Abstract
Both proteolytic and nonproteolytic functions of ubiquitination are essential regulatory mechanisms for promoting DNA repair and the DNA damage response in mammalian cells. Deubiquitinating enzymes (DUBs) have emerged as key players in the maintenance of genome stability. In this minireview, we discuss the recent findings on human DUBs that participate in genome maintenance, with a focus on the role of DUBs in the modulation of DNA repair and DNA damage signaling.
DEUBIQUITINATING ENZYMES
Safeguarding the genome from genotoxic stress is critical for cell survival and for preventing various human diseases, including cancer. DNA repair or DNA damage responses are under exquisite control and must be accurately and rapidly executed when genome integrity is challenged. Understanding the molecular mechanisms and regulation of critical DNA repair and damage response factors in mammalian cells will provide insight into the pathogenesis of human diseases and aid in the development of therapeutics. Ubiquitination is a posttranslational modification event that allows for rapid and dynamic changes in a protein fate or function. The ubiquitin-proteasome system (UPS) is an essential posttranslational regulatory mechanism that permeates into diverse biological processes, including the DNA repair and damage response pathways. While the role of ubiquitin in mediating controlled degradation/proteolysis of specific proteins is well established, nonproteolytic functions of ubiquitination have also emerged as important players in signaling pathways that often exist in parallel with the UPS. Deubiquitinating enzymes (DUBs), responsible for reversing the ubiquitination reaction by removing covalently attached ubiquitin molecules from substrates or polyubiquitinated chains, have recently exploded onto the ubiquitin field as key regulators of both the UPS and of the nonproteolytic functions of ubiquitination. Thus, DUBs exert profound influence on many cellular pathways, including one of the best-studied biological processes involving DNA repair and damage response in mammalian cells.
There are approximately 95 DUBs encoded by the human genome, which fall into one of the five subclasses: ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), Machado-Joseph domain-containing proteins (MJDs), Otubain domain-containing proteases (OTUs), and JAMM (JAB1/MPN/Mov34) proteases (1, 2). Ubiquitin has seven internal lysine residues (Lys6, -11, -27, -29, -33, -48, and -63), each providing sites for the generation of an isopeptide bond with the carboxy terminus of another ubiquitin to form ubiquitin polymers, otherwise known as polyubiquitination. Linear ubiquitin chains can also be generated when the amino-terminal methionine (Met) of ubiquitin (Met1) forms a peptide bond with the carboxy-terminal glycine (Gly) of ubiquitin. As different types of polyubiquitin polymers adopt different conformations (3), it is not surprising that many DUBs show some degree of specificity toward differentially linked polyubiquitination, each of which results in a different physiological outcome. Some DUBs cleave proteasome-targeting K48 or K11 chains (Ub linkages occur via Lys48 or Lys11 residues within ubiquitin itself), thus protecting the substrates from degradation, whereas some DUBs preferentially cleave nondegradative K63-linked chains or monoubiquitinated substrates that usually serve as altered binding or signaling platforms. While many USP family members are promiscuous (4, 5), most OTU or JAMM protease members show inherent specificity, preferring one or a few defined subsets of linkage types. For example, OTUB1 preferentially cleaves K48-linked chains, and Cezanne cleaves K11-linked chains (5), while the JAMM proteases, such as AMSH or BRCC36, preferentially cleave K63-linked substrates (6, 7). Some OTU members such as OTUB1 and OTUD4 act noncatalytically, providing examples of how these enzymes have diverse modes of action.
Consistent with the findings that factors involved in protein ubiquitination (E1, E2, and E3 enzymes) are integral in modulating the DNA damage response and repair (8), increasing evidence demonstrates the critical role of DUBs in these processes as well. In particular, there has been a plethora of discoveries on DUBs regulating the DNA damage response and repair of DNA double-strand breaks (DSBs). In this minireview, we summarize and discuss the known roles of these DUBs in dictating DSB repair (see Table 1 and Fig. 1). We will also discuss the DUBs involved in the repair of other forms of DNA damage (see Table 1 and Fig. 2). Due to the focused nature of this minireview, we will not discuss the DUBs that regulate the cell cycle checkpoints such as p53 regulation or other chromatin-associated events.
TABLE 1.
Types and functions of DUBs that regulate various repair pathways
| DUB | Ub chain type specificitya | Function/targets | Recruitment mechanism | Functional consequence(s) to DSB repair upon genetic ablationb | Reference(s) |
|---|---|---|---|---|---|
| In DSB repair | To DSB sites | ||||
| BRCC36 | K63 | Removes K63-Ub conjugates at DSBs | Part of BRCA-1 complex | Increased HR repair, decreased NHEJ repair | 58–62 |
| POH1 | K63-proximal Ub on substrates | Removes K63-Ub conjugates at DSBs | Part of the 19S lid complex of the proteasome | Decreased HR repair | 64–71 |
| USP3 | Mono-Ub K63? | Removes mono-Ub or possibly K63 chains from H2A and H2AX | Unknown. Forms foci at DSBs | DSB repair defect, increased ϒH2AX and 53BP1 foci, slow replication | 75, 80, 81 |
| OTUB1 | K48, noncatalytic | Inhibits UBC13 E3 enzyme | Directly binds UBC13 | Overexpression suppresses HR repair. Knockdown increases HR repair | 5, 100, 101 |
| OTUB2 | K63 K48? | Removes K63-linked chains at DSB sites. Deubiquitinates LSMBTL1 for removal from chromatin. | Unknown | Increased HR repair, decreased NHEJ repair | 5, 76, 77 |
| USP34 | K48? | Deubiquitinates and stabilizes RNF168 | Directly binds RNF168 | Decreased HR repair; IR sensitivity | 104 |
| USP7 | K48? | Deubiquitinates and stabilizes RNF168 | Binds RNF168 | Reduces IR-induced ubiquitin and BRCA1 foci | 103 |
| USP5 | K63? unanchored | Required for removal of polyubiquitin at DSBs | Unknown | Decrease in HR and NHEJ repair | 72–75 |
| USP44 | Mono-Ub K63? K48? | Removes H2A mono-Ub or possibly K63-chains on H2A | Unknown | Overexpression reduces IR-induced 53BP1 foci | 82, 86 |
| USP11 | Monoubiquitin-SUMO hybrid, K48? | Deubiquitinates H2AX-Ub and BRCA2 | Binds H2AX, binds RNF4 | Decreased HR repair, sensitivity to IR and PARPi | 75, 91–94 |
| USP26 | Mono-Ub? | Deubiquitinates H2A-Ub and H2AX-Ub? | Unknown | DSB repair defect | 75, 90 |
| USP37 | Mono-Ub? | Deubiquitinates H2A-Ub and H2AX-Ub? | Unknown | DSB repair defect | 90 |
| Dub3 | Mono-Ub | Deubiquitinates H2AX-Ub | Binds H2AX | Overexpression prolongs ϒH2AX foci | 84 |
| USP4 | K63, various | Autodeubiquitinates to modulate interaction with CtIP and MRN | Interacts with CtIP and NBS1 | Decreased HR repair, sensitive to IR | 148, 149, 151 |
| UCHL5 | K48? | Deubiquitinates and stabilizes NFRKB | Part of INO80 complex? | Decreased HR repair | 75 |
| BAP1 | Mono-Ub | Deubiquitinates H2A-Ub and H2AX-Ub | Recruitment dependent on PARP. Associates with dPRC1 and H2A-Ub? | Decreased HR repair, reduced BRCA1 and RAD51 foci, IR sensitivity | 115–119 |
| USP16 | Mono-Ub | Deubiquitinates H2A-Ub | Binds H2A | Defects in DSB-associated transcriptional repression | 120, 239 |
| In replication-associated repair | To replication fork lesions | ||||
| USP1 | Mono K48 | Removes monoubiquitination of FANCD2, FANCI, and PCNA | SUMO-like delivery system via binding to UAF1 | ICL repair defects, increased UV-induced mutagenesis, replication fork slowdown | 157, 158, 171–175, 178, 183 |
| BAP1 | K48? | Stabilizes INO80 | Associates with INO80, H2A-Ub | Replication fork slowdown | 197 |
| USP7 | K48? | Deubiquitinates and stabilizes RAD18, Pol eta | Associates with Rad18 | Defect in TLS for UV lesions | 186, 187 |
| In NER | To replication fork lesions | ||||
| USP7 | K48? K63? | Stabilizes CSB and XPC7 | Binds UVSSA, binds XPC7 | Impairs CPD, thymidine dimer repair. Reduced RNA synthesis recovery from UV damage. UV sensitivity | 204–206 |
| USP24 | K48? | Stabilizes DDB2 | Binds DDB2 | UV sensitivity | 212, 213 |
| In DNA alkylation repair | To SSBs | ||||
| OTUD4 | Noncatalytic | Functions as a platform for recruiting USP9x and USP7. Stabilizes ALKBH2 and ALKBH3 | Interacts with ALKBH | Sensitivity to alkylating agent MMS | 229 |
| USP7 | K48 | Deubiquitinates ALKBH2 and ALKBH3 | Interacts with OTUD4 | Sensitivity to alkylating agent MMS | 229 |
| USP9x | K48 | Deubiquitinates ALKBH2 and ALKBH3 | Interacts with OTUD4 | Sensitivity to alkylating agent MMS | 229 |
| USP47 | K48? | Deubiquitinates Polβ | Unknown | Sensitivity to H2O2 | 223 |
| USP7S | K48? | Deubiquitinases Mdm2 and Mule | Interacts with Mule | Unknown | 224 |
A question mark indicates that it is undefined but is only purported.
PARPi, poly(ADP-ribose) polymerase inhibitor; CPD, cyclobutane pyrimidine dimer.
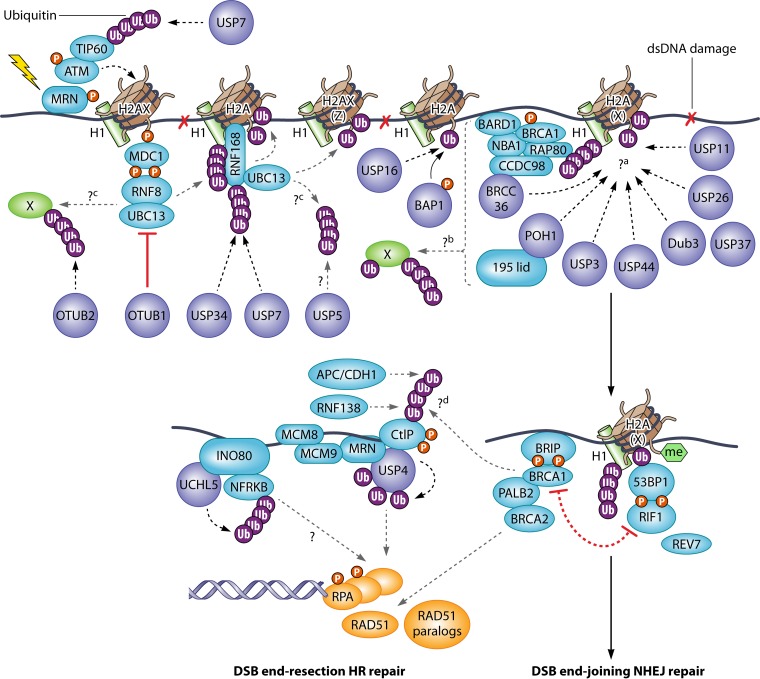
FIG 1.
DUBs that regulate ubiquitin-dependent signaling, leading to DSB repair. The ubiquitination events upon DSB generation are largely mediated by RNF8 and RNF168 on histones. H1, H2A, H2AX, and H2AZ are known to be ubiquitinated by these ligases. Different DUBs counteract these ubiquitinating events by different mechanisms, and some of them appear to act redundantly. The ubiquitination output is read by downstream factors, such as 53BP1, which functionally antagonize HR factors, such as BRCA1, and regulate the choice of DSB repair pathways. BAP1 and USP16 deubiquitinate H2A-Ub (K119; induced by the Polycomb E3 ligase complex). BAP1 regulates HR repair, and USP16 regulates DSB-induced transcriptional silencing (120, 239). Ubiquitination events mediated by other E3 ligases, such as RNF4, RNF138 (ubiquitinating Ku [240]), and HERC2, or other types of modification (e.g., SUMO or PAR) are not shown for simplicity. ?a, for the most part, it is not clear whether these DUBs cleave multimonoubiquitinated forms or polyubiquitinated forms on H2A or H2AX. It is not yet known whether some of these DUBs cleave the K63-linked polyubiquitinated H1. RNF168-induced ubiquitination on H2AZ (241) may also be a substrate. Some DUBs may also act to cleave the RNF168-induced K27-linked chains on H2A(X). ?b, these DUBs may also target other unknown ubiquitinated proteins participating in DSB repair signaling. ?c, RNF8 or RNF168 may also have other substrates, or it is possible that unanchored polyubiquitin chains nearby DSB sites are produced. ?d, both proteolytic and nonproteolytic forms of ubiquitination of CtIP by multiple E3 ligases were reported. The identities of deubiquitinating enzymes that counteract these processes are unknown.
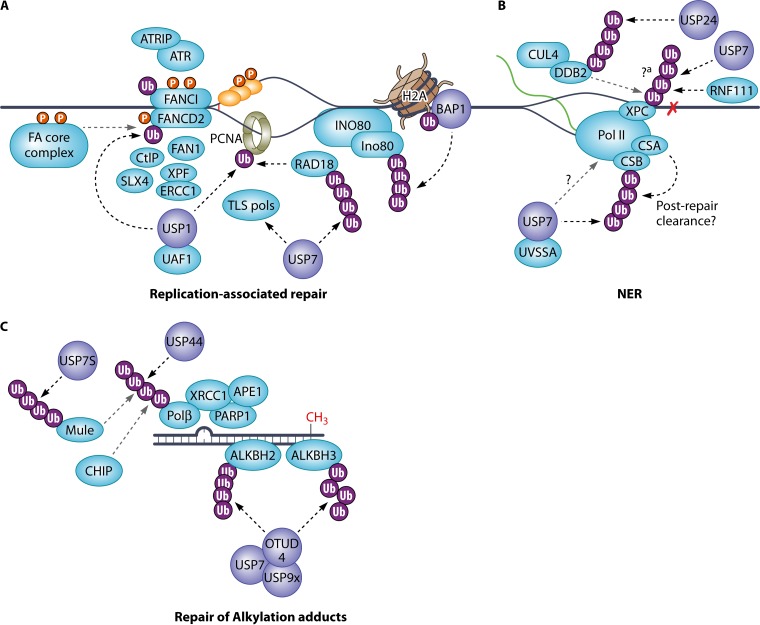
FIG 2.
