Abstract
In the yeast Saccharomyces cerevisiae, the switch from respiratory metabolism to fermentation causes rapid decay of transcripts encoding proteins uniquely required for aerobic metabolism. Snf1, the yeast ortholog of AMP-activated protein kinase, has been implicated in this process because inhibiting Snf1 mimics the addition of glucose. In this study, we show that the SNF1-dependent ADH2 promoter, or just the major transcription factor binding site, is sufficient to confer glucose-induced mRNA decay upon heterologous transcripts. SNF1-independent expression from the ADH2 promoter prevented glucose-induced mRNA decay without altering the start site of transcription. SNF1-dependent transcripts are enriched for the binding motif of the RNA binding protein Vts1, an important mediator of mRNA decay and mRNA repression whose expression is correlated with decreased abundance of SNF1-dependent transcripts during the yeast metabolic cycle. However, deletion of VTS1 did not slow the rate of glucose-induced mRNA decay. ADH2 mRNA rapidly dissociated from polysomes after glucose repletion, and sequences bound by RNA binding proteins were enriched in the transcripts from repressed cells. Inhibiting the protein kinase A pathway did not affect glucose-induced decay of ADH2 mRNA. Our results suggest that Snf1 may influence mRNA stability by altering the recruitment activity of the transcription factor Adr1.
INTRODUCTION
The regulation of gene expression by nutritional conditions in Saccharomyces cerevisiae yeast, particularly the availability of glucose, has been a paradigm of transcriptional control (1–7). Glucose deprivation leads to a global reorganization of transcription (8). Numerous genes are upregulated to allow the cell to switch to a respiratory mode of metabolism, and a large number of genes are downregulated as the cells adapt to a lower rate of growth. Snf1, the yeast ortholog of AMP-activated protein kinase (AMPK) (6), is responsible for upregulating the expression of over 400 genes after glucose depletion (9). Snf1, together with the cyclic AMP (cAMP)-dependent protein kinase A (PKA) and target of rapamycin (TOR) pathways, coordinates many of the nutrient-responsive metabolic pathways in yeast (10). Despite the preponderance of evidence indicating altered transcription as the major factor determining the increase in mRNA abundance when yeast cells are depleted of glucose, there is considerable evidence indicating that posttranscriptional changes, particularly an increase in mRNA stability, are also important in determining gene expression levels (11–15). A recent study demonstrated that promoter sequences influence the subcellular location and efficiency of translation of transcripts upregulated by glucose starvation (16). Thus, promoter sequences appear to have a role in gene expression that includes both transcriptional and posttranscriptional processes.
Glucose-induced mRNA decay is a process that leads to rapid loss of mRNAs encoding enzymes required for efficient aerobic respiration when glucose is replenished. Evidence derived from yeast nuclear genes encoding mitochondrial proteins, particularly SDH1 and SDH2 (17), suggested that this was an example of posttranscriptional regulation of gene expression (18). This is a general process affecting mRNAs encoding gluconeogenic and glyoxylate enzymes (12), enzymes required for the metabolism of nonfermentable carbon sources (19), alternative sugars like maltose (20), galactose (14), and sucrose (21), enzymes of β-oxidation and peroxisome biogenesis, and transporters of amino acids and alternative carbon sources like lactate (19, 22). Regulation of the process may involve multiple signaling pathways, because different genes are affected at different glucose concentrations. For example, the transcripts encoding gluconeogenic enzymes are subject to rapid decay at much lower glucose concentrations than transcripts from genes encoding mitochondrial proteins (13). The biological significance of the decrease in mRNA stability is to ensure a rapid decrease in the synthesis of enzymes involved in aerobic metabolic pathways once the ability to ferment glucose is restored. The rapid clearance of the potential to synthesize these enzymes avoids futile cycling of metabolites.
Transcriptional regulation of SDH1 and SDH2 did not seem sufficient to explain their activation after glucose depletion, and further analyses suggested that an increase in the stability of these mRNAs occurred after the shift from fermentative to respiratory metabolism (11, 17, 21). Lombardo and colleagues concluded that their low abundance during fermentative growth was due to their rapid turnover (11). Thus, these genes seemed to represent a major departure from the regulation of many glucose-repressible genes, whose increased expression can be fully explained by transcriptional upregulation when glucose is exhausted (2, 3, 7).
RNA binding proteins (RBPs) play an important role in posttranscriptional regulation of gene expression (23–25). In the case of nuclear transcripts encoding mitochondrial proteins, the role of the RBP Puf3 is one of the best understood in Saccharomyces cerevisiae. The activity of Puf3, but not its expression, localization, or RNA binding, is regulated by carbon source to ensure that transcripts encoding mitochondrial proteins important for aerobic metabolism have a longer half-life when a fermentable carbon source is absent (15).
Although the mechanism of glucose-induced mRNA decay is poorly understood, analysis of SDH2 suggested that sequences in the 5′ untranslated region (5′-UTR) of the transcript, specifically, the sequences just preceding the translation start site, were necessary to mediate glucose-induced mRNA decay (26). In contrast, studies of glucose-induced decay of JEN1 transcripts suggested an RNA-mediated mechanism (22). Glucose-induced decay of GAL transcripts was attributed not to the presence of glucose per se but to the transition from a respiratory to a fermentable carbon source (14). Thus, there may be multiple, gene-specific mechanisms that participate in glucose-induced mRNA decay.
Signaling pathways that might influence glucose-induced mRNA decay were sought by screening yeast genes whose deletion affected glucose repression or that were known components of mRNA decay pathways (21). Deletion of REG1, encoding an inhibitor of Snf1, or XRN1, encoding a major cytoplasmic exoribonuclease, dramatically increased the stability of SDH2 mRNA during glucose-induced mRNA decay. Although these results suggested a role for Snf1 signaling in glucose-induced mRNA decay, the authors reported that SDH2 expression was SNF1 independent and suggested that REG1 might be acting through a SNF1-independent pathway to influence the process. Deletion of CYC8 (also known as SSN6) and SSN3 (also known as UME5, SRB10, or CDK8), encoding an important transcriptional repressor and a component of yeast Mediator complex, respectively, also stabilized SDH2 mRNA during glucose-induced mRNA decay (21), implicating a role for transcription.
Snf1 was linked to glucose-induced mRNA decay by the observation that inhibiting an analog-sensitive allele of SNF1 (SNF1as) led to instability of the same transcripts that were destabilized by the addition of glucose (19). Inhibiting Snf1 and simultaneously adding glucose had the same effect as either perturbation alone, suggesting that glucose-mediated decay might be due solely to inhibiting Snf1. This conclusion was consistent with the earlier observation that deleting REG1 prevented glucose-induced mRNA decay of SDH2 transcripts (21). SNF1-dependent phosphorylation of components of a cytoplasmic mRNA decay pathway might regulate their activity toward specific transcripts during glucose-induced mRNA decay (19). Xrn1 might be one such component, because mutation of the SNF1-dependent phosphorylation sites in its carboxy-terminal region alleviated glucose-induced decay of several SNF1-dependent mRNAs (27). To explore the mechanism of SNF1-dependent mRNA stabilization, we asked whether there was a link between SNF1-dependent transcription and mRNA stability.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth media.
The S. cerevisiae strains used in this study are listed in Table 1. Strains containing the SNF1as allele (encoding a change of I to G at position 132 [I132G]) were made by integrative transformation (28) at the ura3-1 locus by cutting plasmid pSH47 (URA3) with EcoRV. Strains containing fusions of the ADH2 promoter gene to MAP2 and IDP1 were constructed as follows. The NatMX cassette was amplified from pAG25 using primers KB146-MX-F and KB147-MX-R, and the ADH2 promoter, with a 22-bp region homologous to the 3′ end of NatMX, was amplified from pBGM18 (29) [ADH2(−806 to +109)-lacZ CEN-URA3] using KB148-ADH2-F and KB149-ADH2-R [ADH2(−591 to −101)] or KB148-ADH2-F and KB154-ADH2-R2 [ADH2(−509 to −49)]. The NatMX and ADH2 PCR products were denatured, mixed, and annealed, and the ligated product was amplified after annealing to oligonucleotides complementary to either MAP2 [KB150-MAP2-F and KB151-MAP2-R for ADH2p(−591 to −101)-MAP2 or KB150-MAP2-F and KB155-MAP2-R2 for ADH2p(−591 to −49)-MAP2] or IDP1 [KB152-IDP1-F and KB153-IDP1-R for ADH2p(−591 to −101)-IDP1] and IDP1 [KB152-IDP1-F and KB156-IDP1-R2 for ADH2p(−591 to −49)-IDP1]. Each resulting PCR product was integrated into the wild-type (WT) strain W303-1a and an SNF1as strain (KBY77) upstream from the 5′-UTR of the targeted open reading frame (ORF). All oligonucleotides are listed in Table S1 in the supplemental material. The chimeric ADR1 gene, ADR1-EV, was constructed by gap repair cloning (28). The GAL4 DNA binding domain of pRS313-ADH1pr-Myc-GAL4DBD-ERLBD-VP16-Flag (30), a derivative of the original pGAL4.ER.VP16 (31), was replaced by the DNA binding domain of Adr1 (amino acids 1 to 166). Plasmids pKD16 (32), pKD16H (33), pKD17 (32), pHDY10 (34), pBGM18 (29), and pLGADH2 (35) have been described previously. S. cerevisiae cultures were grown in yeast extract peptone (YP) or in synthetic medium (SM) containing 5% glucose (repressing conditions) and lacking the appropriate amino acid or uracil for plasmid selection. For derepression in low glucose, mid-log-phase cells from repressing medium were harvested by centrifugation, and the pellets were drained well, resuspended in either YP or SM having 0.05% glucose and the required supplements, and allowed to grow for the times indicated in the figure legends. To maintain selection for plasmids containing TRP1 and/or URA3, the synthetic selective medium contained 0.2% Casamino Acids rather than the standard dropout solution. All S. cerevisiae strains were grown at 30°C unless stated otherwise. When cultures containing the plasmid bearing the chimeric ADR1-EV were analyzed, induction was initiated by adding β-estradiol dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 1 nM.
TABLE 1.
Yeast strains
| Strain(s) | Genotype | Reference or source |
|---|---|---|
| W303-1a | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 28 |
| CKY27 | CKY20 adr1Δ::natMX snf1Δ::kanMX | K. Dombek |
| CKY18 | CKY20 snf1Δ::kanMX | K. Dombek |
| CKY19 | W303-1a | K. Dombek |
| CKY20 | W303-1a MATα | K. Dombek |
| KBY77 | CKY19 ura3::YIpSNF1as URA3 snf1Δ::kanMX | This study |
| KBY127 | CKY19 map2::natMX-ADH2p(−591 to −101)-MAP2 | This study |
| KBY131 | CKY19 idp1::natMX-ADH2p(−591 to −101)-IDP1 | This study |
| KBY137 | KBY77 map2::natMX-ADH2p(−591 to −49)-MAP2 | This study |
| KBY140 | KBY77 idp1::natMX-ADH2p(−591 to −49)-IDP1 | This study |
| KBY108 | MATa rpb1-1 (ts) snf1Δ::kanMX ADR1-13MYC::natMX ura3::SNF1as-URA3 LEU+ (mixture of W303 and S288C backgrounds) | This study |
| JBY3622 | TPK1as TPK2as TPK3as his3Δ::HIS3 GAL1-ras2-val19 trp1 ura3 ade2 | 76 |
| TYY1077, TYY1078 | CKY20 snf1Δ::kanMX ura3::YIpSNF1as::URA3 ADR1-Myc13::natMX | This study |
| TYY203 | W303-1a YIpADH2-lacZ | 9 |
| TYY204 | W303-1a YIpADH2-lacZ adr1Δ::URA3 | 9 |
| BY4741 | MATa his3Δ1 leu2ΔO met15ΔO ura3ΔO | Research Genetics |
| BY4741 vts1Δ | BY4741 vts1Δ | Research Genetics |
| BY4742 vts1Δ | MATα vts1Δ | Research Genetics |
mRNA isolation and qRT-PCR.
