Abstract
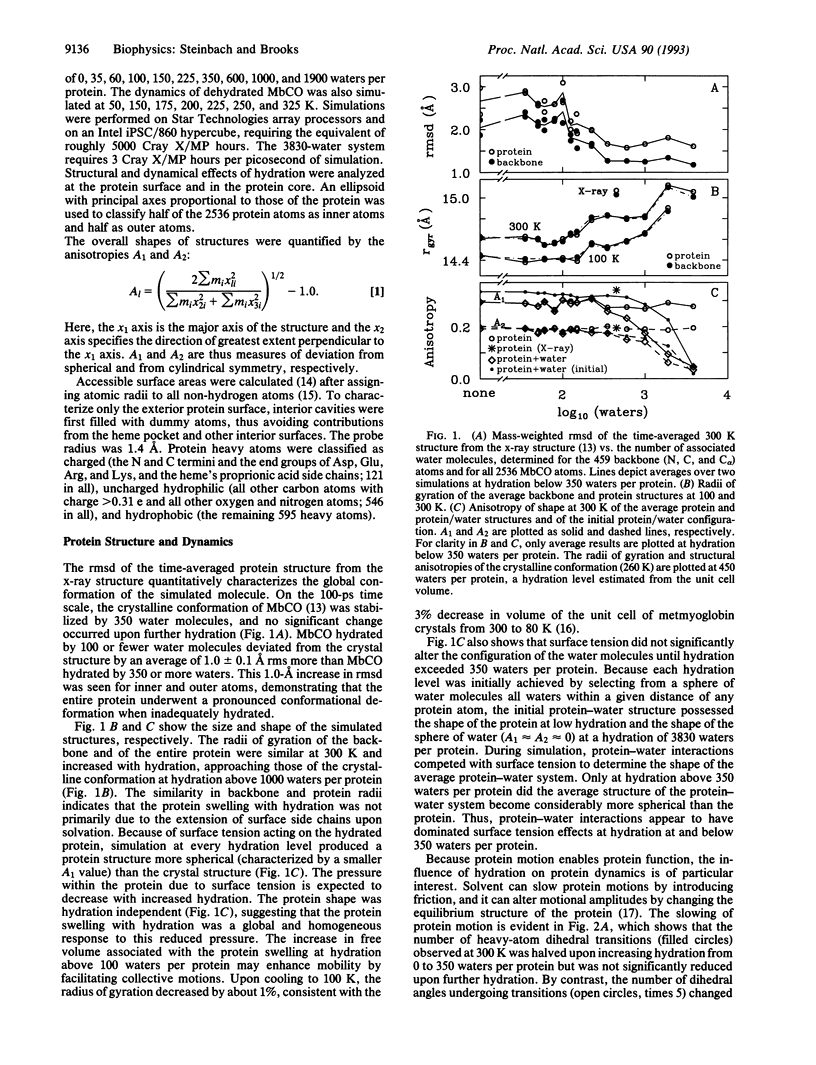
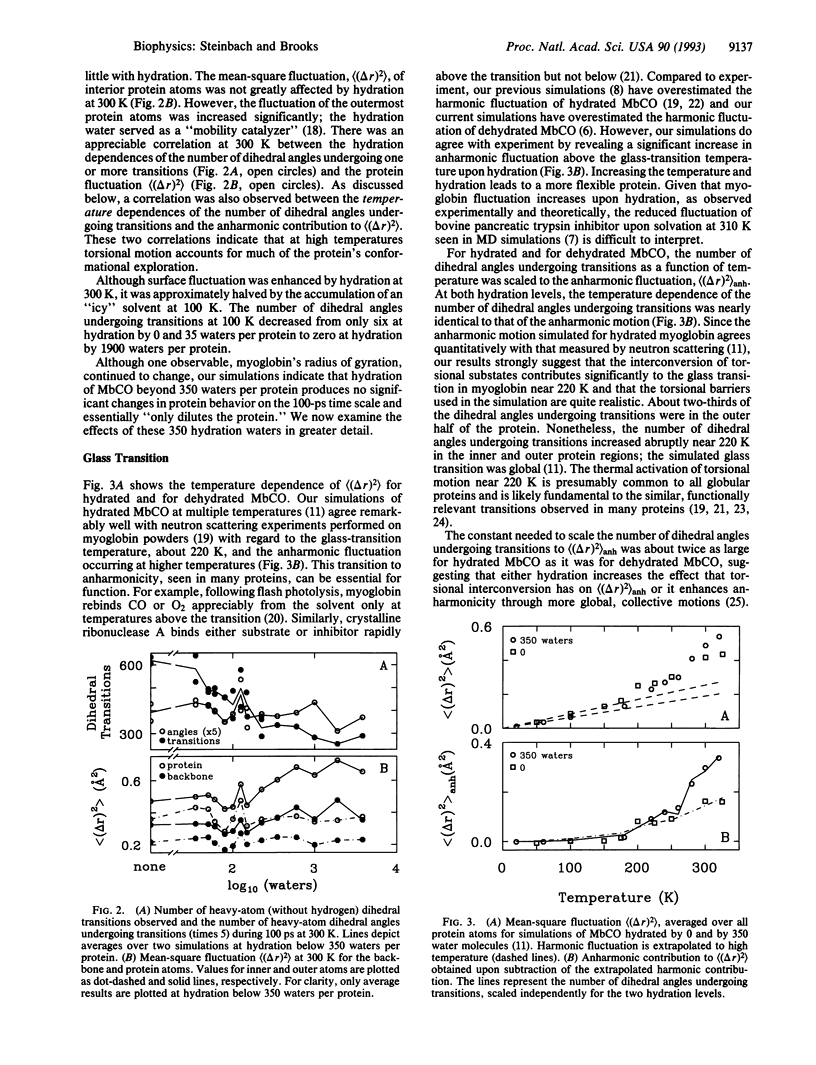
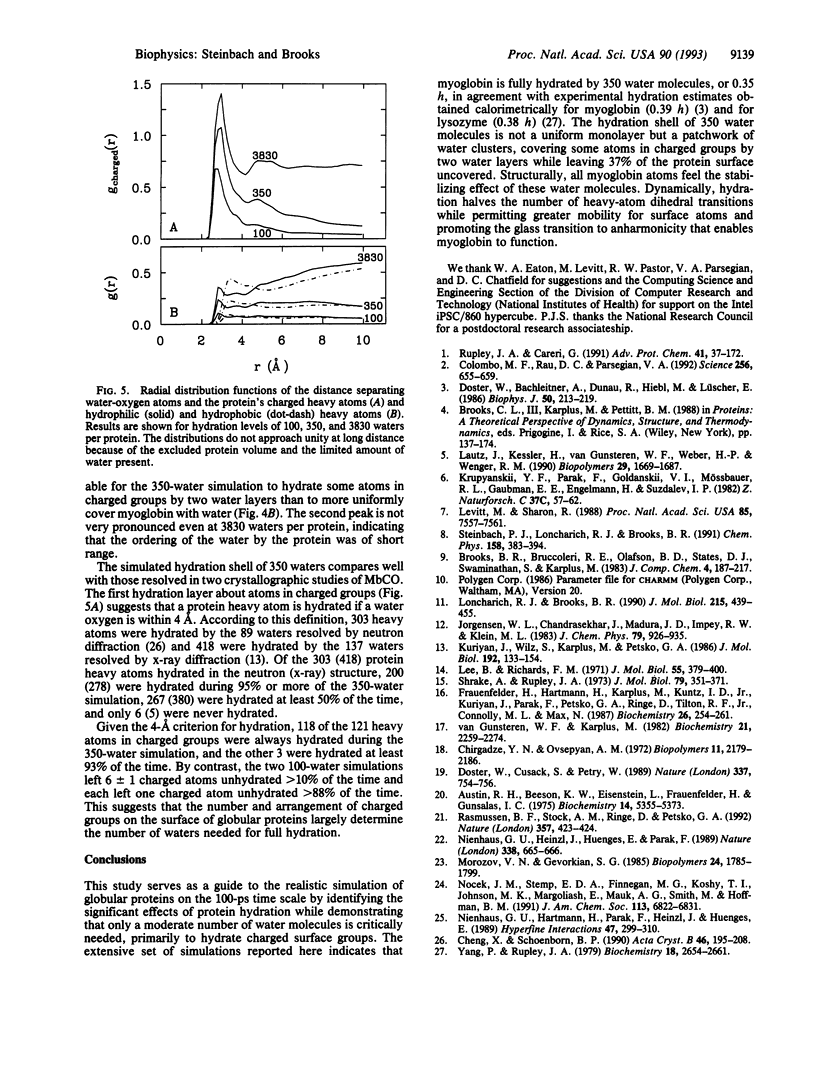
Molecular dynamics (MD) simulation covering a wide range of hydration indicate that myoglobin is fully hydrated by 350 water molecules, in agreement with experiment. These waters, originally placed uniformly about the protein, form clusters that hydrate every charged group throughout the entire simulation. Some atoms in charged groups are hydrated by two water layers while 37% of the protein surface remains uncovered. The locations of the 350 waters are consistent with those of crystallographic waters resolved by x-ray and neutron diffraction. Hydration by 350 waters at 300 K stabilizes the conformation of carboxymyoglobin measured by x-ray diffraction throughout the entire protein, halves the rate of torsional transitions, and promotes alternative conformations for surface atoms. The glass transition observed experimentally in hydrated myoglobin near 220 K is also seen in the simulations and correlates with an increase in the number of dihedral angles undergoing transitions. The anharmonic protein motion above 220 K is enhanced by protein hydration.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Dynamics of ligand binding to myoglobin. Biochemistry. 1975 Dec 2;14(24):5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- Chirgadze Y. N., Ovsepyan A. M. Hydration mobility in peptide structures. Biopolymers. 1972;11(10):2179–2186. doi: 10.1002/bip.1972.360111017. [DOI] [PubMed] [Google Scholar]
