Abstract
Autophagy is a major intracellular degradation system by which cytoplasmic components are enclosed by autophagosomes and delivered to lysosomes. Formation of the autophagosome requires a set of autophagy-related (Atg) proteins. Among these proteins, the ULK1 complex, which is composed of ULK1 (or ULK2), FIP200, Atg13, and Atg101, acts at an initial step. Previous studies showed that ULK1 and FIP200 also function in pathways other than autophagy. However, whether Atg13 and Atg101 act similarly to ULK1 and FIP200 remains unknown. In the present study, we generated Atg13 knockout mice. Like FIP200-deficient mice, Atg13-deficient mice die in utero, which is distinct from most other types of Atg-deficient mice. Atg13-deficient embryos show growth retardation and myocardial growth defects. In cultured fibroblasts, Atg13 deficiency blocks autophagosome formation at an upstream step. In addition, sensitivity to tumor necrosis factor alpha (TNF-α)-induced apoptosis is enhanced by deletion of Atg13 or FIP200, but not by other Atg proteins, as well as by simultaneous deletion of ULK1 and ULK2. These results suggest that Atg13 has both autophagic and nonautophagic functions and that the latter are essential for cardiac development and likely shared with FIP200 but not with ULK1/2.
INTRODUCTION
Macroautophagy (here referred to as autophagy) is an intracellular degradation system by which cytoplasmic components are degraded in the lysosomes (1–3). In the process of autophagy, an isolation membrane sequesters a part of the cytoplasm to form an autophagosome. The autophagosome subsequently fuses with a lysosome to enzymatically degrade sequestered materials. The degradation products, such as amino acids, are released back into the cytoplasm. Although autophagy occurs at low levels under normal conditions, it is highly activated during starvation. Studies in the last decade showed that autophagy is important for various physiological processes, including adaptation to starvation, intracellular quality control of proteins and organelles, preimplantation development, and degradation of intracellular pathogens (3–5).
Autophagosome formation requires a series of autophagy-related (Atg) proteins, which can be classified into several functional units, including (i) the Atg1/ULK complex, (ii) Atg9, (iii) class III phosphatidylinositol 3-kinase (PI3K) complex I, (iv) the Atg2-Atg18/WIPI complex, (v) the Atg12 conjugation system, and (vi) the Atg8/LC3 conjugation system (2, 6, 7). Among these units, the Atg1/ULK complex is one of the most upstream factors of autophagosome formation in both yeast and mammals and is directly regulated by mTORC1 (8, 9). The Saccharomyces cerevisiae Atg1 complex is composed of Atg1, Atg13, Atg17, Atg29, and Atg31. Atg17, Atg29, and Atg31 constitutively form a stable protein complex, with which Atg1 and Atg13 are associated in a starvation-dependent manner. Atg11 can also be included in this complex, particularly within the cytoplasm-to-vacuole pathway (10). This complex is not a simple signal transducer but has a mechanistic role in autophagosome formation (11, 12).
The mammalian ULK complex is composed of ULK1 (or ULK2), Atg13, FIP200 (also known as RB1CC1), and Atg101 (13–19). ULK1 and ULK2 are mammalian homologs of Atg1. FIP200 is hypothesized to act as a hybrid molecule of yeast Atg11 and Atg17 (8). Atg29 and Atg31 are not conserved in higher eukaryotes. In contrast to the yeast Atg1 complex, the mammalian ULK1-Atg13-FIP200-Atg101 complex is stable. It forms an ∼3-MDa complex irrespective of nutrient conditions.
Among these components, FIP200 and ULK1/2 have additional functions in pathways other than autophagy. Mice deficient in FIP200 die during postimplantation embryonic development (20), whereas Atg3−/− (21), Atg5−/− (22), Atg7−/− (23), Atg9a−/− (24), Atg12−/− (25), and Atg16L1−/− (26) mice die shortly after birth (a recent report suggests embryonic lethality of Atg9a−/− mice [27]). Although mice lacking either ULK1 or ULK2 are viable, ULK1/2 double-knockout (DKO) mice die during the neonatal period (28). In addition, FIP200 is involved in various signaling pathways, including those of p53, focal adhesion kinase (FAK), Pyk2, tumor necrosis factor alpha (TNF-α)–Jun N-terminal protein kinase (JNK), tuberous sclerosis complex (TSC)-mTOR, transforming growth factor beta (TGF-β), and β-catenin (29–31). ULK/Atg1 also has a unique role in metazoa, as it is important for neuronal functions. The Caenorhabditis elegans unc-51 (atg1) mutant displays uncoordinated movement and a defect in axonal elongation and transport (32–36). The neuronal function of unc-51 is thought to be independent of autophagy, as other autophagy-deficient mutants, such as epg-1 (atg13) and epg-9 (atg101) mutants, do not show an uncoordinated phenotype or defective axon migration (37, 38). In addition, RNA interference (RNAi)-mediated silencing of bec-1 (atg6), M7.5 (atg7), lgg-1 (atg8), or F41E6.13 (atg18) does not cause defects in axon guidance or axonal transport (35). A neuronal function of ULK has also been reported for Drosophila melanogaster (39–41) and mammals (42–44). It is not understood, however, whether mammalian Atg13 and Atg101 act similarly to ULK or FIP200.
In this study, we generated Atg13-deficient mice and found that Atg13 is essential for autophagy and postimplantation embryonic development. Loss of Atg13 causes growth retardation and myocardial growth defects in developing embryos. Analysis of Atg13-deficient mouse embryonic fibroblasts (MEFs) suggests that, like FIP200 but not like ULK1/2, Atg13 suppresses TNF-α-induced apoptosis. These results suggest that Atg13 has a nonautophagic function, which is required for cardiac development together with FIP200.
MATERIALS AND METHODS
Mice.
Two independent Atg13-deficient mouse lines were used in this study. The first line of Atg13-deficient mice was generated by using the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system (45). The pcDNA3-hCas9 plasmid was linearized by NotI digestion, purified by phenol-chloroform extraction, and transcribed to mRNA by using an mMessage mMachine T7 Ultra kit (Thermo). The synthesized Cas9 mRNA was purified by using an RNeasy minikit (Qiagen). The pT7-Atg13 guide RNA (gRNA) plasmid was linearized by BamHI digestion, purified by phenol-chloroform extraction, and transcribed to mRNA by using a MEGAshortscript T7 transcription kit (Thermo). The synthesized gRNA was purified by using a mirVana microRNA (miRNA) isolation kit (Thermo). Cas9 mRNA (120 ng/μl) and Atg13 gRNA (60 ng/μl) were injected into the cytoplasm of fertilized eggs collected from C57BL/6 mice to obtain heterozygous (Atg13+/−) mutant mice. The second Atg13-deficient mouse line was generated by using the embryonic stem (ES) cell line DD0507 containing an insertion of a gene trap cassette in the Atg13 gene (purchased from Mutant Mouse Regional Resource Centers). ES cells were injected into C57BL/6 blastocysts to obtain chimeric mice, which were crossed with C57BL/6 mice to obtain heterozygous (Atg13+/gt) mutant mice. Caesarean delivery was performed as previously described (22). All animal experiments were approved by the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University and The University of Tokyo.
