ABSTRACT
Cystic fibrosis (CF) is a human genetic disorder which results in a lung environment that is highly conducive to chronic microbial infection. Over the past decade, deep-sequencing studies have demonstrated that the CF lung can harbor a highly diverse polymicrobial community. We expanded our existing in vitro model of Pseudomonas aeruginosa biofilm formation on CF-derived airway cells to include this broader set of CF airway colonizers to investigate their contributions to CF lung disease, particularly as they relate to the antibiotic response of the population. Using this system, we identified an interspecies interaction between P. aeruginosa, a bacterium associated with declining lung function and worsening disease, and Streptococcus constellatus, a bacterium correlated with the onset of pulmonary exacerbations in CF patients. The growth rate and cytotoxicity of S. constellatus 7155 and P. aeruginosa PA14 were unchanged when grown together as mixed biofilms in the absence of antibiotics. However, the addition of tobramycin, the frontline maintenance therapy antibiotic for individuals with CF, to a mixed biofilm of S. constellatus 7155 and P. aeruginosa PA14 resulted in enhanced S. constellatus biofilm formation. Through a candidate genetic approach, we showed that P. aeruginosa rhamnolipids were reduced upon tobramycin exposure, allowing for S. constellatus 7155 biofilm enhancement, and monorhamnolipids were sufficient to reduce S. constellatus 7155 biofilm viability in the absence of tobramycin. While the findings presented here are specific to a biofilm of S. constellatus 7155 and P. aeruginosa PA14, they highlight the potential of polymicrobial interactions to impact antibiotic tolerance in unanticipated ways.
IMPORTANCE Deep-sequencing studies have demonstrated that the CF lung can harbor a diverse polymicrobial community. By recapitulating the polymicrobial communities observed in the CF lung and identifying mechanisms of interspecies interactions, we have the potential to select the best therapy for a given bacterial community and reveal potential opportunities for novel therapeutic interventions. Using an in vitro model of bacterial infection on CF airway cells, we tested how a particular polymicrobial community grows, damages human cells, and responds to antibiotics in single and mixed infections. We describe here the mechanism of an interspecies interaction between two pathogens in the CF lung, P. aeruginosa and S. constellatus, which is potentiated by a commonly prescribed antibiotic, tobramycin.
INTRODUCTION
Cystic fibrosis is a human genetic disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Mutations in the CFTR gene result in a wide array of deleterious effects throughout the body, including chronic lifelong respiratory infections, the primary cause of morbidity and mortality in patients with cystic fibrosis. Historically, chronic lung infections have been attributed to relatively few organisms, including Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and Burkholderia cepacia complex (1–3). However, deep-sequencing studies have revealed that this list of microbes underestimates and oversimplifies the polymicrobial communities that reside in the airways of patients with cystic fibrosis (CF) (4–10). Dozens of bacterial genera have been identified in a single sputum sample, including organisms not generally identified by conventional culturing, such as Streptococcus spp., and anaerobes, including Prevotella (4–14).
Interspecies interactions within polymicrobial communities can be cooperative and competitive and can be influenced by the presence of antibiotics (15–17). Of particular interest in the CF lung is the potential for interspecies interactions between P. aeruginosa and Streptococcus species. P. aeruginosa is the most abundant and prevalent organism identified by deep-sequencing and culture-based methods in sputum samples from adult CF patients (4–10, 18). Colonization by P. aeruginosa is associated with worsening lung function and disease progression (1–3) and thus has received the most attention in studies of microbial pathogenesis in the CF lung.
Interestingly, several recent deep-sequencing studies identified Streptococcus spp. as among the top three most abundant and prevalent organisms in CF sputum, and in a study from our group, Streptococcus spp. were the predominant organisms in the sputum of ∼50% of the patients at our CF center (4, 6, 9). Despite the high abundance and prevalence of Streptococcus spp. in CF sputum, the role that Streptococcus spp. play in disease is unclear. In a cross-sectional study, infection with Streptococcus was reported to be associated with better health (6). However, Sibley et al. (11, 12) and Parkins et al. (19) reported that high numbers of Streptococcus milleri group organisms (S. anginosus, S. constellatus, and S. intermedius) in sputum samples are correlated with the onset of pulmonary exacerbation (11, 12, 19). The S. milleri group can be identified in the sputum samples from comparatively healthy (stable) CF patients (6); therefore, the simple presence or absence of a given microbe does not accurately predict the virulence of a community. Thus, identifying potential pathogenic and/or health-promoting bacterial interactions within the CF sputum community requires more information than survey studies provide.
Here, we sought to analyze how the composition of a bacterial community contributes to its physiology, pathogenesis, and antibiotic response in a mixed-culture model of cystic fibrosis. Given the studies showing that the S. milleri group is associated with the onset of pulmonary exacerbations, we hypothesized that a mixed community of P. aeruginosa and members of the S. milleri group would exhibit increased virulence or increased antibiotic tolerance compared to that with each bacterium alone. To this end, we expanded our previously described coculture system of P. aeruginosa biofilm formation on the surface of CF airway cells (20–22) to include the S. milleri group and other microbes identified by deep sequencing as being highly abundant and prevalent. In addition, we examined mixed communities of S. milleri group organisms and P. aeruginosa and their responses to antibiotic exposure. With this expanded system, we identified and characterized one example of interspecies interaction between P. aeruginosa PA14 and S. constellatus 7155 when treated with the commonly prescribed antibiotic tobramycin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. aeruginosa strains used in this study are listed in Table S1 in the supplemental material and were grown on LB agar or liquid supplemented with 25 μg/ml gentamicin, 100 μg/ml carbenicillin, and where appropriate, shaking at 37°C. S. constellatus 7155 was grown on blood agar and Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) and 20 μl/ml Oxyrase (Oxyrase, Inc.) under static conditions at 37°C in a 5% CO2 atmosphere. For mixed-culture assays, P. aeruginosa was selected on Pseudomonas isolation agar at 37°C, and S. constellatus was enriched on tryptic soy agar (TSA) supplemented with 5% defibrinated sheep's blood (blood agar) and grown anaerobically in GasPak Jars at 37°C. Information relevant to the growth of the other microbes mentioned in the manuscript is detailed in the supplemental material.
Tissue culture cultivation.
The cystic fibrosis bronchial epithelial (CFBE) cell line used in this study overexpresses F508del-cystic fibrosis transmembrane conductance regulator (CFTR) via stable lentiviral transfection of human bronchial epithelial cells (23). The parent cell line, named CFBE41o−, was isolated from a CF patient who was homozygous for the F508del-CFTR mutation and was originally characterized by Bruscia and colleagues (24) and Cozens et al. (25). CFBE cells, here referred to as CF airway cells, were the generous gift of J. P. Clancy. CF airway cells were cultivated as previously described (20, 22). Briefly, CF airway cells were seeded into 24-well plates at 50,000 cells/well and fed every other day with minimal essential medium (MEM) (Life Technologies) supplemented with 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 2 μg/ml puromycin, 5 μg/ml Plasmocin (InvivoGen), and 10% fetal bovine serum until confluent and tight junctions had formed (5 to 7 days).
Growth kinetics of biofilms formed on CF airway cells.
