ABSTRACT
Bordetella pertussis is a bacterium that is considered to be highly adapted to humans, and it has not been isolated from the environment. As this bacterium does not utilize sugars, the abundant supply of glutamate in Stainer Scholte (SS) medium enables B. pertussis to grow efficiently in liquid culture in vitro, and as such, SS medium is a popular choice for laboratory experiments. However, the concentration of glutamate in the in vivo niche of B. pertussis is quite low. We investigated the bacterial response to low concentrations of glutamate to elucidate bacterial physiology via the expression of the type 3 secretion system (T3SS), and we discuss its relationship to the Bvg mode in which the two-component regulator of pathogenesis (BvgAS) is activated. Glutamate limitation induced the expression of both the T3SS apparatus and effector genes at the transcriptional level. (p)ppGpp, a modulator of the stringent response, was necessary for maximum expression of the T3SS genes. These observations indicate that the expression of the T3SS is managed by nutrient starvation. In addition, the autoaggregation ability was high in the absence of glutamate and no autoaggregation was observed in glutamate-replete medium. Taken together, glutamate-limited conditions in Bvg+ mode elicit the high expression of T3SS genes in B. pertussis and promotes its sessile form.
IMPORTANCE Bordetella pertussis is a highly contagious pathogen that causes respiratory infectious disease. In spite of the increasing use of vaccination, the number of patients with pertussis is increasing. The proteins produced in vivo often are different from the protein profile under laboratory conditions; therefore, the development of conditions reflecting the host environment is important to understand native bacterial behavior. In the present study, we examined the effect of glutamate limitation, as its concentration in vivo is much lower than that in the culture medium currently used for B. pertussis experiments. As predicted, the T3SS was induced by glutamate limitation. These results are suggestive of the importance of regulation by nutrient conditions and in the pathogenicity of B. pertussis.
INTRODUCTION
One of the major amino acids that compose the structural and functional biosynthesized proteins, l-glutamate (glutamate) is also utilized as an energy source as a result of its conversion to α-ketoglutarate, one of the intermediate products of the tricarboxylic acid (TCA) cycle. In addition to uptake from the environment, cells are able to utilize cellular enzymes to synthesize glutamate from proline or arginine. While central neurons in animals possess high concentrations (10 to 20 mM) of glutamate in their cells for use as a neurotransmitter, most eukaryotic cells metabolize glutamate rapidly. Even near nerve cells, extracellular glutamate is actively imported via a glutamate transporter to avoid damage to neurons by high concentrations of extracellular glutamate (1). By this mechanism, extracellular concentrations of glutamate are kept much lower than the intracellular concentration at approximately 10 to 100 μM (2).
Bordetella pertussis is known as one of the etiological agents of pertussis (whooping cough), together with B. parapertussis and B. holmesii. B. pertussis utilizes neither glucose nor other sugars, as this bacterium possesses an incomplete TCA cycle, lacking several enzymes in this pathway (3). Since glutamate is a valuable carbon source for B. pertussis, abundant glutamate (63.4 mM) is supplied in the Stainer Scholte (SS) medium used for its liquid culture (4, 5).
In vivo B. pertussis adheres to and persists on the surface of cells, an environment with relatively low glutamate concentrations. Therefore, we investigated the physiology of B. pertussis growing under glutamate-limited conditions and whether this could exact further regulation of pathogenesis in its host.
The type 3 secretion system (T3SS) is a widely distributed secretion system in Gram-negative bacteria and is known as the injectisome/injectosome due to its function of injecting bacterial proteins (effectors) into host cells (6). Although the genes are present and expressed as mRNA, it had been thought that B. pertussis did not produce or utilize the T3SS. Recently, the production of the T3SS proteins was discovered by analysis of fresh clinical isolates (7), and laboratory-adapted strains showed the production of the T3SS in cultures grown under iron-depleted conditions (8).
The pathogenesis of B. pertussis is regulated by the BvgAS two-component system at the transcriptional level (9). In Bvg+ mode, BvgAS is fully activated and all Bvg-dependent virulence factors are expressed. In contrast, bacteria in Bvg− mode do not express any of these. Between Bvg+ and Bvg− modes is Bvgi mode, in which bacteria express only adhesins and a specific gene, bipA. Basically, bacteria are in Bvg+ mode unless specific modulators, such as a high concentration of MgSO4 or nicotinic acid, inactivate this two-component system (10–12). It has been shown that btrS, which encodes an extracellular sigma factor required for the transcription of T3SS genes, is exclusively expressed in Bvg+ mode, thereby activating the production of the T3SS in Bvg+ mode in B. pertussis as well as in Bordetella bronchiseptica (13).
In response to nutrient starvation, concentrations of small nucleotide second messengers, guanosine pentaphosphate or tetraphosphate [pppGpp or ppGpp, respectively; collectively named (p)ppGpp], are increased in bacterial cells, and the expression of many genes is modulated at the transcriptional and posttranscriptional levels (14). This response, called the stringent response, is triggered by various nutrient limitations, such as the depletion of amino acids or carbon sources (15–17). Previously, it was shown that a deficiency of (p)ppGpp synthesis repressed the expression of bsp22, which encodes a component of the T3SS, as well as decreased autoaggregation accompanied by a decreased number of filamentous structures. In the present study, it was determined that T3SS expression was upregulated by glutamate limitation under activation of BvgAS. Based on these findings, we further investigated the effect of glutamate on autoaggregation and suggest that under glutamate-limited conditions, B. pertussis tends to autoaggregate and increase the expression of T3SS genes.
MATERIALS AND METHODS
Bacterial strains and cultivation.
B. pertussis strains used in the present study are listed in Table 1. B. pertussis was routinely cultured on Bordet-Gengou agar supplemented with 15% defibrinated sheep blood or, for nutrient-rich conditions, in SS medium supplemented with Casamino Acids at a final concentration of 0.5% (SSC medium) under vigorous shaking at 35°C.
TABLE 1.
Bordetella pertussis strains and plasmids used in this study
| Strain or plasmid | Relevant genotype/description | Reference |
|---|---|---|
| Strains | ||
| UT25Sm1 | Clinical isolate | 18 |
| PMK21 | Derivative of UT25Sm1, ΔrelA ΔspoT | 19 |
| Tohama I | Clinical isolatea | 20 |
| BP157 | Clinical isolate | 21 |
| BP159 | Clinical isolate | 21 |
| BP163 | Clinical isolate | 21 |
| BP228 | Clinical isolate | 21 |
| BP235 | Clinical isolate | 21 |
| Plasmids | ||
| pRK415 | Cloning vector for B. pertussis, Tetr | 22 |
| pRKTY01 | Derivative of pRK415, relA gene | 19 |
Isolated in 1954 and used for production of acellular vaccine.
