Abstract
Background
Gastric cancer (GC) is a deadly malignancy worldwide. In the past, it has been shown that cellular signaling pathway alterations play a crucial role in the development of GC. In particular, deregulation of the PI3K/AKT/mTOR pathway seems to affect multiple GC functions including growth, proliferation, metabolism, motility and angiogenesis. Targeting alterations in this pathway by microRNAs (miRNAs) represents a potential therapeutic strategy, especially in inhibitor-resistant tumors. The objective of this study was to evaluate the expression of 3 pre-selected miRNAs, miR-101-2, miR-125b-2 and miR-451a, in a series of primary GC tissues and matched non-GC tissues and in several GC-derived cell lines, and to subsequently evaluate the functional role of these miRNAs.
Methods
Twenty-five primary GC samples, 25 matched non-GC samples and 3 GC-derived cell lines, i.e., AGS, MKN28 and MKN45, were included in this study. miRNA and target gene expression levels were assessed by quantitative RT-PCR and western blotting, respectively. Subsequently, cell viability, clone formation, cell death, migration and invasion assays were performed on AGS cells.
Results
miR-101-2, miR-125b-2 and miR-451a were found to be down-regulated in the primary GC tissues and the GC-derived cell lines tested. MiRNA mimic transfections significantly reduced cell viability and colony formation, increased cell death and reduced cell migration and invasion in AGS cells. We also found that exogenous expression of miR-101-2, miR-125b-2 and miR-451a decreased the expression of their putative targets MTOR, PIK3CB and TSC1, respectively.
Conclusions
Our expression analyses and in vitro functional assays suggest that miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in primary GCs as well as in GC-derived AGS cells.
Keywords: Gastric cancer, microRNAs, miR-101-2, miR-125b-2, miR-451a, PI3K/AKT/mTOR pathway
1 Introduction
Gastric cancer (GC) is the fourth most frequent malignant disease and the second cause of cancer mortality worldwide [1, 2]. GC is an aggressive neoplasm associated with an usually late diagnosis, an unsatisfactory treatment and a poor prognosis. An early diagnosis is rarely achieved since the signs and symptoms of GC are not specific and often appear late in the clinical course of the disease [3–5]. Therefore, the diagnosis is generally made when the cancer is already at an advanced stage. So far, co-adjuvant therapy with chemo- and/or radiotherapy has not resulted in a major impact on the survival of patients with advanced GC [6]. Therefore, there is an urgent need for additional therapeutic targets.
Previous studies have revealed that the phosphatidylinositol 3-kinases (PI3K)/AKT/mammalian target of the rapamycin (mTOR) signaling pathway plays an important role in the development of GC and, as such, may provide interesting targets for therapy [7–9]. This pathway plays a crucial role in multiple cellular functions including growth, proliferation, metabolism, survival, and angiogenesis. Growth factor signaling mediated by PI3K through AKT, TSC1 and TSC2 activates mTOR. TSC1 and TSC2 form a dimer (TSC1/TSC2) that regulates mTOR activity indirectly. When upstream signals are activated, TSC1/TSC2 is inhibited by AKT, allowing mTOR activation. Meanwhile, P70S6K1 appears to act as a negative feedback regulator blocking the activity of AKT and other upstream proteins associated with the mTOR complex 1 (mTORC1). In cancer cells, increased PI3K-AKT activity leads to activation of mTORC1 through phosphorylation and a decrease in the feedback activity of P70S6K1-mTORC2. These changes result in increased mitochondrial activity, angiogenesis, ribosome biogenesis for enhanced protein synthesis, cellular growth, proliferation, and autophagy [10, 11]. As such, the PI3K/AKT/mTOR pathway appears to be a good candidate for evaluating the capacity of microRNAs (miRNAs) to regulate specific targets in this pathway and, thereby, to alter the malignant phenotype of GC cells.
MiRNAs are small non-coding RNAs approximately 18–25 nucleotides in length that negatively regulate gene expression [12, 13]. These small molecules often bind to mRNAs in their 3′-UTR regions and, by doing so, can cause a decrease in mRNA expression or translation based on either complete or incomplete complementarity [14–16]. MiRNAs are expressed in a tissue-specific manner and are known to be important in the regulation of essential biological functions including cellular proliferation, differentiation, apoptosis, metabolism and immune responses [17, 18]. Ample reports have shown that functional loss or gain of specific miRNAs may be key events in oncogenesis, since they can act either as oncogenes (in case they are amplified or over-expressed) or as tumor suppressors (in the case they are deleted or repressed) [19, 20]. Many miRNAs are currently under investigation as diagnostic or prognostic biomarkers, as therapeutic targets and as bio-markers for cancer subtypes [21, 22].
Previously, it has been found the miRNAs miR-101-2, miR-125b-2 and miR-451a are deregulated in several types of cancer [23–33], including GC [30, 34–36]. It has also been found these miRNAs may act as tumor suppressors, and that their down-regulation may affect tumorigenesis. However, the cellular signaling pathway(s) regulated by these miRNAs have so far remained unknown.
Here, we identified miR-101-2, miR-125b-2 and miR-451a as down-regulated miRNAs in primary GC samples and in vitro validated their functional effects though exogenous expression in AGS gastric cancer-derived cells. Our findings contribute to a better understanding of the biology underlying GC, and may be instrumental for the design of novel therapeutic applications that complement or enhance the current clinical management of this malignancy.
2 Materials and methods
2.1 Gastric tissues and cancer-derived cell lines
In total 50 gastric tissues were included in this study, encompassing 25 tumor samples from patients with advanced GC (adenocarcinoma) and 25 matched non-tumor gastric samples from the same patients. All gastric samples consisted of frozen tissues collected at the time of diagnosis through an approved tissue collection protocol at the Universidad de La Frontera, Temuco, Chile (see below). In addition, three human GC-derived cell lines (AGS, MKN28, MKN45) were included. One cell line (AGS) was selected for functional assays based on its known aggressiveness. AGS, MKN28 and MKN45 cells were grown in RPMI-1640 medium (Thermo Scientific Hyclone, USA) supplemented with 10 % fetal bovine serum (FBS), 10 units/ml penicillin and 10 mg/ml streptomycin (1 % P/S) (Thermo Scientific Hyclone, USA). All three cell lines were incubated at 37 °C in a humidified atmosphere containing 5 % CO2, and were sub-cultured during their logarithmic phase.
