The short receptor Et/Lat negatively regulates Drosophila JAK/STAT signaling. It binds to intracellular components and the Domeless receptor but cannot bind ligands, thus generating a signaling-incompetent complex. Et/Lat is also more stable than Dome. The study provides insights into how short receptors negatively regulate signaling.
Abstract
Transmembrane receptors interact with extracellular ligands to transduce intracellular signaling cascades, modulate target gene expression, and regulate processes such as proliferation, apoptosis, differentiation, and homeostasis. As a consequence, aberrant signaling events often underlie human disease. Whereas the vertebrate JAK/STAT signaling cascade is transduced via multiple receptor combinations, the Drosophila pathway has only one full-length signaling receptor, Domeless (Dome), and a single negatively acting receptor, Eye Transformer/Latran (Et/Lat). Here we investigate the molecular mechanisms underlying Et/Lat activity. We demonstrate that Et/Lat negatively regulates the JAK/STAT pathway activity and can bind to Dome, thus reducing Dome:Dome homodimerization by creating signaling-incompetent Dome:Et/Lat heterodimers. Surprisingly, we find that Et/Lat is able to bind to both JAK and STAT92E but, despite the presence of putative cytokine-binding motifs, does not detectably interact with pathway ligands. We find that Et/Lat is trafficked through the endocytic machinery for lysosomal degradation but at a much slower rate than Dome, a difference that may enhance its ability to sequester Dome into signaling-incompetent complexes. Our data offer new insights into the molecular mechanism and regulation of Et/Lat in Drosophila that may inform our understanding of how short receptors function in other organisms.
INTRODUCTION
The development of multicellular organisms requires individual cells to coordinate their behavior. Communication events are frequently initiated by secreted ligands, which bind to transmembrane receptors, triggering intracellular signaling cascades. In many cases, these signaling pathways alter target gene expression and modulate cellular behavior. The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway represents one such communication system. The pathway has been evolutionarily conserved and is essential for many developmental processes, including cellular proliferation, innate immune responses, and stem cell maintenance (Arbouzova and Zeidler, 2006; Bausek, 2013; Morin-Poulard et al., 2013; Amoyel et al., 2014). Furthermore, dysregulation of intercellular signaling is frequently associated with human diseases, including immune disorders and cancers (Teng et al., 2014; Thomas et al., 2015; Villarino et al., 2015). Given the importance of appropriate pathway activation, multiple regulatory mechanisms act at many levels to tightly control both the strength and duration of activity (Arbouzova and Zeidler, 2006; Stec et al., 2013).
In vertebrates, the JAK/STAT pathway transduces signaling of multiple transmembrane receptors, which can bind extracellular cytokines, including interleukins (ILs), growth factors, and interferons. Of these, the IL-6 family of vertebrate cytokine receptors, most similar to Drosophila Dome, can be broadly split into two categories: long β-receptors, such as GP130, LIFRβ, and OSMRβ, and shorter α-receptors, such as IL-6Rα, IL-11Rα, and IL-13Rα2 (Rahaman et al., 2002; reviewed in Taga and Kishimoto, 1997; Heinrich et al., 2003). Associating with specific domains within the cytosolic tail of receptor complexes (Kaplan et al., 1996; Taga and Kishimoto, 1997), four JAKs (JAK1–3 and TYK2) play a central role in the transduction of signaling via their tyrosine kinase activity. Phosphorylating themselves and their receptors in response to ligand binding, the JAKs generate sites to which cytosolic STATs are able to bind (Stahl et al., 1995; Klingmüller et al., 1996; May et al., 1996). Once bound to the receptor complex, the STATs are tyrosine phosphorylated before translocating into the nucleus, where they bind DNA and modulate gene expression.
The Drosophila JAK/STAT cascade represents a highly conserved yet lower-complexity system in which to study JAK/STAT signaling. In the fly, only three ligands—Upd, Upd2, and Upd3 (Harrison et al., 1998; Hombria et al., 2005; Wright et al., 2011)—one signaling receptor (Dome; Brown et al., 2001), one JAK (Hopscotch [Hop]; Binari and Perrimon, 1994), and one STAT (STAT92E; Hou et al., 1996; Yan et al., 1996)—make up the complete core signaling cascade. Regulatory mechanisms mediated by factors such as SOCS36E (Stec et al., 2013) and the tyrosine phosphatase PTP61F (Müller et al., 2005) have also been conserved.
Originally identified on the basis of cytokine-binding motifs similar to those present in Dome (Hombria and Brown, 2002), the short receptor Eye Transformer (Et)/Latran (Lat) is located adjacent to the long Dome receptor in the genome and is likely to be the product of an evolutionarily recent duplication event. With a primary structure similar to that of vertebrate α-receptors (Hombria and Brown, 2002), Et/Lat was also identified in two studies that named the protein Eye Transformer (Kallio et al., 2010) and Latran (Makki et al., 2010), respectively, subsequently referred to in this study as Et/Lat. One study focused on the in vivo mutant phenotype of Et/Lat, which demonstrated a requirement in innate immune responses. On parasitic wasp infestation, Et/Lat is up-regulated, whereas Dome expression is down-regulated. This increase in the Et/Lat:Dome ratio reduces pathway activity and thus allows hemocyte precursors within the lymph gland to differentiate into lamellocytes, which in turn attempt to smother the wasp egg (Makki et al., 2010). In the second study, Et/Lat was identified in a cell-based RNA interference (RNAi) screen as a negative regulator of JAK/STAT pathway reporter activity. It was shown to bind to Hop and Dome by immunoprecipitation, and this report suggested that Et/Lat does not affect Dome:Hop or Dome:Dome dimer formation (Kallio et al., 2010).
Here we examine the mechanism of Et/Lat negative regulation. We find that Et/Lat is able to bind to Dome and block the formation of Dome homodimers. We further show that Et/Lat can bind to both JAK and STAT proteins to form a complex that is unable to bind to the pathway ligands and therefore unable to trigger the phosphorylation events necessary for signaling. We find that whereas Et/Lat can be degraded via the lysosome, the protein lasts over a much longer time frame than Dome. Therefore we suggest that Et/Lat is able to sequester JAK/STAT signaling components in an inactive but stable complex.