For glutamate limitation response experiments, decreased amounts of glutamate, at 50% (SSC-0.5E), 20% (SSC-0.2E), or 0% (SSC-0E) of that in SSC medium, were used. To ensure uniform conditions, B. pertussis was cultured in a similar manner except as described for individual experiments. Bacterial cells from overnight cultures were harvested, washed, and resuspended in appropriate medium for each experiment. After adjusting the turbidity to an optical density at 600 nm (OD600) of 0.1, 30 ml of bacterial suspension was subcultured in 250-ml plastic flasks with shaking at 200 rpm. Antibiotics were added as necessary at the following concentrations: 10 μg per ml of gentamicin, 12 μg per ml of tetracycline, and 100 μg per ml of streptomycin.
Quantification of glutamate in the bacterial culture milieu.
To measure concentrations of glutamate, bacterial cells were removed by centrifugation at 12,000 × g for 5 min and passed through a 0.45-μm-pore-size membrane filter. After this, samples were treated with 5 U of l-ascorbic oxidase per ml for 5 min at room temperature to digest vitamin C in the medium (which inhibits the reaction of l-glutamate oxidase in the l-glutamate assay kit II; Yamasa, Co., Choushi, Japan). Samples were diluted 1- to 40-fold with HEPES (pH 7.1) to obtain glutamate concentrations in an appropriate concentration range for measurement (0.05 mM to 3 mM), and 15-μl aliquots of the solutions were added to 96-well flat-bottom plates containing 225 μl of enzyme-reagent mixture. After mixing, they were incubated at room temperature for 20 min. Absorbance at 595 nm of 100 μl of reaction mixture was measured with a microplate reader (iMARK plate reader; Bio-Rad, Hercules, CA). Concentrations of glutamate in the samples were determined by the calibration curve method.
Preparation of protein samples and SDS-PAGE.
Protein samples used for SDS-PAGE analysis were prepared as described previously, with some modifications (23). Briefly, proteins in the culture fluids were precipitated with 15% trichloroacetic acid after removing bacterial cells by centrifugation at 7,500 rpm at 4°C for 10 min and passing through a 0.45-μm-pore-size membrane filter. Bacterial cells were harvested and lysed with CelLytic B cell lysis reagent (Sigma-Aldrich) in the presence of protease inhibitor cocktail (P8849; Sigma-Aldrich). Proteins in the cell-free culture fluids corresponding to B. pertussis culture at an OD600 of 1.2 were analyzed on a 12.5% SDS-PAGE gel except as indicated in the text. Bacterial lysates were normalized with respect to protein concentration, with 40 μg of protein in each sample.
Identification of proteins.
Proteins were separated on a 12.5% SDS-PAGE gel and stained with Coomassie brilliant blue R250, cut out, and treated with trypsin. Digested peptides were separated by a direct nano-liquid chromatography (LC) system (DiNA-KYA Technologies) and analyzed by mass spectrometry (LTQ Orbitrap Velos mass spectrometer). The data were analyzed using the Mascot search engine in the NCBI database.
Immunoblotting.
Proteins separated by SDS-PAGE were transferred to a polyvinylidene difluoride (PVDF) membrane and reacted with diluted antibodies at room temperature for 1 h. Unless described elsewhere, dilution ratios were 1,500-fold for antipertactin (anti-Prn) (24), anti-pertussis toxin (anti-PTX) (Santa Cruz Biotechnology, Inc.), and anti-adenylate cyclase toxin (anti-ACT) (Santa Cruz Biotechnology, Inc.) antibodies from mouse and anti-Bsp22 (kind gift from Akio Abe, Kitasato University) and anti-filamentous hemagglutinin (anti-FHA) antibodies from rabbit (23). Rabbit anti-BspR antibody (kind gift from Akio Abe, Kitasato University) was diluted 800-fold. Appropriate secondary antibodies were diluted 20,000-fold for use. Specific proteins were detected using ECL Prime Western blot detection reagents (GE Healthcare) and visualized with ImageQuant LAS-4000 (GE Healthcare).
Quantitative reverse transcription-PCR (qRT-PCR).
The extraction of total RNA and quantification of specific mRNAs were performed as described previously (19). Briefly, harvested bacterial cells were treated with RNAprotect bacterial reagent (Qiagen), and total RNA was extracted. After treatment with DNase, RNA concentrations of 50 ng per ml were used for the synthesis of cDNA with reverse transcriptase in the presence of RNase inhibitor (PrimeScript RT reagent kit; TaKaRa, Otsu, Japan). The cDNA fractions were used for quantitative PCR by the SYBR green method (SYBR premix Ex Taq GC; TaKaRa, Otsu, Japan) with a real-time PCR system 7500 (Applied Biosystems) using primer pairs described in Table 2. The data were normalized to the endogenous recA level and analyzed by the ΔΔCT method (where CT is threshold cycle) (19). The data were used to calculate relative fold changes in SSC medium compared to the level in medium with a modified glutamate concentration (SSC-0.2E). The error bars represent the standard deviations of the values.
TABLE 2.
PCR primers used for quantitative PCR in this study
| Name | Sequence (5′–3′) | Target gene |
|---|---|---|
| bsp22F rt | CGAACTCCTCACGGCTCAAATG | bsp22 |
| bsp22R rt | GACAGCGCGGACAGGACCTC | bsp22 |
| btrS-F rt | CAGAGATTCATCGCCAAGCACATC | btrS |
| btrS-R rt | GAGTCGCCACGGAACGATTGATAC | btrS |
| btrV-F rt | GGATTTGGGGCGGCTTGACTAC | btrV |
| btrV-R rt | CTTCAGTTCGCACAGCACCAGTTC | btrV |
| bopN-F rt | CATCACTCCGAACGCAAGGTCAC | bopN |
| bopN-R rt | GTCGGGGTCGTGGGTGTGG | bopN |
| bscN-F rt | CAACCGCCATCGCCGAATAC | bscN |
| bscN-R rt | CTGCCGCCAAGCCGATTTC | bscN |
Autoaggregation.
Exponentially growing bacterial cells in SSC medium were harvested and washed with SSC medium or SSC-0E. Two-milliliter aliquots of bacterial suspensions normalized to the same value were dispensed into polystyrene round-bottom tubes and incubated at 35°C for 6 h. The OD600 values for 500-μl samples of the upper portion of each suspension and resuspension following mixing were measured.