2.2 RNA purification and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the gastric tissues and GC-derived cell lines using a mirVana miRNA extraction kit (Ambion, USA) according to the manufacturer's protocol. The RNAs were quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, USA).
The expression of a set of pre-selected miRNAs was assessed in the 50 gastric tissue samples and the three GC-derived cell lines. The relative expression level of each selected miRNA was quantified using a TaqMan MicroRNA Assay (Life Technologies, USA) following the manufacturer's protocol. MiRNA expression data were acquired using the Stratagene Mx3000P qPCR System (Agilent Technologies, USA). The small RNA RNU6B was used as an internal control for input normalization. The cycle number at which the real-time PCR reaction reached a certain threshold (Ct) was recorded for both the miRNAs and RNU6B, and the relative miRNA expression was expressed as 2−ΔΔCt.
The mRNA expression levels of the PIK3CB, AKT1, TSC1, MTOR and TWFL1 genes were assessed by quantitative RT-PCR 48 h after miRNA mimic transfection (see below). Briefly, total RNA was reverse-transcribed with random primers using M-MLV reverse transcriptase (200 U/μl; Promega, USA). The resulting cDNA was subsequently amplified by PCR using a Brilliant II Ultra-Fast SYBR® Green qPCR Master Mix according to the manufacturer's recommendation using a Stratagene Mx-3000p Real-Time PCR System (Agilent Technologies, USA). Relative fold mRNA levels were determined using the 2−ΔΔCt method, with GAPDH as an internal control. Primer sequences (5′-3′) were: for PIK3CB, forward TTGTCTGTCACACTTCTGTAGTT and reverse AACAGTTCCCATTGGATTCAACA, for AKT1, forward TTCTGCAGCTATGCGCAATGTG and reverse TGGCCAGCATACCATAGTGAGGTT, for TSC1, forward GCAGCGTGACACTATGGTAACCAA and reverse AGTTCTATCCGCAGCTCCGCAAT, for MTOR, forward GCTTGATTTGGTTCCCAGGACAGT and reverse GTGCTGAGTTTGCTGTACCCATGT, for TWFL1, forward TGGGGATGAGTTGACTGCAGACTT and reverse TTCCGCTGGGCCCCTAATTAGT, and for GAPDH, forward TGCACCACCAACTGCTTAGC and reverse GGCATGGACTGTGGTCATGAG.
The primers were tested to determine their optimal concentrations for PCR analysis and the resulting products were run on 2 % agarose gels to confirm the appropriate size. The efficiency of the real-time PCR reaction was calculated using standard curves (not shown).
2.3 Transfections and controls
Transfection of AGS cells was performed using a Lipofectamine RNAiMAX Transfection Reagent (Life Technologies, USA) according to the manufacturer's instructions. Briefly, AGS cells were seeded at a concentration of 1× 105 in 6-well culture plates. After 24 h (70 % confluence), the cells were transfected with 50 nM hsa-miR-101-2 (5′UACAGUACUGUGAUAACUGAA′3), hsa-miR-125b-2 (5′UCCCUGAGACCCUAACUUGUGA′3) or hsa-miR-451a (5′AAACCGUUACCAUUACUGAGUUU′3) (mirVana™ mimics & controls; Ambion, USA). Non-specific pre-miR (mirVana™ miRNA Negative Control) was used as a negative control (scrambled) and miR-1 (mirVana™ miRNA Positive Control) and siRNA (BLOCK-iT™ Alexa Fluor® Red Fluorescent) were used as positive controls.
The transfection efficiency was checked by quantitative real-time PCR and visualization of positive controls by confocal microscopy (Olympus FV-1000 Inverted Microscope). The controls were used under the same experimental conditions as the miRNA mimics, thus allowing a transfection efficiency assessment during each assay.
2.4 Cell viability assay
Cell viability was evaluated using a tetrazolium-based MTS assay (CellTiter 96 AQueous One Solution Reagent; Promega, USA). Briefly, transfected AGS cells were seeded in 96-well plates at a density of 2500 cells per well. After 4, 24, 48, 72 and 96 h, 20 μl MTS reagent was added to each well and the cells were incubated for one additional hour. Mitochondrial dehydrogenases of viable cells reduce MTS to a blue formazan dye, the absorbance of which correlates to the number of viable cells. The absorbance was measured at 490 nm using an automated microplate reader (Bio-Rad, USA). Each test was performed in triplicate, from which mean values and standard deviations (SD) were calculated.
2.5 Colony formation assay
For the assessment of colony formation, cells were transfected with miRNA mimics or controls for 24 h, after which 100 cells were seeded in 6-well plates in triplicate. After 10 days of incubation, the plates were gently washed with PBS and stained with 2 % crystal violet in 25 % methanol/PBS for one hour at room temperature. Colonies with>50 cells were assessed using Image J software.
2.6 Cell migration and invasion assays
For the assessment of migration, cells were transfected with miRNA mimics or controls for 24 h. Then 2.5×104 cells in serum-free medium were placed into the upper chamber of an insert (8-μm pore size; BD Bioscience, USA). For the assessment of invasion, 5×105 transfected cells in serum-free medium were placed into the upper chamber of an insert coated with Matrigel (BD Bioscience, USA). For both experiments, media containing 10 % fetal bovine serum were added to the lower chamber. After 24 h of incubation, the cells remaining on the upper membrane were removed with cotton wool, whereas the cells that had migrated or invaded through the membrane were stained with 2 % crystal violet in 25 % methanol/PBS, imaged and counted using an EVOS XL Core inverted microscope (Life Technologies, USA). The experiments were independently repeated three times.
2.7 Cell death assay
For cell death assessment by flow cytometry, AGS cells were seeded in 6-well plates 24 h prior to transfection with mirVana™ mimics and controls (see above). After 48 h, dead cells were detected through double staining with Annexin V/7-Aminoactinomycin D (PE Annexin V Apoptosis Detection Kit I, BD Biosciences, USA). Briefly, cells were harvested in normal media and suspended in 1× binding buffer. The resulting cell suspension (100 μl) was incubated with Annexin V (3 μl) and 7-AAD (7 μl) at room temperature for 20 min in darkness and then analyzed by flow cytometry.
