PR) Cell Stress Chaperones. 2007;12:353–363. doi: 10.1379/CSC-299.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaughan CK, Mollapour M, Smith JR, Truman A, Hu B, Good VM, Panaretou B, Neckers L, Clarke PA, Workman P, Piper PW, Prodromou C, Pearl LH. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31:886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawahara T, Jackson HM, Smith SM, Simpson PD, Lambeth JD. Nox5 forms a functional oligomer mediated by self-association of its dehydrogenase domain. Biochemistry. 2011;50:2013–2025. doi: 10.1021/bi1020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 48.Karagoz GE, Rudiger SG. Hsp90 interaction with clients. Trends Biochem Sci. 2015;40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh A, Stuehr DJ. Soluble guanylyl cyclase requires heat shock protein 90 for heme insertion during maturation of the NO-active enzyme. Proc Natl Acad Sci U S A. 2012;109:12998–13003. doi: 10.1073/pnas.1205854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh A, Chawla-Sarkar M, Stuehr DJ. Hsp90 interacts with inducible NO synthase client protein in its heme-free state and then drives heme insertion by an ATP-dependent process. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2011;25:2049–2060. doi: 10.1096/fj.10-180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Billecke SS, Bender AT, Kanelakis KC, Murphy PJ, Lowe ER, Kamada Y, Pratt WB, Osawa Y. hsp90 is required for heme binding and activation of apo-neuronal nitric-oxide synthase: geldanamycin-mediated oxidant generation is unrelated to any action of hsp90. J Biol Chem. 2002;277:20504–20509. doi: 10.1074/jbc.M201940200. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa WC. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 53.Helmcke I, Heumuller S, Tikkanen R, Schroder K, Brandes RP. Identification of structural elements in Nox1 and Nox4 controlling localization and activity. Antioxid Redox Signal. 2009;11:1279–1287. doi: 10.1089/ars.2008.2383. [DOI] [PubMed] [Google Scholar]
- 54.Martyn KD, Frederick LM, von Loehneysen K, Dinauer MC, Knaus UG. Functional analysis of Nox4 reveals unique characteristics compared to other NADPH oxidases. Cell Signal. 2006;18:69–82. doi: 10.1016/j.cellsig.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 55.Serrander L, Cartier L, Bedard K, Banfi B, Lardy B, Plastre O, Sienkiewicz A, Forro L, Schlegel W, Krause KH. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bechor E, Dahan I, Fradin T, Berdichevsky Y, Zahavi A, Gross Federman A, Rafalowski M, Pick E. The dehydrogenase region of the NADPH oxidase component Nox2 acts as a protein disulfide isomerase (PDI) resembling PDIA3 with a role in the binding of the activator protein p67 (phox.) Front Chem. 3(3):2015. doi: 10.3389/fchem.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayer MP, Le Breton L. Hsp90: Breaking the Symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 58.Papapetropoulos A, Zhou Z, Gerassimou C, Yetik G, Venema RC, Roussos C, Sessa WC, Catravas JD. Interaction between the 90-kDa heat shock protein and soluble guanylyl cyclase: physiological significance and mapping of the domains mediating binding. Mol Pharmacol. 2005;68:1133–1141. doi: 10.1124/mol.105.012682. [DOI] [PubMed] [Google Scholar]
- 59.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takac I, Schroder K, Zhang L, Lardy B, Anilkumar N, Lambeth JD, Shah AM, Morel F, Brandes RP. The E-loop is involved in hydrogen peroxide formation by the NADPH oxidase Nox4. J Biol Chem. 2011;286:13304–13313. doi: 10.1074/jbc.M110.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueyama T, Sakuma M, Ninoyu Y, Hamada T, Dupuy C, Geiszt M, Leto TL, Saito N. The Extracellular A-loop of Dual Oxidases Affects the Specificity of Reactive Oxygen Species Release. J Biol Chem; 2015 doi: 10.1074/jbc.M114.592717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nisimoto Y, Diebold BA, Cosentino-Gomes D, Lambeth JD. Nox4: a hydrogen peroxide-generating oxygen sensor. Biochemistry. 2014;53:5111–5120. doi: 10.1021/bi500331y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Lohneysen K, Noack D, Wood MR, Friedman JS, Knaus UG. Structural insights into Nox4 and Nox2: motifs involved in function and cellular localization. Mol Cell Biol. 2010;30:961–975. doi: 10.1128/MCB.01393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tirone F, Radu L, Craescu CT, Cox JA. Identification of the binding site for the regulatory calcium-binding domain in the catalytic domain of NOX5. Biochemistry. 2010;49:761–771. doi: 10.1021/bi901846y. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto S, Subedi GP, Hanashima S, Satoh T, Otaka M, Wakui H, Sawada K, Yokota S, Yamaguchi Y, Kubota H, Itoh H. ATPase activity and ATP-dependent conformational change in the co-chaperone HSP70/HSP90-organizing protein (HOP) J Biol Chem. 2014;289:9880–9886. doi: 10.1074/jbc.M114.553255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh M, Shah V, Tatu U. A novel C-terminal homologue of Aha1 co-chaperone binds to heat shock protein 90 and stimulates its ATPase activity in Entamoeba histolytica. J Mol Biol. 2014;426:1786–1798. doi: 10.1016/j.jmb.2014.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Measurement of superoxide and hydrogen peroxide levels with L-012 chemiluminescence and amplex red fluorescence in cells and cell membranes. Cells were transfected with a control gene (β-gal) or Nox5, and superoxide (A) and hydrogen peroxide (B) were measured using L-012 or amplex red, respectively, in the presence and absence of SOD (200 U/ml), DPI (3μM), or catalase (200 U/ml). (C) Membrane preparations from COS-7 cells expressing Nox5 were treated with or without RAD (20μM), SOD (200 U/ml) or catalase (200 U/ml) for 30 mins, and superoxide production initiated following the addition of NADPH (200μM). Superoxide was measured using L-012 chemiluminescence. *different from Vehicle, p < 0.05 (n = 5–6).
Supplemental Figure 2. Densitometry analysis of Nox5 expression in COS-7 cells in the presence and absence of vehicle or the Hsp90 inhibitor RAD (5–20μM) for 24h.
Supplemental Figure 3. Mutation of amino acids in and around the distal loop in the Hsp90 binding region (aa490–600) does not alter Hsp90 binding. COS-7 cells expressing RFP, HA-Nox5 WT, or the indicated HA-Nox5 mutants were subject to immunoprecipitation using an anti-HA antibody. Immune complexes were immunoblotted for HA and associated Hsp90. Results are representative of at least 2 separate experiments.
Supplemental Figure 4. The Hsp90 inhibitor RAD potently decreases the interaction between Nox5 and Hsp90. COS-7 cells expressing RFP or Nox5 were treated with or without RAD for 30min. Cell lysates were immunoprecipitated using anti-HA antibody, and immune complexes were immunoblotted for HA-Nox5 and Hsp90. Results are representative of at least 3–5 separate experiments.


