ABSTRACT
Streptococcus gordonii is a commensal inhabitant of the human oral cavity. To maintain its presence as a major component of oral biofilms, S. gordonii secretes inhibitory molecules such as hydrogen peroxide and bacteriocins to inhibit competitors. S. gordonii produces two nonmodified bacteriocins (i.e., Sth1 and Sth2) that are regulated by the Com two-component regulatory system, which also regulates genetic competence. Previously we found that the thiol-disulfide oxidoreductase SdbA was required for bacteriocin activity; however, the role of SdbA in Com signaling was not clear. Here we demonstrate that ΔsdbA mutants lacked bacteriocin activity because the bacteriocin gene sthA was strongly repressed and the peptides were not secreted. Addition of synthetic competence-stimulating peptide to the medium reversed the phenotype, indicating that the Com pathway was functional but was not activated in the ΔsdbA mutant. Repression of bacteriocin production was mediated by the CiaRH two-component system, which was strongly upregulated in the ΔsdbA mutant, and inactivation of CiaRH restored bacteriocin production. The CiaRH-induced protease DegP was also upregulated in the ΔsdbA mutant, although it was not required for inhibition of bacteriocin production. This establishes CiaRH as a regulator of Sth bacteriocin activity and links the CiaRH and Com systems in S. gordonii. It also suggests that either SdbA or one of its substrates is an important factor in regulating activation of the CiaRH system.
IMPORTANCE Streptococcus gordonii is a noncariogenic colonizer of the human oral cavity. To be competitive in the oral biofilm, S. gordonii secretes antimicrobial peptides called bacteriocins, which inhibit closely related species. Our previous data showed that mutation of the disulfide oxidoreductase SdbA abolished bacteriocin production. In this study, we show that mutation of SdbA generates a signal that upregulates the CiaRH two-component system, which in turn downregulates a second two-component system, Com, which regulates bacteriocin expression. Our data show that these systems are also linked in S. gordonii, and the data reveal that the cell's ability to form disulfide bonds is sensed by the CiaRH system.
INTRODUCTION
Oral biofilms are highly competitive, constantly fluctuating environments. The bacteria that colonize this niche contend with a high density of competing bacteria of an estimated 2,000 different taxa and dramatic environmental changes that are dependent on host behavior (1, 2). Streptococcus gordonii is a pioneer colonizer of this environment, where it initiates biofilm formation by binding directly to the acquired salivary pellicle on the tooth surface (2, 3). Once established, S. gordonii persists within the host as part of the oral microbiota. The presence of S. gordonii is associated with oral health (4, 5), and it has been shown to inhibit biofilm formation by cariogenic species (6, 7).
S. gordonii uses several strategies to gain advantage over its competitors and to colonize the oral cavity successfully. These strategies include the production of inhibitory molecules to prevent growth and biofilm formation by related species. For example, S. gordonii produces hydrogen peroxide as a metabolic byproduct, which inhibits the growth of neighboring species that are more sensitive to oxidative stress (8, 9). It also targets closely related species more directly, by secreting small antimicrobial peptides called bacteriocins (10). To defend itself from similar counterattacks, it secretes a protease that degrades the signaling peptide required for bacteriocin production and biofilm formation in a competitor, Streptococcus mutans (11).
S. gordonii DL-1 Challis produces two nonlantibiotic bacteriocins, namely, Sth1 and Sth2, encoded by sthA and sthB, respectively. Sth1 is active against other S. gordonii strains, including C219 and Wicky (12), while Sth1 and Sth2 work in conjunction to target other streptococci, such as Streptococcus mitis and Streptococcus oralis (10). Sth1 and Sth2 are the only known bacteriocins produced by S. gordonii, and they are both regulated by the Com two-component regulatory system, which also controls natural genetic competence (10, 13).