2.8 Western blot analysis
AGS cells were transfected with miRNA mimics and controls for 48 h. Subsequently, protein lysates were harvested using RIPA buffer (50 mM Tris, pH 7.2; 150 mM NaCl; 1 % Triton X-100; and 0.1 % SDS) containing protease (1:100, Roche, USA) and phosphatase (1:100, Sigma-Aldrich, USA) inhibitors. The protein concentrations were determined using a bicinchoninic acid assay (Pierce, Thermo Scientific, USA). Sixty μg of the proteins were separated by SDS-PAGE and transferred (Bio-Rad, USA) to PVDF membranes (Millipore, USA). Human PI3K-β, TSC1 and mTOR protein expression levels were quantified using rabbit polyclonal antibodies specific for each protein (1:1000, Cell Signaling, USA). The expression levels of these three proteins were standardized to human α-actin using a mouse polyclonal anti-α-actin antibody (1:5000, Millipore, USA). Primary antibodies were detected using goat anti-rabbit or goat anti-mouse horse-radish peroxidase (HRP)-conjugated secondary antibodies (1 : 5000, Santa Cruz Biotechnology, USA). Immunoreactive bands were visualized using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer, USA) according to the manufacturer's instructions, and then quantified by densitometry using a ChemiGenius Gel Bio Imaging System (Syngene, USA).
2.9 Statistical analyses
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc., USA). All experiments were repeated at least three times and performed in triplicate. Means were compared using the student's t-test or two-way ANOVA with the corresponding post-test. A p-value≤0.05 was considered statistically significant.
2.10 Ethics statement
The Institutional Review Board of the School of Medicine of Pontificia Universidad Católica approved this study and issued a waiver authorizing the use of archival material without informed consent for samples of more than two years old, thereby preserving the anonymity of the patients.
3 Results
3.1 miR-101-2, miR-125b-2 and miR-451a are down-regulated in primary GC tissues and GC-derived cell lines
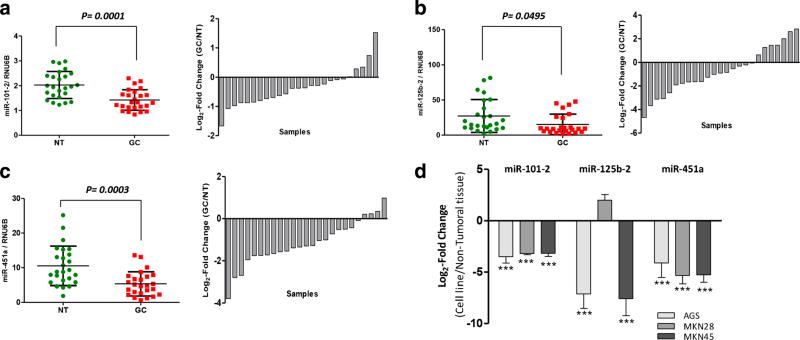
To validate alterations in miRNA expression levels as reported before [30, 34–36], qRT-PCR analyses for the miRNAs miR-101-2, miR-125b-2 and miR-451a were performed in a set of 25 primary gastric cancer (GC) tissues and 25 paired non-tumor gastric tissues from the same patients. Fold changes were normalized to the level of RNU6B expression, and calibrated against the relative expression in the non-tumor tissues from each patient. By doing so, we found that the levels of miR-101-2, miR-125b-2 and miR-451a were lower in the tumor tissues than in the non-tumor tissues (Fig. 1a, b and c). These observations are highly consistent with those reported before, including those in other types of cancer.
Fig. 1.
Expression of miR-101-2, miR-125b-2 and miR-451a in gastric cancer (GC) tissues and GC-derived cell lines. a Expression of miR-101-2 in GC tissues and a sample expression comparison. b Expression of miR-125b-2 in GC tissues and a sample expression comparison. c Expression of miR-451a in GC tissues and a sample expression comparison. The expression of each miRNA in the GC tissues was compared to that in the paired non-tumor tissues (NT). A Wilcoxon paired test was performed for statistical analyses. d Relative expression of miR-101-2, miR-125b-2 and miR-451a in the AGS, MKN28 and MKN45 GC-derived cell lines calibrated with the respective values obtained from a pool of 10 RNA samples from non-tumor tissues samples. The expression of the respective miRNAs in these samples and cell lines was assessed using TaqMan qPCR in triplicate and normalized against the endogenous control RNU6B. *p< 0.05, **p<0.01, ***p<0.001
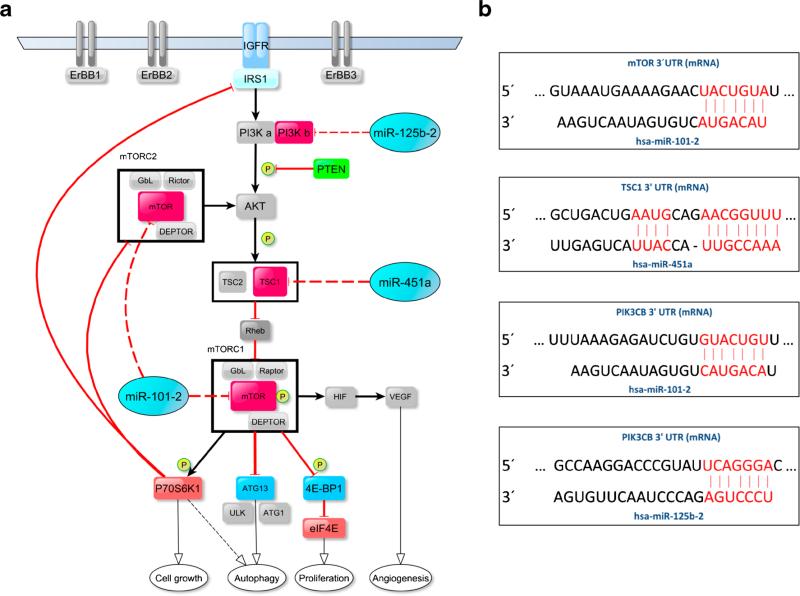
It has previously been suggested that down-regulation of miR-101-2, miR-125b-2 and miR-451a may be associated with a cancerous phenotype in diverse types of neoplasms [23–33]. Therefore, we set out to evaluate the possible functional implications of the down-regulation of these miRNAs in GC. We first examined their expression in three human GC-derived cell lines (AGS, MKN28, MKN45), followed by in vitro assays to determine their potential functional role(s). We found that the miR-101-2, miR-125b-2 and miR-451a expression levels were significantly decreased in the GC-derived cell lines (p<0.001) compared to a non-biological calibrator consisting of a pool of total RNAs from 10 non-tumor gastric tissues. The only exception was the expression of miR-125b-2 in MKN28 cells, where no differences were observed (Fig. 1d). In order to investigate the functional role of these three miRNAs in an in vitro model, an in silico analysis was performed to predict miRNA binding sites within the 3′UTR regions of mRNAs encoded by genes acting in the PI3K/AKT/mTOR pathway, and which are of potential import in GC such as PIK3CB, TSC1 and MTOR (Fig. 2a, b). For these analyses we initially used three algorithms (TargetScan, http://www.targetscan.org; PicTar, http://pictar.bio.nyu.edu; and miRanda, http://www.microrna.org). In order to confirm that these miRNAs effectively regulate the PI3K/AKT/mTOR pathway, a complementary bioinformatics analysis oriented to signaling pathways was performed using the Diana-miRpath-CDS tool. The results were significant for all cases (data not shown).
