Abstract
Purpose
It has been hypothesized that increased predisposition to breast cancer may correlate with radiosensitivity and thus increased risk of toxicity following breast irradiation. This study investigated the relationship between common breast cancer risk variants and radiotherapy toxicity.
Experimental Design
Single nucleotide polymorphism genotypes were determined in female breast cancer patients from the RAPPER (Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy) study using the Illumina CytoSNP12 genome-wide array. A further 15,582,449 genotypes were imputed using the 1000 Genomes Project reference panel. Patient (n=1160) polygenic risk scores were generated by summing risk-allele dosages, both un-weighted and weighted by published effect sizes for breast cancer risk. Regression models were used to test associations of individual variants and polygenic risk scores with acute and late toxicity phenotypes (telangiectasia, breast edema, photographically assessed shrinkage, induration, pigmentation, breast pain, breast sensitivity, overall toxicity).
Results
Genotypes of 90 confirmed breast cancer risk variants were accurately determined and polygenic risk scores were approximately normally distributed. Variant rs6964587 was associated with increased breast edema five years following radiotherapy (beta=0.22; 95% CI=(0.09-0.34); p-value=7 × 10−4). No other associations were found between individual variants or the un-weighted (p>0.17) or weighted (p>0.13) polygenic risk score and radiotherapy toxicity. This study had >87% power to detect an association between the polygenic risk score (relative risk>1.1) and toxicity.
Conclusions
Cancer patients with a high polygenic predisposition to breast cancer do not have an increased risk of radiotherapy toxicity up to five years following radiotherapy but individual variants may increase risk.
Keywords: radiotherapy, toxicity, adverse effects, genetics, breast cancer
Introduction
To date, 94 common breast cancer susceptibility loci have been confirmed (p-value<5 × 10−8) via meta-analyses of 11 genome wide association studies (GWAS; 15,748 cases and 18,084 controls) and 41 studies in the Breast Cancer Association Consortium (BCAC; 46,785 cases and 42,892 controls) (1–3). The effect sizes of each locus are generally modest with odds ratios (ORs) ≤1.34. It is estimated that 16% of the familial risk of breast cancer can be explained by common genetic variation (3). In a recent study, single nucleotide polymorphism (SNP) markers of 77 confirmed breast cancer susceptibility loci were combined to create a polygenic risk score for breast cancer (4). Women in the highest 1% of the risk score were 3.6 times more likely to develop breast cancer than women in the middle quintile (4).
Radiation exposure is a known risk factor for the development of breast cancer (5) and individuals with the disease have been reported to have increased radiosensitivity (6) postulated as due to inherited defects in cellular responses to DNA damage from either endogenous or exogenous sources (7). It has also been suggested that elevated radiosensitivity may be a marker of low penetrance predisposition to cancer (8). Since radiosensitive individuals are more likely to experience toxicity following radiotherapy (9), it has been hypothesized that genetic variants that predispose to breast cancer may also increase the risk of radiation side-effects. This hypothesis was based on the assumption that gene variants involved in cancer pre-disposition may have roles in both tumor formation and in the response to radiation-induced DNA damage.
We have tested this hypothesis using data from the UK RAPPER (Radiogenomics: Assessment of Polymorphisms for Predicting the Effects of Radiotherapy) study (10). Common genetic variants individually have small effects on risk of breast cancer and are likely to have similarly small effects on radiotherapy toxicity. Thus, in order to search for evidence of associations with radiosensitivity, we tested the currently known breast cancer risk variants, both individually and within a more statistically powerful polygenic risk score.
Materials and Methods
Patients
The UK RAPPER (UKCRN1471) study recruits patients from clinical trials and observational studies involving radiotherapy given with curative intent. This study comprised 1160 female breast cancer patients who were included in a GWAS described elsewhere, which provides details on the cohorts included (11): 937 patients from the Cambridge Breast IMRT Trial, 167 from the RACE (Radiation Complications and Epidemiology) study and 56 from a prospective study at the Christie Hospital, Manchester. Briefly, all patients underwent conservative surgery followed by adjuvant radiotherapy with doses of: 40 Gy in 15 fractions over three weeks (Cambridge Breast IMRT trial, Christie Hospital); 50 Gy in 25 fractions or 39.0 Gy/42.9 Gy in 13 fractions over five weeks (RACE study). An alpha/beta ratio of 3.4 (12) was used to calculate equivalent dose in 2Gy fraction for these schedules for inclusion in multivariable analyses. Patient characteristics and treatment factors are summarized in Table 1.
