Normal epithelial cells have an ability to sense and actively eliminate neighboring transformed cells, a process called epithelial defense against cancer (EDAC). Exogenous S1P plays a crucial role in EDAC; the S1P–S1PR2 pathway regulates Rho–Rho kinase–filamin in the surrounding normal cells, promoting apical extrusion of RasV12-transformed cells from epithelia.
Abstract
At the initial step of carcinogenesis, transformation occurs in single cells within epithelia, where the newly emerging transformed cells are surrounded by normal epithelial cells. A recent study revealed that normal epithelial cells have an ability to sense and actively eliminate the neighboring transformed cells, a process named epithelial defense against cancer (EDAC). However, the molecular mechanism of this tumor-suppressive activity is largely unknown. In this study, we investigated a role for the sphingosine-1-phosphate (S1P)–S1P receptor 2 (S1PR2) pathway in EDAC. First, we show that addition of the S1PR2 inhibitor significantly suppresses apical extrusion of RasV12-transformed cells that are surrounded by normal cells. In addition, knockdown of S1PR2 in normal cells induces the same effect, indicating that S1PR2 in the surrounding normal cells plays a positive role in the apical elimination of the transformed cells. Of importance, not endogenous S1P but exogenous S1P is involved in this process. By using FRET analyses, we demonstrate that S1PR2 mediates Rho activation in normal cells neighboring RasV12-transformed cells, thereby promoting accumulation of filamin, a crucial regulator of EDAC. Collectively these data indicate that S1P is a key extrinsic factor that affects the outcome of cell competition between normal and transformed epithelial cells.
INTRODUCTION
At the initial stage of carcinogenesis, it is generally believed that oncogenic transformation occurs in single cells within epithelia. However, it is not clearly understood what happens at the interface between normal epithelial cells and newly emerging transformed cells. In previous studies, we demonstrated that RasV12- or Src-transformed cells are apically extruded when they are surrounded by normal epithelial cells. When transformed cells alone are present, apical extrusion does not occur, indicating that the presence of neighboring normal cells profoundly influences the behavior of the transformed cells (Hogan et al., 2009; Kajita et al., 2010). In addition, it has become clear that normal epithelial cells have an ability to actively eliminate transformed cells from the epithelium (Kajita et al., 2014). When normal and Ras- or Src-transformed cells are copresent within the epithelial monolayer, filamin and vimentin are accumulated in normal cells at the interface with the neighboring transformed cells. Knockdown of filamin or vimentin suppresses apical extrusion of transformed cells, indicating an active role of these proteins in this process. Filamin acts upstream of vimentin and regulates its dynamic accumulation, which produces physical forces for the apical extrusion. Furthermore, we show that the Rho–Rho kinase pathway regulates the accumulation of filamin. Collectively these results indicate that normal epithelial cells have antitumor activity that does not involve immune systems. This process is termed epithelial defense against cancer (EDAC; Kajita et al., 2014). However, the molecular mechanism of how Rho–Rho kinase–filamin is regulated at the interface between normal and transformed cells remains to be elucidated.
Sphingosine-1-phosphate (S1P) is a lipid mediator involved in the regulation of various cellular processes, such as cell proliferation, cell migration, actin cytoskeletal reorganization, and cell adhesion (Kihara et al., 2007; Blaho and Hla, 2014). S1P is secreted from a variety of cell types and binds to its cognate receptors S1P receptors (S1PRs) 1–5 (Blaho and Hla, 2014; Nishi et al., 2014). Gu et al. (2011) showed that S1P-S1PR2 is involved in apical extrusion of apoptotic cells from the epithelial monolayer. At the early phase of apoptosis, dying cells produce S1P, and the secreted S1P binds to S1PR2 in the surrounding normal cells. Then S1PR2 activates the downstream Rho–Rho kinase pathway, leading to the formation of actin–myosin rings that squeeze out apoptotic cells.
In this study, we examined whether the S1P–S1PR2 pathway is also involved in the elimination of transformed cells from the epithelium. Unexpectedly, not endogenous S1P but exogenous S1P plays a major role in this process. S1P–S1PR2 regulates Rho–Rho kinase–filamin in surrounding normal epithelial cells, mediating apical extrusion of RasV12-transformed cells. These data demonstrate that the S1P–S1PR2 pathway is a crucial regulator of EDAC and that cell competition can be substantially influenced by factors from the outer environment.
RESULTS
S1PR2 in the surrounding normal epithelial cells is involved in apical extrusion of RasV12-transformed cells
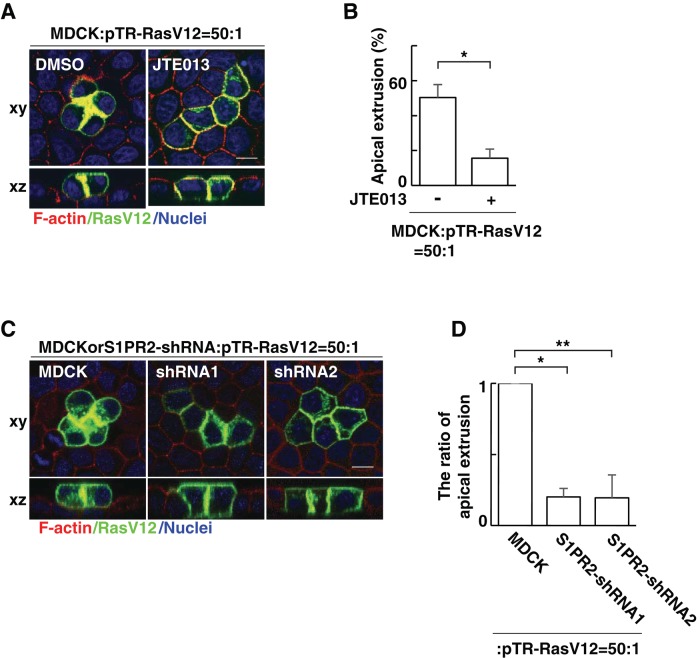
In a previous study, we reported that when Madin–Darby canine kidney (MDCK) cells transformed with human H-RasV12 are surrounded by normal MDCK cells, RasV12 cells are apically extruded from a monolayer of normal epithelial cells (Hogan et al., 2009). To examine the involvement of the S1P–S1PR2 pathway in this process, we analyzed the effect of JTE013, a specific inhibitor of S1PR2. We found that addition of JTE013 profoundly suppressed apical extrusion of RasV12 cells (Figure 1, A and B). According to the previous report, S1PR2 in the surrounding cells is involved in apical extrusion of apoptotic cells (Gu et al., 2011). Therefore we established MDCK cells stably expressing S1PR2 short hairpin RNA (shRNA; Supplemental Figure S1) and examined whether S1PR2 in the surrounding cells is also involved in apical extrusion of transformed cells. Indeed, we found that knockdown of S1PR2 in the surrounding cells significantly suppressed apical extrusion of RasV12 cells (Figure 1, C and D). Collectively these data indicate that S1PR2 in the surrounding normal cells plays a positive role in apical extrusion of RasV12-transformed cells.
