Abstract
RNAi is a powerful tool to target the insect genes involved in host-pest interactions. Key insect genes are the choice for silencing to achieve pest derived resistance where resistance genes are not available in gene pool of host plant. In this study, an attempt was made to determine the effect of dsRNA designed from two genes Cytochrome P450 derivative (CYP6) and Aminopeptidase N (APN) of rice yellow stem borer (YSB) on growth and development of insect. The bioassays involved injection of chemically synthesized 5′ FAM labeled 21-nt dsRNA into rice cut stems and allowing the larvae to feed on these stems which resulted in increased mortality and observed growth and development changes in larval length and weight compared with its untreated control at 12–15 days after treatment. These results were further supported by observing the reduction in transcripts expression of these genes in treated larvae. Fluorescence detection in treated larvae also proved that dsRNA was readily taken by larvae when fed on dsRNA treated stems. These results from the present study clearly show that YSB larvae fed on dsRNA designed from Cytochrome P450 and Aminopeptidase N has detrimental effect on larval growth and development. These genes can be deployed to develop YSB resistance in rice using RNAi approach.
Keywords: yellow stem borer-YSB, double stranded RNA (dsRNA), growth and development, bioassays, cytochrome P450 (CYP6), aminopeptidase N (APN)
Introduction
The rice yellow stem borer (YSB), Scirpophaga incertulas (Lepidoptera: Crambidae), is a widely distributed and destructive insect that causes significant yield losses to an extent of 10–90% by feeding on rice crop at all stages (Bandong and Litsinger, 2005). The YSB larvae that enter inside the stem cause death of the central shoot known as “dead hearts” at vegetative stage and unfilled panicles known as “white heads” at reproductive stage (Pathak and Khan, 1994). There are no resistant sources available for this deadly pest in cultivated rice germplasm (Brar and Khush, 1997). Although the option of expression of cry genes from Bacillus thuringiensis (Bt) is available, development of insect resistance in field has become one of the long standing issue with Bt food crops (Tabashnik et al., 2013). Given these circumstances, silencing of YSB genes through RNA interference (RNAi) by inactivating the key insect genes leading to aberration in pest growth and metabolism (Yang et al., 2011) offers an alternative. RNAi is sequence-specific gene silencing at the post-transcription level, induced by double stranded RNA (dsRNA; Fire et al., 1998; Hamilton et al., 2002). Down regulation of the expression of specific target genes through RNAi has been widely used for genetic research in several insects (Price and Gatehouse, 2008). However, selection of crucial genes which have an important role in insect life cycle for a specific insect is still a challenge (Kola et al., 2015).
Cytochrome P450 monooxygenases (cytochrome P450s) are found in virtually all living organisms. It plays an important role in the metabolism of xenobiotics such as drugs, pesticides, and plant toxins (Scott, 2008). In insects, cytochrome P450s play a predominant role in the metabolism of insecticides, which often results in the development of insecticide resistance in insect populations (Zhou et al., 2010). Most insect cytochrome P450 genes belong to microsomal CYP4, CYP6, CYP9, CYP28, CYP321, and mitochondrial CYP12 families which have been associated with detoxification processes allowing the insect to become tolerant or resistant to insecticides or host plant allelochemicals (Guo et al., 2011). Aminopeptidase N (APN) is a member of metallo enzymes present in larvae midgut of lepidopteran insects. It plays an important physiological role in dietary protein digestion (Marchler-Bauer et al., 2015). Inhibition of its activity in the midgut can result in detrimental effect on larval growth, development, and lead to larval mortality (Reed et al., 1999). APNs are mainly studied for their role as receptors in Cry toxin-induced pathogenesis in insects (Bravo et al., 2007). Expression of APNs was found in midgut and malphigian tubules (Wang et al., 2005). RNAi mediated silencing of CYP6AE14 (Cytochrome P450 derivative) and APN in other lepidopteran insects like Helicoverpa armigera, Plutella xylostella, Achaea janata, and Spodoptera litura resulted in significant down-regulation of corresponding transcript and protein expression causing larval growth arrest and mortality and development of lethal larval-pupal intermediates (Rajagopal et al., 2002; Mao et al., 2007; Sivakumar et al., 2007; Bautista et al., 2009; Ningshen et al., 2013).
The present work was aimed to examine the effect of dsRNA molecules on silencing of YSB genes CYP6 and APN by feeding larvae on dsRNA treated cut stems. The dsRNAs fed larvae were examined to see the effect of silencing of these genes on insect growth and metabolism. Our bioassays simulated the in vivo mechanism of gene silencing and showed that dsRNA molecules can be taken up through the normal dietary path of YSB. Further, the target gene expression level was examined in control and dsRNA fed larvae which suggested silencing of the corresponding gene of YSB which is the first RNAi approach for YSB.