RESULTS AND DISCUSSION
Et/Lat disrupts formation of Dome:Dome dimers
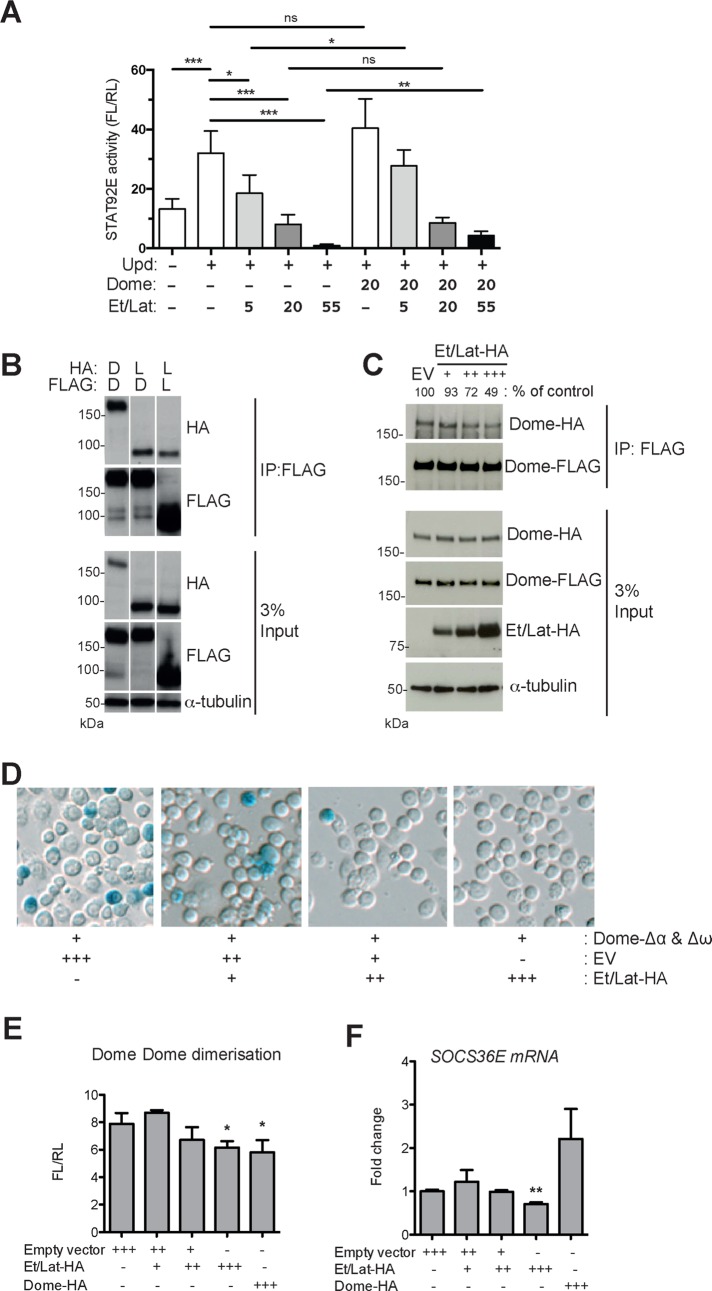
Although it was previously shown that Et/Lat negatively regulates JAK/STAT signaling and binds to the Dome receptor complex, the molecular mechanism underlying this activity is not understood. We first set out to confirm that Et/Lat is indeed a negative regulator of JAK/STAT signaling, using an established STAT92E-responsive luciferase reporter in Drosophila Kc167 cells (Müller et al., 2005). Although increasing concentrations of cotransfected pathway ligand resulted in higher STAT92E activity, coexpression of additional Dome had no effect on signaling. However, expression of Et/Lat-HA in these cells dramatically reduced signaling output in a dose-dependent manner (Figure 1A). Furthermore, expression of additional Dome was sufficient to at least partially relieve the Et/Lat-induced suppression (Figure 1A), findings that are similar to those of Makki et al. (2010). Consistent with a previous report (Kallio et al., 2010), we found that Et/Lat physically associates with Dome, as shown by coimmunoprecipitation of tagged receptors expressed in cells (Figure 1B). Furthermore, we also found that Et/Lat can itself form homodimers (Figure 1B), although the relative affinity of these associations and their potential functional relevance is not clear.
FIGURE 1:
Et/Lat can disrupt formation of Dome homodimers. (A) STAT92E transcriptional reporter activity assay. Pathway activity increases after the cotransfection of Upd ligand. Cotransfection of increasing quantities of Et/Lat blocks signaling capability in a dose-dependent manner. Expression of additional Dome does not significantly alter signaling (column 6) in the absence of Et/Lat but is sufficient to significantly increase signaling inhibited by Et/Lat coexpression (compare column 3 to column 7 and column 5 to column 9). Numbers indicate relative levels of Dome- and Et/Lat-expressing plasmids transfected. (B) Coimmunoprecipitation assays using FLAG-tagged Dome (D) or Et/Lat (L) to pull down HA-tagged Dome or Et/Lat indicate that both Dome and Et/Lat can form both homodimers and heterodimers. Constructs transfected are indicated. α-Tubulin was used as a loading control. (C) Coimmunoprecipitation assays using Dome-FLAG to pull down Dome-HA from cells. Quantification of this blot is shown as percentage of control levels. Increasing the level of Et/Lat cotransfected into cells reduces the amount of Dome-HA that can be coimmunoprecipitated. EV, empty vector. (D) Cells transfected with the Dome-Δα and Dome-Δω dimerization assay and stained with X-gal (blue) to visualize bimolecular complementation. Owing to transient transfection, not every cell expresses both constructs. Increasing amounts of Et/Lat retransfected into previously transfected cells progressively block Dome homodimerization. (E) Dome homodimerization as described in D but quantified using β-Glo reagent to measure β-galactosidase activity levels. (F) Quantitative PCR of the JAK/STAT pathway target gene socs36E. mRNA levels are reduced after increased expression of Et/Lat but not after increased Dome. The analysis used unpaired t tests with Welch’s correction. *p < 0.05, **p < 0.01, and ***p < 0.001. ns, not significant.