Statistical analysis.
The Student t test was used to determine statistical significance, with a P value of less than 0.05 being considered significant.
RESULTS
Decreased glutamate in the medium alters the quantity of T3SS proteins in the culture milieu.
B. pertussis strains were cultured for 30 h in SSC, SSC-0.5E, SSC-0.2E, SSC-0E, or SS medium, and then proteins in the culture fluids were analyzed by SDS-PAGE (Fig. 1). The amounts of several proteins in the culture milieu were altered under lower glutamate concentrations compared to their levels in SSC medium. Proteins with molecular masses of approximately 69 kDa, 38 kDa, 32 kDa, and 24 kDa apparently were increased at low glutamate concentrations, with the maximum amounts of these proteins detected in cultures with SSC-0.2E medium. These proteins were identified by mass spectrometric analysis as BteA (also known as BopC), BopN, BopD, and Bsp22, respectively (Fig. 1). Interestingly, these proteins are T3SS secreted proteins that must be secreted for the construction of the active form of the apparatus. Therefore, these results suggest that the T3SS was upregulated by decreased concentrations of glutamate in the medium.
FIG 1.
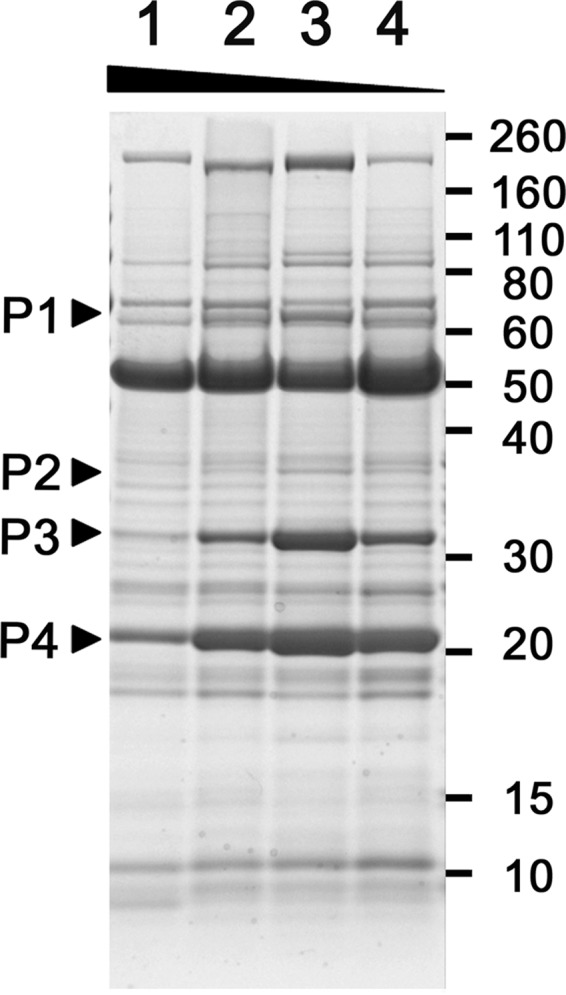
Proteins secreted by wild-type B. pertussis growing in modified SS media. B. pertussis strain UT25Sm1 was cultured in SSC (lane 1), SSC-0.5E (lane 2), SSC-0.2E (lane 3), and SSC-0E (lane 4) medium. Proteins in the cell-free culture fluids were prepared and applied to 12.5% polyacrylamide SDS-PAGE gels. The protein bands indicated by arrowheads were identified as described in Materials and Methods. P1, P2, P3, and P4 are BteA (coverage rate, 91.92%; number of unique peptides, 26; score, 30,179.41), BopN (coverage rate, 65.75%; number of unique peptides, 17; score, 4,058.76), BopD (coverage rate, 62.30%; number of unique peptides, 18; score, 3,416.24), and Bsp22 (coverage rate, 99.51%; number of unique peptides, 8; score, 21,064.88), respectively.
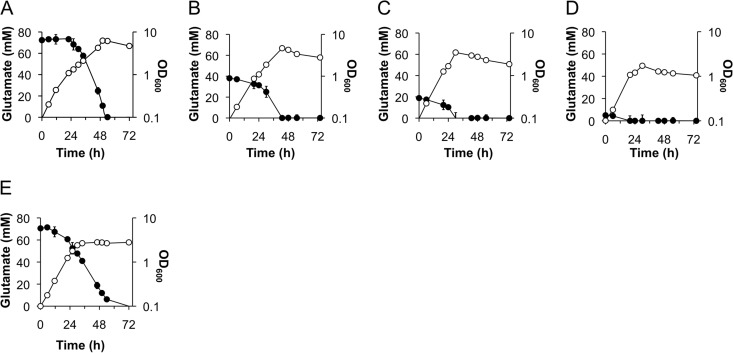
Consumption of glutamate in the medium.
To analyze glutamate consumption by B. pertussis, glutamate in the culture medium was quantified at the indicated times after starting the culture. The initial glutamate concentration in SSC medium was slightly higher than that in SS medium due to the glutamate present in the Casamino Acids (Fig. 2A and E). During bacterial proliferation, the concentration of glutamate decreased, as it was catabolized by B. pertussis, with consumption and depletion under all conditions examined (Fig. 2). Interestingly, bacterial glutamate consumption patterns were different depending on the presence or absence of Casamino Acids (Fig. 2A and E). Glutamate was consumed from the beginning of bacterial growth in the absence of Casamino Acids, and bacterial growth was arrested despite glutamate (approximately 50 mM) remaining in the medium. Additionally, the maximum turbidity of the culture was much lower than that in the presence of Casamino Acids (Fig. 2A and E). Therefore, we can infer that in the absence of Casamino Acids, termination of bacterial growth is due to factors other than glutamate depletion. In cultures in SSC-0.5E or SSC-0.2E, the timing of entry into stationary phase was accelerated (Fig. 2B and C). In the presence of both Casamino Acids and glutamate, a delay of glutamate consumption occurred regardless of glutamate concentration (Fig. 2A to C). B. pertussis still grew modestly in the presence of Casamino Acids even in the absence of glutamate (Fig. 2D). If neither glutamate nor Casamino Acids was supplied, then no bacterial growth was observed (data not shown). These results suggest that amino acids other than glutamate are utilized first in the presence of Casamino Acids. Subsequently, B. pertussis cells consume glutamate efficiently as a carbon source until depletion, finally entering stationary phase (Fig. 2A to D).
