Adaptor complexes are important for cargo sorting in clathrin-coated vesicles. The µ adaptor subunits Apm1 and Apm2 create functionally distinct versions of the yeast AP-1 complex. A novel regulatory protein is identified that selectively binds Apm2-containing complexes and contributes to their membrane recruitment.
Abstract
Heterotetrameric adaptor protein complexes are important mediators of cargo protein sorting in clathrin-coated vesicles. The cell type–specific expression of alternate μ chains creates distinct forms of AP-1 with altered cargo sorting, but how these subunits confer differential function is unclear. Whereas some studies suggest the μ subunits specify localization to different cellular compartments, others find that the two forms of AP-1 are present in the same vesicle but recognize different cargo. Yeast have two forms of AP-1, which differ only in the μ chain. Here we show that the variant μ chain Apm2 confers distinct cargo-sorting functions. Loss of Apm2, but not of Apm1, increases cell surface levels of the v-SNARE Snc1. However, Apm2 is unable to replace Apm1 in sorting Chs3, which requires a dileucine motif recognized by the γ/σ subunits common to both complexes. Apm2 and Apm1 colocalize at Golgi/early endosomes, suggesting that they do not associate with distinct compartments. We identified a novel, conserved regulatory protein that is required for Apm2-dependent sorting events. Mil1 is a predicted lipase that binds Apm2 but not Apm1 and contributes to its membrane recruitment. Interactions with specific regulatory factors may provide a general mechanism to diversify the functional repertoire of clathrin adaptor complexes.
INTRODUCTION
Clathrin-coated vesicles mediate the transfer of membrane proteins between different cellular compartments. Heterotetrameric adaptor protein (AP) complexes bind short linear motifs on cargo proteins and incorporate them into forming vesicles by linking them to the clathrin coat (Edeling et al., 2006). Five AP complexes (AP-1 to AP-5) have been identified that are believed to regulate distinct trafficking pathways at the Golgi, endosome/lysosome, or plasma membrane (Hirst et al., 2013). These AP complexes share the same basic plan, being composed of two large subunits (β1-5 and γ/α/δ/ε/ζ, ∼100 kDa), one medium subunit (μ1-5, ∼50 kDa), and one small subunit (σ1-5, ∼20 kDa). Use of alternate subunits has the potential to further expand the diversity of AP complexes (Mattera et al., 2011). For example, three AP-1 subunits (γ, μ1, and σ1) have isoforms encoded by different genes, and mutations in the three σ1 isoforms AP1S1, AP1S2, and AP1S3 cause different diseases: MEDNIK syndrome, X-linked mental retardation, and pustular psoriasis, respectively (Tarpey et al., 2006; Montpetit et al., 2008; Setta-Kaffetzi et al., 2014).
Different AP subunits are responsible for binding clathrin, cargo-sorting motifs, accessory factors, and membrane lipids (Edeling et al., 2006). Sorting motifs that conform to the YXXΦ consensus (where Φ is a bulky, hydrophobic amino acid) bind a conserved pocket in the medium subunit, whereas dileucine-sorting motifs (D/E)XXXL(L/I) are recognized by the small and non-β large subunits (Canagarajah et al., 2013; Traub and Bonifacino, 2013). In the case of AP-1, Arf1 cooperates with cargo and phosphoinositides to mediate membrane recruitment. Arf1-GTP bridges two AP-1 complexes by binding the β and γ subunits (Ren et al., 2013), whereas the γ subunit of AP-1 binds phosphatidylinositol 4-phosphate at a site similar to that used by the AP-2 α subunit to bind phosphatidylinositol (4,5)-bisphosphate (Heldwein et al., 2004). Finally, clathrin and accessory factors interact with linker and appendage domains of the large subunits, respectively.
The extent to which the use of variant subunit isoforms alters adaptor function is not well understood. The ubiquitous AP-1A and epithelial cell–specific AP-1B complexes, which differ only in the incorporation of μ1A or μ1B, provide one of the best-studied examples (Bonifacino, 2014; Nakatsu et al., 2014). Several studies have suggested that differential lipid binding by the μ subunits directs AP-1A to the Golgi but causes AP-1B to localize to recycling endosomes, presenting a plausible basis for their functional diversity (Fölsch et al., 2003; Fields et al., 2010). However, subsequent work showed AP-1A and AP-1B largely colocalize and that μ1A and μ1B recognize cargo proteins with different affinities (Guo et al., 2013). Thus the basis for the differential functions of these isoforms is unclear (Rodriguez-Boulan et al., 2013), and it is not known whether subunit exchange alters the function of other adaptor complexes.
Of the three AP complexes in the yeast Saccharomyces cerevisiae, only AP-1 uses alternate subunits. The two forms of AP-1 share the same large (Apl2 and Apl4) and small (Aps1) subunits but incorporate different medium subunits (Apm1 or Apm2). Whereas the classical Apm1-containing AP-1 complex transports well-characterized cargo, including chitin synthase III (Chs3) and Sna2 (Valdivia et al., 2002; Renard et al., 2010), no function has been identified for the Apm2-containing complex, which we refer to as AP-1R (AP-1 Related). Early studies suggested that these complexes are biochemically distinct (Stepp et al., 1995; Yeung et al., 1999). Whereas Apm1 and Apm2 were proposed to act redundantly in trafficking the lipid flippase Drs2 (Liu et al., 2008), Apm2 is dispensable for many clathrin-dependent sorting events (Stepp et al., 1995).
Here we show that the Apm2 subunit confers distinct functions on the yeast AP-1 complex. AP-1R and AP-1 sort distinct cargo proteins and exhibit differential sensitivity to cationic lipophilic drugs. These differences are not readily explained by divergent cargo recognition properties because AP-1R is unable to substitute for AP-1 in the sorting of cargo via a dileucine-based motif that is recognized by subunits common to both AP-1 and AP-1R. Strikingly, Apm2 but not Apm1 binds a novel regulatory protein that we call Mil1 (mu-interacting ligand). Mil1 is a predicted serine hydrolase that is required for Apm2-dependent sorting processes and has a role in recruiting Apm2 to membranes. This suggests that binding of specific regulatory factors might be an important mechanism by which subunit exchange confers new functions on clathrin adaptors.
RESULTS
Apm2 is part of a functionally distinct AP-1–related complex
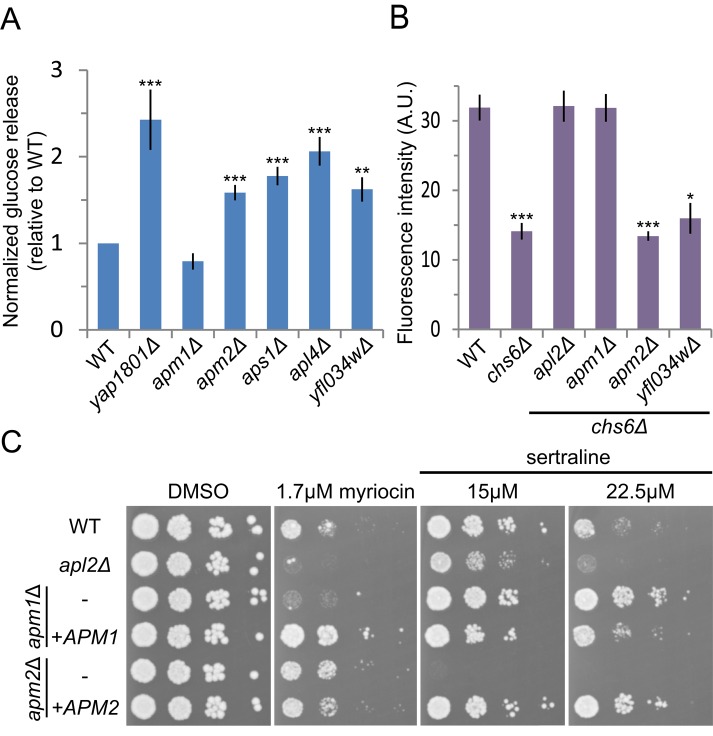
Previously we uncovered genes required for the trafficking of the vesicle soluble N-ethylmaleimide–sensitive factor attachment protein receptor (v-SNARE) Snc1 in a genome-wide screen that assessed changes in the surface levels of the GFP-Snc1-Suc2 (GSS) reporter protein (Burston et al., 2009). Genetic and physical interaction analysis identified a cluster of four genes, representing three AP-1 subunits (Apm2, Apl4, and Aps1) and the uncharacterized Yfl034w, whose deletion increased surface GSS levels. The identification of Apm2, but not the more abundant μ-subunit isoform Apm1, was unexpected and suggested that Apm2-containing AP-1R complex has a specific role in Snc1 sorting. To confirm these observations, we used a quantitative assay that measures the invertase (Suc2) activity of GSS present at the cell surface (Figure 1A; Dalton et al., 2015). Loss of APM2, APL4, and APS1 led to a slight but significant increase in GSS surface levels, comparable to that of the endocytosis mutant yap1801 (Burston et al., 2009). This increase was not observed in the absence of APM1, showing that GSS sorting is an AP-1R–specific function and does not require the Apm1-containing AP-1 complex, which we will refer to here as AP-1.