Total cellular RNA was isolated from yeast cultures using either the hot acidic phenol method described in Collart and Oliviero (36) or an RNeasy kit (Qiagen). Residual DNA in the RNA preparation was reduced by using the Turbo DNA-free kit (Ambion RNA Life Technologies) according to the manufacturer's recommendations. cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad), following the manufacturer's protocol. Reverse transcription quantitative real-time PCR (RT-qPCR) was performed for measuring mRNA levels using SsoFast EvaGreen supermix (Bio-Rad), diluted cDNA, and specific primer pairs. A standard curve was generated with specific primer pairs and used to quantify the mRNA levels.
Analysis of mRNA decay.
The decrease in mRNA abundance was measured after adding glucose (final concentration of 5%), 1,10-o-phenanthroline (final concentration of 100 μg/ml in 100% ethanol), or 2-naphthylmethyl pyrazolopyrimidine (2NM-PP1; final concentration of 10 μM in 100% DMSO) to a 4-h derepressed cell culture. Cells in 10-ml aliquots were collected at the times indicated in the figures into 50-ml Falcon tubes containing 20 g of ice and 200 μg of cycloheximide to stabilize mRNA on the ribosomes. Total RNA isolation and mRNA analysis were performed as described above. Apparent half-lives were calculated from the slope of a graph of log(mRNA) versus time (37). The half-life was calculated from the linear portion of the curve, usually following a 5-min lag.
Labeling and purification of 4-thiouracil-labeled RNA.
The basic protocol for 4-thioruacil labeling of yeast RNA, modification of the thiol group by biotinylation, and purification by streptavidin affinity chromatography was followed (38). 4-Thiouracil (Sigma) was used at a final concentration of 0.2 mM in SM medium containing 0.05% glucose and supplemented with Casamino Acids (0.2%), adenine, tryptophan, and 1/5 the normal concentration of uracil (final concentration, 10 μM). After 60 min in the presence of 4-thiouracil, a 50-fold excess of uracil was added from a 20-mg/ml stock (dissolved by heating) to give a 50-fold excess over the 4-thiouracil. RNA isolated from a 50- to 100-ml culture was purified by hot phenol extraction (36) and biotinylated, and 4-thiouracil-containing RNA was purified by streptavidin affinity chromatography on GE Healthcare magnetic beads and converted to cDNA as described above. Total Escherichia coli 4-thiouracil-containing RNA isolated from E. coli strain DH10B grown in M9 synthetic medium containing Casamino Acids was added to the total yeast RNA prior to the biotinylation reactions as an internal control for biotinylation, purification, and cDNA synthesis.
Primer extension analysis.
Oligonucleotides of noncoding-strand sequences near the 5′ end of ADH2, IDP1, and MAP2 (sequences are in Table S1 in the supplemental material) were labeled with [γ-32P]ATP in kinase buffer (50 mM Tris, pH 7.6, 10 mM MgCl2, 5 mM dithiothreitol) using 25 units of T4 polynucleotide kinase and then purified by centrifugation through a 1-ml syringe barrel containing Sephadex G25 equilibrated in 10 mM Tris, pH 7.5. Approximately 1 ng of primer extension oligonucleotide (typically 200,000 to 400,00 cpm) was annealed with 50 to 100 μg total yeast RNA in 5 mM Tris, pH 8.3, 75 mM KCl, 1 mM EDTA for 45 min at 48°C after heating the annealing solution for 1 min at ∼99°C. Primer extension reactions were carried out in primer extension buffer (10 mM Tris, pH 8.3, 25 mM Tris, pH 8.3, 75 mM KCl, 0.33 mM EDTA, 5 mM MgCl2, 15 mM dithiothreitol, 0.5 mM each deoxynucleoside triphosphate, using 100 units of Moloney murine leukemia virus reverse transcriptase [Gibco-BRL]) and were incubated for 30 min at 37°C. The RNA template and reaction products were ethanol precipitated, washed with 80% ethanol, dried, and resuspended in 4 μl of a solution containing 40 μg/ml of boiled RNase A. After incubating at room temperature for 10 min, 5 μl of formamide gel loading buffer (80% deionized formamide, 0.025% xylene cyanol and bromophenol blue, 30 mM EDTA) was added and the samples were heated for 1 min at 90°C and then rapidly chilled on ice. Samples were loaded onto a Novex precast Tris-boric acid-EDTA (TBE)–10% acrylamide sequencing gel (Invitrogen, Thermo Fisher Scientific) and run in an XCell SureLock minicell apparatus according to the manufacturer's protocol (Invitrogen, Thermo Fisher Scientific). The primers used for these analyses are listed in Table S1.
Polysome gradient analysis of mRNA.
Whole-cell extracts were made and analyzed on polysome gradients as described previously (39). In brief, a 150-ml culture of strain TYY1078 (relevant genotype, SNF1as) was grown to early log phase in YP with 5% glucose and derepressed for 4 h, and then the culture was divided into three 50-ml portions. Glucose (final concentration, 5%) or 2NM-PP1 (final concentration, 5 μM) was added to two aliquots. The third portion had no addition. Five minutes after the addition of glucose or 2NM-PP1, all three cultures were poured onto crushed ice made by freezing YP medium with 5% glucose and 10 μg/ml cycloheximide. Cell extracts were made and fractionated by sucrose density gradient centrifugation as described previously (39). RNA was recovered from the gradient fractions by phenol extraction and concentrated for cDNA synthesis by isopropanol precipitation. E. coli RNA was added prior to phenol extraction to monitor recovery and cDNA synthesis efficiency by RT-qPCR using primers for 16S E. coli RNA.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (40), using 10 μl of anti-Rpb3 antiserum per mg of whole-cell extract. The antiserum was obtained from S. Hahn. Enrichment is expressed as the immunoprecipitated (IP) DNA-to–input ratio of the specific amplicon over the IP DNA-to-input ratio corresponding to the amplicon for a telomeric sequence.
Analysis of sequences in SNF1-dependent mRNAs occupied by RBPs in vivo.
The protein binding sites in nontranslating mRNAs of exponentially growing and glucose-starved cells that were occupied in vivo were obtained from the global photoactivatable ribonucleotide-enhanced coupled immunoprecipitation of RBPs (gPAR-CLIP) data of Freeberg et al. (41). After reading the annotated binding data into an R script (42), unreliable data, i.e., the presence of only one of two replicate data points, were removed. Data were retained if one experimental condition had two data points and the other had none, indicating that binding was below the level of detection under the other condition. Then, the binding site occupancy ratio of the sequence from high-glucose versus glucose-starved cells was calculated for each of the retained binding sites. Sites with an occupancy ratio of 3 or greater or 0.33 or lower were retained for further analyses. The data for the 32 Snf1-dependent mRNAs, as well as for ICL1 and ICL2, were retrieved from this filtered data subset, categorized by binding site location in the transcript, and classified as having an increased level of occupancy if the binding ratio was high or decreased occupancy if the binding ratio was low. The mRNAs having one or more gPAR-CLIP sites were counted in each category for each occupancy level. This data classification and the binding site counting were also performed on 24 subsets of randomly chosen genes with replacement from the filtered-data subset after removal of the SNF1-dependent mRNAs. Finally, a two-sided, one-sample t test comparing these counts for the SNF1-dependent group to those of the randomly chosen mRNA sets was performed for each occupancy level of each mRNA location at a confidence level of 0.95 to determine which region of the Snf1-dependent mRNAs might be enriched over randomness for RNA binding sites with increased occupancy after glucose addition.
To search for motifs recognized by known yeast RNA binding proteins, the sequences of the RNA binding sites identified by gPAR-CLIP were retrieved using an R script that employed the Bioconductor packages Biostrings (43) and BSgenome.Scerevisiae.UCSC.sacCer3 (44) and the binding site coordinates provided by Freeberg et al. in supplementary file 4 of their article (41). Then, a panel of search patterns representing the known sequence motifs for yeast RBPs obtained from Freeberg et al. (41) and Riordan et al. (45) was constructed (see Table S2 in the supplemental material). After combining the lists of motifs from these two sources, the regular expressions were converted into IUPAC nucleotide symbols and duplicate motifs were removed from the combined list. Each motif pattern was searched against the SNF1-dependent subset of binding site sequences identified by gPAR-CLIP and counted using the Biostrings vcountPattern function with the “max.mismatch” parameter set to “1” and the “fixed” parameter set to “FALSE.” The motif counts for RNA binding proteins represented by multiple patterns were summed for each mRNA. Then, the motif counts were quantized to indicate either the presence or absence of an occupied RBP motif in each SNF1-dependent mRNA. These quantized motif counts for each RNA binding protein were summed across all mRNAs in the SNF1-dependent mRNA group to obtain the number of SNF1-dependent transcripts having one or more instances of a binding site occupied by a known RBP.
De novo sequence motif identification in SNF1-dependent mRNA sequences using the MEME suite.
The nucleotide sequences of the 32 mRNAs exhibiting Snf1-dependent turnover upon glucose addition (19) were analyzed using the MEME suite of programs (46) to identify common motifs enriched over 32 SNF1-independent mRNAs. A zero-order Markov model of the yeast genome minus the 32 SNF1-dependent mRNAs was constructed using the fasta-get-markov perl script written by Timothy Bailey. Then, the 32 SNF1-dependent mRNA sequences were loaded into MEME (http://meme-suite.org/tools/meme) and de novo motif finding was performed, looking for zero or one motif occurrence per sequence for 5 different motifs each with a width between 6 and 15 bases on the coding strand, using 32 randomly chosen SNF1-independent mRNA sequences as negative sequences and the Markov model as the background model. Identified motifs with an E value of less than 0.05 were loaded into the Tomtom motif comparison tool (47) and searched against the CISBP-RNA single species database of RNA binding protein recognition sequences (48). Matches with a P value of less than 0.05 were considered significant.
Identification of known RNA binding protein recognition sequences enriched in SNF1-dependent mRNAs over the S. cerevisiae genome.
Sequences of all yeast mRNAs were retrieved using an R script as described above for the qPAR-CLIP data analysis but this time using the transcription start site (TSS) and polyadenylation site (PAS) coordinates obtained from Park et al. (49). Sequences without a TSS coordinate were retrieved beginning 100 bp upstream from the start codon, while sequences without a PAS coordinate were retrieved ending 150 bp downstream from the stop codon. The 5′-UTR sequences included the translation start codon, and the 3′-UTR sequences included the translation stop codon. Each coding sequence (CDS) retrieved began at the start codon and ended at the stop codon. Sequences for each region of the mRNAs were then split into two subsets, one containing the 34 SNF1-dependent mRNAs described above for the gPAR-CLIP data analysis and the other containing the remaining mRNA sequences from the yeast genome for use as a negative-control group. 5′-UTR, CDS, 3′-UTR, and mRNA sequence sets from the SNF1-dependent and negative-control groups were searched with the panel of known yeast RBP binding site motifs (see Table S2 in the supplemental material) and matches counted as described above. The motif counts for RBPs represented by multiple patterns were summed for each mRNA, and the motif counts were quantized to indicate either the presence or absence of a particular RNA binding protein motif in each mRNA of the sequence set searched. Then, these quantized motif counts for each RNA binding protein were summed across all mRNAs of each sequence set for the SNF1-dependent and negative-control groups. Enrichment for RBP motifs in the SNF1-dependent mRNA sequences was determined by comparing the summed, quantized motif counts for the SNF1-dependent mRNAs with those of the negative-control group using Fisher's exact test.
RESULTS
The ADH2 promoter is sufficient to confer glucose-induced mRNA decay.