To better understand the nature of receptor interactions, we tested the effect of Et/Lat on Dome:Dome homodimers. Using coimmunoprecipitation as an assay, we found that increasing levels of Et/Lat can reduce the Dome:Dome interaction (Figure 1C). To confirm the immunoprecipitation results, we also developed a cell-based molecular complementation assay, termed βlue-βlau, previously used to report Dome dimerization in vivo (Brown et al., 2003). In this assay, the dome-coding region is fused to two LacZ mutants termed Δα and Δω. These mutants each produce truncated, enzymatically inactive β-galactosidase (β-gal) proteins (Supplemental Figure S1). However, when brought into close enough proximity (e.g., via interaction of the proteins to which they are fused), they are able to complement one another and thus reconstitute a functional β-gal enzyme (Rossi et al., 1997). We first expressed Dome-Δα and Dome-Δω in Kc167 cells before retransfecting them with increasing concentrations of Et/Lat-hemagglutinin (HA). Visualization of β-gal activity showed that higher levels of Et/Lat were able to reduce β-gal activity and complementation (Figure 1D). This suggests that either Dome:Dome dimer formation is reduced or dimer breakdown is enhanced by Et/Lat, with the approaches used here not able to differentiate between these mechanisms. To quantify this more accurately, we went on to use a substrate that is cleaved by β-gal to release luciferin. This luciferin acts as a substrate in a firefly luciferase reaction, giving a quantifiable measurement of β-gal activity (Hannah et al., 2003). Despite higher levels of background associated with this assay, we found that the signal from Dome-Δα:Dome-Δω dimers was significantly reduced at the highest doses of both Et/Lat-HA and Dome-HA, with the latter as a positive control (Figure 1E).
Consistent with the reduction in Dome:Dome dimers observed, high doses of Et/Lat-HA were also sufficient to reduce significantly the expression of the STAT92E transcriptional target socs36E (Bina et al., 2010; Figure 1F), whereas high Dome-HA levels were not. This indicates that whereas Et/Lat and Dome can both interact with a signaling-competent receptor complex, only Et/Lat has the ability to block downstream signaling.
Et/Lat can bind to intracellular JAK/STAT signaling components
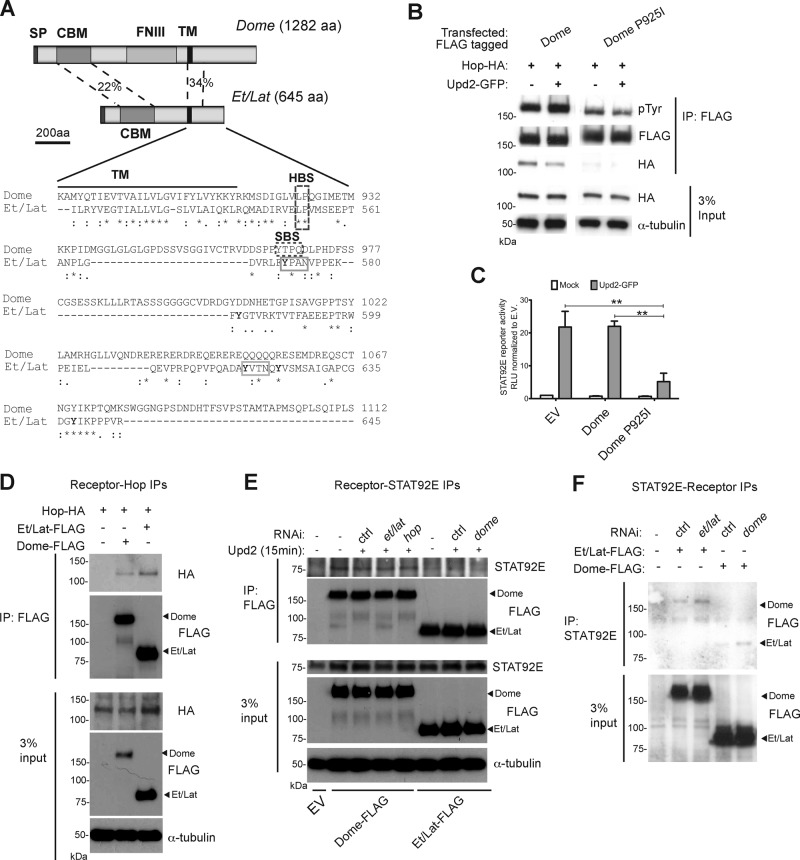
To determine the mechanism underlying how Et/Lat negatively regulates JAK/STAT signaling, we set out to investigate whether Et/Lat could physically associate with other components of the JAK/STAT pathway. Examination of the protein sequence alignments between intracellular Dome and Et/Lat revealed the presence of a conserved potential Hop-binding site located at the same position relative to the transmembrane domain within both molecules (HBS in Figure 2A). This LP motif is similar to the 266LPKS sequence in interferon-γ receptor 1 previously shown to be required for binding to JAK1 (where P is the critical residue required for binding; Kaplan et al., 1996). To test whether this site does indeed confer Hop binding, we mutated the proline at amino acid 925 (P925I). As expected, Dome P925I showed very little binding to Hop even when coexpressed at high levels (Figure 2B). Furthermore, whereas tyrosine phosphorylation of wild-type Dome was increased after stimulation with Upd2-conditioned medium, the basal phosphorylation level of Dome P925I was lower and remained unchanged upon stimulation (Figure 2B). We went on to test the signaling activity of Dome P925I using a STAT92E- responsive reporter assay. We found that increasing the level of Dome did not affect the reporter activity compared with empty vector–transfected cells. (Figure 2C). However, expression of Dome P925I substantially reduced pathway activity, suggesting that Dome P925I is acting as a dominant-negative receptor, possibly via the formation of heterodimers with endogenous Dome (Figure 2C). Taken together, these data suggest that the LP motif within Dome is a Hop-binding site and also suggest that two Hop molecules are required to form a signaling receptor complex. Given that the sequence and the relative position of this site are conserved between Dome and Et/Lat (Figure 2A), we suggest that this site could also act as a binding site for Hop in Et/Lat. To test this possibility, we also tested for physical interactions between Hop and Et/Lat. As predicted, immunoprecipitation of both Dome and Et/Lat is able to coprecipitate Hop (Figure 2D).