Materials and methods
Design of double stranded RNA and prediction of off targets
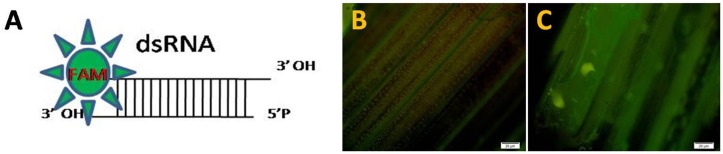
To clone cDNAs of CYP6 and APN genes, a total of 48 corresponding sequences of CYP6 gene were downloaded from lepidopteron database, NCBI. Out of these, 10 sequences could find near homologies which were used for designing three primers. Similarly, to clone APN, total 47 sequences were downloaded from lepidopteron database, NCBI. Out of these, 25 sequences showed near homologies which were used to design two sets of primers from the conserved region through online tool (http://primer3.ut.ee/). Using these primers 750 bp cDNA of CYP6 and 728 bp cDNA for APN were cloned from YSB and submitted to Genbank (CYP6: KC904274, APN: KF290773). These YSB cDNA sequences were used for identification of siRNA hotspots and dsRNAs were designed based on the Reynold rules (Reynolds et al., 2004). The designed dsRNA for CYP6 sense strand was 5′-AGUUGAGAAUGAAAUGACUGA-3′ and the antisense was 3′-UCAACUCUUACUUUACUGACU-5′ which had high Reynolds score of six out of eight. The similar strategy was followed for designing dsRNA from APN gene, the designed dsRNA sense strand sequence was 5′ GACGACGUAUACUUAACUACU-3′ and the antisense was 3′-CUGCUGCAUAUGAAUUGAUGA-5′ which had the high Reynolds score eight out of eight. Care was taken for designed dsRNA not to have any unintended off targets, which was apparent by doing BLAST search. The designed dsRNAs were also tested in in silico to ascertain the fulfillment of different parameters for maximum silencing by using siMAX siRNA design tool (https://www.eurofinsgenomics.eu/en/dna-rna-oligonucleotides/custom-rna-oligos/simax-sirna.aspx). The selected dsRNA were chemically synthesized at Eurofins Genomics GmbH, Germany by attaching FAM (5′carboxy fluorescein) at 5′ position (Figure 1A).
Figure 1.
(A) Schematic representation of dsRNA labeled with 5 FAM, Fluorescence detection of dsRNA in TN1 cut stems under microscope (B) Untreated (C) Treated cut stem.
Bio assay
Adult moths of YSB were collected from the rice field at Indian Institute of Rice Research (IIRR), and released on susceptible cv. TN1 plants in glasshouse for egg laying. These egg masses were collected and transferred into plastic vials for hatching. The neonate larvae that hatched from a single egg mass were released on to ~8 cm long TN1 stems and reared at 25 ± 2°C for 3 days (Padmakumari et al., 2013). For in vitro cut stem bioassay, fresh TN1cut stems (~8 cm) were placed in petri plate with moistened filter paper. For standardization of optimum concentration of dsRNA required for assay, three different concentrations i.e., 10, 20, and 30 pM of dsRNA were chosen. The dsRNA was injected into cut stems using small insulin syringe with volume of 30 μl per stem. Buffer (1 ×) of dsRNAs was used as control. The cut stems were kept at room temperature for 1 h for dsRNA to spread completely within the stem. On each treated cut stem, three larvae of 3 day old were released with the help of a soft paint brush. The insects were reared at 25 ± 2°C, 65%RH and 16:8 h (L: D). The optimized 30 pM concentration of dsRNA was used to determine its effect on larva, initially the experiment was carried out with different dsRNA concentration viz., 10, 20, 30, and 40 pM. (Figures showing the effect of these concentrations on YSB were submitted as Supplementary Material). We found that the response was good at 30 pM. Hence in all the experiments we used 30 μl of 30 pM dsRNA per 8 cm cut stem to evaluate the effect on the target insect. Each bioassay was carried with 15 stems in three replication (N = 45) and each experiment repeated twice to obtain uniform results. Various parameters like insect molting, abnormal morphological changes in the larval growth and length, and survival were observed at 6, 12, 15 days after treatment (DAT). At each time interval, three samples (nine larvae) were selected for observation by opening stems through destructive sampling. The dead larvae were dissected to observe under fluorescent microscope (Olympus CH40, Japan).
Effects of dsRNA on YSB larval growth and development
Larval mortality
The mortality of larvae was recorded by opening the cut stems under a microscope at 3 day intervals before changing on to fresh treated stems. The number of surviving larvae per concentration of dsRNA was recorded at different time intervals of 6, 12, and 15 days after treatment. The mortality of the larvae in both treated and untreated stems were recorded.
Larval weight and length
Reduction in larval weight was obtained by calculating differences in the mean of larval weight in dsRNA treated samples with that of untreated control. Larval weight was measured in milligrams using an electronic balance and larval size with centimeter scale.
Fluorescent detection of dsRNA
To confirm the dsRNA uptake by larvae which were fed on treated stems, fluorescence detection was conducted using a fluorescent microscope (Olympus CH40, Japan) in the range of 460–480 nm. To detect the dsRNA in whole larva as well as in the larval midgut, florescence was observed at 6, 12, and 15 days after treatment.
Statistical analysis
Data on larval length, larval weight and percentage of larval mortality for CYP6 and APN were analyzed by factorial ANOVA using Statistix 8.1 (Analytical Software, 2005). We observed that variance of mortality among the time intervals was slightly unequal, to remove this effect, percent mortality was arc sine transformed, this transformation was made following rules mentioned in Gomez and Gomez (1984).
Determination of silencing
To detect the silencing of CYP6 and APN in treated larvae, total RNA was extracted using Trizol reagent from 4 mg of single larvae. cDNA was synthesized according to manufacturer's instructions by using Prime Script™ cDNA Synthesis Kit (Takara, Japan) from the total RNA extracted at various time intervals. The Quantitative Real time PCR (qRT-PCR) reactions were carried out with SYBR Premix Ex TaqTM (Takara Bio INC, Japan) following the manufacturer's protocol using a Real time PCR LC-96 (Roche LightCycler® 96). The qRT-PCR primers were designed using the online tool Primer3 (http://primer3.ut.ee/). The primers used were CYP6 Forward-GATTTT CGACGTTACCCTCG Reverse- CCGCTGGGTTGGTAATTCC and APN Forward- AGGATTCAAGAGCTGGTCGT Reverse- GATGACTTCGGTGTGAGGCA. For internal control, endogenous genes 18s rRNA from the Lepidopteron database and β-Actin were used (Kumar et al., 2009). Standard qRT-PCR procedures were followed with annealing of 60°C for 30 s. The specificity of the PCR reactions was monitored with melting curve analysis and gel electrophoresis. The relative gene expression data were analyzed by LC96 qPCR and by using the 2−ΔΔCt method as described previously (Schmittgen and Livak, 2008).