FIG 2.
Patterns of glutamate consumption in the culture medium. B. pertussis strain UT25Sm1 was cultured in SSC (A), SSC-0.5E (B), SSC-0.2E (C), SSC-0E (D), and SS (E) medium. Growth was traced by measurement of culture OD600 values (open circles). The concentration of glutamate in the culture was quantified as described in Materials and Methods (closed circles) at the indicated time points.
Since B. pertussis growth is affected by glutamate in the presence of Casamino Acids, bacterial growth in media with different concentrations of glutamate can be considered to show a response to various stages of glutamate limitation. Therefore, cultures grown for 30 h in SSC and SSC-0.2E medium were used to compare glutamate-replete and -limited conditions, respectively, in the ensuing experiments (Fig. 2A and C).
Effect of glutamate limitation on the expression of the T3SS.
The genes encoding the T3SS are located in the bsc and btr loci in B. pertussis, except for bteA (bopC). The expression of Bordetella T3SS genes is regulated hierarchically, similar to those in other bacteria (25). The bsc locus contains the T3SS apparatus and effector genes, and the regulatory proteins for expression of the T3SS are encoded by the btr locus. BtrS, encoded by the btr locus, is an extracellular sigma factor for RNA polymerase, which transcribes the genes of both the btr and bsc loci by binding to specific promoters, regulating T3SS protein expression.
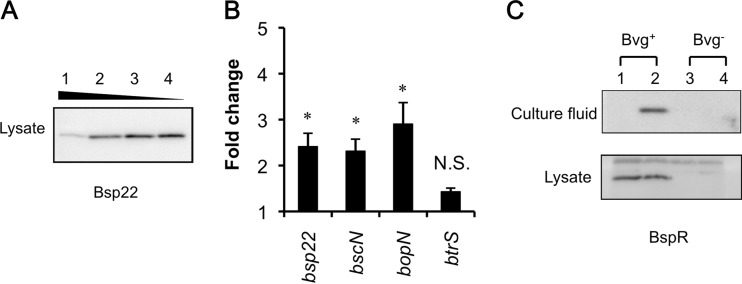
Bsp22, a tip protein of the T3SS, is secreted via the T3SS apparatus and is the most abundant of the T3SS secreted proteins found in the culture. Similar to secreted Bsp22, the amount of Bsp22 in the lysate was found to be altered depending on the glutamate concentration (Fig. 3A). Both bscN, encoding the ATPase essential for secretion activity, and bopN, encoding an effector, are located in the bsc locus. To analyze the regulation of transcription by glutamate limitation, B. pertussis UT25Sm1 was cultured in 10 ml of SSC and SSC-E0.2 for 22 h in a 100-ml flask with shaking at 295 rpm, and total RNA was extracted to quantify bsp22, bscN, and bopN transcripts by qRT-PCR. Glutamate in the cultures used for this experiment was abundant in SSC medium (68.7 mM) and limited (3.4 mM) in SSC-0.2E medium, and T3SS proteins were produced only in the culture growing in SSC-0.2E medium (data not shown). The levels of gene transcripts in bacterial cells grown under glutamate-limited conditions were significantly higher than those in bacteria grown under glutamate-replete conditions (Fig. 3B). Unlike the bsc locus genes, the transcript level of the btrS gene was not significantly induced by glutamate limitation. Similarly, the production of the transcript of the btrV gene in the btr locus showed no significant alteration under glutamate limitation (data not shown).
FIG 3.
Effect of glutamate limitation on the expression of genes encoding the T3SS. (A) Bsp22 production by B. pertussis UT25Sm1. Lysate was prepared using bacteria cultured in SSC (lane 1), SSC-0.5E (lane 2), SSC-0.2E (lane 3), and SSC-0E (lane 4) for 30 h and analyzed by immunoblotting as described in Materials and Methods, except the dilution of anti-Bsp22 antibody was 2,500-fold. (B) Effect of glutamate limitation on transcription of the T3SS genes. Total RNA was extracted from B. pertussis cells, and bsp22-, bscN-, bopN-, and btrS-specific transcripts were quantified. Results presented are averages from three independent experiments. Error bars indicate standard deviations of the means. Asterisks indicate significant differences (P > 0.05) as determined by Student t test. (C) B. pertussis was cultured in SSC or SSC-0.2E medium for 30 h, and protein samples were prepared. The production of BspR under glutamate-replete (odd-numbered lanes) or -depleted (even-numbered lanes) conditions was analyzed by immunoblotting. Bvg− mode bacteria were obtained by the addition of 50 mM MgSO4 to the culture medium.
The bspR gene is present in the btr locus in B. bronchiseptica and is conserved in B. pertussis. Its product, BspR, is one of the T3SS secreted proteins that is translocated into host cell nuclei and is also a negative transcriptional regulator of T3SS genes in the bacterial cytosol (26). To assess the effect of BspR on the expression of the T3SS, the levels of BspR were analyzed by immunoblotting. Glutamate limitation significantly increased BspR levels in the culture fluid, although it did not alter the amount in the lysate (Fig. 3C).
These data suggest that glutamate limitation induces the expression of bsc locus genes at the transcription level but did not affect the expression of btr locus genes.
Induction of the T3SS by glutamate limitation in Tohama I and clinical isolates.
B. pertussis UT25Sm1 is a streptomycin-resistant strain derived from a clinical isolate (18). To investigate whether the glutamate limitation response is conserved in other B. pertussis strains, Bsp22 in the culture fluid of the B. pertussis Tohama I vaccine strain and five clinical isolates was analyzed (Fig. 4). Clinical isolates secreted Bsp22 at detectable levels into their culture milieu, and they were induced by glutamate limitation (Fig. 4A and C). Bsp22 production by the B. pertussis Tohama I strain was not detected under glutamate-replete conditions, but it was induced at detectable levels under glutamate limitation even though the level was significantly lower than that of clinical isolates (Fig. 4A and B). Taken together, these results suggest that the mechanism of regulation of expression of the T3SS by glutamate limitation is conserved in B. pertussis strains.
FIG 4.
Production of T3SS proteins in clinical isolates and a laboratory strain of B. pertussis in response to glutamate limitation. B. pertussis clinical isolates (BP157, BP159, BP162, BP228 and BP235) and a laboratory-adapted strain (Tohama I) were cultured in SSC medium (odd-numbered lanes) or SSC-0.2E medium (even-numbered lanes). At 30 h after starting the culture, proteins in the culture fluid were prepared. (A) The quantities of proteins were normalized with the bacterial cell number corresponding to an OD600 of 1.2 (clinical isolates) or 2.0 (Tohama I) and applied for analysis by immunoblotting using a 2,500-fold dilution of the primary antibody. (B and C) Protein profiles developed in 12.5% polyacrylamide gels by SDS-PAGE were analyzed.