Genotyping of mice and embryos.
For genotyping of the CRISPR/Cas9 line, a heteroduplex mobility assay was performed (46). Primer F (5′-ATGAAGCCTGCAGTTGATCC-3′) and primer R (5′-GAGTGGTCAGAACGCCTCTC-3′) were used for PCR-mediated amplification of a 402-base fragment of the genome DNA by using PrimeSTAR Max DNA polymerase (TaKaRa). The resulting PCR products were electrophoresed on a 6% polyacrylamide gel. Slower-migrating bands represent heteroduplexes. To detect homozygous mutants, the PCR product amplified from each sample was mixed with the PCR product from a wild-type mouse. The mixtures were denatured at 95°C for 3 min, followed by cooling to room temperature. The resulting samples were subjected to polyacrylamide gel electrophoresis. For genotyping of the gene trap line, PCR was performed by using genomic DNA. Primer F (5′-GTGTGGGTGTGTTTGACTAG-3′) and primer R1 (5′-CAGGAGATCTACAGGGCTAAG-3′) were used to amplify a 286-base fragment of the wild-type allele. Primer F (5′-GTGTGGGTGTGTTTGACTAG-3′) and primer R2 (5′-GTGTGAAAATGAGATGGATTG-3′) were used to amplify a 490-base fragment of the gene trap allele.
Cell culture.
Atg13−/− MEF line 1 and Atg13gt/gt MEFs were prepared from Atg13−/− and Atg13gt/gt embryos, respectively, at embryonic day 13.5 (E13.5). Atg13gt/gt MEFs were transformed with pEF321-T, a simian virus 40 (SV40) large T antigen expression vector (kindly provided by S. Sugano), and immortalized cell lines were established. ULK1/2 DKO MEFs (47, 48), Atg3 knockout (KO) MEFs (21), Atg5 KO MEFs (22), Atg9A KO MEFs (24), Atg16L1 KO MEFs (26), FIP200 KO MEFs (20), and Atg14 KO MEFs (49) were generated previously. Atg13−/− MEF line 2 cells were generated as follows. Wild-type MEFs were transfected with a Cas9-encoding plasmid (catalog number 41815; Addgene) and pCR-Blunt II-TOPO-Atg13 gRNA and cultured in the presence of 2 mg/ml Geneticin (G418) for 2 days. Single colonies were isolated, and homozygous Atg13−/− cells were selected. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 μg/ml penicillin, and streptomycin (complete medium) in a 5% CO2 incubator. For starvation treatment, cells were washed twice with phosphate-buffered saline (PBS) and incubated in amino acid-free Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) without FBS. For propidium iodide staining, phenol red-free DMEM was used and the cells were observed with a fluorescence microscope (IX81; Olympus) equipped with a charge-coupled device (CCD) camera (CoolSNAP HQ2; Photometrics).
Retroviral infection and generation of stable cell lines.
Stable cell lines were generated by using a retroviral expression system as previously described (13). Briefly, Plat E cells (kindly provided by T. Kitamura) were transiently transfected with retroviral vectors by using FuGENE HD reagent (Roche). After culture for 3 days, the growth medium containing retrovirus was collected. MEFs were incubated overnight with the collected virus-containing medium with 8 μg/ml Polybrene. Uninfected cells were removed by puromycin (Sigma-Aldrich) selection.
Antibodies and reagents.
Rabbit polyclonal antibodies against LC3 (NM1) (50), Atg16L1 (51), Atg101 (Ab#1) (19), Atg9A (52), and FIP200 (13) were described previously. Antibodies against Atg13 (catalog number SAB4200100; Sigma-Aldrich), ULK1 (catalog number A7481; Sigma-Aldrich), p62 (catalog numbers PM045 [MBL] and GP62-C [Progen]), beclin 1 (catalog number 3738; Cell Signaling), cleaved caspase-3 (catalog number 9664; Cell Signaling), IκBα (catalog number 9242; Cell Signaling), phospho-IκBα (catalog number 9246; Cell Signaling), JNK (catalog number 9252; Cell Signaling), phospho-JNK (catalog number 9251; Cell Signaling), caspase-8 (catalog number ALX-804-447; Enzo), and β-actin (catalog number A2228; Sigma-Aldrich) were used. For immunocytochemistry, Alexa Fluor 488-conjugated anti-rabbit IgG and Alexa Fluor 568-conjugated anti-rabbit IgG or anti-guinea pig IgG secondary antibodies (Molecular Probes) were used. Bafilomycin A1, Hoechst 33342 dye, and propidium iodide were purchased from Wako. Recombinant mouse TNF-α was purchased from Calbiochem. Staurosporine was purchased from Sigma-Aldrich.
Plasmids.
To generate pcDNA3-hCas9, Cas9 cDNA obtained from Addgene (catalog number 41815) was cloned into pcDNA3. To generate pCR-Blunt II-TOPO-Atg13 gRNA, a 20-bp guide sequence (5′-CCGTCTGTAGGGAGATTCTA-3′) was cloned into pCR-Blunt II-TOPO-gRNA obtained from Addgene (catalog number 41824). To generate pT7-Atg13 gRNA, a 20-bp guide sequence (5′-GTAGGGAGATTCTATGGAGT-3′) was cloned into pT7-gRNA, obtained from Addgene (catalog number 46759). The following plasmids were used, as previously described: pMXs-IP-GFP-LC3 (13), pMXs-IP-GFP-Atg14 (53), pMXs-IP-GFP-Atg13 (15), pMXs-IP-GFP-FIP200 (13), pMXs-puro-GFP-WIPI-1, pMXs-puro-VMP1-GFP, and pMXs-puro-GFP-DFCP1 (54).
Immunocytochemistry.
Cells grown on coverslips were fixed with 4% paraformaldehyde (PFA), permeabilized with 50 μg/ml digitonin, and stained with specific antibodies. These cells were observed with a confocal laser microscope (FV1000D IX81; Olympus) using a 60× PlanApoN oil immersion lens (1.42 numerical aperture [NA]; Olympus). Images were processed by using Photoshop Elements 5.0 (Adobe).
Histology.
Images of whole embryos were acquired by using a microscope (SZX12; Olympus) equipped with a digital camera (DP70; Olympus). For histological analysis, embryos were fixed in 4% PFA in PBS and embedded in paraffin. The sections were stained with Mayer's hematoxylin and eosin. Samples were photographed by using a microscope (SZX12 or BX51; Olympus) equipped with a digital camera (DP70; Olympus). Obtained images were processed by using Photoshop Elements 5.0.
Reverse transcriptase PCR (RT-PCR).