FIGURE 1:
APM1 and APM2 deletions have distinct sorting phenotypes. (A) APM2 but not APM1 is required for Snc1 sorting. Cell-surface levels of the Snc1 reporter GSS were quantified in yeast deletion strains by measuring invertase activity of liquid cultures normalized to wild type. Unpaired t test of mutant strains compared with wild type, ***p < 0.0001 and **p < 0.01. Error bars represent SEM (n = 6). (B) Deletion of APM1 but not APM2 bypasses the Chs3 trafficking defect of a chs6 mutant strain. Chs3 surface activity was quantified as the fluorescence intensity of strains plated on 50 μg/ml calcofluor white and reported in arbitrary units (A.U.). Unpaired t test compared with wild type, ***p < 0.0001 and *p < 0.05. Error bars represent SEM (n = 8). (C) To assess drug sensitivity, yeast were spotted in 10× dilution series and grown on YPD with Hyg B plus indicated drugs or DMSO as control.
Previous studies showed that the AP-1 complex is required for the transport of the chitin synthase Chs3 between Golgi and endosomal compartments (Valdivia et al., 2002). Exomer mutants, such as chs6, have low chitin levels because Chs3 is not transported to the surface but instead is retained in an AP-1–dependent intracellular recycling loop. Deletion of the large AP-1 subunit Apl2 or of Apm1 bypasses this intracellular retention, restoring Chs3 transport to the cell surface and chitin production. Previous studies found no role for Apm2 in Chs3 sorting using a qualitative assay (Valdivia et al., 2002). To detect subtle defects, we assessed the requirement for Apm2 in Chs3 transport using a sensitive quantitative assay based on the fluorescent compound calcofluor white (CW; Lam et al., 2006; Burston et al., 2008), which binds chitin produced by cell-surface Chs3. Whereas deletion of AP-1 components (APM1, APL2) restored wild-type cell surface levels of Chs3 in chs6 cells, deletion of APM2 or YFL034W did not (Figure 1B). These results confirm that Apm2 does not play a role in the AP-1–mediated sorting of Chs3.
Because the Snc1 sorting defect in apm2 mutants was relatively mild, we sought alternative phenotypes that distinguish Apm1 and Apm2. Genome-wide chemogenomic studies (Hoepfner et al., 2014; Lee et al., 2014) suggested that Apm1 and Apm2 have differential drug sensitivities. By exposing apm1 and apm2 strains to a subset of these drugs, we found that strains lacking APM1 but not APM2 were sensitive to myriocin, an inhibitor of sphingolipid biosynthesis (Figure 1C). In contrast, apm2 mutants were sensitive to cationic amphiphilic drugs (CADs) such as sertraline, as previously reported (Rainey et al., 2010), whereas apm1 mutants exhibited sertraline resistance (Figure 1C). CADs intercalate in phospholipid bilayers and may interfere with protein function, including lipid binding (Daniel et al., 2015). The differential sensitivities of these adaptor protein mutants may reflect missorting of cargo and be important in responding to stresses imposed by the drug. These differential drug sensitivities, together with the cargo-specific sorting defects, demonstrate that Apm1 and Apm2 have distinct functions.
AP-1R localizes to the late Golgi and early endosomes
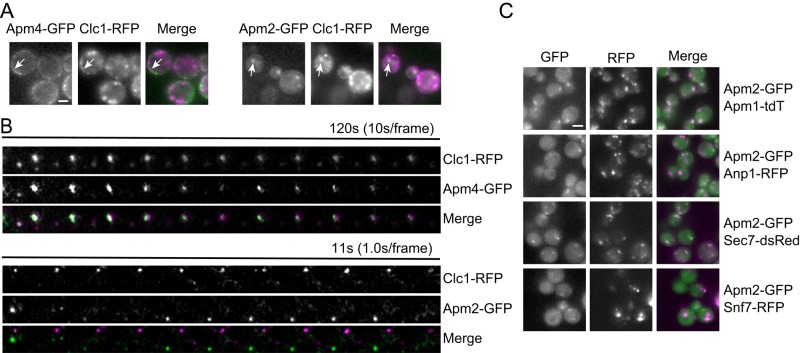
Because Snc1 undergoes a rapid cycle of internalization and recycling to the plasma membrane, increased surface levels in cells lacking AP-1R could reflect reduced endocytosis or an enhanced rate of recycling from intracellular compartments. To determine whether Apm2 is part of the endocytosis machinery, we compared the localizations of Apm2-GFP and the AP-2 medium subunit Apm4-GFP with that of clathrin light chain Clc1-RFP in sla2 strains, which stall endocytosis of clathrin-coated pits (Kaksonen et al., 2003; Newpher et al., 2005). Whereas Apm4-GFP colocalized with Clc1-RFP at the plasma membrane, Apm2-GFP was largely localized to intracellular clathrin compartments (Figure 2A). A small number of Apm2-GFP puncta were observed at or near the plasma membrane. To determine whether these represent sites of endocytosis, we carried out time-lapse imaging using total internal reflection fluorescence microscopy of cells treated with the actin inhibitor latrunculin A (LatA) to block the budding of cell-surface clathrin-coated pits. The AP-2 subunit Apm4-GFP maintained strong colocalization with Clc1–red fluorescent protein (RFP) at immobile patches on the plasma membrane (Figure 2B). In contrast, Apm2-GFP–containing structures near the cell surface were highly mobile and did not colocalize with plasma membrane Clc1-RFP puncta over time. Some lack of colocalization between AP-1 and clathrin was previously observed (Daboussi et al. 2012), suggesting that adaptors can be present at membranes without significant clathrin binding. The absence of AP-1R at the plasma membrane is inconsistent with a role in endocytosis and suggests instead that it functions in an intracellular pathway.
FIGURE 2:
Apm2 localizes to late Golgi/early endosomes. (A) Fluorescence microscopy of live endocytosis-defective sla2 yeast shows that Apm2 is not present at cell-surface clathrin-coated pits. White arrows indicate puncta that colocalize in GFP and RFP channels. (B) Kymograph analysis of Apm4-GFP or Apm2-GFP relative to cortical clathrin patches marked by Clc1-RFP. Cells were incubated with the actin inhibitor latrunculin A and imaged by TIRF microscopy. (C) Localization of Apm2-GFP relative to that of Apm1-tdTomato (AP-1 complex), Anp1-RFP (early Golgi), Sec7-DsRed (late Golgi/endosomes), and Snf7-RFP (late endosomes). Scale bar, 4 μm.