Fig. 2.
The PI3K/AKT/mTOR signaling pathway and a proposed model for its regulation through the evaluated miRNAs. a Diagram showing the targets of miR-101-2, miR-125b-2 and miR-451a within the PI3K/AKT/mTOR pathway. Target genes are in red and miRNAs are in blue. b Potential binding sites of miR-101-2 on the MTOR 3′-UTR sequence, potential binding site of miR-451a on the TSC1 3′-UTR sequence and potential binding sites of miR-101-2 and miR-125b-2 on the PIK3CB 3′-UTR sequence in human
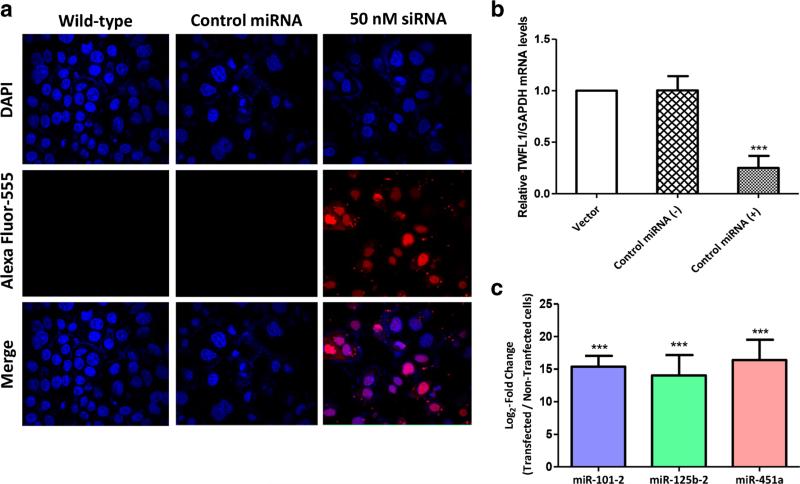
Next, transfections were standardized in AGS cells by measuring miRNA expression levels through qRT-PCR and by visualizing the positive control with BLOCK-iT™ Alexa Fluor® Red Fluorescent. In both cases high transfection efficiencies were observed (Fig. 3a). In addition, the activity of miR-1 (positive miRNA control) in transfected cells was confirmed through expression down-regulation of its target gene TWFL1 (Fig. 3b). The relative expression of the three miRNAs (miR-101-2, miR-125b-2 and miR-451a) was assessed in stably transfected AGS cells at time points up to 72 h post-challenge. As expected, an over 12-fold increase in the expression of all three miRNAs was observed in mimic-transfected cells compared to non-transfected cells (Fig. 3c).
Fig. 3.
Transfection efficiency in AGS cells. a Transfection of a positive control miRNA using ™BLOCK-iTAlexa Fluor Red Fluorescent Oligo® and Lipofectamine RNAiMax. Wild-type AGS cells and negative control miRNA-transfected AGS cells were included as negative controls. Red fluorescence indicates the presence of the control oligonucleotide inside the cells (siRNA, 50 nM). b Relative expression of TWFL1 in AGS cells transfected with a positive control miRNA (miR-1) and controls (non-transfected cells and negative control miRNA). Gene expression analysis revealed a significant reduction in the TWFL1 mRNA level (***p<0.001) in cells expressing miR-1. c Levels of transfected mimics (miR-101-2, miR-125b-2 and miR-451a) 24 h after transfection. Transfected cells were normalized to RNU6B miRNA and calibrated against non-transfected AGS cells
3.2 miR-101-2, miR-125b-2 and miR-451a inhibit cell proliferation and colony formation, increase cell death, and decrease cell migration and invasion in AGS cells
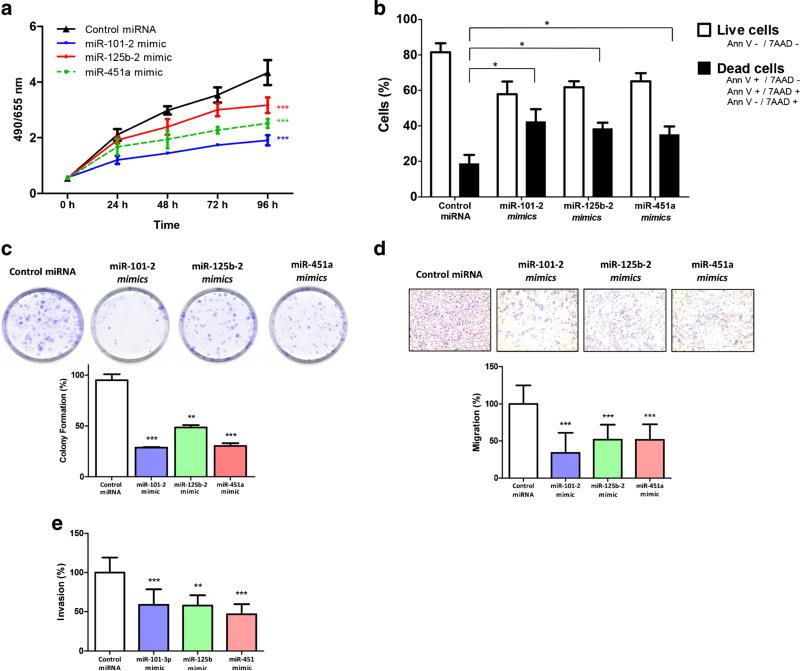
Through MTS and colony forming assays, we found that miR-101-2, miR-125b-2 and miR-451a dramatically reduced AGS cell viability compared to the negative control miRNA (Fig. 4a, p<0.001). In addition, we found that the number of colonies was significantly reduced in cells transfected with miR-101-2 (69.5 %, p<0.001), miR-125b-2 (48.9 %, p<0.05) and miR-451a (68.1 %, p<0.05) mimics (Fig. 4c), compared to cells transfected with control miRNA.