DUBs that regulate replication-associated repair (A), nucleotide excision repair (B), and alkylation adducts repair (C).?a indicates that both proteolytic and nonproteolytic ubiquitination are reported. RNF111 induces K63-linked polyubiquitin chains on SUMO-modified XPC (not shown). RNF111 and CUL4-DDB2 possibly have opposing roles in the chromatin recruitment of XPC but collaboratively regulate the timely recruitment and removal of XPC at the lesions. As USP7 is not known to cleave K63 linkages, there may be more than one deubiquitinating enzymes involved.
DUBs THAT MODULATE DSB REPAIR SIGNALING
Introduction to ubiquitin-mediated signaling at DSB sites.
DNA double-strand breaks (DSBs) are lethal lesions that must be repaired before cell division ensues. Homologous recombination (HR) repair and nonhomologous end joining (NHEJ) repair represent two major forms of DSB repair mechanisms. HR repair operates by duplicating genetic information from opposite sister chromatids and is thus generally considered error-free, whereas NHEJ repair operates by ligating two broken ends with potential mutagenic events arising in the process (9). Mounting evidence suggests that the appropriate choice between HR and NHEJ repair is crucial in preserving genome integrity. One of the key events in initiating HR repair is chromatin loading of RAD51, a single-stranded DNA (ssDNA)-binding protein that facilitates a homology search in the sister chromatid to copy the lost genetic materials. ssDNA loading or recruitment of RAD51 to the DSB lesion requires highly regulated signaling cascades that involve numerous DNA damage response (DDR) factors (10). Among several posttranslational modifications, ubiquitination plays a key role in the recruitment processes (Fig. 1). A key initial signaling event in DSB repair is the phosphorylation of histone variant H2AX by ATM (ataxia and telangiectasia mutated), which then recruits downstream factor MDC1 which binds to phosphorylated H2AX (11, 12). Subsequent phosphorylation of MDC1 by ATM recruits the two RING E3 ubiquitin ligases RNF8 and RNF168 in a sequential manner (13–18). The current model involves RNF8 recruitment to the sites of chromatin damage by binding of its FHA (forkhead associated) domain to the phosphorylated MDC1; RNF8 then induces K63-linked polyubiquitination (polyubiquitinated chains linked through the K63 residue of ubiquitin) on histone H1, which generates a binding site for RNF168 (19). The initial recruitment of RNF168 requires a MIU (motif interacting with ubiquitin) ubiquitin binding motif (binds K48- or K63-linked polyubiquitin [20, 21]), which binds the K63 chain of H1 (19). The RNF168 MIU motif is embedded in surrounding motifs that contain specificity determinants termed LR motifs (22). The recruited RNF168 may further induce K63-linked polyubiquitination on the K15 residue of core histone H2A, as suggested by an in vitro reconstitution study (23). The ubiquitin and acidic patches of H2A may further enhance the recruitment of RNF168 to DSB sites to propagate the signal (19, 23–25). It is possible that RNF8 or RNF168 generates ubiquitination on yet-to-be identified targets at DSB sites (24). A proteomic screen found that RNF168 is also recruited to replication fork-stalled lesions and regulates the recruitment of the SMC6/6 complex (26). The K63-linked polyubiquitination (hereafter referred to as “K63 chains”) by RNF8 and RNF168 is catalyzed by the UBC13 E2 ubiquitin-conjugating enzyme, which is known to form a heterodimer with ubiquitin-conjugating enzyme variant (UEV1a) and assembles the nonproteolytic K63 chains (27). Consistent with the role of UBC13 in K63 chain formation, UBC13-knockout DT40 cells are impaired in the DSB-induced nuclear focus formation of stable polyubiquitin conjugates (as detected by the FK2 polyubiquitin-specific antibody) as well as downstream DNA repair factors such as RAD51 (13, 28). Consequently, the UBC13-depleted cells display increased genome instability and are hypersensitive to DSB inducers, suggesting HR repair defects (28). In addition to the primary role of K63 chains, a recent study also showed that noncanonical K27 chains synthesized on H2A by RNF168 are critical for the DNA damage response (29). One of the key factors immediately recruited to the K63 chains generated at DSB sites is RAP80, a BRCA1-associated factor that contains tandem K63 chain recognizing UIM (ubiquitin-interacting motif) domains (30–33). RAP80-knockout mice are hypersensitive to ionizing irradiation and prone to developing lymphomas, and the cells display prolonged ubiquitin foci at DSB sites (34). RAP80 serves as a scaffold to facilitate recruitment of several factors to DSB sites, including BRCA1. BRCA1, a RING E3 ligase, performs multiple functions in promoting HR repair: it interacts with BRCA2-PALB2 to recruit RAD51 to chromatin (35, 36), and it promotes DSB end resection to generate ssDNA, a key HR-initiating event (37). The role of RAP80 at DSB sites is suggested to sequester BRCA1 from functioning as a HR-promoting factor. The results that RAP80 depletion elevates DSB end resection, RAD51 loading, and HR repair support this model (38, 39). This mechanism may restrict excessive or aberrant HR repair, which can lead to genomic instability. Another key function of RAP80 is to recruit BRCC36, a K63 chain-specific DUB. As we will discuss below, BRCC36 antagonizes K63 chain formation at DSB sites, thereby regulating NHEJ repair. How do K63 chains at DSB sites promote NHEJ repair? A key factor recruited to the DSB sites is 53BP1, which possesses a TUDOR domain (protein structural motif originally identified as a region of 50 amino acids found in the Tudor protein encoded by genes in Drosophila melanogaster that binds methylated histone H3 (40), and an ubiquitination-dependent recruitment (UDR) motif, which enables 53BP1 to interact with the K15-ubiquitinated H2A generated by RNF168 (41). The positioned 53BP1 facilitates NHEJ repair by multiple mechanisms: by suppressing BRCA1 recruitment and subsequent DNA end resection (9, 37, 42, 43), by recruiting anti-DNA end resection factors RIF1 (44–49) and REV7 (50, 51), and by recruiting Artemis nuclease through PTIP (52). Excess RNF168 or 53BP1 increases mutagenic, but not physiological, NHEJ repair (53). The proper regulation of the K63 chain formation and the subsequent recruitment of HR or NHEJ factors are critical in dictating DNA repair and cellular survival in response to DNA-damaging agents. Determining the pathway choice between HR and NHEJ in a given cell cycle stage bears significant biological importance, as activation of an inappropriate repair pathway(s) can lead to DNA mutation, chromosome translocation, and overall genome instability.
Increasing evidence demonstrates the critical role of deubiquitinating enzymes in mediating the aforementioned ubiquitin-mediated DSB repair signaling. As we will discuss, much effort has been put into elucidating how the ubiquitination-dependent DNA repair signaling is negatively or positively regulated by DUBs. Below we discuss the current information on the DUBs that regulate ubiquitin-mediated signaling in the regulation of DNA repair (Table 1 and Fig. 1).
DUBs that modulate the outcome of double-strand break repair. (i) BRCC36 limits K63 chains at DSB sites.
BRCC36 is a K63 linkage-specific DUB that belongs to the metalloprotease family containing the JAMM/MPN+ catalytic domain, unlike the rest of the other DUB family members that are cysteine (Cys)-based proteases with a Cys active site as its catalytic residue (54). A role for BRCC36 in DSB repair was initially suggested by localization studies that found BRCC36 in nuclear foci at DSB sites (or IRIF) and that the localization was dependent on RAP80 (30). Further studies found that BRCC36 forms two distinct subcomplexes: (i) a nuclear “BRCA1-A” complex, which contains BRCC36, BRCA1, BARD1, RAP80, NBA1/MERIT40, BRE/BRCC45, and CCDC98/Abraxas (55–60), and (ii) the cytoplasmic BRISC (BRCC36 isopeptidase complex) complex, which contains BRCC36, KIAA0157/Abro, BRE/BRCC45, and MERIT40/NBA1 (61). A key role of the BRCA1-A complex is to target or sequester BRCA1 to DSB sites to facilitate DNA repair and to regulate the cell cycle checkpoint (30–32). In addition, recruiting BRCC36 to the DSB site as part of the BRCA1-A complex appears to be a crucial event as well. Knockdown of BRCC36 or overexpression of a catalytically inactive form of BRCC36 increases spontaneous ubiquitin foci at DSB sites, which suggests that BRCC36 suppresses the ubiquitination. The study also showed that BRCC36 knockdown restores the ubiquitin foci in RNF8 knockdown cells (RNF8 is believed to be the main ligase for K63 chains at DSB sites), suggesting that BRCC36 antagonizes the RNF8-mediated K63 chain formation (62). Consistent with the restoration of ubiquitin foci, BRCC36 knockdown can restore 53BP1 focus formation and partially restore the ionizing radiation (IR) resistance of RNF8-depleted cells. Thus, BRCC36 serves as a negative regulator that counteracts the K63 chain formation at DSB sites to balance DNA repair. Consistent with this model, BRCC36 can suppress RNF168-mediated chromosome end-to-end fusions via the 53BP1 signaling axis (63), further underscoring that a physiological function of BRCC36 is to limit K63 chains during the duration of DNA repair, which otherwise would promote unnecessary NHEJ events. Consistent with this finding, the formation of excess K63-linked chains upon BRCC36 knockdown is associated with increased DNA end resection, RAD51 loading, HR repair, and IR sensitivity, further suggesting that stabilized K63 linkages lead to an unproductive repair outcome (38, 39). Interestingly, depletion of BRCC36 leads to elevated recruitment of the anti-HR repair factor 53BP1 to chromatin-associated K63 chains, yet these cells still experience a hyper-HR phenotype. One possibility is that 53BP1 recruited to the stabilized K63 chains is not sufficient to suppress HR repair (or enhance NHEJ repair). What is clear is that, based upon increased chromosomal translocation and IR sensitivity phenotype, the increased K63 chains in BRCC36-deficient cells appear to be incapable of supporting DSB repair. Altogether, BRCC36 appears to play a critical role in suppressing excessive K63 chains at DSB sites to balance the signals leading to conditions that are conducive for both HR and NHEJ DNA repair activities.
(ii) POH1 limits excessive NHEJ repair and promotes HR repair.
POH1 (pad one homolog; also called PSMD14, or Rpn11 in Saccharomyces cerevisiae), similar to BRCC36, also possesses a JAMM/MPN+ type metalloprotease activity (64, 65). POH1 is an integral component of the 19S regulatory lid particle of the 26S proteasome. POH1 was suggested to deubiquitinate proteasomal substrates prior to feeding them through the 20S cylindrical core for degradation (64, 65). In addition to disassembling the K48 chains of proteasomal substrates, POH1 can also disassemble K63-linked polyubiquitinated chains (54). Although the exact physiological function and substrates of POH1 are unclear, one purpose of the K63 chain disassembly may be to regulate the DSB repair signaling; similar to BRCC36, POH1 acts to limit the IR-induced formation of K63 chains at DSB sites (66, 67). The diameter of the nuclear foci for ubiquitin (antiubiquitin FK2 and K63-specific antibody) and 53BP1 are larger upon POH1 depletion, suggesting that the DSB repair signal spreads in the absence of the DUB (66). As depletion of RNF8 reduces 53BP1 nuclear foci and NHEJ repair activity, the concomitant reduction of both POH1 and RNF8 can restore the effects of RNF8 partial deficiency. This suggests that POH1 antagonizes the action of RNF8 in K63 chain formation and NHEJ repair (66). Importantly, however, POH1 knockdown in healthy cells does not elevate NHEJ repair activity, again implying that excessive 53BP1 recruitment does not lead to a more functional NHEJ (66). POH1 knockdown also reduces RAD51 focus formation and HR repair activity and sensitizes cells to various DNA-damaging agents via a 53BP1-independent mechanism (66). Although it is not clear how POH1 promotes HR repair, a three-dimensional (3D) imaging study showed that POH1 cooperates with BRCA1 to eliminate 53BP1 from a nuclear focus core and to allow repositioning of replication protein A (RPA) (replication protein A binds to single-stranded DNA) in the focus core, potentially indicating an HR-initiating event (67). Consistent with POH1's role in DNA end resection, POH1-depleted cells lack RPA phosphorylation (a marker for the initiating events of DNA end resection for HR repair). Thus, POH1 appears to act as a decision maker between HR and NHEJ repair by modulating the DNA end resection events. Altogether, these results suggest that POH1 antagonizes NHEJ activity by limiting the abundance of K63 chains, and perhaps repositioning 53BP1 at the DSB sites, to promote HR repair. Why and how is POH1 localized to such sites to regulate DSB repair signaling? Insight could potentially be derived from a structural study of yeast Rpn11 (yeast homolog for POH1) (68). First, the study found that Rpn11 is an unstable protein without its neighboring subunit Rpn8 and that the Rpn8-Rpn11 heterodimer is catalytically active in vitro. This suggests that POH1 may also need to be in complex with its neighboring 19S subunits for activity and that an intact 19S lid complex may be localized to DSB sites as whole, along with POH1. Indeed, PSMC5, a 19S subunit, is observed in the DSB sites (66), and proteasomal subunits have been found to localize to DSB sites using chromatin immunoprecipitation (ChIP) studies in yeast (69). The mechanism by which the 19S lid complex is recruited to the DSB sites is unknown. The N terminus of POH1 has been shown to interact with a transcription factor, Mitf, to regulate its activity (70), which provides an example pf how POH1 can be localized to specific sites in chromatin. Second, the study found that the purified Rpn11-Rpn8 complex lacks specificity toward different ubiquitin chain types tested, which is supported by the fact that Rpn11, unlike other MPN family DUBs, lacks a conserved binding surface for the Ile44 residue on ubiquitin that helps to properly orient the ubiquitin polymer for linkage-specific cleavage. This suggests that Rpn11 is not likely to cleave internal linkages within the K63 chains and that it may instead cleave the first “proximal” ubiquitin attached to substrates (71). This feature of cleaving polyubiquitin chains en bloc off various proteasomal substrates is consistent with a previous study on Rpn11 (65). Perhaps the ability of Rpn11 to cleave a bulk polyubiquitin chain at the proteasome lid suggests that it can perform similarly at the DSB sites; it is possible that POH1 also cleaves the ubiquitin chains en bloc off histones or other unknown substrates at DSB sites, which may rapidly neutralize ongoing DSB repair signaling. This activity may generate free unanchored K63 chains that can be disassembled by other K63 linkage-specific DUBs to completely turn off the signaling pathways. UBP14, a yeast homolog of USP5, is largely responsible for disassembling unanchored free polyubiquitin chains in cells (72). Interestingly, USP5 is localized to DSB sites, and the polyubiquitin chains at DSB sites are sustained longer in USP5-depleted cells (73). USP5 may indeed disassemble any unanchored K63 chains at DSB sites, as a structural study showed that a ZnF-UBP (zinc finger ubiquitin binding domain) domain of USP5 recognizes the C-terminal “Gly-Gly” motif of unanchored ubiquitin, rather than the Ile44 residue of ubiquitin which most DUBs recognize (74). Indeed, mutation in the domain that recognizes the free C terminus of ubiquitin inactivated the polyubiquitin-disassembling activity of USP5. Thus, it is possible that sequential activities of POH1 and USP5 collaborate to turn off DSB repair signaling. The possible role of USP5 in the DSB repair is also supported by a RNA interference (RNAi) screen that found USP5 localized at DSB sites and that USP5-depleted cells exhibit repair defects (75).