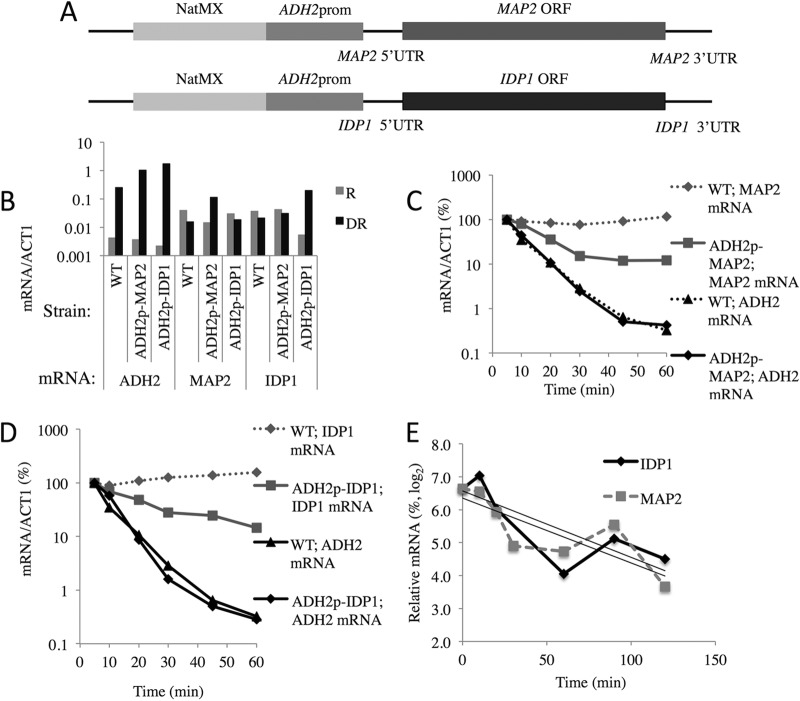
To test whether the ADH2 promoter is sufficient to cause glucose-induced mRNA decay, we replaced the native promoters of two constitutively expressed, SNF1-independent yeast genes with the ADH2 promoter (ADH2p) by integrative transformation (Fig. 1A). The ADH2p-gene fusions retain the native 5′- and 3′-UTRs and ORFs of MAP2 and IDP1. The transcripts of these genes have long half-lives (38, 50, 51) and are nonessential.
FIG 1.
The ADH2 promoter/regulatory region confers glucose-induced mRNA instability on normally stable transcripts. (A) Structures of the MAP2 and IDP1 loci after targeted integration of the ADH2 promoter. (B) ADH2, MAP2, and IDP1 transcript levels under repressed (R) and derepressed (DR) growth conditions. (C and D) Glucose-induced mRNA decay experiments were performed as described in Materials and Methods in strains W303-1a (WT), KBY127 (ADH2p-MAP2), and KBY131 (ADH2p-IDP1). (E) Levels of endogenous IDP1 and MAP2 transcripts after transcription was inhibited by the addition of 100 μg/ml 1,10-o-phenanthroline following 4 h of derepression in strain KBY77. The slopes of the linear regression curves were used to calculate the mRNA half-lives reported in Table 3.
To determine the influence of the ADH2 promoter (residues −591 to −101) on the expression of MAP2 and IDP1, cultures of the WT (W303-1a), ADH2p-MAP2 (KBY127), and ADH2p-IDP1 (KBY131) strains were subjected to a glucose-induced mRNA decay experiment as described in Materials and Methods. RNA was isolated, and mRNAs corresponding to ACT1, ADH2, MAP2, and IDP1 were quantified by RT-qPCR in samples isolated from repressing and derepressing conditions and at various times after glucose repletion. As shown by the results in Fig. 1B, the mRNAs derived from ADH2p-MAP2 and ADH2p-IDP1 increased 9- and 15-fold in abundance upon glucose depletion. The levels of the endogenous MAP2 and IDP1 transcripts in the WT or in strains containing ADH2p-IDP1 and ADH2p-MAP2, respectively, decreased between two- and fourfold under the same conditions, suggesting that the expression of the endogenous genes is downregulated under derepressing growth conditions. The extent of derepression of the ADH2p-driven reporter genes represented a 30- to 40-fold increase over the level of expression of the endogenous genes in the absence of glucose. Thus, ADH2p mediates important increases in the levels of expression of MAP2 and IDP1, as has been shown for other heterologous genes driven by this promoter (34, 35, 52). The increased levels of expression of the heterologous genes are not as great as that shown by ADH2 itself, for unknown reasons that might include chromatin effects on gene expression due to the truncated promoter. MAP2 and IDP1 transcripts under the control of ADH2p decreased rapidly starting 5 min after glucose addition, whereas in WT cells, MAP2 and IDP1 mRNA levels increased by about twofold between 5 and 60 min after glucose addition (Fig. 1C and D). The positive-control ADH2 mRNA showed a rapid decline in mRNA abundance beginning 5 min after glucose repletion in both the WT strain and in the strains bearing the gene fusions (Fig. 1C and D). The rapid decrease in ADH2 transcript levels represents mRNA decay without concomitant transcription because polymerase II (Pol II) is rapidly lost from the promoter and gene body of ADH2 and several other Adr1-dependent genes after glucose repletion (19). We assume that the decrease in MAP2 and IDP1 mRNA levels also represents mRNA decay without concomitant mRNA synthesis. We tested this assumption for IDP1 by performing chromatin immunoprecipitation (ChIP) for RNA Pol II in cells expressing IDP1 from the ADH2 promoter. RNA Pol II was reduced at the 5′ and 3′ ends of the ADH2p-IDP1 gene by 20 min after the addition of glucose to a derepressed culture (Table 2). RNA Pol II was also lost from the 5′ region of the ADH2 gene in these cultures (Table 2). We assume that RNA Pol II is lost from the ADH2p-MAP2 gene in a similar manner. The apparent half-lives of MAP2 and IDP1 mRNAs after glucose addition were 9.0 and 14 min, respectively, when they were expressed under the control of the ADH2 promoter (Table 3). We measured the stability of endogenous MAP2 and IDP1 transcripts expressed from their own promoters in a typical glucose-induced mRNA decay experiment. The nonspecific transcription inhibitor 1,10-o-phenanthroline was added at the same time that glucose was added to a derepressed culture (Fig. 1E). An apparent half-life of about 60 min was determined for both transcripts (Table 3), a value that is similar to those previously reported (38, 50, 53). Thus, the data suggest that ADH2p is sufficient to impart glucose-induced mRNA instability on two heterologous transcripts containing their native 5′- and 3′-UTRs and ORFs.
TABLE 2.
Chromatin immunoprecipitation analysis of RNA Pol II occupancy at the endogenous ADH2 promoter and at the ADH2p-IDP1 genea
| Culture condition | Enrichment factor (IP DNA/input) for: |
|||||
|---|---|---|---|---|---|---|
|
ADH2 |
IDP1 |
|||||
| 3′ end |
5′ end |
|||||
| Mean | Range | Mean | Range | Mean | Range | |
| DR | 12 | 7.3–16 | 12 | 9.4–15 | 11 | 8.7–13 |
| DR→R | 1.9 | 1.1–2.8 | 0.52 | 0.22–0.81 | 0.58 | 0.5–1.5 |
The enrichment factor is a measure of the immunoprecipitated (IP) DNA-to-input ratio at the indicated gene compared to that of a telomeric region under derepressed conditions (DR) and 30 min after adding glucose (DR→R) to 4-h cultures of strain KBY131. The means and ranges of the results from two biological samples are shown.
TABLE 3.
MAP2 and IDP1 mRNA half-lives are reduced after glucose repletion when their native promoters are replaced by the ADH2 promotera
| Genotype | mRNA | Half-life (min)b |
|---|---|---|
| WT | ADH2 | 5.1 |
| ADH2p-MAP2 | ADH2 | 4.7 |
| ADH2p-IDP1 | ADH2 | 4.1 |
| WT | MAP2 | 60 |
| ADH2p-MAP2 | MAP2 | 9.0 |
| WT | IDP1 | 60 |
| ADH2p-IDP1 | IDP1 | 14 |
The strain names and a description of the glucose-dependent mRNA decay experiment are in the legend to Fig. 1.
Half-lives for ADH2, ADH2p-IDP1, and ADH2p-MAP2 mRNAs were estimated from the data shown in Fig. 1C and D. The data for the 5-, 10-, 20-, and 30-min time points were plotted on a semilog scale, and the half-lives were calculated from the slopes of the decay curves obtained using linear regression in excel. For MAP2 and IDP1, the data shown in Fig. 1E were used in a similar manner.
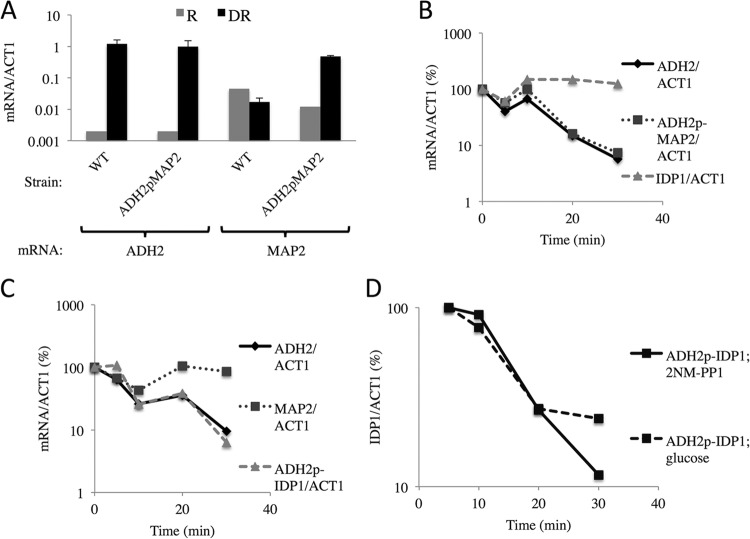
Inhibiting Snf1 imparts instability to mRNAs expressed from ADH2p-MAP2 and ADH2p-IDP1.
To determine the role of Snf1 in the stability of MAP2 and IDP1 transcripts expressed from the chimeric genes, we employed a strain expressing an analog-sensitive allele of Snf1 (SNF1as) (19). The gene fusions used in these experiment had a longer portion of the ADH2 regulatory region (residues −591 to −49) fused to the MAP2 and IDP1 gene than the fusion used in the experiment whose results are shown in Fig. 1 (residues −591 to −101). Cultures of strains containing the SNF1as allele and either the ADH2p-MAP2 (KBY137) or ADH2p-IDP1 (KBY140) strain were grown under repressing conditions and then shifted to derepressing medium and incubated for 4 h, at which time 2NM-PP1 was added. Samples were taken for RNA isolation and analysis at the time of inhibitor addition and at various times thereafter. As shown by the results in Fig. 2A, MAP2 transcripts in the strain containing the ADH2p-MAP2 gene increased more than 10-fold after 4 h under derepressing conditions. IDP1 and MAP2 transcripts expressed from the ADH2 promoter decreased in abundance after the inhibition of Snf1 at approximately the same rate as ADH2 transcripts (Fig. 2B and C). The abundance of IDP1 and MAP2 transcripts expressed from their own promoters was unaffected by the inhibition of Snf1. Thus, Snf1 is important for the stability of IDP1 and MAP2 transcripts when they are expressed from the ADH2 promoter. To directly compare the effects of Snf1 inhibition and glucose addition on the abundance of IDP1 transcripts synthesized under the control of the ADH2 promoter, either glucose or 2NM-PP1 was added to a culture of strain KBY140 after 4 h of derepression. Samples were taken for analysis of IDP1 mRNA levels at 5, 10, 20, and 30 min. IDP1 mRNA levels decreased at similar rates after the addition of either glucose or 2NM-PP1 (Fig. 2D), confirming the glucose sensitivity shown in Fig. 1 for a different ADH2p-IDP1 gene fusion. The apparent half-lives of ADH2, ADH2p-MAP2, and ADH2p-IDP1 after Snf1 was inhibited are listed in Table 4. These results show that glucose addition and Snf1 inhibition caused similar destabilization of MAP2 and IDP1 transcripts when they were expressed from the ADH2 promoter. In summary, the stability of MAP2 and IDP1 transcripts is SNF1 dependent when the genes are expressed under the control of the ADH2 promoter.
FIG 2.
The ADH2 promoter confers SNF1-dependent mRNA expression and stability to normally SNF1-independent transcripts. (A) ADH2 and MAP2 transcript levels under repressed (R) and derepressed (DR) growth conditions. The mean results and ranges for two biological replicates are shown. (B and C) Strains KBY137 and KBY140 with the relevant genotypes [map2::natMX-ADH2p(−591 to −49)-MAP2 and idp1::natMX-ADH2p(−591 to −49)-IDP1 ura3::YIpSNF1as::URA3 snf1Δ::kanMX, respectively] were grown to mid-log phase in YP plus 5% glucose and treated as described in the legend to Fig. 1, except that 2NM-PP1 rather than glucose was added to a final concentration of 10 μM after 4 h of derepression. (D) IDP1 transcript levels in strain KBY140 after the addition of glucose or 2NM-PP1 to a 4-h derepressed culture.
TABLE 4.