FIGURE 1:
S1PR2 in the surrounding normal cells plays a positive role in the apical extrusion of RasV12-transformed cells. (A) Confocal microscopic immunofluorescence images of xy- and xz-sections of MDCK-pTR GFP-RasV12 cells in a monolayer of normal MDCK cells cultured in the absence or presence of JTE013. Twenty-four hours after tetracycline addition, cells were stained with phalloidin (red) and Hoechst (blue). (B) Quantification of the apical extrusion of RasV12 cells. Data are mean ± SD from three independent experiments. *p = 0.0027. (C) Confocal microscopic immunofluorescence images of xy- and xz-sections of MDCK-pTR GFP-RasV12 cells in a monolayer of normal MDCK cells or S1PR2-knockdown MDCK cells. Cells were stained with phalloidin (red) and Hoechst (blue). Scale bars, 10 μm (A, C). (D) Quantification of the apical extrusion of RasV12 cells. Data are mean ± SD from three independent experiments. Values are expressed as a ratio relative to MDCK cells. *p = 2.2 × 10−5, **p = 0.0010.
S1P produced by RasV12-transformed cells or the surrounding normal cells does not play an active role in apical extrusion
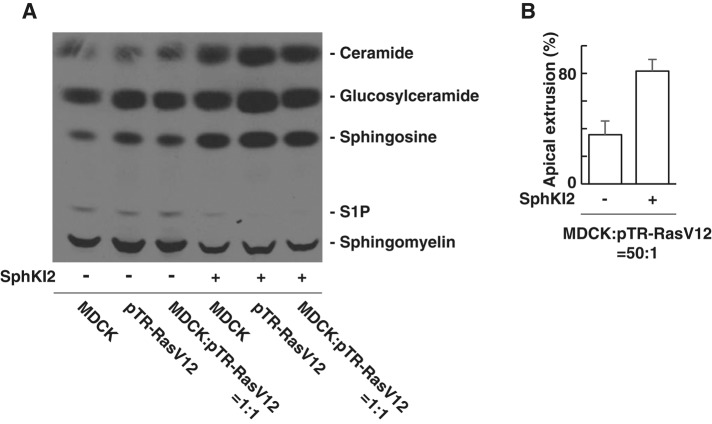
S1P expressed by apoptotic cells or RasV12-transformed cells has been reported to be an important regulator for the elimination of those cells from the epithelium (Gu et al., 2011; Slattum et al., 2014). Sphingosine kinase inhibitor 2 (SphKI2) is a specific inhibitor for sphingosine kinase 1 that catalyzes the production of S1P from sphingosine. We showed that treatment of SphKI2 strongly suppressed production of S1P from normal or RasV12-transformed MDCK cells (Figure 2A). Unexpectedly, addition of SphKI2 did not suppress, but even promoted, apical extrusion of RasV12-transformed cells (Figure 2B). These results suggest that S1P produced by RasV12-transformed cells or the surrounding normal cells does not play an active role in apical extrusion.
FIGURE 2:
S1P produced by RasV12-transformed cells or the surrounding normal cells does not play a positive role in apical extrusion. (A) TLC of sphingolipids extracted from [3H]sphingosine-labeled MDCK cells, MDCK-pTR GFP-RasV12 cells, or the mixture of MDCK and MDCK-pTR GFP-RasV12 cells cultured in the absence or presence of SphKI2. (B) Quantification of the apical extrusion of RasV12 cells. The relatively low frequency of apical extrusion in the absence of SphKI2 may be due to high concentration of dimethyl sulfoxide (0.16%). Data are mean ± SD from three independent experiments.
