In mitotically dividing cells, septin proteins form cytoskeletal filaments that function in cell morphogenesis and division. Gametogenesis in yeast couples meiosis with a fundamentally different form of cytokinesis involving de novo membrane synthesis. Budding yeast septins are critical for spore membrane extension and wall assembly.
Abstract
The highly conserved family of septin proteins has important functions in cytokinesis in mitotically proliferating cells. A different form of cytokinesis occurs during gametogenesis in Saccharomyces cerevisiae, in which four haploid meiotic products become encased by prospore membrane (PSMs) and specialized, stress-resistant spore walls. Septins are known to localize in a series of structures near the growing PSM, but previous studies noted only mild sporulation defects upon septin mutation. We report that directed PSM extension fails in many septin-mutant cells, and, for those that do succeed, walls are abnormal, leading to increased susceptibility to heating, freezing, and digestion by the Drosophila gut. Septin mutants mislocalize the leading-edge protein (LEP) complex required for normal PSM and wall biogenesis, and ectopic expression of the LEP protein Ssp1 perturbs mitotic septin localization and function, suggesting a functional interaction. Strikingly, extra copies of septin CDC10 rescue sporulation and LEP localization in cells lacking Sma1, a phospholipase D–associated protein dispensable for initiation of PSM assembly and PSM curvature but required for PSM extension. These findings point to key septin functions in directing efficient membrane and cell wall synthesis during budding yeast gametogenesis.
INTRODUCTION
When deprived of nutrients, diploid yeast cells exit the mitotic cell cycle and undergo sporulation (Neiman, 2011). Unlike cytokinesis in vegetative (mitotically dividing) cells, in which a single plasma membrane is divided in two, sporulating cells in meiosis II initiate de novo synthesis of four new membranes, each of which encapsulates a haploid nucleus and essential cellular components. As the four “haploid” lobes of the nucleus separate, the process of spore morphogenesis begins at the spindle pole bodies (SPBs) implanted in the nuclear envelope (Lynn and Magee, 1970; Neiman, 1998; Knop and Strasser, 2000). Prospore membrane (PSM) development proceeds through a series of transitions, beginning as SPB-proximal disks and forming spheres upon closure (Neiman, 2011). During subsequent spore wall synthesis, mannan and β-1,3-glucan layers are deposited in the lumen formed by the double-bilayer PSM (Smits et al., 2001). The outermost two layers, rich in chitosan (Coluccio et al., 2004) and dityrosine (Briza et al., 1990b), form after breakdown of the outer PSM leaflet and endow spores with resistance to environmental assaults (Neiman, 2011).
Septins were first identified in Saccharomyces cerevisiae and are found throughout nonplant eukaryotes (Pan et al., 2007; Nishihama et al., 2011). Five septin proteins expressed in vegetative cells (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) assemble into hetero-octamers that interact with the plasma membrane and anneal into a filamentous network (Bertin et al., 2008, 2010, 2012; Garcia et al., 2011; Ong et al., 2014). Filamentous septin rings at the site of vegetative cytokinesis, the mother–bud neck, act as scaffolds to recruit chitin synthases (Larson et al., 2008) and components of the actomyosin ring (Roh et al., 2002; Oh et al., 2013; Feng et al., 2015; Finnigan et al., 2015). Septin rings are also associated with a diffusion barrier function that compartmentalizes regions of the cortex (Dobbelaere and Barral, 2004).
By contrast, roles for septins in sporulation are poorly understood. Expression of CDC3, CDC10, and the two sporulation-specific septins SPR3 and SPR28 is highly induced during sporulation via the transcription factor Ndt80 (Kaback and Feldberg, 1985; Chu and Herskowitz, 1998; Pierce et al., 2003). The products of these genes colocalize with Cdc11 in PSM-associated higher-order structures (Ozsarac et al., 1995; De Virgilio et al., 1996; Fares et al., 1996; McMurray and Thorner, 2008; Pablo-Hernando et al., 2008), whereas Cdc12 and Shs1 are largely evicted from existing complexes and appear to be replaced by Spr3 and Spr28, respectively (McMurray and Thorner, 2008). Septin assemblies probably interact with the inner PSM leaflet, because vegetatively synthesized Cdc10 molecules are inherited by mature spores and are recycled for use upon spore germination (Joseph-Strauss et al., 2007; McMurray and Thorner, 2008); were they on the outer leaflet, they would be left behind in the ascal cytoplasm. Others previously found only mild sporulation defects in septin mutants, leading to the idea that septins are unimportant for this process (Neiman, 2011). Here we carefully analyze PSM dynamics and spore wall composition in mutant cells to identify specific functional requirements for septins during budding yeast sporulation.
RESULTS AND DISCUSSION
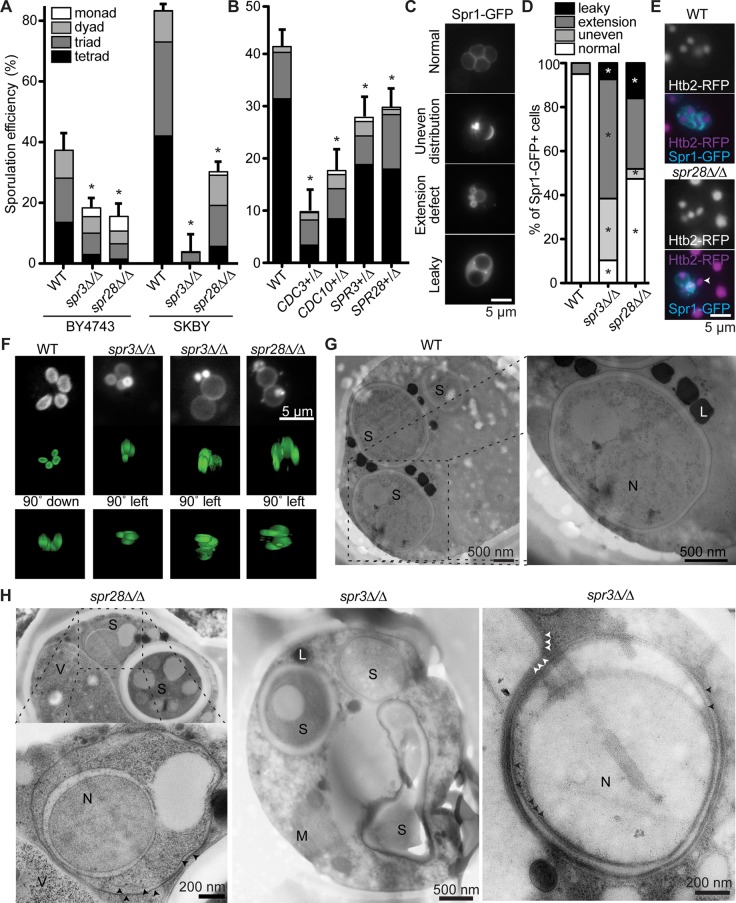
In previous studies, the magnitude of effects of septin mutations on sporulation varied with strain background (De Virgilio et al., 1996; Fares et al., 1996). Overall sporulation efficiency was calculated as the frequency of asci per total number of cells. We constructed homozygous deletions of SPR3 or SPR28 in two nearly isogenic strain backgrounds with very different sporulation efficiencies. BY4743 sporulates with relatively low efficiency (37%), but alteration of single nucleotides at three loci in the BY4743 genome (to create an “SKBY” strain) is able to reconstitute nearly all of the high sporulation efficiency of the SK1 strain background (Kane and Roth, 1974; Deutschbauer and Davis, 2005; Figure 1A). In both strain backgrounds, sporulation efficiency was reduced drastically (up to 10-fold) in spr3∆/∆ mutants and spr28∆/∆ mutants (Figure 1A).