Table 1. Demographic and treatment-related factors of patients in the RAPPER cohort.
| Characteristic | RAPPER (N=1160) | |
|---|---|---|
| Age | N; mean (SD) | 1160; 58 (9.2) |
| Missing (%) | 0 | |
| Diabetesa | Yes (%) | 42 (3.6) |
| No (%) | 925 (79.8) | |
| Missing (%) | 193 (16.6) | |
| Smokera | Yes (%) | 138 (11.9) |
| No (%) | 845 (72.8) | |
| Missing (%) | 177 (15.3) | |
| Tumour size (mm)a | N; mean (SD) | 968; 15.4 (7.6) |
| Missing (%) | 192 (16.6) | |
| Adjuvant chemotherapy | Yes (%) | 198 (17.1) |
| No (%) | 935 (80.6) | |
| Missing (%) | 27 (2.3) | |
| Tamoxifen use | Yes (%) | 785 (67.7) |
| No (%) | 334 (28.8) | |
| Missing (%) | 41 (3.5) | |
| Post-operative breast infectiona | Yes (%) | 182 (15.7) |
| No (%) | 789 (68.0) | |
| Missing (%) | 189 (16.3) | |
| Post-operative hematomaa | Yes (%) | 69 (6.0) |
| No (%) | 759 (65.4) | |
| Missing (%) | 332 (28.6) | |
| Breast boost | Yes (%) | 735 (63.4) |
| No (%) | 424 (36.5) | |
| Missing (%) | 1 (0.1) | |
| Actual volume of breast receiving >107% of radiation dosea | N; mean (SD) | 932; 19.1 (51.5) |
| Missing (%) | 228 (19.7) |
Not available in RACE
Assessment of Radiotherapy Toxicity
Late toxicity data were collected prospectively using standardized scoring systems (Supplementary Table 1). This study used toxicity data collected at two specific time points: two years and five years following radiotherapy. Two-year telangiectasia, breast edema, photographically assessed shrinkage, induration, pigmentation, breast pain and breast sensitivity was recorded for 1134 patients. Five-year telangiectasia, breast edema, photographically assessed shrinkage and induration was collected for 594 patients in the Cambridge Breast IMRT trial. Scale-independent Standardized Total Average Toxicity (STAT) scores were derived from the individual endpoints for overall measures of two-year and five-year toxicity (13). Acute toxicity was recorded for 920 patients in the Cambridge Breast IMRT Trial at 3 weeks following radiotherapy, using the Radiation Therapy Oncology Group (RTOG) scoring system (see Supplementary Table 2).
Genotyping, Quality Control and Imputation
Genotyping and imputation across the whole genome was performed for the RAPPER GWAS. Samples were genotyped using the Illumina CytoSNP12 (Illumina, San Diego, CA, USA). Standard quality control procedures were applied to remove variants that were missing in >5% of samples; had a minor allele frequency (MAF) < 1%; or had MAF<5% and were also missing in >1% samples. Variants were also removed if their genotype frequencies deviated from those expected under Hardy-Weinberg equilibrium (p-value < 10−6). Samples were removed that had >3% of all variants missing. Principle components analysis (PCA) was used to identify and exclude individuals with non-European ancestry. Genome coverage was increased by imputation using SHAPEIT (14) and IMPUTE2 (15) with the 1000 Genomes Phase III reference panel (16). In this study, expected dosages of the alleles of the 94 breast cancer risk variants were extracted from the imputed data.
Statistical Analysis
Polygenic risk scores were created to quantify the patients’ genetic risk of breast cancer. For each patient, genotype dosages for the 94 breast cancer risk-increasing alleles were calculated and then summed across all the variants. Two types of risk score were calculated:
where j = variants 1..94
βj = the per-allele log-odds ratio for risk of breast cancer associated with variant j
G = risk allele dosage
The log-odds ratios used to weight the risk score were taken directly from the report by Mavaddat et al and were estimated by testing the association of each variant with breast cancer risk while adjusting for the effect of other variants (4). Seventeen variants were identified after Mavaddat et al performed their analysis, so log-odds ratios used for these variants were estimates from BCAC (3).
For univariable analysis, ordinal logistic regression was used to test the association between the acute or individual late toxicity endpoints and polygenic risk score. Linear regression was used to test the association between STAT and risk score. Each genetic variant was also tested individually. For multivariable analysis, a residual analysis was performed (17). Residuals from multivariable analyses of the two-year toxicity endpoints with non-genetic covariates, produced as part of the original RAPPER GWAS of late toxicity (11), were used to analyze two-year toxicity. These residuals estimate the risk of toxicity not explained by available patient- and treatment- related factors including baseline symptoms that resulted from surgery and were present before radiotherapy. Residuals for acute toxicity and five-year toxicity were calculated from multivariable regression models including significant non-genetic factors as part of this study. Linear regression was used to evaluate the association between the residuals and both the genetic risk scores and the individual variants. The association between polygenic risk and baseline symptoms was assessed separately to confirm that adjustment for baseline symptoms would not substantially change the results. For the primary analysis, the polygenic risk scores were considered statistically significant if p-value<0.05. To reduce the chance of a false-positive association due to multiple testing, the individual genetic variants were deemed statistically significant if p-value<5 × 10−4.
The study was well powered to detect significant associations between breast cancer polygenic risk score and many radiotherapy toxicity endpoints. Assuming a 20% prevalence of moderate/severe toxicity (grade≥2) in breast cancer patients, the power to detect an association between toxicity (grade≥2) and polygenic risk score at p-value<0.05 would be 87% for a small difference in mean risk score of 0.1. This difference in mean risk would be equivalent to a relative risk of toxicity of 1.1 for the subset of patients with a higher mean polygenic risk of breast cancer. All analyses were performed using statistical software package STATA version 13.1. STrengthening the Reporting Of Genetic Association studies in Radiogenomics (STROGAR) guidelines were followed throughout (18).