Effect of dsRNA on pink stem borer (PSB)
To check the specificity of RNAi effect and off-target effect of dsRNA, the designed dsRNAs were tested on rice pink stem borer (Sesamia inferens) through in vitro bioassay. Bioassays were performed as like YSB and observation were taken on larval length and weight. The data was analyzed statistically by T-test using Statistix 8.1 software (Analytical Software, 2005).
Results
Cloning of cDNAs from YSB and designing dsRNA
In this study, systematic efforts were made to demonstrate the silencing of CYP6 and APN genes in YSB through in vitro studies using rice cut stems injected with corresponding dsRNAs. cDNAs of CYP6 and APN were cloned, CYP6 cDNA of YSB showed 70% identity with Chilo suppressalis, 69% with Spodoptera, 64% with Helicoverpa, and only 69% with Rice Leaf roller Cnaphalocrocis medinalis. Similarly, APN of YSB showed 71% identity with Chilo suppressalis, 89% with Spodoptera, 82 % with Helicoverpa, and very less 69% with Rice Leaf roller Cnaphalocrocis medinalis. Among the siRNA hotspot regions, the region which does not have any homology to the related species was selected to design 21-nucleotide dsRNA. The designed two dsRNA sequences did not match to the any of the related species as well with mammalian species available in Genbank which indicates that it does not have the off targets. The cut stem assay was designed so as to simulate a real time situation wherein the larva bore the stem pieces and enter into stem. As YSB is a monophagous pest of paddy, we used cut stems of TN1 (highly susceptible variety) for standardization of dsRNA concentration. We found 30 pM concentration of dsRNA could effectively silence the CYP6 and APN genes and showed reduced growth and development, whereas, 10 and 20 pM concentration of dsRNA showed very less effect on larval growth (Supplementary Figure 1).
Effect of dsRNA on growth and development
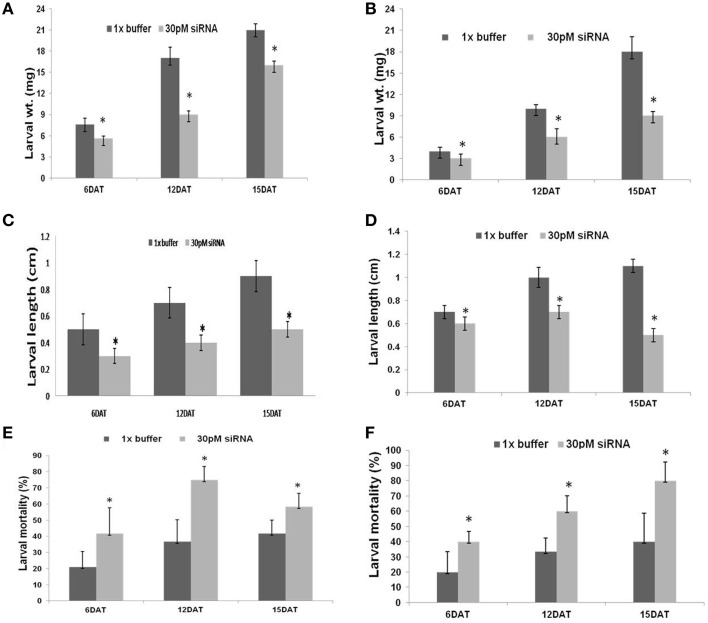
The dsRNA of CYP6 and APN had a pronounced effect on growth and development of the larvae. Larvae fed with dsRNA specific to the CYP6 and APN showed significant inhibition of growth in a time dependent manner. Treated larvae (dsRNA) showed significant reduction in growth and development characters like larval length and weight compared with untreated larvae at regular time intervals. Larvae fed on CYP6 dsRNA showed maximum weight reduction (8 mg) at 12 DAT and minimum was at 6 DAT as compared to its untreated control (Figure 2A). dsRNA designed from APN also showed significant larval weight and length reduction compared to its untreated larvae. The larval weight reduction was maximum at 15 DAT (Figure 2B). There was a significant reduction in larval length with increase in exposure to dsRNAs. Treated larvae showed a mean reduction in length of 0.45 cm at 15 DAT, 0.3 cm at 12 DAT, and 0.25 cm at 6 DAT compared to untreated control larvae (Figures 2C,D).
Figure 2.
(A) Effect of CYP6 30 pM dsRNA on YSB larval weight at different time intervals, *Indicates that larval weight in treatments were significantly different (N = 3, F = 60.27, DF = 11, P < 0.001) from larval weight in control. DAT -days after treatment. (B) Effect of APN 30 pM dsRNA on YSB larval weight at different time intervals, *Indicates that larval weight in treatments were significantly different (N = 3, F = 28, DF = 12, P < 0.002) from control, DAT- days after treatment. (C) Effect of CYP6 30 pM dsRNA on YSB larval length at different time intervals, *Indicates that larval lengths in treatments were significantly different from control larval length. (D) Effect of APN 30 pM dsRNA on YSB larval length at different time intervals, *Indicates that larval length were significantly different (N = 3, F = 35.64, DF = 14, P < 0.0001) from larval length in control. (E) Effect of CYP6 30 pM dsRNA on YSB larval mortality at different time intervals, *Indicates that larval mortality in treatments were significantly different (N = 4, F = 9.13, DF = 18, P < 0.007) from control larval mortality. (F) Effect of Amino 30 pM dsRNA on YSB larval mortality at different time intervals,*Indicates that larval mortality were significantly different (N = 5, F = 12.73, DF = 24, P < 0.0016) from control larval mortality.