With the exception of the Sth bacteriocins, which are unique to S. gordonii, the Com signaling system of S. gordonii is similar to the pathway in Streptococcus pneumoniae (13–15). Competence in S. gordonii occurs during the early exponential growth phase and is activated by competence-stimulating peptide (CSP), a small secreted autoinducer derived from a larger peptide encoded by comC (13, 15). Processing and secretion of CSP are mediated by an ABC transporter, ComAB, which recognizes peptides with a specific double-glycine motif (GG motif) in the N-terminal leader sequence. When the extracellular concentration of CSP surpasses a threshold level, a membrane-bound histidine kinase, ComD, phosphorylates its cognate response regulator, ComE, thereby activating the Com pathway and ultimately modulating the expression of over 150 genes (13).
The genes controlled by the Com system can be divided into two groups, i.e., early genes that are activated directly by ComE, such as comCDE and comAB, and late genes that are regulated by two alterative sigma factors, ComR1 and ComR2, which are homologs of S. pneumoniae ComX (15). Unlike in S. pneumoniae, the S. gordonii comR genes lack an identifiable ComE binding site and are activated by an unknown mechanism (15). Nevertheless, the ComR sigma factors direct expression of the late genes, including the DNA uptake machinery for genetic competence and the bacteriocin genes sthA and sthB (10, 13) (see Fig. 6).
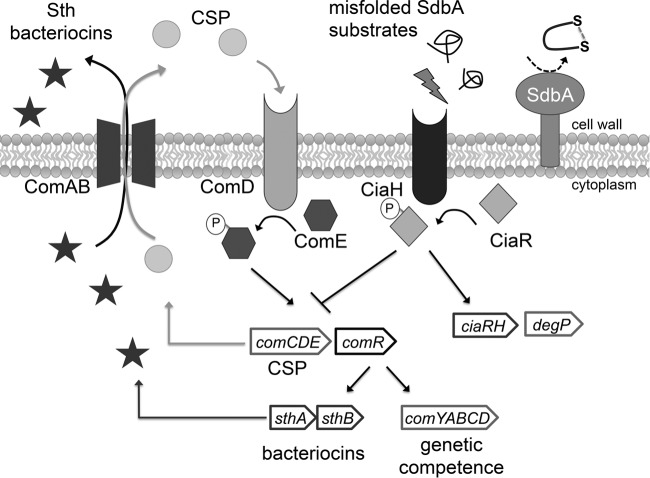
FIG 6.
Summary of the pathway regulating bacteriocin production in S. gordonii. Bacteriocin production in S. gordonii is regulated by the Com quorum-sensing system, which also controls genetic competence. The system is activated by extracellular competence-stimulating peptide (CSP), which is encoded by comC. When ComD, a histidine kinase located at the cell surface, binds to CSP, it becomes active to phosphorylate its cognate response regulator, ComE. ComE drives the expression of the comCDE operon in an autoregulatory loop, as well as the ComAB transporter required for CSP and bacteriocin secretion. In addition to ComE, the Com pathway requires two alternative sigma factors, ComR1 and ComR2, to activate expression of the genes required for genetic competence (comYABCD), as well as the sthA and sthB bacteriocins. Mutation of the disulfide oxidoreductase SdbA generates a signal that results in upregulation of the CiaRH two-component regulatory system. Activation of CiaRH somehow shuts down the Com system, leading to loss of bacteriocin production.
The systems regulating bacteriocin production in other streptococci have been studied in greater detail than those in S. gordonii, and the findings have revealed greater complexity, often involving multiple regulatory systems. Streptococcus pneumoniae produces two bacteriocins encoded by the blp locus, which are controlled by a dedicated quorum-sensing and secretion system; however, the activity of the Blp system is also modulated by at least two additional regulatory systems, namely, ComDE (16) and CiaRH (17). Although the mechanisms involved are not fully understood, the serine protease HtrA (DegP) also appears to play an important role in regulating S. pneumoniae bacteriocin production (17, 18). Similarly, S. mutans produces at least 10 different bacteriocins, which vary by strain, include both lantibiotics and nonlantibiotics (19), and are subject to regulation by complex overlapping systems, including ComDE (20, 21), CiaRH (22), VicRK (23), HdrRM (24), and BrsRM (25). The effects of regulatory systems other than ComDE on bacteriocin production in S. gordonii are not known.