Results
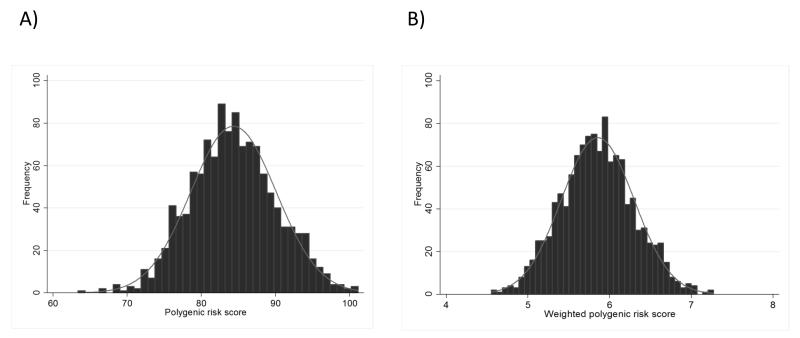
Out of the 94 genetic variants known to increase the risk of breast cancer, 90 were genotyped or imputed successfully (r2>0.66) in the 1160 patients in the RAPPER cohort – details in Table 2. Figure 1 shows the distributions of the un-weighted and weighted polygenic risk scores among all patients with late toxicity data available (N=1134), which approximate to a normal distribution in these samples.
Table 2. Genetic variants known to influence risk of breast cancer.
| Variant | Nearest Gene | Chromf | Position (build 37g) | Breast cancer risk allele | Published odds ratioa | RAPPER risk allele frequency | RAPPER imputation r2c |
|---|---|---|---|---|---|---|---|
| rs616488 | PEX14 | 1 | 10566215 | A | 1.06 | 0.67 | 1 |
| rs11552449 | PTPN22-BCL2L15-AP4B1-DCLRE1B-HIPK1 | 1 | 114448389 | T | 1.08 | 0.20 | 0.98 |
| rs11249433 | None | 1 | 121280613 | G | 1.10 | 0.38 | 0.67 |
| rs12405132 | RNF115 | 1 | 145644984 | C | 1.05b | 0.64 | 1 |
| rs12048493 | OTUD7B | 1 | 149927034 | C | 1.07b | 0.39 | 0.66 |
| rs6678914 | LGR6 | 1 | 202187176 | G | 1.01 | 0.59 | 0.99 |
| rs4245739 | MDM4 | 1 | 204518842 | C | 1.03 | 0.28 | 0.99 |
| rs72755295 | EXO1 | 1 | 242034263 | G | 1.15b | 0.03 | 0.69 |
| rs12710696 | OSR1 | 2 | 19320803 | A | 1.04 | 0.38 | 0.99 |
| rs4849887 | INHBB | 2 | 121245122 | C | 1.09 | 0.90 | 0.99 |
| rs2016394 | METAP1D-DLX1-DLX2 | 2 | 172972971 | G | 1.05 | 0.54 | 1 |
| rs1550623 | CDCA7 | 2 | 174212894 | A | 1.06 | 0.83 | 0.98 |
| rs1045485 | CASP8 d | 2 | 202149589 | G | 1.04 | 0.86 | 0.98 |
| rs13387042 | IGFBP5d | 2 | 217905832 | A | 1.14 | 0.55 | 1 |
| rs16857609 | DIRC3 | 2 | 218296508 | T | 1.07 | 0.29 | 0.98 |
| rs6762644 | ITPR1-EGOT | 3 | 4742276 | G | 1.07 | 0.40 | 0.99 |
| rs4973768 | SLC4A7 | 3 | 27416013 | T | 1.09 | 0.50 | 1 |
| rs12493607 | TGFBR2 | 3 | 30682939 | C | 1.05 | 0.33 | 0.99 |
| rs6796502 | PRSS42 | 3 | 46866866 | G | 1.09b | 0.90 | 0.96 |
| rs1053338 | ATXN7 | 3 | 63967900 | G | 1.08b | 0.15 | 0.98 |
| rs9790517 | TET2 | 4 | 106084778 | T | 1.05 | 0.20 | 0.99 |
| rs6828523 | ADAM29 | 4 | 175846426 | C | 1.10 | 0.89 | 0.99 |
| rs10069690 | TERT d | 5 | 1279790 | T | 1.02 | 0.27 | 1 |
| rs7726159 | TERT d | 5 | 1282319 | A | 1.04 | 0.33 | 0.85 |
| rs2736108 | TERT d | 5 | 1297488 | C | 1.07 | 0.72 | 0.90 |
| rs13162653 | MARCH11 | 5 | 16187528 | G | 1.05b | 0.55 | 0.89 |
| rs2012709 | SUB1 | 5 | 32567732 | T | 1.05b | 0.49 | 1 |
| rs10941679 | None | 5 | 44706498 | G | 1.12 | 0.30 | 0.98 |
| rs889312 | MAP3K1 d | 5 | 56031884 | C | 1.12 | 0.31 | 0.99 |
| rs10472076 | RAB3C | 5 | 58184061 | C | 1.04 | 0.39 | 0.96 |
| rs1353747 | PDE4D | 5 | 58337481 | T | 1.09 | 0.90 | 0.98 |
| rs7707921 | ATG10 | 5 | 81538046 | A | 1.08b | 0.76 | 0.99 |
| rs1432679 | EBF1 | 5 | 158244083 | G | 1.07 | 0.46 | 0.99 |
| rs11242675 | FOXQ1 | 6 | 1318878 | T | 1.06 | 0.63 | 0.94 |
| rs204247 | RANBP9 | 6 | 13722523 | G | 1.05 | 0.45 | 0.99 |
| rs9257408 | None | 6 | 28926220 | C | 1.05b | 0.39 | 0.99 |
| rs17529111 | None | 6 | 82128386 | G | 1.05 | 0.21 | 0.99 |
| rs12662670 | ESR1 e | 6 | 151918856 | G | 1.14 | 0.09 | 0.99 |
| rs2046210 | ESR1 e | 6 | 151948366 | A | 1.05 | 0.37 | 0.99 |
| rs6964587 | AKAP9 | 7 | 91630620 | T | 1.05b | 0.40 | 0.98 |
| rs4593472 | LINC-PINT | 7 | 130667121 | C | 1.05b | 0.66 | 0.92 |
| rs720475 | ARHGEF5-NOBOX | 7 | 144074929 | G | 1.06 | 0.74 | 0.95 |
| rs9693444 | None | 8 | 29509616 | A | 1.07 | 0.34 | 0.99 |