Assays of biofilms formed on CF airway cells were performed as previously described (20–22), with the following modifications. An overnight culture of P. aeruginosa or S. constellatus was washed and resuspended in 1 ml of MEM. A P. aeruginosa inoculum was prepared to an optical density at 600 nm (OD600) of 0.05 (∼5 × 107 CFU/ml), and an S. constellatus inoculum was prepared to an OD600 of 0.1 (∼5 × 107 CFU/ml), both in MEM supplemented with 2 mM l-glutamine. A 1:1 mixture of P. aeruginosa and S. constellatus was prepared from these suspensions. Half a milliliter of each suspension (P. aeruginosa alone, S. constellatus alone, and a 1:1 mixture) was then gently added to each well of CF airway cells that had been washed twice with MEM. The cocultures were incubated 1 h at 37°C in 5% CO2. At 1 h, unattached bacteria were removed by aspiration and the medium replaced with MEM supplemented with 2 mM l-glutamine and 0.4% arginine. Biofilms were allowed to incubate for a total of 1, 3, or 6 h. At each time point, planktonic cells were removed by aspiration, and the remaining biofilms were washed once with MEM supplemented with 2 mM l-glutamine and 0.4% arginine. Fresh MEM supplemented with 2 mM l-glutamine and 0.4% arginine was added to the wells before scraping biofilms with a pipette tip. Biofilm-grown bacteria were serially diluted and plated on Pseudomonas isolation agar (PIA) and blood agar, as described above, to identify P. aeruginosa and Streptococcus spp., respectively. PIA was incubated overnight at 37°C, and blood agar was incubated overnight anaerobically in GasPak jars at 37°C. After overnight incubation, the resulting colonies were counted and the CFU determined.
Biofilm antibiotic assay.
Coculture biofilm assays were performed as previously described (20), with the following modifications. The coculture assays were inoculated as described above in the growth kinetic assays. One hour postinoculation, unattached cells were removed by aspiration and the medium replaced with MEM supplemented with 2 mM l-glutamine and 0.4% arginine. Six hours postinoculation, the unattached bacteria were removed by aspiration; the biofilm fraction was washed once with MEM supplemented with 2 mM l-glutamine and 0.4% arginine and then incubated with or without 5 μg/ml tobramycin in MEM supplemented with 2 mM l-glutamine and 0.4% arginine. Twenty-one hours postinoculation, planktonic fractions were removed and saved for use in a drop collapse assay. MEM supplemented with 2 mM l-glutamine and 0.4% arginine was added to the wells, and biofilms were disrupted by scraping with a pipette tip. The biofilm-grown bacteria were serially diluted and then plated on PIA and blood agar to distinguish the microbes, as described above. PIA was incubated overnight at 37°C, and blood agar was incubated anaerobically overnight at 37°C in GasPak jars. After overnight incubation, the resulting colonies were counted and the CFU determined. Antibiotic assays on plastic were performed exactly as on CF airway cells, except biofilms were formed directly on plastic tissue culture-treated 24-well plates. When indicated, rhamnolipids (50 μg/ml, dissolved in MEM; Sigma) were added to washed preformed biofilms at 6 h postinoculation. When indicated, heat-killed wild-type P. aeruginosa cells were added to the washed preformed biofilms at 6 h postinoculation. Heat-killed P. aeruginosa was prepared from an overnight culture grown in LB that was washed once and resuspended in 1 ml of MEM. The OD600 was calculated for this suspension, and an appropriate volume of cells was heat killed at 80°C for 10 min. The heat-killed bacteria were then resuspended in MEM supplemented with 2 mM l-glutamine and 0.4% arginine with or without tobramycin and added to the washed preformed biofilms.
Microscopy.
MatTek dishes were seeded with 200,000 CF airway cells/dish and grown until tight junctions formed (5 to 7 days), as described previously (20–22). P. aeruginosa PA14 harboring a constitutively expressed gfp gene (26) and S. constellatus 7155 were prepared as described for the coculture assays and added to twice-washed CF airway cells. Unattached bacteria were removed after 1 h, and the medium was replaced with MEM supplemented with 2 mM l-glutamine and 0.4% arginine. At 3 and 4 h postinoculation, the unattached bacteria were removed, and the medium was replaced with MEM supplemented with 2 mM l-glutamine, 0.4% arginine, and 2.5 μl of a 3.125% hexidium iodine solution (which selectively stains Gram-positive bacteria red to stain S. constellatus) and incubated for 15 min at 37°C in 5% CO2. Single-strain and mixed biofilms were visualized on a Nikon Eclipse Ti inverted microscope with a 100× objective, and images were acquired using the Nikon ND acquisition software and a Hamamatsu C11440 Orca-Flash 4.0 digital camera. P. aeruginosa was visualized via its green fluorescence with the green fluorescent protein (GFP) channel of the ND acquisition software; S. constellatus was visualized via its red fluorescence in the mCherry channel of the ND acquisition software.
Drop collapse assay.
Drop collapse assays were performed as previously described (27, 28). Briefly, planktonic fractions from biofilm assays were collected and clarified by centrifugation. The clarified supernatants were serially diluted (1:1) with water in a 96-well plate. Thirty microliters of each dilution was spotted onto the circles on the underside of the lid of the 96-well plate and assessed for surfactant activity, as measured by the spread of the droplet. As surfactant quantities are reduced by dilution, surface tension increases, resulting in the beading up of the droplet. Surfactant scores are equal to the reciprocal of the greatest dilution at which there was surfactant activity (a collapsed drop). A score of 1 is assigned to surfactant activity in the undiluted supernatant of the wild-type strain.
Cytotoxicity assays.
Cytotoxicity was measured as fraction of lactate dehydrogenase (LDH) release compared to total lysis control of uninfected CF airway cells treated with Triton X-100 detergent to completely lyse cells and release all intracellular LDH using the Cyto Tox 96 nonradioactive cytotoxicity kit (Promega), according to the manufacturer's instructions.
RESULTS
Expanded model system of bacterial biofilm formation on CF airway cells.
Our existing model system of bacterial biofilm formation on the surface of CF-derived bronchial epithelial (CFBE) cells was originally designed to study P. aeruginosa biofilms. P. aeruginosa grown in this model system recapitulates several key aspects of chronic biofilm formation, including the formation of biofilm-like microcolonies, expression of genes associated with biofilm growth, induction of quorum sensing, requirement for genes necessary for biofilm formation on abiotic surfaces, and, of importance in the clinic, increased antibiotic tolerance (20, 22). We expanded this system to assess the biofilm-forming ability of seven Streptococcus species and other microbes identified by deep-sequencing studies as highly abundant and/or prevalent in sputum from CF patients, including Prevotella intermedia, S. aureus, Fusobacterium nucleatum, Stenotrophomonas maltophilia, and Gemella species (4–6, 9). Each species was inoculated individually onto CF airway cells for a total of 1, 3, or 6 h and assessed for bacterial viability. Fifteen of the 16 species tested were able to grow on the surface of CF airway cells (see Fig. S1A in the supplemental material). We were unable to recover viable bacteria from obligate anaerobe F. nucleatum cultures on CF airway cells, likely because our culturing conditions were not anoxic (see Fig. S1A).
Given that P. aeruginosa and S. milleri group organisms may influence the overall pathogenesis in the CF lung (1–3, 6, 11, 12, 14, 19), we sought to assess whether P. aeruginosa and Streptococcus interactions might impact cell growth or cytotoxicity to host cells. Pairwise combinations of P. aeruginosa and S. milleri group organisms were grown as biofilms on CF airway cells, assayed for their cytotoxicity and bacterial growth kinetics, and compared with respect to their single-species cytotoxicity and growth kinetics. S. milleri group and P. aeruginosa mixed biofilms on CF airway cells showed a level of cytotoxicity after 1 and 3 h of biofilm formation similar to that seen with single P. aeruginosa biofilms. At 6 h, the two strains of S. constellatus showed similar levels of cytotoxicity toward host cells, while S. anginosus and S. intermedius showed reduced levels of cytotoxicity toward host cells compared to those with single P. aeruginosa biofilms formed on CF airway cells (see Fig. S1B in the supplemental material). P. aeruginosa growth was largely unaffected by growth in the presence of the four streptococcal strains tested (Fig. 1A; see also Fig. S1C in the supplemental material). We did observe a modest but significant reduction of P. aeruginosa growth at 6 h (t test, P < 0.05) when this microbe was cocultured with S. constellatus 7155 (Fig. 1A), but no difference in CFU was observed by 24 h. While coculture with P. aeruginosa did not impact the growth of S. constellatus 7155 (Fig. 1B), S. constellatus C818, or S. anginosus, we did notice a significant impact of growth on a coculture of S. intermedius with P. aeruginosa, as described in Fig. S1D to F in the supplemental material.