To define the intracellular distribution of Apm2-GFP, we performed colocalization experiments with fluorescently tagged marker proteins (n = 3, >125 cells/experiment; Figure 2C). Consistent with a role in Golgi/endosomal sorting, 63 ± 3% of Apm2-GFP puncta colocalized with the late-Golgi/early endosome marker Sec7-dsRed. Apm2-GFP only partially colocalized with the early Golgi marker Anp1-RFP (28 ± 3%) and showed little colocalization with the late endosome marker Snf7-RFP (8 ± 1%). These results indicate that AP-1R localizes primarily to the late Golgi/early endosomes, similar to previous findings placing AP-1 at these compartments (Liu et al., 2008; Daboussi et al., 2012). Of importance, Apm2-GFP showed extensive colocalization with Apm1-tdTomato (77 ± 2%), suggesting that the bulk of AP-1R is present at the same intracellular compartment as AP-1.
Recognition of tyrosine-based signals by Apm1 and Apm2
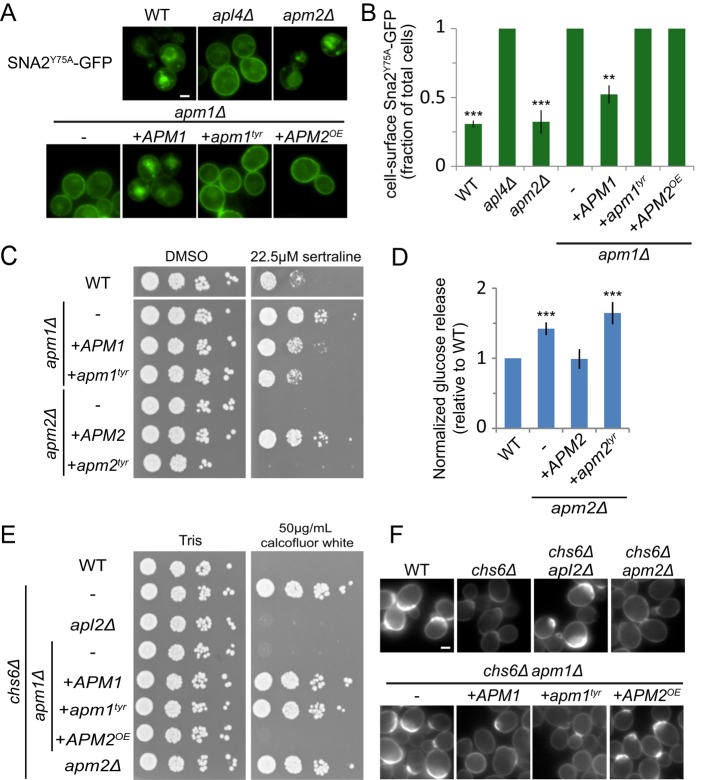
The different phenotypes of apm1 and apm2 mutants suggest that Apm1 and Apm2 recognize different cargo for incorporation into vesicles. The μ subunits of AP complexes interact with tyrosine-based motifs (YxxΦ; Canagarajah et al., 2013) via residues conserved in both Apm1 and Apm2 (Supplemental Figure S1A). The vacuolar protein Sna2 has two tyrosine-based sorting motifs at Y65 and Y75, which govern its trafficking by AP-1 and AP-3, respectively (Renard et al., 2010). We tested sorting of a Sna2 mutant (Sna2Y75A) that lacks the AP-3 signal and whose vacuolar transport depends on AP-1 (Figure 3, A and B). Loss of the AP-1 components Apl4 and Apm1 missorted Sna2Y75A-GFP to the cell surface, consistent with previous studies (Renard et al., 2010). The Sna2 tyrosine-based signal is expected to bind the conserved pocket in Apm1. Indeed, a version of Apm1 containing mutations at this site (APM1tyr: F179A D181S) was present at the same level as wild-type Apm1 (Supplemental Figure S1B) but was unable to rescue the Sna2Y75A-GFP sorting defect of an apm1 mutant.
FIGURE 3:
The predicted YxxΦ-binding pocket is required for some but not all functions of Apm1 and Apm2. (A) Expression of an Apm1 mutant lacking a functional tyrosine signal-binding pocket (apm1tyr) or overexpression of APM2 from the APM1 promoter (APM2OE) is unable to restore the vacuolar localization of the AP-1–specific cargo Sna2Y75A-GFP in apm1 mutant cells. Scale bar, 2 μm. (B) Quantification of surface fluorescence of Sna2Y75A-GFP. Error bars represent SEM (n = 3). Unpaired t test compared with apm1Δ, ***p < 0.0001 and **p < 0.01. (C) An Apm2 mutant lacking a functional tyrosine signal-binding pocket (apm2tyr) and expressed on a plasmid from its native promoter exhibits sertraline sensitivity. Yeast were plated in a 10× dilution series on YPD containing Hyg B and DMSO or 22.5 μM sertraline. (D) The apm2tyr mutant is unable to restore sorting of the Snc1 reporter GSS. Cell-surface GSS levels in the indicated strains were determined by quantifying invertase activity and normalizing to levels in wild-type cells. Unpaired t test compared with wild type, ***p < 0.0001. Error bars represent SEM (n = 6). (E) Wild-type APM1 and apm1tyr, but not overexpressed APM2 (APM2OE), restores the calcofluor resistance of chs6 apm1 mutants. Yeast were plated in a 10× dilution series on buffered YPD containing Hyg B and 50 μg/ml calcofluor white (CW). (F) Representative fluorescence images of calcofluor white–stained strains.
We found that Apm2 is not required for Sna2Y75A-GFP sorting (Figure 3, A and B; Renard et al., 2010). However, it is much less abundant than Apm1 and could have a minor but redundant role that would not be detected in knockout experiments. To determine whether Apm2 can substitute for Apm1 when expressed at similar level, we placed hemagglutinin (HA)-tagged Apm2 under the Apm1 promoter (APM2OE). We confirmed that expression of Apm2 was comparable to that of Apm1 (Supplemental Figure S1) but observed no rescue of Sna2Y75A-GFP sorting in an apm1 strain. In contrast, reintroduction of wild-type Apm1 significantly restored localization of Sna2Y75A-GFP to the cell surface (n = 3, p = 0.0005; Figure 3B). These results suggest that whereas Apm1 sorts Sna2 through a tyrosine-based motif, Apm2 does not. Apm2 could lack a functional YxxΦ-binding pocket. However, wild-type Apm2, but not the Apm2 tyrosine motif-binding mutant (Apm2tyr: F273A D275S), rescued growth on sertraline in an apm2 background (Figure 3C), suggesting that Apm2 may sort a protein required for sertraline resistance via a tyrosine-containing signal. The Snc1 cytosolic domain lacks tyrosine residues and may bind to Apm2 via a noncanonical sorting motif. Of interest, the Apm2tyr mutant also failed to sort GSS in the invertase assay (Figure 3D). This suggests that Apm2 sorts another cargo necessary for the normal recycling of Snc1 or that a noncanonical signal in Snc1 binds the same site in Apm2.
AP-1 binds Chs3 via a dileucine-based motif (Starr et al., 2012). Such motifs are recognized by the γ/σ subunits. Thus mutation of the Apm1 tyrosine-binding pocket is not predicted to affect Chs3 sorting. To test this, we grew serial dilutions of deletion strains on media containing toxic amounts of calcofluor white (Figure 3E). Exomer mutants, such as chs6, that retain Chs3 inside the cell have reduced chitin and are resistant to calcofluor white, whereas AP-1 mutants that bypass chs6 and allow Chs3 to reach the surface restore calcofluor white sensitivity. Expression of either wild-type APM1 or apm1tyr in the chs6 apm1 background resulted in a chs6 phenotype (Figure 3, E and F), confirming that Chs3 sorting does not require the Apm1 YxxΦ-binding pocket.
Because the dileucine-based motif of Chs3 binds the γ/σ subunits present in both AP-1 and AP-1R, Apm2 should substitute for Apm1 if present at the same level. Surprisingly, expression of APM2 from the APM1 promoter did not alter the calcofluor white–sensitive phenotype of chs6 apm1 strains (Figure 3, E and F), indicating that AP-1R is incapable of sorting Chs3. This result suggests that differential cargo recognition is not the reason for the different sorting specificity of AP-1 and AP-1R.