Previously, we found that S. gordonii mutants lacking the thiol-disulfide oxidoreductase SdbA did not exhibit bacteriocin activity (26). SdbA catalyzes disulfide bond formation in secreted proteins, and these bonds are important for protein folding and activity. It is not unusual for bacteriocins to contain disulfide bonds, and all class IIa bacteriocins (pediocin-like), including those produced by Streptococcus uberis and Streptococcus thermophilus, contain a disulfide bond that is essential for activity (27, 28), as do many lantibiotic bacteriocins, such as bovicin produced by Streptococcus bovis HJ50 (29). S. gordonii bacteriocins, however, do not contain cysteines to form a disulfide bond, and the role of SdbA in their production was unclear.
In this study, we aimed to determine how SdbA affects bacteriocin production. Using SdbA active site mutants, we confirmed that bacteriocin production does require the enzyme's disulfide oxidoreductase activity; ΔsdbA mutants did not secrete bacteriocins into the medium, and expression of the bacteriocin-encoding gene sthA was dramatically reduced, compared to the parent, as was that of the gene encoding the CSP autoinducer, comC. The effects of SdbA on the Com pathway and bacteriocin production were indirect and required the CiaRH two-component system, which was upregulated in ΔsdbA mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Experiments were carried out using S. gordonii SecCR1 as the parent strain. S. gordonii SecCR1 is a derivative of S. gordonii DL-1 Challis that secretes a single-chain variable-fragment antibody (scFv) against complement receptor 1 (CR1), which is a protein that requires disulfide bonds for stability (30). This strain was used in our previous studies of disulfide bond formation in S. gordonii (26), and the S. gordonii SecCR1 ΔsdbA mutant has the same phenotype as the ΔsdbA mutant of S. gordonii DL-1 Challis (see Fig. S1 in the supplemental material). Additional strains and mutants are described in Table 1. Streptococcus spp. were grown in brain heart infusion (BHI) medium supplemented with 5% fetal bovine serum (BHIS), at 37°C in 5% CO2 without shaking. Escherichia coli XL-1 Blue was grown in Luria-Bertani (LB) medium, at 37°C with shaking. Antibiotics were used at the following concentrations: for S. gordonii, erythromycin at 10 μg/ml, tetracycline at 10 μg/ml, spectinomycin at 250 μg/ml, kanamycin at 250 μg/ml, and chloramphenicol at 5 μg/ml; for E. coli, ampicillin at 100 μg/ml, tetracycline at 10 μg/ml, and chloramphenicol at 20 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| S. gordonii strains | ||
| SecCR1 | DL-1 Challis, secretes anti-CR1 scFv; Tetr Spcr | 30 |
| ΔsdbA mutant | SecCR1 sdbA::ermAM Tetr Spcr Ermr | 26 |
| SdbA Compl | ΔsdbA; sdbA complemented on chromosome; Tetr Spcr Kanr | 26 |
| ΔsdbAΔdegP mutant | ΔsdbA degP::aphA3 Tetr Spcr Kanr Ermr | This study |
| SdbA C86P/C89A mutant | SecCR1 with SdbA cysteine point mutation; Tetr Spcr Kanr | This study |
| ΔciaRH mutant | SecCR1 ciaRH::aphA3 Tetr Spcr Kanr | This study |
| ΔsdbAΔciaRH mutant | ΔsdbA ciaRH::aphA3 Tetr Spcr Kanr Ermr | This study |
| ΔsdbA CiaRH Compl | ΔsdbA ΔciaRH; ciaRH complemented on chromosome; Tetr Spcr Ermr Cmr | This study |
| E. coli XL-1 Blue | Host for DNA manipulations and expression of recombinant proteins | Stratagene |
Genetic manipulations.