Effect of dsRNA on larval mortality
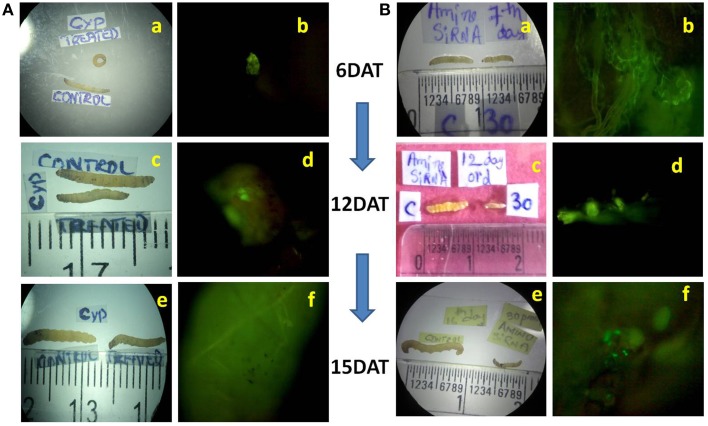
Mortality of treated larvae in both dsRNAs increased from 6 to 15 DAT. The highest mortality rate of 74.95% was observed at 12 DAT in CYP6 (Figure 2E) and 80% at 15 DAT in APN treatment (Figure 2F; Supplementary Tables 1A,B). Florescent microscope observation before and after the release of larvae in the treated and untreated stems confirms that the dsRNA abundance and translocation within the injected stem (Figures 1B,C; Supplementary Figure 2). CYP6 dsRNA fed larvae showed the presence of fluorescence in larval midgut at 6, 12, 15 DAT and it increased with the time course which was also proportional to the increase in larval mortality as well as decrease in larval weight (Figure 3A). Similarly, we found that fluorescence in larval midgut in APN dsRNA fed larvae was proportional to the effect on larval growth (Figure 3B). These observations indicate that the designed dsRNA were systemic in nature, and through oral feeding could reach the targets genes, bind to them, thus affecting the larval growth and development.
Figure 3.
(A) Effect of CYP6 dsRNA on YSB larvae fed on treated TN1 cut stems, Growth reduction in treated larvae at (a) 6th DAT, (c) 12th DAT (e) 15th DAT after feeding on dsRNA treated TN1 cut stems. Presence of fluorescence in larval midgut indicating that dsRNA was ingested by larvae when fed on treated stems at (b) 6th DAT, (d) 12th DAT, (f) 15th DAT. (B) Effect of Amino dsRNA on YSB larvae fed on treated TN1 cut stems. Growth reduction in treated larvae at (a) 6th DAT, (c) 12th DAT (e) 15th DAT after feeding on dsRNA treated TN1 cut stems. Presence of fluorescence in larval midgut at (b) 6th DAT, (d) 12th DAT, (f) 15th DAT.
Confirmation of gene silencing by qRT-PCR
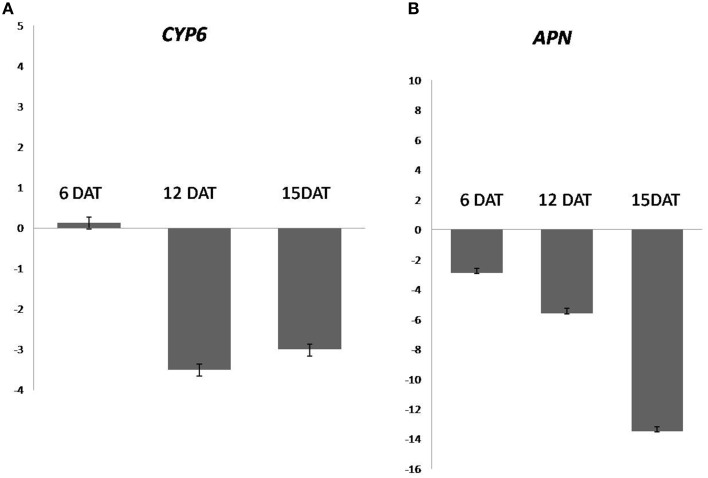
We have observed the reduction in the expression level of CYP6, APN in all the stages of dsRNA treated samples, whereas, maximum silencing was observed at 12 DAT in CYP6, and 15 DAT in case of APN. Quantitative RT–PCR revealed, CYP6 expression in treated larvae was reduced by 3.5, 3.0 fold in 12, 15 DAT, respectively. In case of APN, the fold change in gene expression was increased by reducing its expression 2.9, 5.6, 13.5 fold in 6, 12, and 15 DAT compared to the control larvae (Figures 4A,B). The reduction in transcript levels in treated samples correlated with bioassay data for CYP6 and APN genes. So these results demonstrated that both genes have key roles in maintaining YSB growth and development.
Figure 4.
Relative expression fed on TN1 cut stems treated with CYP6-dsRNA and APN-dsRNA in an individual bioassays. β-actin and 18 s were used as an internal controls (A) expression of CYP6 gene from larvae sampled at 6, 12, 15 days after treatment along with respective controls. (B) Expression of APN in 6, 12, 15 days after treatment along with respective controls.