FIGURE 1:
Defects in ascus production, spore number per ascus, and prospore membrane morphogenesis in septin-mutant cells. (A) Overall sporulation efficiencies and spore number per ascus for WT, spr3∆/∆, or spr28∆/∆ cells in two genetic backgrounds (BY4743 and SKBY). Sporulation efficiency was calculated as the percentage of cells that formed an ascus visible by transmitted light; >200 cells/genotype. Error bars, SE of the proportion. Asterisk, chi-square p < 0.05 compared with WT. (B) As in A, for BY4743-background diploids heterozygous for the indicated septin deletion alleles. (C) Representative images of normal and three types of defective PSMs as visualized by single-focal-plane epifluorescence microscopy. (D) Proportion of asci exhibiting the Spr1-GFP localization patterns shown in C; >50 cells/genotype. (E) Representative images of WT or spr28∆/∆ cells coexpressing a fluorescently tagged histone (Htb2-RFP) and Spr1-GFP. Arrowhead, postmeiotic haploid nucleus lacking an associated PSM. (F) Top, maximum-intensity projections of z-stacks of cells prepared as in C. Bottom, 3D reconstructions of the same cells, viewed from other perspectives, as indicated. (G, H) Negative-stain EM of BY4743-background cells of the indicated genotypes. L, lipid droplet; M, mitochondrion; N, nucleus; S, spore; V, vacuole. Black arrowheads, aberrant PSMs. White arrowheads, presumptive membrane of an unidentified organelle as it abuts a PSM.
Septin-mutant cells that were able to form asci typically did not preserve all four meiotic products in the form of tetrads. Triads, dyads, or monads were the most common classes of mutant asci (Figure 1A). Loss of even a single copy of SPR3, SPR28, CDC3, or CDC10 significantly decreased overall efficiency and spore number per ascus (Figure 1B); by contrast, CDC3 and CDC10 are haplosufficient in vegetative cells (Deutschbauer et al., 2005). These data demonstrate that sporulation is particularly dependent on septin expression and agree with the results of unbiased screening in which BY4743 spr3∆/∆ or cdc10∆/∆ mutants displayed severe decreases in sporulation efficiency (Deutschbauer et al., 2002; Enyenihi and Saunders, 2003). Variations in genetic background among different lab strains are known (BUD4; Wloka et al., 2011) or likely (SSD1; Wanless et al., 2014) to influence the severity of vegetative defects accompanying septin mutation; these or other differences may explain why other studies reported milder sporulation phenotypes. In addition, we used a medium and temperature determined empirically to be optimal for wild-type (WT) BY4743 sporulation, which could involve a particular dependence on septin function. However, unbiased analysis of natural isolates of S. cerevisiae identified as a candidate regulator of sporulation efficiency a polymorphism within the CDC10 promoter (Tomar et al., 2013), which we noticed lies within the Ndt80 binding-site consensus sequence (CACAAAT; Chu and Herskowitz, 1998; polymorphic base in bold). Thus, rather than representing minor players in budding yeast sporulation, alterations in septin gene expression during sporulation may in fact drive evolutionary differences in gametogenesis.
When defects occur early in sporulation, the two haploid nuclei adjacent to the two “newest” SPBs are the most likely to be encased in PSMs and spore walls (Davidow et al., 1980), leading to the preferential inheritance in dyads of one of each of any heterozygous alleles at centromere-linked loci (i.e., “nonsister” dyads [NSDs]). By contrast, defects arising after PSM biogenesis produce dyads with a random pair of meiotic products (Moreno-Borchart et al., 2001; Nickas and Neiman, 2002). We found that NSDs were strongly enriched in spr3∆/∆ and spr28∆/∆ mutant asci (Supplemental Figure S1). Thus defects in septin-mutant cells arise as soon as PSM biogenesis begins.
To visualize PSMs in septin-mutant cells, we used a fusion of green fluorescent protein (GFP) to Spr1, which localizes to the lumen between the two PSM bilayers and eventually resides in the mature spore wall (Suda et al., 2009; Figure 1C). After 24 h in sporulation medium, all PSMs within a given WT cell were similar in size and phase of growth (Figure 1, C and D). In stark contrast, the growth of PSMs in spr3∆/∆ and spr28∆/∆ cultures was severely perturbed. PSMs within a single ascus were differently sized and asynchronous in development, and some PSMs collapsed into bright foci (Figure 1, C and D). A large proportion (∼30%) of spr3∆/∆ mutants had PSMs in which Spr1-GFP was unevenly distributed around the prospore perimeter; this was almost never observed in WT cells (Figure 1, C and D). Another subset of spr3∆/∆ and spr28∆/∆ mutant cells displayed a characteristic of abnormally permeable spore walls (Suda et al., 2009): Spr1-GFP, which normally remains restricted to the spore periphery, “leaked” out into the ascal cytoplasm (Figure 1, C and D). Of importance, although mammalian septins help to coordinate cytokinesis with mitotic chromosome segregation (Spiliotis et al., 2005), in yeast, even severe defects in the fidelity of meiotic chromosome segregation do not perturb PSM biogenesis (Moens et al., 1974; Wagstaff et al., 1982; Gaudet and Fitzgerald-Hayes, 1989). Indeed, coexpression of a fluorescently tagged histone with Spr1-GFP confirmed that four postmeiosis II nuclear “lobes” formed normally in septin mutants, but PSMs often failed to develop around individual lobes (Figure 1E). Finally, monitoring chromosome segregation by fluorescently labeling both a centromere-linked locus (using the tetO/TetR-GFP system; Hediger et al., 2004) and SPBs (with DsRed-tagged Spc110) in spr3∆/SPR3 heterozygous cells revealed that the tagged chromosome segregated properly in each of 34 dyads (24 of which were shown by this assay to be NSDs; unpublished data). Thus defects in meiosis per se cannot explain the PSM defects in septin mutants.