(iii) RNAi screen identifies that OTUB2 antagonizes RNF8-mediated K63 linkages.
A small interfering RNA (siRNA) screen identified that the K63-ubiquitin conjugates at DSB sites are further antagonized by OTUB2, a member of the OTU family of DUBs (76). OTUB2 depletion spontaneously increases focus formation of ubiquitin, 53BP1, RAP80, and RNF168 at DSB sites. While overexpressing OTUB2 eliminated RAP80, RNF168, and ubiquitin foci, OTUB2 did not affect the focus formation of their upstream factors (MDC1 or RNF8); this suggests that OTUB2 inhibits the RNF8 activity or certain downstream functions of RNF8 that are required for RNF168 recruitment. OTUB2 can cleave UBC13/MMS2-induced free K63-linked polyubiquitin conjugates in vitro, supporting the fact that OTUB2 acts to antagonize the K63 chains at DSB sites. Purified OTUB2 indeed shows a preference for cleaving the K63-linked diubiquitin substrates, with some activity toward K48 and K11 linkages (5) (although OTUB2 can cleave both K48- and K63-linked tetraubiquitin substrates as well [77]). As Mattiroli et al. suggested, RNF8/UBC13-mediated K63 chain formation precedes the RNF168 recruitment to nucleosomes (24), and OTUB2 may act to suppress K63 chain formation to inhibit RNF168 recruitment. Consistent with the notion that RAP80 and 53BP1 suppress HR-initiating events such as DNA end resection and RPA foci, HR repair activity is decreased upon OTUB2 depletion. Coknockdown of 53BP1 rescued RAD51 and RPA focus formation in OTUB2 knockdown cells, which suggested that increased 53BP1 is responsible for the suppression of end resection and HR repair. Surprisingly, overexpressing OTUB2 did not reduce the level of multi- or polyubiquitinated H2A (OTUB1 [discussed below] overexpression had the opposite effect). This raises the possibility that there is yet-to-be appreciated complexity among the H2A-ubiquitin types that differentially recruit downstream repair factors. Perhaps related to this point is that BRCC36 and OTUB2, while both antagonize K63 chain formation at DSB sites, lead to very different outcomes: BRCC36 depletion elevates HR repair, whereas OTUB2 depletion suppress HR repair. It may be that recruitment and formation of HR-suppressive RNF168-RAP80-53BP1 components are differently configured depending on the differential actions of DUBs at DSB sites. Alternatively, it is possible that BRCC36 or OTUB2 have additional functions in regulating DNA repair other than modulating the K63 chains. The Kato et al. study (76) found that OTUB2 deubiquitinates LSMBT1, an event which may promote the chromatin loading of 53BP1 recruitment.
(iv) USP3 deubiquitinates H2AX and suppresses genome instability.
USP3 is a homolog of Saccharomyces cerevisiae Ubp8, which deubiquitinates H2B (78, 79). Consistent with its role in chromatin modification, USP3 is localized to the nucleus and chromatin-enriched fractions (80). Ectopic expression of USP3, not the catalytically inactive mutant (active site Cys mutant), leads to a reduction in mono- and diubiquitinated H2A and H2B, whereas knockdown of USP3 increases the ubiquitinated histones (80). The persistent ubiquitination induced by USP3 knockdown is associated with persistent H2A-Ub and H2AX foci. As the authors suggested, this may be due to a defect in attenuation of DDR that is dependent on histone ubiquitination or other functions of USP3 that may be related to DNA replication or damage response.
More recent work further demonstrated that USP3 deubiquitinates H2AX and H2A (81). Knockdown of USP3 increases staining intensity of ubiquitin foci, and overexpressing wild-type USP3 but not the catalytic inactive mutant suppresses UV-induced monoubiquitination of H2A/H2AX and focus formation of BRCA1 and 53BP1. The reduction of H2A-Ub was observed with the FLAG-H2A K118-119R mutant but not with the FLAG-H2A K13-15R mutant, suggesting that USP3 cleaves ubiquitin conjugated to K13-15 residues. Therefore, it was suggested that USP3 counteracts the RNF168-mediated K13-15 ubiquitination on H2A. It is possible that USP3 also regulates H2B-ubiquitin, similar to its yeast homolog Ubp8, which is based upon the reduction of H2B-ubiquitin upon USP3 overexpression observed in another study (82). The in vivo significance of USP3 function was demonstrated by studies with mice deficient in Usp3 (usp3Δ/Δ [83]). The cells from usp3 knockout mice display an increased level of H2A-Ub, and spontaneous focus formation of Ub chains (FK2), 53BP1, and γ-H2AX, suggesting that the DSB signaling is increased and genome integrity is compromised. USP3 depletion causes defects in hematopoietic stem cell homeostasis, and interestingly, the usp3 knockout mice spontaneously developed tumors, which can be attributed to impaired genome integrity and underscores the importance of ubiquitin-mediated signaling in DNA repair.
(v) Overexpression screening studies identify various DUBs as inhibitors of the DSB signaling.
Several studies used human DUB overexpression libraries to identify DUBs that regulate the ubiquitin and DNA damage signaling, as we discuss in this subsection.
In a study that used H2AX monoubiquitination Western blotting as a readout, Dub3 (also known as USP17L2) was found as a major hit (84). Overexpression of Dub3 but not its catalytic inactive mutant reduces the monoubiquitinated forms of H2AX and H2A. The study convincingly showed that Dub3 physically binds to H2AX and that purified Dub3 can deubiquitinate the monoubiquitinated H2AX in vitro, suggesting that the overexpression effect is direct. Overexpressing Dub3 abrogates the damage-induced focus formation of 53BP1, BRCA1, and RNF168 but not H2AX or MDC1, suggesting that the Dub3-induced deubiquitination of H2AX influences canonical DSB signaling. This result is consistent with a finding that 53BP1 is a direct reader of H2A-monoubiquitin (41). Excessive deubiquitination of H2A or H2AX by overexpressing Dub3 resulted in sustained activation of H2AX and MDC1, suggesting that overall DSB signaling is impaired upon Dub3 overexpression.
In a study that used 53BP1 chromatin foci as a screening readout, USP44 was identified as an antagonist of RNF168-mediated Ub conjugates at DSB sites (82). Overexpression of USP44 inhibits focus formation of IR-induced Ub conjugates (FK2), RNF168, RAP80, and 53BP1 at DSB sites. Similar to USP3, USP44 overexpression effectively reduced mono- and multiubiquitinated H2A (and H2B-Ub to a lesser degree). USP44 is recruited to the IR-induced DSB sites, a process dependent on RNF8 and RNF168, suggesting that USP44 recruitment depends on Ub conjugate formation and that USP44 antagonizes IR- and RNF168-induced H2A ubiquitination; whether USP44 specifically cleaves H2AK119-Ub or K13/K15-Ub is not known. The study found only a mild increase in the Ub conjugates and 53BP1 foci upon individual knockdown of USP3, USP44, or OTUB1. This result suggests that these DUBs function redundantly or synergistically and that there is not a single DUB that is solely responsible for removing the bulk ubiquitin conjugates at DSB sites. It was also noted that simultaneous knockdown of these DUBs did not result in further increase in focus formation (82). The reduction in ubiquitinated H2B upon overexpressing USP44 in the study is consistent with another study that links USP44 in deubiquitinating H2B and stem cell differentiation (85).
USP44 also plays an important role during cell division. USP44 inhibits premature activation of APC/C E3 ligase by stabilizing the APC-inhibitory MAD2-CDC20 complex (86) and regulates proper separation and positioning of centrosomes—failure of which can lead to missegregation of sister chromatids during anaphase (87). Indeed, mouse embryo fibroblast (MEF) cells from the USP44 knockout mice display aneuploidy, and the mice are prone to tumor development (87), and overexpressing USP44 also induces chromosome segregation errors and aneuploidy (88). Interestingly, it was found that USP44 is localized to centrosomes via specific interaction with centrin and that wild-type USP44, but not the centrosome localization-defective point mutant or the catalytically inactive mutant, can rescue the chromosome segregation defects, demonstrating that the centrosome pool of USP44 is essential in preventing chromosome missegregation (87). Although the increased aneuploidy can be a significant factor in tumor development, aneuploidy alone may not play a causative role in tumor development (89). In light of the connections between defective DNA repair and mitotic defects, it is possible that the aberrant ubiquitin signaling in DSB repair contributes to the mitotic defects and tumor development upon USP44 ablation.
Another study also used 53BP1 focus formation as a screening readout, and this study found USP26 and USP37 as major antagonists of the IR-induced 53BP1 foci (90). This study found Dub3 and USP44 in the screen, consistent with the other screenings described above. The study showed that overexpressing wild-type USP26 and USP37 but not catalytic inactive mutants of USP26 and USP37 suppressed the 53BP1 and ubiquitin foci at DSB lesions. Overexpression of USP26 significantly suppressed the RNF168-induced H2A ubiquitination, but USP37 did not have much effect. Localization of both USP26 and USP37 at DSB sites was observed, suggesting that the effects are direct. The increased ubiquitin (FK2) and 53BP1 foci upon USP26 or USP37 depletion are associated with downregulation of the HR repair; damage-induced foci of HR factors RAD51, BRCA1, and PALB2 are decreased. Interestingly, codepletion of RAP80 rescued the reduced HR foci. These results suggest that perhaps the role of RNF168-mediated ubiquitination is to control the abundance between the HR-promoting BRCA1-RAP80-PALB2 complex and the HR-suppressing “BRCA1-A complex” BRCA1-RAP80-MERIT40 (NBA1)-Abraxas (CCDC98)-BRCC36; in general, RNF168 would promote the recruitment of the latter complex. This provides another example of how the intricate balance of ubiquitination at DSB sites is important for a proper choice of DSB repair.
In another screen that used a DUB overexpression library, USP11 was shown to deubiquitinate H2AX (91). This study used ubiquitinated H2AX (induced by overexpressing RNF8) coupled with overexpressing 72 DUBs. The primary hits identified in this study included USP26, OTUB1, DUB3, in addition to USP11, validating the results from other screenings. Knockdown of USP11 increased the H2AX-Ub forms, which is consistent with the overexpression results. USP11 knockdown also leads to increases in 53BP1 and ubiquitin (FK2) formation at DSB sites and to cellular sensitization to IR, suggesting that deregulation of H2AX ubiquitination leads to improper DSB signaling and repair. USP11 physically associates with H2AX, further suggesting that H2AX is a direct substrate of USP11. Consistent with this finding, USP11 is one of the hits found to be localized at DSB sites, and cells exhibit repair defects when USP11 is depleted (75). Also consistently, earlier studies found that USP11 knockdown sensitizes cells to PARP inhibition (found in a RNAi screen) and IR (92, 93), suggesting that USP11 regulates the DSB repair. One study showed that USP11 interacts with and deubiquitinates BRCA2, possibly to regulate the proteasomal degradation of BRCA2 (92). USP11 may further regulate the DSB repair by a different mechanism. USP11 is shown to associate with RNF4, an E3 ligase that modulates SUMO-modified targets, and counteracts the RNF4-induced SUMO-ubiquitin hybrid chains (94). Since RNF4 is recruited to DSB sites and is required for DSB repair (95–99), it is possible that USP11 antagonizes some of the RNF4-directed ubiquitination at DSB sites.
(vi) Inhibition of UBC13 E2-conjugating enzyme by OTUB1.