FIG 1.
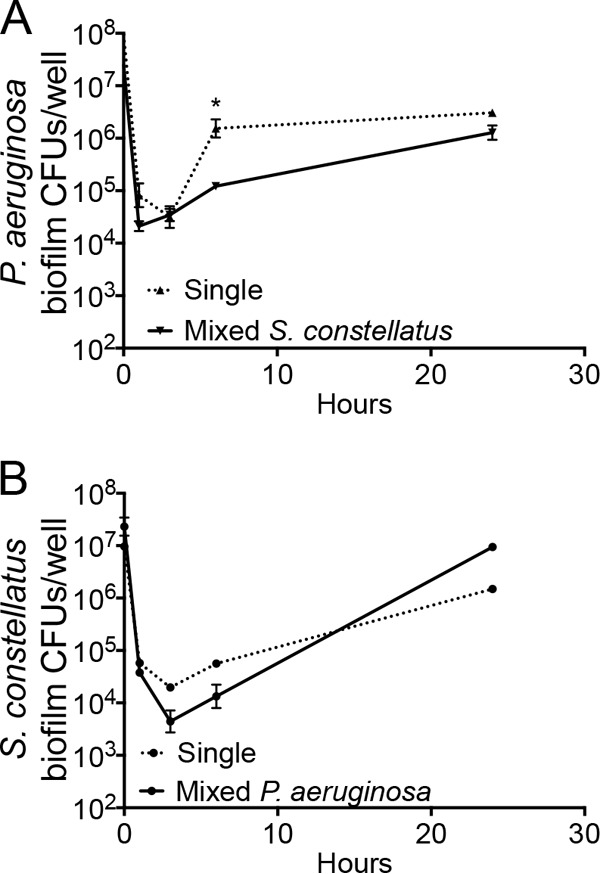
Growth kinetics of P. aeruginosa and S. constellatus in CF airway cells. (A) P. aeruginosa growth kinetics are largely unaffected by the presence of S. constellatus 7155. The dashed line indicates the viability of P. aeruginosa grown as single biofilms on CF airway cells, and the solid line indicates the viability of P. aeruginosa grown as mixed biofilms on CF airway cells with S. constellatus 7155. The only significant difference was noted at 6 h, with no significant difference at 24 h. *, P < 0.05. (B) S. constellatus 7155 growth kinetics are unaffected by the presence of P. aeruginosa PA14. The dashed line indicates viability of S. constellatus 7155 grown as single biofilms on CF airway cells, and the solid line indicates viability of S. constellatus 7155 grown as mixed biofilms on CF airway cells with P. aeruginosa. For all panels, the symbols and error bars indicate the means and standard deviations (SD) of the results from three biological replicates.
Tobramycin treatment enhances growth of S. constellatus 7155 in a mixed biofilm with P. aeruginosa.
Individuals with CF receive aerosolized antibiotics on a regular basis as part of a maintenance therapy regimen and intravenous antibiotics upon admission to the hospital during a pulmonary exacerbation. We therefore hypothesized that antibiotic treatment would affect the physiology and/or pathogenesis of P. aeruginosa or the S. milleri group in mixed biofilms. To test this idea, we established single and mixed biofilms of P. aeruginosa and S. constellatus, S. anginosus, or S. intermedius for 6 h on CF airway cells before treating the established biofilms with 5 μg/ml tobramycin or growth medium as a control, as previously reported (20–22). After 15 h of antibiotic treatment, bacterial biofilm viability (measured as the number of CFU per well) was determined for the mixed community and compared to the bacterial biofilm viability for each organism grown individually under the same conditions.
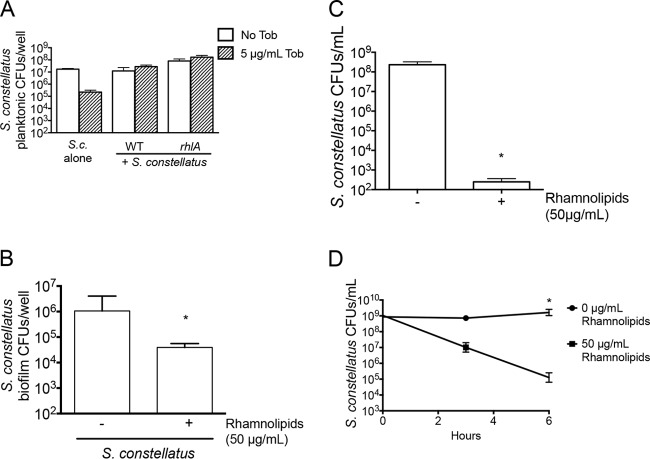
Viable P. aeruginosa levels decreased >200-fold when P. aeruginosa was treated with 5 μg/ml tobramycin and grown alone or in mixed biofilms with S. constellatus 7155 (Fig. 2A). S. constellatus 7155 grown alone on CF airway cells showed a 2.5-fold decline in viability when treated with 5 μg/ml tobramycin. A mixed biofilm of S. constellatus 7155 and P. aeruginosa in the absence of tobramycin resulted in a nonsignificant trend toward enhanced S. constellatus 7155 viability similar to that seen with single S. constellatus 7155 biofilms after 21 h of growth. However, when mixed biofilms of S. constellatus 7155 and P. aeruginosa were treated with tobramycin, the viability of S. constellatus 7155 statistically significantly increased 6-fold compared to that of the mixed community without tobramycin and 292-fold compared to that of single S. constellatus 7155 biofilms in the presence of this antibiotic (Fig. 2A). Thus, the presence of P. aeruginosa PA14 may be beneficial for S. constellatus 7155 when this microbe is grown in biofilms on CF airway cells, and S. constellatus 7155 clearly benefits when the community is treated with tobramycin. Interestingly, we saw no differences in single and mixed cultures with or without tobramycin when we examined the planktonic population (Fig. 2B) or cytotoxicity (Fig. 2C).
FIG 2.