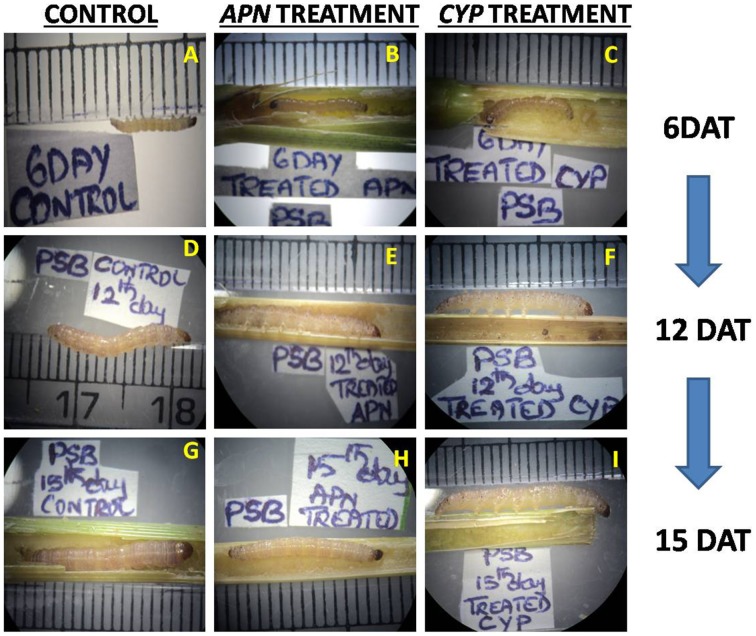
Effect of dsRNA on PSB
The PSB larvae were reared on the rice TN1 cut stems where YSB specific dsRNAs was injected into the stems. Interestingly, no abnormalities were observed in PSB larvae when fed on treated stems of both dsRNA. In APN dsRNA treatment, the PSB larvae lengths were similar to that of untreated control at 6, 12, and 15 DAT. Larval weight (mg) was also similar to the control at 6 and 12 DAT. However, decrease in larval weight was observed at 15 DAT (30 ± 1.0 mg), which is significantly lower (P-value 0.0055) compared to untreated control (49.0 ± 1.0 mg) but larval mortality was not observed. Whereas, in case of CYP6 dsRNA, we did not observe any changes in larval length and weight between control and treatments at three different time intervals. (Supplementary Table 2). Interestingly, larvae were quite active in both the treatments and mortality was not at all observed (Supplementary Data Videos 1–3) and pupae were formed. This clearly indicates that, the designed dsRNAs are specific to the YSB and they did not show any abnormal effect on PSB (Figure 5).
Figure 5.
PSB larvae fed on rice TN1 cut stems, 6th DAT (A) Control, (B) APN treated, (C) CYP treated. 12th DAT (D) Control, (E) APN treated, (F) CYP treated. 15th DAT (G) Control, (H) APN treated, (I) CYP treated.
Discussion
The success of RNAi for insect control depends on identification of suitable genes for RNAi. The candidate gene should not only have insecticidal effects on the target pests, but also safe toward other organisms, natural enemies and human beings (Agarwal et al., 2012; Kola et al., 2015). The present study showed that RNAi technology is an effective strategy for silencing the key genes involved in the metabolic activities of YSB. RNAi-triggered gene suppression through uptake of dsRNAs has been reported for many insect species belonging to Coleoptera, Diptera, Hemiptera, Hymenoptera, Isoptera, Lepidoptera, and Orthoptera (Huvenne and Smagghe, 2010).
In this study, two important genes viz., CYP6 and APN were selected based on their earlier successful reports of silencing in lepidopteron pests (Rajagopal et al., 2002; Mao et al., 2007; Sivakumar et al., 2007; Bautista et al., 2009; Crava et al., 2010; Yang et al., 2010; Zhang et al., 2011; Edi et al., 2014). The Cytochrome P450 monooxygenases are present in almost all organisms including insects. They play vital roles in hormone regulation, metabolism of xenobiotics and in biosynthesis or inactivation of endogenous compounds (Kola et al., 2015; Yu et al., 2015). Similarly, Aminopeptidase N (APNs) is primarily involved in dietary protein digestion majorly located at the midgut epithelium. APNs are known to cleave a single amino acid residue from the N-terminus of oligopeptides, preferentially the neutral amino acids (Wang et al., 2005; Pigott and Ellar, 2007). As, these two genes are known to play very important role in insect growth and development (Mao et al., 2007; Ningshen et al., 2013), we selected them as a candidate for silencing in YSB. Wang et al. (2015) succeeded in silencing CYP6AB14 by injecting dsRNA derived from CYP6AB14 into S. litura and found reduced transcript levels of CYP6AB14 and increased developmental abnormalities and higher mortality rates. Zhang et al. (2013), also observed larval mortality and reduced growth in H. armigera upon larval feeding on dsRNA designed from CYP6B6. Similar results were also reported with transgenic expression of CYP6AE14 in tobacco and cotton (Hodgson et al., 1995; Mao et al., 2007, 2011). Recently, Jin et al. (2015) designed dsRNAs targeted to Chitin synthase (Chi), CYP6AE14, and V-ATPase genes and expressed in the tobacco chloroplasts for control of Helicoverpa and observed reduced transcription of target genes in the insect midgut and stunted larval growth. Silencing of APN1 was reported in Castor Semilooper, A. janata (Ningshen et al., 2013).