S1P from the outer environment positively regulates apical extrusion of RasV12-transformed cells
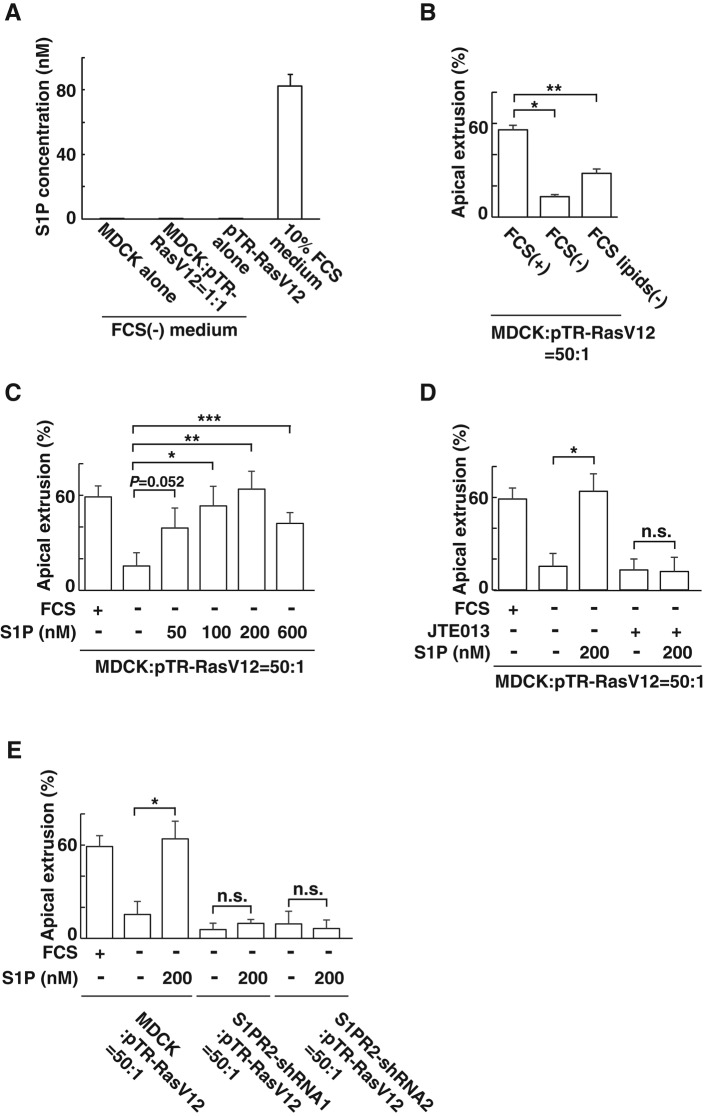
S1P is produced not only from epithelial cells but also from a variety of cell types, including endothelial cells and erythrocytes (Kihara and Igarashi, 2008; Blaho and Hla, 2014). Fetal calf serum (FCS) has been reported to contain a substantial amount of S1P (Edsall and Spiegel, 1999). Indeed, mass spectrometric analysis demonstrated that the S1P concentration in medium containing FCS (∼80 nM) was much higher than that secreted from normal or RasV12-transformed cells (∼0.15 nM; Figure 3A). We then found that apical extrusion of RasV12 cells was profoundly suppressed when the mixture of normal and RasV12 cells was incubated in the absence of FCS (Figure 3B). Furthermore, incubation with FCS depleted of lipids significantly suppressed the apical extrusion (Figure 3B), suggesting a positive role of lipids contained in FCS in this process. Next we examined the effect of exogenously added S1P on apical extrusion of RasV12 cells. When S1P was added in the medium that did not contain FCS, apical extrusion was restored in a dose-dependent manner, with the maximum effect at 200 nM (Figure 3C), suggesting a positive role for exogenous S1P in the apical extrusion. In the presence of JTE013, S1P did not promote apical extrusion (Figure 3D). Furthermore, when RasV12 cells were surrounded by S1PR2-knockdown cells, addition of S1P did not induce apical extrusion of RasV12 cells (Figure 3E). These data suggest that exogenous S1P actively regulates apical extrusion of RasV12-transformed cells via S1PR2 in the surrounding normal cells. In contrast to apical extrusion, basal extrusion of RasV12-transformed cells did not frequently occur (<5%), and neither JTE013 treatment nor S1PR2 knockdown substantially affected the frequency of basal extrusion (unpublished data). Sphingosylphosphorylcholine (SPC) is another ligand for S1PR2 (Gonda et al., 1999). We found that SPC also promoted apical extrusion of RasV12 cells and that the effect of SPC was not observed when JTE013 was added or RasV12 cells were surrounded by S1PR2-knockdown cells (Supplemental Figure S2). The amount of SPC required for induction of apical extrusion was larger than that of S1P (Figure 3C and Supplemental Figure S2), consistent with the fact that the affinity of SPC–S1PR2 is lower than that of S1P–S1PR2 (Gonda et al., 1999). We also showed by quantitative real-time PCR analyses that expression of S1PR2 or the S1P transporter Spns2 was not specifically altered in normal or RasV12-transformed cells under the mixed culture condition (Supplemental Figure S3).
FIGURE 3:
S1P from the outer environment positively regulates the apical extrusion of RasV12-transformed cells. (A) Measurement of endogenously secreted or exogenous S1P by mass spectrometry. MDCK cells and MDCK-pTR GFP-RasV12 cells were cultured alone or cocultured in the absence of FCS. The S1P concentration in the conditioned medium from each condition is compared with that in the 10% FCS–containing medium by mass spectrometry. Data are mean ± SD from three independent experiments. (B) Effect of depletion of lipids from FCS on the apical extrusion of RasV12 cells surrounded by normal MDCK cells. Data are mean ± SD from two independent experiments. *p = 0.0027, **p = 0.010. (C) Effect of exogenously added S1P on the apical extrusion of RasV12 cells surrounded by normal MDCK cells. Data are mean ± SD from three independent experiments. *p = 0.012, **p = 0.0039, ***p = 0.012. (D) Effect of exogenously added S1P in the absence or presence of JTE013 on the apical extrusion of RasV12 cells surrounded by normal MDCK cells. Data are mean ± SD from three independent experiments. *p = 0.0039. (E) Effect of exogenously added S1P on the apical extrusion of RasV12 cells surrounded by normal MDCK cells or S1PR2-knockdown MDCK cells. Data are mean ± SD from three independent experiments. *p = 0.0039. n.s., not significant (D, E).