Fig. 4.
Ectopic expression of miR-101-2, miR-125b-2 and miR-451a in AGS cells affects cell viability, colony formation, apoptosis, migration, and invasion in vitro. a AGS cells expressing miR-101-2, miR-125b-2 and miR-451a show a decreased viability compared to cells expressing the negative miRNA control (scramble). b Over-expression of miR-101-2, miR-125b-2 and miR-451a significantly promotes cell death in AGS cells. c The percentage of colonies show a significantly decrease in cells expressing miR-101-2, miR-125b-2 and miR-451a (representative image of colony formation). d miR-101-2, miR-125b-2 and miR-451a transfected AGS cells show both a decrease in migration and e a decrease in invasion in (Matrigel coated) transwell assays. For each assay values were compared to control miRNA using a two-way ANOVA test. The error bars indicate the standard deviation. *p<0.05, **p<0.01, ***p<0.001
Cell death was assayed by flow cytometry in miRNA mimic-transfected AGS cells. We found that exogenous over-expression of miR-101-2, miR-125b-2 and miR-451a significantly increased cell death compared to control miRNA-transfected cells (Fig. 4b). The average percentage of dead cells was 42.2 % for miR-101-2 (p<0.05), 38.2 % for miR-125b-2 (p<0.05), 35 % for miR-451a (p<0.05) and 18.3 % in cells transfected with control miRNA. In summary, these data confirm that miR-101-2, miR-125b-2 and miR-451a play a key role in increasing cell death, and decreasing cell viability and colony formation in AGS cells.
Chamber-based assays were used to determine whether miR-101-2, miR-125b-2 and miR-451a are involved in GC cell migration and/or invasion. For these assays we used AGS cells transfected with the respective mimics. By doing so, we found that miR-101-2, miR-125b-2, and miR-451a inhibited both cell migration and invasion in AGS cells compared to controls (Fig. 4d, e).
3.3 miR-101-2, miR-125b-2 and miR-451a expression is inversely correlated with MTOR, PIK3CB and TSC1 expression
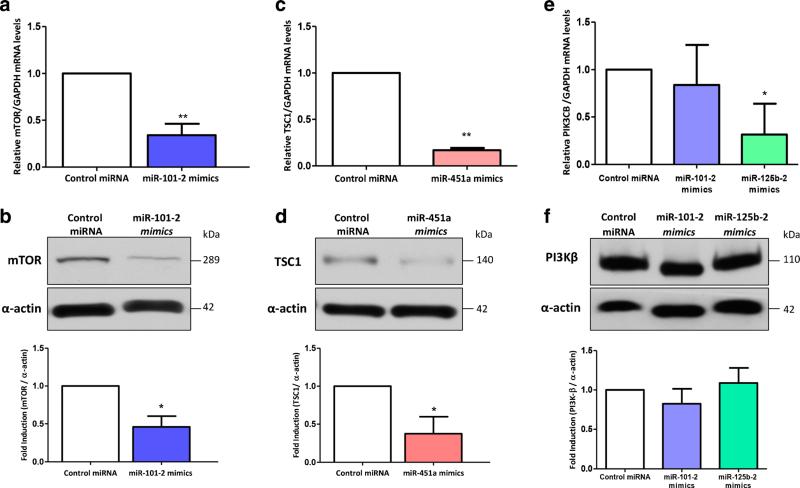
Using the bioinformatics-based predictions (see above), we set out to examine the effect of miR-101-2, miR-125b-2 and miR-451a on the mRNA and protein expression levels of targets from the PI3K/AKT/mTOR pathway (i.e., MTOR, PIK3CB and TSC1, respectively). The experiments were conducted in AGS cells previously transfected with the respective miRNA mimics or scrambled control miRNA. We found that the mRNA and protein expression levels of MTOR were dramatically decreased in miR-101-2-transfected cells compared to control transfected cells (p<0.01; p<0.05, respectively) (Fig. 5a, b). Additionally, our results showed that the mRNA and protein expression levels of the TSC1 gene were significantly decreased in cells transfected with a miR-451a mimic compared to scrambled control-transfected cells (p<0.01; p<0.05, respectively) (Fig. 5c, d). Finally, we found that AGS cells transfected with a miR-125b-2 mimic showed a knockdown of PIK3CB mRNA expression compared to that of control cells (p<0.05). However, at the protein level (PI3Kβ, p110) the expression was not affected. The miR-101-2 mimic had no effect on either PIK3CB mRNA or protein expression levels (Fig. 5e, f), even when this mimic was co-transfected with miR-125b mimics-2 (data not shown).
Fig. 5.