FIGURE 2:
Et/Lat can bind to intracellular JAK/STAT components. (A) Visualization of Dome and Et/Lat protein domain structure. Clustal sequence alignment for intracellular portion of Dome and Et/Lat. Transmembrane domain (TM; black bar), putative binding sites for Hop (HBS), and STAT92E (SBS) are highlighted. Tyrosine residues in Et/Lat are shown with bold letters. SP, signal peptide; CBM, cytokine-binding motif; FNIII, fibronectin 3 domains; TM, transmembrane domain; HBS, Hop-binding site; SBS, STAT binding site; aa, amino acids). Percentage identity between the indicated domains. (B) Dome-FLAG can coimmunoprecipitate Hop-HA, but this is greatly reduced when the putative Hop-binding site is mutated (Dome P925I). Tyrosine phosphorylation (pTyr) of Dome is also reduced in the Dome-P925I. (C) STAT92E transcriptional reporter activity assay. JAK/STAT pathway signaling is greatly reduced in cells expressing Dome P925I even in the presence of Upd2 ligand, **p < 0.01. (D) Immunoprecipitation assays show that both Dome-FLAG and Et/Lat-FLAG can coprecipitate Hop-HA. (E) Immunoprecipitation assays after transfection of the indicated Dome-FLAG and Et/Lat-FLAG proteins show that both proteins can coprecipitate endogenous STAT92E. dsRNA targeting the labeled mRNAs was added as indicated; ctrl, a nontargeting dsRNA. Knockdown of endogenous Dome (final lane) did not disrupt the ability of Et/Lat to bind to STAT92E. Upd2-conditioned medium was added for 15 min before protein harvesting. EV, empty vector. (F) Immunoprecipitation of endogenous STAT92E from cells expressing FLAG-tagged Dome or Et/Lat shows that both receptors are coprecipitated. Treatment with dsRNAs targeting the endogenous receptors suggests that interactions detected are independent of potential heterodimer formation.
Previous studies of mammalian receptors identified STAT-binding sites within the intracellular domain of multiple cytokine receptors. STAT5 was shown to bind to a YXXL motif in EPO-R (Klingmüller et al., 1996), and STAT3 can associate with GP130 via a YXXQ motif (Stahl et al., 1995). Consistent with these vertebrate receptors, Dome also contains a YXXQ motif, which may represent a STAT92E-binding site (SBS in Figure 2A; Chen et al., 2002). Neither STAT consensus binding motif is present within the Et/Lat intracellular domain; however, five tyrosine residues (highlighted in Figure 2A), including two YXXN sites (solid gray boxes in Figure 2A), are present. Somewhat surprisingly, we find that Et/Lat is able to bind to endogenously expressed STAT92E, an interaction that we detect in reciprocal precipitation experiments of both receptor and STAT92E (Figure 2, E and F). Furthermore, we find that the interaction is not destabilized upon Dome RNAi treatment and is not modulated by addition of ligand (Figure 2, E and F), suggesting that STAT92E is binding directly to Et/Lat in this system. Given the similarity of the glutamine residue within the Dome YXXQ STAT-binding motif and the amide side chain of asparagine, it is tempting to suggest that the two YXXN motifs present in the cytosolic tail of Et/Lat may represent the STAT92E binding sites in this molecule.
Et/Lat is unable to bind Upd ligands
Because Et/Lat is able to bind the key intracellular signaling pathway components Hop and STAT92E, it is possible that an inability to bind to extracellular ligands may explain its negative pathway activity. Despite clear homology between the cytokine-binding motifs of both Dome and Et/Lat (Makki et al., 2010), we investigated whether this molecule was in fact able to bind to any of the Upd family of ligands (Upd, Upd2, Upd3; Harrison et al., 1998; Hombria et al., 2005; Wright et al., 2011). First, we cotransfected cells with green fluorescent protein (GFP)–tagged versions of the Upd, Upd2, and Upd3 cytokines previously shown to be active (Wright et al., 2011), along with HA-tagged Dome or Et/Lat. We find that Et/Lat is unable to bind to any of the pathway ligands at detectable levels under conditions in which interactions with Dome are clearly detected (Figure 3A). We next sought to confirm that the interactions between Dome and pathway ligands were not arising from their coexpression and therefore repeated this experiment using Upd2-conditioned medium. We found that Dome coprecipitated with GFP-tagged ligand present in the conditioned medium, suggesting a stable interaction at the plasma membrane. However, under the same conditions, no interaction was detected between Upd2-GFP and Et/Lat. In addition, RNAi targeting the untranslated region (UTR) of endogenous dome mRNA had no effect on this result, indicating that endogenous Dome is not affecting receptor/ligand interactions in this assay and suggesting that Dome:Et/Lat heterodimers are also incapable of binding ligand. This failure to interact with Upd2-GFP suggests that the shorter receptor is either unable to bind ligand or that any interaction that does occur is weaker than the Dome:ligand interaction and is disrupted under the experimental conditions used (Figure 3B).