ΔciaRH mutants were constructed by creating a clean deletion of ciaRH and replacing the genes with a kanamycin resistance cassette (aphA3) amplified from the plasmid pDL276 (31). PCR was carried out using Phusion high-fidelity DNA polymerase (New England BioLabs, Whitby, ON, Canada) to amplify 425 bp of the upstream gene sgo_1071 and 525 bp of the downstream gene sgo_1074, using the SL1178/SL1222 and SL1220/SL1221 primer pairs, respectively (see Table S1 in the supplemental material). The PCR products were digested with restriction enzymes, as indicated in Table S1 in the supplemental material, and ligated with T4 DNA ligase (New England BioLabs). The ligation product was amplified using the outside primers SL1178 and SL1221, and the resulting construct was used to transform S. gordonii SecCR1 or S. gordonii DL1 Challis, as described previously (30). Transformants were selected on BHI medium containing the appropriate antibiotics, and insertion of aphA3 and deletion of ciaRH were confirmed by PCR. The ΔdegP mutants were prepared using the same strategy, with the primers indicated in Table S1 in the supplemental material.
Complementation of ciaRH in the ΔsdbA ΔciaRH mutant was achieved by introducing functional ciaRH genes back into the chromosome, under their native promoter, as follows. The ciaRH genes and the upstream 130-bp intergenic region were amplified using the SL1180/SL1221 primer pair. The resulting PCR fragment was digested with KpnI and ligated to a chloramphenicol acetyltransferase (cat) resistance cassette cut from pCopCAT/pUC18 using KpnI and HindIII. pCopCAT/pUC18 was constructed by subcloning the 1.6-kb PstI-BamHI DNA fragment containing the cat gene under the control of a S. mutans cop promoter from pHSL2/pUC (32) into the same sites on pUC18. Next, a 425-bp fragment of the gene located upstream of ciaRH, sgo_1071, was amplified by PCR using the SL1178/SL1179 primer pair and digested with HindIII. The three fragments were ligated together with T4 DNA ligase and amplified by PCR using the primers SL1178 and SL1221. The resulting construct was used to transform ΔsdbA ΔciaRH cells by homologous recombination, replacing aphA3 with the cat-ciaRH construct. Transformants were selected on BHI medium with chloramphenicol, and replica plating was used to identify kanamycin-sensitive, chloramphenicol-resistant colonies. Complementation was confirmed by PCR.
To test for polar effects, expression of the upstream and downstream genes, sgo_1071 and sgo_1074, respectively, was tested by reverse transcription (RT)-PCR. The results indicated that the genetic manipulations did not create a polar effect in the strains (see Fig. S2 in the supplemental material).
Site-directed mutagenesis.
Point mutation of the SdbA cysteines at the active site (C86PDC89) was used to generate a catalytically inactive SdbA mutant. The cysteine at codon 86 was mutated first. The upstream portion of sdbA amplified with the SL756/SL975 primer pair and the downstream portion of sdbA amplified with the SL974/SL803 primer pair were cloned into pBluescript. Primers SL974 and SL975 contained the cysteine (TGT)-to-proline (CCA) substitution. The resulting pBluescriptsdbAC86P was digested with BamHI and ligated to aphA3 and a downstream portion of sdbA. The ligated DNA was amplified using primers SL762 and SL759 and transformed into the ΔsdbA mutant to generate the SdbA C86P mutant. This strategy was used because it created a unique MscI site (TGGTGT to TGGCCA) that could be used as a second approach, in addition to DNA sequencing, to confirm the mutation quickly.
The second cysteine of the active site was mutated by overlapping PCR (33). Overlapping PCR with the SL756/SL1039 and SL1038/SL759 primer pairs was used to construct a cysteine (TGT)-to-alanine (GCT) substitution at the cysteine codon at position 89. The fragments were combined and amplified with SL756 and SL803. The resulting construct was then ligated to aphA3 and a downstream portion of sdbA. The ligated DNA was amplified using the primers SL756 and SL759. This PCR product was used to transform an SdbA C86P mutant to produce a C86P/C89A double mutant. Point mutations were confirmed by DNA sequencing (McGill University and Génome Québec Innovation Centre).
Bacteriocin activity assay.
The activity of Sth bacteriocins was tested as described by Heng et al. (10), using S. mitis I18 or S. oralis 34 as the target strain. Briefly, overnight starter cultures of S. gordonii were diluted 1:40 into prewarmed BHIS medium and were grown to an optical density at 600 nm (OD600) of ∼0.200. The culture supernatant fluids were filter sterilized using 0.22-μm filters and were mixed 1:1 with fresh BHIS medium. The conditioned medium was warmed at 37°C for 15 min prior to inoculation with a 1:100 dilution of the target strain. Cultures were grown for 6 h (S. mitis) or 10 h (S. oralis), and the OD600 was read using a spectrophotometer (Shimadzu UV-1700; Shimadzu, Kyoto, Japan).
