Nuclear villin regulates the expression and activity of Slug, a key transcriptional regulator of epithelial–mesenchymal transition, by directly interacting with its transcriptional corepressor, ZBRK1. Villin accumulates in the nucleus during wound repair, and altering the cellular microenvironment by hypoxia increases the nuclear villin.
Abstract
Villin is a tissue-specific, actin-binding protein involved in the assembly and maintenance of microvilli in polarized epithelial cells. Conversely, villin is also linked with the loss of epithelial polarity and gain of the mesenchymal phenotype in migrating, invasive cells. In this study, we describe for the first time how villin can switch between these disparate functions to change tissue architecture by moonlighting in the nucleus. Our study reveals that the moonlighting function of villin in the nucleus may play an important role in tissue homeostasis and disease. Villin accumulates in the nucleus during wound repair, and altering the cellular microenvironment by inducing hypoxia increases the nuclear accumulation of villin. Nuclear villin is also associated with mouse models of tumorigenesis, and a systematic analysis of a large cohort of colorectal cancer specimens confirmed the nuclear distribution of villin in a subset of tumors. Our study demonstrates that nuclear villin regulates epithelial–mesenchymal transition (EMT). Altering the nuclear localization of villin affects the expression and activity of Slug, a key transcriptional regulator of EMT. In addition, we find that villin directly interacts with a transcriptional corepressor and ligand of the Slug promoter, ZBRK1. The outcome of this study underscores the role of nuclear villin and its binding partner ZBRK1 in the regulation of EMT and as potential new therapeutic targets to inhibit tumorigenesis.
INTRODUCTION
The epithelium is the first tissue that appears during ontogenesis, and epithelial cells have fundamental roles in embryogenesis and organ development (Bryant and Mostov, 2008). Epithelial cells are distinguished from other cell types by their organization into adherent cells that maintain a distinct apicobasal polarization. This apicobasal polarization guides tissue morphogenesis and is required to perform crucial vectorial transport functions by epithelial cells. The tight association of epithelial cells with each other and the extracellular matrix also prevents them from moving when in their apicobasal polarized state. Epithelial cells undergo epithelial–mesenchymal transition (EMT) to lose cell polarity and cell–cell adhesion and to gain the migratory and invasive property of a mesenchymal stem cell. EMT reduces epithelial organization locally, disrupts intercellular junctions, and enhances migration, but it also promotes stem cell–like properties that facilitate metastatic colonization and cancer cell resistance to treatment (Kalluri and Weinberg, 2009). More than 90% of malignant human cancers are derived from epithelial cells. Thus the benefit of understanding the molecular mechanisms that guide the regulation of the EMT is quite significant (McCaffrey and Macara, 2011; Muthuswamy and Xue, 2012).
The villin gene family encodes a number of actin-binding proteins, which function in the cytoplasm by severing, capping, nucleating, and bundling actin filaments (Khurana, 2006). Villin is expressed in very significant amounts in epithelial cells with well-developed and extensive microvilli, particularly of the gastrointestinal (GI), urogenital, and respiratory tracts (Ferrary et al., 1999; Khurana, 2006; Khurana and George, 2008; Revenu et al., 2012). Furthermore, villin participates in the assembly and maintenance of the microvilli in these polarized differentiated epithelial cells (Ferrary et al., 1999; Khurana, 2006; Revenu et al., 2012; Ubelmann et al., 2013). Despite this association of villin with the polarized epithelial phenotype, villin is also recruited both in vitro and in vivo to promote EMT (Athman et al., 2003; Ubelmann et al., 2013). In addition, the absence of villin impairs the ability of epithelial cells to respond to signals that regulate EMT, resulting in deficiencies in apicobasal polarity, cell migration, and wound repair (Ferrary et al., 1999; Athman et al., 2003; Ubelmann et al., 2013). The phenotypic markers of the onset of EMT, including changes in apicobasal polarity, increased capacity for migration and cell invasion, and resistance to apoptosis, have all been linked to villin (Khurana, 2006; Khurana and George, 2008; Wang et al., 2008, 2012; Ubelmann et al., 2013). Villin’s function in EMT depends on activation of receptor tyrosine kinases (EGFR/c-Met), villin’s phosphorylation by the tyrosine kinase c-Src, and villin’s direct association with PLC-γ1 (Tomar et al., 2004, 2006; Wang et al., 2007; Khurana et al., 2008; Mathew et al., 2008; Lhocine et al., 2015). Note that pp60c-Src-Met signaling and PLC-γ are up-regulated in metastatic cancers and have independently been shown to promote metastases. Relevant to that is the fact that villin expression is maintained in most GI adenocarcinomas and adenocarcinomas of the bladder, cervix, endometrium, gall bladder, kidney, liver, lung, and pancreas, medullary carcinoma of the thyroid, and Merkel cell tumors (Khurana, 2006). Villin is also expressed in intestinal metaplasia and is associated with Barrett’s esophagus and chronic atrophic gastritis, even though normal gastric and esophageal tissue do not express villin (Khurana, 2006). How villin actuates these disparate functions to regulate epithelial plasticity remains to be characterized.
In this study, we provide for the first time a molecular basis for how villin could maintain or alternatively disperse the epithelial phenotype. Using fluorescence recovery after photobleaching (FRAP), fluorescence loss in photobleaching (FLIP), and live-cell imaging, we demonstrate the dynamic trafficking of villin between the cytoplasm and the nucleus. Our studies show that the shuttling of villin between these two compartments can be regulated by posttranslational modification (tyrosine phosphorylation) of villin, as well as by changes in the cellular microenvironment (hypoxia). Nuclear localization of villin was noted in epithelial cells lines that express endogenous or ectopic villin, in vivo in xenograft experiments performed in severe combined immunodeficiency (SCID) mice using colorectal cancer cell lines, and in human colorectal cancer specimens. Functional studies allow us to confirm that the nuclear trafficking of villin is required for villin’s effects on EMT. Of greater importance, we demonstrate that nuclear villin regulates the expression of the transcriptional factor and regulator of EMT, Slug. Villin’s effect on Slug is likely mediated by villin’s direct interaction with the transcriptional corepressor and ligand of the Slug promoter, zinc finger and BRCA1-interaction protein with a KRAB domain 1 (ZBRK1; also called ZNF350). Villin’s function in modifying the actin cytoskeleton when in the cytoplasm and its ability to moonlight in the nucleus to regulate Slug expression suggest that it is a key contributor to the spatial and temporal regulation of EMT. Most notably, our study demonstrates that this moonlighting function of villin could be converted into a pathological one by promoting tumorigenesis, thus predicting that increased nuclear localization of villin could be detrimental for patients with fibrosis or carcinomas.
RESULTS
Nuclear localization of villin in epithelial cell lines
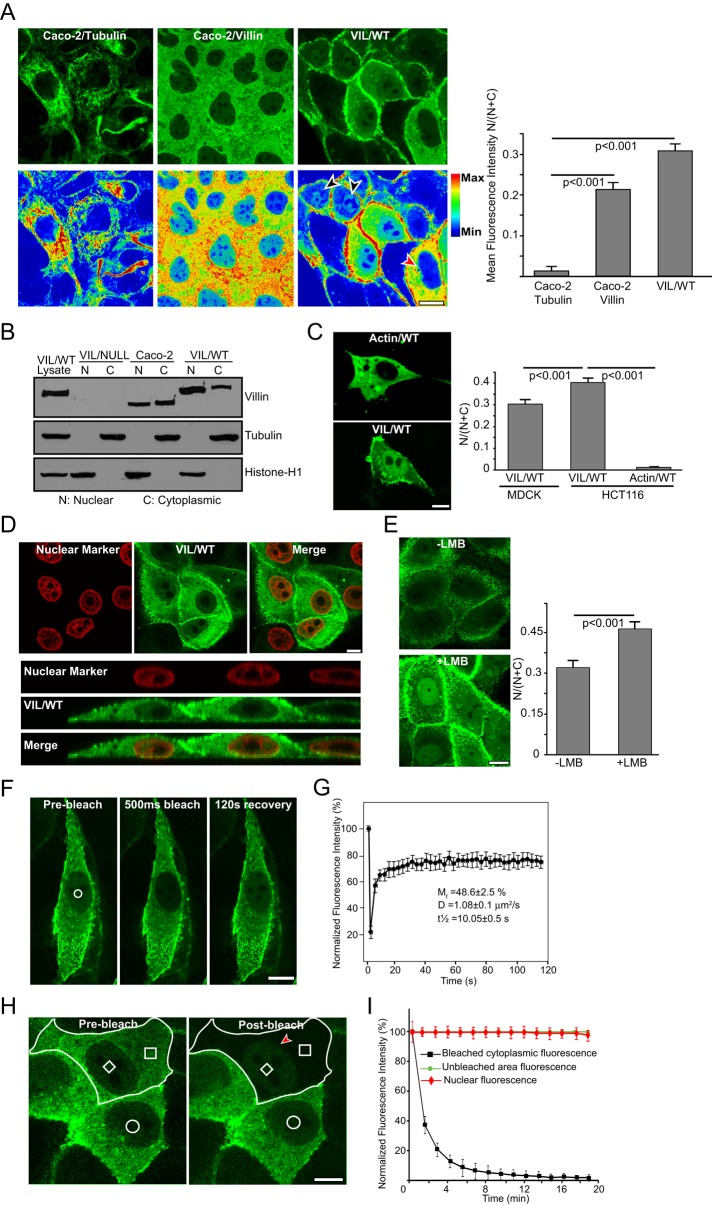
To characterize the nuclear localization of villin in epithelial cells, we examined the subcellular distribution of villin in cells that ectopically (HCT-116 or Madin–Darby canine kidney [MDCK] cells) or endogenously (Caco-2 or HT-29 cells) express human villin. We used the nontransformed epithelial cell line MDCK Tet-Off and the villin-null colon adenocarcinoma cell line HCT-116 to ectopically express superenhanced yellow fluorescent protein (seYFP)–tagged full-length human villin as described previously (Tomar et al., 2006). We used two complementary methods to characterize the cytoplasmic–nuclear distribution of villin—immunofluorescence of live and fixed cells and subcellular fractionation—to characterize the nuclear and cytoplasmic extracts of these cells. To quantify the enrichment of villin in the nucleus, we calculated fluorescence intensities within regions of interest for 40–60 cells across four or five independent experiments for each cell type. We noted that, under basal conditions villin is distributed throughout the cell, with varying levels in the nucleus (Figure 1A). Both endogenous (Caco-2) and ectopically expressed villin (VIL/WT) accumulate significantly in the nucleus (p < 0.001 compared with the negative control, tubulin; Figure 1A). Subcellular fractionation confirmed the nuclear localization of villin in cells expressing both ectopic (VIL/WT) and endogenous (Caco-2) villin (Figure 1B). For these studies, tubulin and histone-H1 were used as cytoplasmic and nuclear markers, respectively. Of interest, we noted that ectopic expression of villin in the colon cancer cell line, HCT-116, resulted in significantly more nuclear accumulation of villin than in the nontransformed epithelial cell line, MDCK (Figure 1C; quantitative analysis done by comparison of the ratio N/(N + C) of VIL/WT in HCT-116 with that in MDCK cells). Control HCT-116 cells were transfected with green fluorescent protein (GFP)–actin (Actin/WT; Figure 1C). It is possible that metastatic tumor cells have molecular mechanisms to either traffic or retain more nuclear villin, and there may be a correlation between nuclear distribution of villin and tumorigenesis (Kau et al., 2004). Similar data were obtained in multiple clones of transfected cells and with a population of mixed clones (unpublished data). The nuclear localization of villin was also not cell-type or species specific (human vs. dog epithelial cells). Nuclear localization of villin was validated using the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI; Figure 1D and Supplemental Figure S1A).