For successful design of dsRNAs, there is a need to identify the specific regions or motif of key pest genes. To attain this, for the first time partial length of two candidate genes, CYP6 and APN were cloned from the YSB through PCR based strategy. Though YSB belongs to lepidopteron family, CYP6 showed 69% identity and APN 82% identities with Helicoverpa indicating significant differences. The unique regions of two YSB genes were selected for designing dsRNA, following Reynolds rules (Reynolds et al., 2004). The higher scores with these principles suggest having more stability and greater silence effects (Horn et al., 2010). Much of the commercial software currently available for designing dsRNAs follow Reynolds rule, Ui-Tei rule, Amarzguioui rule, and Tuschl rule (Naito and Ui-Tei, 2012). The dsRNAs designed in this study of CYP6 and APN genes had the highest score based on Reynolds rule, which might have positive influence of their higher expression. There were some reports of using long dsRNA for effective RNAi than small dsRNA for insects (Miller et al., 2012; Li et al., 2015), however, recent reports indicated the more effectiveness of small dsRNAs insect control (Kumar et al., 2009; Naito and Ui-Tei, 2012; Gong et al., 2013). Our designed dsRNA from CYP6 lies in the principle domain of CYP450 group of enzymes, which belongs to haem-thiolate proteins involved in the oxidative degradation of various compounds. This class of proteins has domains viz., haem-binding loop (with an absolutely conserved cysteine that serves as the 5th ligand for the haem iron), the proton-transfer groove and the absolutely conserved EXXR motif in helix K forms a principle domain. Similarly APN (zinc-dependent metallopeptidases) is a Type II integral membrane protease belongs to Gluzincin family (thermolysin-like peptidases or TLPs), this family consists of several zinc-dependent metallopeptidases including M1 peptidases and know to consist of a small N-terminal cytoplasmic domain, a single transmembrane domain and a large extracellular ectodomain that contains the active site. The dsRNA designed is lies in the part of active site of this enzyme so we expect that it binds to the target effectively.
Generally insect bioassays have been carried out through introducing dsRNA into an organism by microinjection (Ghanima et al., 2007), soaking and oral feeding through artificial diet (Mao et al., 2007; Chen et al., 2008). Since, YSB is a monophagous pest and does not have artificial diet; we resorted to rearing on rice cut stems, so as to simulate a real time situation where larva bore the stem pieces and enter into stem. In earlier studies cut stem assay were used to assess the toxicity of various toxins and transgenics against YSB (Nayak et al., 1997; Nguyen Huu Ho et al., 2001; Ramesh et al., 2004; Padmakumari et al., 2013). We found lower concentration of dsRNA (30 pM) was effective for silencing of target genes. Kumar et al. (2009) also found that 50 nM concentrations were effective for silencing Acetyl cholinesterase (Ache) gene in H. armigera. Similarly, Bautista et al. (2009) reported that 250 ng dsRNA targeting CYP6BG1 delivery through droplet feeding to 4th instar larvae of Helicoverpa greatly reduced the CYP6BG1 expression. To track accurately the presence of dsRNA in various insect tissues, FAM was used to label the dsRNA and confirmed their systemicity in insect tissue. Earlier reports have indicated the use of fluorescent dyes like Fluoroscein isothiocyanate (FITC), Cyanine Cy-3, Cy-5, and FAM to track the movement and binding of dsRNA in the tissues (Urwin et al., 2002; Karim et al., 2010; Hui et al., 2011; Wuriyanghan et al., 2011; Bolognesi et al., 2012; Li et al., 2015).
Our results indicated the increase of YSB larval mortality rates in feeding assay with dsRNA of CYP6 and APN genes. Change in growth and developmental characters were observed from very early larval developmental stages by showing reduced larval growth (larval weight and length), delayed molting and led to death. Interestingly, none of the larvae entered into pupae form. We observed an increase in fluorescence signal with increase in time of exposure which indicates the signal amplification. These observations were correlated with qRT-PCR experiments data.
The designed dsRNA sequences did not match with mammals or related species which are present in GenBank and beneficial insects which suggests that the designed dsRNA molecules may be specific to target insect and may not have unintended effects. Further to make sure of specificity of these molecules, bioassays were carried out in PSB, another stem borer of rice. The results indicated that dsRNAs designed from CYP6 and APN did not showed significant effect on larval length and weight except in case of 15 DAT with APN treatment. Further investigations are required to fully understand the delayed response of APN treatment in PSB larval weight. There was no morality in both the cases, larvae were quite active completed their instars and entered in to the pupal stage. These results suggest that the YSB dsRNAs of CYP6 and APN genes could not find targets in PSB which further confirms that the designed dsRNAs were more specific to YSB.
In our study, we conclusively proved the key functions of Cytochrome P450 and Aminopeptidase N enzymes in various metabolic pathways of YSB such as normal growth and also its involvement in many stages of development. The present results strongly suggest that these CYP6 and APN genes can be potential targets for insect-control, and insect-resistant transgenic plants may be obtained through RNAi-mediated silencing of insect genes. The off targeting study through in silico and in vivo showed that the designed dsRNA from these genes are highly specific to insects and do not have any unintended effects on other organisms.
Author contributions
Conceived and designed the experiment: MSM, SKM, KVSR, PR, APP, SMB, and VRB.
Conflict of interest statement
The authors declare that the research was conducted by the support of DBT, Govt of India.