FIGURE 3:
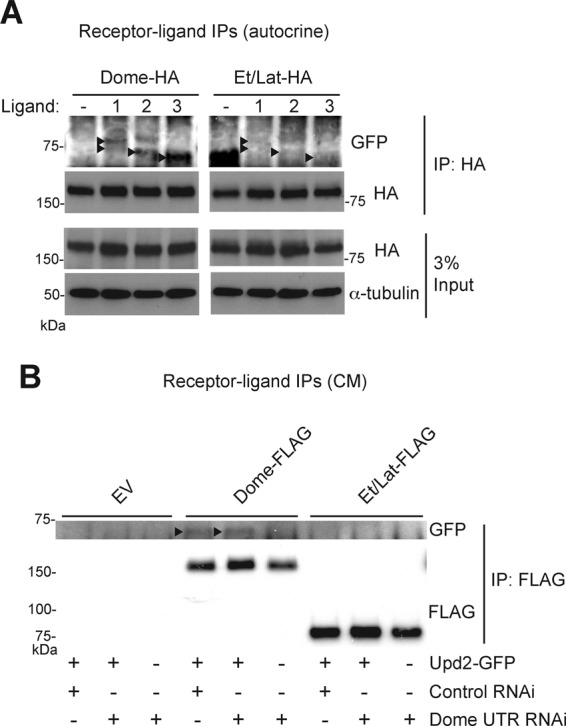
Et/Lat cannot bind to pathway ligands. (A) Immunoprecipitation assays show that Dome-FLAG can coprecipitate cotransfected GFP-tagged pathway ligands (black arrowheads). By contrast, no interaction is detected after precipitation of Et/Lat-FLAG. 1, Upd; 2, Upd2; 3, Upd3. Diffuse banding pattern of ligand-GFP fusions are believed to be affected by posttranslational glycosylation of these secreted proteins. (B) Immunoprecipitation assays show that Upd2-GFP (arrowheads) can be coprecipitated from conditioned medium by cells expressing Dome-FLAG but not by cells expressing empty vector (EV) or Et/Lat-FLAG. Addition of dsRNA targeting the UTR of the dome mRNA was used to deplete endogenous Dome from cells, ensuring that the interaction observed is via the transfected FLAG-tagged proteins.
Although independently identified as a negative regulator of JAK/STAT signaling in a number of assays, the mechanism underlying how Et/Lat expression reduces pathway activity has been unclear. It was previously shown that Dome homodimerization is required to form a functional signaling complex, whereas monomeric Dome is not active (Brown et al., 2003). By inference, a single molecule of Dome is either not capable of binding to ligand or unable to transduce signaling in response to binding. Here we demonstrate that Dome can effectively form heterodimers with Et/Lat and that the cytosolic domains of both receptor molecules are capable of binding to Hop and STAT92E. Furthermore, we demonstrate that whereas Dome homodimers are able to bind to each of the three pathway ligands, no such binding could be detected between Et/Lat and Upd, Upd2, or Upd3 (Figure 3).
Our data suggest two potential mechanisms via which Et/Lat negatively regulates downstream pathway activity. First, Et/Lat could act as a negative regulator by forming heterodimers with Dome. This generates receptor complexes containing only one molecule capable of binding to extracellular ligands, an arrangement likely to be incapable of transducing pathway activation to intracellular components. The second potential mechanism is that Et/Lat homodimers may simply bind to positively acting components of the JAK/STAT pathway such as Hop and STAT92E, sequestering these into signaling incompetent complexes. It is most likely that both mechanisms contribute toward negative regulation of the pathway at some level. However, the ability of additional Dome to partly relieve Et/Lat-induced pathway suppression (Figure 1A) argues against a “squelching” mechanism and instead suggests that Et/Lat is likely to negatively regulate JAK/STAT signaling via formation of inactive Dome:Et/Lat heterodimers.
Et/Lat has increased protein stability compared to Dome
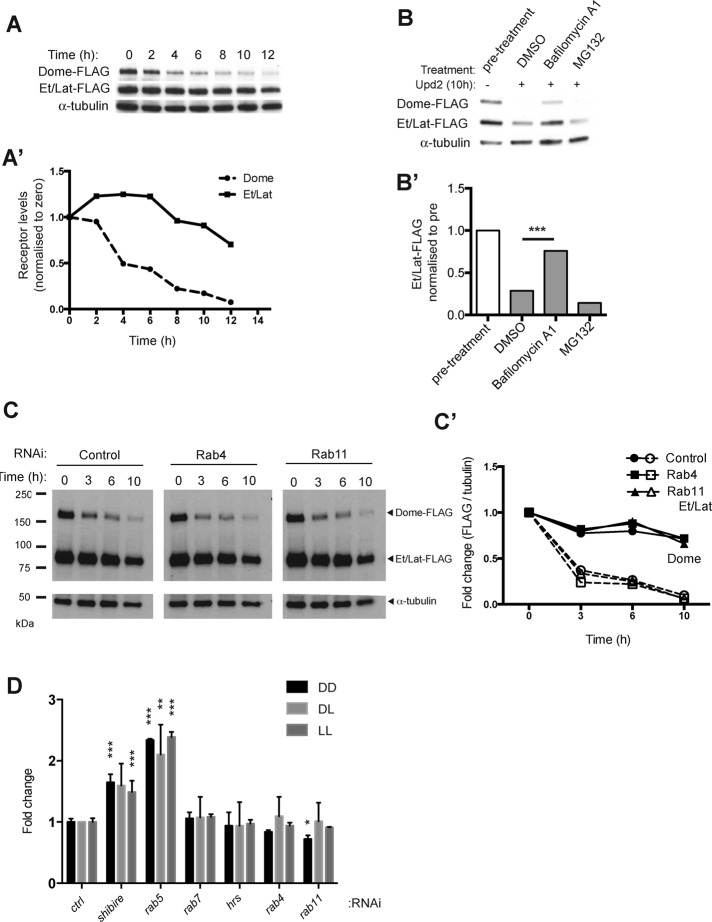
We and others previously demonstrated that Dome is rapidly internalized and targeted for lysosomal degradation even in the absence of ligand stimulation, a trafficking process that results in attenuation of signaling (Vidal et al., 2010; Stec et al., 2013; Ren et al., 2015). We therefore investigated whether the turnover dynamics of Et/Lat was similar to that of Dome, using a time-course experiment that involved ligand stimulation in the presence of cyclohexamide (CHX) to block new protein translation. Consistent with previous findings (Stec et al., 2013), we observed a rapid decrease in Dome protein levels (Figure 4, A and A′). By contrast, Et/Lat appeared to be much more stable, showing only a modest decrease in protein levels even after 10 h of stimulation (Figure 4, A and A′). To determine whether this difference was the result of alternative degradation mechanisms, we examined the effect of the proteosomal inhibitor MG132 and the lysosomal inhibitor bafilomycin A1 on the stability of both receptors. As is also the case for Dome, Et/Lat degradation occurs within the lysosome, as shown by the increased stability of the receptor after treatment with bafilomycin A1 (Figure 4, B and B′; Stec et al., 2013).