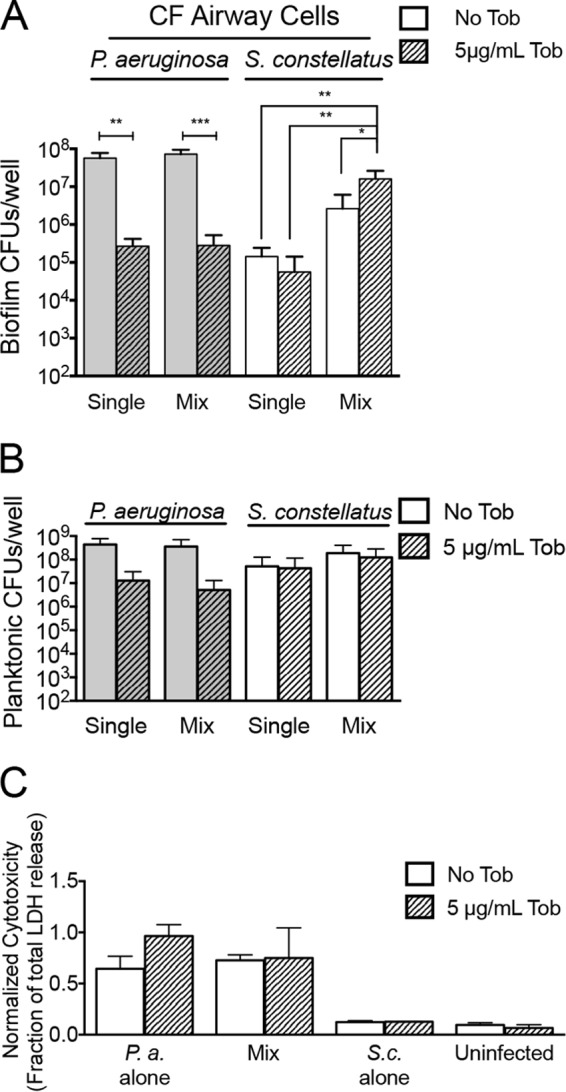
Tobramycin (Tob) treatment of P. aeruginosa and S. constellatus mixed biofilms enhances S. constellatus viability. The data represent the viability of bacteria grown as biofilms at 21 h on CF airway cells from assays of single or mixed biofilms of P. aeruginosa and S. constellatus 7155 treated with no tobramycin or with 5 μg/ml tobramycin. (A) Viability of bacteria in the biofilm fraction. (B) Viability of bacteria in the planktonic fraction. (C) Cytotoxicity of indicated mixed biofilms on CF airway cells. Cytotoxicity was normalized to total cell lysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by analysis of variance (ANOVA) with Tukey's posttest for multiple comparisons. P.a., P. aeruginosa; S.c., S. constellatus. For all panels, the bars and error bars indicate the means and SD of the results from three biological replicates.
The coculture of S. constellatus 7155 with P. aeruginosa PAO1 did not show a significant enhancement in S. constellatus 7155 growth in the presence or absence of tobramycin (see Fig. S2A in the supplemental material). We also found no evidence of a tobramycin-dependent interaction with P. aeruginosa PA14 when tested with two other members of the S. milleri group, S. intermedius and S. anginosus, or with S. constellatus C818 (see Fig. S2B in the supplemental material), indicating that the observed phenotype was specific for this strain of S. constellatus (see Discussion).
Characterization of tobramycin-dependent enhancement of S. constellatus 7155 in mixed biofilms with P. aeruginosa PA14.
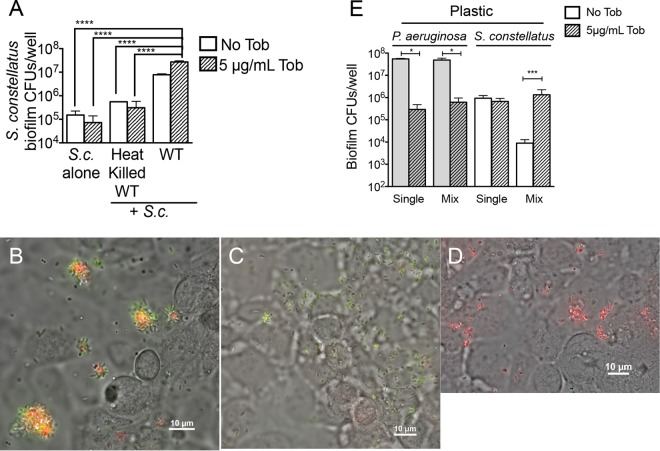
One explanation for the increase in S. constellatus 7155 CFU in mixed biofilm with P. aeruginosa treated with tobramycin may be enhanced nutrient availability. That is, tobramycin reduced viable P. aeruginosa levels in this assay by 256-fold; it is possible that these dead P. aeruginosa serve as a source of nutrients for S. constellatus 7155, thus allowing for their increased viability. To address this possibility, heat-killed P. aeruginosa was added to single S. constellatus 7155 biofilms grown on CF airway cells at the same time as the addition of antibiotic to mimic the availability of nutrients from dying P. aeruginosa. S. constellatus 7155 biofilms that received heat-killed P. aeruginosa had significantly lower viable counts than those of S. constellatus 7155 grown as mixed biofilms with live P. aeruginosa (Fig. 3A). Additionally, the viability of S. constellatus 7155 that received heat-killed P. aeruginosa did not increase upon tobramycin treatment (Fig. 3A).
FIG 3.
Characterization of tobramycin enhancement of S. constellatus in mixed biofilms with P. aeruginosa. (A) Viability of S. constellatus 7155 grown as biofilms on CF airway cells from assays of single or mixed biofilms of heat-killed P. aeruginosa PA14 or non-heat-killed P. aeruginosa PA14, as indicated, treated with no tobramycin or with 5 μg/ml tobramycin. ****, P < 0.0001, as determined by ANOVA, with Tukey's posttest for multiple comparisons. WT, wild type. (B) P. aeruginosa and S. constellatus colocalize in vitro. Shown is a representative image of a mixed-species biofilm of P. aeruginosa (green, expressing GFP) and S. constellatus 7155 (red, stained with hexidium iodide) on the surface of CF airway cells (gray) at ∼3 h postinoculation with P. aeruginosa and S. constellatus 7155. (C) Representative image of a single-species biofilm of P. aeruginosa (green, expressing GFP) on the surface of CF airway cells (gray) at ∼3 h postinoculation with P. aeruginosa alone. Red staining indicates the nonspecific staining levels of hexidium iodide for CF airway cells. (D) Representative image of S. constellatus 7155 (red, stained with hexidium iodide) on the surface of CF airway cells (gray) at ∼3 h postinoculation with S. constellatus 7155 alone. (E) Tobramycin treatment of P. aeruginosa and S. constellatus 7155 mixed biofilms enhances S. constellatus biofilm formation on plastic. The data represent the viability of bacteria grown as biofilms on plastic from biofilm assays on single or mixed biofilms of P. aeruginosa and S. constellatus 7155 treated with no tobramycin or 5 μg/ml tobramycin. *, P < 0.05; ***, P < 0.001, as determined by the Mann-Whitney test. For panels A and E, the bars and error bars indicate the means and SD of the results from three biological replicates.
We next examined P. aeruginosa PA14 and S. constellatus 7155 growing as biofilms on CF airway cells via epifluorescence microscopy. We reasoned that understanding the physical proximity of S. constellatus 7155 and P. aeruginosa in mixed biofilms on CF airway cells would be informative in understanding the mechanism of this interspecies interaction. In these studies, P. aeruginosa PA14 was visualized by its green fluorescence from a GFP constitutively expressed from a plasmid (26), while S. constellatus 7155 was visualized by its red fluorescence after staining with hexidium iodide, which preferentially stains Gram-positive bacteria (Fig. 3C and D) (29). When examined by phase contrast and fluorescence microscopy, P. aeruginosa and S. constellatus 7155 were found to colocalize on the surface of CF airway cells in vitro (Fig. 3B). Colocalization was not required for biofilm formation, as single-species biofilms of P. aeruginosa or S. constellatus 7155 were also visualized in mixed biofilms (Fig. 3B) and single-species biofilms (Fig. 3C and D).
To determine if the impact of tobramycin on S. constellatus 7155 when grown in mixed biofilms with P. aeruginosa on CF airway cells was dependent upon the presence of host cells, the biofilm assays described above were repeated for biofilms grown on the plastic surface of 24-well plates rather than a monolayer of CF airway cells. Levels of P. aeruginosa grown alone or in mixed biofilms on plastic decreased by 187-fold and 78-fold with tobramycin treatment (Fig. 3E), respectively, recapitulating the findings obtained from CF airway cell-grown biofilms (Fig. 2). Similarly, the growth and antibiotic response of plastic-grown S. constellatus 7155 single-species biofilms were comparable to those observed for CF airway cell-grown S. constellatus 7155 single-species biofilms, decreasing by 1.4-fold (Fig. 3E). In contrast, mixed biofilms of S. constellatus 7155 and P. aeruginosa on plastic resulted in a sharp 105-fold decline in the numbers of S. constellatus 7155 without tobramycin treatment compared to untreated single S. constellatus 7155 biofilms. Interestingly, a 150-fold increase in S. constellatus 7155 viability was observed in the presence of P. aeruginosa during tobramycin treatment of these mixed-species plastic-grown biofilms (Fig. 3E), a finding similar to that shown for CF airway cell-grown biofilms (Fig. 2A).