Mil1(Yfl034w) specifically binds the Apm2 C-terminal subdomain B
One explanation for the functional differences between Apm1 and Apm2 is that they bind different regulatory proteins that direct them into specific pathways. The uncharacterized protein Yfl034w clustered with AP-1R subunits in our genome-wide analysis (Burston et al., 2009) and, like Apm2, was required for sorting of the Snc1 reporter but not Chs3 (Figure 1, A and B), suggesting that it shares a closely related function. On the basis of results described later, we refer to Yfl034w as Mil1 (medium adaptin-interacting ligand).
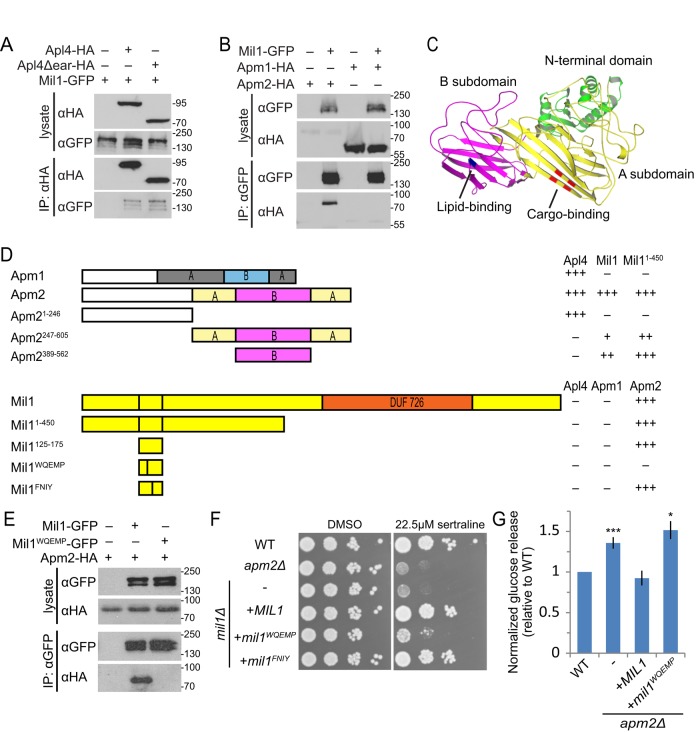
Physical interactions between Mil1 and components of the AP-1 complex were reported in several proteomic studies (Gavin et al., 2006; Krogan et al., 2006; Collins et al., 2007; Babu et al., 2012). We confirmed this interaction, showing that Mil1-GFP copurifies with Apl4-HA immunoprecipitated from yeast cell lysates (Figure 4A). Like all heterotetrameric clathrin adaptors, AP-1 consists of a core complex with two appendages (“ears”) projecting from the large subunits by flexible linkers (Edeling et al., 2006). Although the Apl4 (γ) appendage recruits many regulatory proteins, Mil1 was able to bind a truncated form of Apl4 lacking this appendage (Apl4Δear; Figure 4A). We reasoned that if Mil1 is an AP-1R regulator, it might associate preferentially with Apm2, which is part of the core complex. Apm2-HA was present at low levels in the cell lysate, yet we observed a clear enrichment of Apm2-HA upon immunoprecipitation of Mil1-GFP, whereas there was little if any pull down of Apm1-HA, showing that the physical interaction between Apm2 and Mil1 is highly specific (Figure 4B).
FIGURE 4:
Mil1 interacts with Apm2 through its WQEMP motif. (A) Mil1-GFP copurified with immunoprecipitated Apl4-3HA and with a truncated version lacking the γ appendage (Apl4Δear-HA) known to bind several AP-regulatory proteins. Loading of lysate relative to immunoprecipitate was 1:9. All proteins were genomically tagged. (B) Pull down of Mil1-GFP in strains coexpressing either Apm1-3HA or Apm2-3HA shows that Mil1 binds specifically to Apm2. All proteins were genomically tagged. (C) Phyre2 homology model of Apm2, colored to indicate the N-terminal AP-binding domain (green) and a C-terminal region composed of A (yellow) and B (magenta) subdomains. Key residues predicted to be involved in YxxΦ binding (red) or lipid binding (blue) based on alignment with regions of AP-1 and AP-2 μ subunits are indicated. (D) Yeast two-hybrid mapping of Apm2-Mil1–binding domains. Full-length or truncated Apm2 constructs were fused to the GAL4 DNA-binding domain (GBD), and full-length, truncated, or mutated Mil1 constructs were fused to the GAL4-activating domain (GAD). Qualitative interaction strengths are indicated. Mil1WQEMP represents the W143QEMP>AAEAA mutant, and Mil1FNIY represents the F152NIY>ANAA mutant. (E) Anti-GFP immunoprecipitation of plasmid-expressed wild-type Mil1-GFP or Mil1WQEMP-GFP from strains coexpressing genomically tagged Apm2-3HA. (F) Sertraline sensitivity was assessed by plating strains in a 10× dilution series on YPD containing 22.5 μM sertraline or DMSO as a control. (G) The mil1WQEMP mutant is unable to restore sorting of the Snc1 reporter GSS. Cell-surface GSS levels in the indicated strains were determined by quantifying invertase activity and normalizing to levels in wild-type cells. Unpaired t test compared with wild type, ***p < 0.0001 and *p < 0.05. Error bars represent SEM (n = 10).
A yeast two-hybrid assay was used to map interacting regions. Based on structural homology to other μ adaptins, Apm2 has an N-terminal domain required for its incorporation into the AP complex, whereas the C-terminus consists of two subdomains: an A domain, containing the tyrosine-binding pocket, and a B domain, which may contribute to membrane recruitment but does not contain known cargo-binding sites (Figure 4C; Fields et al., 2010; Canagarajah et al., 2013). The minimal Mil1-binding region was mapped to residues 389–562, which correspond to the B domain (Figure 4D).
Additional mutations were used to define the Apm2-binding site on Mil1. First, the interaction was narrowed to a region between residues 125 and 175. Point mutations that alter conserved residues in this 50–amino acid fragment revealed that a F152NIY>ANAA mutant retained the yeast two-hybrid interaction with Apm2, but a W143QEMP>AAEAA mutant did not. Furthermore, introducing the W143QEMP>AAEAA mutation into the full-length Mil1 protein resulted in a Mil1WQEMP mutant that was stably expressed but did not coprecipitate with Apm2 (Figure 4E).
Having identified an Apm2 binding–deficient Mil1 mutant, we tested whether the Apm2-Mil1 interaction was important for Apm2 function. Sensitivity of MIL1 deletion strains could be rescued by a plasmid expressing Mil1 or the full-length F152NIY>ANAA (Mil1FNIY) mutant, whereas the Mil1WQEMP mutant showed only weak complementation (Figure 4F). In addition, only wild-type Mil1 was able to complement GSS sorting (Figure 4G). Thus the Apm2-Mil1 interaction is functionally important, suggesting that Mil1 plays a key role in AP-1R–mediated sorting processes.
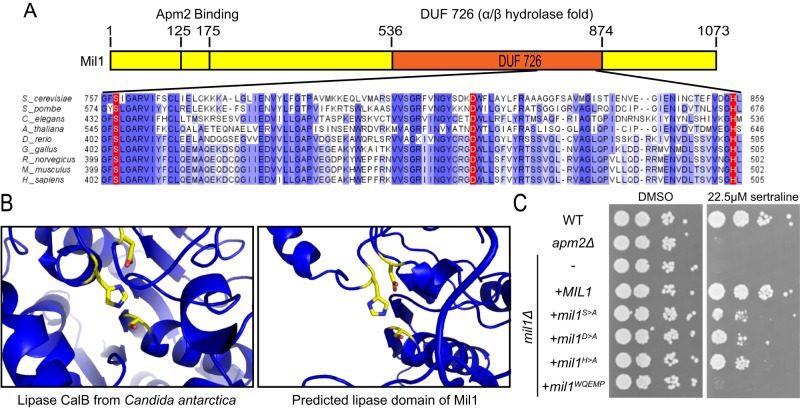
Mil1 has a conserved α/β-hydrolase catalytic motif required for its function
Mil1, which contains a C-terminal DUF726 domain, is predicted to be a serine hydrolase with an α/β hydrolase fold (Baxter et al., 2004). Serine hydrolases constitute a large protein family that uses the conserved serine nucleophile to hydrolyze amide, ester, and thioester bonds in a variety of substrates (Lenfant et al., 2013). Alignment of Mil1 and its homologues revealed conserved residues that match the Ser-Asp-His catalytic triad of many serine hydrolases, consisting of S759 within a GxSxG motif, D817, and H858 (Figure 5A). Structural modeling using Phyre2 (Kelley and Sternberg, 2009) suggested the DUF726 domain of Mil1 is most similar to lipases and that residues S759, D817, and H858 are oriented in the necessary geometry to form a catalytic triad (Figure 5B). The active site of the well-characterized lipase CalB from Candida antarctica is shown for comparison; CalB was one of the highest template matches used for three-dimensional (3D) homology modeling.