FIGURE 4:
Et/Lat is trafficked and degraded in the lysosome via a slower mechanism than that for Dome. (A) Time course of receptor degradation over the indicated time in hours. Cyclohexamide to block new translation and Upd2-conditioned medium were present throughout the assay. (A′) Quantification of data shown in A after normalization of FLAG to α-tubulin (Dome-FLAG, dashed line; Et/Lat-FLAG, solid line). (B) Levels of Dome-FLAG and Et/Lat-FLAG 10 h after the addition of cyclohexamide and Upd2 ligand stimulation. By comparison to the dimethyl sulfoxide carrier control, incubation with 0.2 μM bafilomycin A1 greatly reduced ligand-induced degradation of Dome and Et/Lat, whereas treatment with 10 μM MG132 did not. (B′) Quantification of data shown in B. (C) Time course of Dome-FLAG and Et/Lat-FLAG degradation over the indicated time course after prior treatment with dsRNA targeting the indicated mRNAs. (C′) Quantification of receptor levels normalized to α-tubulin and shown by solid lines (Et/Lat-FLAG) and dashed lines (Dome-FLAG). (D) Steady-state levels of Dome:Dome (DD), Dome:Et/Lat (DL), and Et/Lat:Et/Lat (LL) dimers as reported by the βlue-βlau bimolecular complementation and β-Glo assay after 5 d of RNAi treatment. Results are shown as fold changes to control. Control levels were averaged from two wells treated with rh5 dsRNA. Significant results are shown; all unlabeled bars were not significantly different from controls. *p < 0.05, **p < 0.01, ***p < 0.001.
We next examined whether the greater stability of Et/Lat might be a consequence of its recycling back to the plasma membrane after endocytic uptake. We therefore knocked down Rab4 and Rab11, two molecules key to the recycling of previously endocytosed proteins (Jones et al., 2006), and measured the degradation rate of both Dome and Et/Lat over time (Figure 4C). Even after quantification and normalization for loading differences, we found no change in the stability of either receptor compared with controls (Figure 4, C and C′), suggesting that endocytic recycling does not play a significant role in the differing stability of the Dome and Et/Lat receptors.
Finally, we returned to our βlue-βlau complementation assay to examine the steady-state circumstances under which Dome and Et/Lat homodimers and Dome:Et/Lat heterodimers accumulate in cells. We found that knockdown of shibire and Rab5, which encode components required for the formation of the early endosome, resulted in increased levels of each receptor dimer combination (Figure 4D). By contrast, knockdown of rab7 or hrs, two molecules involved in the formation of the late endosome and multivesicular bodies, respectively, or rab4 and rab11, required for recycling, have no effect on the steady-state levels of receptor complexes (Figure 4D). In the absence of antibodies, we are unable to confirm complete loss of these proteins; however, if taken at face value, this result suggests that blocking the formation of early endosomes is sufficient to cause an increase in the levels of receptor dimers at the plasma membrane. By contrast, blocking downstream aspects of endocytic trafficking has no effect on the levels of receptor dimers, suggesting that receptors are no longer dimerized in endosomal compartments.
We suggest that Et/Lat inhibits JAK/STAT signaling via the formation of signaling-incompetent Dome:Et/Lat heterodimers. If Et/Lat were to strongly down-regulate pathway signaling via competitive binding with Dome, this would require higher levels of Et/Lat in relation to Dome. Evidence from previous in vivo studies, as well as from our analysis of Et/Lat and Dome stability, suggests that this is indeed the case. In vivo, Et/Lat acts within the lymph gland to down-regulate pathway signaling after infestation by the parasitic wasp Leptopilina boulardi. Strikingly, infestation not only triggers the up-regulation of Et/Lat expression, but also simultaneously down-regulates dome mRNA levels, a combination that is likely to change the relative abundance of receptors present in vivo (Makki et al., 2010). In addition to changes in relative gene expression levels, we demonstrated that Et/Lat has a significantly longer protein half-life than Dome. As a result, expression of long-lived Et/Lat protein could produce strong and lasting effects with relatively small changes in transcription.
In vertebrates, multiple ligands, including IL-6 and IL-11, first bind to their respective α-receptors, forming ligand:receptor complexes, which then associate with long β-receptors to form functional signaling complexes (Heinrich et al., 2003). In this scenario, expression of α-receptors sensitizes cells to ligand-induced signaling and thus positively regulates downstream signaling. By contrast, a handful of short receptors, including IL-13Rα2, soluble forms of oncostatin M receptor (OSMR), and short forms of the prolactin receptor, act negatively (Perrot-Applanat et al., 1997; Diveu et al., 2006; Chandriani et al., 2014). In the case of IL-13Rα2 and soluble OSMR, short receptors do not interact with JAKs or STATs, but instead act as decoys that compete for ligand binding (Diveu et al., 2006; Chandriani et al., 2014). By contrast, the short, 291–amino acid form of the prolactin receptor is identical to the full-length form and can bind to JAK1 but is missing cytoplasmic domains required for STAT binding (Lebrun et al., 1995; Perrot-Applanat et al., 1997). This short receptor cannot activate downstream signaling and acts in a dominant-negative manner to reduce prolactin-mediated JAK/STAT pathway activation (Perrot-Applanat et al., 1997; Devi et al., 2009). Similarly to Dome and Et/Lat, prolactin receptors also predimerize in the absence of ligand to form both long:long homodimers and long:short heterodimers (Qazi et al., 2006; Tan and Walker, 2010). Whereas homodimers signal through JAK2 and STAT5 after ligand stimulation, heterodimers are able to bind ligand but do not activate downstream JAK/STAT signaling (reviewed in Devi and Halperin, 2014).
Although they share some mechanistic similarities with the short form of the human prolactin receptor, we show that Et/Lat is unable to associate with extracellular ligands but is able to bind to both the JAK Hop and STAT92E, as well as dimerize with the long receptor Dome. In both examples, the inability to bind to one key component of the pathway results in a strong dominant-negative effect and the down-regulation of pathway signaling. As such, it is possible that insights into molecular mechanisms of short receptor antagonists in Drosophila may provide clues to similar mechanisms in vertebrate systems.