| rs13365225 | KCNU1 | 8 | 36858483 | A | 1.05b | 0.85 | 0.98 |
| rs6472903 | CASC9 | 8 | 76230301 | T | 1.10 | 0.84 | 0.94 |
| rs2943559 | HNF4G | 8 | 76417937 | G | 1.13 | 0.09 | 0.98 |
| rs13267382 | None | 8 | 117209548 | A | 1.05b | 0.36 | 0.96 |
| rs13281615 | MYC d | 8 | 128355618 | G | 1.10 | 0.44 | 0.99 |
| rs11780156 | MYC d | 8 | 129194641 | T | 1.07 | 0.17 | 0.99 |
| rs1011970 | CDKN2A/B | 9 | 22062134 | T | 1.05 | 0.18 | 1 |
| rs10759243 | KLF4 d | 9 | 110306115 | A | 1.05 | 0.29 | 0.75 |
| rs865686 | KLF4 d | 9 | 110888478 | T | 1.11 | 0.62 | 0.99 |
| rs2380205 | ANKRD16 | 10 | 5886734 | C | 1.02 | 0.59 | 0.99 |
| rs7072776 | MLLT10-DNAJC1 | 10 | 22032942 | A | 1.06 | 0.27 | 0.91 |
| rs11814448 | DNAJC1 | 10 | 22315843 | C | 1.22 | 0.02 | 0.84 |
| rs10995190 | NRBF2 d | 10 | 64278682 | G | 1.17 | 0.87 | 0.99 |
| rs704010 | ZMIZ1 | 10 | 80841148 | T | 1.07 | 0.43 | 0.97 |
| rs7904519 | TCF7L2 | 10 | 114773927 | G | 1.06 | 0.48 | 0.99 |
| rs11199914 | FGFR2 d | 10 | 123093901 | C | 1.06 | 0.70 | 0.73 |
| rs2981579 | FGFR2 d | 10 | 123337335 | A | 1.25 | 0.47 | 0.99 |
| rs3817198 | LSP1 | 11 | 1909006 | C | 1.07 | 0.33 | 0.92 |
| rs3903072 | DKFZp761e198-OVOLI-SNX32-CFL1-MUS81 | 11 | 65583066 | G | 1.06 | 0.55 | 0.99 |
| rs78540526 | CCND1 d | 11 | 69331418 | T | 1.18 | 0.08 | 0.95 |
| rs554219 | CCND1 d | 11 | 69331642 | G | 1.12 | - | - |
| rs75915166 | CCND1 d | 11 | 69379161 | A | 1.024 | 0.06 | 0.86 |
| rs11820646 | BARX2 | 11 | 129461171 | C | 1.05 | 0.60 | 0.98 |
| rs12422552 | None | 12 | 14413931 | C | 1.03 | - | - |
| rs10771399 | PTHLH | 12 | 28155080 | A | 1.16 | - | - |
| rs17356907 | NTN4 | 12 | 96027759 | A | 1.10 | - | - |
| rs1292011 | None | 12 | 115836522 | A | 1.08 | 0.59 | 1 |
| rs11571833 | BRCA2-N4BP2LI-N4BP2L2 | 13 | 32972626 | T | 1.26 | 0.01 | 0.87 |
| rs2236007 | PAX9-SLO25A21 | 14 | 37132769 | G | 1.09 | 0.82 | 1 |
| rs2588809 | RAD51L1 | 14 | 68660428 | T | 1.07 | 0.17 | 0.97 |
| rs999737 | RAD51L1 | 14 | 69034682 | C | 1.08 | 0.77 | 0.97 |
| rs941764 | CCDC88C | 14 | 91841069 | G | 1.06 | 0.36 | 0.95 |
| rs11627032 | RIN3 | 14 | 93104072 | T | 1.06b | 0.74 | 0.93 |
| rs3803662 | TOX3 | 16 | 52586341 | A | 1.23 | 0.29 | 1 |
| rs17817449 | MIRI972-2-FTO | 16 | 53813367 | T | 1.08 | 0.60 | 0.99 |
| rs11075995 | FTO | 16 | 53855291 | T | 1.04 | 0.76 | 0.99 |
| rs13329835 | CDYL2 | 16 | 80650805 | G | 1.08 | 0.24 | 0.98 |
| rs146699004 | TEFM | 17 | 29230520 | GGT | 1.08b | 0.74 | 0.99 |
| rs6504950 | COX11 e | 17 | 53056471 | G | 1.07 | 0.72 | 0.97 |
| rs745570 | CBX8 | 17 | 77781725 | A | 1.05b | 0.50 | 0.97 |
| rs527616 | None | 18 | 24337424 | G | 1.04 | 0.66 | 0.98 |
| rs1436904 | CHST9 | 18 | 24570667 | T | 1.06 | 0.63 | 0.99 |
| rs6507583 | SETBP1 | 18 | 42399590 | A | 1.10b | 0.93 | 0.99 |
| rs8170 | ABHD8/ANKLE1 e | 19 | 17389704 | A | 1.03 | 0.20 | 0.99 |
| rs2363956 | ABHD8/ANKLE1 e | 19 | 17394124 | T | 1.03 | 0.51 | 0.98 |
| rs4808801 | SSBP4-ISYNA1-ELL | 19 | 18571141 | A | 1.07 | 0.67 | 0.99 |
| rs3760982 | C19orf61-KCNN4-LYPD5-ZNF283 | 19 | 44286513 | A | 1.06 | 0.49 | 1 |
| rs2823093 | NRIP1 | 21 | 16520832 | G | 1.08 | 0.75 | 0.96 |
| rs17879961 | CHEK2 | 22 | 29121087 | G | 1.36 | 0.001 | 0.69 |
| rs132390 | EMID1-RHBDD3-EWSR1 | 22 | 29621477 | C | 1.11 | 0.04 | 1 |
| rs6001930 | MKL1 | 22 | 40876234 | C | 1.13 | 0.11 | 0.99 |
adjusted odds ratios from Mavaddat et al. 2015
unadjusted odds ratios from Michailidou et al. 2013 or Michailidou et al. 2015