The S1P–S1PR2 pathway acts upstream of Rho–Rho kinase–filamin in EDAC
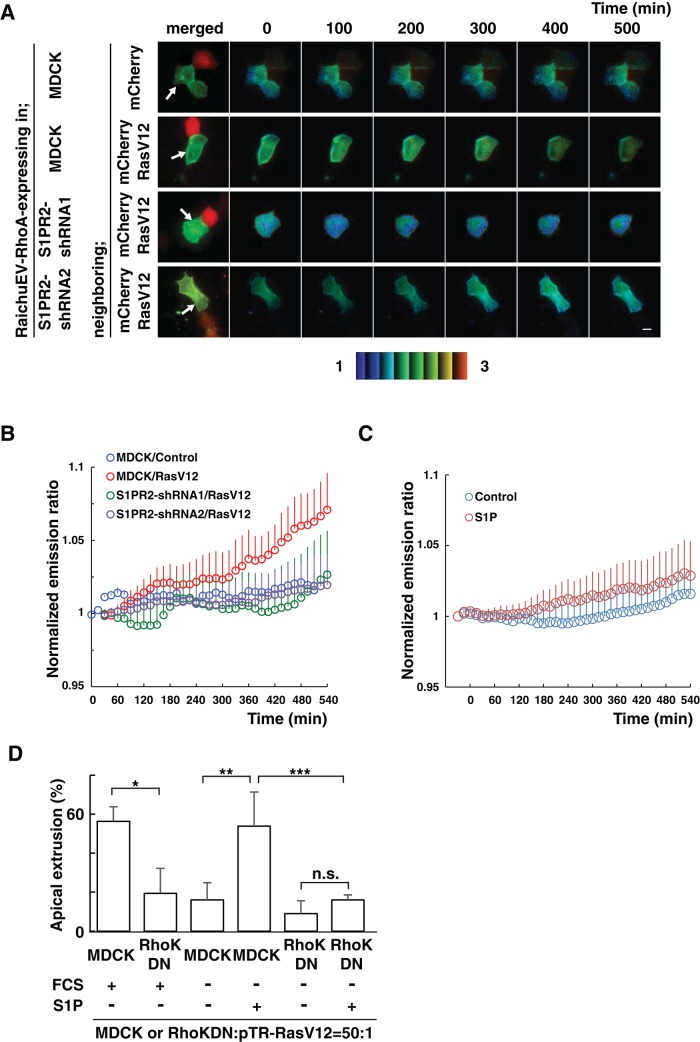
In a previous study, we reported that filamin is accumulated in the surrounding normal cells at the interface with RasV12-tranformed cells and positively regulates apical extrusion. In addition, Rho–Rho kinase functions upstream of filamin in this process (Kajita et al., 2014). It was also reported that binding of S1P to S1PR2 leads to activation of the Rho–Rho kinase pathway (Lepley et al., 2005). Therefore we investigated the relationship between the S1P–S1PR2 and Rho–Rho kinase–filamin pathways. Using the Raichu–fluorescence resonance energy transfer (FRET) system, we first examined the activity of Rho. In normal cells neighboring RasV12/mCherry-expressing cells, activity of Rho gradually increased (Figure 4, A and B). In contrast, Rho activity did not change in cells that were adjacent to cells expressing mCherry alone (Figure 4, A and B), indicating the specific activation of Rho in normal cells abutting the transformed cells. We then found that the activation of Rho did not frequently occur in S1PR2-knockdown cells neighboring RasV12/mCherry-expressing cells (Figure 4, A and B). These data suggest that S1PR2 regulates the activation of Rho in normal cells neighboring RasV12-transformed cells. When normal cells were treated with S1P, Rho activity was slightly elevated (Figure 4C). However, the extent of S1P-mediated Rho activation was less than that of Rho activation in normal cells neighboring RasV12-transformed cells (Figure 4, B and C), suggesting the presence of additional signaling pathways that function together with S1P–S1PR2. When RasV12 cells were surrounded by cells expressing the Rho kinase dominant-negative mutant, apical extrusion was significantly suppressed (Figure 4D; Kajita et al., 2014). We also showed that addition of S1P did not promote apical extrusion of RasV12 cells when they were surrounded by Rho kinase dominant–negative–expressing cells (Figure 4D), suggesting that S1P-mediated apical extrusion requires the activity of Rho kinase in the surrounding cells.
FIGURE 4:
The S1P–S1PR2 pathway acts upstream of Rho–Rho kinase in normal cells neighboring RasV12-transformed cells. (A) MDCK cells or S1PR2-knockdown MDCK cells were transfected with an expression vector for the FRET-based biosensor RaichuEV-RhoA. The cells were further transfected with an expression vector for mCherry alone or together with that for RasV12 and then seeded on the collagen gel. After 12 h, the sample was screened under a fluorescence microscope for a pair of adjacent cells, one of which expressed RaichuEV-RhoA and the other mCherry/RasV12. The Raichu-expressing cells were then monitored by dual-emission fluorescence microscopy. FRET/CFP ratio images were generated to represent FRET efficiency. Representative images at the indicated time points. The upper and lower limits of the ratio range are indicated at the bottom. Arrows indicate Raichu-expressing cells neighboring mCherry-expressing cells. Scale bar, 10 μm. (B, C) Traces for three-point moving average of the FRET/CFP emission ratio. (B) Quantification of A. p = 5.2 × 10−5 between MDCK/control and MDCK/RasV12; p = 4.0 × 10−8 between MDCK/RasV12 and S1PR2-shRNA1/RasV12; p = 2.0 × 10−6 between MDCK/RasV12 and S1PR2-shRNA2/RasV12. n = 7, MDCK/control; n = 5, MDCK/RasV12; n = 8, S1PR2-shRNA1/RasV12; and n = 8, S1PR2-shRNA2/RasV12. (C) Effect of S1P on the Rho activity in normal cells. p = 1.7 × 10−9 between MDCK/control and MDCK/RasV12. n = 6, control (fatty acid–free BSA); and n = 11, 200 nM S1P. Error bars indicate SEM. Time 0 min is set when the microscopic observations start (A, B) or S1P is added (C). (D) Effect of exogenously added S1P on the apical extrusion of RasV12 cells surrounded by normal MDCK cells or MDCK cells expressing the Rho kinase dominant-negative mutant. Data are mean ± SD from three (first, second, fifth, and sixth bars) or six (third and fourth bars) independent experiments. *p = 0.013, **p = 0.00082, ***p = 0.0088. n.s., not significant.