Acknowledgments
The work was carried out with the grant (Grant BT/PR2468/AGr/36/701/2011 dt 23.05.2012) from Department of Biotechnology, Government of India. The authors are thankful to Director, IIRR for providing research facilities, Dr. P.C Latha, Scientist-IIRR for providing the fluorescent microscope and Dr. M. Sampath Kumar, Scientist-IIRR for providing the larvae of PSB.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2016.00020
References
- Agarwal S., Mohan M., Mangrauthia S. K. (2012). RNAi: machinery and role in pest and disease management, in Crop Stress and its Management: Per- spectives and Strategies, eds Venkateshwarlu B., Shankar A. K., Shankar C., Maheshwari M. (Springer; ), 447–469. [Google Scholar]
- Analytical Software (2005). Statistix Version 8.1: User's Manual. Tallahassee, FL: Analytical Software. [Google Scholar]
- Bandong J. P., Litsinger J. A. (2005). Rice crop stage susceptibility to the rice yellow stem borer Scirpophaga incertulas (Walker) (Lepidoptera: Pyralidae). Int. J. Pest. Manag. 51, 37–43. 10.1080/09670870400028276 [DOI] [Google Scholar]
- Bautista M. A., Miyata T., Miura K., Tanaka T. (2009). RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem Mol. Biol. 39, 38–46. 10.1016/j.ibmb.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Bolognesi R., Ramaseshadri P., Anderson J., Bachman P., Clinton W., Flannagan R., et al. (2012). Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE. 7:e47534. 10.1371/journal.pone.0047534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar D. S., Khush G. S. (1997). Alien introgression in rice. Plant Mol. Bio. 35, 35–47. [PubMed] [Google Scholar]
- Bravo A., Gill S. S., Soberón M. (2007). Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49, 423–435. 10.1016/j.toxicon.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Tian H., Zou L., Tang B., Hu J., Zhang W. (2008). Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull. Entomol. Res. 98, 613–619. 10.1017/S0007485308005932 [DOI] [PubMed] [Google Scholar]
- Crava C. M., Bel Y., Lee S. F., Manachini B., Heckel D. G., Escriche B. (2010). Study of the aminopeptidase N gene family in the lepidopterans Ostrinia nubilalis (Hübner) and Bombyx mori (L.): sequences, mapping and expression. Insect Biochem. Mol. Biol. 40, 506–515. 10.1016/j.ibmb.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Edi C. V., Djogbénou L., Jenkins A. M., Regna K., Muskavitch M. A. T. (2014). CYP6 P450 Enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 10:e1004236. 10.1371/journal.pgen.1004236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Ghanima M., Kontsedalov S., Czosnek H. (2007). Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem Mol Biol. 37, 732–738. 10.1016/j.ibmb.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Gomez K. A., Gomez A. A. (1984). Statistical Procedures for Agricultural Research, 2nd Edn. NewYork, NY: John wiley and sons. [Google Scholar]
- Gong L., Chen Y., Hu Z., Hu M. (2013). Testing insecticidal activity of novel chemically synthesized siRNA against Plutella xylostella under laboratory and field conditions. PLoS ONE 8:e62990. 10.1371/journal.pone.0062990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Q., Zhang J. Z., Yang M. L., Yan L. Z., Zhu K. Y., Guo Y. P., et al. (2011). Comparative analysis of cytochrome P450-like genes from Locusta migratoria manilensis (Meyen): expression profiling and response to insecticide exposure. Insect Sci. 19, 75–85. 10.1111/j.1744-7917.2011.01450.x [DOI] [Google Scholar]
- Hamilton A., Voinnet O., Chappell L., Baulcombe D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. 10.1093/emboj/cdf464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E., Rose R. L., Ryu D. Y., Falls G., Blake B. L., Levi P. E. (1995). Pesticide-metabolizing enzymes. Toxicol. Lett. 82–83, 73–81. 10.1016/0378-4274(95)03469-2 [DOI] [PubMed] [Google Scholar]
- Horn T., Sandmann T., Boutros M. (2010). Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol. 11, R61. 10.1186/gb-2010-11-6-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui X. M., Yang L. W., He G. L., Yang Q. P., Han Z. J., Li F. (2011). RNA interference of ace1 and ace2 in Chilo suppressalis reveals their different contributions to motor ability and larval growth. Insect Mol. Biol. 20, 507–518. 10.1111/j.1365-2583.2011.01081.x [DOI] [PubMed] [Google Scholar]
- Huvenne H., Smagghe G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. 10.1016/j.jinsphys.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Jin S., Singh N. D., Li L., Zhang X., Daniell H. (2015). Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnol. J. 13, 435–446. 10.1111/pbi.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S., Troiano E., Mather T. N. (2010). Functional genomics tool: gene silencing in Ixodes scapularis eggs and nymphs by electroporated dsRNA. BMC Biotech. 10:1. 10.1186/1472-6750-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola V. S. R., Renuka P., Madhav M. S., Mangrauthia S. K. (2015). Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front. Physiol. 6:119. 10.3389/fphys.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Gupta G. P., Rajam M. V. (2009). Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J. Insect Physiol. 55, 273–278. 10.1016/j.jinsphys.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Li Z., Zeng B., Ling L., Xu J., You L., Aslam A. F., et al. (2015). Enhancement of Larval RNAi Efficiency by Over-expressing Argonaute2 in Bombyx mori. Int. J. Biol. Sci. 11, 176–185. 10.7150/ijbs.10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. B., Cai W. J., Wang J. W., Hong G. J., Tao X. Y., Wang L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. 10.1038/nbt1352 [DOI] [PubMed] [Google Scholar]