FIGURE 5:
Mil1 has a conserved serine hydrolase catalytic triad. (A) BLAST alignment of the conserved DUF726 domain in Mil1. Intensity of blue-highlighted residues corresponds to percentage identity. Conserved residues corresponding to the predicted Ser-Asp-His catalytic triad are highlighted in red. (B) Structural model showing the catalytic triad of the well-characterized lipase B from C. antarctica (left; Protein Data Bank ID: 1TCA) and the Phyre2 homology model showing the predicted catalytic triad of Mil1 (right). (C) Sertraline sensitivity was assessed by plating strains in a 10× dilution series on YPD containing 22.5 μM sertraline or DMSO as a control.
To investigate whether Mil1 is likely to have an enzymatic activity important for Apm2-dependent sorting pathways, we tested the sertraline resistance of mutants carrying an alanine substitution of each predicted catalytic residue. A plasmid expressing wild-type Mil1 fully complemented the sertraline sensitivity of a mil1 strain, but the S759A (S>A), D817A (D>A), and H858A (H>A) mutants were only partially functional (Figure 5C), despite being expressed at similar levels (unpublished data). Each catalytic mutant displayed a similar level of sertraline sensitivity, suggesting that the putative catalytic activity of Mil1 contributes to Apm2 function.
Mil1 is a peripheral membrane protein that promotes Apm2 recruitment
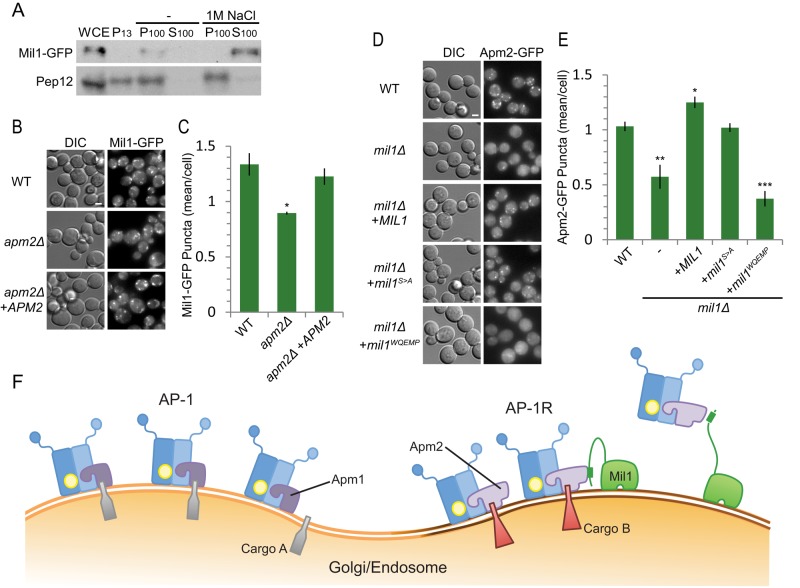
The transmembrane prediction algorithms Phobius (Käll et al., 2004) and TMHMM (Krogh et al., 2001) suggest that Mil1 contains four transmembrane domains. However, two of these predicted TMDs overlap the DUF726 α/β hydrolase domain, which is expected to be soluble. Subcellular fractionation was used to examine Mil1 membrane association under conditions that can dislodge peripheral, but not integral, membrane proteins (Figure 6A). In the presence of high salt concentrations, the transmembrane protein Pep12 was found in the pellet (P100) fraction as expected, whereas Mil1 was present exclusively in the soluble (S100) fraction, indicating that Mil1 is peripherally associated with membranes.
FIGURE 6:
Mil1 is a peripheral membrane protein that promotes Apm2 membrane recruitment. (A) Subcellular fractionation of Mil1 and the transmembrane protein Pep12. WCE, whole-cell extract. P13, low-speed pellet fraction; P100, high-speed pellet fraction; S100, high-speed soluble fraction. Samples were resolved by 15% SDS–PAGE and detected by immunoblotting. (B) Representative maximum-intensity z-projection images from nine slices at 0.3-μm increments. Scale bar, 2 μm. (C) Localization of Mil1-GFP in wild-type or apm2 mutant strains quantified in a single slice. Unpaired t test, *p < 0.05. Error bars represent SEM (n = 3). (D) Representative maximum-intensity z-projection images from nine slices at 0.3-μm increments. Scale bar, 2 μm. (E) Localization of Apm2-GFP expressed in wild-type or mil1 mutant strains and quantified in a single slice. Unpaired t test, ***p < 0.0001, **p < 0.01, and *p < 0.05. Error bars represent SEM (n = 4). (F) Proposed model: Mil1 helps to recruit Apm2 to a distinct membrane region.
We hypothesized that binding of Mil1 to the B domain of Apm2 could be responsible for its membrane recruitment. Accordingly, we examined the localization of Mil1-GFP in wild-type and apm2 mutants (Figure 6B). We observed a slight but significant decrease in the number of Mil1-GFP puncta in apm2 cells compared with wild type, which could be complemented by introduction of APM2 on a plasmid (Figure 6C). Thus Mil1 localization seems to be largely Apm2 independent.
We also considered the hypothesis that Mil1 promotes Apm2 membrane recruitment. We saw a significant decrease in the number of Apm2-GFP puncta in mil1 cells compared with wild-type strains (Figure 6D). Loss of Mil1 reduced the number of Apm2 puncta by ∼50% (Figure 6E) but did not significantly change the number of Apm1-GFP or Sec7-dsRed puncta (Supplemental Figure S2, A and B). Of note, the Mil1S>A catalytic-site mutant complemented this defect almost as well as wild-type Mil1, but the Mil1WQEMP deficient in Apm2 binding failed to complement, suggesting that direct physical interaction between Mil1 and Apm2, but not Mil1 catalytic activity, is important for recruitment of Apm2, but not Apm1, to membranes.
DISCUSSION
Subunit exchange may represent a general mechanism for creating functionally distinct adaptor complexes. How the incorporation of distinct but highly homologous μ subunits alters the function of the two AP-1 complexes of mammalian epithelial cells, AP-1A and AP-1B, is the subject of debate (Rodriguez-Boulan et al., 2013). Although AP-1A and AP-1B were shown to localize to different intracellular compartments in early studies, recent work suggested that these adaptor complexes are present in the same vesicles but recognize different cargo (Guo et al., 2013). Our work provides an example of how subunit exchange can create functionally distinct AP-1 adaptor complexes in yeast and highlights the importance of unique interactions with regulatory partners in this process.
We find that inclusion of either Apm1 or Apm2 changes the cargo specificity of AP-1 complexes: whereas the classical Apm1-containing AP-1 complex is involved in sorting Chs3 and Sna2, consistent with previous studies (Valdivia et al., 2002; Renard et al., 2010), the Apm2-containing AP-1R complex works in pathways that sort the v-SNARE Snc1 and give resistance to sertraline. The lipid flippase Drs2 is the only cargo reported to require both Apm1 and Apm2 for its localization (Liu et al., 2008). Because Drs2 regulates the formation of AP-1/clathrin-coated vesicles by generating membrane curvature, it may constitute an essential component of vesicles in both AP-1– and AP-1R–dependent pathways.