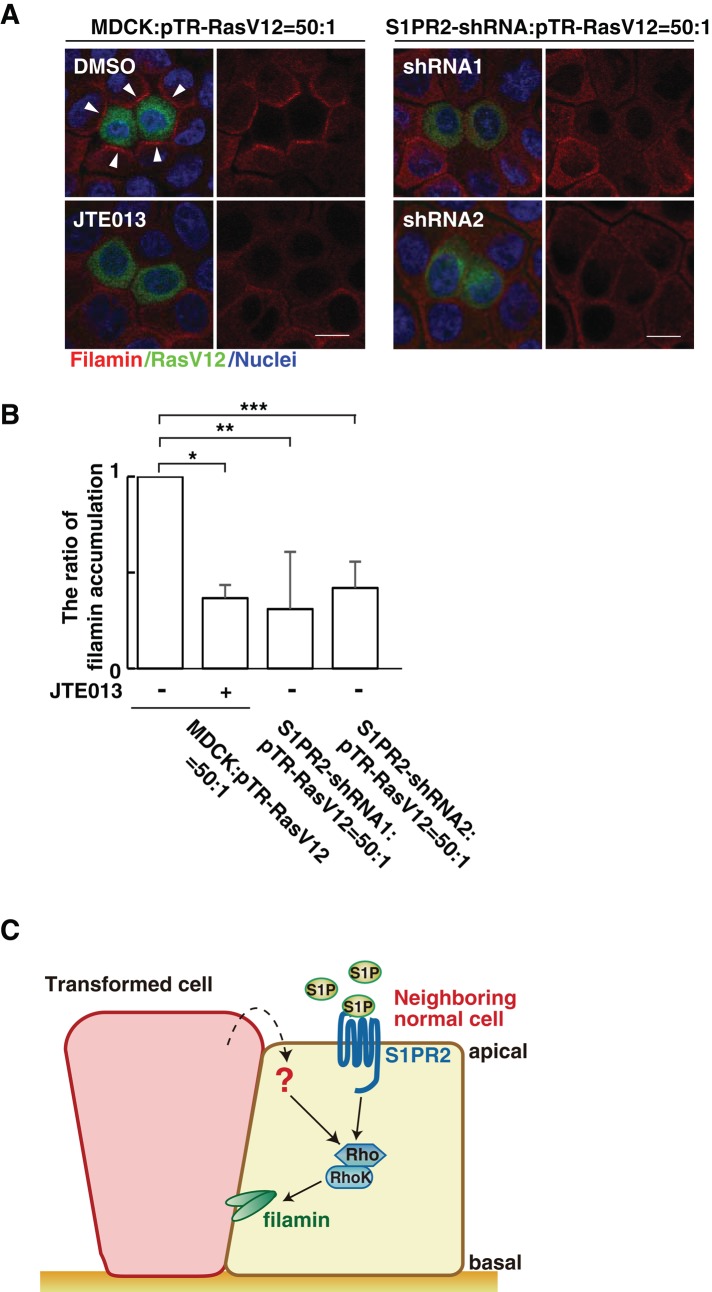
Furthermore, we showed that incubation with JTE013 substantially suppressed filamin accumulation between normal and RasV12 cells (Figure 5, A and B). Similarly, when RasV12 cells were surrounded by S1PR2-knockdown cells, the ratio of filamin accumulation was decreased (Figure 5, A and B). Collectively these results suggest that the S1P–S1PR2 pathway acts upstream of Rho–Rho kinase–filamin in the elimination of transformed cells from the epithelium.
FIGURE 5:
The S1P–S1PR2 pathway regulates filamin accumulation. (A) Confocal microscopic immunofluorescence images of filamin (red) in normal MDCK cells or S1PR2-knockdown MDCK cells at the interface with MDCK-pTR GFP-RasV12 cells. Arrowheads indicate filamin accumulation. Left, bottom, mixture of normal and RasV12 cells incubated with JTE013. Scale bars, 10 μm. (B) Quantification of the filamin accumulation by confocal microscopic analyses. Several confocal xy-section images were examined for the presence of filamin accumulation. Data are mean ± SD from three independent experiments. Values are expressed as a ratio relative to MDCK cells: pTR-RasV12 = 50:1 (JTE013 –). *p = 8.8 × 10−5, **p = 0.016, ***p = 0.0019. (C) Model for the functional role of S1P–S1PR2 in the interaction between normal and transformed epithelial cells. 1) S1P from the outer environment binds to S1PR2 in the surrounding normal cells, which 2) leads to activation of Rho; then 3) Rho–Rho kinase mediates filamin accumulation, resulting in the apical extrusion of the neighboring transformed cells.
DISCUSSION
Based on data obtained in this study, our current model for the involvement of S1P in apical extrusion of RasV12-transformed cells is demonstrated in Figure 5C. S1P from the outer environment binds to S1PR2 in the surrounding normal cells, affecting the activity of Rho. The Rho–Rho kinase pathway plays a positive role in the accumulation of filamin at the interface with transformed cells, which eventually results in the apical extrusion of the transformed cells. Collectively all this implies that S1P is a key extrinsic factor of EDAC; this is the first study demonstrating that an extrinsic factor can profoundly affect the competitive interaction between normal and transformed epithelial cells. Addition of exogenous S1P slightly activates Rho in normal cells (Figure 4C) but to a lesser extent than with the Rho activity in normal cells surrounding RasV12 cells, suggesting that there are additional molecular mechanisms from the neighboring transformed cells that mediate Rho activation in normal cells (a red question mark in Figure 5C) together with the S1P–S1PR2 pathway. Molecules and/or signaling pathways involving those mechanisms should be identified in future studies.
We and other groups have demonstrated that RasV12-transformed cells are frequently extruded from the apical surface of the normal epithelial monolayer (Hogan et al., 2009; Wu et al., 2014). In contrast, Slattum et al. (2014) showed that RasV12-transformed cells are basally extruded when cocultured with normal epithelial cells. This apparent discrepancy may result from the different experimental conditions. We used an inducible system in which RasV12-expression is induced in a mosaic manner within a monolayer of normal epithelial cells that form tight cell–cell adhesions, which is believed to reflect the events occurring at the initial stage of carcinogenesis. In contrast, Slattum et al. (2014) used a noninducible transformed cell system with ultraviolet irradiation in which RasV12-expressing cells were mixed and cocultured with normal epithelial cells. Future studies should examine whether apical or basal extrusion of RasV12-transformed cells predominantly occurs under physiological conditions, using mouse in vivo model systems (Yamauchi et al., 2015). Slattum et al. (2014) also showed that S1P secreted from RasV12-transformed cells binds to S1PR2 in neighboring normal cells, causing basal delamination of RasV12 cells. Unexpectedly, we found that the blockage of endogenous S1P production does not suppress apical extrusion of RasV12 cells (Figure 2). Instead, apical extrusion is strongly suppressed by depleting FCS in the culture medium of lipids, which is rescued by addition of exogenous S1P (Figure 3, B and C). Moreover, mass spectrometric analysis demonstrates that the amount of S1P in culture medium containing 10% FCS is much higher than that secreted from normal or RasV12-transformed cells (Figure 3A). Collectively these data strongly suggest that exogenous S1P, not endogenous S1P, is mainly involved in apical extrusion, although the possibility cannot be completely ruled out that endogenously secreted S1P may have a role by modulating its local concentration around the cells.