FIGURE 1:
Dynamic nuclear localization of villin in epithelial cell lines. (A) Localization of endogenous villin in Caco-2 cells and ectopically expressed seYFP villin (VIL/WT) in MDCK cells. Tubulin was used as a cytoplasmic control in these studies. Statistically significant accumulation of villin was noted in both cell types compared with control cells stained for α-tubulin (p < 0.001, n = 6). Fluorescence intensities are shown in pseudocolor (increases from blue to red). Black arrowhead shows nuclear villin expression in MDCK cells expressing exogenous villin. Red arrowhead shows lack of nuclear villin in MDCK cells overexpressing exogenous villin. Nuclear localization of villin is not dependent on level of exogenous villin expression in cells. (B) Subcellular fractionation of MDCK cells expressing seYFP-tagged VIL/WT and Caco-2 cells expressing endogenous villin shows both nuclear and cytoplasmic localization of villin. Tubulin and histone H1 were used as cytoplasmic and nuclear markers, respectively. Whole-cell lysate from seYFP-villin–transfected MDCK cells (VIL/WT) were used as a positive control. Data are representative of three independent experiments. (C) Localization of ectopically expressed seYFP-villin in the colon cancer cell line HCT-116 shows strong nuclear distribution. Quantification of mean fluorescence intensity shows that nearly 40% of villin is localized to the nucleus of HCT-116 cells compared with control cells transfected with GFP-actin (p < 0.001, n = 3). Ectopic expression of GFP-actin (Actin/WT) was used as a control for these studies. The nuclear accumulation of villin in the transformed cell line HCT-116 cells was also significantly more than in the nontransformed MDCK cells (p < 0.001, n = 6). (D) Colocalization of villin in Caco-2 cells with nuclear dye DAPI. The xy-plane and xz-orthologue of cells expressing nuclear villin. (E) Representative images of seYFP-tagged villin (VIL/WT) in cells treated with 20 nM leptomycin B (+LMB) or vehicle control (–LMB) for 8 h. A significant accumulation of nuclear villin was noted in cells treated with LMB compared with vehicle- treated cells (p < 0.001, n = 6). (F) FRAP analysis of seYFP-tagged villin in the nucleus before (prebleach), immediately after 500-ms bleaching (500-ms bleach), and 120 s after bleaching (120-s recovery). The bleached region is indicated with a white circle. (G) Quantitative analysis of FRAP recovery of seYFP villin in the nucleus with Mf, D, and t1/2 as indicated. (H, I) FLIP analysis of seYFP villin. (H) The imaged cell is outlined in white, and the circular region outlines an unbleached region inside the nucleus of an adjoining cell. The smaller white box in the nucleus is unbleached region, and the larger white box is the bleached region in the cytoplasm. (I) Quantification of fluorescence intensity of the three regions indicated in H over time. Bars, 5 μm.
Dynamic cytoplasmic–nuclear shuttling of villin
Because the molecular mass of villin is too large (95 kDa) to diffuse passively through the nuclear pore, we measured the kinetics of cytoplasmic–nuclear shuttling of villin. Proteins that are actively transported through the nuclear pore complex contain signals that are recognized by the nuclear transport receptors, importins and exportins (Cook et al., 2007). Nuclear trafficking of villin was characterized using the nuclear export receptor (CRM1/exportin) inhibitor leptomycin B (LMB). MDCK cells were used for these studies because of their low basal nuclear villin expression. Treatment of MDCK cells with 20 nM LMB caused a significant accumulation of seYFP-villin in the nucleus, confirming the regulated nuclear–cytoplasmic shuttling of villin via a CRM1-dependent nuclear export signal (p < 0.001, n = 50; Figure 1E). LMB treatment did not change total villin levels (Supplemental Figure S1B). LMB treatment had no effect on the subcellular distribution of seYFP alone (unpublished data). An increase in nuclear villin was also noted in LMB-treated Caco-2 cells expressing endogenous villin (Supplemental Figure S1C).
FRAP was used to selectively bleach the nuclear seYFP-villin and monitor the fluorescence recovery of this fraction, determine the characteristic diffusion time of villin molecules into the bleached regions in the nucleus, and measure the mobility of villin molecules (Figure 1F and Supplemental Movie S1). We noted that the recovery of villin reached a plateau at ∼80% and remained stable after that, suggesting that the majority of the villin can freely diffuse into the nucleus and also identifying a small fraction of nuclear villin that is not exchanged (Figure 1G). The mobile fraction (Mf) value confirmed that ∼50% of nuclear villin molecule can diffuse freely into the bleached region during the time course of the experiment, implying that at least half of the villin protein remains bound or immobilized or consists of an extremely slowly exchanging fraction. The effective diffusion coefficient in the nucleus for villin was estimated as 1.08 μm2/s. For comparison, the diffusion coefficient of Alexa 488 is 200–400 μm2/s, and that of nuclear CapG is 0.27 μm2/s (Renz and Langowski, 2008). The FRAP bleach protocols were kept short to limit photodamage and did not result in any cytotoxicity. For comparison, we measured the kinetics of seYFP, which passively diffuses through the nuclear pores of all cells. The FRAP data were normalized for loss of total fluorescence during the bleaching and recovery periods. FLIP analysis showed that reducing cytosolic seYFP-villin had no effect on nuclear seYFP-villin (Figure 1, H and I), indicating differences in the dynamic properties of villin molecules within the two compartments. Lack of changes in nuclear villin after cytosolic bleaching also suggests that a low basal level of nuclear villin is maintained in unstimulated epithelial cells (Figure 1A). Cells expressing seYFP alone were used as controls for these studies. Together with the data shown earlier, these findings demonstrate that villin dynamically shuttles to maintain a basal level of nuclear villin in epithelial cells.