For assays that included synthetic CSP (DVRSNKIRLWWENIFFNKK) (Biomatik, Cambridge, ON, Canada), cultures were grown in BHIS medium to an OD600 of 0.150, followed by the addition of 10 ng/ml CSP (from a 1-mg/ml stock solution in MilliQ water). The cultures were then incubated for an additional 30 min at 37°C, to allow induction and protein expression. Antibiotics were omitted from all cultures. Assays were performed in triplicate and repeated at least three times.
Detection of DegP.
Cells from BHIS cultures with OD600 values of 0.2 were boiled in SDS-PAGE sample buffer (250 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol, 10% 2-mercaptoethanol, 0.01% bromphenol blue), and the protein extracts were electrophoresed on 12.5% SDS-PAGE gels. The proteins were transferred to nitrocellulose membranes (Bio-Rad Laboratories) using standard techniques (34), blocked with 5% skim milk, and reacted with either rabbit anti-HtrA (S. pneumoniae) antiserum (1:500 dilution; a gift from Jeffrey Weiser, University of Pennsylvania) (35) or mouse anti-PrsA antiserum (36). The membranes were then reacted with goat anti-rabbit IgG–alkaline phosphatase (1:30,000 dilution; Sigma-Aldrich) or goat anti-mouse IgG–alkaline phosphatase (1:30,000 dilution; Sigma-Aldrich).
Purification of Sth from culture supernatants.
Antibodies were raised in New Zealand White rabbits (37) and BALB/c mice (36) against Sth1 conjugated to keyhole limpet hemocyanin (Biomatik), using methods similar to those described previously. Sth1-specific antibodies were affinity purified from rabbit sera using Sth1 peptides cross-linked to CNBr-activated Sepharose 4B beads (GE Healthcare Life Sciences, Mississauga, ON, Canada), according to the manufacturer's instructions. The purified antibodies were then irreversibly cross-linked to protein A-Sepharose beads (Sigma-Aldrich) with dimethyl pimelimidate, using standard techniques (38).
To purify secreted bacteriocins, overnight starter cultures were diluted 1:40 into prewarmed BHIS medium and grown to an OD600 of 0.200, unless otherwise noted. Cells were removed by centrifugation (5,000 × g for 10 min at 4°C), and the supernatant was passed through a 10-ml column packed with 1 ml of anti-Sth1-protein A-Sepharose. The column was washed with 10 ml of 10 mM Tris (pH 7.5), followed by 10 ml of 10 mM Tris (pH 7.5) containing 500 mM NaCl. Sth1 was eluted in 200-μl fractions with 100 mM glycine (pH 2.5) and was immediately neutralized with 20 µl of 1 M Tris (pH 8.0). Microtiter plates (Maxisorp; Fischer Scientific, Ottawa, ON, Canada) were coated with 100-μl aliquots of the fractions and incubated overnight at 4°C. The plates were then blocked with 200 μl of 1% (wt/vol) gelatin in phosphate-buffered saline with 0.1% Tween 20 (PBST) at room temperature for 1 h. After blocking, mouse anti-Sth1 antiserum (1:1,000) was added to the wells and incubated overnight at 4°C. Sth1 was detected using goat anti-mouse IgG–biotin (1:20,000; Sigma-Aldrich), followed by ExtrAvidin-alkaline phosphatase (1:60,000; Sigma-Aldrich). The plates were developed with p-nitrophenyl phosphate (1 mg/ml; Bioshop Canada Inc., Burlington, ON, Canada) in diethanolamine buffer, and the absorbance at 405 nm was read using a microplate reader.
Reverse transcription-PCR and quantitative real-time PCR.