imputation r2 calculated by the mean info score from IMPUTE2. The info score represents the certainty with which the SNP has been imputed and lies between 0 (no certainty) and 1 (high certainty; r2=1 for genotyped SNPs)
published target gene
known target gene, not yet published
chromosome
human genome build 37 (GRCh37) by the Genome Reference Consortium, released 3 March 2009. Available from: http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/
Figure 1.
Distribution of polygenic risk scores in the 1134 RAPPER patients for whom late radiotherapy toxicity data were available
A) Un-weighted polygenic risk score
B) Weighted polygenic risk score
The distributions of the nine different acute and late-arising toxicity endpoints (acute toxicity, telangiectasia, breast edema, photographically assessed shrinkage, induration, pigmentation, breast pain, breast sensitivity and a standardized total average toxicity (STAT) score) measured in this study are shown in Table 3. Baseline symptoms that resulted from surgery and were present before radiotherapy were not associated with any of the genetic variants either individually or in the polygenic risk score. The significant non-genetic factors associated with acute toxicity and the five-year toxicity endpoints are shown in Supplementary Table 3.
Table 3. Distributions of toxicity endpoints in RAPPER patient sample.
| Number of patients (%) | |||||
|---|---|---|---|---|---|
| Acute toxicity | Grade 0 | Grade 1 | Grade 2a | Grade 2b | Grade 3 |
| RTOG acute toxicity | 30 (3.3) | 549 (59.7) | 328 (35.7) | 12 (1.3) | 1 (0.1) |
| 2 year toxicity | Grade 0 | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
| Telangiectasia | 738 (79.7) | 116 (12.5) | 53 (5.7) | 18 (1.9) | 1 (0.1) |
| Breast edema | 522 (56.4) | 261 (28.2) | 110 (11.9) | 33 (3.6) | 0 |
| Shrinkage assessed photographically | NA | 593 (63.5) | 285 (30.5) | 56 (6.0) | NA |
| Induration | 177 (18.3) | 444 (45.8) | 276 (28.5) | 70 (7.2) | 3 (0.3) |
| Pigmentation | 629 (83.0) | 62 (8.2) | 67 (8.8) | 0 | 0 |
| Breast pain | NA | 416 (49.9) | 362 (43.4) | 45 (5.4) | 11 (1.3) |
| Breast sensitivity | NA | 507 (60.7) | 278 (33.3) | 41 (4.9) | 10 (1.2) |
| Overall toxicity (STATa) | N=1134, mean (standard deviation) = 0.003 (0.66) | ||||
| 5 year toxicity | Grade 0 | Grade 1 | Grade 2 | Grade 3 | Grade 4 |
| Telangiectasia | 493 (83.1) | 48 (8.1) | 30 (5.1) | 22 (3.7) | 0 |
| Breastedema | 487 (82.1) | 73 (12.3) | 27 (4.6) | 6 (1.1) | 0 |
| Shrinkage assessed photographically | NA | 337 (58.7) | 201 (35.0) | 36 (6.3) | NA |
| Induration | 174 (29.3) | 267 (45.0) | 130 (21.9) | 22 (3.7) | 0 |
| Overall toxicity (STATa) | N=593, mean (standard deviation) = 0.01 (0.64) | ||||
Standardised total average toxicity
None of the individual genetic variants were associated, at the pre-specified significance level of p<5 × 10−4, with acute toxicity or two-year toxicity in either univariable or multivariable analyses (Supplementary Table 4, Supplementary Figures 1-9). Univariable analysis of five-year toxicity identified one SNP, rs6964587, to be significantly associated with increased risk of breast edema five years following radiotherapy (OR=1.75, 95%CI=(1.30-2.49), p-value=4 × 10−5). The association remained strong after adjustment for non-genetic factors (residual beta=0.22, 95% CI=(0.09-0.34), p-value=7 × 10−4). No other individual variants were associated with five-year toxicity on univariable or multivariable analysis (Supplementary Table 5).