Identification of the nuclear targeting sequences in human villin
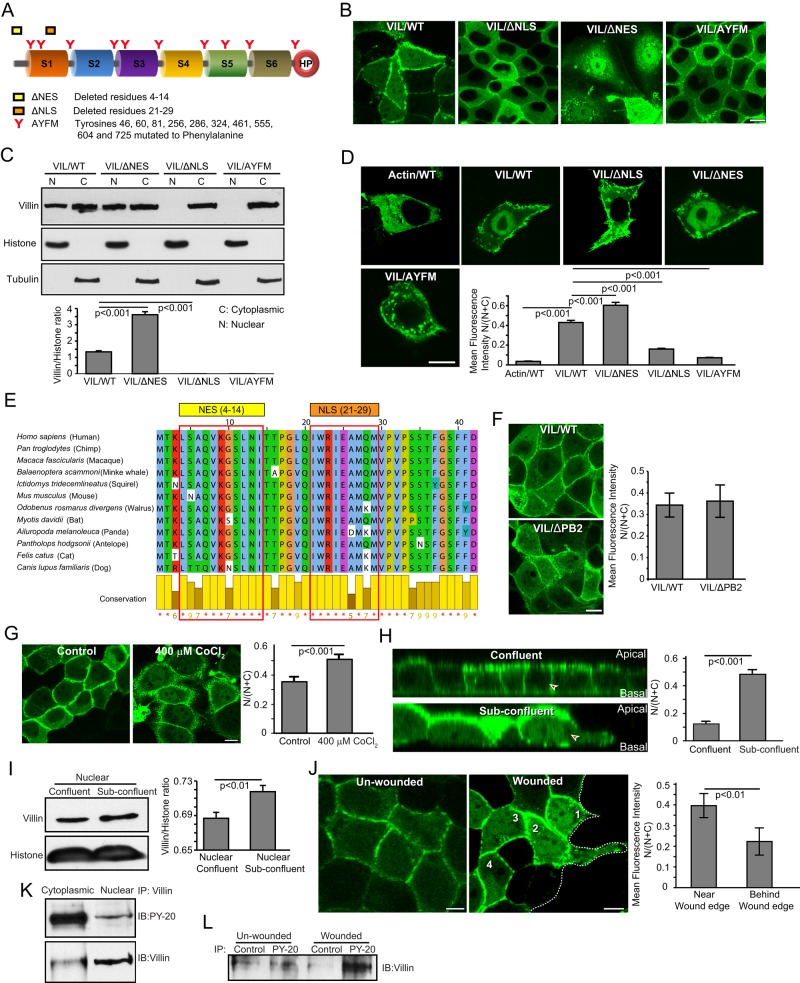
Import of proteins into the nucleus is mediated by a nuclear localization signal (NLS), which consists of a cluster of basic amino acids (Dingwall et al., 1988). In addition, phosphorylation within or around an NLS is a common strategy to regulate nuclear transport of proteins. Export is determined by a nuclear export signal (NES), which is a short sequence of four hydrophobic residues (la Cour et al., 2004). We analyzed the sequence of human villin and identified a motif resembling a putative NLS (21IWRIEAMQM29) and a single sequence that fits the putative CRM1-dependent NES consensus (4L-SAQ-V-KGS-L-N-I14; Figure 2A). Using site-directed mutagenesis, we deleted the NLS and NES sites and noted a significant loss in the nuclear accumulation of villin with the NLS (VIL/ΔNLS) and a significant gain in the nuclear accumulation of villin with the NES (VIL/ΔNES) deletion mutants expressed in MDCK cells (Figure 2, B and C, and Supplemental Figure S2A). We further confirmed that the same sites also direct the import and export of villin in the colon cancer cell line HCT-116 (Figure 2D). Both the NLS and the NES sequences are well conserved among villin proteins from different species, highlighting villin’s relevance as a regulator of intranuclear functions (Figure 2E and Supplemental Figure S2B). In addition, we noted that a villin mutant that lacks 10 previously identified tyrosine phosphorylation sites in human villin (VIL/AYFM) also failed to localize to the nucleus (Figure 2, A–D). We previously showed that these tyrosine phosphorylation sites in villin are regulated by c-Src kinase and the tyrosine phosphatases SHP-2 and PTP-PEST (Mathew et al., 2008). In addition, we reported that the villin mutant AYFM prevents villin’s effects on wound healing (Tomar et al., 2006). These data demonstrate that not only is villin actively trafficked between the cytoplasm and the nucleus, but also this dynamic shuttling of villin could be regulated by signaling mechanisms that involve upstream nonreceptor tyrosine kinases and phosphatases. Because the villin mutant VIL/AYFM lacks the actin-severing function of wild-type villin, we used the villin mutant VIL/ΔPB2, which lacks actin severing but can be tyrosine phosphorylated like wild-type villin (Kumar et al., 2004; Tomar et al., 2006). As shown in Figure 2F, VIL/ΔPB2 localizes to the nucleus like VIL/WT, suggesting that tyrosine phosphorylation of villin likely regulates its trafficking to the nucleus. The villin mutants VIL/ΔPB2 and VIL/AYFM were described in our previous studies (Kumar et al., 2004; Tomar et al., 2006). In addition, we previously showed that tyrosine phosphorylation of villin is required for its effect on EMT (Tomar et al., 2006). Villin mutants VIL/ΔNLS and VIL/AYFM also did not accumulate in the nucleus in response to LMB treatment (Supplemental Figure S2C). The villin mutant proteins are stable and maintain their actin-modifying properties (Supplemental Figure S3, A–D). Similar data were obtained in multiple clones of transfected cells and with a population of mixed clones (unpublished data).
FIGURE 2:
Characterization of nuclear villin. (A) Schematic representation of the NLS (ΔNLS) and NES (ΔNES) sites mutated in human villin. The individual homology domains are listed as S1–S6; headpiece is shown as HP. An additional mutant AYFM that lacks the 10 previously identified tyrosine-phosphorylated sites in human villin was used in these studies. (B) MDCK Tet-Off cells expressing seYFP-tagged wild-type (VIL/WT) or mutant villin (VIL/ΔNLS; VIL/ΔNES; VIL/AYFM) were transfected by using Lipofectamine 2000. Live-cell confocal images of MDCK cells expressing wild-type (VIL/WT) and mutant (VIL/ΔNES, VIL/ΔNLS, and VIL/AYFM) villin proteins. Bar, 5 μm. (C) Subcellular distribution of villin in MDCK cells expressing wild-type (VIL/WT) and mutant (VIL/ΔNES, VIL/ΔNLS, and VIL/AYFM) villin proteins. Tubulin and histone H1 were used as cytoplasmic and nuclear markers, respectively. Data are representative of three independent experiments. (D) Live-cell confocal images of HCT-116 cells expressing GFP-tagged actin (Actin/WT) or seYFP-tagged wild-type (VIL/WT) and mutant (VIL/ΔNES, VIL/ΔNLS, and VIL/AYFM) villin proteins. Bar, 5 μm. Quantification of nuclear wild-type and mutant villin proteins expressed in HCT-116 cells. There is significant accumulation of nuclear villin in cells expressing VIL/WT and VIL/ΔNES compared with cells expressing the mutant protein VIL/ΔNLS or VIL/AYFM (p < 0.001, n = 30). (E) The NLS (amino acids 21–29) and NES (amino acids 4–14) sequences identified in human villin are well conserved in villin from different species. Sequence conservation is shown at the bottom of the. (F) Nuclear localization of villin in MDCK cells expressing full-length (VIL/WT) or mutant (VIL/ΔPB2) villin. VIL/ΔPB2 lacks the actin severing activity of full-length villin but can be tyrosine phosphorylated like full-length villin. (G) To induce hypoxia, cells were incubated with 400 μM CoCl2 for 16 h. Representative images of seYFP-tagged villin (VIL/WT) in MDCK cells treated with 400 μM CoCl2 or vehicle control for 16 h. Bar, 5 μm. Quantification of mean fluorescence intensity shows significant nuclear accumulation of villin in hypoxic cells compared with normoxic control cells (p < 0.001, n = 20). (H) Representative images of seYFP-tagged villin (VIL/WT) in subconfluent and confluent monolayers of MDCK Tet-Off cells grown on filters. A significant increase in nuclear accumulation of villin is noted in subconfluent monolayers compared with confluent monolayers of MDCK cells expressing VIL/WT (compare cells indicated with arrowheads; p < 0.001, n = 20). (I) Western analysis of total and subcellular fractions of confluent and subconfluent cells, showing increased nuclear accumulation of villin in subconfluent cells compared with confluent monolayers of MDCK cells (p < 0.001, n = 3). Tubulin and histone were run as cytoplasmic and nuclear fraction controls, respectively. (J) Representative cross-section across the xz-plane, showing intense nuclear villin accumulation at the wound edge (cell 1) compared with cells further away (cells 2–4). Also shown is the nuclear villin distribution in unwounded monolayers. Data are representative of three independent experiments and show significant accumulation in cells near the wound edge compared with behind it (p < 0.01, n = 20). (K) Western analysis shows distribution of tyrosine-phosphorylated villin between cytoplasmic and nuclear fractions of cells. (L) Western analysis shows nuclear accumulation of tyrosine-phosphorylated villin in wounded monolayers. Phosphorylated proteins were immunoprecipitated (IP) with anti-phosphotyrosine antibody (PY-20) from nuclear extracts from wounded monolayers, and Western analysis done with villin antibody. Data are representative of three independent experiments.
Nuclear accumulation of villin can be regulated
To determine whether we can alter the nuclear accumulation of villin by altering the microenvironment of the cells similar to what may occur within tumors, we subjected MDCK cells expressing seYFP-villin to hypoxia. The subcellular localization of villin was analyzed in control normoxic cells and hypoxic cells (treated with 400 μM CoCl2). Hypoxia caused a significant increase in the amount of nuclear villin accumulation compared with normoxic control cells (p < 0.001, n = 30; Figure 2G). An increase in nuclear villin in response to hypoxia was also seen in Caco-2 cells (Supplemental Figure S4A). Tubulin distribution in normoxic and hypoxic cells was used as an additional control for these studies, demonstrating that not all cytoplasmic proteins are trafficked into the nucleus in response to hypoxia (Supplemental Figure S4B). Hypoxia also did not alter total villin levels (Supplemental Figure S1B). In addition, we noted that confluent monolayers expressed lower levels of nuclear villin than subconfluent cells (Figure 2H; compare cells labeled with arrowheads). Western analysis confirmed the nuclear accumulation of villin in subconfluent monolayers compared with that in confluent monolayers (Figure 2I). Other cell lines studied showed a similar increase in nuclear villin in subconfluent compared with confluent monolayers (unpublished data). Moreover, in a wounded monolayer, we noted a significant increase in nuclear villin distribution in cells closer to the wound edge than further away from it (compare cell 1 with cells 2–4 and with cells in unwounded monolayer; Figure 2J). Villin is tyrosine phosphorylated in both the cytoplasmic and nuclear fractions (Figure 2K). In addition, we noted an increase in the tyrosine phosphorylation of nuclear villin in the wounded monolayer compared with the unwounded cells (Figure 2L). Increased nuclear accumulation of villin is associated with tyrosine phosphorylation of villin, and because mutation of the tyrosine residues involved in this process (VIL/AYFM) also prevents the nuclear accumulation of villin (Figure 2, A–D), these data strongly suggest that tyrosine phosphorylation of villin may regulate its nuclear trafficking. Together these studies indicate that changing the microenvironment, local geometry, apicobasal polarity, cellular communities, or microregions in vivo could alter the nuclear distribution of villin.