Three-dimensional (3D) renderings created from serial images through the z-plane revealed that in cells with the “membrane extension” defect, a significant degree of PSM extension occurred before collapse, creating dense, torpedo-shaped structures (Figure 1F). Negative-stain electron microscopy (EM) confirmed that in septin mutants, PSMs developed asynchronously and were frequently misdirected in ways that clearly corresponded to the distinct defects seen by light microscopy. PSMs that had either collapsed onto or grown in a direction away from the nucleus (Figure 1H) likely correspond to the Spr1-GFP signals that appeared as small foci in two-dimensional images and as torpedo-shaped structures in 3D reconstructions (Figure 1, C and F). Prospores in which multiple layers of membrane were stacked on one side, and others in which the width of the lumen varied widely, likely correspond to the “uneven distribution” Spr1-GFP category (Figure 1, C and H). Together, these images indicate that septins are critical for proper PSM growth—particularly for ensuring a constant lumen size and maintaining normal PSM distance from the nuclear envelope—and are consistent with the observed decreases in sporulation efficiency and spore number per ascus. These defects are also highly reminiscent of defects in forespore membrane (equivalent to the PSM) orientation in septin-mutant fission yeast cells (Onishi et al., 2010).
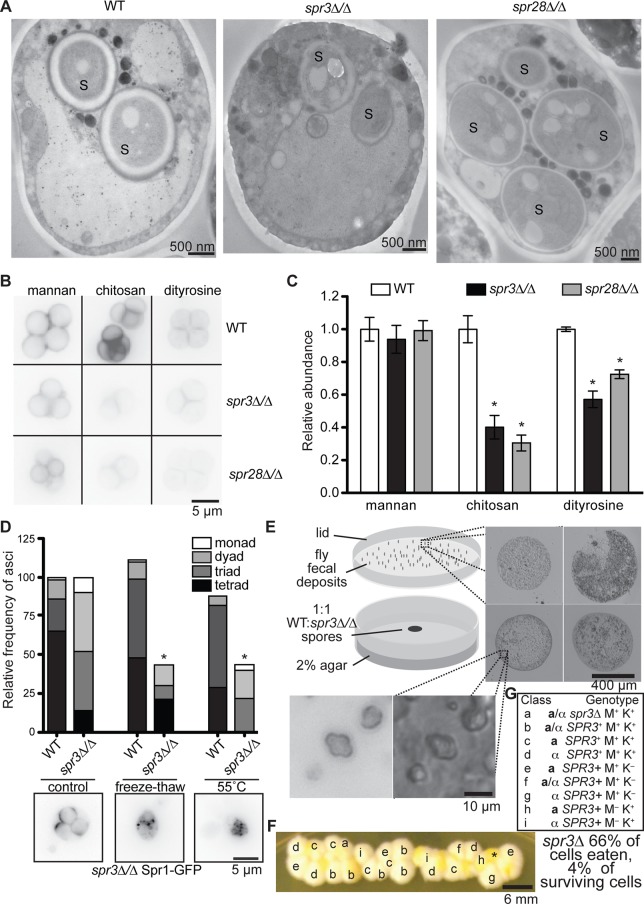
Many spr3∆/∆ and spr28∆/∆ prospores that had apparently succeeded in PSM morphogenesis nonetheless had spore walls that lacked the trilaminar appearance of WT spores (Rij, 1978; Figure 2A). PSM biogenesis is necessary but not sufficient to ensure proper spore wall deposition. Indeed, spr3∆/∆ and spr28∆/∆ spores within tetrads contained significantly less dityrosine and chitosan than WT but deposited equivalent amounts of inner spore wall layer mannan (Figure 2, B and C). The acquisition of spore refractility in light microscopy occurs just before chitosan synthesis (Neiman, 2005); because the spores we analyzed were refractile, this places the earliest spore wall defect in septin-mutant cells at the chitosan stage. Thus septin-mutant prospores inefficiently deposit the outer spore wall layers, which are primarily responsible for resistance to environmental insults (Briza et al., 1990a). We found that, whereas WT spores retained their refractility to transmitted light, spore walls in spr3∆/∆ and spr28∆/∆ asci disappeared after exposure to either of two ecologically relevant stressors, freeze–thaw and high heat (Figure 2D). Examination of Spr1-GFP revealed that wall disappearance was accompanied by apparent PSM collapse (Figure 2E). Thus impaired wall biogenesis in the absence of septins is manifested at the organismal level as increased susceptibility to harsh environmental conditions.
FIGURE 2:
Spore wall defects in septin-mutant cells. (A) EM micrographs as in Figure 1F. (B) Representative images of spores of the indicated genotypes stained with fluorescein–Con A (mannan), eosin Y (chitosan), or autofluorescence in unstained cells visualized with a DAPI filter (dityrosine; Briza et al., 1986). (C) Quantification via line scans of fluorescence signals as in B for at least 9, 14, or 34 cells/genotype. Error bars, SEM. *p < 0.0001 for unpaired t test compared with WT. (D) Asci of the indicated genotypes were analyzed as in Figure 1A (control) and then exposed to either a freeze–thaw cycle or 55°C for 2 h and analyzed again. Values were normalized to pretreatment values for each genotype. Bottom, representative images of Spr1-GFP fluorescence in spr3∆/∆ cells after the indicated treatments. (E) WT and spr3∆/∆ sporulation cultures were mixed to create an equal proportion of total spores of each genotype and fed to starved adult WT Drosophila for 18 h in a Petri dish. Top right, four representative fecal deposits on the lid visualized directly with a 10× objective. Bottom left, transmitted light and fluorescence image taken with a GFP filter cube and 40× objective. (F) Colonies on rich medium produced by resuspended fecal deposits (E). Asterisk, nonyeast colony. Other labels correspond to genotypes shown in G; see Supplemental Figure S2 for details. K, lysine; M, methionine.