Apical extrusion of transformed cells from epithelia occurs in a stochastic manner, and the frequency of this event can be influenced by various factors. Previous studies revealed that at the interface between normal and transformed epithelial cells, expression and/or activity of various molecules are modulated within both cells, which profoundly regulates the occurrence of apical extrusion. Our results suggest that not only intrinsic factors but also extrinsic factors can influence the outcome of the cell competition between normal and transformed cells. S1P is expressed by a variety of cell types, including endothelial and immune cells. Thus it is likely that within tissues containing higher concentration of S1P, elimination of transformed cells is actively promoted. In contrast, under pathological conditions in which S1P production from the surrounding cells is reduced, EDAC is weakened, leading to cancer progression. In future studies, the functional role of S1P in cell competition needs to be examined in vivo. In addition, other environmental cell competition–regulating factors should be further explored.
MATERIALS AND METHODS
Antibodies and materials
Mouse anti-filamin (F6682) antibody was from Sigma-Aldrich (Gillingham, United Kingdom). Alexa Fluor 568–conjugated phalloidin (Life Technologies, Paisley, United Kingdom) was used at 1.0 U/ml. Alexa Fluor 568– and 647–conjugated secondary antibodies were from Life Technologies. Hoechst 33342 (Life Technologies) was used at a dilution of 1:5000. For immunofluorescence, primary antibody was used at 1:100, and all secondary antibodies were used at 1:200. Type I collagen (Cellmatrix Type I-A) was obtained from Nitta Gelatin (Osaka, Japan) and neutralized on ice to a final concentration of 2 mg/ml according to the manufacturer’s instructions. S1P, SPC, and bovine serum albumin (BSA) were from Sigma-Aldrich.
Cell culture
MDCK cells, MDCK-pTR GFP-human H-RasV12 cells, and MDCK cells stably expressing the Rho kinase dominant-negative mutant were cultured as previously described (Hogan et al., 2009; Kajita et al., 2014). S1PR2 shRNA oligonucleotides (S1PR2-shRNA1: 5′-GATCCCCGCAAGTTCCACTCAGCCATGTTTCAAGAGAACATGGCTGAGTGGAACTTGCTTTTTC-3′ and 5′-TCGAGAAAAAGCAAGTTCCACTCAGCCATGTTCTCTTGAAACATGGCTGAGTGGAACTTGCGGG-3′; or S1PR2-shRNA2: 5′-GATCCCCGCTCAAGACGGTCACCATTGTTTCAAGAGAACAATGGTGACCGTCTTGAGCTTTTTC-3′ and 5′-TCGAGAAAAAGCTCAAGACGGTCACCATTGTTCTCTTGAAACAATGGTGACCGTCTTGAGCGGG-3′) were cloned into the BglII and XhoI sites of pSUPERIOR.neo + gfp (Oligoengine, Seattle, WA). MDCK cells were transfected with pSUPERIOR.neo + gfp S1PR2-shRNA using Lipofectamine 2000 (Life Technologies), followed by selection in the medium containing 800 μg/ml G418 (Life Technologies). For tetracycline-inducible MDCK cell lines, 2 μg/ml tetracycline (Sigma-Aldrich) was used to induce expression of RasV12. All inhibitors were added at the beginning of the 24-h incubation, except for Figure 5, A and B, for which JTE013 was added for 16 h. JTE013 (20 μM) was from TOCRIS Bioscience (Ellisville, MO), and SphKI2 (80 μM) was from Cayman Chemical (Ann Arbor, MI). To remove lipids from FCS, 100 ml of FCS was incubated with activated charcoal (10 g; Sigma-Aldrich) at 4°C overnight. After centrifugation (13,000 × g for 40 min), the supernatant was filtered through a 0.22-μm polyethersulfone filter (Corning, New York, NY). S1P was dissolved in ethanol to make 1 mM stock solution. S1P was mixed with fatty acid-free BSA (40 μg BSA per 100 pmol S1P) and added to the FCS-free culture medium.
Immunofluorescence
For immunofluorescence, MDCK-pTR GFP-RasV12 cells were mixed with MDCK cells at a ratio of 1:50 and cultured on the collagen matrix as previously described (Hogan et al., 2009). The mixture of cells was incubated for 8–12 h, followed by tetracycline treatment for 16 h for analysis of filamin accumulation or for 24 h for analysis of apical extrusion. Cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline, except that for Figure 5, A and B, cells were fixed in methanol for 2.5 min at −20°C. After PFA fixation, cells were permeabilized as previously described (Kajita et al., 2010). Immunofluorescence images were analyzed by confocal microscopy, using the Olympus FV1000 system and Olympus FV10-ASW software (Olympus, Tokyo, Japan).
[3H]Sphingosine labeling assay
Sphingosine labeling assay was performed as previously described (Nakahara et al., 2012). Cells were incubated for 12 h at 37°C with DMEM containing charcoal-treated 10% FCS and for another 12 h with DMEM. Cells were then labeled with 0.6 μCi of [3H]sphingosine (PerkinElmer Life Sciences, Waltham, MA) for 4 h at 37°C. Lipids were extracted, separated by normal-phase TLC on Silica Gel 60 high-performance TLC plates (Merck Millipore, Billerica, MA) with 1-butanol/acetic acid/water (3:1:1 [vol/vol]), and visualized by fluorography.
Quantitative real-time PCR
For Supplemental Figure S1, total RNA was isolated from MDCK and MDCK cells stably expressing S1PR2 shRNA using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. Isolated RNA was treated with deoxyribonuclease I, amplification grade (Life Technologies) and reverse transcribed using ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan) according to the manufacturer’s guidelines. In brief, PCR amplification of the targeted fragments was performed with 45 cycles of denaturation at 95°C (30 s) and annealing and extension at 60°C (30 s). For Supplemental Figure S3, MDCK cells, MDCK pTR GFP-RasV12 cells, or a 1:1 mix culture of MDCK and MDCK pTR GFP-RasV12 cells were cultured at 9 × 106 cells into 100-mm dishes. After incubation with tetracycline for 16 h, green fluorescent protein GFP)–negative MDCK cells and GFP-positive RasV12 cells were separated using FACSAria II (BD Biosciences, Franklin Lakes, NJ). Total RNA was extracted from isolated cells with TRIzol (Thermo Fisher Scientific, Waltham, MA), purified using RNeasy Mini Kit (Qiagen, Venlo, Netherlands), and reverse transcribed using a QuantiTect Reverse Transcription Kit (Qiagen). GeneAce SYBR qPCR Mix (Nippon Gene) was used to perform quantitative PCR using the StepOnePlus system (Thermo Fisher Scientific). For both figures, relative mRNA expression was quantified using the comparative threshold method with the content of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as internal control. The primer sequences used were as follows: GAPDH, 5′-ATTCTATCCACGGCAAATCC-3′ and 5′-GGACTCCACAACATACTCAG-3′; S1PR2, 5′-GCCTCGTGGCTCATCTCG-3′ and 5′-GGTGACCGTCTTGAGCAGG-3′; and Spns2, 5′-CTGCATCTTTGTTGGGGAGA-3′ and 5′-CCGAGATAAAGCCGATGAGG-3′.