- Colombo M. F., Rau D. C., Parsegian V. A. Protein solvation in allosteric regulation: a water effect on hemoglobin. Science. 1992 May 1;256(5057):655–659. doi: 10.1126/science.1585178. [DOI] [PubMed] [Google Scholar]
- Doster W., Bachleitner A., Dunau R., Hiebl M., Lüscher E. Thermal properties of water in myoglobin crystals and solutions at subzero temperatures. Biophys J. 1986 Aug;50(2):213–219. doi: 10.1016/S0006-3495(86)83455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doster W., Cusack S., Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989 Feb 23;337(6209):754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Hartmann H., Karplus M., Kuntz I. D., Jr, Kuriyan J., Parak F., Petsko G. A., Ringe D., Tilton R. F., Jr, Connolly M. L. Thermal expansion of a protein. Biochemistry. 1987 Jan 13;26(1):254–261. doi: 10.1021/bi00375a035. [DOI] [PubMed] [Google Scholar]
- Krupyanskii YuF, Parak F., Goldanskii V. I., Mössbauer R. L., Gaubman E. E., Engelmann H., Suzdalev I. P. Investigation of large intramolecular movement within metmyoglobin by Rayleigh scattering of Mössbauer radiation (RSMR). Z Naturforsch C. 1982 Jan-Feb;37(1-2):57–62. doi: 10.1515/znc-1982-1-211. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Wilz S., Karplus M., Petsko G. A. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 A resolution. J Mol Biol. 1986 Nov 5;192(1):133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M., Sharon R. Accurate simulation of protein dynamics in solution. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7557–7561. doi: 10.1073/pnas.85.20.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncharich R. J., Brooks B. R. Temperature dependence of dynamics of hydrated myoglobin. Comparison of force field calculations with neutron scattering data. J Mol Biol. 1990 Oct 5;215(3):439–455. doi: 10.1016/s0022-2836(05)80363-8. [DOI] [PubMed] [Google Scholar]
- Rasmussen B. F., Stock A. M., Ringe D., Petsko G. A. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature. 1992 Jun 4;357(6377):423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- Rupley J. A., Careri G. Protein hydration and function. Adv Protein Chem. 1991;41:37–172. doi: 10.1016/s0065-3233(08)60197-7. [DOI] [PubMed] [Google Scholar]
- Shrake A., Rupley J. A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol. 1973 Sep 15;79(2):351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Yang P. H., Rupley J. A. Protein--water interactions. Heat capacity of the lysozyme--water system. Biochemistry. 1979 Jun 12;18(12):2654–2661. doi: 10.1021/bi00579a035. [DOI] [PubMed] [Google Scholar]
- van Gunsteren W. F., Karplus M. Protein dynamics in solution and in a crystalline environment: a molecular dynamics study. Biochemistry. 1982 May 11;21(10):2259–2274. doi: 10.1021/bi00539a001. [DOI] [PubMed] [Google Scholar]