In summary, we demonstrated that heat-killed P. aeruginosa cannot recapitulate the tobramycin-dependent enhancement of S. constellatus 7155 growth in mixed biofilms with P. aeruginosa and that S. constellatus 7155 colocalizes with P. aeruginosa on CF airway cells. Studies of biofilms grown on plastic indicate that P. aeruginosa inhibited the growth of S. constellatus 7155 on plastic, but this inhibition could be relieved by the addition of tobramycin. The growth of biofilms on CF airway cells is somewhat more complex. In the absence of tobramycin, S. constellatus 7155 shows a stimulation of growth in the presence of P. aeruginosa which is further enhanced upon treatment with tobramycin, indicating a potential multifactorial interaction between these two microbes when grown on CF airway cells. In the subsequent studies described below, we focus on the ability of tobramycin to enhance the population of S. constellatus 7155 when this microbe is grown in the presence of P. aeruginosa PA14, thereby allowing us to perform our assays on plastic or on CFBE cells.
Screening for P. aeruginosa genes responsible for tobramycin-dependent S. constellatus biofilm growth enhancement using a candidate gene approach.
We hypothesized that the increase in S. constellatus 7155 biofilm viability when treated with tobramycin in mixed biofilms with P. aeruginosa PA14 was due to a tobramycin-dependent change in gene expression by P. aeruginosa, consistent with several studies demonstrating that P. aeruginosa gene expression changes in response to tobramycin treatment (20, 30–36). Specifically, we hypothesized that tobramycin treatment activates one or more P. aeruginosa genes that enhance S. constellatus 7155 biofilm viability, or alternatively, that tobramycin treatment could repress one or more P. aeruginosa genes that diminish S. constellatus 7155 biofilm viability. From a list of candidate genes chosen for their reported change in expression in response to tobramycin treatment, role in biofilm formation, and/or antibiotic tolerance (see Table S2 in the supplemental material), we performed a screen with mutant P. aeruginosa PA14 strains and wild-type S. constellatus 7155 to identify P. aeruginosa genes responsible for enhancing S. constellatus 7155 survival in mixed biofilms on plastic when treated with tobramycin. We performed the first round of this screen on plastic because growth on plastic served as a facile experimental tool, as this assay allowed us to focus on the P. aeruginosa-mediated growth inhibition of S. constellatus 7155 and the relief of that inhibition upon treatment with tobramycin. Forty-three mutant P. aeruginosa strains were tested in the screen with wild-type S. constellatus 7155 grown as biofilms on plastic (see Table S2); a mixture of the wild-type P. aeruginosa with wild-type S. constellatus 7155 served as the control.
Several genes related to rhamnolipid production were identified as candidates for mutations capable of enhancing S. constellatus 7155 growth in the absence of tobramycin treatment; that is, the viable counts of S. constellatus 7155 were equally high with or without tobramycin when cultured with P. aeruginosa strains carrying these mutations on plastic. Rhamnolipids are produced by the rhlA, rhlB, and rhlC gene products, which are responsible for the biosynthesis of β-hydroxyalkanoyl–β-hydroxyalkanoic acids (HAAs), monorhamnolipids, and dirhamnolipids, respectively, all of which are surfactants secreted by P. aeruginosa (Fig. 4A). The rhlA and rhlB genes are organized in an operon and transcriptionally induced by the activated quorum-sensing regulator RhlR. The rhlC gene is located elsewhere on the chromosome and is also activated by RhlR (27, 37–40).
FIG 4.
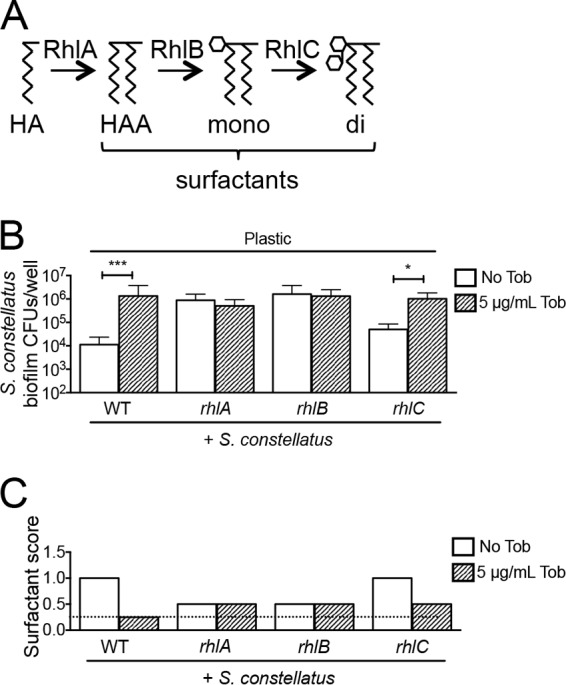
Monorhamnolipids prevent S. constellatus biofilm formation in mixed biofilms with P. aeruginosa on plastic. (A) Diagram of rhamnolipid biosynthetic pathway and the proteins required for each step in the biochemical pathway. Mutating rhlA would result in an accumulation of hydroxyacyl-ACP (HA), mutating rhlB results in the accumulation of β-hydroxyalkanoyl–β-hydroxyalkanoic acid (HAA), and the loss of rhlC causes the accumulation of monorhamnolipids (mono). di, dirhamnolipid. (B) Viability of S. constellatus 7155 grown as mixed biofilms on plastic from biofilm assays of S. constellatus 7155 with the wild type or the rhlA, rhlB, or rhlC mutant of P. aeruginosa, not treated with tobramycin or treated with 5 μg/ml tobramycin. The bars and error bars indicate the means and SD of the results from three biological replicates. ***, P < 0.001; *, P < 0.05, as determined by the Mann-Whitney test. (C) Representative surfactant score from assays of mixed biofilms of S. constellatus 7155 with the wild type or the rhlA, rhlB, or rhlC mutant of P. aeruginosa on plastic, not treated with tobramycin or treated with 5 μg/ml tobramycin. The dashed line indicates the limit of detection.
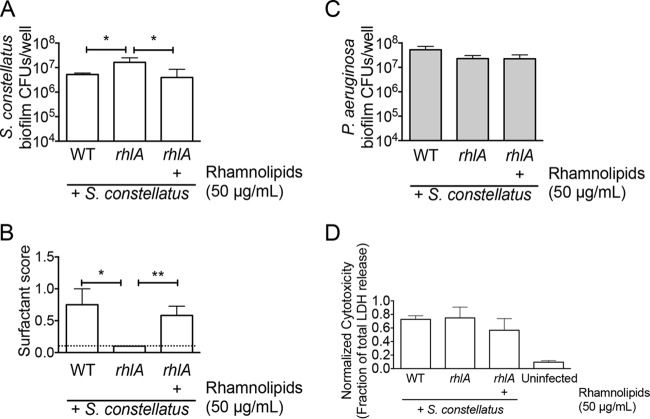
Mixed biofilms of S. constellatus 7155 with P. aeruginosa PA14 strains carrying mutations in the rhlA or rhlB genes resulted in high S. constellatus 7155 viability without the addition of tobramycin on plastic (Fig. 4B), a phenotype suggesting that tobramycin repressed the production of HAAs and monorhamnolipids. Consistent with this finding, mixed biofilms of S. constellatus 7155 and P. aeruginosa PA14 mutants with defects in the transcriptional activators of rhlA and rhlB (the lasR, rhlR, and lasR rhlR mutants) also resulted in high S. constellatus 7155 biofilm viability without the addition of tobramycin (see Table S2 in the supplemental material). Mixed biofilms of S. constellatus 7155 and a P. aeruginosa PA14 strain carrying a mutation in the rhlC gene did not result in an increase in the level of S. constellatus 7155 without the addition of tobramycin (Fig. 4B), phenocopying mixed biofilms of S. constellatus 7155 and wild-type P. aeruginosa PA14. This result suggested that HAAs and/or monorhamnolipids produced by the rhlC P. aeruginosa strain were sufficient to repress S. constellatus 7155.