Nuclear localization of villin is required for villin’s effect on EMT
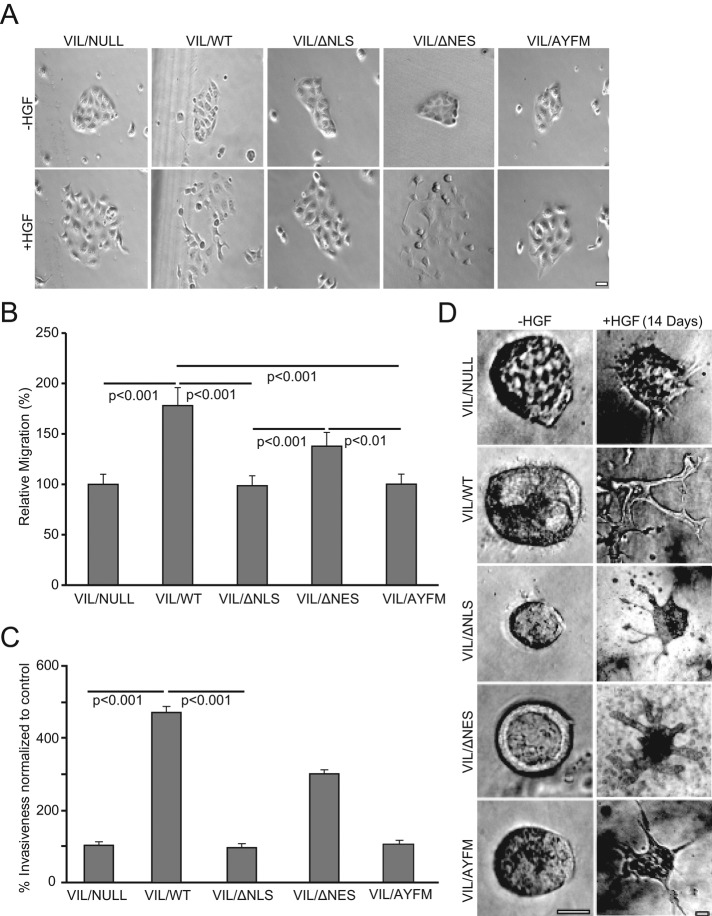
Functional studies demonstrated that villin mutants that fail to localize to the nucleus fail to regulate EMT. As shown in Figure 3A and consistent with previous reports, hepatocyte growth factor (HGF) induced cell scatter in epithelial cells expressing full-length villin (VIL/WT) but not in VIL/NULL cells. Preventing the cytoplasmic–nuclear trafficking of villin (mutant VIL/ΔNLS and VIL/AYFM) prevented HGF-induced cell scatter, whereas promoting nuclear accumulation of villin (VIL/ΔNES) enhanced cell scatter relative to VIL/NULL cells. We and others previously reported that ectopic expression of villin increases cell migration compared with control cells that lack villin (Athman et al., 2003; Tomar et al., 2004, 2006). We now report that ectopic expression of villin mutants that fail to localize to the nucleus fail to increase cell migration rates compared with VIL/WT cells or VIL/ΔNES (Figure 3B). The nuclear localization of villin also regulates villin’s function in cell invasion (Figure 3C). The effect of nuclear villin on morphogenesis was assayed using a standard protocol in which MDCK cells are grown in three-dimensional collagen in the absence or presence of 100 ng/ml HGF to promote tubulogenesis (Figure 3D). We found a significant increase in the number of tubules formed with cells expressing full-length villin (VIL/WT) compared with villin-null cells (VIL/NULL; p < 0.01, n = 280). In contrast, there was a significant decrease in the number of tubules formed with VIL/ΔNLS compared with VIL/WT (p < 0.001). No significant difference was noted between the number of tubules formed by VIL/ΔNES and that formed by VIL/WT. After 10 d in collagen matrix, cells lacking villin (VIL/NULL) were still undergoing tubulogenesis, showing mostly cysts with extensions but no tubules. In contrast, full-length villin–expressing cells induced potent morphogenesis, which started as early as 10 d and lasted >20 d, forming large, branched tubular structures, demonstrating that tubulogenesis is much more efficient in these cells than in control cells that lack villin (Figure 3D and Supplemental Figure S4C; Athman et al., 2003). A similar observation was made with VIL/ΔNES. Cell expressing villin mutant VIL/ΔNLS or VIL/AYFM, in contrast, behaved like villin-null cells, forming cysts and extensions but no tubules (p < 0.01, n = 280). Together these studies demonstrate a distinct role for nuclear villin in the regulation of EMT.
FIGURE 3:
Nuclear villin regulates EMT. (A) Scatter assay with VIL/NULL cells and cells expressing wild-type (VIL/WT) or mutant (VIL/ΔNLS, VIL/ΔNES, or VIL/AYFM) villin proteins. We plated 5 × 104 cells in six-well culture dishes and allowed them to grow as discrete colonies. Cells were serum starved overnight, followed by addition of medium containing 100 ng/ml HGF to initiate scattering. Phase contrast image show cell scatter with VIL/WT but not control VIL/NULL or the mutant VIL/ΔNLS, VIL/ΔNES, or VIL/AYFM cells. Images taken before (0 h, –HGF) and 9 h after the addition of HGF. Bar, 10 μm. This is a representative of six independent experiments. (B) Migration rates of VIL/NULL cells or cells expressing VIL/WT, VIL/ΔNLS, VIL/ΔNES, or VIL/AYFM. Migration rates were compared with control cells that lack villin (VIL/NULL). The error bars are the measured SE (n = 36). (C) Cell invasion was measured using a modified Boyden chamber assay. Cells expressing VIL/WT were significantly more invasive than VIL/NULL or the mutant VIL/ΔNLS, VIL/ΔNES, or VIL/AYFM cells. The error bars are the measured SE (n = 12). (D) Tubulogenesis assay with VIL/NULL, VIL/WT, VIL/ΔNLS, VIL/ΔNES, and VIL/AYFM cells. We dispensed 1 × 104 cells embedded in a Vitrogen type I collagen gel into 0.02-mm tissue culture inserts in 24-well plates and allowed them to form cysts. Tubulogenesis was initiated by the addition of HGF (100 ng/ml; renewed every 3 d). Images were collected every day (20 cysts/tubules every day for a maximum of 20 d). Representative phase contrast images are shown before the addition of HGF and 14 d after HGF treatment. Quantitative analysis of number of cysts, extensions, and tubules was done using one-way repeated-measure analysis of variance and Tukey’s modified t test. This is a representative of four independent experiments. Bar, 500 μm.
Nuclear localization of villin in xenografts generated in SCID mice
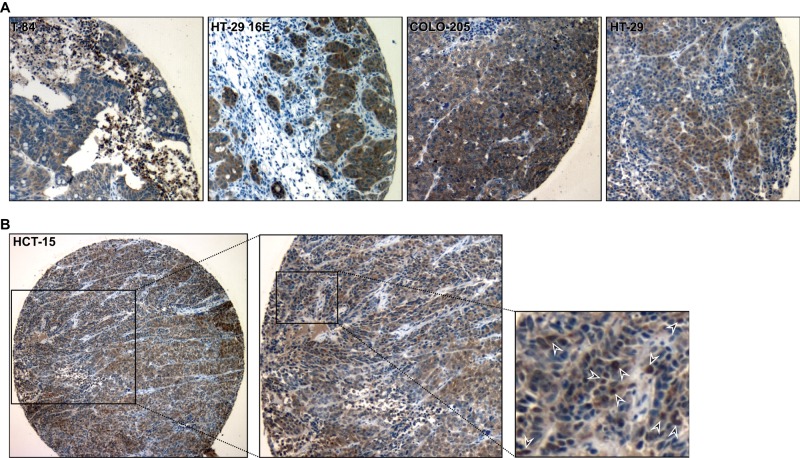
To test our hypothesis that nuclear localization of villin occurs during tumorigenesis, we stained xenografts derived from 30 colon cancer cell lines with villin antibodies. With five of these cell lines, tumors develop within 21 d at 100% frequency in SCID mice inoculated subcutaneously with 107 cells. Xenografts derived from these five cell lines—T-84, HT-29 16E, Colo205, HT-29 19A, and HCT-15—show strong nuclear accumulation of villin (Figure 4). The remainder show primarily cytoplasmic staining (unpublished data). Higher magnification of HCT-15 cells shows strong localization of villin in individual nuclei (indicated with arrowheads; Figure 4B). Together these data demonstrate that villin is mislocalized to the nucleus in vivo in mice during tumorigenesis.
FIGURE 4:
Nuclear localization of villin in colon cancer cell xenografts. Colon cancer cell lines endogenously expressing villin—T-84, HT-29 16E, COLO-205, HT-29 19A, and HCT-15 cells—were grown as xenografts in SCID mice. The tumors were excised and Formalin fixed. Villin expression was determined by immunohistochemistry using a commercially available anti-villin antibody. (A) Representative images of T-84, HT-29 16E, COLO-205, and HT-29 19A. (B) Higher magnification of HCT-15 xenograft shows localization of villin in individual nuclei (arrowheads).
Nuclear localization of villin in colorectal cancers
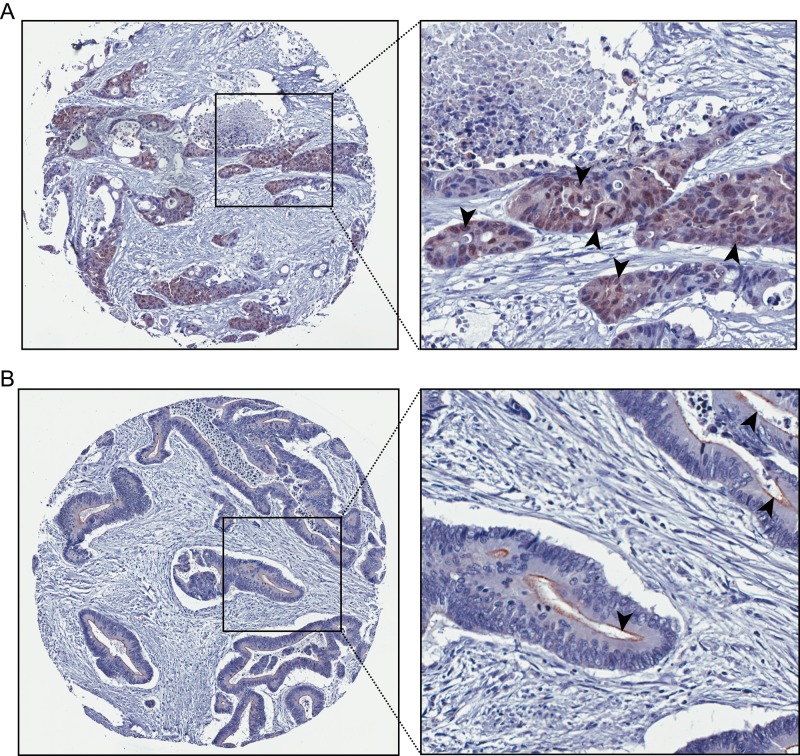
Previous studies indicated that mislocalization of villin away from the brush border is associated with poor prognosis in patients with colorectal cancer (Arango et al., 2012). However, no systematic study had been performed to describe the subcellular localization of villin in tumors. Because a small number of proteins of the villin superfamily have been shown to traffic to the nucleus and regulate gene transcription, our hypothesis was that mislocalization of villin to the nucleus could be linked to tumorigenesis. Additional support for our hypothesis came from our in vitro studies (Figure 1) and studies showing nuclear localization of villin in mouse xenografts with colon cancer cell lines (Figure 4). We analyzed 533 Dukes’ B and C colorectal cancers to characterize the nuclear localization of villin in colorectal cancers. Tissue microarrays were stained with anti-villin antibodies, and tumors were classified as having exclusive membrane staining or some nuclear staining. A comparison of these tumors demonstrated that villin is localized to the nucleus (Figure 5A) in a significant number (5.3%) of colon tumors compared with others in which villin is distributed like the normal tissue to the epithelial brush border (Figure 5B).