For yeast cells, the crops of insects represent a refuge from the environment (Stefanini et al., 2012), but reemergence requires passage through the intestinal tract. Previous research suggested that whereas vegetative S. cerevisiae cells excreted by Drosophila are mostly inviable, spores with properly formed walls are resistant to this killing process (El-Tabey Awab Shihata and Mrak, 1951; Reuter et al., 2007; Coluccio et al., 2008). To ask whether the wall defects we observed in septin-mutant spores compromise survival during insect digestion, we fed starved, WT Drosophila a mix of WT and spr3∆/∆ sporulation cultures (adjusted to 50% total spr3∆/∆ spores) and examined the cells deposited in their feces. As expected, each fecal deposit was primarily composed of yeast, among which asci were clearly visible (Figure 2F and Supplemental Figure S2B).
To determine whether the survivors were vegetative cells or spores, we tested clones arising from resuspended feces spread on a plate (Figure 2F) for heterozygosity at the MAT, MET17, and LYS2 loci, genes that were heterozygous in both starting diploid strains. Whereas spontaneous mitotic loss of heterozygosity is rare, meiosis ensures that each spore inherits a single allele. Seventy percent (16 of 23) of surviving clones had a single allele at one or more of these loci (Figure 2, F and G, and Supplemental Figure S2). Twenty-two percent (2 of 9) of MATa/α diploids had only met17∆ or lys2∆, similar to the 25% expected from interascus mating. Thus nearly all survivors were spores. Crucially, although ∼60% of cells and 50% of spores entering the Drosophila gut carried the spr3∆ allele, only one of the 23 surviving clones did (4%) (Figure 2, F and G), indicating strong selection during insect digestion against spores produced by spr3∆/∆ cells.
The leading-edge protein (LEP) complex found at the “lip” of the growing PSM is believed to guide proper orientation of PSM extension (Moreno-Borchart et al., 2001; Nickas and Neiman, 2002). Among known LEP components, Ssp1 is essential for PSM closure and recruitment of the LEP Ady3 (Moreno-Borchart et al., 2001). Defects in ssp1∆/∆ mutants are reminiscent of those we describe for septin mutants, including torpedo-shaped prospores, PSMs that extend in the wrong directions, and incompletely synthesized walls (Moreno-Borchart et al., 2001). Ssp1 has even been proposed to be a “divergent septin” that performs septin-like functions during sporulation (Maier et al., 2007). Septins are able to form higher-order structures in ssp1∆/∆ cells (Moreno-Borchart et al., 2001), but their association with PSMs is perturbed (Supplemental Figure S3).
Whether septins are required for efficient LEP localization was unknown. Ssp1 does not tolerate standard GFP fusions (Maier et al., 2007), so we used Ady3-GFP as a reporter for LEP localization. In WT cells, Ady3-GFP localized primarily to rings, usually four per ascus (Figure 3A), as expected (Moreno-Borchart et al., 2001; Nickas and Neiman, 2002). In cells lacking SPR3 or SPR28, on the other hand, Ady3-GFP was most frequently found in numerous small puncta (Figure 3A). Indistinguishable aberrant Ady3 “precursor structures” have been described for other mutants (Moreno‐Borchart et al., 2001; Riedel et al., 2005). Of note, dyads formed in ady3∆ mutants incorporate a random pair of meiotic products, consistent with a primary defect in wall production, not PSM biogenesis (Moreno-Borchart et al., 2001; Nickas and Neiman, 2002). The NSDs and more severe PSM defects that we found in septin mutants thus suggest that proper localization of Ssp1 (which does not require Ady3; Moreno-Borchart et al., 2001) is also perturbed when septins are mutated, providing a plausible rationale for at least some of the PSM morphogenesis and spore wall defects we observed in septin-mutant cells.
FIGURE 3:
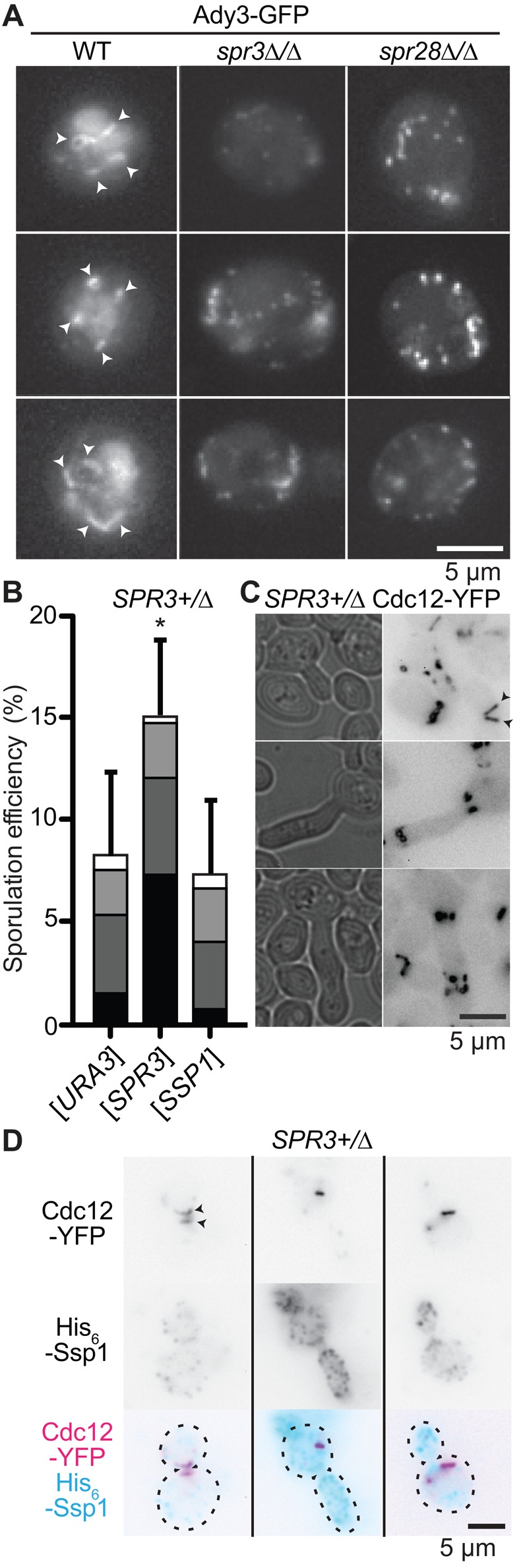
LEP complex mislocalization in septin mutants and septin mislocalization upon ectopic LEP expression. (A) Representative images of cells of the indicated genotypes expressing Ady3-GFP. Arrowheads, presumptive rings. (B) As in Figure 1A, for SPR3+/∆ cells carrying plasmids encoding the indicated genes. URA3, negative control. (C) Transmitted light and fluorescence micrographs (yellow fluorescent protein filter) of representative vegetative cells ectopically expressing GST-His6-tagged Ssp1. Arrowheads, normal septin rings in cells with normal, round morphology. (D) Representative cells as in C, immunostained with anti-His6 mAb and Alexa Fluor 594–labeled secondary antibodies.