To address whether tobramycin did indeed reduce the production of surfactants by P. aeruginosa, bulk surfactant production was measured by a drop collapse assay, as previously described (27, 28), to measure the effect of tobramycin on surfactant secretion into the supernatant. In a representative assay, mixed biofilms of S. constellatus 7155 and wild-type P. aeruginosa PA14 resulted in a surfactant score of 1.0 without tobramycin treatment and results below the limit of detection with tobramycin treatment (Fig. 4C). Mixed biofilms of S. constellatus 7155 and the P. aeruginosa PA14 rhlA mutant or the rhlB mutant resulted in a surfactant score of 0.5 with and without tobramycin treatment (Fig. 4C). A mixed biofilm of S. constellatus 7155 and the P. aeruginosa PA14 rhlC mutant showed surfactant scores of 1.0 without tobramycin treatment and 0.5 with tobramycin treatment (Fig. 4C). These results suggested that tobramycin interferes with rhamnolipid production and that the surfactants produced by the P. aeruginosa PA14 rhlC mutant are sufficient to inhibit S. constellatus 7155 biofilms. Thus, we propose that rhamnolipids negatively impact S. constellatus 7155 viability in a biofilm formed on plastic, monorhamnolipids are sufficient for this effect, and the loss of rhamnolipid production by P. aeruginosa PA14 upon tobramycin treatment allows for the increased S. constellatus 7155 observed in our study.
P. aeruginosa-produced monorhamnolipids inhibit S. constellatus biofilms on airway cells.
We next asked if rhamnolipids reduced S. constellatus 7155 CFU in mixed biofilms with P. aeruginosa PA14 formed on the more clinically relevant CF airway cells. Mixed biofilms of S. constellatus 7155 and the P. aeruginosa PA14 strains with mutations in the rhlA or rhlB genes formed on airway cells resulted in a level of S. constellatus 7155 viability without tobramycin (Fig. 5A) which is typically observed for this organism only when grown in the presence of P. aeruginosa PA14 with tobramycin. Mixed biofilms of S. constellatus 7155 and the P. aeruginosa PA14 rhlC mutant formed on airway cells did not result in increased S. constellatus 7155 CFU without tobramycin (Fig. 5A). Similar to what we observed on plastic, when grown as mixed biofilms on airway cells, S. constellatus 7155 CFU were the highest in mixed biofilms in which surfactant levels were the lowest (Fig. 5A and B). These results suggest that the surfactants produced by the P. aeruginosa rhlC mutant are sufficient to repress S. constellatus 7155 growth on airway cells.
FIG 5.
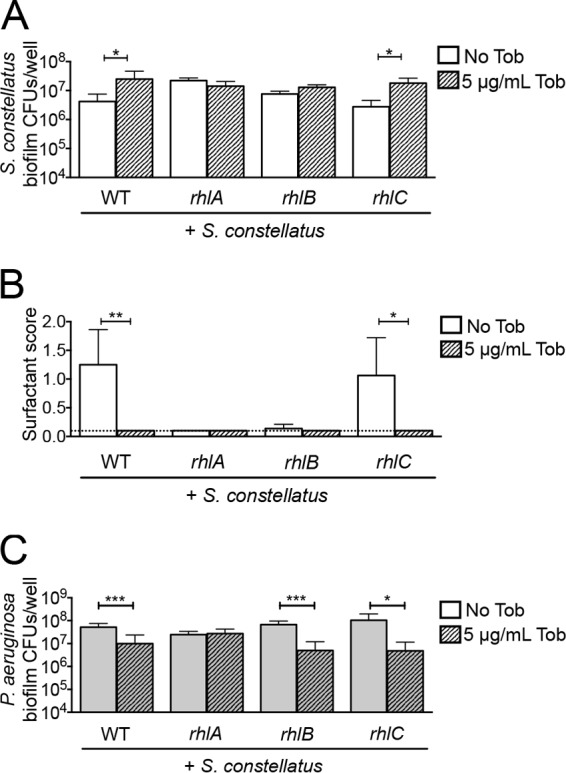
Monorhamnolipids reduce S. constellatus biofilm viability in mixed culture with P. aeruginosa on CF airway cells. (A) Viability of S. constellatus 7155 grown as mixed biofilms on CF airway cells with the wild type or the rhlA, rhlB, or rhlC mutant of P. aeruginosa not treated with tobramycin or treated with 5 μg/ml tobramycin. (B) Surfactant score from assays of mixed biofilms of S. constellatus 7155 with the wild type or the rhlA, rhlB, or rhlC mutant of P. aeruginosa on CF airway cells not treated with tobramycin or treated with 5 μg/ml tobramycin. The dashed line indicates the limit of detection. (C) Viability of the wild type or the rhlA, rhlB, or rhlC mutant of P. aeruginosa grown as mixed biofilms on CF airway cells with S. constellatus 7155 not treated with tobramycin or treated with 5 μg/ml tobramycin. For all panels, the bars and error bars indicate the means and SD of the results from three biological replicates. *, P < 0.05; **, P < 0.01, as determined by an unpaired t test.
It is possible that the tobramycin-dependent increase in S. constellatus 7155 viability in mixed biofilms formed on airway cells was due to an increased opportunity to colonize and/or additional nutrients that were available due to the reduced population of P. aeruginosa in the tobramycin-treated mixed biofilm. Several lines of evidence argue against this idea. First, single S. constellatus 7155 biofilms grown on CF airway cells, which should have the most available colonization surface and nutrients, have smaller amounts of viable S. constellatus 7155 than mixed biofilms of S. constellatus 7155 and P. aeruginosa (Fig. 2A). Second, adding heat-killed P. aeruginosa to an S. constellatus 7155 biofilm grown on CF airway cells did not result in increased S. constellatus 7155 viability (Fig. 3A). Third, the rhlA mutant used in these studies harbors a gentamicin resistance cassette in place of the rhlA gene, conferring cross-resistance to tobramycin to this mutant. In mixed biofilms of S. constellatus 7155 and the P. aeruginosa rhlA mutant, there are high levels of S. constellatus 7155 and P. aeruginosa with and without the addition of tobramycin (Fig. 5A and C). These results suggest that the growth enhancement of S. constellatus 7155 is not promoted by the absence of P. aeruginosa. Instead, these genetic studies support the model that P. aeruginosa-secreted rhamnolipids inhibit S. constellatus 7155 biofilm formed on CF airway cells and that surfactant production can be repressed by the addition of tobramycin.
Exogenous rhamnolipids inhibit S. constellatus biofilm formation on airway cells.