We showed that Apm2 could not substitute for Apm1 in Sna2 or Chs3 sorting even when expressed at similar levels, suggesting that these μ adaptins share little functional overlap. Some of these differences could result from differential cargo binding. The μ adaptins of AP-1, AP-2, and AP-3 bind YxxΦ motifs at a conserved site (Owen and Evans, 1998; Jia et al., 2012; Mardones et al., 2013). We found that mutations in Apm1 that disrupt this site block the YxxΦ-dependent AP-1 sorting of Sna2, whereas corresponding mutations in Apm2 cause sertraline sensitivity, suggesting that resistance to this drug requires correct sorting of a YxxΦ-containing cargo by Apm2. Thus, whereas both Apm1 and Apm2 are likely to recognize YxxΦ motifs, small variations at this site could alter the affinity for different cargo, as reported for AP-1 and AP-3 (Mardones et al., 2013).
Apm1 and Apm2 may have additional sites for binding noncanonical signals, as demonstrated for AP-4 (Boll et al., 2002; Burgos et al., 2010). Apm1 binds the Ste13 cytoplasmic via an 11–amino acid peptide that lacks recognized motifs (Foote and Nothwehr, 2006). Snc1 also lacks consensus YxxΦ or dileucine sorting signals, suggesting that Apm2 recognizes a divergent sorting signal in the Snc1 cytoplasmic domain or acts indirectly to sort a protein that is itself needed for Snc1 transport. Further work will be needed to define the relative affinities of Apm1 and Apm2 for different sorting signals and determine whether the signals are bound at distinct sites on the μ chain.
Although differential recognition of sorting signals might explain some apm1 and apm2 phenotypes, Chs3 sorting, which depends solely on Apm1, requires a dileucine motif (Starr et al., 2012). Such motifs are typically recognized by the γ/σ subunits present in both AP-1 and AP-1R (Doray et al., 2007; Kelly et al., 2008; Mattera et al., 2011). Indeed, we found the Apm1 tyrosine-binding pocket required for Sna2 transport was dispensable for Chs3 sorting. The fact that Apm2 is unable to substitute for Apm1 in this pathway, even at comparable levels of expression, suggests that something other than cargo binding affinity distinguishes the AP-1 and AP-1R complexes. It is possible that the Chs3 dileucine signal binds noncanonically to the AP-1 μ subunit or that Apm2 alters the conformation or accessibility of a dileucine-binding site in the γ/σ subunits. However, we favor an alternative model: that AP-1R is located in a separate compartment or microdomain, where it does not encounter Chs3 (Figure 6F). Although we found that Apm2 and Apm1 are largely present at the same Golgi/endosomal structures, our data are consistent with the model that AP-1 and AP-1R sort cargo into separate vesicle populations.
We identified an Apm2-interacting protein, Mil1, which was required specifically for Apm2-mediated protein trafficking. We hypothesize that Mil1 plays a key role in recruiting Apm2 into a distinct pathway in which Apm1 does not participate. Mil1 is predicted to be structurally similar to fungal lipases and phospholipases and contains conserved residues that match a Ser-Asp-His catalytic triad and GxSxG motif (Ha et al., 2012). Phospholipases A1 and A2 (PLA1 and PLA2) have been implicated in vesicle formation at the Golgi, either by producing cone-shaped lipids that promote membrane curvature or by creating particular phospholipid species that recruit trafficking components (Bechler et al., 2012; Ha et al., 2012). The cytosolic phosphatidic acid phospholipase A1 (PAPLA1) is important in the COPII-dependent transport of rhodopsin 1 in flies, but this function does not require its active site GxSxG residues (Kunduri et al., 2014). Similarly, the lipid-binding activity of the phospholipase A1 family member p125A, but not its enzymatic activity, is suggested to maintain COPII residency at endoplasmic reticulum exit sites (Klinkenberg et al., 2014). We found that Mil1 mediates the efficient recruitment of Apm2 to membranes, and this requires Mil1-Apm2 interaction but not Mil1’s presumed enzymatic activity. Because Mil1 is largely targeted to membranes in the absence of Apm2, it could bind a particular class of lipid and reinforce the recruitment of Apm2 to a specific microdomain, similar to the role proposed for p125A in COPII recruitment (Figure 6F).
We found that a conserved sequence in a predicted flexible, extended N-terminal region of Mil1 interacted with the B lobe of the Apm2 medium chain, distal to known cargo-recognition sites. Thus Mil1 binding is unlikely to interfere with cargo recognition by Apm2, consistent with a regulatory role for Mil1. Of interest, other lipid-modifying proteins have been found to associate with AP μ subunits. AP-1 μ1B binds and activates the phosphatidylinositol 4-phosphate (PI(4)P) 5-kinase PIPKIγ, which may contribute to the formation of a phosphatidylinositol (3,4,5)-trisphosphate–enriched domain that favors further AP-1B recruitment (Ling et al., 2007; Fields et al., 2010). All three (PI(4)P) 5-kinase isoforms bind the C-terminal domain of AP2 μ subunit, and this does not require the AP2 μ subunit’s tyrosine-binding pocket (Krauss et al., 2006). We found that predicted active-site residues in Mil1 were not required for Apm2 recruitment but were partially required for Apm2-mediated processes, suggesting that Mil1 has a catalytic activity whose role is downstream of adaptor recruitment. Our discovery that Mil1 regulates vesicle trafficking in yeast indicates that its uncharacterized human homologue, TMCO4, may act in a similar process. The Apm2-binding region of Mil1 is not conserved in TMCO4, suggesting that this protein regulates other trafficking pathways in higher organisms. Further studies are required to determine the extent to which lipid-modifying proteins interact with different adaptor protein complexes and other coat proteins to regulate vesicle transport.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains from this study were made by homologous recombination as described (Longtine et al., 1998; Janke et al., 2004; Sheff and Thorn, 2004). BY4741 and BY4742 yeast strains were purchased from GE Dharmacon (Lafayette, CO). Strains were propagated in rich medium (YPD: 1% yeast extract, 2% peptone, 2% dextrose) or SD minimal medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% synthetic complete mix, 2% dextrose) supplemented with the appropriate amino acids for plasmid selection. Plasmids were made by homologous recombination in yeast, rescued in Escherichia coli, and confirmed by sequencing. pHPH was made by PCR amplification of the hygromycin resistance gene HPH from pFA6a-hphNT1 and cotransformation of the PCR product with linearized pRS416. Proteins were tagged with either the bright GFP variant GFP+ (Scholz et al., 2000) or GFP(envy) (Slubowski et al., 2015), a gift from C. Slubowski (University of Massachusetts, Boston, MA), by PCR amplification of the GFP::HIS cassette and transformation into yeast. Strains and plasmids are described in Supplemental Tables S1 and S2.
Chemical compounds
A calcofluor white (fluorescent brightener 28; Sigma Aldrich, St. Louis, MO) stock solution was made in 0.5 M Tris-HCl, pH 9.6, to a final concentration of 10 mg/ml. Stock solutions of sertraline hydrochloride (Cedarlane Laboratories, Burlington, Canada) and myriocin (Cayman Chemical Company, Ann Arbor, MI) of 25 and 1 mg/ml, respectively, were made in dimethyl sulfoxide (DMSO) and stored at −20°C until use. Myriocin was purchased from the Cayman Chemical Company, resuspended to a final concentration of 1 mg/ml in DMSO, and treated similarly. Hygromycin B (Hyg B; Roche Diagnostics, Indianapolis, IN) was purchased as a 50 mg/ml stock in phosphate-buffered saline. All other chemicals were purchased from Sigma-Aldrich and stored according to supplier’s recommendations.
Invertase assay
Strains expressing the GSS construct were grown for 20 h in 2 ml of YP-fructose (final OD600 of ∼10). Cultures were diluted to 3 OD600/ml, and the liquid invertase assay was performed as described (Dalton et al., 2015). Results are reported as nanomoles of glucose produced per 1 OD600 of culture, normalized to wild-type readings from the day. Glucose standards (ranging from 5 to 50 nM) and blanks were included in each experiment to ensure that measurements were within the linear range.