Overnight cultures of the parent strain and the ΔsdbA, ΔciaRH, and ΔsdbA ΔciaRH mutants grown in BHIS medium were diluted 1:40 and grown to an OD600 of 0.200 in fresh BHIS medium, with or without exogenous CSP (100 ng/ml). Total RNA was isolated using the hot acid phenol method, as described previously (39). The RNA (1 μg) was treated with 1 unit of amplification-grade DNase I (Life Technologies, Burlington, ON, Canada) for 15 min at room temperature, and removal of DNA was confirmed by PCR with 16S rRNA primers (SL525 and SL697). cDNA synthesis was carried out using random primers and SuperScript II reverse transcriptase (Life Technologies), according to the manufacturer's directions.
Quantitative real-time PCR (qPCR) to amplify comC, comE, sthA, ciaR, and degP was carried out using the primers listed in Table S1 in the supplemental material and iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Mississauga, ON, Canada), according to the manufacturer's directions. The reactions were performed using a 7900 HT Fast real-time PCR system (Applied Biosystems) at 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The cycle threshold (CT) was calculated using SDS 2.2.2 software (Applied Biosystems). Relative expression levels were calculated using the comparative CT method (40), using 16S rRNA as an internal control. Each reaction was performed in duplicate, using cDNA from at least three biological replicates.
Statistical analysis.
Results were analyzed by one-way analysis of variance, with Tukey's posttests, using GraphPad Prism version 6 (GraphPad Software, Inc., La Jolla, CA).
RESULTS
The thiol-disulfide oxidoreductase SdbA is required for bacteriocin production.
Previously, we found that ΔsdbA mutants were defective in bacteriocin activity (26). To investigate how SdbA affects bacteriocin production, we started by constructing a catalytically inactive SdbA active site mutant, to determine whether bacteriocin production requires the oxidoreductase activity of SdbA. The active site of thiol-disulfide oxidoreductases contains a CXXC motif, where X is any amino acid, and the two cysteines are required for activity (41). To eliminate SdbA oxidase activity, the N- and C-terminal cysteines were changed to proline and alanine, respectively, and the loss of enzyme activity was confirmed using an RNase A folding assay (see Fig. S3 in the supplemental material).
Bacteriocin production by the active site mutant was tested in an activity assay using the target strains S. mitis and S. oralis (10, 12). Consistent with our previous results, supernatants obtained from early-exponential-phase cultures of the S. gordonii parent strain contained active bacteriocins that inhibited the growth of the target strains (Fig. 1A and B). In contrast, supernatants from the ΔsdbA mutant and the SdbA active site mutant failed to inhibit growth. When a functional sdbA gene was reintroduced into the chromosome of the ΔsdbA mutant, bacteriocin activity was restored, confirming that the enzyme activity of SdbA is required for normal bacteriocin activity.
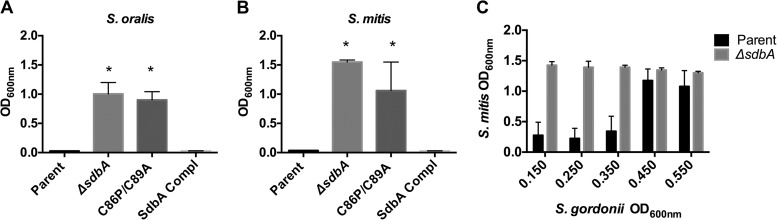
FIG 1.
SdbA is required for bacteriocin activity. The inhibitory activity of S. gordonii bacteriocins secreted into the medium was tested against two target strains. (A and B) Growth of S. oralis 34 (A) and S. mitis I18 (B) in the presence of filter-sterilized culture supernatants from the S. gordonii parent strain, the ΔsdbA mutant, the SdbA active site cysteine mutant (C86P/C89A), and the sdbA-complemented mutant (SdbA Compl), grown to an OD600 of ∼0.200. (C) Growth of S. mitis in the presence of culture supernatants obtained from the S. gordonii parent strain or the ΔsdbA mutant at different time points. Results are means ± standard deviations (SDs) from three experiments. *, P < 0.001, compared with the parent strain.