MATERIALS AND METHODS
Cell culture
Drosophila Kc167 cell lines were cultured, and conditioned medium was produced as previously described (Vidal et al., 2010). Cell transfections were performed using Effectene (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Pathway activity was measured using the 6×2xDrafLuc reporter assay, as previously described (Müller et al., 2005; Fisher et al., 2012), and is presented as the ratio of Firefly (reporter) to Renilla (constitutively expressed cell density control) luciferase. RNAi experiments, pharmacological treatments, and socs36E quantitative PCR were carried out as previously described (Stec et al., 2013). Templates for double-strand RNA (dsRNA) generation were provided by the Sheffield RNAi Screening Facility, using designs previously described (Horn et al., 2010; Fisher et al., 2012).
βlue-βlau and β-Glo assays
Kc167 cells were batch-transfected with pAc-Dome-LacZ-Δα, pAc-Dome-LacZ-Δω, and pAc-RLuc and incubated for 24 h. Cells were either split into 12-well plates for retransfection with Et/Lat-HA or control plasmid or changed into serum-free medium and split into 384-well white plates with 250 ng of dsRNA. In RNAi experiments, serum-containing medium was added after 1 h. After 5 d, cells were assayed for β-gal activity using β-Glo Assay System (Promega), followed by measurement of Renilla luciferase activity. For whole-cell assays, cells were fixed in 2% gluteraldehyde/phosphate-buffered saline before development using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), using standard techniques (Brown et al. 2003).
Constructs
The previously described plasmids were pAc-Dome-FLAG, pAc-Dome-HA, and pAc-Hop-HA (Stec et al., 2013), and pAc-Upd-GFP, pAc-Upd2-GFP, and pAc-Upd3-GFP (Wright et al., 2011). A full-length sequenced Et/Lat cDNA clone (MIP10547; a kind gift of the Susan Celniker lab [Berkeley, CA] and the Berkeley Drosophila Genome Project [BDGP]) was used as template for amplification. The PCR-amplified fragment was inserted into a Gateway System Entry vector using a pENTR Directional TOPO Cloning Kit (Invitrogen, Carlsbad, CA) and subsequently cloned into destination vector pAWH (Drosophila Gateway Vector Collection) using Gateway LR Clonase II Enzyme Mix (Invitrogen), according to manufacturer’s instructions. Gateway destination vectors were obtained from the Drosophila Genomics Resource Center (Bloomington, IN). Single point mutation in Dome was introduced using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) using the following primers: Dome-P925I-F, GATATCGGTCTAGTGCTGattCAGGGAATCATGGAGACC; and Dome-P925I-R, GGTCTCCATGATTCCCTGaatCAGCACTAGACCGATATC.
Dome-LacZ-Δα and Dome-LacZ-Δω were cut out from pUAST vectors previously described using standard methods (Brown et al., 2003) and cloned into pAc5.1 (Invitrogen). Et/Lat was subcloned into pCR-2.1-TOPO (Invitrogen) and then used to replace Dome to create pAc-Et/Lat-Δα and pAc-Et/Lat-Δω.
Immunoprecipitation, Western blotting, and antibodies
Immunoprecipitation and Western blotting were carried out as described previously (Stec et al., 2013). The 3% of the input used is shown in each experiment. Tubulin was used as a loading control on all westerns. Kilodalton markers are shown for each Western panel in every figure. Western densitometric analysis was carried out using ImageJ (National Institutes of Health, Bethesda, MD) and analyzed in Prism. One-way analysis of variance with multiple comparisons was used to calculate statistical significance. The following antibodies were used: 1:2500 monoclonal M2 anti-FLAG (Sigma-Aldrich, St. Louis, MO), 1:2500 anti-HA High Affinity (Roche, Basel, Switzerland), 1:400 anti-pTyrosine PY20 (Calbiochem, San Diego, CA), 1:5000 monoclonal anti–α-tubulin (Sigma-Aldrich), 1:200 anti-STAT92E (Santa Cruz Biotechnology, Dallas, TX), and 1:500 anti-GFP (Abcam, Cambridge, UK).
Protein alignment
Alignment of amino acid sequences was carried out using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2/). Transmembrane and intracellular portions were aligned for Dome (amino acids 888–1112) and Et/Lat (amino acids 517–645).
Supplementary Material
Acknowledgments
We thank Lucie N’Koy, Lindsay Farrell, and Kirsty Johnstone for technical assistance and James Castelli-Gair Hombría, the Celniker lab, and the BDGP for reagents. M.P.Z. was a Cancer Research-UK Senior Cancer Research Fellow, W.S. was a Cancer Research-UK PhD fellow, and K.F. was supported by the Framework 7 Project Cancer Pathways.
Abbreviations used:
- β-gal
β-galactosidase
- CHX
cyclohexamide
- ET
Eye Transformer
- IL
interleukin
- JAK
Janus kinase
- Lat
Latran
- STAT
signal transducer and activator of transcription.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0546) on December 10, 2015.