Calcofluor white assay
The calcofluor assay was performed as described (Burston et al., 2008). Briefly, knockout strains were plated in 1536-array format onto YPD plates containing 50 μg/ml calcofluor white, using a Virtek automated colony arrayer (Bio-Rad Laboratories, Hercules, CA). After incubation at 30°C for 3 d, white-light images were acquired using a model 2400 flat-bed scanner (Epson, Nagano, Japan), and fluorescent-light images were captured with a Fluor S Max MultiImager (Bio-Rad Laboratories) using the 530DF60 filter and Quantity One software (version 4.2.1; Bio-Rad Laboratories). The open-source spot-finding program GridGrinder was used for densitometry of digital images.
Small-molecule inhibitor assays
We spotted 1 OD600/ml of log-phase yeast grown under selective conditions to maintain plasmids (either SD-Leu or YPD + Hyg B) in 10× serial dilution on selective plates containing inhibitors or DMSO as a control, incubated them at 30°C for 3 d, and imaged them with a Canon Rebel T3I camera (Canon, Tokyo, Japan).
Fluorescence microscopy
Log-phase yeast in minimal selective media were imaged at room temperature with an Axioplan 2 fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a Plan-Apochromat 100×/1.40 numerical aperture (NA) oil immersion objective lens (Zeiss) and a CoolSNAP camera (Roper Scientific, Tucson, AZ) using MetaMorph 7.7 software (MDS Analytical Technologies, Sunnyvale, CA). Alternatively, in Figures 2C and 6, images were acquired with a DMi8 microscope (Leica Microsystems, Wetzlar, Germany) using an HC PL APO 63×/1.30 Glyc CORR CS or an HC PL APO 100×/1.40 OIL STED WHITE objective (Leica) and an ORCA-Flash4.0 digital camera (Hamamatsu Photonics, Hamamatsu City, Japan). Images were adjusted using MetaMorph and Photoshop CS5 (Adobe, San Jose, CA). Maximum-intensity z-projections were based on z-stacks of nine planes with a 0.3-μm step size and made using MetaMorph after photobleaching correction based on histogram matching in Fiji (www.fiji.sc/Fiji). Cellular features were quantified by manually scoring images, except the Sec7-dsRed quantification, which was done by automated counting using in-house MetaMorph-based algorithms. For total internal reflection fluorescence (TIRF) microscopy experiments, cells from early log-phase cultures were incubated in a final concentration of 200 μM LatA dissolved in DMSO for 30 min at 30°C. We spotted 200 μl of treated cells onto concanavalin A–coated eight-well Lab-Tek dishes (Nalge Nunc International, Rochester, NY) and collected TIRF images with a 3i Marianas microscope (Intelligent Imaging Innovations, Denver, CO) equipped with an α-Plan-Fluor 100× 1.45 NA objective lens and a Zeiss TIRF slider (Carl Zeiss). Images were acquired with 488-nm and/or 561-nm laser excitation, with GFP and RFP emission split between two Cascade II 512 EM cameras (Photometrics, Tucson, AZ) with an Optical Insights Dual Cam (Photometrics) with an exposure time of 750 ms. TetraSpeck 100-nm beads (Life Technologies, Carlsbad, CA) were used to align the two channels and for subsequent registration in software. Slide-Book 4.2 software (Intelligent Imaging Innovations) was used for image acquisition and dual-channel image registration. Montages were created using ImageJ (National Institutes of Health, Bethesda, MD) with the Kymograph plug-ins installed (www.embl.de/eamnet/html/kymograph.html).
Coimmunoprecipitation and Western blotting
Log-phase cells in YPD medium were converted into spheroplasts by digesting with Zymolyase (MJS BioLynx, Brockville, Canada), and stored at −85°C until use, as described (Conibear and Stevens, 2000). We resuspended 40 OD600 of cells in coimmunoprecipitation lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 100 mM NaCl, and 2 mM EDTA, with 1% 3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1-propanesulfonate and 1 mM phenylmethylsulfonyl fluoride added before use) and centrifuged them for 15 min at 4°C. Supernatant fractions were incubated at 4°C for 1 h with rabbit anti-HA (Santa Cruz Biotechnology, Dallas, TX) or rabbit anti-GFP (Molecular Probes, Carlsbad, CA), followed by protein A–Sepharose beads (GE Healthcare, Little Chalfont, United Kingdom) for 2.5 h. The beads were washed, and proteins were eluted by heating at 95°C for 5 min. Samples were run on 10% SDS–PAGE gels. Proteins were transferred overnight to nitrocellulose membranes and blotted with either mouse anti-GFP (Roche) or mouse anti-HA (Covance, Princeton, NJ) and then with goat anti-mouse antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA). Blots were developed with the enhanced chemiluminescent West Pico (Pierce, Rockford, IL) and West Femto (Pierce) and exposed to Amersham Hyperfilm (GE Healthcare).
Yeast two-hybrid assay
PJ694a strains carrying pGBDU-C2-based plasmids were mated to PJ694α strains carrying pGAD-C2-based plasmids, each expressing the indicated full-length or truncation proteins (James et al., 1996). Positive two-hybrid interactions were scored on minimal medium (SD-Ura-Leu-His+Ade) containing 3, 5, or 10 mM 3-amino-1,2,4-triazole.
Bioinformatic analyses
All sequences are from Uniprot (www.uniprot.org). Homologues in other organisms were found using the Panther classification system (www.pantherdb.org). Visualization of MUSCLE-aligned sequences (Edgar, 2004) used Jalview (www.jalview.org). For 3D structure modeling, Phyre2 with “intensive” settings was used (Kelley and Sternberg, 2009). Apm2 tyrosine-motif recognition pocket residues were labeled based on sequence conservation with known tyrosine-pocket residues of AP complex μ subunits (Owen and Evans, 1998; Heldwein et al., 2004), and the predicted region involved in lipid binding was extrapolated based on alignment with human μ1B (Fields et al., 2010).
Membrane fractionation
The membrane fractionation protocol was adapted from Conibear and Stevens (2000). Log-phase cells at 60 OD600 were spheroplasted, lysed, and spun at 500 × g for 5 min at 4°C. The P13 fraction was pelleted at 13,000 × g for 10 min at 4°C and resuspended in SDS sample buffer (5% SDS, 50 mM Tris, pH 6.8, 0.4 mg/ml bromophenol blue, 1% β-mercaptoethanol). The supernatant was centrifuged at 100,000 × g in a table-top ultracentrifuge for 60 min to generate the P100 pellet, which was resuspended in 1 ml of lysis buffer as the P100 fraction, and the supernatant was kept as the S100 fraction. For high-salt treatment of the P100 and S100 fractions, NaCl was added to a final concentration of 1 M before the 100,000 × g spin. Samples were trichloroacetic acid precipitated, resuspended in SDS sample buffer, and subjected to SDS–PAGE and immunoblotting.
Supplementary Material
Acknowledgments
We thank P. Morsomme and C. Slubowski for reagents and E. Perlstein for helpful discussions. This work was supported by funding from the Canadian Institutes of Health Research (64394 to E.C.), the Natural Sciences and Engineering Council of Canada (371618-09 to E.C.), the Canada Foundation for Innovation (7664 and 30636 to E.C.), and the National Institutes of Health (R01 GM-60979 and NIGMS T32 007231 to B.W.). Additional support was provided by Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship and Canada Graduate Scholarship awards (to H.E.B.) and a Ford Foundation Diversity Fellowship (to L.M.B.).
Abbreviations used:
- AP
adaptor protein
- GFP
green fluorescent protein
- GSS
GFP:Snc1:Suc2 reporter
- HA
hemagglutinin
- Hyg B
hygromycin B.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0621) December 10, 2015.