OTUB1 is an OTU family deubiquitinating enzyme that has high specificity toward K48-linked chains (5). OTUB1 was identified as a negative regulator of RNF168-mediated Ub conjugate formation in a siRNA screen; knockdown of OTUB1 leads to spontaneous and persistent Ub conjugates and 53BP1 foci (100). Intriguingly, overexpressing a catalytically inactive mutant of OTUB1 inhibited 53BP1 focus formation as efficiently as overexpressing the wild-type enzyme, which suggested that the catalytic activity of OTUB1 is not required (although the whole OTU domain must be present), and together with the fact that OTUB1 is known to be specific for K48 linkages, it was concluded that OTUB1 may not directly deubiquitinate the K63 linkages at DSB sites. Further analysis revealed that OTUB1 physically interacts with the UBE2D/E subfamily of E2-conjugating enzymes, including UBC13 which assembles the K63 chains at DSB sites. Interestingly, OTUB1 binds preferentially to UBC13 that are charged with ubiquitin through the OTUB1's unique N-terminal extension that bears a ubiquitin binding motif (100). The interaction leads to disruption of RNF168/UBC13-mediated polyubiquitination through a mechanism independent of deubiquitinating activity. A structural study revealed that the binding of the N terminus of OTUB1 to the Ub-charged UBC13 is allosterically regulated by the binding of a free ubiquitin to a second site in OTUB1; it triggers conformational changes in the ubiquitin-binding helix in the N terminus, thereby enhancing affinity to the E2-charged ubiquitin. The interaction ultimately disrupts the ability of the E2-charged ubiquitin thioester bond to be attacked by an acceptor ubiquitin (on the substrate), which can explain the inhibition of K63 chain synthesis. Consistent with the model, OTUB1 inhibits UBC13-dependent diubiquitin synthesis but not the initial monoubiquitination (100). It was suggested that the configuration of the E2-charged ubiquitin and the free ubiquitin moieties at the OTUB1 catalytic site mimic cleaved K48 linkages, which may be a basis for OTUB1 inhibiting the E2 enzyme (101). By inhibiting UBC13 and K63 chain synthesis, OTUB1 regulates DNA repair; overexpression of OTUB1 suppresses HR repair activity, and interestingly, knockdown of OTUB1 leads to nearly complete restoration of the HR repair defect that is caused by ATM inhibition (100). OTUB1 is also capable of inhibiting other E2 enzyme family members UBE2D and UBE2E, suggesting that it also exerts other physiological functions outside the DNA repair signaling through similar mechanisms (100, 101).
OTUB1 is also subject to reciprocal regulation by an E2 enzyme UBCH5B. It was shown that UBCH5B directly binds to and stabilize the N-terminal ubiquitin-binding helix of OTUB1, thus increasing the affinity to a proximal ubiquitin moiety and stimulating the di-K48-ubiquitin chain cleavage activity (102). It is possible that the OTUB1-UBC13 interaction may be dynamically regulated during the DNA damage response.
(vii) Stabilization of RNF168 E3 enzyme by USP34 and USP7.
Two USPs have been reported to directly associate with RNF168 and prevent it from undergoing proteasome-mediated destruction (UPS pathway). RNF168 is the aforementioned E3 ubiquitin ligase that induces mono- and polyubiquitination of H2A/H2AX. USP7 (103) and USP34 (104) directly bind, deubiquitinate, and stabilize RNF168. Consistent with the role of RNF168 in DNA damage-induced formation of monoubiquitin and K63-linked polyubiquitin conjugates, depletion of USP7 or USP34 reduces the level of H2A-ubiquitin and overall Ub conjugates at DSB sites (104). Thus, USP7 and USP34 promote Ub conjugate formation at DSB sites, which in turn promotes recruitment of downstream DDR mediators 53BP1 and BRCA1, and confer resistance to DNA-damaging agents (103, 104). Maintaining the balance of RNF168 abundance at DSBs may have an important consequence, as failure to do so results in improper spreading of ubiquitin conjugates to undamaged chromatin (105). It is not known how USP7 and USP34 are recruited to the damaged sites or whether they form stable complexes with RNF168; however, USP34 forms IR-inducible foci at DSBs in a manner that is dependent on RNF168 and UBC13. This suggests that USP34 recruitment may be dependent on the local ubiquitinated proteins or direct binding to RNF168 (104). In yeast, Rsp5 HECT E3 ligase forms a stable complex with Ubp2 deubiquitinating enzyme to promote assembly and disassembly of K63-linked polyubiquitin chains on their substrates as well as on Rsp5 itself (106–108). Similarly, it is possible that USP7 or USP34 is recruited to the damaged sites by forming stable complexes with RNF168 to support the stability of RNF168. It is not known whether USP7 or USP34 regulates RNF168 in different contexts or whether they serve redundant roles. Interestingly, RNF168 is itself polyubiquitinated and destabilized upon IR treatment, which is reversed by USP34, suggesting that USP34 specifically stabilizes RNF168 when DNA becomes damaged (104). It is not clear by what mechanism RNF168 is induced for polyubiquitination and degradation, although two HECT E3 ubiquitin ligases UBR5 and TRIP12 have been shown to negatively regulate the abundance and spreading of RNF168 foci at DSB sites (105). Whether the two HECT E3 enzymes directly catalyze polyubiquitination of RNF168 or whether RNF168 self-catalyzes by coupling with K48 linkage-specific E2 enzymes is unknown. Overall, these studies indicate that maintaining the balance of RNF168 abundance at DSBs is important in promoting DSB repair signaling. USP7 is also shown to stabilize TIP60 histone acetyltransferase (109), which acetylates ATM and promotes DNA damage signaling (110); it also promotes the stability of BMI1 (103), a component of the Polycomb-repressive complex (PRC1) that induces ubiquitination of H2A at the K119 residue that induces transcriptional repression (111). BMI1 localizes to IR-induced foci and promotes DSB repair signaling (112–114). Thus, USP7 appears to exert multiple functions at chromatin to promote DSB repair signaling and perhaps organize damage-inducible local transcriptional repression.
(viii) BAP1 promotes HR repair.
BAP1 (BRCA1-associated protein 1) is a nuclear deubiquitinating enzyme with a ubiquitin carboxy-terminal hydrolase (UCH) domain. BAP1 was initially reported as a tumor suppressor that suppresses breast cancer growth in a BRCA1-dependent manner (115). Cancer-associated BAP1 mutants are linked to deficiency in deubiquitinating activity, and the catalytic mutant cannot suppress tumor growth in mice (116). Studies found that BAP1 is a regulator of DSB repair, in particular by promoting HR repair (117, 118). BAP1 is rapidly recruited to DSB sites, and BAP1 depletion leads to reduced BRCA1, RAD51, and RPA foci. Consequently, BAP1 depletion leads to reduced HR repair and sensitivity to IR. Overexpressing BAP1 reduces ubiquitinated forms of H2A and H2AX, while depletion increases them, suggesting that BAP1 is a DUB for the ubiquitinated H2A(X) (117). The ubiquitinated site for H2A(X) was suggested to be the K119 residue, and this is consistent with the fact that a BAP1 homolog in Drosophila, Calypso, is associated with PRC1 and regulates H2AK119 ubiquitination (119). Therefore, BAP1 may engage in a dynamic regulation of this modification. Whether the role of BAP1 is redundant or cooperative with USP16, another DUB that deubiquitinates H2AK119 (120), is unknown.
(ix) UCHL5 and USP4 regulate the HR repair by modulating DSB end resection.
A crucial regulatory event during the DSB repair is the generation of a stretch of single-stranded DNA (ssDNA) at the DSB sites, or so-called DSB end resection. The ssDNA generated at the DSB lesions is subsequently coated by heterotrimeric RPA complex. The coated RPA performs many important functions.
Studies with yeast showed that the coated RPAs prevent the activation of MMEJ (microhomology-mediated end joining), an error-prone form of NHEJ (121), and the formation of DNA secondary structures that induce genome instability (122). RPAs also recruit the ATR-ATRIP kinase complex (123) and activate DNA damage checkpoint and cell cycle arrest (123–125). As a prochoice mechanism for HR repair, RPA-coated ssDNA recruits RAD51, which initiates strand invasion, homology search, and subsequent HR repair (126, 127).
The DSB end resection during mitotic DSB repair is mediated by a concerted action of several nucleases. Current models suggest that the MRN (MRE11-RAD50-NBS1) complex collaborates with CtIP endonuclease (Sae2 in budding yeast, Ctp1 in fission yeast) to initially trim the DSB ends (128–131). Recruitment of the MRN complex and the end resection require the MCM8-MCM9 complex (132). Subsequent reactions requiring the nucleases Exo1, Sgs1 (BLM in human) and Dna2 function redundantly or collaboratively to induce extensive ssDNA, onto which RPA binds (131, 133–135). CtIP knockout (KO) mice is embryonic lethal, and CtIP is essential for suppressing genome aberration (136, 137). CtIP/Sae2 is also highly regulated by posttranslational modifications, including phosphorylation (138–141), acetylation (142, 143), ubiquitination (both proteolytic and nonproteolytic) (144–147), and deregulation of these processes lead to DSB repair deficiency, evidence that the DSB end resection is subject to a dynamic regulation.
Studies identified that two DUBs, UCHL5 and USP4, regulate DSB end resection. Two studies found that DNA end resection is positively regulated by USP4 in response to DNA damage (148, 149). Both studies found that USP4 interacts with the DNA end resection factor CtIP, as well as with the MRN complex, through two distinct regions within USP4. USP4 knockdown reduced the recruitment of CtIP, RPA (reduced RPA2 S4-S8 phosphorylation), and RAD51 to sites of DSBs, suggestive of impaired DNA end resection. Unexpectedly, both studies found that CtIP is not a direct ubiquitin-modified substrate of USP4; rather, USP4 is itself ubiquitinated, and its deubiquitination is dependent on its own catalytic activity. These studies found that the catalytically inactive USP4 mutant cannot efficiently interact with CtIP and MRN components compared to the wild-type form, implying that the increased ubiquitination of USP4 interferes with these interactions. Expression of the catalytic mutant forms cannot fully support HR, underscoring the significance of the autodeubiquitination activity of USP4 to promote proper CtIP interaction and localization to enable DNA end resection and HR activity. Interestingly, the Wijnhoven et al. study found that the ability of USP4 to undergo autodeubiquitination to mediate protein-protein interactions may be shared by other USPs, such as the structurally related USP11 and USP15 (149). As USP15 and USP4 may have arisen from genome duplication (150), USP15 may also participate in DSB repair. Additionally, it was purported that the zinc-coordinating Cys residues on USP4, not lysines, are the targets of ubiquitin conjugation. It is unclear whether ubiquitination of Cys residues is a prevalent mode of regulation of Cys-based DUBs in general. Although the ubiquitin linkage type targeted by USP4 for autodeubiquitination remains elusive, USP4 has been shown to deubiquitinate K63-linked proteins and regulate the spliceosome assembly (151), providing another example that USP4 functions to modulate protein interactions.
Nishi et al. (75) used a combination of green fluorescent protein (GFP) localization and RNAi screenings of the human DUBs to identify that UCHL5 (also called UCH37) is a regulator of DSB end resection. UCHL5 is localized to DSB lesions, and UCHL5 knockdown leads to increased DSBs and cellular sensitization to DSB-inducing drugs. Further analysis showed that UCHL5 knockdown selectively impaired HR repair but not NHEJ and specifically that DSB end resection was impaired upon UCHL5 knockdown. The fact that UCHL5 is a component of both the proteasome lid as well as the INO80 chromatin-remodeling complex prompted the authors to sort out which macrocomplex is involved in the end resection regulation; they found that rather than collaborating with the proteasome, UCHL5 suppresses ubiquitination and proteasomal degradation of NFRKB (nuclear factor related to kB-binding protein), a factor known to mediate the association of UCHL5 with INO80 (152). Indeed, INO80 is known to directly associate with DSB ends and is required for DSB end resection and overall DSB repair (153, 154).
DUBs THAT REGULATE DNA REPLICATION-ASSOCIATED DNA DAMAGE RESPONSE
USP1 deubiquitinates FANCD2 and FANCI to regulate DNA ICL repair.
DNA interstrand cross-links (ICLs) are complex DNA lesions that can be generated by various endogenous and exogenous sources that include aldehyde and platinum-based drugs such as cisplatin or mitomycin C. ICLs can be lethal if unrepaired, leading to blockade of DNA replication and transcription. In vertebrates, ICL repair is largely coupled to DNA replication due to the activation of a dedicated coordinator of replication-coupled ICL repair called the Fanconi anemia (FA) pathway. The FA pathway facilitates ICL repair by coordinating multiple types of repair pathways, including nucleotide excision repair (NER), homologous recombination (HR), and translesion synthesis (TLS) (155). One of the key steps that leads to FA pathway activation is the monoubiquitination of FANCD2 and FANCI proteins on Lys561 and Lys523, respectively (human sequence), which is mediated by the multisubunit FA core E3 ligase complex (156–158) (Fig. 2). Monoubiquitination of FANCD2 and FANCI peaks during S phase of the cell cycle, which is consistent with the timing of HR and TLS events in the cell. The monoubiquitinated FANCD2 performs multiple functions in coordinating the downstream repair activities by recruiting the following: (i) FAN1 and XPF-ERCC nucleases to facilitate the incision process and unhooking of the cross-linked DNA segment (159–164); (ii) TLS polymerase to facilitate replicative bypass of the unhooked ICL lesion (165); and (iii) exonuclease CtIP to induce 3′-to-5′ resection of DSB DNA ends to generate ssDNA to channel the broken DNA ends to HR-mediated repair (166–168). Although the requirement for monoubiquitinated FANCD2 is well established in replication-coupled ICL repair, the receptor that interacts specifically with the monoubiquitinated form remains controversial and has yet to be identified. The role of FANCI monoubiquitination is less well understood, but it is minimally required for the efficient monoubiquitination of FANCD2 and is thought to stabilize the FANCD2-FANCI heterodimer (157). There are also monoubiquitination-dependent and -independent functions of FANCD2 and FANCI in protection against general DNA replication stress (169, 170) that will not be discussed in this minireview due to space constraints.