Total RNA was extracted from MEFs by using Isogen (Nippon Gene) and treated with RQ1 DNase (Promega) to remove genomic DNA contamination. cDNA was synthesized from total RNA by using ReverTra Ace and random primers (Toyobo). Subsequent PCR was performed by using the following primers: Arhgap1 primer F (5′-AAAGCAGCTCTCCAGAACCT-3′) and primer R (5′-CCGGCAAGCACTGAATACAA-3′) and β-actin primer F (5′-CTGGGTATGGAATCCTGTGG-3′) and primer R (5′-GTACTTGCGCTCAGGAGGAG-3′).
Southern blotting.
Genomic DNAs were digested with EcoRI, separated by electrophoresis on an agarose gel, transferred onto Biodyne nylon transfer membranes (Pall Corporation), and hybridized with a digoxigenin (DIG; Roche)-labeled probe. The probe was detected by anti-DIG antibody labeled with alkaline phosphatase (Roche) and visualized with CDP-Star (Roche). A 0.6-kb fragment of the Atg13 gene, which is upstream of exon 1, was labeled with DIG by PCR using primers 5′-CAGCCATTCACTTGTTAACTC-3′ and 5′-CTTCACAGGCAAACCCTGTGC-3′ and used as a probe.
Immunoblotting.
Cells were lysed with regular lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM NaF, 0.4 mM Na3VO4, 10 mM sodium pyrophosphate, and a protease inhibitor cocktail [Complete EDTA-free protease inhibitor; Roche]). The lysates were centrifuged at 17,400 × g for 15 min, and the supernatants were boiled in sample buffer. Samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Immobilon-P polyvinylidene difluoride membranes (Millipore). Immunoblot analysis was performed, and signals were visualized with the SuperSignal West Pico chemiluminescent substrate (Pierce) or Immobilon Western substrate (Millipore). The signal intensities were analyzed by using an LAS-3000mini imaging analyzer and Multi Gauge software version 3.0 (Fujifilm). Contrast and brightness adjustments were applied to the whole images by using Photoshop Elements 5.0. To detect protein phosphorylation by Phos-tag, cells were lysed with lysis buffer without EDTA. Polyacrylamide gels containing 50 μM Phos-tag acrylamide (Wako) and 50 μM MnCl2 were used for SDS-PAGE. After electrophoresis, Phos-tag acrylamide gels were washed with transfer buffer containing 2 mM EDTA for 10 min and then subjected to transfer.
Gel filtration analysis.
Cells were homogenized in hypotonic buffer (40 mM Tris-HCl [pH 7.5] and a protease inhibitor cocktail) by repeated passage (15 times) through a 1-ml syringe with a 27-gauge needle. The homogenates were centrifuged at 13,000 × g for 15 min, and the supernatants were further centrifuged at 100,000 × g for 60 min. The supernatant fraction was filtered with an Ultrafree-MC 0.45-μm filter unit (Millipore) and then applied to a Superose 6 column (GE Healthcare). Next, 0.5-ml fractions were collected at a flow rate of 0.5 ml/min with elution buffer (40 mM Tris-HCl [pH 7.5] and 150 mM NaCl). The fractions were then analyzed by Western blotting. The column was calibrated with thyroglobulin (669 kDa; Wako), ferritin (440 kDa; Sigma-Aldrich), catalase (240 kDa; Wako), and ovalbumin (43 kDa; Wako).
Caspase activity assay.
The activity of caspase-8 was measured by using a fluorometric assay kit (MBL). Briefly, cell lysates were incubated with a 7-amido-4-methylcoumarin (AMC)-conjugated substrate specific for caspase-8. Substrate cleavage was monitored by measuring the fluorescence of AMC (excitation at 380 nm and emission at 460 nm).
RESULTS
Establishment of Atg13-deficient mice and cells.
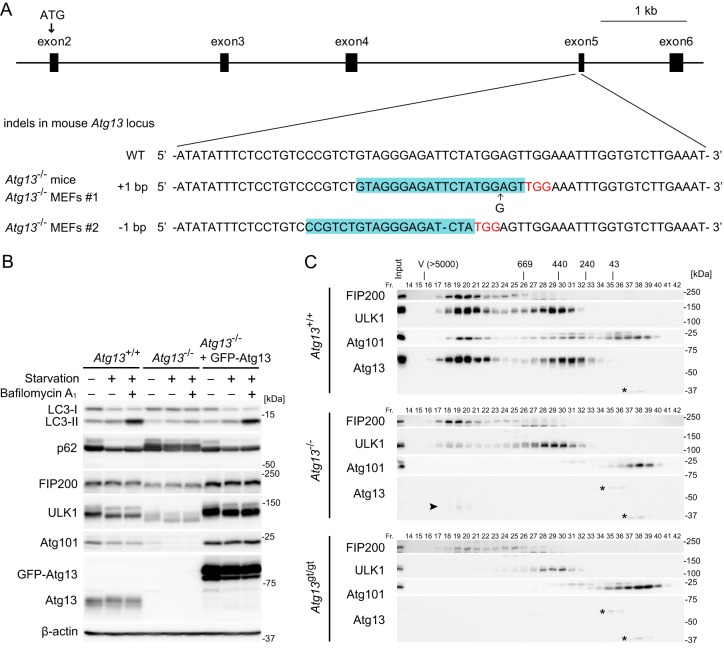
Two independent Atg13-deficient mouse lines were generated. The first line was generated by using the CRISPR/Cas9 system (referred to as Atg13−/− mice here). A frameshift mutation was introduced into exon 5 of the Atg13 gene (Fig. 1A). The second line was generated by using an ES cell line carrying a gene trap cassette containing a splicing acceptor and β-galactosidase–neomycin phosphotransferase fusion gene (β-geo) downstream of exon 1 of the Atg13 gene (referred to as Atg13gt/gt mice here) (see Fig. S1A in the supplemental material). Insertion of the gene trap cassette was confirmed by Southern blotting and PCR analysis (see Fig. S1B in the supplemental material). Expression of Atg13 was abolished in Atg13−/− MEF line 1 derived from Atg13−/− embryos, Atg13−/− MEF line 2 (prepared from wild-type MEFs by using the CRISPR/Cas9 system), and Atg13gt/gt MEFs (Fig. 1B; see also Fig. S1C and D in the supplemental material). In Atg13gt/gt MEFs, however, we found that expression of Arhgap1, which is a gene next to Atg13, was also abolished, suggesting that the gene trap cassette destroyed the Arhgap1 gene (see Fig. S1E in the supplemental material). In contrast, the expression of Arhgap1 was not affected in Atg13−/− MEF lines 1 and 2, which were generated by using the CRISPR/Cas9 system (see Fig. S1E). Thus, in the following experiments, we primarily used Atg13−/− mice and MEFs and also used Atg13gt/gt mice and cells to confirm phenotype specificity and rule out the possibility of potential off-target effects caused by CRISPR-mediated gene editing.