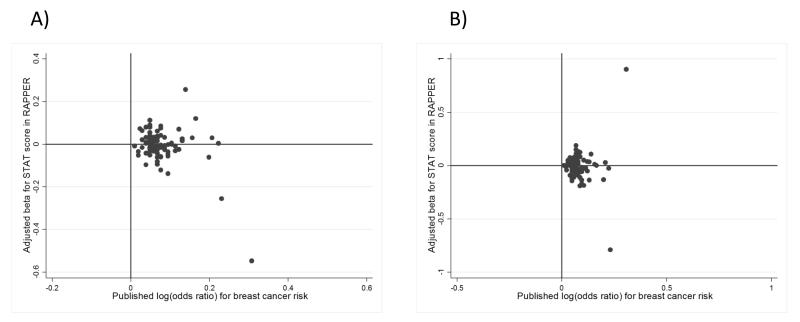
Scatter plots comparing each variant’s effect on breast cancer risk with their effects on STAT (overall toxicity score) at two years and five years are shown in Figure 2. Scatter plots for all other toxicity endpoints can be found in Supplementary Figures 10-17. Points in the top right hand quarters of the plots represent alleles associated with both increased breast cancer and radiation toxicity risk. Conversely for points in the bottom right hand quarter, the allelic effects are opposite for breast cancer and radiation toxicity risk. The plots reveal no correlations between these confirmed breast cancer risk alleles and increased radiotherapy toxicity risk in this RAPPER cohort.
Figure 2.
Scatter plot of individual breast cancer risk variants comparing their effect on breast cancer risk with their effect on STAT (Standardized Total Average Toxicity) score
A) STAT at two years
B) STAT at five years
When the more statistically powerful polygenic risk scores were tested, there were no associations with acute toxicity (weighted p-values>0.30; un-weighted p-values>0.39) or any of the two-year toxicity endpoints (weighted p-values ≥0.13; un-weighted p-values≥0.17) or five-year endpoints (weighted p-values≥0.19; un-weighted p-values≥0.39) (Table 4).
Table 4. Association of risk scores with toxicity endpoints from residual analyses.
| Toxicity | betaa (95% confidence interval) & p-value | |
|---|---|---|
|
| ||
| Un-weighted risk score | Weighted risk score | |
| Acute toxicity | ||
| RTOG grade | −0.01 (−0.02-0.01) p=0.39 | −0.08 (−0.23-0.07) p=0.30 |
| 2 year toxicity | ||
| Telangiectasia | −0.01 (−0.02-0.01) p=0.40 | −0.05 (−0.20-0.10) p=0.54 |
| Breast edema | 0.001 (−0.01-0.01) p=0.80 | 0.02 (−0.13-0.17) p=0.80 |
| Shrinkage assessed photographically | −0.01 (−0.02-0.01) p=0.41 | −0.08 (−0.22-0.07) p=0.30 |
| Induration | −0.003 (−0.01-0.01) p=0.64 | −0.05 (−0.20-0.10) p=0.51 |
| Pigmentation | 0.01 (−0.004-0.02) p=0.17 | 0.13 (−0.04-0.29) p=0.13 |
| Breast pain | 0.003 (−0.01-0.01) p=0.67 | 0.01 (−0.14-0.17) p=0.85 |
| Oversensitivity | −0.001 (−0.01-0.01) p=0.85 | −0.05 (−0.21-0.10) p=0.49 |
| Overall toxicity (STATb) | 0.002 (−0.01-0.01) p=0.72 | 0.02 (−0.13-0.16) p=0.81 |
| 5 year toxicity | ||
| Telangiectasia | −0.0004 (−0.02-0.01) p=0.96 | −0.05 (−0.24-0.14) p=0.61 |
| Breast edema | −0.003 (−0.02-0.01) p=0.72 | −0.05 (−0.23-0.13) p=0.58 |
| Shrinkage assessed photographically | 0.001 (−0.01-0.02) p=0.92 | 0.05 (−0.14-0.25) p=0.58 |
| Induration | −0.01 (−0.03-0.01) p=0.19 | −0.09 (−0.28-0.11) p=0.39 |
| Overall toxicity (STATb) | −0.01 (−0.02-0.01) p=0.47 | −0.07 (−0.26-0.12) p=0.48 |
beta estimates are for the association between polygenic risk score and change in residual toxicity; a positive beta represents an increased risk of toxicity and a negative beta represents reduced risk of toxicity
Standardised total average toxicity
Discussion
The hypothesis behind this study was that common genetic variants known to increase the risk of breast cancer may also increase the likelihood of developing toxicity following radiotherapy for breast cancer. We found no conclusive evidence supporting this hypothesis. One SNP, rs6964587, was identified as associated with increased breast edema at 5 years (OR=1.75; 95% CI=(1.30-2.49)). This SNP is a missense SNP lying in a coding region of the AKAP9 gene, which encodes the A kinase (PRKA) anchor protein 9 - a key component of signal transduction that contributes to carcinogenesis (2,19,20). The SNP is predicted to generate a M643I substitution in the AKAP9 protein sequence. Of note, a study by Han et al suggested that breast tumor cells secrete the AKAP9 protein in response to ionizing radiation (20). This SNP association merits further investigation in independent radiotherapy toxicity cohorts.