FIGURE 5:
Nuclear localization of villin in human colorectal cancers. Dukes’ B and C colorectal tumor samples were analyzed for villin expression and classified as either having exclusive membrane staining or some nuclear staining. Villin was found to be localized in a small but significant number of colon tumors (5.3% of the tumors; p < 0.01, n = 533). Representative images of nuclear villin staining (A) and brush border villin staining (B). Arrowheads highlight villin staining in individual nuclei (A) and brush border (B).
Nuclear villin regulates the expression of the transcriptional factor Slug
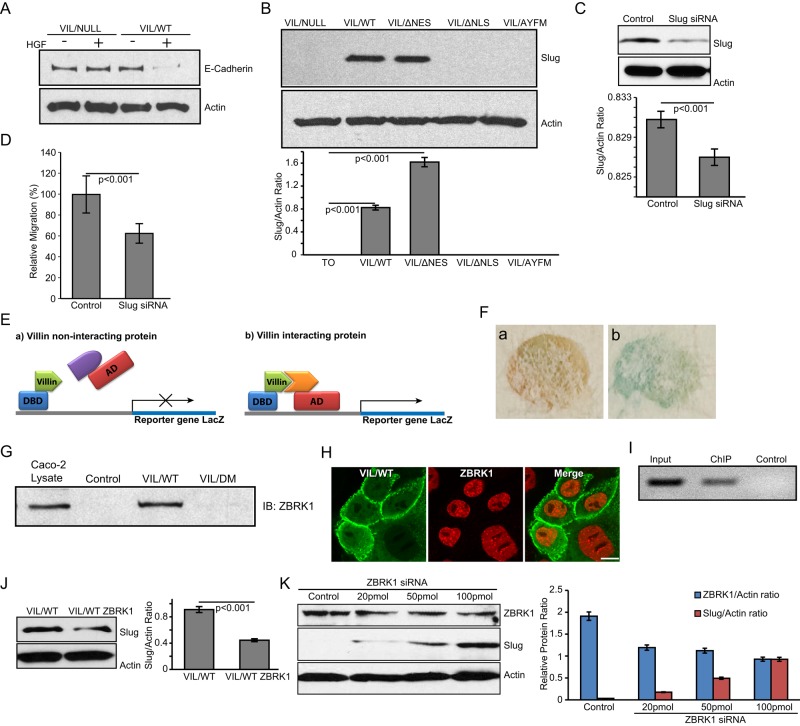
Consistent with previous studies, we report that the expression of villin regulates EMT (Athman et al., 2003; Revenu et al., 2007; Ubelmann et al., 2013). MDCK cells expressing full-length human villin or null cells were treated with HGF (100 ng/ml for up to 72 h) to induce EMT. As shown in Figure 6A, villin-expressing cells show down-regulation of E-cadherin, but villin-null cells or villin-expressing cells treated with vehicle (–HGF) do not, as early as 24 h after HGF treatment. To determine the targets of villin that mediate this effect on EMT, we analyzed multiple transcriptional regulators of EMT. Our studies indicate that villin’s effects on EMT are mediated through the expression and activation of the transcriptional factor Slug (Figure 6B). Of greater note notably, villin mutant cells that fail to localize to the nucleus showed no activation of Slug expression (Figure 6B). Slug was up-regulated in cells expressing VIL/WT and VIL/ΔNES but not in VIL/NULL cells or cells expressing the villin mutant VIL/ΔNLS or VIL/AYFM. Consistent with this, knockdown of endogenous Slug using small interfering RNA (siRNA; Figure 6C) suppresses villin-induced increase in cell migration (Figure 6D). No change was noted in Slug knockdown villin-null cells (Supplemental Figure S5, A and B). In addition, we noted no change in several other transcriptional markers of EMT, including Snail and TCF-8 (unpublished data).
FIGURE 6:
Nuclear villin regulates Slug expression. (A) Whole-cell lysates from VIL/NULL and VIL/WT cells treated with HGF (100 ng/ml for 0–72 h) or vehicle control were analyzed by Western analysis for E-cadherin expression. Actin was detected as a loading control. Data are representative of three independent experiments showing E-cadherin expression at 24 h posttreatment. (B) Whole-cell lysates from VIL/NULL, VIL/WT, VIL/ΔNES, VIL/ΔNLS, and VIL/AYFM cells treated with HGF (100 ng/ml for 0–72 h) were analyzed by Western analysis for Slug expression. Actin was used as a loading control in these studies. Data are representative of three independent experiments. (C) Knockdown of Slug using siRNA in MDCK cells. Actin was used as a loading control in these studies. (D) Cell migration was measured using a wound-healing assay in MDCK cells expressing VIL/WT with control siRNA or VIL/WT with Slug knocked down using siRNA. (E) A model of the yeast two-hybrid screen performed using villin fused to the DNA-binding domain of GAL4 (DBD). Proteins that are able to interact with villin activate the expression of LacZ. AD refers to activation domain of GAL4. (F) Positive clone (b) identified by LacZ expression (blue) compared with control (a) shows direct interaction of villin with ZBRK1. (G) GST (Control) and VIL/WT recombinant proteins bound to agarose beads were used to pull down ZBRK-1 from Caco-2 whole-cell lysates. A villin mutant, VIL/DM, lacking the NH2-terminal residues 21–67/112–118 showed loss of ZBRK1 binding. Total Caco-2 whole-cell lysate was also used as a positive control for the ZBRK-1 antibody. Data are representative of three independent experiments. (H) MDCK cells expressing seYFP-tagged villin (VIL/WT) and mCherry-tagged ZBRK-1 were imaged using fluorescence microscopy to characterize the intracellular distribution of both proteins. Data are representative of three independent experiments. Merged images show colocalization of villin and ZBRK1 in the nucleus. Bar, 10 μm. (I) ChIP assay showing the association of ZBRK1 with the Slug promoter. A 300–base pair, PCR-amplified product was generated from ChIP with anti-ZBRK1antibody (ChIP) but not from beads (Control). (J) Slug expression in MDCK cells overexpressing villin or overexpressing villin and ZBRK1. In the presence of villin and endogenous ZBRK1, Slug expression is activated. Overexpression of ZBRK1 in these cells even in the presence of villin inhibits Slug expression. Data are representative of three independent experiments. (K) Knockdown of ZBRK1 increases Slug expression in MDCK cells. Actin was run as a loading control.
A yeast two-hybrid screen using the human villin in-frame with GAL4 DNA-binding domain was used to identify the direct binding partners of villin (Figure 6E). Using this approach, we identified the zinc finger protein and transcriptional coregulator ZBRK1 as a direct binding partner of villin (Figure 6F). We identified no direct association between villin and Slug using this approach (unpublished data). Using recombinant human villin protein (VIL/WT), we pulled down ZBRK1 from Caco-2 cell lysates, thus validating the interaction of ZBRK1 with villin (Figure 6G). We identified no direct interaction between villin and Slug using similar pulldown assays (unpublished data). In addition, we identified a domain in the amino terminus of human villin protein (VIL/DM: 21–67/112–118) that fails to associate with ZBRK1 (Figure 6G). Additional details on the villin mutant DM are described in our previous study (George et al., 2007). Using live-cell assays, we also identified in MDCK cells the nuclear localization of both villin and ZBRK1 (Figure 6H). Because villin can directly associate with ZBRK1, as shown by our yeast two-hybrid studies, we suggest that at least some of this nuclear ZBRK1 could be colocalized with villin (Figure 6H). A similar observation was made in the other cell lines studied (unpublished data). Chromatin immunoprecipitation (ChIP) assay confirmed the interaction of ZBRK1 with human Slug promoter (Figure 6I), indicating that Slug expression could be regulated by ZBRK1. Endogenous expression of ZBRK1 in MDCK cells is shown in Supplemental Figure S3C. Overexpression of villin in these cells activates Slug expression (Figure 6B). However when ZBRK1 is also in excess, it down-regulates villin-induced Slug expression (Figure 6J). In addition, we noted that knockdown of ZBRK1 using siRNA results in a concomitant increase in Slug expression (Figure 6K). Taken together, these data indicate that villin does not regulate Slug activity by directly associating with Slug. These data also reveal that ZBRK1 is a corepressor of Slug expression and that the villin-ZBRK1 complex could prevent the corepressor function of ZBRK1, allowing for Slug expression and activation.