FIG 1.
Establishment of Atg13-deficient mice and cells. (A) Sequences of mutant alleles from Atg13−/− mice (and Atg13−/− MEF line 1) and Atg13−/− MEF line 2 generated by the CRISPR/Cas9 system. The gRNA-targeting sequence used to generate each mutation is highlighted (blue), and the protospacer-adjacent motif sequence is indicated (red). (B) Wild-type (WT) MEFs, Atg13−/− MEF line 2, and Atg13−/− MEF line 2 stably expressing GFP-Atg13 were cultured in regular or starvation medium in the presence or absence of 100 nM bafilomycin A1 for 2 h. (C) The cytosolic fractions (Fr.) of wild-type MEFs, Atg13−/− MEF line 2, and Atg13gt/gt MEFs were subjected to gel filtration analysis using a Superose 6 column. Positions of the molecular mass standards (in kilodaltons) are shown at the top. V, void fraction. Asterisks indicate nonspecific immunoreactive bands. The arrowhead indicates a potential truncated form of Atg13.
As we previously reported, using Atg13 small interfering RNA (siRNA)-treated cells (15, 19), disassembly of the ULK1 complex (Fig. 1C) and destabilization of the ULK1 complex components (i.e., ULK1, FIP200, and Atg101) were observed in Atg13 KO MEFs (Fig. 1B; see also Fig. S1C and D in the supplemental material). Although ULK1 was completely absent in the 3-MDa complex in Atg13gt/gt MEFs, ULK1 partially remained in the 3-MDa complex in Atg13−/− MEF line 2 (Fig. 1C) as well as in MEFs described previously by other groups (55, 56). This is presumably because of the expression of a truncated Atg13 fragment (downstream of exon 5), which, even though not functional, could link ULK1 to FIP200 (Fig. 1C). To measure autophagic activity in Atg13 KO MEFs, we performed an LC3 turnover assay (57). LC3 is one of the mammalian Atg8 homologs and is present in two forms: LC3-I (the cytosolic form) and LC3-II (the phosphatidylethanolamine-conjugated form). Because LC3-II is present on both outer and inner autophagosomal membranes, LC3-II is degraded after autophagosome-lysosome fusion. In wild-type MEFs, induction of autophagy by nutrient starvation converted LC3-I to LC3-II, and LC3-II was further increased by treatment with the vacuolar H+ ATPase inhibitor bafilomycin A1 (Fig. 1B; see also Fig. S1C and D in the supplemental material). In Atg13 KO MEFs, on the other hand, the increase in the level of LC3-II caused by starvation and lysosomal inhibition was suppressed (Fig. 1B; see also Fig. S1C and D). We also examined starvation-induced degradation of p62 (also known as SQSTM1), a selective substrate of autophagy (57). In Atg13 KO MEFs, the starvation-induced decrease in p62 levels, which was observed in wild-type MEFs, was abolished (Fig. 1B; see also Fig. S1C and D). The stability of the ULK1 complex components and turnover of LC3 and p62 were restored by exogenously expressed green fluorescent protein (GFP)-Atg13 (Fig. 1B; see also Fig. S1C). These results confirm that Atg13 is essential for autophagy.
Impaired recruitment of Atg proteins in Atg13-deficient cells.
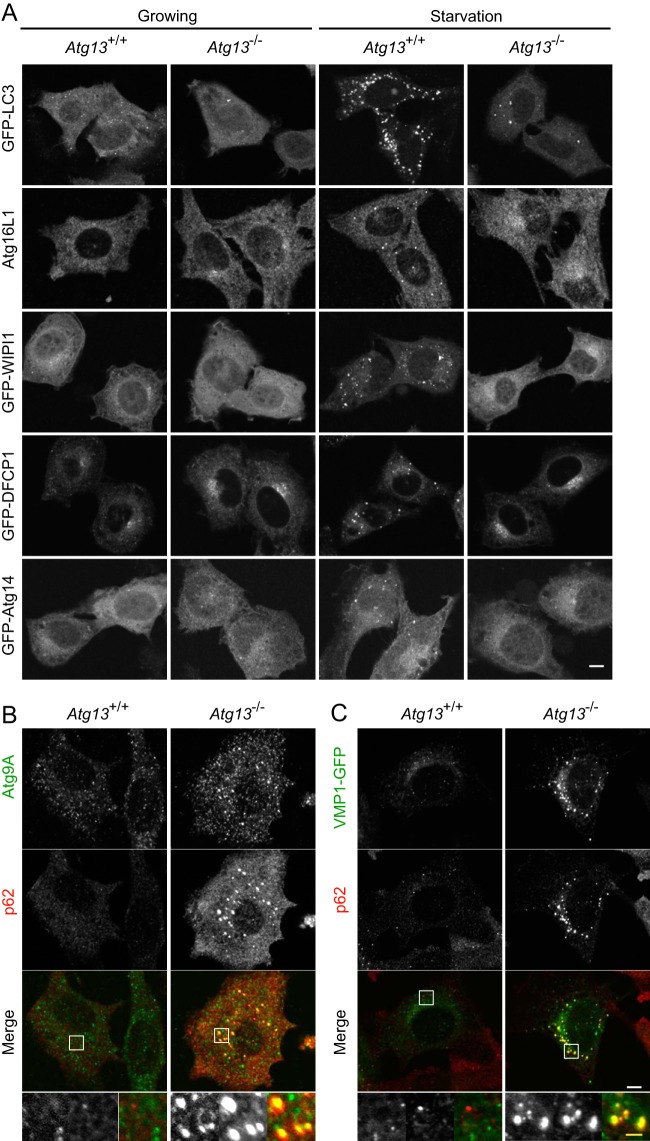
The ULK1 complex is considered to be one of the most upstream units among mammalian Atg proteins. This is based on the observation that punctum formation of LC3, Atg16L1, WIPI-1, DFCP1, and Atg14 is impaired in FIP200 KO MEFs (54). In wild-type MEFs, LC3 Atg16L1, WIPI-1, DFCP1, and Atg14 formed puncta after starvation (Fig. 2A; see also Fig. S2A in the supplemental material). In Atg13 KO MEFs (Atg13−/− MEF line 2 and Atg13gt/gt MEFs), on the other hand, punctum formation of these Atg proteins was almost completely suppressed (Fig. 2A; see also Fig. S2A). LC3 puncta were occasionally observed even in Atg13 KO MEFs, as in FIP200 KO MEFs, which could represent aggregation of LC3 proteins (Fig. 2A; see also Fig. S2A) (13, 54). These results indicate that Atg13 functions upstream of these proteins, as reported previously for FIP200.
FIG 2.