If Ssp1 mislocalization underlies PSM and wall defects in septin-mutant cells, then Ssp1 overexpression might drive proper Ssp1 localization in septin-mutant cells and improve sporulation. When overexpressed in SPR3+/∆ heterozygotes, Spr3 increased sporulation efficiency and spore number per ascus (Figure 3B), as expected, but Ssp1 expressed in an identical manner had no effect on sporulation (Figure 3B) and instead induced severe defects in vegetative septin localization and function (i.e., elongated cellular morphologies; Figure 3C). Although Ssp1 can form rare cytosolic rings in vegetative cells (Maier et al., 2007), it did not colocalize with the ectopic septin structures in our Ssp1-expressing cells (Figure 3D). Thus the requirement for septins in Ssp1 function cannot be overcome solely by increasing Ssp1 levels, and functional Ssp1–septin interactions are likely indirect.
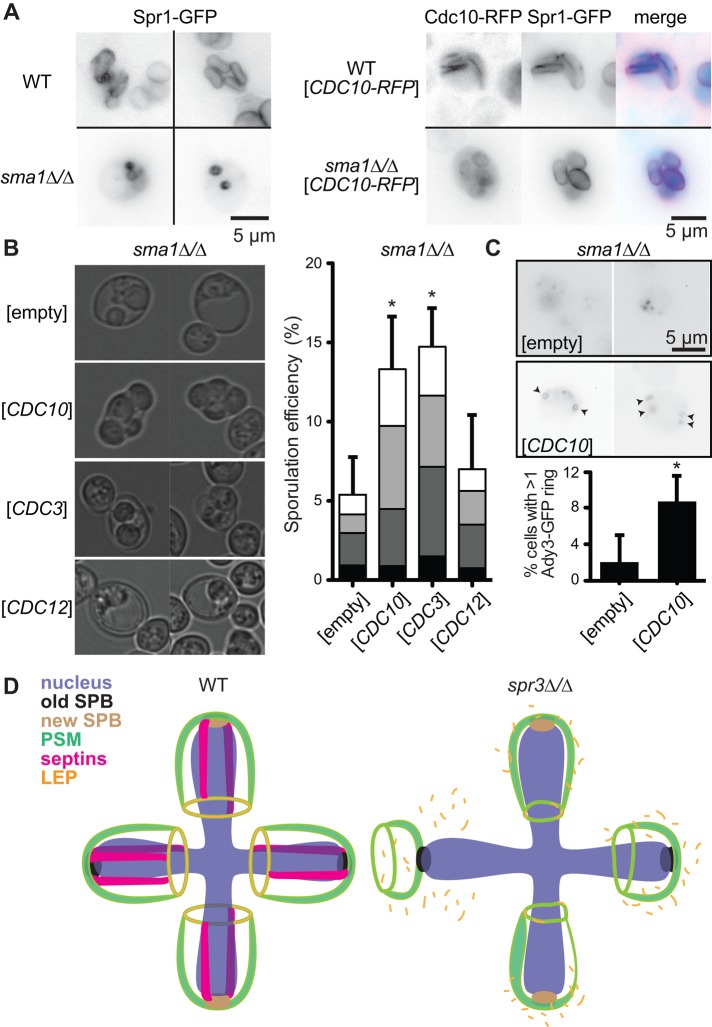
Cells lacking the PSM-associated protein Sma1 form few spores because PSM extension does not proceed far beyond the SPB (Enyenihi and Saunders, 2003; Riedel et al., 2005). As in septin mutants, LEPs mislocalize in sma1∆/∆ cells (Rabitsch et al., 2001; Riedel et al., 2005). To our surprise, in sma1∆/∆ diploids carrying Spr1-GFP and Cdc10–red fluorescent protein (RFP) plasmids, a large proportion of cells achieved PSM extension, proper Cdc10-RFP localization, and formation of mature spores (Figure 4A). However, within each culture, there was also a population of cells that exhibited the expected PSM extension defect, and these cells always lacked Cdc10-RFP (Figure 4A). This result suggested that the presence of extra, plasmid-encoded Cdc10 in sma1∆/∆ cells promoted extension of the otherwise stunted PSMs. Experiments with distinct plasmids and controls showed that extra copies of CDC3 or CDC10, but not of CDC12 or the empty vector, restored spore-forming ability to sma1∆/∆ mutants (Figure 4B) but had no effect on sporulation in mutants lacking SSP1 or any of a number of other genes implicated in this process (Supplemental Figure S3; unpublished data). A CDC10 plasmid failed to improve sma1∆/∆ sporulation in the absence of SPR3 (no visible spore among 280 apparent asci), indicating that the CDC10 rescue required assembly of Spr3-containing septin complexes. Additional studies will be required to understand at a mechanistic level how septin function can be altered by increasing the expression of only a single subunit of a complex believed to function as a stoichiometric hetero-oligomer.
FIGURE 4:
Septin overexpression rescues LEP localization and sporulation in PSM-extension-deficient sma1∆/∆ cells. (A) As in Figure 1C, but from cultures of WT or sma1∆/∆ cells expressing Spr1-GFP with or without Cdc10-RFP. (B) Left, representative transmitted light images of asci in sma1∆/∆ cultures with plasmids carrying the indicated septin gene or an empty vector. Right, as in Figure 1A, for at least 500 cells within cultures in B. (C) Top, as in Figure 3A, but with sma1∆/∆ cells and a plasmid encoding untagged Cdc10 or an empty vector. Bottom, quantification of cells with multiple Ady3-GFP rings among >1000 cells/genotype. Error bars, SE of the proportion. Asterisk, chi-square p < 0.05. (D) Model illustrating the defects in PSM biogenesis in spr3∆/∆ mutants.