- Mao Y. B., Tao X. Y., Xue X. Y., Wang L. J., Chen X. Y. (2011). Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Trans Res. 20, 665–673. 10.1007/s11248-010-9450-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. C., Miyata K., Brown S. J., Tomoyasu Y. (2012). Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE. 7:e47431. 10.1371/journal.pone.0047431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Ui-Tei K. (2012). siRNA design software for a target gene-specific RNA interference. Front. Genet. 3:102. 10.3389/fgene.2012.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak P., Debabrata B., Sampa D., Asitava B., Dipankar G., Neeliyath R., et al. (1997). Transgenic elite indica rice plants expressing CryIAc -endotoxin of Bacillus thuringiensis are resistant against yellow stem borer (Scirpophaga incertulas). Proc. Natl. Acad. Sci. U.S.A. 94, 2111–2116. 10.1073/pnas.94.6.2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Huu Ho Nguyen Van Uyen, Karabi Datta, Swapan Kumar Datta. (2001). Production of transgenic rice plants resistant to yellow stem borer and herbicide in two Vietnamese varieties via Agrobacterium tumefaciens. Omonrice 9, 30–35. [Google Scholar]
- Ningshen T. J., Aparoy P., Ventaku V. R., Dutta-Gupta A. (2013). Functional Interpretation of a Non-Gut Hemocoelic Tissue Aminopeptidase N (APN) in a lepidopteran insect pest Achaea janata. PLoS ONE 8:e79468. 10.1371/journal.pone.0079468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumari A. P., Katti G., Sailaja V., Padmavathi C. H., Jhansi Lakshmi V., Prabhakar M., et al. (2013). Delineation of larval instars in field populations of rice yellow stem borer, Scirpophaga incertulas (Walk.). Oryza 50, 259–267. [Google Scholar]
- Pathak M. D., Khan Z. R. (1994). Insect Pests of Rice. Manila: International Rice Research Institute. [Google Scholar]
- Pigott C. R., Ellar D. J. (2007). Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71, 255–281. 10.1128/MMBR.00034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. R. G., Gatehouse J. A. (2008). RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. 10.1016/j.tibtech.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Rajagopal R., Sivakumar S., Agrawal N., Malhotra P. M., Bhatnagar R. K. (2002). Silencing of midgut aminopeptidase N of Spodoptera litura by double stranded RNA establishes its role as Bacillus thuringiensis receptor. J. Biol. Chem. 277, 46849–46851. 10.1074/jbc.C200523200 [DOI] [PubMed] [Google Scholar]
- Ramesh S., Nagadhara D., Pasalu I. C., PadmaKumari A. P., Sarma N. P., Reddy V. D., et al. (2004). Development of stem borer resistant transgenic parental lines involved in the production of hybrid rice. J. Biotech. 111, 131–141. 10.1016/j.jbiotec.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Reed B. J., Chandler D. S., Sandeman R. M. (1999). Aminopeptidases as potential targets for the control of the Australian sheep blowfly, Lucilia cuprina. Int. J. Parasitol. 29, 839–850. 10.1016/S0020-7519(99)00043-0 [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khorova A. (2004). Rational siRNA design for RNA interference. Nat. Biotech. 22, 326–330. 10.1038/nbt936 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C (T) method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Scott J. G. (2008). Insect cytochrome P450s: thinking beyond detoxification. Recent Adv. Insect Physiol. Toxicol. Mol. Biol. 1, 117–124. [Google Scholar]
- Sivakumar S., Rajagopal R., Venkatesh G. R., Srivastava A., Bhatnagar R. K. (2007). Knockdown of aminopeptidase -N from Helicoverpa armigera larvae and in transfected SF21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J. Biol. Chem. 282, 7312–7319. 10.1074/jbc.M607442200 [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E., Brévault T., Carrière Y. (2013). Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31, 510–521. 10.1038/nbt.2597 [DOI] [PubMed] [Google Scholar]
- Urwin P. E., Lilley C. J., Atkinson H. J. (2002). Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 15, 747–752. 10.1094/MPMI.2002.15.8.747 [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang X., Zhang J. (2005). Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 35, 611–620. 10.1016/j.ibmb.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Wang R. L., Xia Q. Q., Baerson S. R., Ren Y., Wang J., Su Y. J., et al. (2015). A novel cytochrome P450 CYP6AB14 gene in Spodoptera litura (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J. Insect Physiol. 75, 54–62. 10.1016/j.jinsphys.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Wuriyanghan H., Rosa C., Falk B. W. (2011). Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PLoS ONE 6:e27736. 10.1371/journal.pone.0027736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., You M., Vasseur L., Zhao Y., Liu C. (2011). Development of RNAi in insects and rnai-based pest control, pesticides, in The Modern World - Pests Control and Pesticides Exposure and Toxicity Assessment, ed Margarita Stoytcheva (Shanghai: Intechopen; ), 27–38. [Google Scholar]
- Yang Y., Zhu Y. C., Ottea J., Husseneder C., Leonard B. R., Abel C., et al. (2010). Molecular characterization and RNA interference of three midgut aminopeptidase N isozymes from Bacillus thuringiensis-susceptible and -resistant strains of sugarcane borer, Diatraea saccharalis. Insect Biochem. Mol. Biol. 40, 592–603. 10.1016/j.ibmb.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Yu L., Tang W., He W., Ma X., Vasseur L., Baxter S. W., et al. (2015). Characterization and expression of the cytochrome P450 gene family in diamond back moth, Plutella xylostella (L.). Sci. Rep. 5, 8952 10.1038/srep08952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu X., Ma J., Zhao J. (2013). Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. Bull. Entomol. Res. 103, 584–591. 10.1017/S0007485313000151 [DOI] [PubMed] [Google Scholar]
- Zhang Y. L., Kulye M., Yang F. S., Xiao L., Zhang Y. T., Zeng H. M., et al. (2011). Identification, characterization, and expression of a novel P450 gene encoding CYP6AE25 from the Asian corn borer, Ostrinia furnacalis. J. Insect Sci. 11, 37. 10.1111/j.1744-7917.2004.tb00178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. J., Ma C. X., Li M., Sheng C. F., Liu H. X., Qiu X. H. (2010). CYP9A12 and CYP9A17 in the cotton bollworm, Helicoverpa armigera: sequence similarity, expression profile and xenobiotic response. Pest Manag. Sci. 66, 65–73. 10.1002/ps.1832 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.