Impairment of Atg protein recruitment in Atg13-deficient cells. (A) Wild-type MEFs and Atg13−/− MEF line 2 stably expressing GFP-LC3, GFP-WIPI1, GFP-DFCP1, or GFP-Atg14 were cultured in regular or starvation medium for 1 h. Cells were fixed and analyzed by fluorescence microscopy. Atg16L1 was immunostained with anti-Atg16L1 antibody. (B) Wild-type MEFs and Atg13−/− MEF line 2 were cultured in regular medium. Cells were fixed and analyzed by immunofluorescence microscopy using anti-Atg9A and anti-p62 antibodies. (C) Wild-type MEFs and Atg13−/− MEF line 2 stably expressing VMP1-GFP were cultured in regular medium. Cells were fixed and analyzed by immunofluorescence microscopy using anti-p62 antibody. Bars, 10 μm (white) and 2 μm (yellow).
We also examined the phosphorylation of beclin 1, which is a substrate of ULK1 (58). As a result, phosphorylation of beclin 1 was slightly but clearly reduced in Atg13 KO MEFs as well as in FIP200 KO MEFs and ULK1/2 DKO MEFs, suggesting an impairment of ULK1 activity (see Fig. S3 in the supplemental material).
We previously reported that p62, VMP1-GFP, and Atg9A accumulate at the autophagosome formation site when early steps of autophagosome formation are blocked (52, 54, 59). Consistently, Atg9A and VMP1-GFP accumulated on enlarged puncta, which were colocalized with p62, in Atg13-deficient but not in wild-type MEFs (Fig. 2B and C; see also Fig. S2B and C in the supplemental material). Taken together, these results indicate that Atg13 functions at an initial step of autophagosome formation together with FIP200.
Atg13 is essential for postimplantation embryonic development.
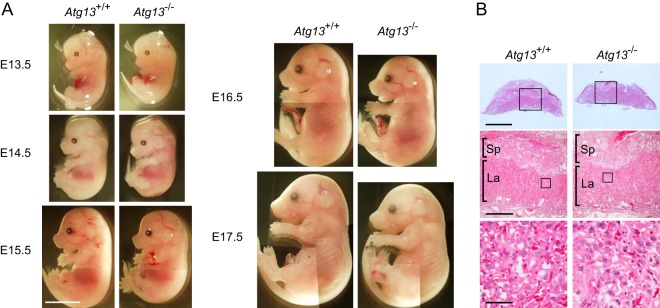
Although heterozygous Atg13+/− and Atg13+/gt mice are healthy and fertile, we obtained no live-born homozygous offspring from heterozygous breeding, indicating that Atg13-deficient mice died during embryogenesis (Table 1; see also Table S1 in the supplemental material). Mating of Atg13+/− mice produced Atg13 KO embryos at almost the expected Mendelian frequency at E16.5, but the survival rate was decreased thereafter, and no live pups were obtained (Table 1). This phenotype appeared slightly earlier for Atg13gt/gt embryos, which may reflect an additive effect of the deletion of Arhgap1 (see Table S1 in the supplemental material). These results suggest that Atg13 is essential for postimplantation embryonic development. Gross examination of embryos revealed growth retardation of Atg13-deficient embryos (Fig. 3A; see also Fig. S4A in the supplemental material). The body sizes of both Atg13−/− and Atg13gt/gt embryos were smaller than that of wild-type embryos. Because placental failure could be a cause of postimplantation embryonic lethality (60), we examined the placentas of Atg13−/− and Atg13gt/gt embryos. Although the vast majority of the placental phenotypes showed defects in the establishment of the labyrinth layer (61), this layer was normally observed in both wild-type and Atg13-deficient placentas, suggesting that growth retardation and embryonic lethality of Atg13-deficient mice are not derived from placental failure (Fig. 3B; see also Fig. S4B in the supplemental material).
TABLE 1.
Total numbers of embryos and neonates obtained from mating between Atg13+/− mice
| Stageb | No. of offspring obtained of genotype: |
Total no. of offspring obtained | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| E13.5 | 5 | 8 | 1 | 14 |
| E14.5 | 1 | 2 | 4 | 7 |
| E15.5 | 1 | 6 | 2 | 9 |
| E16.5 | 5 | 7 | 4 | 16 |
| E17.5 | 5 | 13 | 1 | 19 |
| E18.5 | 3 | 3 | 0 | 6 |
| P0a | 9 | 16 | 0 | 25 |
Neonates were obtained by natural birth.
E, embryonic day; P, postnatal day.
FIG 3.
Loss of Atg13 causes growth retardation. (A) Wild-type (Atg13+/+) and Atg13−/− embryos at the indicated stages. Bar, 5 mm. (B) Paraffin sections of placentas from wild-type (Atg13+/+) and Atg13−/− embryos at E16.5 were subjected to staining with Mayer's hematoxylin and eosin. Sp, spongiotrophoblast; La, labyrinth layer. Bars, 2 mm (top), 500 μm (middle), and 50 μm (bottom).
Loss of Atg13 causes myocardial growth defects in the heart.
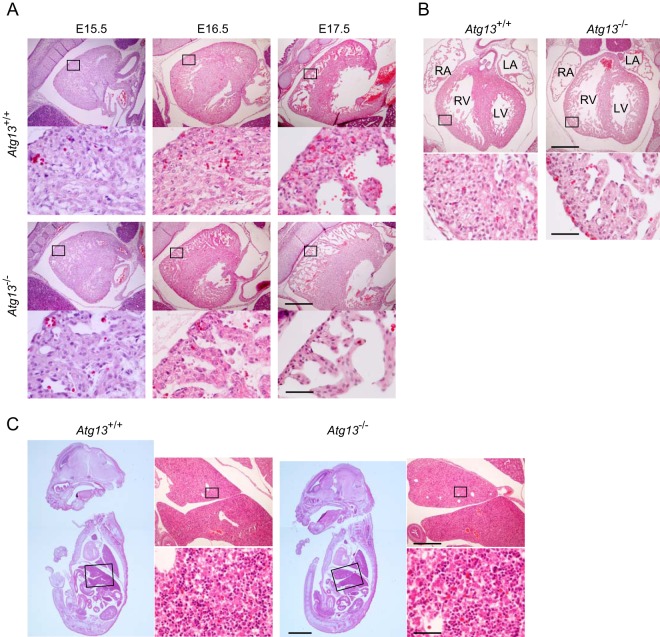
Histological analysis of the embryos showed defects in the heart. Thinning of the ventricular wall was observed in the hearts of both Atg13−/− and Atg13gt/gt embryos after E13.5, suggesting that the growth of the compact layer was impaired (Fig. 4A and B; see also Fig. S5 in the supplemental material). This was apparent during late embryogenesis in Atg13−/− embryos (Fig. 4A and B) but was observed as early as E14.5 or E15.5 in Atg13gt/gt embryos and in Atg13−/− embryos (see Fig. S5). No remarkable abnormality was observed in organs other than the heart (Fig. 4C). Although FIP200 KO mice show a liver defect, it was not clear in Atg13-deficient embryos (Fig. 4C) (20). These results suggest that Atg13 is essential for myocardial growth in developing embryos. The embryonic lethality of Atg13 KO mice is likely due to heart failure.