MAP2 and IDP1 mRNA half-lives are reduced after Snf1 inhibition when their native promoters are replaced by the ADH2 promotera
| Genotype | mRNA | Half-life (min) (SD or range)b |
|---|---|---|
| WT | ADH2 | 5.0 |
| ADH2p-MAP2 | ADH2 | 7.1 (6.0–8.2) |
| ADH2p-IDP1 | ADH2 | 7.1 (0.81) |
| ADH2p-MAP2 | MAP2 | 9.5 (5–14) |
| ADH2p-IDP1 | IDP1 | 5.5 (5–6) |
The strain names and a description of the Snf1-inhibited mRNA decay experiment are presented in the legend to Fig. 2.
IDP1 transcripts synthesized using the ADH2 promoter have the same 5′ ends as endogenous transcripts.
To determine whether the 5′ ends of IDP1 transcripts synthesized using the ADH2 promoter are the same as those present on endogenous transcripts, we performed primer extension analysis. As shown by the results in Fig. 3A, the 5′ extension products synthesized using RNA isolated from a WT strain were the same size as those synthesized using RNA isolated from a strain expressing IDP1 from the ADH2 promoter, and they were close to the expected size of 68 nucleotides. The endogenous transcripts were less abundant than the transcripts synthesized from the ADH2 reporter after 4 and 7 h of derepression, as expected from the data shown in Fig. 1A. We conclude that the different stabilities of the endogenous and ADH2p-driven IDP1 transcripts are unlikely to be due to using different start sites for transcription.
FIG 3.
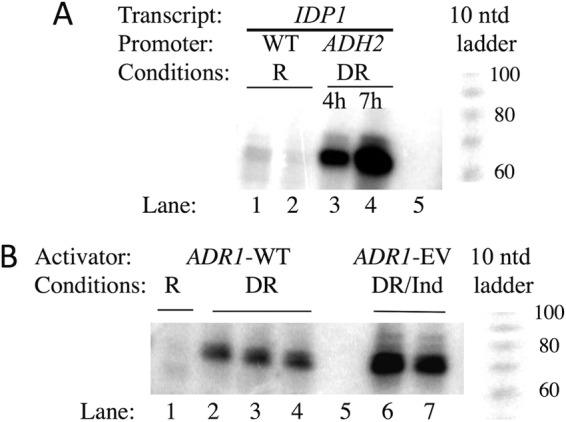
Primer extension analysis. (A) The 5′ ends of IDP1 mRNA were determined as described in Materials and Methods. The strains used were KBY137 (WT IDP1) and BY4741 (WT IDP1) grown under repressing (R) conditions (lanes 1 and 2, respectively) and strain KBY140 (ADH2p-IDP1) grown under derepressing (DR) conditions for 4 and 7 h (lanes 3 and 4, respectively). Lane 5, no RNA during primer extension. A 10-nucleotide (ntd) ladder is shown to the right. (B) 5′ ends of ADH2. Lane 1, RNA isolated from strain KBY140 grown under repressing conditions; lanes 2 to 4, three replicates using RNA from strain KBY140 grown for 4 h under derepressing conditions; lane 5, no RNA during the primer extension; lanes 6 and 7, two RNA preparations from strain TYY204 containing pRR01(ADR1-EV) grown under derepressing conditions for 3 h and then induced with 1 nM β-estradiol for 1 h (DR/Ind). A 10-nucleotide ladder is shown to the right.
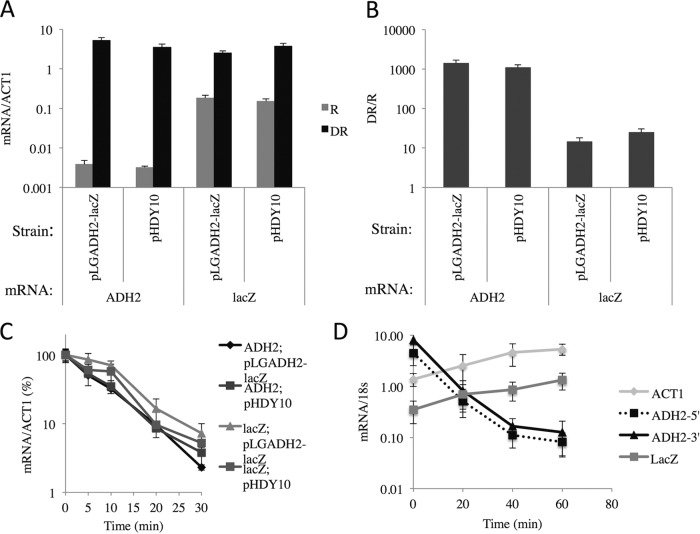
Adr1 binding sites are sufficient to confer glucose-induced mRNA decay.
To determine whether Adr1 binding sites are sufficient to confer glucose-induced mRNA decay, we used a reporter plasmid, pHDY10, in which the CYC1 UAS region of the lacZ reporter plasmid pLG669Z (54) has been replaced by multiple Adr1 binding sites. These regulatory region-gene fusions are under the control of ADR1, SNF1, and glucose (34). As a control to determine whether the lacZ ORF was susceptible to glucose-induced mRNA decay when the entire ADH2 regulatory region was fused upstream from CYC1 TATA-lacZ, plasmid pLGADH2 was also tested (35). The expression levels and glucose-induced mRNA decay of the reporter gene and endogenous ADH2 were measured as described above. The endogenous ADH2 gene was tightly repressed and activated about 1,000-fold under derepessing conditions (Fig. 4A and B). The lacZ reporter gene was repressed and derepressed about 20-fold by glucose depletion. Glucose-induced mRNA instability was observed for lacZ transcripts derived from pHDY10 and pLGADH2. The lacZ mRNA decayed with apparent half-lives of 6.6 and 7.3 min in strains with pHDY10 and pLGADH2, respectively (Fig. 4C). The apparent half-lives of ADH2 transcripts in the two strains were 6.2 and 5.6 min. To measure the stability of lacZ mRNA transcribed independently of SNF1, we used a lacZ reporter gene under the control of the CDC19 (also known as PYK1) promoter that was carried on a centromeric plasmid. Under conditions of glucose-induced mRNA decay with 1,10-o-phenenthroline added to inhibit transcription at the time of glucose addition, there were increases in the lacZ and ACT1 mRNA levels following the addition of glucose to a derepressed culture. Figure 4D shows the results of a representative experiment of the four performed using different transformants and two different batches of 1,10-o-phenanthroline. As expected, ADH2 mRNA levels decreased rapidly after glucose addition (Fig. 4D). The increases in lacZ and ACT1 mRNA levels were unexpected. Increased transcription of stress-related genes was observed after the inhibition of transcription with thiolutin, a different nonspecific inhibitor of transcription (55). We have no explanation for this observation, but it cautions against the exclusive use of nonspecific inhibitors of biosynthesis to study mRNA decay (32). Previous studies using a similar reporter gene measured an apparent half-life of 25 to 26 min for lacZ transcripts (56). Both our analysis and the results of previous studies indicate that lacZ transcripts made independently of SNF1 are more stable than lacZ transcripts whose synthesis is dependent on ADR1 and SNF1. Thus, we conclude that binding sites for Adr1 are sufficient to confer glucose-induced mRNA instability on heterologous transcripts.
FIG 4.
Binding sites for Adr1 are sufficient to promote glucose-induced mRNA decay. (A and B) ADH2 and lacZ transcript levels under repressed (R) and derepressed (DR) growth conditions (A), and the ratio of expression levels under derepressed and repressed growth conditions (B). (C and D) Strain W303-1a (WT) was transformed with pHDY10 (34) or pLGADH2 (35), and cultures were grown to mid-log phase in synthetic complete (SC) medium lacking uracil plus 5% glucose. (C) After 21 h of derepression, a typical glucose-induced mRNA decay experiment was performed. (D) lacZ, ACT1, and ADH2 transcript levels in strain CKY19 transformed with the lacZ expression plasmid YCpPKG2 (PYK1lacZ). After 4 h of derepression, glucose and 1,10-o-phenanthroline were added. The values for mRNA were normalized to the abundance of 18S RNA. The average results and standard deviations for four transformants of CKY19 transformed with YCpPKG2 are shown.
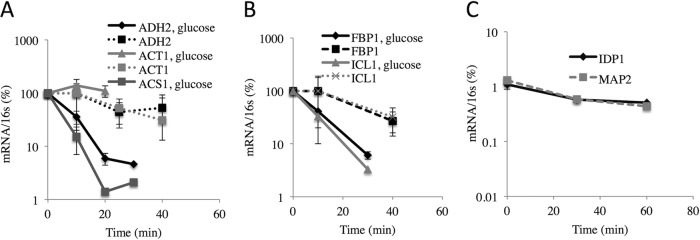
Pulse-chase experiments with 4-thiouracil confirm the stability of SNF1-dependent transcripts during derepression.
Because inhibiting transcription can affect the balance between mRNA synthesis and decay (57–59), we measured the rate of decay of SNF1-dependent transcripts synthesized during derepressing conditions without inhibition of transcription using a 4-thiouracil pulse-chase protocol (37, 38). RNA was labeled with 4-thiouracil for 60 min starting after 3 h of derepression. At the end of the labeling period, either uracil or uracil and glucose was added to duplicate cultures and samples were removed at various times for purification and quantitation of the abundance of specific 4-thiouracil-containing transcripts. E. coli RNA containing 4-thiouracil was added to the samples prior to modification and purification to serve as a control for the efficiency of modification and purification. The results in Fig. 5A show that 4-thiouracil-containing ADH2 mRNA decreased at a rate similar to that of ACT1 mRNA when the cells were maintained under derepressing conditions. The rates of decay suggest half-lives of 30 and 22 min for ADH2 and ACT1 transcripts, respectively, similar to values reported previously (19). In contrast, the apparent rates of decay of 4-thiouracil-containing ADH2 and ACS1 transcripts after glucose addition were 5 and 3 min, respectively (Fig. 5A), which are similar to the rates of decay measured using total RNA preparations (19). ACT1 transcripts did not decrease in abundance during the 20 min after glucose addition, suggesting a longer half-life under repressing conditions. The stability of 4-thiouracil-containing IDP1 and MAP2 transcripts synthesized from their endogenous promoters was analyzed in an analogous experiment. (Fig. 5B). Their apparent half-lives were about 60 min, similar to the half-lives measured under repressing conditions in another study using a 4-thiouracil pulse-chase protocol (38) and somewhat shorter than was measured using the transcription inhibitor 1,10-o-phenthroline (Fig. 1E). Thus, the data confirm the stability of these transcripts synthesized under derepressing conditions after the addition of glucose. In summary, the glucose-induced instability of ADH2 and ACS1 transcripts was confirmed using a pulse-chase protocol.
FIG 5.
Glucose-induced mRNA instability assessed by pulse-chase using 4-thiouracil as described in Materials and Methods. Strain TYY203 was grown in SM with limiting uracil and 5% glucose and then shifted to derepressing medium with 0.05% glucose and limiting uracil. After the addition of a 50-fold excess of normal uracil with or without glucose, samples were removed at the indicated times and 4-thiouracil-containing RNA was purified. Specific mRNA levels were measured by RT-qPCR and normalized to the amount of reference 16S RNA that was added prior to the biotinylation reactions. (A) ADH2, ACT1, and ACS1 mRNAs. (B) FBP1 and ICL1 mRNAs. (C) IDP1 and MAP2 transcript levels were measured in an analogous experiment conducted using the same strain. (A and B) The values represent the means and ranges of the results for two replicate cultures.
SNF1-dependent transcripts can be stable when synthesized in the presence of glucose.