Without extra copies of CDC10, Ady3-GFP in sma1∆/∆ cells localized almost exclusively to aberrant “cap”-like PSM structures near the SPBs (Riedel et al., 2005; Figure 4C). By contrast, a CDC10 plasmid restored Ady3-GFP localization to the characteristic LEP rings in a sizeable subset of sma1∆/∆ cells (Figure 4C). sma1∆/∆ rescue by CDC10 could reflect improved ability of the PSM to extend in an LEP-independent manner. However, we believe that these findings are more consistent with a model (Figure 4D) in which septins promote PSM extension by facilitating proper LEP localization.
In summary, we conclude that septins are critical factors in budding yeast gametogenesis. Without them, PSM biogenesis is severely perturbed, and walls that are produced do not provide the stress resistance that is key for this stage of the yeast life cycle. Intriguingly, septins are also required for gametogenesis in mammals (Kuo et al., 2012), but the precise roles that septins play remain unclear. Our study provides a foundation for investigations of additional mechanistic details of septin functions during gametogenesis in yeast and beyond.
MATERIALS AND METHODS
Strains and media
Yeast strains and plasmids are listed in Supplemental Tables S1 and S2, respectively. Rich (YP) and synthetic defined media (Sherman, 1991) contained dextrose, galactose, and/or raffinose to 2% total sugar. Drosophila Oregon-R flies were reared on cornmeal, molasses, and yeast medium before starvation.
Sporulation conditions
Diploid strains were grown in liquid yeast extract/peptone/dextrose (YPD) medium to saturation. A 250-μl amount of the culture was pelleted, washed twice with water, and resuspended in 2 ml of sporulation medium (1% potassium acetate, 0.05% dextrose, 20 mg/ml leucine, 40 mg/ml uracil). Cells carrying plasmids were grown in synthetic defined dextrose medium lacking appropriate nutrients for 1 d and then switched to YPD for 18 h before washing and transferring to sporulation medium. The CDC3, CDC12, and non–RFP-tagged CDC10 plasmids used in the sma1∆/∆ rescue experiments were pMVB100, pMVB66, and pLA10, respectively; the empty vector was pRS316. Cells carrying plasmids with glutathione S-transferase (GST)–hexahistidine (His6)-tagged open reading frames under control of the GAL1/10 promoter were prepared the same, way except they were switched to YP medium with 2% raffinose for 5 h and then to YP with 1% galactose and 1% raffinose for 18 h before being washed and transferred to sporulation medium. Cultures were incubated at room temperature (∼22°C). Depending on strain background (SKBY or BY4743), cultures were sporulated for 2–5 d before examination.
Sporulation efficiency and spore counting
Mature spore walls are refractile when viewed by transmitted light. Spores were thus identified as refractile spheres 1–5 μm in diameter. The accuracy of this scoring method was validated in two ways: 1) using a strain expressing a fluorescently tagged histone to identify the four-lobed meiosis II nucleus (unpublished data) and 2) using fluorescent signals generated from the Spr1-GFP plasmid, whose expression is restricted to sporulating cells (Suda et al., 2009; unpublished data). At least 200 cells were scored for each culture.
Microscope image acquisition
All single-focal-plane images were acquired using an EVOSfl inverted fluorescence microscope (ThermoFisher Scientific, Waltham, MA) with 10× dry, 40× dry, and 60× oil objectives. Filter cubes were as follows: 4′,6-diamidino-2-phenylindole (DAPI; excitation 357/44 nm, emission 447/60 nm), Texas red (excitation 585 nm, emission 624 nm), yellow fluorescent protein (excitation 500 nm, emission 542 nm), and GFP (excitation 470/22 nm, emission 510/42 nm). Images were cropped, contrast adjusted, and inverted in Photoshop (Adobe, San Jose, CA). Magenta-cyan merged images were generated in ImageJ (Schneider et al., 2012) by preparing a standard red-green merged image from raw white-on-black images and then converting to RGB and inverting the image. For z-stack and 3D projections, images were acquired using a Nikon (Melville, NY) Ti-E microscope with a 1.45 numerical aperture 100× CFI Plan Apo objective, piezoelectric stage (Physik Instrumente, Auburn, MA), spinning-disk confocal scanner unit (CSU10; Yokogawa, Tokyo, Japan), 488- and 561-nm lasers (Agilent Technologies, Santa Clara, CA), and an electron-multiplying charge-coupled device camera (iXon Ultra 897; Andor Technology, Belfast, United Kingdom) using NIS Elements software (Nikon). Image z-stacks were created by capturing consecutive images at 0.3-μm intervals. The z-stack and 3D stack projections were created using Fiji software (Schindelin et al., 2012). Image stacks were imported as 16-bit files, cropped, and compiled to make either a z-stack projection or 3D renderings. The z-stacks modeled as 3D renderings were cropped to size, filtered with a gaussian blur of 1.0, converted to an 8-bit image, and projected using the 3D viewer plug-in for Fiji.
Electron microscopy
BY4743-background cells were grown to saturation in 5 ml of YPD. A 1-ml amount of saturated culture was pelleted, washed twice with water, and inoculated into 25 ml of sporulation medium. Cultures were sporulated for 30 h and prepared as described previously (Giddings et al., 2001). Cells were collected by filtration, and the resulting paste was rapidly frozen under high pressure in a BAL-TEC HPM-010 high-pressure freezer (Technotrade International, Manchester, NH). The frozen cells were freeze substituted into acetone containing 2% osmium tetroxide and 0.1% uranyl acetate at –80°C and then warmed to room temperature. Samples were embedded in Spurr’s resin per manufacturer’s instructions (Electron Microscopy Sciences, Port Washington, PA). Embedded cells were serially sectioned at a thickness of 50 nm, and the sections were stained with 2% uranyl acetate in 70% methanol and lead citrate. Sections were viewed on a Philips CM10 electron microscope (Philips, Eindhoven, Netherlands), and images were collected with a Gatan BioScan digital camera by using the Digital Micrograph software package (Gatan, Pleasanton, CA).
Analysis of cell wall composition
Relative dityrosine fluorescence was calculated from images taken using the DAPI filter. Images were acquired at 70% intensity for 1 s. For chitosan analysis, spores were pelleted, washed twice with McIlvaine’s buffer (0.2 M sodium phosphate, 0.1 M citric acid, pH 6.0) and resuspended in McIlvaine’s buffer and 30 μl of eosin Y (5 mg/ml in water; VWR 22816; Baker et al., 2007). Samples were incubated for 10 min in darkness at room temperature. After incubation, spores were pelleted, washed twice, and resuspended in the buffer before imaging with the GFP filter. For mannan analysis, spores were pelleted and resuspended in 50 μl of 5 mg/ml 20T Zymolyase (0832092; MP Biomedicals, Santa Ana, CA) to remove the cell wall. Spores were incubated at 30°C for 10 min. Samples were again pelleted and resuspended in water with 0.1 mg/ml concanavalin A (Con A) coupled to fluorescein (C827; Life Technologies, Carlsbad, CA) before imaging with the GFP filter. For all images, using the line scan function in Fiji, an 8 pixel–wide line was drawn from outside of the ascus, across two spores, and back out of the cell. Maximum fluorescence (the spore walls) was subtracted from minimum fluorescence (slide background) to yield a background-corrected value.