Role of BvgAS in the induction of T3SS genes by glutamate.
The BvgAS system positively regulates the transcription of T3SS genes as well as most virulence factors in B. pertussis. To clarify the role of glutamate limitation in BvgAS-mediated regulation, the production of Bsp22 in Bvg− mode generated by addition of 50 mM MgSO4 to the culture medium was analyzed. The results show that B. pertussis cells in Bvg− mode did not produce detectable levels of Bsp22 even under glutamate limitation (Fig. 5). Therefore, glutamate limitation regulation is thought to occur exclusively via BvgAS activation. The response of other BvgAS-dependent virulence factors to glutamate limitation also was assessed by immunoblotting. FHA, ACT, PT, and Prn are Bvg-dependent virulence factors repressed in Bvg− mode and are not produced under Bvg− in vitro modulating conditions by addition of 50 mM MgSO4 in the culture medium. The production of similar levels of FHA, ACT, and PT regardless of glutamate concentration indicates that the expression of these genes is not affected by glutamate limitation (Fig. 5B and D). This control mechanism therefore is likely to be specific to genes encoding the T3SS proteins and dependent on BvgAS activity.
FIG 5.
Production of Bvg-dependent proteins in B. pertussis. B. pertussis strain UT25Sm1 was cultured in SSC medium (odd-numbered lanes) or SSC-0.2E medium (even-numbered lanes). Bacteria cultured in the presence of 50 mM MgSO4 were used as Bvg− mode bacteria. At 30 h after starting the culture, bacterial protein samples were prepared as described in Materials and Methods. SDS-PAGE gels of protein in cell lysates and culture fluids were visualized by staining with Coomassie brilliant blue (A and C). Bsp22, FHA, ACT, and PTX in protein samples were analyzed by immunoblotting (B and D). For the detection of Bsp22 in the cell-free culture fluids, the primary antibody was diluted 2,500-fold.
(p)ppGpp is necessary for the maximum production of the T3SS proteins.
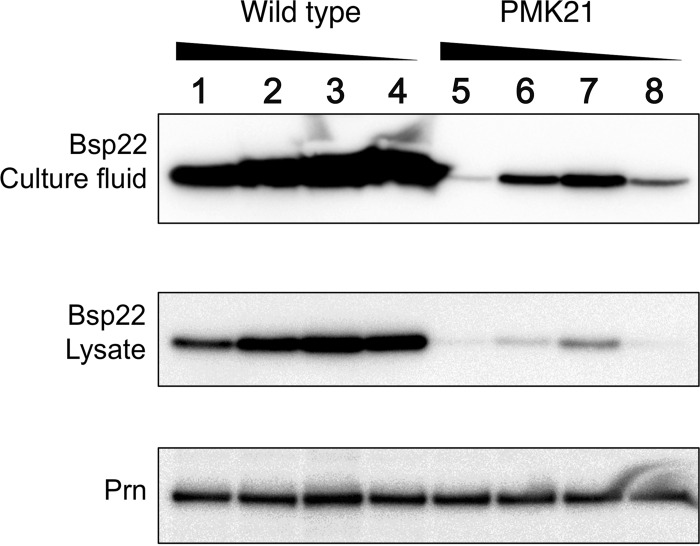
Cells accumulate (p)ppGpp, which is synthesized by RelA/SpoT homologues (RSH) (27), in response to nutrient starvation due to the activation of the stringent response. Previously, we demonstrated a significant reduction of the T3SS in strain PMK21, a (p)ppGpp-deficient mutant in which both the relA and spoT genes have been deleted, even though the expression of other virulence factors was not affected (19). Therefore, it was anticipated that glutamate limitation would elicit a stringent response in B. pertussis. Using B. pertussis strain PMK21, we examined the involvement of the stringent response in T3SS induction by glutamate limitation. Prn production was not affected by either (p)ppGpp depletion or glutamate limitation (Fig. 6). In contrast, Bsp22 was significantly reduced by the depletion of (p)ppGpp, similar to previous results (19). Moreover, Bsp22 was slightly induced by glutamate limitation in both cell-free culture fluids and lysates (Fig. 6). These results not only indicate that (p)ppGpp is essential for full expression of the T3SS but also imply that T3SS gene expression is regulated by other factors via glutamate limitation.
FIG 6.
Production of Bsp22 and pertactin (Prn) in B. pertussis strains UT25Sm1 and PMK21. B. pertussis strains were cultured in SSC (lanes 1 and 5), SSC-0.5E (lanes 2 and 6), SSC-0.2E (lanes 3 and 7), and SSC-0E (lanes 4 and 8) medium. At 30 h after starting the culture, protein samples were prepared and separated by 12.5% polyacrylamide SDS-PAGE gels. Prn and Bsp22 were detected by immunoblotting as described in Materials and Methods.
Autoaggregation is affected by glutamate in B. pertussis strains.