USP1 was initially identified as a regulator of FANCD2 monoubiquitination in a DUB family siRNA genetic screen, which showed that depletion of USP1 increases spontaneous monoubiquitination of FANCD2 (171). A later study found that USP1 is also required for deubiquitination of FANCI (157). Intriguingly, the USP1 knockout is associated with defects in DNA repair and increased cellular sensitivity to DNA ICL-inducing agents in mice and DT40 cells, suggesting that deubiquitination of FANCD2-FANCI is also an essential event in DNA ICL repair (172, 173). How can this phenomenon be interpreted? It is speculated that perhaps FANCD2 (and FANCI) monoubiquitination is dynamically regulated by USP1 during S phase, undergoing multiple rounds of reversible monoubiquitination events. Disruption of this dynamic regulation in USP1-depleted cells led to reduced overall FANCD2 recruitment to the damaged sites (localization of FANCD2 and FANCI nuclear foci was initially thought to be monoubiquitination dependent). Thus, perhaps a role for USP1 is to regulate the turnover of FANCD2-FANCI monoubiquitination at the damaged sites to facilitate multiple rounds of repair during S phase, rather than for USP1 to act only once just as cells exit S phase for repair recovery. The fact that the USP1 level is higher while FANCD2-FANCI monoubiquitination is also elevated (S-G2 phases) further supports the idea that USP1 activity is needed for the FANCD2-mediated ICL repair (174).
How does USP1 directly deubiquitinate FANCD2 and FANCI? The ability of USP1 to gain access to FANCD2-FANCI proteins may be mediated through its WD40 repeat-containing catalytic cofactor, UAF1 (WDR48). UAF1 was identified as a stoichiometric binding partner of USP1 (175). Similar to USP1, depletion or knockout of UAF1 also results in hypermonoubiquitinated FANCD2 and FANCI (176, 177), suggesting that UAF1 is a positive regulator of USP1. Importantly, UAF1 binding to purified USP1 increases the catalytic turnover (kcat) but does not increase the affinity of USP1 for the substrate (Km) ubiquitin-7-amido-4-methylcoumarin (Ub-AMC) in an intrinsic DUB activity measurement (177). In addition to its role as a catalytic cofactor, UAF1 may also recognize and bind directly to USP1 substrates, serving as a bridge or adaptor for USP1 to recognize its ubiquitinated substrates. The carboxy terminus of UAF1 harbors SUMO-like sequences (termed the SUMO-like domain [SLD]), which can be recognized by a SUMO-interacting motif (SIM) present in FANCI (178). Thus, USP1 may be recruited to the FANCD2 and FANCI (inhibitor of DNA binding [ID]) complex through the SUMO-like delivery system between UAF1 and FANCI proteins. However, the physiological relevance of this interaction has yet to be formally demonstrated. Adding to this piece of the puzzle, UAF1 also serves as a catalytic cofactor and binding partner for two other DUBs, USP12 and USP46 (175). To date, these DUBs do not appear to function in the FA pathway. It is unclear whether these DUBs can also be recruited to FANCI via the SUMO-like delivery system on UAF1.
USP1 deubiquitinates PCNA to regulate translesion synthesis (TLS) for DNA damage tolerance.
USP1 also plays an important role in regulating TLS events in cells via the deubiquitination of PCNA (proliferating cell nuclear antigen). PCNA normally functions as a DNA replicative polymerase sliding clamp that ensures the replicative DNA polymerase's processivity during DNA synthesis. It also plays a key role in mediating DNA damage tolerance through the recruitment of the more error-prone Y-family TLS polymerases (Pols), such as Pol eta, kappa, iota, and Rev1, to allow for replicative bypass of DNA lesions during replication, otherwise known as postreplication repair. PCNA is known to be monoubiquitinated on Lys164 (conserved from yeast to mammals) by various DNA-damaging agents (hydroxyurea [HU], aphidicolin, UV, and MMS) that cause the slowing or stalling of replication forks. Interestingly, USP1 also utilizes the SUMO-like delivery system to recognize monoubiquitinated PCNA, similar to FANCD2. Instead of recognizing the SIMs on FANCI, the SLD sequence on UAF1 recognizes the SIM on ELG1, a PCNA-binding RFC-like protein which serves as a substrate adaptor for PCNA deubiquitination by the USP1-UAF1 complex (178, 179). While Y-family DNA Pol eta, kappa, and iota are recruited to PCNA through a weak PCNA-interacting protein (PIP) box, Rev1 utilizes its BRCT domain and/or its PAD domain for localization. All Y-family TLS Pols have one or two ubiquitin-binding domains (UBD), which adds to their affinity for monoubiquitinated PCNA at the sites of stalled forks. Another mechanism that facilitates TLS Pol localization to damaged DNA is through the direct recruitment to Rev1, which can serve as a scaffolding protein for the TLS Pols (180). Conversely, it has been postulated that the deubiquitination of PCNA by USP1 facilitates the removal of TLS Pols from the replication fork in exchange for the replicative Pols (Epsilon and Delta) (181).
Similar to FANCD2, elevated levels of PCNA monoubiquitination (caused by USP1 depletion in unperturbed cycling cells) can be disruptive to its normal error-free DNA replication kinetics, as excessive and/or aberrant recruitment of TLS Pols, such as Pol kappa, to the replication fork leads to reduced fork speeds and is a contributing factor to genomic instability (182). In contrast, DNA damage leads to the reduction of USP1 levels via transcriptional and posttranslational mechanisms. It was shown that UV damage causes the proteasomal degradation of USP1 (183). The degradation of USP1 is initiated through an autocleavage event, coupled to recognition by the N-end rule pathway enzymes that subsequently target it for polyubiquitination and degradation (184). How USP1 autocleavage is regulated in a signal-dependent manner is still unknown.
In summary, USP1 negatively regulates several key ubiquitination processes in response to DNA damage. In addition to the well-characterized ubiquitinated substrates described above, USP1 was more recently shown to promote the stability of inhibitors of DNA binding (IDs) (family of transcriptional repressor proteins) and rescue them from the UPS (185). The ID proteins antagonize basic-helix-loop-helix (bHLH) transcription factors to regulate differentiation and maintenance of stem cell fate. ID proteins are known to inhibit the transcription of the cyclin kinase (CDK) inhibitor, p21. p21 has a strong PIP box, and low levels of p21 are sufficient to prevent TLS onset. Whether USP1 loss can indirectly lead to upregulation of p21 transcriptional levels due to the rapid turnover of ID proteins remains to be tested. Thus, USP1 may alter S-phase DNA repair events by targeting the deubiquitination of multiple substrates, including FANCI, FANCD2, PCNA, and ID proteins. There are likely other substrates of USP1 that directly regulate DNA repair and DNA replication kinetics during S phase that have yet to be identified. While USP1 is the major DUB that regulates TLS, USP7 has been implicated in the deubiquitination of Rad18 and Pol eta to prevent their degradation via the UPS (186, 187). Loss of USP7 destabilizes both Rad18 and Pol eta proteins and compromises UV-induced PCNA monoubiquitination and Pol eta recruitment to stalled replication forks, respectively. As more ubiquitinated substrates are implicated in TLS, there will likely be more DUBs involved in the regulation of the DNA damage tolerance pathway.
Regulation of DUBs by oxidative stress.
Bursts of reactive oxygen species (ROS), such as H2O2, can be produced both by environmental carcinogens and/or endogenous sources. While it was once previously considered to be just a unwanted by-product of aerobic respiration, accumulating evidence indicates that H2O2 also serves as a critical signaling molecule in cell proliferation and survival (188). While there is a wealth of research demonstrating the general adverse effect of oxidative stress on DNA damage and cell death, much less is known about the signaling targets of ROS that regulate cell growth and DNA repair in a more direct manner. Protein tyrosine phosphatases (PTPs) are a family of redox-regulated proteins that are mediators of multiple signaling pathways in essential cellular processes such as proliferation, differentiation, and migration (189). PTPs can be directly inactivated by ROS through the oxidation of the catalytic Cys residue (190–192), and this inactivation is typically transient and reversible under physiological conditions. Thus, the reversibility of the reaction fine-tunes the signaling pathways emanating from receptor tyrosine kinases.
In a series of studies initiated by multiple groups, it was demonstrated that members of the Cys protease family of DUBs, similar to PTPs, were susceptible to redox regulation (193–195). Strikingly, they found that these DUBs can be rapidly targeted by ROS both in vitro and in cells. USP1, a DUB critical for both the FA pathway and TLS (as mentioned above), is transiently oxidized and inactivated in response to oxidative stress in cells, suggesting that USP1 is responsible for fine-tuning the monoubiquitination levels of PCNA during DNA lesion bypass (damage tolerance) for oxidative DNA damage in replicating cells (193, 194, 196). USP7, an important regulator of the nucleotide excision repair (NER) and Mdm2-p53 tumor suppressor pathway (see below), can also be reversibly oxidized and inactivated in cells (193). Using mass spectrometry and X-ray crystallography, the catalytic Cys residue was shown to be the target of oxidative modification (193, 195). There are likely many other redox-regulated DUBs that play a role in genome stability pathways. The molecular mechanisms that govern DUB redox regulation and how dysregulation can impact multiple DNA repair pathways and genomic instability in human cells are still active areas of study.
Regulation of replication fork progression by BAP1.
While BAP1 was shown to be involved in HR repair as discussed above, a more recent study showed that BAP1 is also required for DNA replication fork progression by deubiquitinating and stabilizing the INO80 ATPase subunit of the large INO80 complex, a chromatin remodeler that is involved in DNA replication and transcription (197). BAP1 is visible at the PCNA-containing replication fork foci, and by associating with H2A-Ub, it recruits INO80 to the replication foci (197). Similar to the effect of INO80 depletion, BAP1 depletion leads to impairment of replication fork progression, aneuploidy, chromosomal aberration, and developmental defects of mice (197). The role of BAP1 in DNA replication progression is partly attributed to stabilizing INO80 at replication forks, but there may be another unknown function of BAP1 in supporting DNA replication and alleviating replication stress (197). INO80 was also shown to be recruited to DSB sites through interacting with H2A and H2AX in yeast, and it promotes DSB end resection possibly by clearing out nucleosomes (154, 198, 199). Therefore, it is possible that BAP1 also affects DSB repair efficiency by modulating INO80 stability at DSB sites. Indeed, BAP1 is implicated in promoting DSB repair and conferring resistance to DNA-damaging agents (117). However, the INO80-stabilizing role of BAP1 may be specific only in the context of replication fork progression, as evidenced by S-phase-specific costaining of BAP1 and INO80 at the PCNA-containing replication foci (197).
DUBs THAT REGULATE REMOVAL OF BULKY ADDUCTS ON DNA
USP7 regulates nucleotide excision repair.
USP7 (also known as HAUSP [herpesvirus-associated ubiquitin-specific protease]) regulates cellular DNA damage response by modulating many substrates including MDM2 and p53, but probably the most direct role of USP7 in DNA repair is in regulating nucleotide excision repair (NER). NER is a repair mechanism that removes various types of bulky DNA adducts, such as UV-induced thymidine dimers. Recognition of the bulky lesions leads to removal of the short stretch of single-stranded DNA that contains the lesion, while the undamaged strand remains intact and serves as a template for DNA synthesis by DNA polymerases. Two distinct subpathways exist for NER: global genomic NER (GG-NER), which operates throughout the genome, and transcription-coupled NER (TC-NER), which removes DNA adducts at the transcriptionally active sites (200, 201). A major difference between these two pathways lies in the way damaged sites are recognized; GG-NER utilizes DDB2 (forms a complex with DDB1) or XPC (forms a complex with human RAD23 [hRAD23]) as the lesion recognition factor, while TC-NER appears to be initiated by RNA polymerase II stalling at the lesion. Genetic defects in TC-NER can lead to disorders such as Cockayne syndrome (CS) and UV-sensitive syndrome (UVSS), whose common feature is increased UV sensitivity. The current model suggests that stalled RNA polymerase II at damaged sites triggers recruitment of TC-NER-specific factors CSA (or ERCC8) and CSB (or ERCC6). CSB may rearrange the nucleosome structure around the stalled Pol II, and together with CSA, it facilitates recruitment of the TC-NER repair complex that includes repair/transcription complex including TFIIH, XPA, XPG, and ERCC-XPF1 nuclease (200–202). CSA and CSB play essential roles in TC-NER by temporarily displacing (or allowing backtracking) RNA Pol II from the damaged sites. Upon completion of repair, it is necessary to remove the TC-NER repair complex, and polyubiquitination and degradation of CSB were suggested to be one such post-TC-NER recovery mechanism (203) (Fig. 2). Whether deubiquitination of CSB is an essential event for the integrity of the TC-NER pathway was unknown. In theory, the deubiquitination activity may preserve or enhance the CSB-mediated displacement of RNA Pol II at damaged sites during the repair time frame. It was indeed shown that USP7 stabilizes CSB to regulate the efficiency of the TC-NER pathway (204, 205). Recruitment of USP7 to the damaged sites and to the RNA Pol II complex is mediated by a scaffold protein UVSSA (UV-stimulated scaffold protein A), one of the three complementation groups identified for UVSS patients (the other two groups being CSA and CSB). The protein level of CSB is lower in patient-derived UVSSA-deficient cells, and consistently, knockdown of UVSSA or USP7 leads to accelerated degradation of CSB by the UPS, UV sensitivity, and reduced RNA synthesis recovery from UV (204–206). Whether USP7 directly deubiquitinates CSB was not demonstrated in biochemical assays; however, a model in which USP7 deubiquitinates and stabilizes CSB/ERCC6 to facilitate the completion of TC-NER repair has been suggested (207). Of note was the observation that overexpressing CSB did not rescue the TC-NER defect of UVSS cells, which suggested that there are other mechanisms/substrates regulated by the UVSSA/USP7 complex (204). The steady-state level of RNA Pol II was also slightly lower in USP7 knockdown cells, suggesting that USP7 also deubiquitinates and stabilizes RNA Pol II, although this has not been formally tested (207).