NSD identification
A 100-μl amount of sporulation culture was pelleted and resuspended in 50 μl of 5 mg/ml Zymolyase, and 5 μl of the resuspension was spread on a YPD agar plate. After 30–45 min at 30°C to allow for digestion, dyads were isolated and dissected using a Singer MSM microscope (Singer Instruments, Watchet, United Kingdom).
Spore integrity in extreme temperatures
We calculated frequency of asci and spore number per ascus for sporulation cultures of BY4743 and made a diploid by mating MMY0184 and MMY0151 (>500 cells). We exposed 500-μl aliquots to extreme temperatures (55°C heat block for 3 h or three rounds of freeze–thaw in a dry ice/ethanol bath) in 1.5-ml microcentrifuge tubes and then recalculated efficiency and spore number values.
Spore survival during passage through Drosophila
A 25-ml amount of molten 2% agar made in water was poured into a 100 mm × 15 mm Petri dish (FB0875712; Fisherbrand; Fisher Scientific, Pittsburgh, PA). The sporulation efficiency of cultures of WT (strain BY4743 carrying plasmid pSPR1-GFP) or spr3∆/∆ cells (strain MMY0200) was calculated by examining aliquots microscopically (WT, 67 spores/157 cells; spr3∆/∆, 39 spores/197 cells) and used to prepare a mixture of cultures containing 50% WT and 50% spr3∆ spores (66% spr3∆/∆ culture, 33% WT culture). A 100-μl amount of this mixture was spotted in the center of the polymerized, cooled agar. Approximately 50 adult male flies were transferred to a vial containing a half-circle of 7.0-cm Whatman Qualitative #3 filter paper (1003070) and 500 μl of sterile water. After 6.5 h at 25°C, 30 flies were transferred directly to the Petri dish containing agar and yeast. After overnight incubation at 25°C, flies were anesthetized with CO2 gas and removed. The lid of the plate was scanned with a Typhoon imager (600 PMT, 532-nm laser, no emission filter; GE Healthcare, Pittsburgh, PA) to determine the overall number of fecal deposits, several of which were then visualized with 10× and 40× objectives on the EVOSfl microscope. Each of the deposits was resuspended in 1 μl of sterile water, and these were pooled together. An aliquot of 5 μl was spread on the surface of a YPD agar plate. Note that of 50 individual asci that were micromanipulated to defined positions in a grid pattern on the plate, only one could be dissected, and none of these isolated spores or the 49 undissected asci produced a colony after 7 d incubation at 30°C, suggesting that survival and ascus wall removal are very inefficient. Of the ∼25 colonies that emerged from the unmanipulated fecal spread, 23 were picked and analyzed for marker phenotypes on appropriate media. In addition, multiplex colony PCR (Huxley et al., 1990) was performed to determine the genotype at MAT. For the two reactions that failed, mating type was determined by mixing the clones with other clones of known, complementary genotypes at MET17 and LYS2 and selecting for diploids prototrophic for methionine and lysine.
Immunofluorescence
Immunofluorescence was performed as described elsewhere (Babst et al., 1998), with the following modifications. Diploid cells made by mating MMY0184 and BY4742 and carrying plasmids for SPR3, SSP1, or URA3 overexpression were grown in synthetic dropout medium lacking uracil overday and switched to YP medium with 2% raffinose for 5 h and then to YP with 1% galactose and 1% raffinose for 18 h before spheroplasting. Spheroplasts were fixed in 4% formaldehyde for 15 min at 30°C and mounted on glass coverslips coated with poly-l-lysine (P8920; Sigma-Aldrich, St. Louis, MO). After blocking, cells were incubated for 1 h with anti-His6 monoclonal antibody (mAb; Y1011; Ubiquitin-Proteasome Biotechnologies, Aurora, CO) diluted 1:1000 in 50 mM Tris, pH 8, 150 mM NaCl, 1% nonfat dry milk, 0.5 mg/ml bovine serum albumin, and 0.1% Tween 20 (buffer B). Coverslips were washed five times in buffer B and then incubated for 1 h with Alexa Fluor 594 goat anti-mouse mAb (1:2000). Cells were washed five times with buffer B and mounted onto slides with mounting medium (90% glycerol).
Supplementary Material
Acknowledgments
We thank Fred Winston, JoAnne Engebrecht, Michael Knop, Mark Johnston, and Jeremy Thorner for sharing reagents, Jeff Moore for reagents and use of the confocal microscope, Tom Giddings for electron microscopy, members of Tania Reis’s lab for help with the Drosophila experiments, and Courtney Johnson and Rachel Schaefer for technical assistance. Research reported here was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Awards R00GM086603 (to M.A.M.) and T32GM008730 (to L.R.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- LEP
leading-edge protein
- NSD
nonsister dyad
- PSM
prospore membrane
- SPB
spindle pole body
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0721) on December 17, 2015.