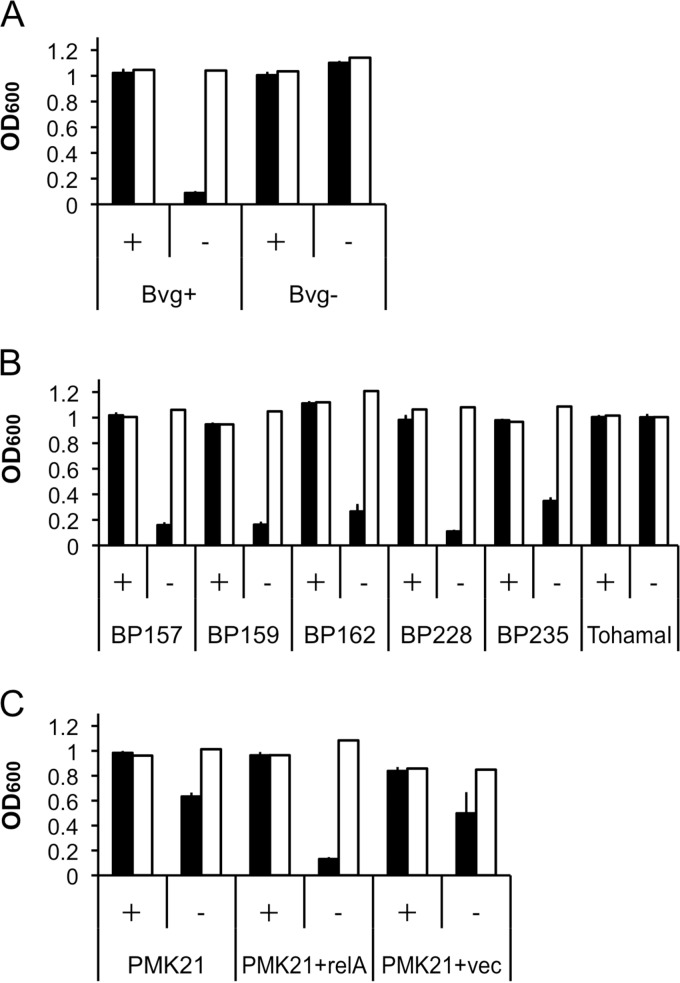
Since filamentous structures such as the T3SS often are involved in cell aggregation, we investigated the correlation between T3SS production and the ability to autoaggregate under glutamate-limited conditions. In SSC medium with abundant glutamate, B. pertussis in Bvg+ mode did not aggregate at all during 6 h of incubation, but autoaggregation was significantly increased under glutamate-depleted conditions (Fig. 7A). Under Bvg-modulated conditions with 50 mM MgSO4, bacterial cells did not aggregate regardless of the presence of glutamate (Fig. 7A). On the other hand, Bsp22 was already induced 6 h after transferring bacterial cells to SSC-0E or SS medium (Fig. 8). In particular, B. pertussis responded by altering protein production by 6 h after exposure to glutamate limitation. As shown in Fig. 7B, all clinical isolates examined showed no autoaggregation under glutamate-replete conditions and high autoaggregation under glutamate-limited conditions. On the other hand, B. pertussis Tohama I did not autoaggregate at all (Fig. 7B). The (p)ppGpp-deficient strain B. pertussis PMK21 partially autoaggregated under glutamate-limiting conditions, but full autoaggregation was restored when the relA gene was provided on pRKT01 (Fig. 7C). These results suggest a role for (p)ppGpp in the induction of autoaggregation under glutamate-limited conditions; however, other factors likely are involved.
FIG 7.
Autoaggregation of B. pertussis strains under various nutritional conditions. (A) B. pertussis strains were cultured in SSC medium for 24 h, and autoaggregation was evaluated by measurement of OD600 values from the upper portion of the culture (closed bars) and following resuspension by vortexing (open bars) after incubation in SSC medium (+) or SSC-0E medium (−) or in the absence (Bvg+) or presence (Bvg−) of 50 mM MgSO4. (B) The autoaggregation of B. pertussis clinical isolates and Tohama I strains was analyzed, and the effect of (p)ppGpp production on autoaggregation is shown. (C) B. pertussis strains PMK21 and PMK21, carrying pRKT01 (relA+) or pRKT415 (vec+), were cultured and autoaggregation was investigated. OD600 values are the averages from three independent experiments. Error bars indicate standard deviations of the means. Representative results from three independent experiments are indicated in the figures.
FIG 8.
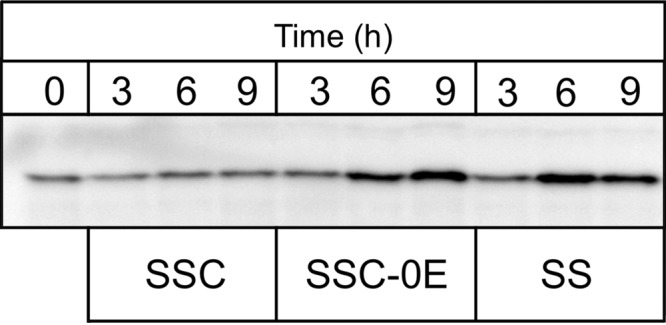
Production of cellular Bsp22 in response to nutrient limitation. Exponential-phase B. pertussis UT25Sm1 cells cultured in SSC medium were collected and resuspended in fresh SSC medium, SSC-E0 medium, or SS medium. At 3, 6, and 9 h after changing the medium, bacterial cells were lysed and Bsp22 was visualized by immunoblotting using a 2,500-fold dilution of anti-Bsp22 antibody. Before replacement of the medium, a portion of the bacterial cells was stored and used for preparation of lysate at 0 h.
DISCUSSION
The construction of the T3SS apparatus or activation of the secretion machinery often is stimulated by specific culture conditions, presumably corresponding to the strain's required microenvironments. Modifications of culture conditions therefore are required to analyze the T3SS by in vitro experiments. Many bacterial strains cultured in nutrient-poor media express high levels of the T3SS while producing undetectable levels under nutrient-rich conditions (28–30). In the case of both enterohemorrhagic and enteropathogenic Escherichia coli, bacteria that are cultured in Dulbecco's modified Eagle's medium with low glucose and grown overnight at 37°C without agitation produce T3SS proteins (31–33). This obviously indicates that the expression of the T3SS is activated under nutrient limitation; therefore, there is tight control in response to the microenvironment.
In the present study, we investigated the effect of glutamate limitation on B. pertussis. The production of the T3SS proteins was significantly increased when glutamate was depleted from the medium. Conversely, FHA, ACT, PTX, and Prn, which also are BvgAS-dependent virulence factors, were not regulated by glutamate. The unaltered production of other Bvg-dependent virulence factors suggests that the function of BvgAS itself is not directly modified by glutamate limitation. BvgAS activation is necessary for the transcription of T3SS genes but is insufficient for maximum expression of T3SS proteins. We assessed the possibility of an effect on BspR; however, the level in the lysate remained the same despite glutamate limitation (Fig. 3C). Thus, the transcription of genes of the bsc locus, which is BtrS dependent, was upregulated by glutamate limitation, whereas the expression of the btr locus genes was not significantly altered. Therefore, it seemed that the upregulation of the bsc genes occurs after the transcription of btr locus genes.
The effect of glutamate limitation on gene expression in B. pertussis first was investigated by DNA microarray analysis (34). Contrary to the case in the present study, the amount of T3SS transcripts was decreased under glutamate limitation. Moreover, decreased transcript levels were observed during the transition from exponential to stationary phase (34). It is notable that Nicholson et al. demonstrated that B. bronchiseptica expressed higher levels of T3SS genes and cytotoxicity mediated by T3SS in late exponential phase and stationary phase than during mid-exponential phase (35). At the entry into stationary phase, the nutrient limitation response is an important aspect of growth phase-dependent gene regulation in addition to other factors, such as the effect of metabolite accumulation. The reason why these results are inconsistent with those of the present study is not clear but may be due to culture conditions or differences between analyses of transcripts and gene products.