Construction of the expression vector for RaichuEV-RhoA
The coding sequence for the Rho-binding domain (RBD) of protein kinase N was amplified by PCR, followed by subcloning into the XhoI and Asp718 sites of the pEKAREV-nes vector (Komatsu et al., 2011). The coding sequence for the N-terminus of RhoA (amino acids 1–189) was also amplified by PCR, and the resulting PCR products were digested with AgeI and NotI and inserted into the Aor13H and NotI sites of the aforementioned pEKAREV-nes–derived vector. The tandem coding sequences for RBD, EV linker, and RhoA were cleaved out from this vector by digestion with XhoI and NotI and then inserted into the XhoI and NotI sites of a pCSII-derived vector harboring YPet, Turquoise, the CAAX box of K-Ras4B, and blasticidin S resistance genes (Komatsu et al., 2011) to obtain the expression vector for RaichuEV-RhoA.
Fluorescence microscopy
Cells were imaged with an Olympus IX-81 inverted microscope equipped with BioPoint MAC5000 excitation and emission filter wheels (Ludl Electronic Products, Hawthorne, NY), a motorized XY stage (Chuo Precision Industrial, Tokyo, Japan), and a CoolSNAP MYO cooled charge-coupled device camera (Nippon Roper, Tokyo, Japan). The following filters were used in this study: an XF1071 excitation filter, XF3075 and XF3079 emission filters for cyan fluorescent protein (CFP) and FRET, respectively, and an XF2034 dichroic mirror (Omega Optical, Brattleboro, VT). Cells were illuminated with a SOLA light engines illuminator (Lumencor, Beaverton, OR) through a 25% neutral density filter. Exposure times for 4 × 4 binning were 1 s and 30 ms for fluorescence images and differential interference contrast images, respectively. MetaMorph software (Universal Imaging, West Chester, PA) was used for control of the charge-coupled device camera, motorized stage, and filter wheels, as well as for analysis of cell-imaging data. FRET imaging and data analyses were performed essentially as previously described (Fujioka et al., 2013). In brief, MDCK cells or S1PR2-knockdown MDCK cells were transfected with an expression vector for RaichuEV-RhoA for 24 h. The cells were further transfected with pcDNA4/TO/mCherryN1 alone or together with pRK5-myc-RasV12 and cultured on a collagen gel in 35-mm-diameter, glass-bottom dishes (Matsunami Glass, Osaka, Japan). After 12 h, the cells were placed in a Chamlide stage-top incubator (Live Cell Instrument, Seoul, Korea) on a microscope in which the temperature was maintained at 37°C. A differential interference contrast (DIC) image and CFP and FRET fluorescence images were recorded every 15 min for 10 h. DIC images were used for focusing the planes and checking the appearance of cells. FRET efficiency was calculated as a quotient of background-subtracted FRET and CFP images (emission ratio) and is presented in an intensity-modified display mode with MetaMorph software. In the intensity-modified display mode shown here, eight colors from red to blue are used to represent the emission ratio, with the intensity of each color indicating the mean intensity of FRET and CFP as indicated. Alternatively, whole-cell fluorescence intensities of FRET and CFP at each time point were exported to Excel software (Microsoft, Redmond, WA), and emission ratio (FRET/CFP) was calculated. The three-point moving average of emission ratio at time t [= (FRET/CFPt-2 + FRET/CFPt-1 + FRET/CFPt)/3] was further calculated and plotted over time (Figure 4, B and C). In Figure 4C, MDCK cells transfected with RaichuEV-RhoA were starved in FCS-free medium for 12 h. At 1 h after the start of CFP and FRET imaging, S1P was added to be 200 nM at the final concentration.
Measurement of S1P levels by mass spectrometry
S1P levels in culture media were quantified by ultraperformance liquid chromatography (UPLC) coupled with electrospray ionization (ESI) tandem triple quadrupole mass spectrometry (Xevo TQ-S; Waters, Milford, CT). MDCK cells, MDCK pTR GFP-RasV12 cells, or 1:1 mix culture of MDCK and MDCK pTR GFP-RasV12 cells were placed on 60-mm dishes at 3.2 × 106 cells and incubated in DMEM without FCS but containing 2 μg/ml tetracycline at 37°C for 16 h. The culture media (4 ml) were transferred to glass tubes, and lipids were extracted from them by successive addition and mixing of 15 ml of chloroform/methanol/12 M formic acid (100:200:1 [vol/vol]), 5 ml of chloroform, and 5 ml of water. Lipids were also extracted from DMEM containing 10% FCS. After phase separation by centrifugation (2600 × g, room temperature, 3 min), the organic phase was recovered and dried. Lipids were suspended in 200 μl of methanol, separated by UPLC on a reverse-phase column (ACQUITY UPLC BEH C18 column, length 150 mm; Waters) at 45°C, and analyzed by mass spectrometry. UPLC was conducted using the binary solvent gradient system of mobile phase A (methanol/water/formic acid, 74:25:1 [vol/vol], containing 5 mM ammonium formate) and mobile phase B (methanol/formic acid, 99:1 [vol/vol], containing 5 mM ammonium formate) as follows: 0–2.4 min, 20% B; 2.4–9.6 min, gradient to 100% B; 9.6–12 min, 100% B; and 12–12.5 min, gradient to 20% B; 12.5–15 min, 20%. Samples were injected at the volume of 5 μl and flowed at 0.2 ml/min. The ESI capillary voltage was set at 3.0 kV. The sampling cone was set at 30 V, and source offset was at 50 V in positive-ion mode. S1P was detected by multiple reaction monitoring by selecting the m/z 380.3 at Q1 and the m/z 264.4 at Q3 with the collision energy setting at 20 V. S1P levels were quantified using a standard curve plotted from serial dilutions of S1P (d18:1; Avanti Polar Lipids, Alabaster, AL) standard. Data were analyzed and quantified using MassLynx software (Waters).