Expression of miR-101-2, miR-125b-2 and miR-451a are inversely correlated with the expression of MTOR, PIK3CB and TSC1. a miR-101-2 down-regulates MTOR mRNA expression in AGS cells. b Western blot and densitometry results showing mTOR protein down-regulation in AGS cells transfected with miR-101-2. c miR-451a down-regulates TSC1 mRNA expression in AGS cells. d Western blot and densitometry results showing TSC1 protein down-regulation in AGS cells transfected with miR-451a. e The mRNA expression level of PIK3CB mRNA is down-regulated after miR-125b-2 mimic, but not after miR-101-2 mimic, transfection. f Western blot and densitometry results of PI3Kβ (p110β) protein in AGS cells transfected with either miR-101-2 or miR-125b-2, or control miRNA. The gene and protein analyses were performed 48 h post transfection. Baseline mRNA and protein expression levels of control miRNA transfected cells were used to calibrate the qRT-PCR and densitometry values, respectively. GAPDH was used to normalize mRNA expression values and α-actin was used as a loading control in the western blots. Each assay was performed in triplicate and for comparison of the values student's t-test was used. The error bars indicate the standard deviation. *p<0.05, **p<0.01, ***p<0.001
4 Discussion
MiRNAs are known to regulate many key biological processes through post-transcriptional regulation of the expression of genes involved in both normal (developmental/homeostatic) and disease-related processes. Regulatory cellular processes such as inflammation, cell cycle progression, stress response, differentiation, apoptosis and invasion may directly affect tumor development. As a result, miRNAs have been afforded great potential as biomarkers and putative therapeutic targets. It has e.g., been found that they can restore normal gene expression levels and, by doing so, reduce the growth of tumor cells or even completely eliminate tumors in animal model systems.
Since the PI3K/AKT/mTOR pathway has previously been reported to be up-regulated in gastric cancer (GC) [7, 8], we focused our study on miR-101-2, miR-125b-2 and miR-451a, which are according to TargetScan, PicTar and miRanda related to MTOR, PIK3CB and TSC1 expression, respectively. miR-101-2 is encoded by a gene located in 9p21, a chromosomal region exhibiting a high frequency of allelic loss in several cancers including liver, colon, pancreas and breast cancer, and in neuroblastoma [23–26]. MiR-125b-2 is encoded by a gene located on chromosome 21 and is commonly down-regulated in breast and liver cancer, and in osteosarcoma [27–29]. MiR-451a is encoded by a gene located on chromosome 17, and is frequently down-regulated in glioma and colon cancer cells [30–32]. In addition, miR-451a has been proposed as a potential prognostic biomarker for relapse in GC [33]. Here, the expression levels of these three human miRNAs (i.e., miR-101-2, miR-125b-2 and miR-451a) were assessed in 25 advanced GC tissue samples and their 25 matched non-tumor tissues. We found that the levels of all three miRNAs were lower in the GC tissues compared to the non-tumor tissues from the same patients. Subsequently, the expression of these miRNAs was assessed in three GC-derived cell lines (AGS, MKN45 and MKN28). Our results confirmed a significant decrease in the expression of these three miRNAs in the GC-derived cell lines compared to a pool of non-tumor gastric tissues, the only exception being miR-125b-2 in MKN28 cells. These results are consistent with previous studies that reported a decrease in the expression of these miRNAs in other human tumors, and other GC tissues and GC-derived cell lines not included in this study [30, 34–36].
Additionally, we examined the effect of exogenous miR-101-2, miR-125b-2 and miR-451a expression on several in vitro growth parameters and the expression of potential targets (MTOR, PIK3CB and TSC1) in AGS, a cell line known for its high proliferation rate. We found that exogenous miR-101-2 expression inhibited AGS cell proliferation by decreasing its viability and inhibiting its colony forming capacity more efficiently than the other miRNA mimics. We also found that AGS cell death increased significantly after transfection with miR-101-2 mimics. The decreased cell viability could be related to the silencing of MTOR mRNA and protein expression. When mTOR is down-regulated its downstream effectors, such as P70S6K1 and 4E-BP1, which activate proliferation through the release of the transcription factor proteins RpS6 and eIF4E, respectively, remain inactivated [37]. The increased cell death can, at least partly, be explained by a block in mTOR activity, which may induce 4E-BP1 de-phosphorylation, leading to ASK-1 up-regulation and, consequently, phosphorylation of c-Jun and cell death [38]. On the other hand, we found that exogenous miR-101-2 expression decreased cell migration and invasion, suggesting that the miR-101-2 effect on MTOR mRNA may be important for interrupting AGS cell motility and invasion. The effect on cell migration is attributable to the subsequent inhibition of both the mTORC1/P70S6K and mTORC1/4E-BP1 interactions [39]. P70S6K interacts with thrombin [40] and epidermal growth factor (EGF), and affects actin arc formation in the cytoskeleton, a functional indicator of cell migration [41]. Additionally, phosphorylated P70S6K can trigger fibronectin-induced migration [42] or can act on metalloproteinase-9 (MMP-9), an important endopeptidase involved in extracellular matrix degradation, which is required for tumor invasion and metastasis [43]. The inhibitory effect on mTOR may also decrease HIF-1a and FAK activity, thereby provoking inhibition of cancer cell migration and invasion as has been observed in rapamycin-treated GC cells in vitro [44]. Furthermore, downstream regulators of mTOR complex 2 (mTORC2), such as AKT, GTPases, PKCα and SGK1, have also been implicated in migration and invasion [39]. These data confirm the crucial role of miR-101-2 in disrupting mTOR signaling and attenuation of the cancerous phenotype.