The stringent response is triggered by the accumulation of (p)ppGpp in bacterial cells by RelA/SpoT homologues (RSH proteins), which have a (p)ppGpp synthase domain (27). Since other genes encoding RSH proteins except for relA and spoT have not been found in B. pertussis (19), strain PMK21 did not synthesize detectable (p)ppGpp even in the presence of serine hydroxamate, a strong (p)ppGpp inducer, and it showed significantly decreased production of Bsp22 (19). (p)ppGpp is required for SPI-1 activation under low oxygen conditions and positively regulates PI-2 genes in Salmonella enterica serovar Typhimurium (36). Recently, the regulation of the T3SS by (p)ppGpp was demonstrated in Erwinia amylovora via a sigma factor cascade (37). In B. pertussis, the production of the T3SS was significantly decreased by a deficiency of (p)ppGpp (19), and the level still was low even though the amount of glutamate was limited (Fig. 6). Taking these results together, (p)ppGpp is necessary for maximum induction of the T3SS under conditions of BvgAS activation. Additionally, these results strongly suggest that glutamate limitation is one of the triggers for induction despite the lack of direct evidence for induction of (p)ppGpp by glutamate limitation.
The accumulation of (p)ppGpp is induced by depletion of amino acids and iron (14, 17). The induction of (p)ppGpp accumulation by carbon starvation initially was shown in E. coli (15) and subsequently demonstrated in several other bacteria (16, 38–41). It was reported that T3SS production in both B. pertussis and B. bronchiseptica was upregulated when bacteria were cultured in iron-depleted medium (8, 42). We cultured strain PMK21 under iron-limited conditions to assess the role of the stringent response in the iron limitation response, but it did not grow in iron-depleted SSC medium (data not shown). Meanwhile, SS medium is relatively low in amino acids because only 0.03 mM cysteine and 2 mM proline are supplied in addition to glutamate, and SSC medium is more amino acid rich because of the inclusion of Casamino Acids. Bsp22 in the bacterial lysate was induced by exposure to the amino-acid-poor conditions of SS medium (Fig. 8).
Autoaggregation was stimulated in response to glutamate limitation in B. pertussis strains except for Tohama I, and the stringent response seemed partially related to this stimulation. FHA, which is one of the adhesins for B. pertussis and an important virulence factor, is localized on the cell surface and also released into the medium. In the absence of cyclodextrin, FHA associates with the cell surface and causes strong autoaggregation via FHA-FHA binding, probably due to its hydrophobicity (43). This is likely the reason for the decreased autoaggregation in Bvg−-mode bacteria, but it is not clear why the Tohama I strain was unable to autoaggregate, because this strain evidently produces FHA, which was utilized as a vaccine component. Moreover, glutamate limitation did not alter the amounts of FHA in either the cell lysate or the culture fluid. Therefore, additional relationships involving other factors that are induced by glutamate limitation are likely.
The functions of the T3SS in B. pertussis have not been fully elucidated, although the facilitation of colonization and suppression of proinflammatory cytokine production have been reported in a murine model (28). There is both direct and indirect evidence for in vivo expression of the T3SS in B. pertussis. For example, Bsp22 was detected in lung homogenate of B. pertussis-infected mouse (7, 44, 45). In addition, some clinical isolates that did not produce BteA (one of the effectors of the T3SS) due to an insertion sequence (IS) in the promoter of bteA still secreted BopD and produced a functional T3SS apparatus (21). According to these results, it is possible that the T3SS has an alternative role in colonization or persistence in addition to the roles of T3SS effectors in modulating immunity and regulating pathogenesis. Also, the expression of the T3SS genes might be tightly controlled depending on the environment in the host.
The expression of Bsp22 was slightly increased by glutamate limitation in strain PMK21, although the level was much lower than that in the wild-type strain under glutamate-replete conditions. Even when glutamate is the main carbon source, it is also an amino acid that could be an important nitrogen source; therefore, its limitation will affect other aspects of metabolism, such as the glutamate-glutamine cycle. Thus, the glutamate limitation response is probably a component of the complex stringent response as well as other regulatory systems. Increased production of T3SS proteins and enhanced autoaggregation ability in response to glutamate limitation provides an adhesive phenotype that tends to persist in vivo. Future studies will unveil further details regarding the role of glutamate limitation in pathogenicity.
ACKNOWLEDGMENTS
We thank Akio Abe (Kitasato University) for the gift of antibodies to Bsp22 and BspR, Yoshie Matsubara (Division of Microscopic Anatomy, Kyorin University) for the supporting electron microscopy study, Tomoko Nozaki for technical assistance, and Pedro Peixoto (University of Lisbon) for advice during his stay in Japan.
This work was supported by JSPS KAKENHI grant number 26460537.