We further confirmed the role of rhamnolipids in reducing S. constellatus viability by adding purified commercially available rhamnolipids (a mixture of mono- and dirhamnolipids) to a mixed biofilm of S. constellatus 7155 and the P. aeruginosa rhlA mutant. Rhamnolipids were added at 50 μg/ml, a concentration that has been measured in broth-grown P. aeruginosa (41). The addition of exogenous rhamnolipids to the mixed biofilm of S. constellatus 7155 and the P. aeruginosa rhlA mutant represses S. constellatus 7155 CFU to the same extent as wild-type P. aeruginosa without tobramycin treatment, effectively complementing the rhlA mutant phenotype (Fig. 6A). Drop collapse assays on the supernatants from these biofilms revealed that 50 μg/ml exogenous rhamnolipids is comparable to the amount of surfactant produced by mixed biofilms of S. constellatus 7155 and P. aeruginosa (Fig. 6B). The addition of exogenous rhamnolipids did not impact P. aeruginosa viability (Fig. 6C) or cytotoxicity toward host cells (Fig. 6D). Taken together, our studies are consistent with the model that rhamnolipids produced by P. aeruginosa, or added exogenously, can reduce S. constellatus 7155 viability.
FIG 6.
Exogeneous rhamnolipids reduce S. constellatus viability in biofilms on CF airway cells. (A and B) Shown are the viability (A) and surfactant scores (B) of S. constellatus 7155 grown as single and mixed biofilms on CF airway cells with the wild type or the rhlA mutant of P. aeruginosa treated with 50 μg/ml rhamnolipids, as indicated. The dashed line indicates the limit of detection. (C) Viability of the wild type or the rhlA mutant P. aeruginosa grown as mixed biofilms on CF airway cells with S. constellatus 7155 treated with 50 μg/ml rhamnolipids, as indicated. (D) Cytotoxicity of indicated mixed biofilms on CF airway cells. Cytotoxicity was normalized to total cell lysis. For all panels, the bars and error bars indicate the means and SD of the results from three biological replicates. *, P < 0.05; **, P < 0.01, as determined by an unpaired t test.
Rhamnolipids likely inhibit S. constellatus 7155 in mixed biofilms by direct killing.
Rhamnolipids can disrupt biofilms by directly killing bacteria (42) and/or stimulating dispersion (41, 43). To test the hypothesis that rhamnolipids were dispersing S. constellatus 7155 biofilms, we measured the amount of S. constellatus 7155 in the planktonic fraction of the mixed biofilms formed on airway cells. In mixed biofilms of S. constellatus 7155 and wild-type P. aeruginosa or the P. aeruginosa rhlA mutant, we found no evidence of increased dispersion in mixed biofilms that produced rhamnolipids (S. constellatus 7155 and wild-type P. aeruginosa) compared to those that did not (S. constellatus 7155 and the P. aeruginosa rhlA mutant), as measured by viable S. constellatus 7155 levels in the planktonic fraction of CF airway cell-grown biofilms (Fig. 7A). Interestingly, we saw no statistically significant impact of the rhlA mutant on the viability of planktonic S. constellatus 7155, consistent with the data in Fig. 2B, indicating that the rhamnolipid-mediated reduction in the viability of S. constellatus 7155 by P. aeruginosa PA14 may be a biofilm-specific effect.
FIG 7.
Rhamnolipids directly kill S. constellatus. (A) Viability of the planktonic fraction of S. constellatus 7155 single and mixed biofilms with the wild type or the rhlA mutant of P. aeruginosa PA14 on CF airway cells not treated with tobramycin or treated with 5 μg/ml tobramycin. (B) Viability of S. constellatus 7155 grown as single biofilms on CF airway cells treated or not treated with 50 μg/ml rhamnolipids, as indicated. *, P value < 0.05, as determined by Mann Whitney test. (C) Viability of S. constellatus grown planktonically overnight in THY medium treated or not treated with 50 μg/ml rhamnolipids, as indicated. *, P value < 0.05, as determined by an unpaired t test. (D) Viability of planktonic S. constellatus 7155 incubated in phosphate-buffered saline (PBS) with or without 50 μg/ml rhamnolipids, as indicated. *, P value < 0.05, as determined by one-way ANOVA, followed by Tukey's posttest for multiple comparisons. The bars (A to C) and data points (D) indicate the means and the error bars indicate the SD of the results from three biological replicates.
To test whether S. constellatus 7155 grown in a biofilm on CF airway cells was susceptible to direct killing by rhamnolipids, 50 μg/ml rhamnolipids was added to preformed single S. constellatus 7155 biofilms grown on CF airway cells. Adding rhamnolipids to single S. constellatus 7155 biofilms significantly reduced the viability of S. constellatus 7155 by 27-fold after 15 h of treatment of biofilm-grown bacteria (Fig. 7B). Addition of rhamnolipids to the initial inoculum of a planktonically grown S. constellatus 7155 culture in THY resulted in an approximately 83,000-fold reduction in recovered CFU after 15 h of incubation (Fig. 7C). The kinetics of rhamnolipid-mediated killing is relatively rapid, with a decrease in the viability of S. constellatus 7155 planktonically grown cells noted as early as 3 h after the addition of surfactant and an 8,200-fold reduction by 6 h postaddition (Fig. 7D). Thus, the concentration of rhamnolipids produced by P. aeruginosa in broth culture (41) and in a mixed biofilm with S. constellatus 7155 on CF airway cells (Fig. 6B) is sufficient to significantly reduce the viability of S. constellatus 7155 grown as a biofilm on CF airway cells.
DISCUSSION
Here, we have shown an example of an interspecies interaction between two predominant organisms in the CF airway, P. aeruginosa and S. constellatus, potentiated by tobramycin, the frontline maintenance therapy antibiotic used to combat pulmonary infections in individuals with CF. We found that physiologically relevant concentrations (41) of P. aeruginosa-produced rhamnolipids can directly kill S. constellatus biofilms formed on CF airway cells, and our genetic studies suggest that monorhamnolipid production is sufficient for this effect. Furthermore, tobramycin treatment reduced the amount of rhamnolipids produced by P. aeruginosa, thus allowing enhanced growth of S. constellatus in mixed biofilms with P. aeruginosa on plastic and CF airway cells.
It is important to note that the interaction we observed here is specific to a biofilm of S. constellatus 7155 and P. aeruginosa PA14. We tested other Streptococcus strains and other isolates of P. aeruginosa but did not observe a similar interaction. At this stage, the basis of this specificity is not understood, but it might result, for example, from the differential production of rhamnolipids by P. aeruginosa or from S. constellatus 7155 being particularly sensitive to this surfactant. Despite the strain specificity of the infection, this work does illustrate the potential of microbial interactions in the context of host cells to impact antibiotic tolerance in unanticipated ways. It will be of interest if other similar interactions are revealed in the future.
Rhamnolipid production in the lung is varied (44); P. aeruginosa strains defective in quorum sensing have been isolated from the lungs of CF patients (45). These isolates would presumably reduce the production of rhamnolipids, thereby diminishing a potentially protective response against S. constellatus in vivo. However, Duan and colleagues (14) reported that autoiducer-2 produced by a clinical isolate of Streptococcus from an individual with CF could activate P. aeruginosa genes, including those required for rhamnolipid production (14). This result suggests that the presence of Streptococcus may increase the production of rhamnolipids in vivo, increasing a potentially protective response against S. constellatus unless tobramycin is also administered, which we predict would mitigate rhamnolipid-dependent killing. Thus, the regulation of rhamnolipid production in the lung is complex, and further study is warranted to understand the local and global concentrations of rhamnolipids in particular microbial communities in individual patients.
Interactions among P. aeruginosa and other members of the CF microbial community have been described in vitro and in vivo (14, 15, 46–48). P. aeruginosa has been shown to utilize S. aureus as an iron source, contributing to S. aureus lysis in mixed culture (46). Further, P. aeruginosa can sense and respond to peptidoglycan shed from Gram-positive bacteria (47). A recent study of volatile metabolites collected from breath samples of individuals with CF suggests that P. aeruginosa produces phenazines, small secreted molecules that can serve as alternative electron acceptors (49) in response to 2,3-butanedione (48). The authors hypothesize that 2,3-butanedione is produced by the S. milleri group, of which S. constellatus is a member, supporting the hypothesis that P. aeruginosa and S. constellatus may interact in vivo in the context of CF.