USP7 may also regulate GG-NER by deubiquitinating XPC. XPC, in complex with hRAD23, plays a key role in the GG-NER pathway by inducing the formation of the preincision complex and senses a broad spectrum of lesions. Regulating XPC loading onto chromatin by polyubiquitination appears to be a key event in the GG-NER. XPC is ubiquitinated by two E3 ubiquitin ligases, CUL4-DDB2 (208) and RNF111 (209, 210). The CUL4-DDB2 E3 ligase polyubiquitinates XPC and does not lead to proteasomal degradation but rather promotes increased binding affinity of XPC to DNA (208). XPC is also ubiquitinated by RNF111, a SUMO-targeted E3 ubiquitin ligase that induces polyubiquitination on presumoylated XPC. Ubiquitination is mediated by UBC13/MMS2 E2-conjugating enzyme, suggesting that it is likely a nonproteolytic K63-linked polyubiquitination event. Interestingly, RNF111-mediated polyubiquitination induces the release of XPC from the lesions (209, 210), which is the opposite effect of CUL4-DDB2 mediated polyubiquitination. Both DDB2 and RNF111 depletion impair the UV-induced DNA repair, suggesting a model in which two sequential polyubiquitination events on XPC regulate the recruitment and release of XPC during NER. The RNF111-induced release of XPC from chromatin is required for recruitment of downstream NER factors and completion of repair (210). A recent study found that USP7 directly binds to and deubiquitinates XPC and that UV-induced XPC degradation is accelerated in USP7−/− HCT116 cells (211). The USP7−/− cells are defective in repairing UV-induced photoadducts, confirming that USP7's role in deubiquitinating and stabilizing XPC is important in NER. It is unclear whether USP7 serves to counteract polyubiquitination induced by CUL4/DDB2 or RNF111, but this seems unlikely as these ubiquitination mechanisms are nonproteolytic. It is possible that there is a yet-to-be identified ubiquitin E3 ligase-mediated polyubiquitination event that induces XPC degradation, which may be a necessary step upon completion of NER. It is also possible that an unknown DUB is responsible for counteracting nonproteolytic ubiquitination of XPC that regulates its chromatin loading at damage sites. Altogether, USP7 serves as an important regulator of both subpathways of NER by regulating stability of CSB/ERCC6 and perhaps both stability and chromatin loading of XPC, thus overall orchestrating UV-induced NER repair and cell survival.
A study found that USP24 is also involved in the ubiquitination events during the UV response. USP24 binds to, deubiquitinates, and stabilizes the DDB2 component of CUL4 E3 ligase (212). As CUL4-DDB2 is known to polyubiquitinate XPC, whether USP24 influences the XPC polyubiquitination or overall TC-NER is not known, although USP24 depletion increases the UV sensitivity of cells (213).
DUBs THAT REGULATE REMOVAL OF NONBULKY ADDUCTS ON DNA
USP47 regulates base excision repair by stabilizing DNA polymerase beta.
Base excision repair (BER) is an essential cellular process required for genome stability. Loss of BER function has been linked to premature aging, increased rate of mutagenesis, and cancer (214, 215). BER is active throughout the cell cycle and is critical for the removal of DNA lesions due to deamination, oxidation, and alkylation. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming abasic sites (AP sites). These sites are then cleaved by an AP endonuclease, the resulting single-strand break (SSB) can then be processed by either short-patch BER (where a single nucleotide is replaced) or long-patch BER (where 2 to 10 nucleotides are newly synthesized). The SSB is repaired by a DNA repair complex that includes DNA polymerase beta (Polβ), XRCC1, and DNA ligase. Polβ plays a central role in BER with both an AP lyase and DNA polymerase activity. The importance of Polβ is highlighted by the fact that a knockout of the polβ gene in mice results in early embryonic lethality and hypersensitivity to DNA-damaging agents (216, 217). Furthermore, the regulation of cellular Polβ levels is vital, since both under- and overproduction of Polβ lead to deficient repair or an increased rate of mutagenesis, respectively, and both have been linked to increased cancer susceptibility (218–220). Parsons and colleagues have previously shown that the steady-state level of BER enzymes, including Polβ, is tightly regulated and directly linked to the amount of endogenous DNA lesions (221). For example, if the level of Polβ exceeds the level of DNA lesions, then the excess or newly synthesized Polβ becomes ubiquitinated and degraded by the UPS (Fig. 2). This is achieved by sequential ubiquitination of Polβ by the E3 ubiquitin ligases Mule (ARF-BP1) and CHIP, whereby Mule first targets Polβ for monoubiquitination, which then seeds the protein for polyubiquitination by CHIP and subsequent targeting to the proteasome for degradation (222). The same group has also identified USP47 as the major DUB responsible for deubiquitinating Polβ to rescue it from proteasomal degradation (223). Consistent with this role, loss of USP47 increased the level of ubiquitinated Polβ, decreased Polβ levels, and caused a defect in BER. The defect in BER is reflected by the accumulation of DNA strand breaks and decreased cell viability in response to DNA damage. The E3 ubiquitin ligase Mule is also subjected to regulation via a phosphorylated-Ser18-containing isoform of USP7 (noted as USP7S). USP7S controls the protein stability of Mule by preventing the ubiquitin ligase from autoubiquitination and subsequent proteasomal degradation (224). USP7S was previously shown to be stabilized by CK2 protein kinase-dependent phosphorylation in unstressed cells (225). Upon DNA damage, the protein phosphatase PPM1G is activated by ATM, resulting in the dephosphorylation and subsequent destabilization of USP7S. Thus, DNA damage-dependent downregulation of USP7S leads to the reduction of Mule and likely stabilization of BER proteins. Other BER enzymes, including DNA glycosylases, have been implicated as targets of different E3 ubiquitin ligases (226). The function of ubiquitination in some of these substrates still needs to be worked out, and whether USP47 or USP7S plays a role in their deubiquitination remains to be determined.
The OTUD4-USP7-USP9X complex regulates the reversal of alkylated DNA.
Alkylation of DNA is one of the most frequent mutagenic events the cell encounters. DNA alkylation can occur by endogenous alkylating agents (e.g., S-adenosylmethionine [SAM]), as well as exogenous sources such as chloromethane or MMS (methyl methanesulfonate) (227). Alkylating agents such as MMS are mostly methylating agents. DNA alkylation can be repaired by at least three repair mechanisms: BER (base excision repair), direct base repair by methyltransferases, or base repair by oxidative demethylases. Much of the knowledge on alkylation repair came from studies on AlkB, an oxidative demethylase from Escherichia coli, which removes methyl groups from various sites on bases. In humans, there are nine AlkB family members. Of these family members, ALKBH2 (human ABH2 [hABH2]) and ALKBH3 (hABH3) are considered two human functional homologs of the E. coli AlkB (228). Evidence suggests that ALKBH2 preferentially removes methyl groups from double-stranded DNA and that ALKBH3 prefers single-stranded DNA or RNA as the substrate (228). How this class of DNA repair enzymes is functionally regulated has not been well studied. A recent study demonstrated that the stability of both ALKBH2 and ALKHB3 is regulated by the UPS (229). OTUD4, a member of the OTU DUB family with little known function, has been shown to interact with and stabilize ALKBH3 and ALKBH2 (229). Intriguingly, overexpression of not only the wild type but also a catalytic mutant form of OTUD4 can suppress ubiquitination and degradation of ALKBH3, suggesting that it is not dependent on the deubiquitinating activity of OTUD4. Characterization of ALKBH3-interacting proteins revealed that two USP family DUBs, USP7 and USP9x, are associated with OTUD4, and overexpression of these USPs (wild type but not catalytic mutant) can suppress the ubiquitination and degradation of ALKBH3 in the presence of OTUD4. This led to a model that OTUD4 serves as a scaffolding factor that recruits USP7 and USP9x to ALKBH3 for deubiquitination. Consistent with the role of ALKBH2 and ALKBH3, the knockdown of OTUD4, USP7, or USP9x sensitizes cells to DNA-alkylating agents, which is correlated with reduced ALKBH3. Why and how ALKBH2 and ALKBH3 are ubiquitinated and degraded is unknown, but it is possible that these repair enzymes are kept at low levels under normal conditions, and DNA alkylation events may require rapid stabilization by the DUBs. Thus, there must be a mechanism to inhibit DUB activity upon recovery from damage. Given the increasing evidence for the role of ALKBH2 and ALKBH3 in cancer development and chemoresistance, more information on these DUBs will be necessary for developing effective therapeutic strategies (229). The processing of alkylation products can lead to secondary lesions such as single- and double-strand breaks, and OTUD4-USP7-USP9x may also be essential in repairing more complicated DNA repair processes.
FUTURE PERSPECTIVES
Specific functions or functional redundancy?
Despite greater understanding of DUBs in DNA repair, key questions remain. Why are different DUBs with seemingly redundant roles needed to regulate certain DNA repair processes? Clearly, functional redundancy provides a better safety net to ensure genome integrity. To ensure tight regulation, each DUB may function at temporally distinct stages during DDR. For instance, different kinetics for Ub conjugate and DDR mediator recruitment were seen upon OTUB1 and OTUB2 depletion under the same experimental settings (76). This suggested that OTUB2 may function in the early phases of DDR (<10 min), whereas OTUB2 functions at later phases. It is also possible that different DUBs act sequentially to cooperate together to remove polyubiquitin chains from target substrates. For instance, POH1 may act to cleave the first proximal ubiquitin moiety on H2A/H2AX to release polyubiquitin chains en bloc, which can then be followed by USP5 or other DUBs that may disassemble the unanchored polyubiquitin chains. Perhaps different DUBs acting on the same polyubiquitinated substrate may be subjected to ubiquitin chain editing, leading to diverse functions for the individually modified substrate. A recent study demonstrated the functional relevance of branched ubiquitin chains for cell cycle regulation (242). Whether the formation of branched ubiquitin chains plays a role in the DNA repair pathway remains to be formally tested, but the likelihood is high. Another outstanding question is whether we have completely exhausted the search for ubiquitinated substrates involved in DNA repair at damaged sites (the polyubiquitin signal in DNA damage foci marked by the FK2 antibody). Identifying and generating a comprehensive list of damage-inducible ubiquitinated substrates that localize to damaged sites will enable researchers to determine whether some of these substrates are true targets of these DUBs or whether some of these DUBs are functionally redundant (Table 1). With the advent of quantitative mass spectrometry-based proteomics (243), it may soon be easier to identify all the ubiquitinated substrates for a specific DUB by coupling DUB inhibition (using a chemical inhibitor, RNAi, or inducible knockout strategy) to the enrichment of ubiquitin conjugates using Lys-ε-Gly-Gly (diGly)-based proteomics.
Regulation of DUBs.
How are different DUBs recruited to different types of DNA lesions? There are only a handful of DUBs that have localization signals to specific compartments. Furthermore, DUBs typically do not contain modular substrate-binding domains (akin to the F-box proteins for the SCF ubiquitin E3 ligase complex) that specifically recognize substrates for deubiquitination. Thus, DUB activity must be recruited by its associated factor(s) to specific substrates or cellular compartments for deubiquitination. The critical roles of DUB binding partners in dictating DUB subcellular localization are well demonstrated by BRCC36 and its binding proteins CCDC98/Abraxas (part of the BRCA1-A complex) and KIAA0157 (part of the BRISC complex). Knockdown of CCDC98/Abraxas or overexpressing KIAA0157 increases the cytosolic pool of BRCC36, whereas knockdown of KIAA0157 or overexpressing CCDC98 increases the nuclear pool of BRCC36 (58, 230). The components of the BRCA1-A complex are essential for the catalytic activity and stability of BRCC36 (230, 231). Thus, the activity and localization may be regulated at multiple levels to ensure the proper localization of the HR-limiting BRCC36 activity. How a single DUB, such as USP7, can participate in different types of DNA repair processes remains to be understood. USP7 is localized to UV-induced lesions by a scaffolding protein, UVSSA, which in turn associates with NER factors. A different pool of USP7 may be recruited to the OTUD4 scaffold to regulate repair of alkylated DNA or to associate with RNF168 to regulate DSB repair. It is possible that the differential recruitment of a DUB to different sites is regulated by distinct damage response types. Conversely, a single cofactor can activate different DUBs by simply binding to these DUBs, as shown by UAF1 in forming three distinct DUB complexes (175, 232). It is also intriguing to imagine how DUBs that are part of large complexes are spatially coordinated at the sites of damage. BRCC36 is part of a multisubunit BRCA1 complex, and POH1 is part of the 16-subunit 19S proteasome lid complex. The fact that the catalytic activity and protein stability of BRCC36 and POH1 require their binding subunits or partners suggests that the larger complexes may be present at DSB sites to disassemble the ubiquitin chains. It will be interesting to find out whether these larger complexes are necessary to regulate substrate specificity and/or localization and recruitment processes. Better understanding of the recruitment modules may further elucidate the specific biological functions of each DUB and perhaps explain the complexity of DUB toolkits in DNA repair.
DUBs as therapeutic targets.
As it has become clear that inhibiting selective DUBs impairs the ability to respond and repair DNA lesions, DUBs are appreciated as attractive clinical targets in order to suppress “DNA repair addictive” tumors (233, 234). Given that the activity of these DUBs can be locally inhibited in tumor cells, pharmacologic targeting of these DUBs could be useful for sensitizing the cancer cells to chemotherapeutics in clinical settings. However, without the complete understanding of the molecular basis and biology of these targeted DUBs, it remains a challenge to predict whether DUBs make good therapeutic targets for cancer and other human diseases.