The effect of exogenous miR-125b-2 expression on decreased AGS cell viability and increased AGS cell death may be explained by its binding to PIK3CB mRNA, which encodes PI3Kβ, an important member in the PI3K/AKT pathway. Alterations in PIK3CB expression are responsible for PI3Kβ activation, which may lead to an increase in cellular proliferation and survival [45]. Therefore, an up-regulation of miR-125b-2 may induce lower growth and division rates in cancer cells. The non-significant decrease in PI3Kβ (p110β) protein level 24 h after transfection with miR-125b-2 mimics may be due to a slow degradation rate of this protein within AGS cells. This minor change, however, seemed to be enough to induce a modification of the phenotype. On the other hand, miR-125b-2 over-expression may also significantly inhibit AGS cell migration and invasion, as has previously been reported in other cancers [27, 46]. In this case, PI3Kβ may be directly involved in cell migration and invasion through interactions with integrins and clathrin, which play key roles in governing cell motility [47–49]. Moreover, PI3Kβ blockage could lead to a lower AKT1/AKT2 activity and a decreased function of its downstream modulators PKB, NF-κB, IGF-1R, and others. These events might then trigger a reduced activity of matrix metalloproteinase-2 (MMP-2) and MMP-9 in human cells to decrease invasion [50, 51]. According to our results, miR-451a plays an important role in cell viability, cell death, and cell migration and invasion. Previously, the effect of miR-451a has been related to a direct or indirect reduction of cell proliferation and an alteration in cell cycle progression in cancer cells by targeting MIF, cyclin D1 and p27 expression [30, 31, 52]. Furthermore, miR-451a may induce apoptosis by inhibiting BCL-2, an anti-apoptotic protein involved in the intrinsic apoptotic pathway [31]. Other studies performed in glioma cells have linked the effect of miR-451a to LKB1 complex formation, indicating that mTOR complex 1 (mTORC1) and its downstream targets are activated in these cells [32]. Here, we suggest that miR-451a affects mTOR pathway activation by regulating the mRNA and protein expression levels of TSC1. The TSC1/TSC2 complex acts as an inhibitor of mTORC1, but the exact role of these proteins, especially TSC1, remains unclear. Because of this negative regulation, we initially expected to observe an increased cell proliferation and clone formation after TSC1 inhibition by miR-451a mimics. Our results, however, revealed the contrary, suggesting that another regulatory system is operational within the AKT/TSC1/mTOR axis. Our observations could be due the action of overlapping mechanisms similar to those outlined above (MIF, p27, cyclin D1 or the interaction between LKB1 and mTORC1) or, alternatively, to a compensating interaction between other modulators of the mTOR pathway that could induce activation of mTORC1, despite lower levels of TSC1. Moreover, studies on glioblastoma have shown that miR-451a can reduce cell migration and proliferation via MMP-2 and MMP-9, which are regulators of these characteristics [31, 45]. Here, we found a decrease in cell migration and invasion after transfection with miR-451a mimics compared to control miRNA. This result can likely be explained through a complementary regulating system in the AKT/TSC1/mTOR pathway, which resembles that previously described. However, more work is needed to fully uncover the complex relationship(s) between TSC1, cell migration and invasion.
MiRNAs have emerged as important molecules with promising prospects as biomarkers and as therapeutic targets. Accumulating evidence indicates that miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors involved in a wide range of cellular processes that can be deregulated in several neoplasms, including GC. Loss of function of these miRNAs may result from several mechanisms. Clearly, more work is needed to fully unravel these mechanisms, for example whether the respective miRNA-mRNA interactions are direct or indirect, and to understand the effects of alterations in downstream signaling. MiRNAs are generally located in cancer-associated genomic regions (CAGRs), i.e., regions prone to deletion, mutation, epigenetic silencing and/or miRNA processing alteration [53]. Therefore, exogenous expression of the aforementioned miRNAs could serve as a therapeutic strategy for the treatment of this complex and refractory disease. To this end, validation by in vivo assays is a prerequisite. Moreover, its efficacy and safety should be carefully evaluated, since the ultimate response may depend on the genetic and epigenetic make-up specific to each individual.
Acknowledgments
This study was supported by the Chilean National Fund for Scientific and Technological Development (FONDECYT NO. 1090171), the Chilean National Commission for Scientific and Technological Research (CONICYT) through a PhD scholarship and financial support for a doctoral thesis (NO. 24121456) and a Postdoctoral Scholarship from the Universidad de La Frontera.
The FV-1000 microscope experiments/data analyses were performed in part through use of the VUMC Cell Imagining Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126).
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Yousuf A, Bhat MY, Pandith AA, Afroze D, Khan NP, Alam K, Shah P, Shah MA, Mudassar S, Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D, Gene MGMT. Silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell. Oncol. 2014;37:245–52. doi: 10.1007/s13402-014-0179-3. [DOI] [PubMed] [Google Scholar]
- 3.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, Van Cutsem E. Gastric cancer. Crit. Rev. Oncol. Hematol. 2009;71:127–64. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Hartgrink HH, Jansen EPM, van Grieken NCT, van de Velde CJH. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labots M, Buffart TE, Haan JC, van Grieken NCT, Tijssen M, van de Velde CJH, Grabsch HI, Ylstra B, Carvalho B, Fijneman RJA, Verheul HMW, Meijer GA. High-level copy number gains of established and potential drug target genes in gastric cancer as a lead for treatment development and selection. Cell. Oncol. 2014;37:41–52. doi: 10.1007/s13402-013-0162-4. [DOI] [PubMed] [Google Scholar]
- 6.Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann. Surg. 2005;241:27–39. doi: 10.1097/01.sla.0000149300.28588.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Batran S-E, Ducreux M, Ohtsu A. mTOR as a therapeutic target in patients with gastric cancer. Int. J. Cancer. 2012;130:491–6. doi: 10.1002/ijc.26396. [DOI] [PubMed] [Google Scholar]
- 8.Tapia O, Riquelme I, Leal P, Sandoval A, Aedo S, Weber H, Letelier P, Bellolio E, Villaseca M, Garcia P, Roa JC. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014;465:25–33. doi: 10.1007/s00428-014-1588-4. [DOI] [PubMed] [Google Scholar]