To determine whether SNF1-dependent transcripts are inherently unstable in the presence of glucose, we used two ADR1 alleles that constitutively activate gene expression independent of glucose and SNF1: overexpressed ADR1 (oeADR1) (pKD17) (60, 61) and ADR1-GAL11 (pAG11), a gene encoding a fusion protein in which the DNA binding domain of Adr1 is fused to Gal11, a component of yeast Mediator (62). We used a strain (KBY108) containing a temperature-sensitive allele of RPB1, rpb1-1, the largest subunit of RNA Pol II to measure mRNA stability (37). Plasmids carrying oeADR1 and ADR1-GAL11 were introduced into KBY108, and cultures were grown under repressing conditions at 25°C and then shifted to the nonpermissive temperature for 60 min. WT ADR1 (pKD16) was also introduced into KBY108 to test the effect of the gene expression level on mRNA stability. ADH2 mRNA levels were quantified by RT-qPCR from samples taken at the time of the temperature shift and 60 min later. As expected, ADH2 mRNA levels were low when WT ADR1 was expressed from its own promoter and they were 70- and 90-fold higher when ADH2 was activated by oeAdr1 or the Adr1-Gal11 fusion protein, respectively (Table 5). After 60 min at 37°C, ADH2 transcripts were reduced in abundance by about threefold when activated by either WT Adr1 or oeAdr1, suggesting that they are relatively stable, independent of the level of activation, when synthesized under repressing conditions. In contrast, ADH2 transcript levels were reduced about 100-fold after 60 min at the restrictive temperature when the fusion protein, Adr1-Gal11, activated transcription. The apparent half-lives of the ADH2 transcripts, based on an endpoint assay, were 65, 56, and 10 min when activated by WT Adr1, oeAdr1, and Adr1-Gal11, respectively (Table 5). ADH2 was activated to similar high levels by oeAdr1 and Adr1-Gal11, but the apparent stabilities of the transcripts were very different, indicating that high mRNA abundance did not cause the greater reduction in ADH2 mRNA levels in the strain with ADR1-GAL11. In summary, ADH2 mRNA made in the presence of glucose is not inherently unstable and its mode of expression appears to influence its stability.
TABLE 5.
The half-life of ADH2 mRNA made in glucose-containing medium is affected by the activatora
| Plasmid | Activator | Half-life (min)b | Mean relative abundance (SD) of ADH2 mRNA atc: |
|
|---|---|---|---|---|
| 25°C | 36.5°C | |||
| pKD16 | Adr1 | 65 | 0.74 (0.36) | 0.33 (0.13) |
| pKD17 | oeAdr1 | 56 | 21 (7.5) | 10 (8.3) |
| pAG11 | Adr1-Gal11 | 10 | 28 (19) | 0.5 (0.14) |
Strain KBY108 (relevant genotype, rpb1-1 trp1) was transformed with the TRP1-containing plasmids pKD16, pKD17, and pAG11. Three transformants of each type were grown in synthetic complete (SC) medium lacking tryptophan and containing 5% glucose at 25°C to an A600 of 0.8. Samples were taken for RNA isolation and analysis, and the temperature of the cultures was raised to 36.5°C by adding an equal volume of 50°C medium. After 60 min of vigorous shaking, the cultures were collected for RNA isolation and analysis as described in Materials and Methods.
The apparent half-life of ADH2 mRNA was estimated as described in the legend to Table 3 from a graph of log(ADH2 mRNA/18S rRNA) versus time using the 0- and 60-min time points.
The abundance of ADH2 mRNA and 18s rRNA was determined by RT-qPCR as described in Materials and Methods. Each value represents the mean ADH2/18S RNA level (multiplied by 1,000) and standard deviation from three replicate cultures.
The transition from respiratory to fermentative conditions is not sufficient to trigger glucose-induced mRNA decay.
It was proposed that GAL1 transcripts were destabilized by the transition from a respiratory to a fermentative carbon source (14). To test this explanation for glucose-induced instability of Adr1-dependent transcripts and to assess a possible role for Snf1 in the process, we designed, constructed, and tested a chimeric activator, Adr1-EV. It contains the DNA binding and nuclear localization domains of Adr1 (63, 64), the ligand binding domain of the estrogen receptor (EBD) (30), and the activation domain of VP16. The DNA binding domain of Adr1 allows it to bind ADR1-specific genes, the EBD tethers the protein in the cytoplasm until the inducer, β-estradiol, is added, and the VP16 portion encodes a strong constitutive activation domain. The chimeric activator was expressed under the control of the constitutive ADH1 promoter. We hypothesized that the chimeric activator would be glucose and SNF1 independent because it lacks the major regulatory region of Adr1, the 14-3-3 (also known as Bmh in yeast) binding domain (40, 65), and is highly expressed from the ADH1 promoter. To test the glucose and SNF1 dependence of gene activation by Adr1-EV, we measured the expression of three ADR1-dependent genes (ADH2, ACS1, and POX1) in a strain lacking ADR1 and SNF1 (CKY27) and grown under either repressing or derepressing conditions. As predicted, their expression was dependent on the inducer but independent of glucose and SNF1 (Fig. 6A). The ADH2 mRNA levels increased more than 100-fold in the presence of the inducer under both repressing and derepressing growth conditions. The other ADR1-dependent genes behaved similarly, despite a 100-fold range in expression levels. As a control for the specificity of activation, we measured the levels of transcripts whose synthesis is SNF1 dependent but ADR1 independent. Two SNF1- and CAT8-dependent but ADR1-independent genes, FBP1 and ICL1, were expressed at very low levels in the snf1Δ adr1Δ strain, regardless of the presence of the inducer (Fig. 6A), showing that activation by Adr1-EV is promoter specific. To confirm that the inducer acted through the chimeric activator, we performed a similar experiment in the same strain carrying an empty vector or transformed with plasmids expressing WT ADR1 or ADR1-EV. There was very low expression of ADR1-dependent genes (ADH2, ACS1, and POX1) in the presence of WT ADR1 (because SNF1 was absent) or a vector lacking ADR1 and a high level of expression when ADR1-EV was present (Fig. 6B). These results confirm the Adr1-EV dependence and SNF1 independence of gene expression. The CAT8-dependent genes FBP1 and ICL1 were expressed at very low levels, which was expected because SNF1 was absent (Fig. 6B). Thus, Adr1-EV specifically and efficiently activates ADR1-dependent genes in the absence of SNF1 under repressing and derepessing growth conditions.
FIG 6.
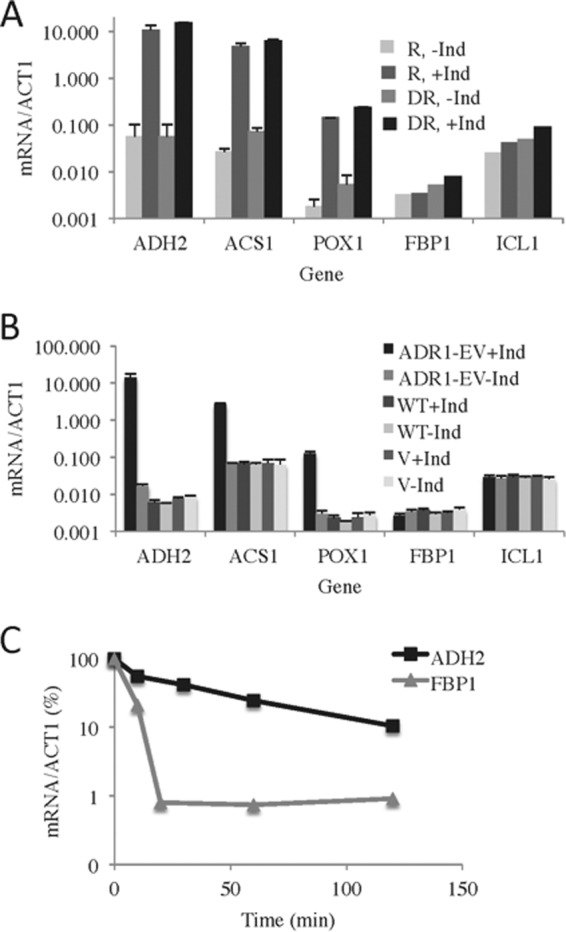
Transcript stability is influenced by the activator, not the transition from respiration to fermentation. (A) Strain CKY27 (adr1Δ snf1Δ) carrying the chimeric gene ADR1-EV on the centromeric plasmid pRR01 was grown in synthetic medium lacking histidine but with 5% glucose to an A600 of ∼1. Different portions of the culture were maintained under repressing conditions or subjected to derepression, in each case with or without induction with 1 nM β-estradiol. The data represent the average results and standard deviations from three replicate experiments. (B) β-Estradiol induction of gene expression requires the chimeric Adr1-EV activator. Cultures of strain CKY27 (adr1Δ snf1Δ) carrying the chimeric gene ADR1-EV on pRR01 (ADR1-EV), a WT ADR1 gene carried on pKD16H (WT), or pRS313, a HIS3 vector with no ADR1 (vector [V]), were grown to mid-log phase in SC medium without histidine but with 5% glucose. Thereafter, the cells were treated as described for panel A. The values shown are the average results and standard deviations from three replicate experiments. (C) ADH2 mRNA made in derepressing conditions under the control of the chimeric Adr1-EV activator is not subject to glucose-induced mRNA decay. Cultures of strain TYY204 (adr1Δ) carrying the chimeric gene ADR1-EV were grown, derepressed, and induced as described for panel A. After 60 min of induction, glucose and 1,10-o-phenanthroline were added to final concentrations of 5% and 100 μg/ml, respectively. Aliquots were removed at the times indicated for RNA isolation and analysis. The results shown are those of a representative example from three replicate experiments.
Primer extension analysis of ADH2 transcripts activated by WT Adr1 (SNF1 dependent) and Adr1-EV (SNF1 independent) was also performed. ADH2 transcripts synthesized by these activators had indistinguishable 5′ ends of the expected size, 77 nucleotides (66). Figure 3B shows a representative gel. Thus, the different stabilities of the WT Adr1- and Adr1-EV-activated transcripts are not caused by using different start sites, at least within the resolution of the analysis.
ADR1-EV allowed us to determine whether the transition from derepressing to repressing growth conditions would trigger glucose-induced decay of ADH2 transcripts synthesized independently of Snf1. Their stability was measured in the SNF1 strain TYY204 carrying the ADR1-EV expression plasmid pRR01. A typical glucose-induced mRNA decay experiment was performed in which the transcription inhibitor 1,10-o-phenanthroline was added at the same time that glucose was restored to a 4-h derepressed culture that had been induced for 60 min with β-estradiol. Thus, transcription would be inhibited at the time of glucose addition, allowing us to measure the stability of transcripts synthesized uniquely under derepressing conditions. The results of a representative experiment are shown in Fig. 6C. In three replicate experiments, the mean decay rate of ADH2 mRNA was 42 min (standard deviation, 26 min), similar to the decay rate that was measured under derepressing conditions when WT Adr1 was the activator (Fig. 5A). As a control to show that glucose-induced decay occurred for SNF1-dependent but ADR1-independent transcripts, we measured the amount of FBP1 mRNA. FBP1 transcripts decayed with an apparent half-life of 2 to 3 min (Fig. 6C), indicating that SNF1-dependent transcripts were still subject to glucose-induced mRNA decay. Thus, ADH2 transcripts synthesized under derepressing conditions under the control of the chimeric activator were not subject to glucose-induced decay. It is likely that ADH2 transcripts synthesized under repressing conditions under the control of Adr1-EV are also relatively stable, because their steady-state level was similar to that under derepressing conditions. We conclude that ADH2 transcripts that are synthesized under derepressing conditions and then exposed to repressing conditions are not inherently unstable. Therefore, their mode of activation and/or synthesis appears to influence their rate of decay.
Identification of RBP binding sites in SNF1-dependent mRNA sequences.
To identify binding site motifs for RBPs that potentially act downstream from Snf1 to influence transcript stability, we compared the sequences of 32 SNF1-dependent transcripts showing rapid glucose-induced decay to those of 32 random transcripts, using MEME as described in Materials and Methods. Of the enriched motifs with E values greater than 0.05, 3 motifs were identified using Tomtom (47) as being similar to the RNA recognition sequence for Vts1. These were the only motifs for a yeast RBP found. In a more targeted approach, we used the known binding motifs for 20 yeast RBPs (see Table S2 in the supplemental material) to search for matches in 34 SNF1-dependent transcript sequences (the 32 described above plus two others, ICL1 and ICL2). The SNF1-dependent transcripts were enriched over the total yeast transcriptome for motifs matching the binding site motifs of Vts1 and Nsr1 (at least one match in the ORFs for 33 and 12 genes, respectively) (Table 6) and Khd1, Nrd1, and Pab1 (at least one match in the 5′-UTR for 26, 15, and 20 genes, respectively) (Table 6). The SNF1-dependent subset of mRNAs was not enriched over the genome for 3′-UTRs having at least one instance of any of the RBP motif sets.