REFERENCES
- 1.Hertz L. 1979. Functional interactions between neurons and astrocytes. I. Turnover and metabolism of putative amino acid transmitters. Prog Neurobiol 13:277–323. [DOI] [PubMed] [Google Scholar]
- 2.Maragakis NJ, Rothstein JD. 2001. Glutamate transporters in neurologic disease. Arch Neurol 58:365–370. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong SK, Gross R. 2007. Primary metabolism and physiology of Bordetella species, p 165–190. In Locht C. (ed), Bordetella: molecular microbiology. Horizon Bioscience, Wymondham, Norfolk, United Kingdom. [Google Scholar]
- 4.Rowatt E. 1955. Amino acid metabolism in the genus Bordetella. J Gen Microbiol 13:552–560. doi: 10.1099/00221287-13-3-552. [DOI] [PubMed] [Google Scholar]
- 5.Stainer DW, Scholte MJ. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 6.Galan JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fennelly NK, Sisti F, Higgins SC, Ross PJ, van der Heide H, Mooi FR, Boyd A, Mills KH. 2008. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect Immun 76:1257–1266. doi: 10.1128/IAI.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman TJ, Cummings CA, Liew SY, Relman DA, Armstrong SK. 2011. Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J Bacteriol 193:4798–4812. doi: 10.1128/JB.05136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol 12:274–288. doi: 10.1038/nrmicro3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupre E, Herrou J, Lensink MF, Wintjens R, Vagin A, Lebedev A, Crosson S, Villeret V, Locht C, Antoine R, Jacob-Dubuisson F. 2015. Virulence regulation with Venus flytrap domains: structure and function of the periplasmic moiety of the sensor-kinase BvgS. PLoS Pathog 11:e1004700. doi: 10.1371/journal.ppat.1004700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupre E, Lesne E, Guerin J, Lensink MF, Verger A, de Ruyck J, Brysbaert G, Vezin H, Locht C, Antoine R, Jacob-Dubuisson F. 2015. Signal transduction by BvgS sensor kinase: binding of modulator nicotinate affects the conformation and dynamics of the entire periplasmic moiety. J Biol Chem 290:23307–23319. doi: 10.1074/jbc.M115.655720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrou J, Bompard C, Wintjens R, Dupre E, Willery E, Villeret V, Locht C, Antoine R, Jacob-Dubuisson F. 2010. Periplasmic domain of the sensor-kinase BvgS reveals a new paradigm for the Venus flytrap mechanism. Proc Natl Acad Sci U S A 107:17351–17355. doi: 10.1073/pnas.1006267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuk MH, Harvill ET, Miller JF. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol Microbiol 28:945–959. doi: 10.1046/j.1365-2958.1998.00850.x. [DOI] [PubMed] [Google Scholar]
- 14.Cashel M, Kalbacher B. 1970. The control of ribonucleic acid synthesis in Escherichia coli. V. Characterization of a nucleotide associated with the stringent response. J Biol Chem 245:2309–2318. [PubMed] [Google Scholar]
- 15.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 16.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. 2002. Dissection of the mechanism for the stringent factor RelA. Mol Cell 10:779–788. [DOI] [PubMed] [Google Scholar]
- 17.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 18.Brickman TJ, Armstrong SK. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol 178:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugisaki K, Hanawa T, Yonezawa H, Osaki T, Fukutomi T, Kawakami H, Yamamoto T, Kamiya S. 2013. Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiology 159:1379–1389. doi: 10.1099/mic.0.066597-0. [DOI] [PubMed] [Google Scholar]
- 20.Kasuga T, Nakase Y, Ukishima K, Takatsu K. 1954. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med 27:57–62. [PubMed] [Google Scholar]
- 21.Han HJ, Kuwae A, Abe A, Arakawa Y, Kamachi K. 2011. Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. PLoS One 6:e17797. doi: 10.1371/journal.pone.0017797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 23.Hanawa T, Yonezawa H, Kawakami H, Kamiya S, Armstrong SK. 2 July 2013. Role of Bordetella pertussis RseA in the cell envelope stress response and adenylate cyclase toxin release. Pathog Dis doi: 10.1111/2049-632X.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otsuka N, Han HJ, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, Shibayama K, Kamachi K. 2012. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One 7:e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattoo S, Yuk MH, Huang LL, Miller JF. 2004. Regulation of type III secretion in Bordetella. Mol Microbiol 52:1201–1214. doi: 10.1111/j.1365-2958.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- 26.Kurushima J, Kuwae A, Abe A. 2012. The type III secreted protein BspR regulates the virulence genes in Bordetella bronchiseptica. PLoS One 7:e38925. doi: 10.1371/journal.pone.0038925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee VT, Mazmanian SK, Schneewind O. 2001. A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J Bacteriol 183:4970–4978. doi: 10.1128/JB.183.17.4970-4978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallis AJ, Yahr TL, Barbieri JT, Frank DW. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67:914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dijk K, Fouts DE, Rehm AH, Hill AR, Collmer A, Alfano JR. 1999. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol 181:4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, Gruenheid S, Strynakda NC, Puente JL, Finlay BB. 2005. Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 73:2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J 17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Frey E, Mackenzie AM, Finlay BB. 2000. Human response to Escherichia coli O157:H7 infection: antibodies to secreted virulence factors. Infect Immun 68:5090–5095. doi: 10.1128/IAI.68.9.5090-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura MM, Liew SY, Cummings CA, Brinig MM, Dieterich C, Relman DA. 2006. Growth phase- and nutrient limitation-associated transcript abundance regulation in Bordetella pertussis. Infect Immun 74:5537–5548. doi: 10.1128/IAI.00781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson TL, Buboltz AM, Harvill ET, Brockmeier SL. 2009. Microarray and functional analysis of growth phase-dependent gene regulation in Bordetella bronchiseptica. Infect Immun 77:4221–4231. doi: 10.1128/IAI.00136-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson A, Rolfe MD, Lucchini S, Schwerk P, Hinton JC, Tedin K. 2006. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J Biol Chem 281:30112–30121. doi: 10.1074/jbc.M605616200. [DOI] [PubMed] [Google Scholar]
- 37.Ancona V, Lee JH, Chatnaparat T, Oh J, Hong JI, Zhao Y. 2015. The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora. J Bacteriol 197:1433–1443. doi: 10.1128/JB.02551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosse AM, Greenway DL, England RR. 2000. Accumulation of ppGpp and ppGp in Staphylococcus aureus 8325-4 following nutrient starvation. Lett Appl Microbiol 31:332–337. doi: 10.1046/j.1472-765x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- 39.Das B, Pal RR, Bag S, Bhadra RK. 2009. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol Microbiol 72:380–398. doi: 10.1111/j.1365-2958.2009.06653.x. [DOI] [PubMed] [Google Scholar]
- 40.Flardh K, Axberg T, Albertson NH, Kjelleberg S. 1994. Stringent control during carbon starvation of marine Vibrio sp. strain S14: molecular cloning, nucleotide sequence, and deletion of the relA gene. J Bacteriol 176:5949–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesley JA, Shapiro L. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol 190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurushima J, Kuwae A, Abe A. 2012. Iron starvation regulates the type III secretion system in Bordetella bronchiseptica. Microbiol Immunol 56:356–362. doi: 10.1111/j.1348-0421.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 43.Menozzi FD, Boucher PE, Riveau G, Gantiez C, Locht C. 1994. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun 62:4261–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero RV, Osicka R, Sebo P. 2014. Filamentous hemagglutinin of Bordetella pertussis: a key adhesin with immunomodulatory properties? Future Microbiol 9:1339–1360. doi: 10.2217/fmb.14.77. [DOI] [PubMed] [Google Scholar]
- 45.Gaillard ME, Bottero D, Castuma CE, Basile LA, Hozbor D. 2011. Laboratory adaptation of Bordetella pertussis is associated with the loss of type three secretion system functionality. Infect Immun 79:3677–3682. doi: 10.1128/IAI.00136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]