FIG 4.
Loss of Atg13 causes myocardial growth defects. (A) Sagittal paraffin sections from hearts of wild-type (Atg13+/+) and Atg13−/− embryos at the indicated embryonic stages. Bars, 500 μm (top) and 50 μm (bottom). (B) Coronal paraffin sections from hearts of wild-type (Atg13+/+) and Atg13−/− embryos at E16.5. RA, right atrium; RV, right ventricle; LA, left atrium; LV, left ventricle. Bars, 500 μm (top) and 50 μm (bottom). (C) Sagittal paraffin sections from livers of wild-type (Atg13+/+) and Atg13−/− embryos at E16.5. Bars, 2 mm (left), 500 μm (top right), and 50 μm (bottom right).
Atg13-deficient cells are sensitive to TNF-α-induced apoptosis.
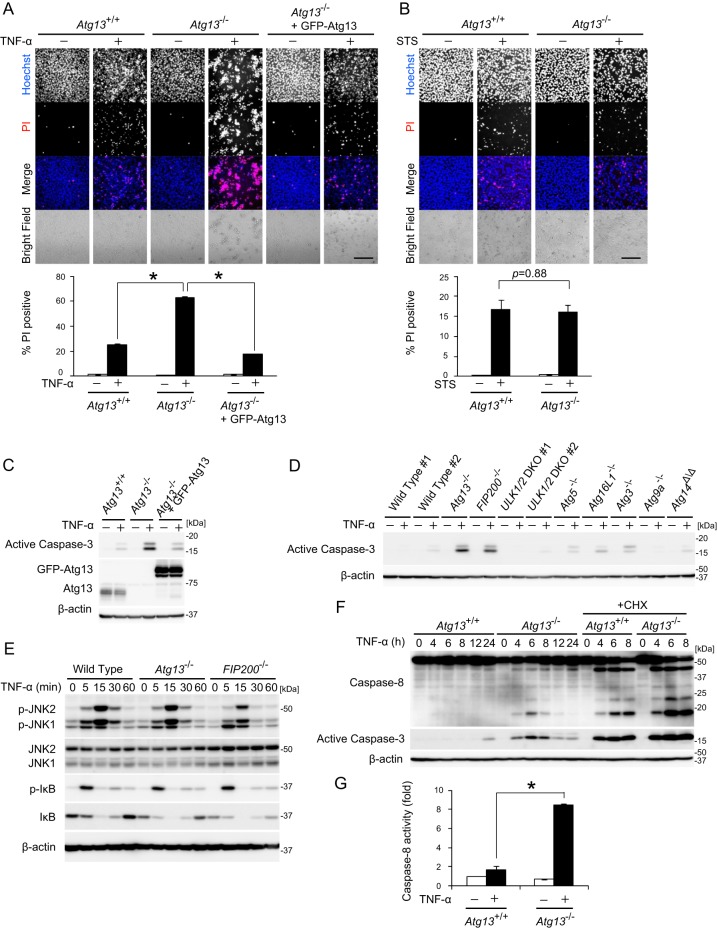
The phenotype of Atg13 KO embryos is similar to that of FIP200 KO embryos (20). This suggests that Atg13 functions together with FIP200 in both autophagic and nonautophagic pathways. Because FIP200 is known to suppress TNF-α-induced apoptosis in MEFs (20), we examined whether Atg13 is also involved in this function. The number of dead cells after 24 h of TNF-α treatment was increased in both Atg13−/− MEF line 2 and Atg13gt/gt MEFs compared to wild-type and Atg13 KO MEFs stably rescued with GFP-Atg13 (Fig. 5A; see also Fig. S6A in the supplemental material). On the other hand, staurosporine-induced cell death was not enhanced in Atg13 KO MEFs, indicating that Atg13 specifically suppresses TNF-α-induced cell death (Fig. 5B; see also Fig. S6B in the supplemental material). TNF-α-induced caspase-3 activation was significantly increased in Atg13−/− MEF line 2, Atg13gt/gt MEFs, and FIP200 KO MEFs compared to that in wild-type and rescued Atg13 KO MEFs (Fig. 5C and D; see also Fig. S6C and D in the supplemental material), suggesting that Atg13 negatively regulates TNF-α-induced apoptosis. The enhanced sensitivity of Atg13 KO MEFs to TNF-α-induced apoptosis is not simply because of decreased FIP200 protein levels because overexpression of GFP-FIP200 did not restore cell death and caspase-3 activation (see Fig. S7 in the supplemental material). The sensitivity to TNF-α-induced caspase-3 activation was not significantly increased in ULK1/2 (DKO), Atg5, Atg16L1, Atg3, Atg9A, or Atg14 KO MEFs. This suggests that Atg13 and FIP200 suppress TNF-α-induced apoptosis independent of autophagy and even of ULK1/2 (Fig. 5D; see also Fig. S6D).
FIG 5.
Atg13 suppresses TNF-α-induced apoptosis independent of autophagy. (A) Wild-type MEFs, Atg13−/− MEF line 2, and Atg13−/− MEF line 2 stably expressing GFP-Atg13 were cultured with or without 20 ng/ml TNF-α for 24 h. Cells were stained with 1 μg/ml Hoechst 33342 dye and propidium iodide (PI) and then analyzed by fluorescence microscopy. Bar, 200 μm. The percentage of propidium iodine-positive cells of at least 300 cells was determined, and the means ± standard errors from three independent experiments are shown. *, P < 0.01, as determined by an unpaired Student t test. (B) Wild-type MEFs and Atg13−/− MEF line 2 were cultured with or without 50 ng/ml staurosporine (STS) for 24 h and analyzed as described above for panel A. (C) Wild-type MEFs, Atg13−/− MEF line 2, and Atg13−/− MEF line 2 stably expressing GFP-Atg13 were cultured with or without 20 ng/ml TNF-α for 12 h. (D) MEFs of the indicated genotypes were cultured with or without 20 ng/ml TNF-α for 12 h. (E) Wild-type MEFs, Atg13−/− MEF line 2, and FIP200 KO MEFs were cultured with 20 ng/ml TNF-α for the indicated times. (F) Wild-type MEFs and Atg13−/− MEF line 2 were cultured with 20 ng/ml TNF-α in the presence or absence of 50 μg/ml cycloheximide (CHX) for the indicated times. (G) Wild-type MEFs and Atg13−/− MEF line 2 were cultured with or without 20 ng/ml TNF-α for 12 h. The activity of caspase-8 was measured. Means ± standard errors (n = 3) are shown. *, P < 0.01, as determined by an unpaired Student t test.