TABLE 6.
RNA-binding proteins having motifs enriched in SNF1-dependent mRNAs over their occurrence in the yeast genome
| RBPa | Location | P valueb | No. of transcripts in: |
|||
|---|---|---|---|---|---|---|
| Snf1-dependent genes |
Whole genome |
|||||
| Matchesc | Totald | Matches | Total | |||
| Khd1 | 5′-UTR | 3.96E−02 | 26 | 34 | 3,566 | 6,550 |
| Nrd1 | 5′-UTR | 3.91E−02 | 15 | 34 | 1,484 | 6,550 |
| Pab1 | 5′-UTR | 3.63E−02 | 20 | 34 | 2,189 | 6,550 |
| Nsr1 | ORF | 4.10E−03 | 12 | 34 | 736 | 6,550 |
| Vts1 | ORF | 7.41E−03 | 33 | 34 | 4,927 | 6,550 |
Each RNA-binding protein is represented by one or more distinct motifs.
The probability that the number of Snf1-dependent genes containing one or more RBP binding sites is not enriched over the whole yeast genome. Values have been corrected for multiple hypothesis testing using the method of Benjamini and Hochberg (94).
The number of genes coding for mRNAs with one or more instances of an RBP motif.
The total number of mRNA sequences searched.
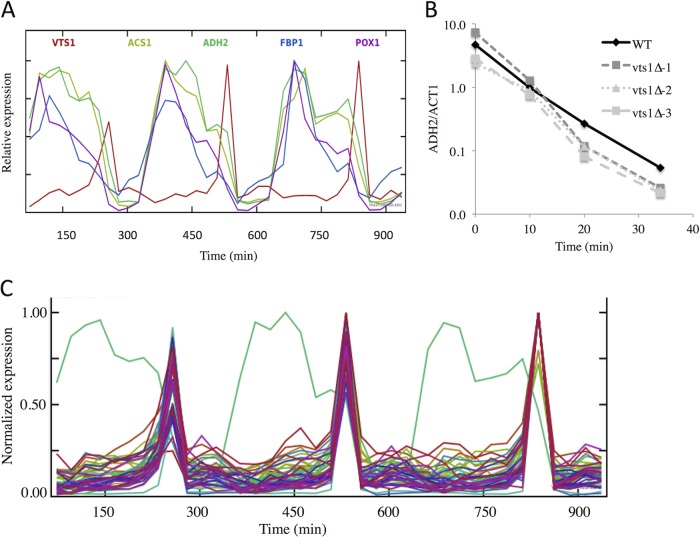
Vts1 is an RBP involved in mRNA decay and repression (67, 68). Because the Vts1 motif was the enriched match most frequently found in the SNF1-dependent transcript sequences, we examined its expression in published data sets to determine whether its expression might be correlated with their decay. VTS1 expression is enhanced 2.5-fold in the presence of glucose when SNF1-dependent transcripts are absent or reduced in abundance (41). A more pronounced expression bias was observed during the yeast metabolic cycle (YMC) (69). VTS1 expression was observed during a brief period of time at the end of the respiratory phase, when the abundance of SNF1-dependent mRNAs (such as ADH2 and others) begins a rapid decline (Fig. 7A and data not shown). This result suggested that Vts1 might be involved in the degradation or the repression of SNF1-dependent transcripts during the fermentative/reductive building phase of the metabolic cycle.
FIG 7.
(A) VTS1 expression is regulated during the metabolic cycle. The abundance of ADH2 and VTS1 transcripts during three turns of the metabolic cycle was retrieved from the data in Tu et al. (69) as described in Materials and Methods. (B) Glucose-induced mRNA decay in WT and vts1 deletion mutants. A typical glucose-induced mRNA decay experiment was performed and analyzed as described in Materials and Methods. The WT strain is BY4741, and it and the three deletion mutants are from the Research Genetics collection. The vts1Δ-1 and vts1Δ-3 strains are MATa, and the vts1Δ-2 strain is MATα. The vts1Δ-1 and vts1Δ-3 strains are derived from two different strain collections. (C) Expression of genes whose timing is correlated with that of VTS1 during the yeast metabolic cycle. Transcript abundance was obtained as described in Materials and Methods.
To assess a possible role of Vts1 during glucose-induced mRNA decay, we compared the rate of decay of ADH2 mRNA in a WT strain with the decay rates in three vts1Δ mutants (two MATa and one MATα). As shown by the results in Fig. 7B, the rates of decay were indistinguishable. Thus, if Vts1 is involved in glucose-induced mRNA decay, it must function redundantly with other proteins. To identify other RBPs that could function redundantly with Vts1, we analyzed the data of Tu et al. (69) for mRNAs whose abundance correlated with that of VTS1. We found 44 mRNAs whose expression correlated with that of VTS1 with a correlation coefficient greater than 0.95. These are plotted together with the expression data for ADH2 in Fig. 7C using SCEPTRANS (http://moment.utmb.edu/cgi-bin/sceptrans.cgi?locus = YOR359W) as described by Kudlicki et al. (70). Yeast GO Slim Mapper (71) identified 7 of the 44 mRNAs as coding for proteins that have an RNA binding function. These are Bud21, Naf1, Ppr43, Pus1, Ssf1, Trm11, and Vts1. Of these, only Vts1 is known to have a role in mRNA decay. These RBPs might act redundantly to promote decay of SNF1-dependent transcripts during the YMC and during glucose-induced mRNA decay, or they might have other functions associated with the end of the respiratory phase. RBPs whose activity rather than expression is regulated could also participate in these processes and would not be identified by this analysis.
Glucose addition to a derepressed culture causes rapid dissociation of ADH2 mRNA from polysomes.
Glucose deprivation in yeast causes a rapid loss of translation initiation and a shift of mRNAs from polysomes to P bodies and stress granules (72). It is not known whether glucose repletion leads to a similar shift of mRNAs synthesized under derepressing conditions that might be causally related to glucose-induced mRNA decay. To test this possibility, we analyzed whole-cell extracts isolated from SNF1as yeast cells (TYY1078) cultured under the following three conditions: derepressed for 4 h or derepressed for 4 h and then treated with either glucose or 2NM-PP for 5 min. A portion of the whole-cell extract was sedimented through a sucrose density gradient to analyze the distribution of polysomes and ADH2 and ADH1 mRNAs. The absorbance was monitored continuously during the collection of nine fractions from each gradient (Fig. 8A). The top and bottom three fractions, representing soluble proteins/ribosomal subunits and large polysomes, respectively, were pooled. The middle three fractions represent 80S monosomes and small- and medium-sized polysomes, respectively. As expected, the extracts from derepressed cells contained a small number of polysomes due to inhibition of translation initiation (72–74). ADH2 mRNA was found primarily in the region of the gradient containing small polysomes in extracts of derepressed cells (Fig. 8B). Five minutes after the addition of glucose, it sedimented more slowly, suggesting that the addition of glucose caused a rapid dissociation of ADH2 mRNA from polysomes. Inhibiting Snf1 did not change the distribution of ADH2 mRNA. ADH1 mRNA, a glucose-induced transcript (75), showed the inverse behavior to ADH2 mRNA, being shifted from a slower-migrating peak of the gradient in extracts of derepressed cells to the polysomal region 5 min after adding glucose (Fig. 8C). These results suggest that glucose-induced mRNA decay is associated with and is perhaps preceded by rapid dissociation of SNF1-dependent transcripts from polysomes. Inhibiting Snf1 was not sufficient to induce this relocation with the same rapid kinetics.
FIG 8.
ADH2 mRNA rapidly dissociates from polysomes after glucose repletion. Cell extracts were made and fractionated by sucrose density gradient centrifugation as described in Materials and Methods and in more detail in reference 39. E. coli RNA was added to fractions 1 to 9 to serve as a reference for the efficiency of RNA purification and cDNA synthesis. (A) A260 values of the material eluting from a 10-to-30% sucrose gradient on which an extract from derepressed cells was sedimented. (B and C) ADH2 (B) and ADH1 (C) transcript abundance normalized to the amount of reference 16S RNA.
Analysis of sequences in SNF1-dependent mRNAs occupied by RNA binding proteins in vivo.
To determine whether RNA binding proteins (RBPs) acting downstream from SNF1 might be involved in the relocation of ADH2 mRNA after glucose repletion, we analyzed the RBP binding sites that were identified in a recent study of the nontranslating portion of the S. cerevisiae transcriptome (41). This analysis used 4-thiouracil-promoted RNA-protein cross-linking and RNA sequencing to identify RBP sites on a global scale, a procedure termed gPAR-CLIP (global photoactivatable ribonucleotide-enhanced coupled immunoprecipitation of RBPs). These experiments characterized sequences derived from cells growing exponentially under repressing conditions and after 2 h of glucose depletion. We determined whether the number of protein binding sites identified for each region (5′-UTR, ORF, 3′-UTR, and intron) of the 34 SNF1-dependent mRNAs increased or decreased threefold or more under repressing conditions compared to the number identified under glucose-depleted conditions. This analysis indicated an enrichment over randomness of bound sites in the 5′- and 3′-UTRs of SNF1-dependent transcripts after the addition of glucose (Table 7). The number of sites with decreased occupancy after glucose depletion did not change, and the bound sites in the introns and ORFs did not show such large differences. The increased occupancy of RBP binding sites in the 5′- and 3′-UTRs of SNF1-dependent transcripts observed in the gPAR-CLIP RNA binding site data is consistent with loading of the RNA decay machinery onto these mRNAs released from ribosomes when glucose is abundant.
TABLE 7.
Occupancy of protein binding sites in Snf1-dependent mRNAs after the addition of glucose to starved cells
| Region | Occupancya | P valueb | No. of bound sites in: |
||
|---|---|---|---|---|---|
| Snf1-dependent genes | Random genesd |
||||
| Countc | Avg count | SD | |||
| 5′-UTR | Increased | 3.45E−16 | 18 | 6.2 | 2.9 |
| Decreased | 6.22E−01 | 1 | 1.1 | 1.2 | |
| 3′-UTR | Increased | 3.14E−09 | 22 | 11 | 5.9 |
| Decreased | 1.22E−01 | 2 | 2.6 | 1.9 | |
| Coding sequence | Increased | 3.81E−02 | 73 | 67 | 14 |
| Decreased | 9.42E−05 | 19 | 14 | 5.6 | |
| Intron | Increased | 1.95E−04 | 0 | 1.5 | 1.6 |
| Decreased | 8.30E−02 | 0 | 0.25 | 0.68 | |
Protein binding sites exhibiting a threefold increase or decrease in binding site reads per million normalized to gene reads per kilobase per million, as defined in Freeberg et al. (41).
The probability that the number of Snf1-dependent genes containing one or more RBP binding sites occurred by random chance. Significant enrichment is seen at P values of less than 1 × 10−6.
The number of Snf1-dependent genes coding for mRNAs with sequences identified by gPAR-CLIP as occupied.
Statistics for RBP binding sites in 24 sets of 34 random Snf1-independent mRNAs.
The binding site sequences with increased occupancy in the 34 SNF1-dependent genes were searched for known RNA binding motifs in the 5′- and 3′-UTRs of the transcript using the known binding motifs for 20 RBPs described above. Although the UTRs of many SNF1-dependent transcripts did have one or more binding sites for known RBPs, the sequences that were bound did not identify a single prevalent RBP known to be involved in mRNA turnover (data not shown).
The cAMP-dependent protein kinase pathway is not required for glucose-induced mRNA decay.