Riedele and Reichl (15) reported differential effects of the β-lactam antibiotic ceftazidime on broth-grown single and mixed cultures of the CF pathogens P. aeruginosa, B. cepacia, and S. aureus; furthermore, S. aureus was protected from ceftazidime treatment in mixed culture. Coupled with the data presented here, the unintended consequence of increasing the viability of a pathogen after antibiotic treatment in mixed culture (with examples using two different classes of antibiotics) underscores the importance of studying the effects of antibiotics in the context of a polymicrobial infection.
In this work, we demonstrate a complex interspecies relationship in a simple two-member community and reveal the molecular mechanism of at least one aspect of the interaction. While the work presented here focuses on one specific interaction, as we better understand how polymicrobial interactions might impact antibiotic tolerance, such findings might impact approaches to clinical care. And while identifying and understanding the network of interactions among CF microbes that contribute to disease with and without antibiotic perturbation are significant challenges, employing emerging CF sputum microbiomic, metagenomic, transcriptomic, and metabolomic studies as a guide to inform hypotheses, we may be able to leverage our expanded model system to discover interspecies interactions and their molecular mechanisms. We suggest that understanding these mechanisms may allow us to better predict how a given sputum community will respond to current antibiotics and may expose opportunities for novel therapeutic interventions. Ultimately, it may be possible to use this model system as a platform to select the best antibiotic(s) for a given microbial community, personalizing treatment plans based on microbial composition in the lungs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01 2 R37 AI83256-06 and P20 GM103413-10 to G.A.O., the Cystic Fibrosis Foundation Carol Basbaum Memorial Postdoctoral Fellowship PRICE13F0 to K.E.P., and a Cystic Fibrosis Foundation Research Development Grant (STANTO07R0).
We thank Russell Monds for constructing the phoB mutant used in this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00705-15.
REFERENCES
- 1.Costerton JW. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol 9:50–52. doi: 10.1016/S0966-842X(00)01918-1. [DOI] [PubMed] [Google Scholar]
- 2.Heijerman H. 2005. Infection and inflammation in cystic fibrosis: a short review. J Cyst Fibros 4(Suppl 2):S3–S5. [DOI] [PubMed] [Google Scholar]
- 3.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filkins LM, Hampton TH, Gifford AH, Gross MJ, Hogan DA, Sogin ML, Morrison HG, Paster BJ, O'Toole GA. 2012. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 194:4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim YW, Schmieder R, Haynes M, Willner D, Furlan M, Youle M, Abbott K, Edwards R, Evangelista J, Conrad D, Rohwer F. 2013. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros 12:154–164. doi: 10.1016/j.jcf.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP, Hoffman L, Daniels TW, Patel N, Forbes B, Bruce KD. 2012. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax 67:867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- 9.Price KE, Hampton TH, Gifford AH, Dolben EL, Hogan DA, Morrison HG, Sogin ML, O'Toole GA. 2013. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zemanick ET, Harris JK, Wagner BD, Robertson CE, Sagel SD, Stevens MJ, Accurso FJ, Laguna TA. 2013. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One 8:e62917. doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibley CD, Grinwis ME, Field TR, Parkins MD, Norgaard JC, Gregson DB, Rabin HR, Surette MG. 2010. McKay agar enables routine quantification of the 'Streptococcus milleri' group in cystic fibrosis patients. J Med Microbiol 59:534–540. doi: 10.1099/jmm.0.016592-0. [DOI] [PubMed] [Google Scholar]
- 12.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A 105:15070–15075. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field TR, Sibley CD, Parkins MD, Rabin HR, Surette MG. 2010. The genus Prevotella in cystic fibrosis airways. Anaerobe 16:337–344. doi: 10.1016/j.anaerobe.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Duan K, Dammel C, Stein J, Rabin H, Surette MG. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 15.Riedele C, Reichl U. 2011. Interspecies effects in a ceftazidime-treated mixed culture of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus: analysis at the single-species level. J Antimicrob Chemother 66:138–145. doi: 10.1093/jac/dkq394. [DOI] [PubMed] [Google Scholar]
- 16.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A 111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tunney MM, Klem ER, Fodor AA, Gilpin DF, Moriarty TF, McGrath SJ, Muhlebach MS, Boucher RC, Cardwell C, Doering G, Elborn JS, Wolfgang MC. 2011. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 66:579–584. doi: 10.1136/thx.2010.137281. [DOI] [PubMed] [Google Scholar]
- 19.Parkins MD, Sibley CD, Surette MG, Rabin HR. 2008. The Streptococcus milleri group–an unrecognized cause of disease in cystic fibrosis: a case series and literature review. Pediatr Pulmonol 43:490–497. doi: 10.1002/ppul.20809. [DOI] [PubMed] [Google Scholar]
- 20.Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O'Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreau-Marquis S, Bomberger JM, Anderson GG, Swiatecka-Urban A, Ye S, O'Toole GA, Stanton BA. 2008. The DeltaF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am J Physiol Lung Cell Mol Physiol 295:L25–L37. doi: 10.1152/ajplung.00391.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentchel-Franks K, Lozano D, Eubanks-Tarn V, Cobb B, Fan L, Oster R, Sorscher E, Clancy JP. 2004. Activation of airway Cl− secretion in human subjects by adenosine. Am J Respir Cell Mol Biol 31:140–146. doi: 10.1165/rcmb.2004-0012OC. [DOI] [PubMed] [Google Scholar]
- 24.Bruscia E, Sangiuolo F, Sinibaldi P, Goncz KK, Novelli G, Gruenert DC. 2002. Isolation of CF cell lines corrected at DeltaF508-CFTR locus by SFHR-mediated targeting. Gene Ther 9:683–685. doi: 10.1038/sj.gt.3301741. [DOI] [PubMed] [Google Scholar]
- 25.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. 1994. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol 10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 26.Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol 63:4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deziel E, Comeau Y, Villemur R. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183:1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caiazza NC, Shanks RM, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason DJ, Shanmuganathan S, Mortimer FC, Gant VA. 1998. A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl Environ Microbiol 64:2681–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 31.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 32.Marr AK, Gooderham WJ, Hancock RE. 2006. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol 6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Linares JF, Gustafsson I, Baquero F, Martinez JL. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A 103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kindrachuk KN, Fernández L, Bains M, Hancock RE. 2011. Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1874–1882. doi: 10.1128/AAC.00935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjarnsholt T, Jensen PO, Burmolle M, Hentzer M, Haagensen JA, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Hoiby N, Givskov M. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 36.Babić F, Venturi V, Maravić-Vlahoviček G. 2010. Tobramycin at subinhibitory concentration inhibits the RhlI/R quorum sensing system in a Pseudomonas aeruginosa environmental isolate. BMC Infect Dis 10:148. doi: 10.1186/1471-2334-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochsner UA, Fiechter A, Reiser J. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J Biol Chem 269:19787–19795. [PubMed] [Google Scholar]
- 38.Ochsner UA, Koch AK, Fiechter A, Reiser J. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol 176:2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol Microbiol 40:708–718. doi: 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 40.Ochsner UA, Reiser J. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boles BR, Thoendel M, Singh PK. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 42.Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A. 2003. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng 81:316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- 43.Davey ME, Caiazza NC, O'Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kownatzki R, Tümmler B, Döring G. 1987. Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet i:1026–1027. [DOI] [PubMed] [Google Scholar]
- 45.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R, Blake DR, Conrad D, Rohwer F. 2014. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J 8:1247–1258. doi: 10.1038/ismej.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol 42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.