Many DUBs exert pleiotropic functions by regulating multiple factors that are not necessarily in the same pathway or even in the same subcellular compartment. However, an upside of this could be that the pleiotropic nature of DUBs may allow them to be more-effective targets, as targeting a single DUB may sensitize cancer cells to chemotherapy due to the pleiotropic functions of the DUB in several DNA repair pathways. For instance, disrupting USP7 causes toxicity to cancer cells by inducing p53-mediated apoptosis (235), but the other roles of USP7 in multiple DSB repair pathways (e.g., DSB repair signaling, NER, TLS, alkylation repair) and in cell cycle checkpoints and transcriptional silencing may potentiate the cell-killing effect further by allowing irreparable DNA lesions to cause mitotic catastrophe. Similarly, USP9x inhibition cannot induce apoptosis only by destabilizing antiapoptotic MCL1 (236), but the apoptotic effect can also be further potentiated by combining with a DNA-alkylating agent based upon its role in stabilizing ALKBH3 (229). As for many other therapeutic approaches of targeting DNA repair pathways, potential complications arising with spontaneous tumor development in the long run should be taken into consideration when inhibiting DUBs in DNA repair; for example, USP3 and USP44 knockout mice were born healthy, but they ultimately develop tumors (83, 87).
A selective USP1-UAF1 inhibitor called ML323 has been developed recently and has shown promise in inhibiting the FA pathway and TLS to potentiate cisplatin cytotoxicity in non-small cell lung cancer and osteosarcoma cells in in vitro studies (237). The idea that targeting USP1 could be useful in overcoming resistance to platinum-based anticancer drugs may be a logical strategy for introducing new types of drugs to deal with chemoresistance. Importantly, USP1 knockout mice are resistant to tumorigenesis and prone to cellular senescence (238), suggesting that USP1 inhibition in the long run may be beneficial in treating certain types of cancers. Further understanding of the DUB function and regulation in DNA repair should equip us with better logistics in developing new strategies for treating various human diseases.
ACKNOWLEDGMENTS
We thank M. Nanjundan and K. Burns-Huang for critical reading of the manuscript. We also thank the reviewers of the manuscript for their invaluable suggestions. We apologize for any related work that may not have been included for discussion due to space constraints.
This work was supported in part by NIH grant R15HL126113 to Y.K. and NIH (GM084244) and American Cancer Society (ACS) (RSG-12-158-DMC) grants to T.T.H.
Biographies

Younghoon Kee received his Ph.D. from the University of Texas—Austin working under Jon Huibregtse and did his postdoctoral fellowship at the Dana-Farber Cancer Institute, Boston, MA, with Alan D'Andrea. He is currently an assistant professor at the University of South Florida. He has been studying the function and regulation of yeast and human deubiquitinating enzymes for more than 10 years, with a current focus on DUBs in genome stability.

Tony T Huang received his Ph.D. from the University of Wisconsin—Madison working under Shigeki Miyamoto and did his postdoctoral fellowship at the Dana-Farber Cancer Institute, Boston, MA, with Alan D'Andrea. In 2007, he began his own independent research group at the New York University School of Medicine working on elucidating molecular mechanisms of Fanconi anemia and translesion synthesis pathways in mammalian cells. His current research group is actively working on the role of deubiquitinases in genome stability pathways in health and disease model systems.
ADDENDUM IN PROOF
Orthwein and colleagues (A. Orthwein et al., Nature 528:422–426, 2015, http://dx.doi.org/10.1038/nature16142) recently reported a cell cycle-regulated mechanism to prohibit HR in G1 by controlling the interaction of BRCA1 with PALB2-BRCA2 through PALB2 ubiquitination that is counteracted by USP11.
REFERENCES
- 1.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D. 2012. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faesen AC, Luna-Vargas MP, Geurink PP, Clerici M, Merkx R, van Dijk WJ, Hameed DS, El Oualid F, Ovaa H, Sixma TK. 2011. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol 18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SM, Ovaa H, Komander D. 2013. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 7.Komander D. 2010. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem 54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 8.Huang TT, D'Andrea AD. 2006. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 9.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 10.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. doi: 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 12.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Elledge SJ. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A 104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 17.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 18.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N. 2015. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 20.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. 2006. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Panier S, Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 8:436–443. doi: 10.1016/j.dnarep.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH, Durocher D. 2012. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell 47:383–395. doi: 10.1016/j.molcel.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 23.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. 2012. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D, Miller KM. 2014. Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS Genet 10:e1004178. doi: 10.1371/journal.pgen.1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, Storchova Z, Cox J, Bekker-Jensen S, Mailand N, Mann M. 2015. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann RM, Pickart CM. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645–653. doi: 10.1016/S0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell 25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L. 2015. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep 10:226–238. doi: 10.1016/j.celrep.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 30.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Chen J, Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 33.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. 2007. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Liu C, Chen J, Yu X. 2012. RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J Biol Chem 287:22919–22926. doi: 10.1074/jbc.M112.351007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sy SM, Huen MS, Chen J. 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A 106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Fan Q, Ren K, Andreassen PR. 2009. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res 7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. 2011. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman KA, Greenberg RA. 2011. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 286:13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 41.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell 35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC. 2011. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmermann M, de Lange T. 2014. 53BP1: pro choice in DNA repair. Trends Cell Biol 24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Feng L, Fong KW, Wang J, Wang W, Chen J. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem 288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M, Jaspers JE, Watanabe K, Pieterse M, Kersbergen A, Sol W, Celie PH, Schouten PC, van den Broek B, Salman A, Nieuwland M, de Rink I, de Ronde J, Jalink K, Boulton SJ, Chen J, van Gent DC, Bartek J, Jonkers J, Borst P, Rottenberg S. 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, Wevers BA, Orthwein A, Durocher D, Jacobs JJ. 2015. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521:537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Aroumougame A, Lobrich M, Li Y, Chen D, Chen J, Gong Z. 2014. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev 28:2693–2698. doi: 10.1101/gad.252478.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zong D, Callen E, Pegoraro G, Lukas C, Lukas J, Nussenzweig A. 2015. Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining. Nucleic Acids Res 43:4950–4961. doi: 10.1093/nar/gkv336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. 2009. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J 28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Wu J, Yu X. 2007. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol 14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 56.Kim H, Huang J, Chen J. 2007. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 57.Wang B, Hurov K, Hofmann K, Elledge SJ. 2009. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev 23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng L, Wang J, Chen J. 2010. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem 285:30982–30988. doi: 10.1074/jbc.M110.135392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu X, Kim JA, Castillo A, Huang M, Liu J, Wang B. 2011. NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes. J Biol Chem 286:11734–11745. doi: 10.1074/jbc.M110.200857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. 2003. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell 12:1087–1099. doi: 10.1016/S1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 61.Zheng H, Gupta V, Patterson-Fortin J, Bhattacharya S, Katlinski K, Wu J, Varghese B, Carbone CJ, Aressy B, Fuchs SY, Greenberg RA. 2013. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep 5:180–193. doi: 10.1016/j.celrep.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. 2009. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A 106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR III, Denchi EL. 2013. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 65.Yao T, Cohen RE. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 66.Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. 2012. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 31:3918–3934. doi: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA. 2013. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res 41:10298–10311. doi: 10.1093/nar/gkt802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Worden EJ, Padovani C, Martin A. 2014. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol 21:220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 69.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. 2004. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 70.Schwarz T, Sohn C, Kaiser B, Jensen ED, Mansky KC. 2010. The 19S proteasomal lid subunit POH1 enhances the transcriptional activation by Mitf in osteoclasts. J Cell Biochem 109:967–974. doi: 10.1002/jcb.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wauer T, Komander D. 2014. The JAMM in the proteasome. Nat Struct Mol Biol 21:346–348. doi: 10.1038/nsmb.2800. [DOI] [PubMed] [Google Scholar]
- 72.Amerik AY, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. 1997. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J 16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakajima S, Lan L, Wei L, Hsieh CL, Rapic-Otrin V, Yasui A, Levine AS. 2014. Ubiquitin-specific protease 5 is required for the efficient repair of DNA double-strand breaks. PLoS One 9:e84899. doi: 10.1371/journal.pone.0084899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. 2006. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 75.Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, Clague MJ, Urbe S, Jackson SP. 2014. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol 16:1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kato K, Nakajima K, Ui A, Muto-Terao Y, Ogiwara H, Nakada S. 2014. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell 53:617–630. doi: 10.1016/j.molcel.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 77.Altun M, Walter TS, Kramer HB, Herr P, Iphofer A, Bostrom J, David Y, Komsany A, Ternette N, Navon A, Stuart DI, Ren J, Kessler BM. 2015. The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages. PLoS One 10:e0115344. doi: 10.1371/journal.pone.0115344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sloper-Mould KE, Eyre HJ, Wang XW, Sutherland GR, Baker RT. 1999. Characterization and chromosomal localization of USP3, a novel human ubiquitin-specific protease. J Biol Chem 274:26878–26884. doi: 10.1074/jbc.274.38.26878. [DOI] [PubMed] [Google Scholar]
- 80.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol 17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 81.Sharma N, Zhu Q, Wani G, He J, Wang QE, Wani AA. 2014. USP3 counteracts RNF168 via deubiquitinating H2A and gammaH2AX at lysine 13 and 15. Cell Cycle 13:106–114. doi: 10.4161/cc.26814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. 2013. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J Biol Chem 288:16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lancini C, van den Berk PC, Vissers JH, Gargiulo G, Song JY, Hulsman D, Serresi M, Tanger E, Blom M, Vens C, van Lohuizen M, Jacobs H, Citterio E. 2014. Tight regulation of ubiquitin-mediated DNA damage response by USP3 preserves the functional integrity of hematopoietic stem cells. J Exp Med 211:1759–1777. doi: 10.1084/jem.20131436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Delgado-Diaz MR, Martin Y, Berg A, Freire R, Smits VA. 2014. Dub3 controls DNA damage signalling by direct deubiquitination of H2AX. Mol Oncol 8:884–893. doi: 10.1016/j.molonc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, Feldmesser E, Brik A, Yu X, Hanna J, Aberdam D, Domany E, Oren M. 2012. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell 46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER III, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, Harper JW, Elledge SJ. 2007. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. 2012. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest 122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, van Deursen J, Galardy PJ. 2011. Overexpression of ubiquitin specific protease 44 (USP44) induces chromosomal instability and is frequently observed in human T-cell leukemia. PLoS One 6:e23389. doi: 10.1371/journal.pone.0023389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holland AJ, Cleveland DW. 2012. The deubiquitinase USP44 is a tumor suppressor that protects against chromosome missegregation. J Clin Invest 122:4325–4328. doi: 10.1172/JCI66420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Typas D, Luijsterburg MS, Wiegant WW, Diakatou M, Helfricht A, Thijssen PE, van de Broek B, Mullenders LH, van Attikum H. 2015. The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Res 43:6919–6933. doi: 10.1093/nar/gkv613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu M, Liu K, Mao Z, Luo J, Gu W, Zhao W. 27 October 2015. USP11 is a negative regulator to gammaH2AX ubiquitylation by RNF8/RNF168. J Biol Chem doi: 10.1074/jbc.M114.624478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. 2004. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol 24:7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wiltshire TD, Lovejoy CA, Wang T, Xia F, O'Connor MJ, Cortez D. 2010. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem 285:14565–14571. doi: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hendriks IA, Schimmel J, Eifler K, Olsen JV, Vertegaal AC. 2015. Ubiquitin-specific protease 11 (USP11) deubiquitinates hybrid small ubiquitin-like modifier (SUMO)-ubiquitin chains to counteract RING finger protein 4 (RNF4). J Biol Chem 290:15526–15537. doi: 10.1074/jbc.M114.618132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, Hendriks IA, Radaelli E, Hochepied T, Blanpain C, Sablina A, van Attikum H, Olsen JV, Jochemsen AG, Vertegaal AC, Marine JC. 2013. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ 20:490–502. doi: 10.1038/cdd.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guzzo CM, Berndsen CE, Zhu J, Gupta V, Datta A, Greenberg RA, Wolberger C, Matunis MJ. 2012. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal 5:ra88. doi: 10.1126/scisignal.2003485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo K, Zhang H, Wang L, Yuan J, Lou Z. 2012. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 31:3008–3019. doi: 10.1038/emboj.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O'Donnell L, Kumakubo A, Munro M, Sicheri F, Gingras AC, Natsume T, Suda T, Durocher D. 2010. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 101.Juang YC, Landry MC, Sanches M, Vittal V, Leung CC, Ceccarelli DF, Mateo AR, Pruneda JN, Mao DY, Szilard RK, Orlicky S, Munro M, Brzovic PS, Klevit RE, Sicheri F, Durocher D. 2012. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell 45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, Wolberger C. 2013. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol 20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu Q, Sharma N, He J, Wani G, Wani AA. 2015. USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell Cycle 14:1413–1425. doi: 10.1080/15384101.2015.1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sy SM, Jiang J, O WS, Deng Y, Huen MS. 2013. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res 41:8572–8580. doi: 10.1093/nar/gkt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J, Lukas C. 2012. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 106.Kee Y, Lyon N, Huibregtse JM. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J 24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kee Y, Munoz W, Lyon N, Huibregtse JM. 2006. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem 281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- 108.Lam MH, Emili A. 2013. Ubp2 regulates Rsp5 ubiquitination activity in vivo and in vitro. PLoS One 8:e75372. doi: 10.1371/journal.pone.0075372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dar A, Shibata E, Dutta A. 2013. Deubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathway. Mol Cell Biol 33:3309–3320. doi: 10.1128/MCB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 112.Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol 31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ismail IH, Andrin C, McDonald D, Hendzel MJ. 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol 191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pan MR, Peng G, Hung WC, Lin SY. 2011. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem 286:28599–28607. doi: 10.1074/jbc.M111.256297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ III. 1998. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 116.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. 2008. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ismail IH, Davidson R, Gagne JP, Xu ZZ, Poirier GG, Hendzel MJ. 2014. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res 74:4282–4294. doi: 10.1158/0008-5472.CAN-13-3109. [DOI] [PubMed] [Google Scholar]