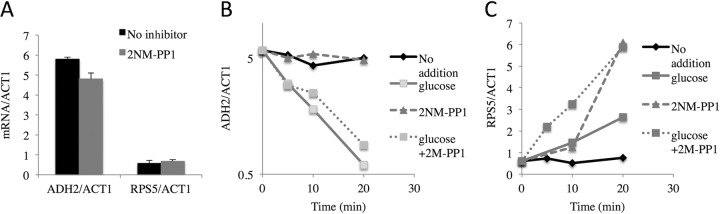
The observation that glucose but not Snf1 inhibition caused a rapid dissociation of ADH2 from polysomes suggested that a glucose-dependent signaling pathway might be required for this process. cAMP-dependent protein kinase (protein kinase A [PKA]) is a major regulator of pathways active during fermentation (5, 76), and low PKA activity inhibits translation during glucose starvation (73). Whether PKA plays a role in glucose-induced mRNA decay is unknown. We tested whether inhibiting PKA would protect ADR1-dependent transcripts from glucose-induced decay. Cultures of a strain carrying analog-sensitive alleles of all three catalytic subunits of PKA, TPK1as, TPK2as, and TPK3as (76), were derepressed for 4 h either in the presence or in the absence of the inhibitor 2NM-PP1. Then, samples of both cultures were removed for RNA isolation and quantitation. ADH2 mRNA was present at the same level whether derepression occurred in the presence or in the absence of 2NM-PP1, indicating that PKA activity is not required for ADH2 derepression (Fig. 9A). The expression of RPS5, encoding a subunit of the small ribosomal subunit, was induced about twofold by 2NM-PP1 (Fig. 9A). The former result is consistent with those of previous studies in which a strain carrying a single weakly active TPK allele was shown to derepress normally (77). To assess a possible role for PKA in glucose-induced mRNA decay, 2NM-PP1, glucose, or 2NM-PP1 plus glucose was added to portions of a culture that had been derepressed for 4 h. A fourth portion of the culture received DMSO, the solvent for 2NM-PP1, as a no-inhibitor control. Samples were taken for RNA isolation and quantitation of ACT1, ADH2, and RPS5 transcripts at 5, 10, 20, and 40 min. Glucose-induced mRNA decay was observed for ADH2, but inhibiting PKA had no effect, and the decrease in ADH2 mRNA abundance after glucose addition was unaffected by simultaneously inhibiting PKA (Fig. 9B). In contrast, glucose, 2NM-PP1, and glucose plus 2NM-PP1 stimulated the expression of RSP5 (Fig. 9C). The increased abundance of RSP5 transcripts due to the presence of 2NM-PP1 under derepressing conditions was unexpected. However, the observation that the inhibitor of PKA had an effect on RSP5 expression demonstrates the effectiveness of 2NM-PP1 in this strain. We conclude that PKA signaling does not play an important role in glucose-induced mRNA decay.
FIG 9.
Activity of the cyclic AMP-dependent protein kinase is not required for glucose-induced mRNA decay. (A) ADH2 and RSP5 mRNA levels after 4 h of derepression in strain JBY3622 (relevant genotype, TPK1as TPK2as TPK3as). 2NM-PP1 or DMSO (No inhibitor) was added at the time derepression was initiated by pelleting the cells and resuspending them in low-glucose medium. (B and C) A typical glucose-induced mRNA decay experiment was performed in strain JBY3622 (described above). 2NM-PP1, glucose, 2NM-PP1 and glucose, or DMSO (No addition) was added after 4 h of derepression, and ADH2, ACT1, and RSP5 transcripts were analyzed by RT-qPCR.
DISCUSSION
Glucose-induced mRNA decay appears to be an example of transcription-coupled mRNA decay (58, 59) that is regulated by Snf1 and acts through SNF1-dependent transcription factors. Transcripts (including the 5′- and 3′-UTRs and ORFs) not normally dependent on SNF1 for their synthesis or stability can be rendered SNF1 dependent and subject to glucose-induced mRNA decay by fusing their transcription units, including the 5′ and 3′-UTRs and ORFs, to the ADH2 regulatory region. Adr1 binding sites were sufficient to confer glucose-induced mRNA decay and SNF1-dependent stability on a heterologous transcript. These results suggest that Snf1-dependent transcription, acting through promoter sequences, and specifically, transcription factor binding sites, is sufficient to confer regulated mRNA decay. Because Adr1 binding sites are sufficient, recruitment by Adr1 of factors that influence decay is implicated.
RBPs play important roles in posttranscriptional regulation of gene expression (24, 57, 78) and are likely to influence glucose-induced mRNA. Because the ADH2 promoter was sufficient to impart SNF1-dependent stability to IDP1 and MAP2 mRNAs containing their own 5′- and 3′-UTRs, these regions of the ADH2 transcript are not necessary for glucose-induced mRNA decay, but they could act downstream from SNF1 to influence decay. The binding site for Vts1 was enriched in the ORFs of 33 of 34 SNF1-dependent transcripts and, thus, appears to be a good candidate for affecting these transcripts posttranscriptionally. Vts1 is a member of the Smaug (Smg) family of posttranscriptional RBPs which bind RNA through a conserved sterile alpha motif (SAM) that interacts with Smg recognition element (SRE)-containing transcripts (79, 80). Vts1 recruits the CCR4-NOT deadenylase complex to SRE-containing mRNAs to initiate their degradation (67). In addition, Vts1 interacts with an eIF4E-interacting protein, Eap1, which represses mRNA translation in S. cerevisiae (68). Thus, Vts1 stimulates both mRNA decay and mRNA repression. The expression of VTS1 is temporally correlated with the decline in abundance of SNF1-dependent transcripts during the reductive/charging phase of the ultradian yeast metabolic cycle (the results in Fig. 9A illustrate this point for ADH2, a typical SNF1-dependent transcript; see the POX cluster of genes in Fig. 3B in reference 69, for other examples). If the activity of Vts1 is regulated in a similar manner, it could be important for the rapid loss of the POX cluster transcripts and inhibition of metabolic activity at this point in the metabolic cycle. Vts1 was not identified as a Snf1 target in phosphoproteomic studies (27). However, two Vts1-interacting proteins, Eap1 and Ccr4, are Snf1 targets for phosphorylation and dephosphorylation, respectively (27). CCR4 deletion protects SNF1-dependent transcripts from glucose-induced mRNA decay (27), but whether dephosphorylation is important for this activity is unknown. In addition, CCR4 is important for transcription-coupled mRNA decay (59). However, deletion of VTS1 did affect glucose-induced ADH2 mRNA decay (Fig. 9B). One possible explanation for this lack of an effect of Vts1 on ADH2 mRNA decay is that it acts redundantly with other RBPs in the process. For example, binding sites for other RBPs, such as Nsr1, Khd1, and Pab1, were enriched in SNF1-dependent transcripts. One or more of these proteins, or RBPs whose motifs were abundant but not enriched or not identified in SNF1-dependent transcripts because their sequence specificity has yet to be defined, might act redundantly with Vts1 in glucose-induced mRNA decay. Another possibility is that Vts1 acts to repress ADH2 mRNA translation rather than to stimulate its decay and, thus, might be involved in the loss of ADH2 mRNA from polysomes after glucose repletion (Fig. 8).
Because we focused on RBP binding sequences that were enriched in the SNF1-dependent mRNAs, RBPs whose activity rather than binding was regulated by SNF1 would not be selected. Puf3 is one such RBP in S. cerevisiae. Puf3 is important for the decay of numerous nuclear transcripts encoding mitochondrial proteins involved in respiration (15). How its activity, but not binding, is regulated is unknown. Puf3-Ser203 is phosphorylated in a SNF1-dependent fashion, and the kinetics of phosphorylation after glucose depletion (see supplementary file 1 in reference 27) suggest that this modification could be important for regulating its activity. It will be interesting to mutate this site and determine whether it has an effect on the activity of Puf3.
It is becoming increasingly evident that how a transcript is synthesized can influence posttranscriptional events in mRNA metabolism (57, 58). That promoter sequences can control the fate of mRNA has been documented for several yeast genes controlled by Rap1, an important transcription factor for genes encoding ribosomal proteins and glycolytic enzymes (81), and also on a genome-wide scale (82). A recent study implicated the promoter in determining the translation efficiency of an mRNA, as well as influencing its cytosolic location (16). Evidence suggests that the dissociable Pol II subunits Rpb4/Rpb7 (83, 84) and the cell cycle kinase Dbf2 (85) remain associated with transcripts after they exit the nucleus and, thus, can influence their fate in the cytoplasm. This led Goler-Baron et al. to propose that mRNA degradation in the cytoplasm and its synthesis in the nucleus were coupled by RNA binding proteins that associated with mRNAs cotranscriptionally and accompanied them to the cytoplasm, where they could influence their fate (86), essentially forming a closed regulatory loop to balance mRNA synthesis and decay (58, 59). However, a direct role for Rpb4/Rbp7 in the process of cytoplasmic mRNA decay has been questioned, because tethering Rpb4 in the nucleus by fusion to a large subunit of RNA polymerase II still rescues the mRNA stability defect found in an rpb4 deletion mutant (87).
Our studies implicate SNF1-dependent transcription as an important contributor to glucose-induced mRNA decay. ADR1-dependent transcripts that are subject to glucose-induced mRNA decay are resistant to this process if they are synthesized in a SNF1-independent fashion. The SNF1-independent, ADR1-dependent transcripts appeared to have the same 5′ ends as the transcripts synthesized in a SNF1-dependent fashion, apparently ruling out a role for different initiation sites in the process. However, other modifications of the 5′ ends of the transcript would not have been detected, nor would changes in their termination or polyadenylation sites.
ADH2 mRNA rapidly dissociated from polysomes when glucose was added to a culture of derepressed cells. Snf1 inhibition was not associated with the same transition. One possibility is that a glucose-induced signaling pathway, such as the PKA or TOR cascade, is required to respond to glucose repletion. A reduction in PKA signaling is required to respond to glucose starvation by repressing translation (73), but whether increased PKA signaling is required to promote transcript instability in response to glucose repletion is not known. Our results suggest that the PKA pathway is not required for glucose-induced decay of SNF1-dependent transcripts, because ADH2 transcripts underwent rapid decay after inhibiting PKA in the presence of glucose.
We hypothesize that transcripts synthesized in a SNF1-dependent fashion are marked during transcription in a way that allows them to be distinguished from SNF1-independent transcripts. The mark could cause them to be targeted for degradation in the absence of active Snf1, such as under repressing growth conditions or after inhibition of Snf1. The mark might also cause them to be dissociated from polysomes. According to this hypothesis, if the same transcript were synthesized in an SNF1-independent fashion, it would not acquire that mark, would not show glucose-induced mRNA decay, and would not be translationally repressed. The mark could be an RBP whose activity is SNF1 dependent. Fourteen RNA binding proteins were identified as Snf1 targets in our recent phosphoproteomics analysis, and four of them, Ccr4, Dhh1, Scp160, and Xrn1, reduced the rate of glucose-induced mRNA decay (27). Xrn1, a major cytoplasmic mRNA-degradative enzyme, is essential for glucose-induced mRNA decay (21). It has an important global role in mRNA synthesis and decay (58, 59), but its role in coupling these processes is controversial (discussed in reference 57). Mutation of the Snf1-dependent phosphorylation sites in Xrn1 made the transcripts partially resistant to glucose-induced decay (27), an observation that is consistent with Snf1 acting through Xrn1 to influence mRNA decay. The other three Snf1 target RBPs that influence glucose-induced mRNA decay have annotations suggesting a role in mRNA decay, mRNA repression, or both. Our results with transcripts expressed from chimeric ADH2 promoter genes suggest that if an RBP protein is important for glucose-induced mRNA decay, its function is downstream from Snf1 in the process. Alternatively, the mark could be a SNF1-dependent covalent modification of the mRNA that was carried out by an enzyme whose activity was SNF1 dependent. Several enzymes of mRNA cap metabolism were targets of Snf1 (27), and one of these might influence glucose-induced mRNA decay and or mRNA repression.
Either of these mechanisms is compatible with an influence of SNF1-dependent transcription on mRNA stability. Snf1 is recruited to the GAL1 promoter by Gal4 (88) and to the INO1 promoter (89). A Snf1 complex containing the activating subunit Gal83 is found at the SNF1- and ADR1-dependent ADY2 promoter (90). The mammalian ortholog of Snf1, AMPK, is found at the promoters and in the ORFs of several mammalian genes (91). Thus, Snf1 is in a location where it might phosphorylate enzymes involved in modifying mRNA or RBPs that were recruited cotranscriptionally by SNF1-dependent transcription factors, such as Adr1. Xrn1 might be one such protein, because it is found in the nucleus (92; but see reference 59 for a conflicting report) and its activity appears to be regulated by Snf1-dependent phosphorylation (27). Although the known nuclear role of AMPK and Snf1 is to modify chromatin through activation of histone acetyltransferases (89, 90, 93), they could also influence posttranscriptional gene regulation by one of the mechanisms described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Broach for pSH47 and JBY3622, S. Hahn for the use of laboratory facilities, R. Rossi for constructing pRR01, P. Parua for help constructing TYY1077, V. MacKay for help with the polysome analysis, S. Gruenberg and Rafał Donczew for help with the primer extension analysis, and C. Zhang for the gift of 2NM-PP1.