We next tested which signals downstream of the TNF receptor were affected in Atg13 KO MEFs. It was reported previously that FIP200 KO MEFs are defective in ASK1/TRAF2-mediated JNK activation, which might explain the higher TNF-α sensitivity (20). However, we could not detect the defect in either Atg13 KO or FIP200 KO MEFs (Fig. 5E; see also Fig. S6E in the supplemental material), suggesting that there is another explanation for the enhanced TNF-α-induced apoptosis in these KO MEFs. Although NF-κB activation is a major survival event in response to TNF-α stimulation (62, 63), phosphorylation of IκB was not impaired in Atg13 KO MEFs (Fig. 5E; see also Fig. S6E). It is known that TNF-induced apoptosis is mediated mainly by caspase-8 and is augmented by treatment with the protein synthesis inhibitor cycloheximide (CHX), which eliminates the endogenous caspase-8 inhibitor c-FLIP (62–64). In the absence of c-FLIP, caspase-8 was activated by autocatalytic cleavage upon TNF-α treatment. Upon treatment with TNF-α in the presence or absence of CHX, caspase-8 cleavage and caspase-3 activation were observed more clearly in Atg13−/− MEF line 2 than in wild-type MEFs (Fig. 5F). To further evaluate caspase-8 activation, we measured caspase-8 activity in TNF-α-treated MEFs and found that it was higher in Atg13−/− MEF line 2 than in wild-type MEFs (Fig. 5G). This tendency was also observed for Atg13gt/gt MEFs (see Fig. S6F and G in the supplemental material). Taken together, these results suggest that Atg13 negatively regulates TNF-α-induced caspase-8 activation to suppress apoptosis.
DISCUSSION
In this study, we showed that Atg13 has a nonautophagic function essential for postimplantation embryonic development in addition to its known role in autophagy. Atg13 and FIP200 seem to act in the same pathway, as evidenced by the fact that the phenotype of Atg13 deficiency is similar to that of FIP200 deficiency to some extent (20). It was reported previously that FIP200 KO embryos show lethality at around E14.5 to E15.5, pale appearance, generalized edema, and thinning and disorganization of the heart ventricular wall, with increased numbers of apoptotic cells (20). Embryonic lethality and thinning of the ventricular wall are also observed in Atg13 KO embryos. These defects were not reported for Atg KO mice other than FIP200 KO mice. A recent report suggested an important function of autophagy in the development of the ventricular septum and valves in Atg5 KO mouse neonates, but the thickness of the ventricular wall appears to be normal in Atg5 KO mouse neonates (65). Furthermore, enhanced TNF-α sensitivity was also observed for both FIP200 KO and Atg13 KO MEFs but not for other Atg KO MEFs. These data suggest that the developmental defects observed for Atg13 KO and FIP200 KO mice are likely derived from defects in a pathway other than autophagy. However, the phenotype of FIP200 KO mice seems to be more severe than that of Atg13 KO mice. Considering that FIP200 is involved in various signaling pathways, FIP200 might have autophagy-independent and even Atg13-independent function. Alternatively, it is still possible that the relative necessities of these Atg factors in the autophagy pathway itself are different, which would result in different phenotypes. More comprehensive studies will be required in the future.
We realized that the Arhgap1 gene is also targeted in Atg13gt/gt mice. Arhgap1 has GTPase-activating activity toward Cdc42 and Rho and regulates cell survival, migration, and motility (66, 67). Simultaneous deletion of the Arhgap1 gene may explain why Atg13gt/gt mice showed some defects that were more severe (e.g., earlier lethality and heart thinning) than those in Atg13−/− mice, although Arhgap1-deficient mice are not embryonic lethal (67).
Unexpectedly, our results and data from recent reports suggest that the nonautophagic functions of Atg13-FIP200 and ULK are different. First, Atg13 KO MEFs and FIP200 KO MEFs show enhanced TNF-α-induced apoptosis, in contrast to ULK1/2 DKO MEFs (Fig. 5D; see also Fig. S6D in the supplemental material). Second, the phenotype of ULK1/2 DKO mice is milder than that of Atg13 KO and FIP200 KO mice: ULK1/2 DKO mice die after birth (28). These milder effects of deletion of ULK1 and ULK2 may indicate that they are not essential for the function of Atg13 and FIP200 in certain settings. In chicken cells, ULK1 and ULK2 are not essential even for autophagy, whereas Atg13 and FIP200 are essential (68). On the other hand, ULK has FIP200- and Atg13-independent functions. In C. elegans, the uncoordinated movement observed for the unc-51 mutant is not observed for the epg-1 (atg13) (37) and epg-9 (atg101) mutants (38). Similarly, in Drosophila melanogaster, neuronal defects observed in the unc-51 mutant are not observed in the Fip200 mutant (69). In addition to effects on the nervous system, the findings of a recent study suggest that ULK1 but not Atg13 and FIP200 regulates melanogenesis in cultured cells (70).
In this study, we also found that Atg13 negatively regulates TNF-α-induced caspase-8 activation to suppress apoptosis. It is possible that the increased sensitivity of death receptor signaling accounts for the defects observed in Atg13 KO embryos because deletion of c-FLIP, which results in death receptor-induced caspase activation, also leads to growth retardation and thinning of the heart wall (71, 72). However, the molecular mechanisms are currently unknown. Considering that survival signaling mediated by IκB phosphorylation is not affected in Atg13 KO MEFs and FIP200 KO MEFs (Fig. 5E; see also Fig. S6E in the supplemental material), Atg13 and FIP200 may regulate a factor(s) downstream of NF-κB and upstream of caspase-8. Accumulation of p62 due to autophagy suppression might contribute to elevated caspase-8 activity (73), although caspase-3 hyperactivation is observed in Atg13 KO and FIP200 KO MEFs but not in other ATG KO MEFs (Fig. 5D; see also Fig. S6D in the supplemental material). Further experiments will provide more insights into the nonconventional roles of Atg13 and FIP200.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nao Hosokawa for the preparation of ES cells, Jun-Lin Guan for FIP200 KO MEFs, Masaaki Komatsu for Atg3 KO MEFs, Tatsuya Saitoh and Shizuo Akira for Atg9A, Atg14, and Atg16L1 KO MEFs, Sharon Tooze and Craig Thompson for ULK1/2 DKO MEFs, Toshio Kitamura and Shoji Yamaoka for retroviral vectors and Plat-E cells, Sumio Sugano for the pEF321-T plasmid, Akiko Kuma, Hideaki Morishita, and Keiko Igarashi for technical instruction, and Rieko Asai and Hiroki Kurihara for technical instruction and helpful discussion.
Funding Statement
This work was supported by the Funding Program for Next Generation World-Leading Researchers (LS043), JSPS KAKENHI grants-in-aid for scientific research on innovative areas (grant number 25111005 to Noboru Mizushima), and a grant-in-aid for JSPS fellows (grant number 23*4974 to Takeshi Kaizuka).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01005-15.