REFERENCES
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Baxter SM, Rosenblum JS, Knutson S, Nelson MR, Montimurro JS, Di Gennaro JA, Speir JA, Burbaum JJ, Fetrow JS. Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Mol Cell Proteomics. 2004;3:209–225. doi: 10.1074/mcp.M300082-MCP200. [DOI] [PubMed] [Google Scholar]
- Bechler ME, de Figueiredo P, Brown WJ. A PLA1-2 punch regulates the Golgi complex. Trends Cell Biol. 2012;22:116–124. doi: 10.1016/j.tcb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll W, Rapoport I, Brunner C, Modis Y, Prehn S, Kirchhausen T. The μ2 subunit of the clathrin adaptor AP-2 binds to FDNPVY and YppØ sorting signals at distinct sites. Traffic. 2002;3:590–600. doi: 10.1034/j.1600-0854.2002.30808.x. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. Adaptor proteins involved in polarized sorting. J Cell Biol. 2014;204:7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos PV, Mardones GA, Rojas AL, Dasilva LLP, Prabhu Y, Hurley JH, Bonifacino JS. Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev Cell. 2010;18:425–436. doi: 10.1016/j.devcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston HE, Davey M, Conibear E. Genome-wide analysis of membrane transport using yeast knockout arrays. Methods Mol Biol. 2008;457:29–39. doi: 10.1007/978-1-59745-261-8_3. [DOI] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Báez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol. 2009;185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canagarajah BJ, Ren X, Bonifacino JS, Hurley JH. The clathrin adaptor complexes as a paradigm for membrane-associated allostery. Protein Sci. 2013;22:517–529. doi: 10.1002/pro.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Payne GS. Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol. 2012;14:239–248. doi: 10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton L, Davey M, Conibear E. Large-scale analysis of membrane transport in yeast using invertase reporters. Methods Mol Biol. 2015;1270:395–409. doi: 10.1007/978-1-4939-2309-0_27. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Chau N, Abdel-Hamid MK, Hu L, von Kleist L, Whiting A, Krishnan S, Maamary P, Joseph SR, Simpson F, et al. Phenothiazine-derived antipsychotic drugs inhibit dynamin and clathrin-mediated endocytosis. Traffic. 2015;16:635–654. doi: 10.1111/tra.12272. [DOI] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu G, Kornfeld S. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell. 2007;18:1887–1896. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Smith C, Owen D. Life of a clathrin coat: insights from clathrin and AP structures. Nat Rev Mol Cell Biol. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields IC, King SM, Shteyn E, Kang RS, Fölsch H. Phosphatidylinositol 3,4,5-trisphosphate localization in recycling endosomes is necessary for AP-1B-dependent sorting in polarized epithelial cells. Mol Biol Cell. 2010;21:95–105. doi: 10.1091/mbc.E09-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote C, Nothwehr SF. The clathrin adaptor complex 1 directly binds to a sorting signal in Ste13p to reduce the rate of its trafficking to the late endosome of yeast. J Cell Biol. 2006;173:615–626. doi: 10.1083/jcb.200510161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Guo X, Mattera R, Ren X, Chen Y, Retamal C, González A, Bonifacino JS. The Adaptor Protein-1 μ1B subunit expands the repertoire of basolateral sorting signal recognition in epithelial cells. Dev Cell. 2013;27:353–366. doi: 10.1016/j.devcel.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KD, Clarke BA, Brown WJ. Regulation of the Golgi complex by phospholipid remodeling enzymes. Biochim Biophys Acta. 2012;1821:1078–1088. doi: 10.1016/j.bbalip.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Irving C, Borner GHH. Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia. Traffic. 2013;14:153–164. doi: 10.1111/tra.12028. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, et al. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res. 2014;169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1426. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol. 2012;19:701–706. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL. A Combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kelly B, McCoy A, Späte K, Miller S, Evans P, Höning S, Owen D. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976– 979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg D, Long KR, Shome K, Watkins SC, Aridor M. A cascade of ER exit site assembly that is regulated by p125A and lipid signals. J Cell Sci. 2014;127:1765–1778. doi: 10.1242/jcs.138784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, Kukhtina V, Pechstein A, Haucke V. Stimulation of phosphatidylinositol kinase type I-mediated phosphatidylinositol (4,5)-bisphosphate synthesis by AP-2mu-cargo complexes. Proc Natl Acad Sci USA. 2006;103:11934–11939. doi: 10.1073/pnas.0510306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kunduri G, Yuan C, Parthibane V, Nyswaner KM, Kanwar R, Nagashima K, Britt SG, Mehta N, Kotu V, Porterfield M, et al. Phosphatidic acid phospholipase A1 mediates ER-Golgi transit of a family of G protein-coupled receptors. J Cell Biol. 2014;206:79–95. doi: 10.1083/jcb.201405020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KKY, Davey M, Sun B, Roth AF, Davis NG, Conibear E. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, et al. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 2014;344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol. 2007;176:343–353. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Surendhran K, Nothwehr S, Graham T. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell. 2008;19:3526–3535. doi: 10.1091/mbc.E08-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Mckenzie A, III, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mardones GA, Burgos PV, Lin Y, Kloer DP, Magadán JG, Hurley JH, Bonifacino JS. Structural basis for the recognition of tyrosine-based sorting signals by the μ3A subunit of the AP-3 adaptor complex. J Biol Chem. 2013;288:9563–9571. doi: 10.1074/jbc.M113.450775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. Conservation and diversification of dileucine signal recognition by Adaptor Protein (AP) complex variants. J Biol Chem. 2011;286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit A, Côté S, Brustein E, Drouin CA, Lapointe L, Boudreau M, Meloche C, Drouin R, Hudson TJ, Drapeau P, et al. Disruption of AP1S1, causing a novel neurocutaneous syndrome, perturbs development of the skin and spinal cord. PLoS Genet. 2008;4:e1000296. doi: 10.1371/journal.pgen.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Hase K, Ohno H. The role of the clathrin adaptor AP-1: polarized sorting and beyond. Membranes (Basel) 2014;4:747–763. doi: 10.3390/membranes4040747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey MM, Korostyshevsky D, Lee S, Perlstein EO. The antidepressant sertraline targets intracellular vesiculogenic membranes in yeast. Genetics. 2010;185:1221–1233. doi: 10.1534/genetics.110.117846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard H-F, Demaegd D, Guerriat B, Morsomme P. Efficient ER exit and vacuole targeting of yeast Sna2p require two tyrosine-based sorting motifs. Traffic. 2010;11:931–946. doi: 10.1111/j.1600-0854.2010.01070.x. [DOI] [PubMed] [Google Scholar]
- Ren X, Farías GG, Canagarajah BJ, Bonifacino JS, Hurley JH. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell. 2013;152:755–767. doi: 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Perez-Bay A, Schreiner R, Gravotta D. Response: the “tail” of the twin adaptors. Dev Cell. 2013;27:247–248. doi: 10.1016/j.devcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz O, Thiel A, Hillen W, Niederweis M. Quantitative analysis of gene expression with an improved green fluorescent protein. p6. Eur J Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- Setta-Kaffetzi N, Simpson MA, Navarini AA, Patel VM, Lu HC, Allen MH, Duckworth M, Bachelez H, Burden AD, Choon SE, et al. AP1S3 mutations are associated with pustular psoriasis and impaired toll-like receptor 3 trafficking. Am J Hum Genet. 2014;94:790–797. doi: 10.1016/j.ajhg.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Slubowski CJ, Funk AD, Roesner JM, Paulissen SM, Huang LS. Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins, Envy and Ivy. Yeast. 2015;32:379–387. doi: 10.1002/yea.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TL, Pagant S, Wang C-W, Schekman R. Sorting signals that mediate traffic of chitin synthase III between the TGN/endosomes and to the plasma membrane in yeast. PLoS One. 2012;7:e46386. doi: 10.1371/journal.pone.0046386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon SK. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Stevens C, Teague J, Edkins S, O’Meara S, Avis T, Barthorpe S, Buck G, Butler A, Cole J, et al. Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet. 2006;79:1119–1124. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Bonifacino JS. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harbor Perspect Biol. 2013;5:a016790. doi: 10.1101/cshperspect.a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Yeung BG, Phan HL, Payne GS. Adaptor complex-independent clathrin function in yeast. Mol Biol Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.