- 9.The Cancer Genome Atlas Research Network Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H. A comprehensive map of the mTOR signaling network. Mol. Syst. Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, Wang T, Zhao G. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell. Oncol. 2013;36:385–94. doi: 10.1007/s13402-013-0144-6. [DOI] [PubMed] [Google Scholar]
- 14.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Nagadia R, Pandit P, Coman WB, Cooper-White J, Punyadeera C. miRNAs in head and neck cancer revisited. Cell. Oncol. 2013;36:1–7. doi: 10.1007/s13402-012-0122-4. [DOI] [PubMed] [Google Scholar]
- 17.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Cho WCS. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M. miRNAs as molecular biomarkers of cancer. Expert Rev. Mol. Diagn. 2010;10:435–44. doi: 10.1586/erm.10.27. [DOI] [PubMed] [Google Scholar]
- 22.Chan E, Prado DE, Weidhaas JB. Cancer microRNAs: from subtype profiling to predictors of response to therapy. Trends Mol. Med. 2011;17:235–43. doi: 10.1016/j.molmed.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G, Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res. 2009;315:1439–47. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Batchu RB, Gruzdyn O, Qazi AM, Bouwman D, Gruber SA, Weaver DW. MicroRNA-101 (miR-101) enhances chemosensitivity of pancreatic ductal adenocarcinoma (PDAC) cells by inhibition of MTOR signaling Via PRAS40. J. Surg. Res. 2012;172:233. [Google Scholar]
- 25.Buechner J, Tømte E, Haug BH, Henriksen JR, Løkke C, Flægstad T, Einvik C. Tumour-suppressor microRNAs Let-7 and Mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang C-W, Huang Y, Leong K-W, Chen L-C, Chen H-C, Chen S-J, Chou C-K. PKCalpha mediated induction of miR-101 in human hepatoma HepG2 cells. J. Biomed. Sci. 2010;17:35. doi: 10.1186/1423-0127-17-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang L, Wong C-M, Ying Q, Fan DN-Y, Huang S, Ding J, Yao J, Yan M, Li J, Yao M, Ng IO-L, He X. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–40. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Li H, Li J, Zhong H, Zhang H, Chen J, Xiao T. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem. Biophys. Res. Commun. 2011;416:31–8. doi: 10.1016/j.bbrc.2011.10.117. [DOI] [PubMed] [Google Scholar]
- 29.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J. Biol. Chem. 2007;282:1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 30.Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009;15:2281–90. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- 31.Nan Y, Han L, Zhang A, Wang G, Jia Z, Yang Y, Yue X, Pu P, Zhong Y, Kang C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010;1359:14–21. doi: 10.1016/j.brainres.2010.08.074. [DOI] [PubMed] [Google Scholar]
- 32.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA, Lawler SE. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell. 2010;37:620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H, Guo J-M, Lou Y-R, Zhang X-J, Zhong F-D, Jiang Z, Cheng J, Xiao B-X. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J. Mol. Med. (Berl.) 2010;88:709–17. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang H-J, Ruan H-J, He X-J, Ma Y-Y, Jiang X-T, Xia Y-J, Ye Z-Y, Tao H-Q. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur. J. Cancer. 2010;46:2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z-X, Lu C-Y, Yang Y-L, Dou K-F, Tao K-S. MicroRNA-125b expression in gastric adenocarcinoma and its effect on the proliferation of gastric cancer cells. Mol. Med. Rep. 2013;7:229–232. doi: 10.3892/mmr.2012.1156. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Chen L, Cao X, Ha S, Xia X. Expression profiling and functional analysis of hsa-miR-125b and its target genes in drug-resistant cell line of human gastric cancer. Hereditas. 2014;36:119–128. doi: 10.3724/sp.j.1005.2014.00119. [DOI] [PubMed] [Google Scholar]
- 37.Clohessy JG, Reschke M, Pandolfi PP. Found in translation of mTOR signaling. Cell Res. 2012;22:1315–8. doi: 10.1038/cr.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–59. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Huang S. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr. Protein Pept. Sci. 2011;12:30–42. doi: 10.2174/138920311795659407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crouch MF. Regulation of thrombin-induced stress fibre formation in swiss 3T3 cells by the 70-kDa S6 kinase. Biochem. Biophys. Res. Commun. 1997;233:193–9. doi: 10.1006/bbrc.1997.6419. [DOI] [PubMed] [Google Scholar]
- 41.Berven LA, Willard FS, Crouch MF. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell Res. 2004;296:183–95. doi: 10.1016/j.yexcr.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 42.Sakakibara K, Liu B, Hollenbeck S, Kent KC. Rapamycin inhibits fibronectin-induced migration of the human arterial smooth muscle line (E47) through the mammalian target of rapamycin. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2861–H2868. doi: 10.1152/ajpheart.00561.2004. [DOI] [PubMed] [Google Scholar]
- 43.Zhou HY, Wong AST. Activation of p70S6K induces expression of matrix metalloproteinase 9 associated with hepatocyte growth factor-mediated invasion in human ovarian cancer cells. Endocrinology. 2006;147:2557–66. doi: 10.1210/en.2005-1404. [DOI] [PubMed] [Google Scholar]
- 44.Lang SA, Gaumann A, Koehl GE, Seidel U, Bataille F, Klein D, Ellis LM, Bolder U, Hofstaedter F, Schlitt H-J, Geissler EK, Stoeltzing O. Mammalian target of rapamycin is activated in human gastric cancer and serves as a target for therapy in an experimental model. Int. J. Cancer. 2007;120:1803–10. doi: 10.1002/ijc.22442. [DOI] [PubMed] [Google Scholar]
- 45.Akinleye A, Avvaru P, Furqan M, Song Y, Liu D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J. Hematol. Oncol. 2013;6:88. doi: 10.1186/1756-8722-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P, Pan Z, Hu X, Zhao Y, Xie H, Jiang G, Chen T, Wang J, Zheng S, Cheng J, Wan D, Yang S, Li Y, Gu J. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int. J. Cancer. 2008;123:1616–22. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 47.Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J. Cell Biol. 2009;187:733–47. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dbouk HA, Backer JM. A beta version of life: p110β takes center stage. Oncotarget. 2010;1:729–33. doi: 10.18632/oncotarget.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, Kim S, Koh H, Yoon SO, Chung AS, Cho KS, Chung J. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. FASEB J. 2001;15:1953–62. doi: 10.1096/fj.01-0198com. [DOI] [PubMed] [Google Scholar]
- 51.Chin YR, Toker A. Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell. Signal. 2009;21:470–6. doi: 10.1016/j.cellsig.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner B, Hoshen MB, Purim O, Ben David M, Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern M, Rosenfeld N, Chajut A, Niv Y, Kushnir M. MicroRNAs as a potential prognostic factor in gastric cancer. World J. Gastroenterol. 2011;17:3976–85. doi: 10.3748/wjg.v17.i35.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jansson MD, Lund AH. MicroRNA and cancer. Mol. Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]