DISCUSSION
Villin is best known for its association with the microvilli, where it has been shown to regulate the assembly, morphology, and function of the epithelial cell brush border (Ferrary et al., 1999; Revenu et al., 2012; Ubelmann et al., 2013). Villin is also uniquely adapted to disassemble the microvilli and the apicobasal polarized phenotype of the epithelial cell by regulating EMT (Tomar et al., 2006; Khurana and George, 2008; Ubelmann et al., 2013). The molecular basis for the regulation of villin’s divergent functions in assembling the epithelial, as well as in the mesenchymal phenotype, remains to be determined. We provide the first evidence that in addition to the cytoplasm, villin moonlights in the nucleus and that this adaptability of villin to alter its subcellular localization is intimately linked with repurposing villin for different cellular functions. Furthermore, this functional adaptability of villin due to its moonlighting property is closely linked with the plasticity of the epithelial cell. In the cytoplasm, villin serves to modify actin dynamics (Khurana, 2006; Khurana and George, 2008; George et al., 2013). By moonlighting in the nucleus, it regulates gene transcription to promote the mesenchymal phenotype. Our studies show that the nuclear distribution of villin is balanced by the nuclear import and export activities, which maintain a basal level of nuclear villin in epithelial cells. The conserved NLS and NES in villin from different species and the fact that nuclear villin is noted in multiple cell types and different tumors indicate a broad mechanism to regulate EMT in all epithelial cells that express villin. Because the villin NLS is also conserved between plants and mammalian cells and plant villin is ubiquitously expressed, our study also suggests a more general role for villin in the regulation of gene expression.
Our study demonstrates that tyrosine phosphorylation could be a significant regulator for the nuclear accumulation of villin. Previous studies showed that tyrosine phosphorylation of villin is required for its effects on EMT (Athman et al., 2003; Tomar et al., 2006). We now show that nuclear villin is tyrosine phosphorylated and that a villin mutant that cannot be phosphorylated does not localize to the nucleus and has no effect on EMT. It was shown previously that negative charge enhances nuclear import by increasing NLS recognition by importin subunits and/or modulating the affinity of NLS for the importin (Zhang et al., 2000). Tyrosine phosphorylation of villin could be a regulatory step in villin’s exchange between the two cellular compartments, which would drastically modify cellular behavior. Imposing such a requirement could be particularly important in preventing unrestrained activity of nuclear villin on EMT. Tyrosine phosphorylation and dephosphorylation of villin provides an effective mechanism to rapidly activate or switch off EMT in response to loss of cellular homeostasis and/or receptor activation. Maintaining low basal levels of nuclear villin in unstimulated cells might also support such a rapid activation of EMT. FRAP and FLIP analysis confirm the dynamic trafficking of villin between the two cellular compartments. Nuclear villin could also be regulated by how much free cytoplasmic villin is available. We previously showed that tyrosine phosphorylation of villin prevents its association with the cytoplasmic actin cytoskeleton (Khurana et al., 1997). This suggests that phosphorylation of villin alters the equilibrium of soluble and actin-bound villin to influence nuclear accumulation of villin. Dissociation of tyrosine-phosphorylated villin from the microvillar actin would also favor the disruption of the microvilli. The fate of microvilli during EMT is relatively unknown. Our study suggests that increased microvilli dynamics might be a normal physiological response to EMT, and proteins like villin could alter the microvilli morphology while promoting EMT.
Tyrosine phosphorylation of villin is regulated by c-Src, SHP-2, and PTP-PEST (Mathew et al., 2008; Lhocine et al., 2015). Aberrant expression and activation of Src occurs in several cancers in which a strong correlation between c-Src levels and advances in stages of cancer, size of tumors, and the metastatic potential of tumors have been noted (Aligayer et al., 2002). Similarly, SHP-2 and PTP-PEST have been linked to tumorigenesis (Grossmann et al., 2010). In this study, we make the novel observation that nuclear accumulation of villin can be regulated by pathophysiology linked to tumorigenesis, including hypoxia. Our study demonstrates the accumulation of nuclear villin in a mouse model of tumorigenesis and in a subset of colorectal cancer patients. Although our study is the first systematic analysis of nuclear villin in tumor samples, a careful review of the literature shows that villin is frequently directed to the nucleus in colon, gastric, and other poorly differentiated, more aggressive metastatic cancers (Goldstein and Thomas, 2001; Lau et al., 2002). However, before our report, no study characterized the nuclear pool of villin.
We identify ZBRK1 as a direct binding partner of villin. Altered expression of ZBRK1 is frequent in several human cancers, including colorectal cancer (Garcia et al., 2005; Furuta et al., 2006). Increased ZBRK1 levels diminish, and loss of ZBRK1 augments, malignant growth, metastasis, and invasion in cancers, indicating that ZBRK1 acts as a tumor suppressor (Lin et al., 2010, 2013). Although the nuclear–cytoplasmic trafficking of ZBRK1 has not been examined, a careful analysis of the primary sequence of ZBRK1 demonstrates that it contains both a putative NLS and NES. Our study demonstrates the direct interaction of ZBRK1 with the Slug promoter and highlights the potential corepressor functions of ZBRK1 in EMT (Figure 7). ZBRK1 has also been implicated in the regulation of EMT functioning as a transcriptional repressor (Zheng et al., 2000; Lin et al., 2013). Sequestration of transcriptional coregulators is increasingly being recognized as a mechanism to regulate gene transcription (e.g., NF-κB regulation by sequestration of SIMPL; Kwon et al., 2004). Slug is capable of repressing the transcription of several polarity factors and the cell adhesion gene E-cadherin (Medici et al., 2008). Slug expression has been correlated with cancer stem cell formation, cell cycle regulation, and apoptosis, as well as with invasion and metastasis in multiple cancers, including colorectal cancer, and is an independent prognostic factor in colorectal cancer (Shioiri et al., 2006; Hotz et al., 2007).
FIGURE 7:
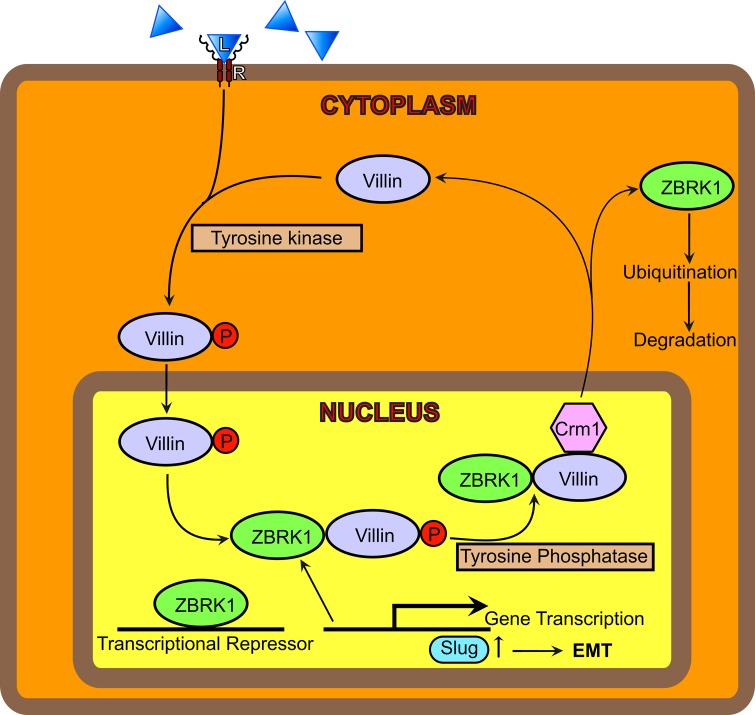
Working model for the role of villin in EMT. We suggest that in response to receptor activation (R), villin is tyrosine phosphorylated and trafficked into the nucleus. In the nucleus, villin interacts with the transcriptional cofactor ZBRK1, which prevents the interaction of ZBRK1 with Slug. Disruption of the ZBRK1-Slug interaction results in the activation and expression of the transcriptional factor Slug. This, we suggest, regulates EMT. The role of ZBRK1 in the regulation of Slug remains to be characterized. We hypothesize that nonreceptor tyrosine kinases and tyrosine phosphatases regulate the cytoplasmic–nuclear shuttling of villin in epithelial cells.
We suggest that proteins like villin, which can alter both cell architecture by modifying the microfilament structure when in the cytoplasm and gene transcription when moonlighting in the nucleus, may be a new class of proteins. This could include other proteins, such as junction-mediating and -regulatory proteins (JMYs). JMY was shown initially to accumulate in the nucleus to function as a transcriptional cofactor that increases p53/TP53 response via its interaction with p300/EP300 (Coutts et al., 2007, 2011). More recent studies have shown that JMY also nucleates actin assembly and colocalizes with actin filaments in the lamellipodia (Zuchero et al., 2009). This is indistinguishable from villin’s discrete functions in the cytoplasm and the nucleus (Tomar et al., 2004, 2006; George et al., 2013). Furthermore, our study suggests that epithelial cell–specific actin-associated proteins may have the unique characteristic to rapidly sense changes in cellular environment and modify simultaneously the cellular architecture and gene transcription by simply moonlighting between the cytoplasm and the nucleus. Like villin, other epithelial cell–specific actin-binding proteins, such as β-catenin, are known to traffic between the cytoplasm and the nucleus and simultaneously modify the epithelial cell structure and have a profound effect on EMT. Future studies will allow us to determine the molecular basis of ZBRK1-induced changes in Slug expression and how the two pools of villin cooperate or maintain distinct roles to regulate cellular behavior as a polarized epithelial cell or as a depolarized mesenchymal cell.
MATERIALS AND METHODS
Cell lines and cell culture
MDCK and HeLa Tet-Off cells were a kind gift from Keith Mostov (University of California, San Francisco, San Francisco, CA). All other cell lines were purchased from the American Type Culture Collection (Rockville, MD). MDCK Tet-Off and HCT-116 cells expressing seYFP-tagged wild-type (VIL/WT) or mutant villin (VIL/ΔNLS; VIL/ΔNES; VIL/AYFM; VIL/ΔPB2; VIL/DM) were transfected by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Transfected cells were cultured in DMEM containing 100 μg/ml G418 sulfate, 100 μg/ml hygromycin B, and 10% fetal calf serum. To repress the expression of villin gene in MDCK Tet-Off transfected cells, cells were cultured in the presence of 10 ng/ml doxycycline. For LMB treatment, cells were incubated in medium containing 20 nM LMB for 8 h. Cells were washed twice with Dulbecco’s PBS and then analyzed. To induce hypoxia, cells seeded on glass-bottom dishes were incubated with 400 μM CoCl2 for 16 h.