The nearest gene to each individual breast cancer risk locus studied is listed in Table 2. For some of these, (marked with d or e), the target gene and mode of action of the causal variant has been elucidated, but for the majority this process is still on-going. A few putative target breast cancer genes on the list (for example, CHEK2 and RAD51b) have known roles in DNA damage repair, but it is unlikely that the majority will prove to have similar functions. In this study of common low-penetrance breast cancer loci, the rare, more highly penetrant mutations in genes such as ATM, BRCA1, BRCA2 and TP53 were not genotyped and could not be included in this analysis. Investigation of these genes requires complete sequencing of the relevant genes and different statistical methods and is on-going. The link between radiosensitivity and cancer risk has been exemplified by ATM gene mutations, causing ataxia-telangiectasia, a cancer predisposing immunodeficiency syndrome characterized by extreme sensitivity to radiation and life-threatening radiotherapy toxicity (21). Carriers of ATM gene mutations, including relatives of people with ataxia-telangiectasia, have an increased risk of breast cancer(22). An in vitro radiosensitivity assay revealed that around 40% of breast cancer patients tested displayed evidence of increased radiosensitivity (7,8,23). Another study demonstrated the heritability of enhanced chromosomal radiosensitivity in breast cancer families (6).
This RAPPER study had a relatively small sample size. The original studies confirming the associations of the 94 genetic variants with increased breast cancer risk were performed in BCAC with over 100,000 study participants. In contrast, the RAPPER breast cohort comprises just fewer than 1,200 patients and the study had low power to detect a true association between individual variants and radiotherapy toxicity. Despite this, a single variant associated with an ~75% increased odds of breast edema five years following radiotherapy has been detected at a stringent statistical significance threshold. Future larger studies with greater power will be able to identify other individual SNPs that may be associated with radiotherapy toxicity. These studies could also explore whether the effect sizes of individual variants on breast cancer risk differ from those for radiation toxicity risk. Although this study was generally underpowered to detect individual variants, the study was well powered (87%) to detect an association between radiotherapy toxicity and a polygenic risk score. The 90 confirmed breast cancer variants, used in this study, have been estimated to explain 16% of the familial risk of breast cancer (3) and an earlier polygenic risk profile, which included a subset of 77 of these SNPs, demonstrated that women in the highest 1% of the risk score were 3.6 times more likely to develop breast cancer than women in the middle quintile (4). We have extended the polygenic risk profile to include 90 currently assayable breast cancer risk loci and have shown that they have no detectable predictive value for risk of radiation toxicity in women being treated for breast cancer. Our analysis was limited to women of European ancestry only and therefore the findings may not be generalizable to women of other ancestries.
In summary, our study found no evidence to support a suggestion that patients with a high polygenic risk of breast cancer have an increased likelihood of developing acute or late radiotherapy toxicity. The conclusion from this work is that it appears to be safe for breast cancer patients with a high polygenic risk of breast cancer to receive standard radiotherapy for breast cancer.
Supplementary Material
Translational Statement.
Common germline genetic variation impacts on the development of cancer and 94 common variants have now been identified, which together explain almost 20% of the variance in breast cancer risk. Up to 75% of breast cancer patients are likely to have radiotherapy and, depending on the country of treatment, up to 80% of patients survive breast cancer. For over 20 years clinicians have been concerned that the biological mechanisms underlying genetic predisposition to breast cancer risk may also increase radiosensitivity, such that some patients receiving breast tumor radiotherapy may be at increased risk of normal tissue toxicity. We have tested the breast cancer risk variants, both individually and as a multi-SNP risk profile, for association with acute and late radiotherapy toxicity phenotypes in 1,160 breast cancer patients. Our study showed that those with a high polygenic risk of breast cancer do not have increased risk of radiotherapy toxicity.
Acknowledgements
The authors would like to acknowledge the Cambridge Centre for Cancer Genetic Epidemiology (CCGE) computing cluster at Strangeways Research Laboratory at the University of Cambridge for providing supercomputing infrastructure and Michael Lush for his technical support. We thank Nasim Mavaddat for early access to pre-published details of the breast cancer risk variants. We acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR.
Funding
LD is funded by the Medical Research Council (UK). CCC and NGB are supported by the Cambridge National Institute for Health Research (NIHR) Biomedical Research Centre. JY is supported by the Institute of Cancer Research & Royal Marsden NIHR Biomedical Research Centre. PDPP is funded by Cancer Research UK (C490/A16561). CMW is supported by Experimental Cancer Medicine Centre (ECMC) funding. The collaborative group (RAPPER) is funded by Cancer Research UK (C1094/A18504). EU FP7 Health-F2-2009-223175 “COGS” funded the discovery or confirmation of the breast cancer risk variants used in this study.