This research was supported by grant GM26079 from the NIH to E.T.Y.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00436-15.
REFERENCES
- 1.Carlson M. 1999. Glucose repression in yeast. Curr Opin Microbiol 2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 2.Schuller H-J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 3.Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gancedo JM. 2008. The early steps of glucose signalling in yeast. FEMS Microbiol Rev 32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- 5.Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu Rev Genet 42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 6.Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front Biosci 13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn S, Young ET. 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Young ET, Dombek KM, Tachibana C, Ideker T. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem 278:26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 10.Broach JR. 2012. Nutritional control of growth and development in yeast. Genetics 192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardo A, Cereghino GP, Scheffler IE. 1992. Control of mRNA turnover as a mechanism of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol 12:2941–2948. doi: 10.1128/MCB.12.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercado JJ, Smith R, Sagliocco FA, Brown AJ, Gancedo JM. 1994. The levels of yeast gluconeogenic mRNAs respond to environmental factors. Eur J Biochem 224:473–481. doi: 10.1111/j.1432-1033.1994.00473.x. [DOI] [PubMed] [Google Scholar]
- 13.Yin Z, Hatton L, Brown AJ. 2000. Differential post-transcriptional regulation of yeast mRNAs in response to high and low glucose concentrations. Mol Microbiol 35:553–565. [DOI] [PubMed] [Google Scholar]
- 14.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. 2011. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell 22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MA, Russo J, Fischer AD, Lopez Leban FA, Olivas WM. 2014. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res 42:3954–3970. doi: 10.1093/nar/gkt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zid BM, O'Shea EK. 2014. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514:117–121. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cereghino GP, Atencio DP, Saghbini M, Beiner J, Scheffler IE. 1995. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell 6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto S, de la Cruz BJ, Scheffler IE. 2000. Glucose-regulated turnover of mRNA and the influence of poly(A) tail length on half-life. J Biol Chem 275:14155–14166. doi: 10.1074/jbc.275.19.14155. [DOI] [PubMed] [Google Scholar]
- 19.Young ET, Zhang C, Shokat KM, Parua PK, Braun KA. 2012. The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae. J Biol Chem 287:29021–29034. doi: 10.1074/jbc.M112.380147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federoff HJ, Eccleshall TR, Marmur J. 1983. Carbon catabolite repression of maltase synthesis in Saccharomyces carlsbergensis. J Bacteriol 156:301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cereghino GP, Scheffler IE. 1996. Genetic analysis of glucose regulation in saccharomyces cerevisiae: control of transcription versus mRNA turnover. EMBO J 15:363–374. [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade RP, Kotter P, Entian KD, Casal M. 2005. Multiple transcripts regulate glucose-triggered mRNA decay of the lactate transporter JEN1 from Saccharomyces cerevisiae. Biochem Biophys Res Commun 332:254–262. doi: 10.1016/j.bbrc.2005.04.119. [DOI] [PubMed] [Google Scholar]
- 23.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 25.Mansfield KD, Keene JD. 2009. The ribonome: a dominant force in co-ordinating gene expression. Biol Cell 101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Cruz BJ, Prieto S, Scheffler IE. 2002. The role of the 5′ untranslated region (UTR) in glucose-dependent mRNA decay. Yeast 19:887–902. doi: 10.1002/yea.884. [DOI] [PubMed] [Google Scholar]
- 27.Braun KA, Vaga S, Dombek KM, Fang F, Palmisano S, Aebersold R, Young ET. 2014. Phosphoproteomic analysis identifies proteins involved in transcription-coupled mRNA decay as targets of Snf1 signaling. Sci Signal 7:ra64. doi: 10.1126/scisignal.2005000. [DOI] [PubMed] [Google Scholar]
- 28.Rothstein RJ. 1983. One-step gene disruption in yeast. Methods Enzymol 101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 29.Young ET, Sloan J, Miller B, Li N, van Riper K, Dombek KM. 2000. Evolution of a glucose-regulated ADH gene in the genus Saccharomyces. Gene 245:299–309. doi: 10.1016/S0378-1119(00)00035-4. [DOI] [PubMed] [Google Scholar]
- 30.Nalley K, Johnston SA, Kodadek T. 2006. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442:1054–1057. doi: 10.1038/nature05067. [DOI] [PubMed] [Google Scholar]
- 31.Louvion JF, Havaux-Copf B, Picard D. 1993. Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 131:129–134. doi: 10.1016/0378-1119(93)90681-R. [DOI] [PubMed] [Google Scholar]
- 32.Dombek KM, Kacherovsky N, Young ET. 2004. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J Biol Chem 279:39165–39174. doi: 10.1074/jbc.M400433200. [DOI] [PubMed] [Google Scholar]
- 33.Ratnakumar S, Kacherovsky N, Arms E, Young ET. 2009. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics 182:735–745. doi: 10.1534/genetics.109.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J, Donoviel MS, Young ET. 1989. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol Cell Biol 9:34–42. doi: 10.1128/MCB.9.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloan JS, Dombek KM, Young ET. 1999. Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J Biol Chem 274:37575–37582. doi: 10.1074/jbc.274.53.37575. [DOI] [PubMed] [Google Scholar]
- 36.Collart MA, Oliviero S. 2001. Preparation of yeast RNA. Curr Protoc Mol Biol Chapter 13:Unit 13.12. doi: 10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Ortin JE, Alepuz P, Chavez S, Choder M. 2013. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol 425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 38.Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L, Martin DE, Tresch A, Cramer P. 2011. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. 2004. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics 3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Parua PK, Dombek KM, Young ET. 2014. Yeast 14-3-3 protein functions as a comodulator of transcription by inhibiting coactivator functions. J Biol Chem 289:35542–35560. doi: 10.1074/jbc.M114.592287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeberg MA, Han T, Moresco JJ, Kong A, Yang YC, Lu ZJ, Yates JR, Kim JK. 2013. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol 14:R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team. 2014. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 43.Pages H, Aboyoun P, Gentleman R, DebRoy S. 2014. Biostrings: string objects representing biological sequences, and matching algorithms. R package, version 2.34.0. http://www.bioconductor.org/. [Google Scholar]
- 44.The Bioconductor Dev Team. 2014. BSgenome.Scerevisiae.UCSC.sacCer3: Saccharomyces cerevisiae (yeast) full genome (UCSC version sacCer3). R package, version 1.3.99 http://www.bioconductor.org/. [Google Scholar]
- 45.Riordan DP, Herschlag D, Brown PO. 2011. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res 39:1501–1509. doi: 10.1093/nar/gkq920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park D, Morris AR, Battenhouse A, Iyer VR. 2014. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res 42:3736–3749. doi: 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. 2002. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A 99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price VL, Taylor WE, Clevenger W, Worthington M, Young ET. 1990. Expression of heterologous proteins in Saccharomyces cerevisiae using the ADH2 promoter. Methods Enzymol 185:308–318. doi: 10.1016/0076-6879(90)85027-L. [DOI] [PubMed] [Google Scholar]
- 53.Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. 2014. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guarente L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 55.Adams CC, Gross DS. 1991. The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol 173:7429–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purvis IJ, Loughlin L, Bettany AJ, Brown AJ. 1987. Translation and stability of an Escherichia coli beta-galactosidase mRNA expressed under the control of pyruvate kinase sequences in Saccharomyces cerevisiae. Nucleic Acids Res 15:7963–7974. doi: 10.1093/nar/15.19.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braun KA, Young ET. 2014. Coupling mRNA synthesis and decay. Mol Cell Biol 34:4078–4087. doi: 10.1128/MCB.00535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haimovich G, Choder M, Singer RH, Trcek T. 2013. The fate of the messenger is pre-determined: a new model for regulation of gene expression. Biochim Biophys Acta 1829:643–653. doi: 10.1016/j.bbagrm.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. 2013. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell 52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 60.Irani M, Taylor WE, Young ET. 1987. Transcription of the ADH2 gene in Saccharomyces cerevisiae is limited by positive factors that bind competitively to its intact promoter region on multicopy plasmids. Mol Cell Biol 7:1233–1241. doi: 10.1128/MCB.7.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biddick RK, Law GL, Young ET. 2008. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS One 3:e1436. doi: 10.1371/journal.pone.0001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young ET, Tachibana C, Chang HW, Dombek KM, Arms EM, Biddick R. 2008. Artificial recruitment of mediator by the DNA-binding domain of Adr1 overcomes glucose repression of ADH2 expression. Mol Cell Biol 28:2509–2516. doi: 10.1128/MCB.00658-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thukral SK, Tavianini MA, Blumberg H, Young ET. 1989. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol 9:2360–2369. doi: 10.1128/MCB.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blumberg H, Eisen A, Sledziewski A, Bader D, Young ET. 1987. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. Nature 328:443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- 65.Parua PK, Young ET. 2014. Binding and transcriptional regulation by 14-3-3 (Bmh) proteins requires residues outside of the canonical motif. Eukaryot Cell 13:21–30. doi: 10.1128/EC.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell DW, Smith M, Williamson VM, Young ET. 1983. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem 258:2674–2682. [PubMed] [Google Scholar]
- 67.Rendl LM, Bieman MA, Smibert CA. 2008. S. cerevisiae Vts1p induces deadenylation-dependent transcript degradation and interacts with the Ccr4p-Pop2p-Not deadenylase complex. RNA 14:1328–1336. doi: 10.1261/rna.955508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rendl LM, Bieman MA, Vari HK, Smibert CA. 2012. The eIF4E-binding protein Eap1p functions in Vts1p-mediated transcript decay. PLoS One 7:e47121. doi: 10.1371/journal.pone.0047121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tu BP, Kudlicki A, Rowicka M, McKnight SL. 2005. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science 310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 70.Kudlicki A, Rowicka M, Otwinowski Z. 2007. SCEPTRANS: an online tool for analyzing periodic transcription in yeast. Bioinformatics 23:1559–1561. doi: 10.1093/bioinformatics/btm126. [DOI] [PubMed] [Google Scholar]
- 71.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simpson CE, Ashe MP. 2012. Adaptation to stress in yeast: to translate or not? Biochem Soc Trans 40:794–799. doi: 10.1042/BST20120078. [DOI] [PubMed] [Google Scholar]
- 73.Ashe MP, De Long SK, Sachs AB. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol 21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denis CL, Ferguson J, Young ET. 1983. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem 258:1165–1171. [PubMed] [Google Scholar]
- 76.Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. 2009. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol 5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dombek KM, Young ET. 1997. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol Cell Biol 17:1450–1458. doi: 10.1128/MCB.17.3.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerber AP, Herschlag D, Brown PO. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. 2003. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- 80.Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH. 2006. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol 13:160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 81.Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 82.Dori-Bachash M, Shalem O, Manor YS, Pilpel Y, Tirosh I. 2012. Widespread promoter-mediated coordination of transcription and mRNA degradation. Genome Biol 13:R114. doi: 10.1186/gb-2012-13-12-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lotan R, Goler-Baron V, Duek L, Haimovich G, Choder M. 2007. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol 178:1133–1143. doi: 10.1083/jcb.200701165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trcek T, Larson DR, Moldon A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. 2008. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schulz D, Pirkl N, Lehmann E, Cramer P. 2014. Rpb4 subunit functions mainly in mRNA synthesis by RNA polymerase II. J Biol Chem 289:17446–17452. doi: 10.1074/jbc.M114.568014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J 24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 90.Abate G, Bastonini E, Braun KA, Verdone L, Young ET, Caserta M. 2012. Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochim Biophys Acta 1819:419–427. doi: 10.1016/j.bbagrm.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. 2010. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haimovich G, Medina DA, Causse SZ, Garber M, Millan-Zambrano G, Barkai O, Chavez S, Perez-Ortin JE, Darzacq X, Choder M. 2013. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y, Xu X, Kuo MH. 2010. Snf1p regulates Gcn5p transcriptional activity by antagonizing Spt3p. Genetics 184:91–105. doi: 10.1534/genetics.109.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. 1995. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses 45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.