Antibodies and reagents
Monoclonal antibodies for villin and actin were purchased from BD Transduction Laboratories (San Jose, CA), tubulin and histone H1 antibody were from Abcam (Cambridge, UK), and ZBRK1 and Slug antibodies were from Sigma-Aldrich (St. Louis, MO). HGF, CoCl2 and LMB and Slug siRNA and control siRNA were purchased from Sigma-Aldrich. A QuikChange Site-Directed Mutagenesis Kit was purchased from Agilent Technologies (Santa Clara, CA). Purified bovine collagen (PureCol) solution was purchased from Advanced Biomatrix (Carlsbad, CA). The yeast three-hybrid pBridge vector was purchased from Clontech (Mountain View, CA).
Cloning of cerulean-tagged ZBRK1
Cerulean tag cloning into the NotI and KpnI sites of pBudCE4.1 vector was done as described previously (George et al., 2007). ZBRK1 was cloned into the XhoI and BstBI site of this vector. Human ZBRK1 was a kind gift from Wen-Hwa Lee (Department of Biological Chemistry, University of California, Irvine, CA). Full-length ZBRK1 was amplified using the primers (forward) 5′-GCGCTCGAGATCCAGGCCCAGGAAT-3′ and (reverse) 5′-CAGACTTCGAATGGGTTTTCTGTAACAT-3′, which contain the XhoI and BstBI sites. The PCR products and cerulean-pBudCE4.1 were restricted using XhoI and BstBI enzymes, ligated, and transformed in Escherichia coli XL-10 Gold Ultracompetent cells. The sequence and frame of the inserts were checked by sequencing. VIL/ΔNLS and VIL/ΔNES mutants were made by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis Kit as described (George et al., 2007).
Cell motility assay
Cell migration was measured as described previously (George et al., 2007). Briefly, confluent monolayers were wounded with a sharp blade across the diameter of the well. Cells were rinsed and images obtained at the initial time of wounding and 7 h postwounding. Images were collected with a Nikon Eclipse TE2000-U inverted microscope equipped with a CoolSnap ES charge-coupled device camera and an Optiscan motorized stage system. Images were analyzed using MetaMorph image analysis software as described previously (George et al., 2007).
Cell invasion assay
Invasion assay was performed as described previously (Wang et al., 2007). We plated 1 × 105 cells in six-well invasion chambers coated with Matrigel. HGF (100 ng/ml) was added to the lower chamber. The invasion chambers were incubated for 24 h at 37°C. The transmigrated cells were visualized using the DIFF-Quick staining kit (IMEB, San Marcos, CA).
Cell scatter assay
We plated 5 × 104 MDCK Tet-Off cells expressing wild-type or mutant villin proteins in six-well culture dishes and allowed them to grow as discrete colonies. Cells were transferred into serum-free medium containing 100 ng/ml HGF to initiate scattering. Nine hours later, phase contrast images were collected using the Nikon Eclipse TE2000-U inverted microscope.
Tubulogenesis assay
MDCK Tet-Off cells expressing wild-type or mutant villin proteins were suspended at 1 × 104 cells/ml in Vitrogen type I collagen and dispensed into 0.02-mm tissue culture inserts in 24-well plates. After the collagen solution had gelled, complete culture medium containing HGF (100 IU/ml) was added and renewed every 3 d. Images were collected every day (20 cysts/tubules each day for a maximum of 20 d) using a Nikon Eclipse TE2000-U inverted microscope.
Live-cell confocal imaging
Live cell images were obtained using the FluoView FV1200 Laser Scanning Confocal Microscope with a 60×/numerical aperture 1.35 objective. FRAP and FLIP measurements were performed using a 405-nm solid-state laser with 40% power and 100% transmission. The bleaching was performed for 200 ms after five prebleach frames were captured at 512 × 512 at a scan speed of 8. Postbleaching images were acquired at the same resolution and power as bleaching with 0.3% transmission at an interval of 3 s. A 3-μm circular region of interest (ROI) was used for these studies and a recovery period of 120 s. Recovery curves were generated by comparing the intensity ratio in ROIs inside and outside the bleached area before bleach and during recovery. The intensities were normalized to correct for total loss of fluorescence due to overall photobleaching as described previously (Snapp et al., 2003). The resulting images were analyzed for calculating t1/2 (time for 50% fluorescence plateau) using Olympus FluoView software. The Mf and diffusion coefficient (D) were calculated using 10 data sets as described previously (Snapp et al., 2003). FLIP was performed by bleaching with 40% transmission at regular intervals of 20 s for 60 cycles, followed by imaging at 0.3% transmission. Bleaching was performed in the nucleus and cytoplasm using 3-mm circular ROIs. Calculations were done by normalizing with nonbleached ROIs using 10 data sets.
Yeast two-hybrid screen
Human villin was used as a bait protein in the yeast GAL4-based, two-hybrid screening of a Matchmaker Pre-transformed Human Kidney cDNA Library (Clontech Laboratories). Specifically, two-hybrid vector pBridge containing villin S1–S3 domains (amino acids 1–300) in-frame with GAL4 DNA-binding domain was used to perform library screening in Saccharomyces cerevisiae strain Y187. Villin expression was driven by the constitutive alcohol dehydrogenase 1 promoter. The bait strain S. cerevisiae AH109 was established by transformation with pBridge-Src-villin using the lithium acetate method in which villin expression was confirmed by reverse transcription PCR and Western blotting, and no significant cell toxicity and autotranscriptional activity were detected. This AH109-pBridge-villin bait strain was mated with library strain Y187(MATα), which contained ∼3.5 × 106 clones of the human cDNA. The AH109-Y187 diploid cells were selected on the (synthetic dropout) SD/-Ade-Leu-His-Trp (quadrodropout) medium for the positive protein–protein interactions by transcription activation of ADE2, MEL1, LacZ, and HIS3 four reporter genes. β-Galactosidase activities were qualitatively monitored by the blue color development with X-gal filter assays. Library cDNA inserts fused with pACT2 GAL4 activation domain from positive clones were rescued by transforming into E. coli KC8, purified, and sequenced.
ChIP assay
ChIP analysis was performed using the ab500 ChIP kit (Abcam) following the manufacturer’s protocol. Briefly, HT29-19A cells were lysed using the lysis buffer provided in the kit. Lysates were treated with 1.1–1.5% formaldehyde to cross-link proteins to DNA. Genomic DNA was sheared using Branson sonicator for 2 min at the 10% amplitude to acquire optimal DNA fragment size of 500–1000 base pairs. Samples were then subjected to immunoprecipitation using ZBRK1 (ChIP grade) antibody, followed by DNA purification. Only beads were used as a negative control for the experiment. Immunoprecipitated chromatin was de-cross-linked and subjected to PCR using primer sets for promoter regions of SLUG. Primer sequences for amplifying the SLUG promoter region are forward, 5′-AGGCAAACCTCTCCAGATGC-3′; and backward, 5′=CCTGGCTTCCAGATGTGGTG-3′). We loaded 3% of input for final assay.
Nuclear and cytoplasmic extract preparation
Nuclear and cytoplasmic extracts were prepared as described previously (Abmayr et al., 2006). Briefly, cells containing wild-type or mutant villin proteins were grown and harvested after a brief washing with ice-cold phosphate-buffered saline (PBS). Cells were treated with hypotonic buffer before 10 shots of homogenization using a glass Dounce homogenizer and pestle B. The nuclear and S-100 fraction were separated by centrifugation at 3210 × g at 4°C. The nuclear and cytoplasmic fractions were cleared by several rounds of centrifugation and dialysis and analyzed by Western blot analysis.
Xenograft experiments
Colon cancer xenografts were generated as described previously (Arango et al., 2012). We mixed 5 × 107 colon cancer cell lines in a volume of 100 μl of PBS with an equal volume of Matrigel and injected them subcutaneously into the right flank of SCID mice. Tumors were excised, fixed in Formalin, and processed for immunohistochemistry. Nuclear localization was quantitated by counting the number of nuclei with primarily nuclear villin localization. Averages of 100 cells randomly selected in 25 distinct areas of the slide were counted for each of the cell lines shown.
Tissue microarrays and immunohistochemistry
Tissue microarray (TMA) generation from 577 human Dukes’ B and C colon cancers has been described previously (Arango et al., 2012). The microarrays were stained with anti-villin antibody obtained from Novacastra (Newcastle on Tyne, United Kingdom) at a dilution of 1:50. The scoring of patient samples for villin expression was described previously (Arango et al., 2012). Each sample was scored for villin staining intensity as 0 (no staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). For each sample, the percentage of positive cells was also scored and the subcellular localization of villin (membrane, membrane and cytoplasmic, cytoplasmic, or nuclear) recorded. The specificity of the antibody was validated previously (Arango et al., 2012) in colon cancer cells in which only those lines that expressed villin mRNA and protein (by Western blot) stained positively for villin by immunohistochemistry when grown as xenografts.
Statistical analysis
Statistical analysis was performed using the two-tailed Student’s t test; p was based on unpaired samples and unequal variance. Quantitative analysis of number of cysts, extensions, and tubules was done using one-way repeated measure analysis of variance and Tukey’s modified t test.
Supplementary Material
Acknowledgments
We thank Langzhu Tan for assistance with the yeast two-hybrid studies, Leon Chatman for assistance with the tubulogenesis assays, and Rizwan Siddiqui for assistance with the E-cadherin assays. This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-98120 (to S.K.) and Public Health Service Grant DK-56338.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- EMT
epithelial–mesenchymal transition
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- LMB
leptomycin B
- NES
nuclear export signal
- NLS
nuclear localization signal
- seYFP
superenhanced yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-06-0453) December 10, 2015.