Data analysis
For data analyses, two-tailed Student’s t tests were used to determine p values, except for Figure 4, B and C, for which p values concerning all the values from the consecutive time points were determined by Friedman’s test and Bonferroni correction (Figure 4B) or one-way analysis of variance (Figure 4C). For quantification of apical extrusion or filamin accumulation, a group of two to four RasV12 cells that were surrounded by the indicated cells was analyzed. For quantification of apical extrusion of RasV12 cells and filamin accumulation using confocal microscopy (Figures 1, B and D, 2B, 3, B–E, 4D, and 5B and Supplemental Figure S2), 100 cells were analyzed for each experimental condition. Cells that were completely detached from the basal matrix were defined as apically extruded cells. Filamin accumulation in the surrounding normal cells at the interface with RasV12-transformed cells was quantified per transformed cells.
Supplementary Material
Acknowledgments
We thank Hideo Satsu for materials. Ya.F. is supported by Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas 26114001, Grant-in-Aid for Scientific Research (A) 26250026, and AMED Strategic Japanese-Swiss Cooperative Program. Ya.F. is also supported by the Takeda Science Foundation. Y.Y. is supported by Japan Society for the Promotion of Science Research Fellowship for Young Scientists (DC1) 26.2366.
Abbreviations used:
- EDAC
epithelial defense against cancer
- MDCK
Madin–Darby canine kidney
- shRNA
short hairpin RNA
- SphKI
sphingosine kinase inhibitor
- S1P
sphingosine-1-phosphate
- S1PR
sphingosine-1-phosphate receptor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-03-0161) December 2, 2015.
REFERENCES
- Blaho VA, Hla T. An update on the biology of sphingosine 1-phosphate receptors. J Lipid Res. 2014;55:1596–1608. doi: 10.1194/jlr.R046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall LC, Spiegel S. Enzymatic measurement of sphingosine 1-phosphate. Anal Biochem. 1999;272:80–86. doi: 10.1006/abio.1999.4157. [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Tsuda M, Nanbo A, Hattori T, Sasaki J, Sasaki T, Miyazaki T, Ohba Y. A Ca(2+)-dependent signalling circuit regulates influenza A virus internalization and infection. Nat Commun. 2013;4:2763. doi: 10.1038/ncomms3763. [DOI] [PubMed] [Google Scholar]
- Gonda K, Okamoto H, Takuwa N, Yatomi Y, Okazaki H, Sakurai T, Kimura S, Sillard R, Harii K, Takuwa Y. The novel sphingosine 1-phosphate receptor AGR16 is coupled via pertussis toxin-sensitive and -insensitive G-proteins to multiple signalling pathways. Biochem J. 1999;337:67–75. [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Forostyan T, Sabbadini R, Rosenblatt J. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J Cell Biol. 2011;193:667–676. doi: 10.1083/jcb.201010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C, Dupre-Crochet S, Norman M, Kajita M, Zimmermann C, Pelling AE, Piddini E, Baena-Lopez LA, Vincent JP, Itoh Y, et al. Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- Kajita M, Hogan C, Harris AR, Dupre-Crochet S, Itasaki N, Kawakami K, Charras G, Tada M, Fujita Y. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci. 2010;123:171–180. doi: 10.1242/jcs.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, Sugimura K, Ohoka A, Burden J, Suganuma H, Ikegawa M, Shimada T, Kitamura T, Shindoh M, Ishikawa S, et al. Filamin acts as a key regulator in epithelial defence against transformed cells. Nat Commun. 2014;5:4428. doi: 10.1038/ncomms5428. [DOI] [PubMed] [Google Scholar]
- Kihara A, Igarashi Y. Production and release of sphingosine 1-phosphate and the phosphorylated form of the immunomodulator FTY720. Biochim Biophys Acta. 2008;1781:496–502. doi: 10.1016/j.bbalip.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Kihara A, Mitsutake S, Mizutani Y, Igarashi Y. Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog Lipid Res. 2007;46:126–144. doi: 10.1016/j.plipres.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepley D, Paik JH, Hla T, Ferrer F. The G protein-coupled receptor S1P2 regulates Rho/Rho kinase pathway to inhibit tumor cell migration. Cancer Res. 2005;65:3788–3795. doi: 10.1158/0008-5472.CAN-04-2311. [DOI] [PubMed] [Google Scholar]
- Nakahara K, Ohkuni A, Kitamura T, Abe K, Naganuma T, Ohno Y, Zoeller RA, Kihara A. The Sjogren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell. 2012;46:461–471. doi: 10.1016/j.molcel.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Nishi T, Kobayashi N, Hisano Y, Kawahara A, Yamaguchi A. Molecular and physiological functions of sphingosine 1-phosphate transporters. Biochim Biophys Acta. 2014;1841:759–765. doi: 10.1016/j.bbalip.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Slattum G, Gu Y, Sabbadini R, Rosenblatt J. Autophagy in oncogenic K-Ras promotes basal extrusion of epithelial cells by degrading S1P. Curr Biol. 2014;24:19–28. doi: 10.1016/j.cub.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SK, Gomez GA, Michael M, Verma S, Cox HL, Lefevre JG, Parton RG, Hamilton NA, Neufeld Z, Yap AS. Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat Cell Biol. 2014;16:167–178. doi: 10.1038/ncb2900. [DOI] [PubMed] [Google Scholar]
- Yamauchi H, Matsumaru T, Morita T, Ishikawa S, Maenaka K, Takigawa I, Semba K, Kon S, Fujita Y. The cell competition-based high-throughput screening identifies small compounds that promote the elimination of RasV12-transformed cells from epithelia. Sci Rep. 2015;5:15336. doi: 10.1038/srep15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.