REFERENCES
- 1.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tooze SA, Yoshimori T. 2010. The origin of the autophagosomal membrane. Nat Cell Biol 12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Cecconi F, Levine B. 2008. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N. 2010. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Wong PM, Puente C, Ganley IG, Jiang X. 2013. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy 9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch-Day MA, Klionsky DJ. 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett 584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheong H, Nair U, Geng J, Klionsky DJ. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 19:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan EYW, Longatti A, McKnight NC, Tooze SA. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol Cell Biol 29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1-ATG13-FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer CA, Kaliappan A, Dennis PB. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 19.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. 2009. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 20.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC-mTOR signaling pathways. J Cell Biol 175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malhotra R, Warne JP, Salas E, Xu AW, Debnath J. 2015. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy 11:145–154. doi: 10.1080/15548627.2014.998917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 27.Kojima T, Yamada T, Akaishi R, Furuta I, Saitoh T, Nakabayashi K, Nakayama KI, Nakayama K, Akira S, Minakami H. 2015. Role of the Atg9a gene in intrauterine growth and survival of fetal mice. Reprod Biol 15:131–138. doi: 10.1016/j.repbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, Lindsten T. 2014. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy 10:45–56. doi: 10.4161/auto.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan B, Guan JL. 2008. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal 20:787–794. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi JD, Ryu M, Ae Park M, Jeong G, Lee JS. 2013. FIP200 inhibits β-catenin-mediated transcription by promoting APC-independent beta-catenin ubiquitination. Oncogene 32:2421–2432. doi: 10.1038/onc.2012.262. [DOI] [PubMed] [Google Scholar]
- 31.Koinuma D, Shinozaki M, Nagano Y, Ikushima H, Horiguchi K, Goto K, Chano T, Saitoh M, Imamura T, Miyazono K, Miyazawa K. 2011. RB1CC1 positively regulates transforming growth factor-β signaling through the modulation of Arkadia E3 ubiquitin ligase activity. J Biol Chem 286:32502–32512. doi: 10.1074/jbc.M111.227561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev 8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 33.Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. 1997. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev 11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- 34.Lai T, Garriga G. 2004. The conserved kinase UNC-51 acts with VAB-8 and UNC-14 to regulate axon outgrowth in C. elegans. Development 131:5991–6000. doi: 10.1242/dev.01457. [DOI] [PubMed] [Google Scholar]
- 35.Ogura K, Goshima Y. 2006. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development 133:3441–3450. doi: 10.1242/dev.02503. [DOI] [PubMed] [Google Scholar]
- 36.Ogura K, Okada T, Mitani S, Gengyo-Ando K, Baillie DL, Kohara Y, Goshima Y. 2010. Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development 137:1657–1667. doi: 10.1242/dev.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian E, Wang F, Han J, Zhang H. 2009. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5:608–615. doi: 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- 38.Liang Q, Yang P, Tian E, Han J, Zhang H. 2012. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy 8:1426–1433. doi: 10.4161/auto.21163. [DOI] [PubMed] [Google Scholar]
- 39.Toda H, Mochizuki H, Flores R III, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. 2008. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev 22:3292–3307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochizuki H, Toda H, Ando M, Kurusu M, Tomoda T, Furukubo-Tokunaga K. 2011. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One 6:e19632. doi: 10.1371/journal.pone.0019632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahantarig A, Chadwell LV, Terrazas IB, Garcia CT, Nazarian JJ, Lee HK, Lundell MJ, Cassill JA. 2009. Molecular characterization of Pegarn: a Drosophila homolog of UNC-51 kinase. Mol Biol Rep 36:1311–1321. doi: 10.1007/s11033-008-9314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24:833–846. doi: 10.1016/S0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 43.Tomoda T, Kim JH, Zhan C, Hatten ME. 2004. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev 18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Babu JR, da Silva S, Shu Q, Graef IA, Oliver T, Tomoda T, Tani T, Wooten MW, Wang F. 2007. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci U S A 104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ota S, Hisano Y, Muraki M, Hoshijima K, Dahlem TJ, Grunwald DJ, Okada Y, Kawahara A. 2013. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 18:450–458. doi: 10.1111/gtc.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. 2011. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A 108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAlpine F, Williamson LE, Tooze SA, Chan EY. 2013. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy 9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N. 2013. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 14:284–291. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quy PN, Kuma A, Pierre P, Mizushima N. 2013. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J Biol Chem 288:1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 52.Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. 2012. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci 125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- 53.Itakura E, Kishi C, Inoue K, Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itakura E, Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. 2011. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A 108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hieke N, Loffler AS, Kaizuka T, Berleth N, Bohler P, Driessen S, Stuhldreier F, Friesen O, Assani K, Schmitz K, Peter C, Diedrich B, Dengjel J, Holland P, Simonsen A, Wesselborg S, Mizushima N, Stork B. 2015. Expression of a ULK1/2 binding-deficient ATG13 variant can partially restore autophagic activity in ATG13-deficient cells. Autophagy 11:1471–1483. doi: 10.1080/15548627.2015.1068488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. 2013. ULK1 induces autophagy by phosphorylating beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itakura E, Mizushima N. 2011. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192:17–27. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papaioannou VE, Behringer RR. 2012. Early embryonic lethality in genetically engineered mice: diagnosis and phenotypic analysis. Vet Pathol 49:64–70. doi: 10.1177/0300985810395725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watson ED, Cross JC. 2005. Development of structures and transport functions in the mouse placenta. Physiology 20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 62.Varfolomeev EE, Ashkenazi A. 2004. Tumor necrosis factor: an apoptosis JuNKie? Cell 116:491–497. doi: 10.1016/S0092-8674(04)00166-7. [DOI] [PubMed] [Google Scholar]
- 63.Oberst A, Green DR. 2011. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat Rev Mol Cell Biol 12:757–763. doi: 10.1038/nrm3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kreuz S, Siegmund D, Scheurich P, Wajant H. 2001. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol 21:3964–3973. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee E, Koo Y, Ng A, Wei Y, Luby-Phelps K, Juraszek A, Xavier RJ, Cleaver O, Levine B, Amatruda JF. 2014. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy 10:572–587. doi: 10.4161/auto.27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, Wang L, Zheng Y. 2006. Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia induction, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol Biol Cell 17:4675–4685. doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Yang L, Burns K, Kuan CY, Zheng Y. 2005. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc Natl Acad Sci U S A 102:13484–13489. doi: 10.1073/pnas.0504420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alers S, Loffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, Stork B. 2011. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 7:1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim M, Park HL, Park HW, Ro SH, Nam SG, Reed JM, Guan JL, Lee JH. 2013. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy 9:1201–1213. doi: 10.4161/auto.24811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalie E, Razi M, Tooze SA. 2013. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS One 8:e75313. doi: 10.1371/journal.pone.0075313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, Mak TW. 2000. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 12:633–642. doi: 10.1016/S1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 72.Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T, Mak TW, Wallach D, Green DR. 2012. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep 1:401–407. doi: 10.1016/j.celrep.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. 2009. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.