REFERENCES
- Amoyel M, Anderson AM, Bach EA. JAK/STAT pathway dysregulation in tumors: a Drosophila perspective. Semin Cell Dev Biol. 2014;28:96–103. doi: 10.1016/j.semcdb.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Bausek N. JAK-STAT signaling in stem cells and their niches in Drosophila. JAKSTAT. 2013;2:e25686. doi: 10.4161/jkst.25686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 2010;11:201–207. doi: 10.1038/embor.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Novel level of signalling control in the JAK/STAT pathway revealed by in situ visualisation of protein-protein interaction during Drosophila development. Development. 2003;130:3077–3084. doi: 10.1242/dev.00535. [DOI] [PubMed] [Google Scholar]
- Chandriani S, DePianto DJ, N’Diaye EN, Abbas AR, Jackman J, Bevers J, 3rd, Ramirez-Carrozzi V, Pappu R, Kauder SE, Toy K, et al. Endogenously expressed IL-13Ralpha2 attenuates IL-13-mediated responses but does not activate signaling in human lung fibroblasts. J Immunol. 2014;193:111–119. doi: 10.4049/jimmunol.1301761. [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–398. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi YS, Halperin J. Reproductive actions of prolactin mediated through short and long receptor isoforms. Mol Cell Endocrinol. 2014;382:400–410. doi: 10.1016/j.mce.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Devi YS, Shehu A, Halperin J, Stocco C, Le J, Seibold AM, Gibori G. Prolactin signaling through the short isoform of the mouse prolactin receptor regulates DNA binding of specific transcription factors, often with opposite effects in different reproductive issues. Reprod Biol Endocrinol. 2009;7:87. doi: 10.1186/1477-7827-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diveu C, Venereau E, Froger J, Ravon E, Grimaud L, Rousseau F, Chevalier S, Gascan H. Molecular and functional characterization of a soluble form of oncostatin M/interleukin-31 shared receptor. J Biol Chem. 2006;281:36673–36682. doi: 10.1074/jbc.M607005200. [DOI] [PubMed] [Google Scholar]
- Fisher KH, Wright VM, Taylor A, Zeidler MP, Brown S. Advances in genome-wide RNAi cellular screens: a case study using the Drosophila JAK/STAT pathway. BMC Genomics. 2012;13:506. doi: 10.1186/1471-2164-13-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah R, Stroke I, Betz N. The Beta-Glo® Assay System provides a sensitive luminescent reagent for detecting and quantifying β-galactosidase activity in a homogeneous assay format. Cell Notes. 2003;6:16–18. [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–R575. doi: 10.1016/s0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Horn T, Sandmann T, Boutros M. Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol. 2010;11:R61. doi: 10.1186/gb-2010-11-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Jones MC, Caswell PT, Norman JC. Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol. 2006;18:549–557. doi: 10.1016/j.ceb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Kallio J, Myllymaki H, Gronholm J, Armstrong M, Vanha-aho LM, Makinen L, Silvennoinen O, Valanne S, Ramet M. Eye transformer is a negative regulator of Drosophila JAK/STAT signaling. FASEB J. 2010;24:4467–4479. doi: 10.1096/fj.10-162784. [DOI] [PubMed] [Google Scholar]
- Kaplan DH, Greenlund AC, Tanner JW, Shaw AS, Schreiber RD. Identification of an interferon-gamma receptor alpha chain sequence required for JAK-1 binding. J Biol Chem. 1996;271:9–12. doi: 10.1074/jbc.271.1.9. [DOI] [PubMed] [Google Scholar]
- Klingmüller U, Bergelson S, Hsiao JG, Lodish HF. Multiple tyrosine residues in the cytosolic domain of the erythropoietin receptor promote activation of STAT5. Proc Natl Acad Sci USA. 1996;93:8324–8328. doi: 10.1073/pnas.93.16.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun JJ, Ali S, Ullrich A, Kelly PA. Proline-rich sequence-mediated Jak2 association to the prolactin receptor is required but not sufficient for signal transduction. J Biol Chem. 1995;270:10664–10670. doi: 10.1074/jbc.270.18.10664. [DOI] [PubMed] [Google Scholar]
- Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, Krzemien´ J, Bourbon HM, Zhou R, Vincent A, Crozatier M. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010;8:e1000441. doi: 10.1371/journal.pbio.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Gerhartz C, Heesel B, Welte T, Doppler W, Graeve L, Horn F, Heinrich PC. Comparative study on the phosphotyrosine motifs of different cytokine receptors involved in STAT5 activation. FEBS Lett. 1996;394:221–226. doi: 10.1016/0014-5793(96)00955-6. [DOI] [PubMed] [Google Scholar]
- Morin-Poulard I, Vincent A, Crozatier M. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT. 2013;2:e25700. doi: 10.4161/jkst.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Perrot-Applanat M, Gualillo O, Pezet A, Vincent V, Edery M, Kelly PA. Dominant negative and cooperative effects of mutant forms of prolactin receptor. Mol Endocrinol. 1997;11:1020–1032. doi: 10.1210/mend.11.8.9954. [DOI] [PubMed] [Google Scholar]
- Qazi AM, Tsai-Morris CH, Dufau ML. Ligand-independent homo- and heterodimerization of human prolactin receptor variants: inhibitory action of the short forms by heterodimerization. Mol Endocrinol. 2006;20:1912–1923. doi: 10.1210/me.2005-0291. [DOI] [PubMed] [Google Scholar]
- Rahaman SO, Sharma P, Harbor PC, Aman MJ, Vogelbaum MA, Haque SJ. IL-13R(alpha)2, a decoy receptor for IL-13 acts as an inhibitor of IL-4-dependent signal transduction in glioblastoma cells. Cancer Res. 2002;62:1103–1109. [PubMed] [Google Scholar]
- Ren W, Zhang Y, Li M, Wu L, Wang G, Baeg GH, You J, Li Z, Lin X. Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet. 2015;11:e1005180. doi: 10.1371/journal.pgen.1005180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JEJ, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- Stec W, Vidal O, Zeidler MP. Drosophila SOCS36E negatively regulates JAK/STAT pathway signaling via two separable mechanisms. Mol Biol Cell. 2013;24:3000–3009. doi: 10.1091/mbc.E13-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- Tan D, Walker AM. Short form 1b human prolactin receptor down-regulates expression of the long form. J Mol Endocrinol. 2010;44:187–194. doi: 10.1677/JME-09-0101. [DOI] [PubMed] [Google Scholar]
- Teng Y, Ross JL, Cowell JK. The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT. 2014;3:e28086. doi: 10.4161/jkst.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113:365–371. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal OM, Stec W, Bausek N, Smythe E, Zeidler MP. Negative regulation of Drosophila JAK-STAT signalling by endocytic trafficking. J Cell Sci. 2010;123:3457–3466. doi: 10.1242/jcs.066902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino AV, Kanno Y, Ferdinand JR, O’Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright VM, Vogt KL, Smythe E, Zeidler MP. Differential activities of the Drosophila JAK/STAT pathway ligands Upd, Upd2 and Upd3. Cell Signal. 2011;23:920–927. doi: 10.1016/j.cellsig.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JEJ. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.