- 118.Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, Barbour H, Corbeil L, Hebert J, Drobetsky E, Masson JY, Di Noia JM, Affar EB. 2014. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A 111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. 2007. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 121.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. 2014. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol 21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng SK, Yin Y, Petes TD, Symington LS. 2015. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell 60:500–508. doi: 10.1016/j.molcel.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 124.Longhese MP, Neecke H, Paciotti V, Lucchini G, Plevani P. 1996. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res 24:3533–3537. doi: 10.1093/nar/24.18.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399–409. doi: 10.1016/S0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 126.Sugiyama T, Zaitseva EM, Kowalczykowski SC. 1997. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem 272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 127.Sung P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 128.Takeda S, Nakamura K, Taniguchi Y, Paull TT. 2007. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 129.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mimitou EP, Symington LS. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst) 8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee KY, Im JS, Shibata E, Park J, Handa N, Kowalczykowski SC, Dutta A. 2015. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat Commun 6:7744. doi: 10.1038/ncomms8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A 105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH. 2005. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol 25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Polato F, Callen E, Wong N, Faryabi R, Bunting S, Chen HT, Kozak M, Kruhlak MJ, Reczek CR, Lee WH, Ludwig T, Baer R, Feigenbaum L, Jackson S, Nussenzweig A. 2014. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J Exp Med 211:1027–1036. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X, Paull TT. 2014. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell 54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barton O, Naumann SC, Diemer-Biehs R, Kunzel J, Steinlage M, Conrad S, Makharashvili N, Wang J, Feng L, Lopez BS, Paull TT, Chen J, Jeggo PA, Lobrich M. 2014. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol 206:877–894. doi: 10.1083/jcb.201401146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J. 2013. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol Cell 49:657–667. doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaidi A, Weinert BT, Choudhary C, Jackson SP. 2010. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 143.Rajendran P, Kidane AI, Yu TW, Dashwood WM, Bisson WH, Lohr CV, Ho E, Williams DE, Dashwood RH. 2013. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics 8:612–623. doi: 10.4161/epi.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yu X, Fu S, Lai M, Baer R, Chen J. 2006. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Steger M, Murina O, Huhn D, Ferretti LP, Walser R, Hanggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak P, Gerrits B, Del Sal G, Zerbe O, Sartori AA. 2013. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell 50:333–343. doi: 10.1016/j.molcel.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 146.Schmidt CK, Galanty Y, Sczaniecka-Clift M, Coates J, Jhujh S, Demir M, Cornwell M, Beli P, Jackson SP. 2015. Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat Cell Biol 17:1458–1470. doi: 10.1038/ncb3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lafranchi L, de Boer HR, de Vries EG, Ong SE, Sartori AA, van Vugt MA. 2014. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J 33:2860–2879. doi: 10.15252/embj.201489017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, Jiang P, Wang T, Guo W, Chen Y, Li X, Zhang P, Pei H. 2015. The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep 13:93–107. doi: 10.1016/j.celrep.2015.08.056. [DOI] [PubMed] [Google Scholar]
- 149.Wijnhoven P, Konietzny R, Blackford AN, Travers J, Kessler BM, Nishi R, Jackson SP. 2015. USP4 auto-deubiquitylation promotes homologous recombination. Mol Cell 60:362–373. doi: 10.1016/j.molcel.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vlasschaert C, Xia X, Coulombe J, Gray DA. 2015. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol Biol 15:230. doi: 10.1186/s12862-015-0511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. 2010. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. 2008. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell 31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gospodinov A, Vaissiere T, Krastev DB, Legube G, Anachkova B, Herceg Z. 2011. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol Cell Biol 31:4735–4745. doi: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van Attikum H, Fritsch O, Gasser SM. 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kee Y, D'Andrea AD. 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7:249–262. doi: 10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 157.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sims AE, Spiteri E, Sims RJ III, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol 14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 159.MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DM, Rouse J. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 161.Liu T, Ghosal G, Yuan J, Chen J, Huang J. 2010. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 162.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. 2014. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Scharer OD, Patel KJ. 2014. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell 54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M, Wang H, Fei P. 2013. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle 12:803–809. doi: 10.4161/cc.23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Murina O, von Aesch C, Karakus U, Ferretti LP, Bolck HA, Hanggi K, Sartori AA. 2014. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep 7:1030–1038. doi: 10.1016/j.celrep.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 167.Unno J, Itaya A, Taoka M, Sato K, Tomida J, Sakai W, Sugasawa K, Ishiai M, Ikura T, Isobe T, Kurumizaka H, Takata M. 2014. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep 7:1039–1047. doi: 10.1016/j.celrep.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 168.Yeo JE, Lee EH, Hendrickson EA, Sobeck A. 2014. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet 23:3695–3705. doi: 10.1093/hmg/ddu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ III, Rothenberg E, Jallepalli PV, Huang TT. 2015. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58:323–338. doi: 10.1016/j.molcel.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chaudhury I, Sareen A, Raghunandan M, Sobeck A. 2013. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res 41:6444–6459. doi: 10.1093/nar/gkt348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 172.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. 2007. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. 2011. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 194:177–186. doi: 10.1083/jcb.201101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park E, Kim JM, Primack B, Weinstock DM, Moreau LA, Parmar K, D'Andrea AD. 2013. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol Cell Biol 33:4360–4370. doi: 10.1128/MCB.00870-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 178.Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D'Andrea AD. 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 25:1847–1858. doi: 10.1101/gad.17020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K. 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J Biol Chem 285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. 2008. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci U S A 105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jones MJ, Colnaghi L, Huang TT. 2012. Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. EMBO J 31:908–918. doi: 10.1038/emboj.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8:339–347. [DOI] [PubMed] [Google Scholar]
- 184.Piatkov KI, Colnaghi L, Bekes M, Varshavsky A, Huang TT. 2012. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell 48:926–933. doi: 10.1016/j.molcel.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. 2011. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 186.Zlatanou A, Sabbioneda S, Miller ES, Greenwalt A, Aggathanggelou A, Maurice MM, Lehmann AR, Stankovic T, Reverdy C, Colland F, Vaziri C, Stewart GS. 11 May 2015. USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene doi: 10.1038/onc.2015.149. [DOI] [PubMed] [Google Scholar]
- 187.Qian J, Pentz K, Zhu Q, Wang Q, He J, Srivastava AK, Wani AA. 2015. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 34:4791–4796. doi: 10.1038/onc.2014.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Poole LB, Hall A, Nelson KJ. 2011. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr Protoc Toxicol Chapter 7:Unit 7.9. doi: 10.1002/0471140856.tx0709s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tonks NK. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 190.Tonks NK. 2005. Redox redux: revisiting PTPs and the control of cell signaling. Cell 121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 191.Salmeen A, Barford D. 2008. Methods for preparing crystals of reversibly oxidized proteins: crystallization of protein tyrosine phosphatase 1B as an example. Methods Mol Biol 476:101–116. [DOI] [PubMed] [Google Scholar]
- 192.den Hertog J, Groen A, van der Wijk T. 2005. Redox regulation of protein-tyrosine phosphatases. Arch Biochem Biophys 434:11–15. doi: 10.1016/j.abb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 193.Cotto-Rios XM, Bekes M, Chapman J, Ueberheide B, Huang TT. 2012. Deubiquitinases as a signaling target of oxidative stress. Cell Rep 2:1475–1484. doi: 10.1016/j.celrep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee JG, Baek K, Soetandyo N, Ye Y. 2013. Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat Commun 4:1568. doi: 10.1038/ncomms2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. 2013. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun 4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. 2011. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol Cell 43:649–662. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 197.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. 2014. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun 5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- 198.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 199.van Attikum H, Fritsch O, Hohn B, Gasser SM. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 200.Fousteri M, Mullenders LH. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res 18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 201.Hanawalt PC, Spivak G. 2008. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol 9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 202.Saijo M. 2013. The role of Cockayne syndrome group A (CSA) protein in transcription-coupled nucleotide excision repair. Mech Ageing Dev 134:196–201. doi: 10.1016/j.mad.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 203.Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. 2006. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev 20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schwertman P, Lagarou A, Dekkers DH, Raams A, van der Hoek AC, Laffeber C, Hoeijmakers JH, Demmers JA, Fousteri M, Vermeulen W, Marteijn JA. 2012. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet 44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 205.Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S, Tahara H, Neilan EG, Honma M, Nohmi T, Yasui A, Tanaka K. 2012. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet 44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- 206.Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, Takahashi Y, Ohyama K, Ito K, Mishima H, Nomura M, Kinoshita A, Ono S, Takenaka K, Masuyama R, Kudo T, Slor H, Utani A, Tateishi S, Yamashita S, Stefanini M, Lehmann AR, Yoshiura K, Ogi T. 2012. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet 44:586–592. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- 207.Schwertman P, Vermeulen W, Marteijn JA. 2013. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma 122:275–284. doi: 10.1007/s00412-013-0420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Hanaoka F. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 209.Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA, Bekker-Jensen S, Mailand N. 2013. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201:797–807. doi: 10.1083/jcb.201212075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.van Cuijk L, van Belle GJ, Turkyilmaz Y, Poulsen SL, Janssens RC, Theil AF, Sabatella M, Lans H, Mailand N, Houtsmuller AB, Vermeulen W, Marteijn JA. 2015. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat Commun 6:7499. doi: 10.1038/ncomms8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.He J, Zhu Q, Wani G, Sharma N, Han C, Qian J, Pentz K, Wang QE, Wani AA. 2014. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. J Biol Chem 289:27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhang L, Lubin A, Chen H, Sun Z, Gong F. 2012. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell Cycle 11:4378–4384. doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhang L, Nemzow L, Chen H, Lubin A, Rong X, Sun Z, Harris TK, Gong F. 2015. The deubiquitinating enzyme USP24 is a regulator of the UV damage response. Cell Rep 10:140–147. doi: 10.1016/j.celrep.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Parsons JL, Dianov GL. 2013. Co-ordination of base excision repair and genome stability. DNA Repair (Amst) 12:326–333. doi: 10.1016/j.dnarep.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 215.Dianov GL, Parsons JL. 2007. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 216.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. 2008. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res 18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ochs K, Sobol RW, Wilson SH, Kaina B. 1999. Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res 59:1544–1551. [PubMed] [Google Scholar]
- 218.Cabelof DC, Guo Z, Raffoul JJ, Sobol RW, Wilson SH, Richardson A, Heydari AR. 2003. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res 63:5799–5807. [PubMed] [Google Scholar]
- 219.Canitrot Y, Frechet M, Servant L, Cazaux C, Hoffmann JS. 1999. Overexpression of DNA polymerase beta: a genomic instability enhancer process. FASEB J 13:1107–1111. [DOI] [PubMed] [Google Scholar]
- 220.Chan K, Houlbrook S, Zhang QM, Harrison M, Hickson ID, Dianov GL. 2007. Overexpression of DNA polymerase beta results in an increased rate of frameshift mutations during base excision repair. Mutagenesis 22:183–188. doi: 10.1093/mutage/gel070. [DOI] [PubMed] [Google Scholar]
- 221.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. 2008. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell 29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 222.Parsons JL, Tait PS, Finch D, Dianova II, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL. 2009. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J 28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. 2011. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol Cell 41:609–615. doi: 10.1016/j.molcel.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 224.Khoronenkova SV, Dianov GL. 2013. USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res 41:1750–1756. doi: 10.1093/nar/gks1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. 2012. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 45:801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Edmonds MJ, Parsons JL. 2014. Regulation of base excision repair proteins by ubiquitylation. Exp Cell Res 329:132–138. doi: 10.1016/j.yexcr.2014.07.031. [DOI] [PubMed] [Google Scholar]
- 227.De Bont R, van Larebeke N. 2004. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 228.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. 2007. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 229.Zhao Y, Majid MC, Soll JM, Brickner JR, Dango S, Mosammaparast N. 2015. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J 34:1687–1703. doi: 10.15252/embj.201490497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA. 2010. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem 285:30971–30981. doi: 10.1074/jbc.M110.135319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. 2009. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev 23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D'Andrea AD. 2010. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J Biol Chem 285:11252–11257. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lim KH, Baek KH. 2013. Deubiquitinating enzymes as therapeutic targets in cancer. Curr Pharm Des 19:4039–4052. doi: 10.2174/1381612811319220013. [DOI] [PubMed] [Google Scholar]
- 234.Sippl W, Collura V, Colland F. 2011. Ubiquitin-specific proteases as cancer drug targets. Future Oncol 7:619–632. doi: 10.2217/fon.11.39. [DOI] [PubMed] [Google Scholar]
- 235.Nicholson B, Suresh Kumar KG. 2011. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys 60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 236.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, Yue P, Dornan D, Huang DC, Dixit VM. 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 237.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. 2014. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol 10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Park E, Kim H, Kim JM, Primack B, Vidal-Cardenas S, Xu Y, Price BD, Mills AA, D'Andrea AD. 2013. FANCD2 activates transcription of TAp63 and suppresses tumorigenesis. Mol Cell 50:908–918. doi: 10.1016/j.molcel.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ismail IH, Gagne JP, Genois MM, Strickfaden H, McDonald D, Xu Z, Poirier GG, Masson JY, Hendzel MJ. 2015. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol 17:1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- 241.O'Connor HF, Lyon N, Leung JW, Agarwal P, Swaim CD, Miller KM, Huibregtse JM. 2015. Ubiquitin-activated interaction traps (UBAITs) identify E3 ligase binding partners. EMBO Rep 16:1699–1712. doi: 10.15252/embr.201540620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Meyer HJ, Rape M. 2014. Enhanced protein degradation by branched ubiquitin chains. Cell 157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Silva GM, Finley D, Vogel C. 2015. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol 22:116–123. doi: 10.1038/nsmb.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]