REFERENCES
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park S-S, Garcia G, 3rd, Patanwala I, Ng H-L, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Pierson J, Thai L, McDonald KL, Zehr EA, García G, Peters P, Thorner J, Nogales E. Three-dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23:423–432. doi: 10.1091/mbc.E11-10-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Thai L, Garcia G, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P, Breitenbach M, Ellinger A, Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990a;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Briza P, Ellinger A, Winkler G, Breitenbach M. Characterization of a DL-dityrosine-containing macromolecule from yeast ascospore walls. J Biol Chem. 1990b;265:15118–15123. [PubMed] [Google Scholar]
- Briza P, Winkler G, Kalchhauser H, Breitenbach M. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J Biol Chem. 1986;261:4288–4294. [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio AE, Rodriguez RK, Kernan MJ, Neiman AM. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One. 2008;3:e2873. doi: 10.1371/journal.pone.0002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics. 1980;94:581–595. doi: 10.1093/genetics/94.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, DeMarini DJ, Pringle JR. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology. 1996;142:2897–2905. doi: 10.1099/13500872-142-10-2897. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- El-Tabey Awab Shihata AM, Mrak EM. The fate of yeast in the digestive tract of Drosophila. Am Nat. 1951:85, 381–383. [Google Scholar]
- Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Okada S, Cai G, Zhou B, Bi E. Myosin‑II heavy chain and formin mediate the targeting of myosin essential light chain to the division site before and during cytokinesis. Mol Biol Cell. 2015;26:1211–1224. doi: 10.1091/mbc.E14-09-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Booth EA, Duvalyan A, Liao EN, Thorner J. The carboxy-terminal tails of septins Cdc11 and Shs1 recruit myosin-II binding factor Bni5 to the bud neck in Saccharomyces cerevisiae. Genetics 200, 843–862. 2015 doi: 10.1534/genetics.115.176503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G, 3rd, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195:993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet A, Fitzgerald-Hayes M. Mutations in CEN3 cause aberrant chromosome segregation during meiosis in Saccharomyces cerevisiae. Genetics. 1989;121:477–489. doi: 10.1093/genetics/121.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, O’Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–42. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Taddei A, Neumann FR, Gasser SM. Methods for visualizing chromatin dynamics in living yeast. Methods Enzymol. 2004;375:345–365. doi: 10.1016/s0076-6879(03)75022-8. [DOI] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N. Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks. Genome Biol. 2007;8:R241. doi: 10.1186/gb-2007-8-11-r241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback DB, Feldberg LR. Saccharomyces cerevisiae exhibits a sporulation-specific temporal pattern of transcript accumulation. Mol Cell Biol. 1985;5:751–761. doi: 10.1128/mcb.5.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y-C, Lin Y-H, Chen H-I, Wang Y-Y, Chiou Y-W, Lin H-H, Pan H-A, Wu C-M, Su S-M, Hsu C-C, et al. SEPT12 mutations cause male infertility with defective sperm annulus. Hum Mutat. 2012;33:710–719. doi: 10.1002/humu.22028. [DOI] [PubMed] [Google Scholar]
- Larson JR, Bharucha JP, Ceaser S, Salamon J, Richardson CJ, Rivera SM, Tatchell K. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:3040–3051. doi: 10.1091/mbc.E08-02-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn RR, Magee PT. Development of the spore wall during ascospore formation in saccharomyces cerevisiae. J Cell Biol. 1970;44:688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier P, Rathfelder N, Finkbeiner MG, Taxis C, Mazza M, Le Panse S, Haguenauer-Tsapis R, Knop M. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 2007;26:1843–1852. doi: 10.1038/sj.emboj.7601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr Biol. 2008;18:1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Esposito RE, Esposito MS. Aberrant nuclear behavior at meiosis and anucleate spore formation by sporulation-deficient (SPO) mutants of Saccharomyces cerevisiae. Exp Cell Res. 1974;83:166–174. doi: 10.1016/0014-4827(74)90700-9. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart AC, Strasser K, Finkbeiner MG, Shevchenko A, Shevchenko A, Knop M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 2001;20:6946–6957. doi: 10.1093/emboj/20.24.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189:737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Neiman AM. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics. 2002;160:1439–1450. doi: 10.1093/genetics/160.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Onishi M, Pringle JR. New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol Chem. 2011;392:681–687. doi: 10.1515/BC.2011.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell. 2013;24:1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K, Wloka C, Okada S, Svitkina T, Bi E. Architecture and dynamic remodelling of the septin cytoskeleton during the cell cycle. Nat Commun. 2014;5:5698. doi: 10.1038/ncomms6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Koga T, Hirata A, Nakamura T, Asakawa H, Shimoda C, Bähler J, Wu J-Q, Takegawa K, Tachikawa H, et al. Role of septins in the orientation of forespore membrane extension during sporulation in fission yeast. Mol Cell Biol. 2010;30:2057–2074. doi: 10.1128/MCB.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsarac N, Bhattacharyya M, Dawes IW, Clancy MJ. The SPR3 gene encodes a sporulation-specific homologue of the yeast CDC3/10/11/12 family of bud neck microfilaments and is regulated by ABFI. Gene. 1995;164:157–162. doi: 10.1016/0378-1119(95)00438-c. [DOI] [PubMed] [Google Scholar]
- Pablo-Hernando ME, Arnaiz-Pita Y, Tachikawa H, del Rey F, Neiman AM, Vázquez de Aldana CR. Septins localize to microtubules during nutritional limitation in Saccharomyces cerevisiae. BMC Cell Biol. 2008;9:55. doi: 10.1186/1471-2121-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Malmberg RL, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol. 2003;23:4814–4825. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Tóth A, Gálová M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol. 2007;17:R81–R83. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Mazza M, Maier P, Körner R, Knop M. Differential requirement for phospholipase D/Spo14 and its novel interactor Sma1 for regulation of exocytotic vesicle fusion in yeast meiosis. J Biol Chem. 2005;280:37846–37852. doi: 10.1074/jbc.M504244200. [DOI] [PubMed] [Google Scholar]
- Rij NJWK-V. Electron microscopy of germinating ascospores of Saccharomyces cerevisiae. Arch Microbiol. 1978;117:73–77. doi: 10.1007/BF00689354. [DOI] [PubMed] [Google Scholar]
- Roh D-H, Bowers B, Schmidt M, Cabib E. The septation apparatus, an autonomous system in budding yeast. Mol Biol Cell. 2002;13:2747–2759. doi: 10.1091/mbc.E02-03-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Smits GJ, van den Ende H, Klis FM. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras J-L, Calabretta A, Di Paola M, De Filippo C, Viola R, Capretti P, Polsinelli M, Turillazzi S, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci USA. 2012;109:13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y, Rodriguez RK, Coluccio AE, Neiman AM. A screen for spore wall permeability mutants identifies a secreted protease required for proper spore wall assembly. PLoS One. 2009;4:e7184. doi: 10.1371/journal.pone.0007184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar P, Bhatia A, Ramdas S, Diao L, Bhanot G, Sinha H. Sporulation genes associated with sporulation efficiency in natural isolates of yeast. PLoS One. 2013;8:e69765. doi: 10.1371/journal.pone.0069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff JE, Klapholz S, Esposito RE. Meiosis in haploid yeast. Proc Natl Acad Sci USA. 1982;79:2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanless AG, Lin Y, Weiss EL. Cell morphogenesis proteins are translationally controlled through UTRs by the Ndr/LATS target Ssd1. PLoS One. 2014;9:e85212. doi: 10.1371/journal.pone.0085212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauss M, Bi E. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392:813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.