REFERENCES
- Abmayr SM, Yao T, Parmely T, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb1201s75. Chapter 12, Unit 12.11. [DOI] [PubMed] [Google Scholar]
- Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE. Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis. Cancer. 2002;94:344–351. doi: 10.1002/cncr.10221. [DOI] [PubMed] [Google Scholar]
- Arango D, Al-Obaidi S, Williams DS, Dopeso H, Mazzolini R, Corner G, Byun DS, Carr AA, Murone C, Togel L, et al. Villin expression is frequently lost in poorly differentiated colon cancer. Am J Pathol. 2012;180:1509–1521. doi: 10.1016/j.ajpath.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athman R, Louvard D, Robine S. Villin enhances hepatocyte growth factor-induced actin cytoskeleton remodeling in epithelial cells. Mol Biol Cell. 2003;14:4641–4653. doi: 10.1091/mbc.E03-02-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Mostov KE. From cells to organs: building polarized tissue. Nat Rev Mol Cell Biol. 2008;9:887–901. doi: 10.1038/nrm2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2007;8:84–90. doi: 10.1038/sj.embor.7400855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Pires IM, Weston L, Buffa FM, Milani M, Li JL, Harris AL, Hammond EM, La Thangue NB. Hypoxia-driven cell motility reflects the interplay between JMY and HIF-1alpha. Oncogene. 2011;30:4835–4842. doi: 10.1038/onc.2011.188. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Robbins J, Dilworth SM, Roberts B, Richardson WD. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol. 1988;107:841–849. doi: 10.1083/jcb.107.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrary E, Cohen-Tannoudji M, Pehau-Arnaudet G, Lapillonne A, Athman R, Ruiz T, Boulouha L, El Marjou F, Doye A, Fontaine JJ, et al. In vivo, villin is required for Ca(2+)-dependent F-actin disruption in intestinal brush borders. J Cell Biol. 1999;146:819–830. doi: 10.1083/jcb.146.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Wang JM, Wei S, Jeng YM, Jiang X, Gu B, Chen PL, Lee EY, Lee WH. Removal of BRCA1/CtIP/ZBRK1 repressor complex on ANG1 promoter leads to accelerated mammary tumor growth contributed by prominent vasculature. Cancer Cell. 2006;10:13–24. doi: 10.1016/j.ccr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Garcia V, Garcia JM, Pena C, Silva J, Dominguez G, Rodriguez R, Maximiano C, Espinosa R, Espana P, Bonilla F. The GADD45, ZBRK1 and BRCA1 pathway: quantitative analysis of mRNA expression in colon carcinomas. J Pathol. 2005;206:92–99. doi: 10.1002/path.1751. [DOI] [PubMed] [Google Scholar]
- George SP, Chen H, Conrad JC, Khurana S. Regulation of directional cell migration by membrane-induced actin bundling. J Cell Sci. 2013;126:312–326. doi: 10.1242/jcs.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SP, Wang Y, Mathew S, Srinivasan K, Khurana S. Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J Biol Chem. 2007;282:26528–26541. doi: 10.1074/jbc.M703617200. [DOI] [PubMed] [Google Scholar]
- Goldstein NS, Thomas M. Mucinous and nonmucinous bronchioloalveolar adenocarcinomas have distinct staining patterns with thyroid transcription factor and cytokeratin 20 antibodies. Am J Clin Pathol. 2001;116:319–325. doi: 10.1309/550P-QLJX-D84W-F6DN. [DOI] [PubMed] [Google Scholar]
- Grossmann KS, Rosario M, Birchmeier C, Birchmeier W. The tyrosine phosphatase Shp2 in development and cancer. Adv Cancer Res. 2010;106:53–89. doi: 10.1016/S0065-230X(10)06002-1. [DOI] [PubMed] [Google Scholar]
- Hotz B, Arndt M, Dullat S, Bhargava S, Buhr HJ, Hotz HG. Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin Cancer Res. 2007;13:4769–4776. doi: 10.1158/1078-0432.CCR-06-2926. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- Khurana S. Structure and function of villin. In: Khurana S, editor. Aspects of the Cytoskeleton. New York: Elsevier; 2006. pp. 89–115. [Google Scholar]
- Khurana S, Arpin M, Patterson R, Donowitz M. Ileal microvillar protein villin is tyrosine-phosphorylated and associates with PLC-gamma1. Role of cytoskeletal rearrangement in the carbachol-induced inhibition of ileal NaCl absorption. J Biol Chem. 1997;272:30115–30121. doi: 10.1074/jbc.272.48.30115. [DOI] [PubMed] [Google Scholar]
- Khurana S, George SP. Regulation of cell structure and function by actin-binding proteins: villin’s perspective. FEBS Lett. 2008;582:2128–2139. doi: 10.1016/j.febslet.2008.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Tomar A, George SP, Wang Y, Siddiqui MR, Guo H, Tigyi G, Mathew S. Autotaxin and lysophosphatidic acid stimulate intestinal cell motility by redistribution of the actin modifying protein villin to the developing lamellipodia. Exp Cell Res. 2008;314:530–542. doi: 10.1016/j.yexcr.2007.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Zhao P, Tomar A, Galea CA, Khurana S. Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J Biol Chem. 2004;279:3096–3110. doi: 10.1074/jbc.M308878200. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Breese EH, Vig-Varga E, Luo Y, Lee Y, Goebl MG, Harrington MA. Tumor necrosis factor alpha induction of NF-kappaB requires the novel coactivator SIMPL. Mol Cell Biol. 2004;24:9317–9326. doi: 10.1128/MCB.24.21.9317-9326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lau SK, Prakash S, Geller SA, Alsabeh R. Comparative immunohistochemical profile of hepatocellular carcinoma, cholangiocarcinoma, and metastatic adenocarcinoma. Hum Pathol. 2002;33:1175–1181. doi: 10.1053/hupa.2002.130104. [DOI] [PubMed] [Google Scholar]
- Lhocine N, Arena ET, Bomme P, Ubelmann F, Prevost MC, Robine S, Sansonetti PJ. Apical invasion of intestinal epithelial cells by Salmonella typhimurium requires villin to remodel the brush border actin cytoskeleton. Cell Host Microbe. 2015;17:164–177. doi: 10.1016/j.chom.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LF, Chuang CH, Li CF, Liao CC, Cheng CP, Cheng TL, Shen MR, Tseng JT, Chang WC, Lee WH, Wang JM. ZBRK1 acts as a metastatic suppressor by directly regulating MMP9 in cervical cancer. Cancer Res. 2010;70:192–201. doi: 10.1158/0008-5472.CAN-09-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LF, Li CF, Wang WJ, Yang WM, Wang DD, Chang WC, Lee WH, Wang JM. Loss of ZBRK1 contributes to the increase of KAP1 and promotes KAP1-mediated metastasis and invasion in cervical cancer. PLoS One. 2013;8:e73033. doi: 10.1371/journal.pone.0073033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew S, George SP, Wang Y, Siddiqui MR, Srinivasan K, Tan L, Khurana S. Potential molecular mechanism for c-Src kinase-mediated regulation of intestinal cell migration. J Biol Chem. 2008;283:22709–22722. doi: 10.1074/jbc.M801319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Xue B. Cell polarity as a regulator of cancer cell behavior plasticity. Annu Rev Cell Dev Biol. 2012;28:599–625. doi: 10.1146/annurev-cellbio-092910-154244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M, Langowski J. Dynamics of the CapG actin-binding protein in the cell nucleus studied by FRAP and FCS. Chromosome Res. 2008;16:427–437. doi: 10.1007/s10577-008-1234-6. [DOI] [PubMed] [Google Scholar]
- Revenu C, Courtois M, Michelot A, Sykes C, Louvard D, Robine S. Villin severing activity enhances actin-based motility in vivo. Mol Biol Cell. 2007;18:827–838. doi: 10.1091/mbc.E06-05-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Ubelmann F, Hurbain I, El-Marjou F, Dingli F, Loew D, Delacour D, Gilet J, Brot-Laroche E, Rivero F, et al. A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell. 2012;23:324–336. doi: 10.1091/mbc.E11-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp EL, Altan N, Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr Protoc Cell Biol. 2003 doi: 10.1002/0471143030.cb2101s19. Chapter 21, Unit 21.21. [DOI] [PubMed] [Google Scholar]
- Tomar A, George S, Kansal P, Wang Y, Khurana S. Interaction of phospholipase C-gamma1 with villin regulates epithelial cell migration. J Biol Chem. 2006;281:31972–31986. doi: 10.1074/jbc.M604323200. [DOI] [PubMed] [Google Scholar]
- Tomar A, Wang Y, Kumar N, George S, Ceacareanu B, Hassid A, Chapman KE, Aryal AM, Waters CM, Khurana S. Regulation of cell motility by tyrosine phosphorylated villin. Mol Biol Cell. 2004;15:4807–4817. doi: 10.1091/mbc.E04-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubelmann F, Chamaillard M, El-Marjou F, Simon A, Netter J, Vignjevic D, Nichols BL, Quezada-Calvillo R, Grandjean T, Louvard D, et al. Enterocyte loss of polarity and gut wound healing rely upon the F-actin-severing function of villin. Proc Natl Acad Sci USA. 2013;110:E1380–E1389. doi: 10.1073/pnas.1218446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, George SP, Srinivasan K, Patnaik S, Khurana S. Actin reorganization as the molecular basis for the regulation of apoptosis in gastrointestinal epithelial cells. Cell Death Differ. 2012;19:1514–1524. doi: 10.1038/cdd.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Srinivasan K, Siddiqui MR, George SP, Tomar A, Khurana S. A novel role for villin in intestinal epithelial cell survival and homeostasis. J Biol Chem. 2008;283:9454–9464. doi: 10.1074/jbc.M707962200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tomar A, George SP, Khurana S. Obligatory role for phospholipase C-gamma(1) in villin-induced epithelial cell migration. Am J Physiol Cell Physiol. 2007;292:C1775–C1786. doi: 10.1152/ajpcell.00420.2006. [DOI] [PubMed] [Google Scholar]
- Zhang F, White RL, Neufeld KL. Phosphorylation near nuclear localization signal regulates nuclear import of adenomatous polyposis coli protein. Proc Natl Acad Sci USA. 2000;97:12577–12582. doi: 10.1073/pnas.230435597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.