Footnotes
The authors declare no conflict of interest.
References
- 1.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milne RL, Burwinkel B, Michailidou K, Arias-Perez J-I, Zamora MP, Menéndez-Rodríguez P, et al. Common non-synonymous SNPs associated with breast cancer susceptibility: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2014;23:6096–111. doi: 10.1093/hmg/ddu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47:373–80. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavaddat N, Pharoah PDP, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of Breast Cancer Risk Based on Profiling With Common Genetic Variants. JNCI J Natl Cancer Inst. 2015;107:djv036. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozasa K, Shimizu Y, Suyama A, Kasagi F, Soda M, Grant EJ, et al. Studies of the Mortality of Atomic Bomb Survivors , Report 14 , 1950 – 2003 : An Overview of Cancer and Noncancer Diseases. Radiat Res. 2012;243:229–43. doi: 10.1667/rr2629.1. [DOI] [PubMed] [Google Scholar]
- 6.Roberts SA, Spreadborough AR, Bulman B, Barber JB, Evans DG, Scott D. Heritability of cellular radiosensitivity: a marker of low-penetrance predisposition genes in breast cancer? Am J Hum Genet. 1999;65:784–94. doi: 10.1086/302544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baria K, Warren C, Roberts SA, West CM, Scott D. Chromosomal radiosensitivity as a marker of predisposition to common cancers? Br J Cancer. 2001;84:892–6. doi: 10.1054/bjoc.2000.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott D. Chromosomal radiosensitivity and low penetrance predisposition to cancer. Cytogenet Genome Res. 2004;104:365–70. doi: 10.1159/000077517. [DOI] [PubMed] [Google Scholar]
- 9.Hoeller U, Borgmann K, Bonacker M, Kuhlmey A, Bajrovic A, Jung H, et al. Individual radiosensitivity measured with lymphocytes may be used to predict the risk of fibrosis after radiotherapy for breast cancer. Radiother Oncol. 2003;69:137–44. doi: 10.1016/j.radonc.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Burnet NG, Elliott RM, Dunning A, West CML. Radiosensitivity, Radiogenomics and RAPPER. Clin Oncol. 2006;18:525–8. doi: 10.1016/j.clon.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Barnett GC, Thompson D, Fachal L, Kerns S, Talbot C, Elliott RM, et al. A genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicity. Radiother Oncol. 2014;111:178–85. doi: 10.1016/j.radonc.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Trialists TS. The UK Standardisation of Breast Radiotherapy (START) Trial A of radiotherapy hypofractionation for treatment of early breast cancer: a randomised trial. Lancet Oncol. 2008;9:331–41. doi: 10.1016/S1470-2045(08)70077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnett GC, West CML, Coles CE, Pharoah PDP, Talbot CJ, Elliott RM, et al. Standardized Total Average Toxicity score: a scale- and grade-independent measure of late radiotherapy toxicity to facilitate pooling of data from different studies. Int J Radiat Oncol Biol Phys. 2012 Mar 1;82:1065–74. doi: 10.1016/j.ijrobp.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012 Feb;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 15.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011 Nov 1;:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentzen SM, Overgaard J. Patient-to-patient variability in the expression of radiation-induced normal tissue injury. Semin Radiat Oncol. 1994;4:68–80. doi: 10.1053/SRAO00400068. [DOI] [PubMed] [Google Scholar]
- 18.Kerns SL, De Ruysscher D, Andreassen CN, Azria D, Barnett GC, Chang-Claude J, et al. STROGAR - STrengthening the Reporting of Genetic Association studies in Radiogenomics. Radiother Oncol. Elsevier Ireland Ltd. 2014;110:182–8. doi: 10.1016/j.radonc.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank B, Wiestler M, Kropp S, Hemminki K, Spurdle AB, Sutter C, et al. Association of a common AKAP9 variant with breast cancer risk: A collaborative analysis. J Natl Cancer Inst. 2008;100:437–42. doi: 10.1093/jnci/djn037. [DOI] [PubMed] [Google Scholar]
- 20.Han NK, Kim BC, Lee HC, Lee YJ, Park MJ, Chi SG, et al. Secretome analysis of ionizing radiation-induced senescent cancer cells reveals that secreted RKIP plays a critical role in neighboring cell migration. Proteomics. 2012;12:2822–32. doi: 10.1002/pmic.201100419. [DOI] [PubMed] [Google Scholar]
- 21.Gatti RA, Berkel I, Boder E, Braedt G, Charmley P, Concannon P, et al. Localization of an ataxia telangiectasia gene to chromosome 11q22-23. Nature. 1988;336:577–80. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 22.Milne RL. Variants in the ATM gene and breast cancer susceptibility. Genome Med. 2009;1:12. doi: 10.1186/gm12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryabchenko NM, Glavin OA, Shtefura VV, Anikusko MF. Chromosomal Radiosensitivity in Ukranian Breast Cancer Patients and Healthy Individuals. Exp Oncol. 2012;34:121–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.