Because bacteriocin production is a transient, growth phase-dependent phenomenon (12, 42), it was possible that the window of growth in which bacteriocin production occurred was altered in the ΔsdbA mutant and bacteriocins were being produced but at a different time point than in the parent strain. Bacteriocin expression peaks 15 min after exposure to CSP (13), and bacteriocin activity can be detected in culture supernatants during early exponential growth (2 h) but not in the mid-exponential to late exponential phase (6 h) (12). To determine whether the ΔsdbA mutant produced bacteriocins at a later growth stage than the parent strain, we assayed bacteriocin activity against S. mitis using S. gordonii supernatants harvested at different time points. Under the conditions used in this study, there was no difference in growth rates between the parent strain and the ΔsdbA mutant (data not shown). Bacteriocin activity in the parent strain was no longer detected as the optical density of the culture increased from an OD600 of 0.350 to an OD600 of 0.450, and the growth of S. mitis was no longer inhibited (Fig. 1C). In contrast, the ΔsdbA mutant showed no indication of bacteriocin activity at any of the time points tested, even as the culture reached the mid-exponential phase of growth.
An alternative explanation for the lack of bacteriocin activity in the ΔsdbA mutant was that the peptides were being produced but were not being processed to their active form. S. gordonii bacteriocins are produced as small peptides that are processed from larger proteins during secretion. Like the CSP autoinducer, Sth bacteriocins contain a GG motif that directs their secretion via ComAB, which couples transport across the membrane with cysteine protease activity that cleaves at the GG motif of the signal sequence (43, 44). ComAB is the only transporter of this type encoded by S. gordonii (45). Thus, it was possible that the ΔsdbA mutant has a defect in bacteriocin processing and/or secretion that would result in loss of biological activity. To determine whether the ΔsdbA mutant secreted an inactive form of Sth, we tested for the presence of bacteriocins in culture supernatants by immunoaffinity chromatography. Sth bacteriocins were isolated from supernatants obtained from the parent strain and the sdbA-complemented mutant but were not detected in supernatants from the ΔsdbA mutant (Fig. 2A). The signal from the ΔsdbA mutant was the same as that from the negative control with medium alone. Sth was not detected above the background absorbance even when the volume of supernatant was increased 4-fold, and thus the bacteriocin was produced either at very low levels or not at all (Fig. 2B).
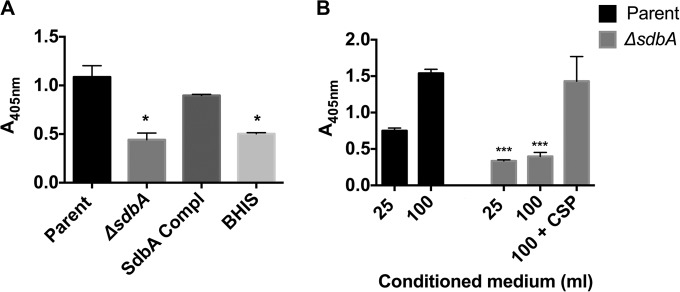
FIG 2.
The ΔsdbA mutant does not secrete Sth1 bacteriocins. Secreted bacteriocins were isolated from culture supernatants using Sth1-specific rabbit IgG-protein A-Sepharose beads and were detected with a mouse anti-Sth1 antibody in an enzyme-linked immunosorbent assay (ELISA). (A) Detection of Sth1 captured from 25 ml of culture supernatant prepared from the parent strain, the ΔsdbA mutant, the sdbA-complemented mutant (SdbA Compl), or uninoculated BHI medium with 5% serum (BHIS) (negative control). (B) Detection of Sth1 captured from 25 or 100 ml of culture supernatant from the parent strain, the ΔsdbA mutant, or the ΔsdbA mutant induced with exogenous CSP for 30 min. Results are means ± SDs from three experiments. ***, P < 0.001, compared with the parent strain; *, P < 0.05, compared with the parent strain.
Finally, we used quantitative real-time PCR (qPCR) to test the expression of the bacteriocin gene sthA in the ΔsdbA mutant. The expression of sthA was markedly lower in the ΔsdbA mutant than in the parent strain, with an average level >1,500-fold lower than that of the parent strain, which indicated that the mutant lacked bacteriocin activity because the gene was not transcribed (Fig. 3A). Complementation of sdbA restored sthA expression.
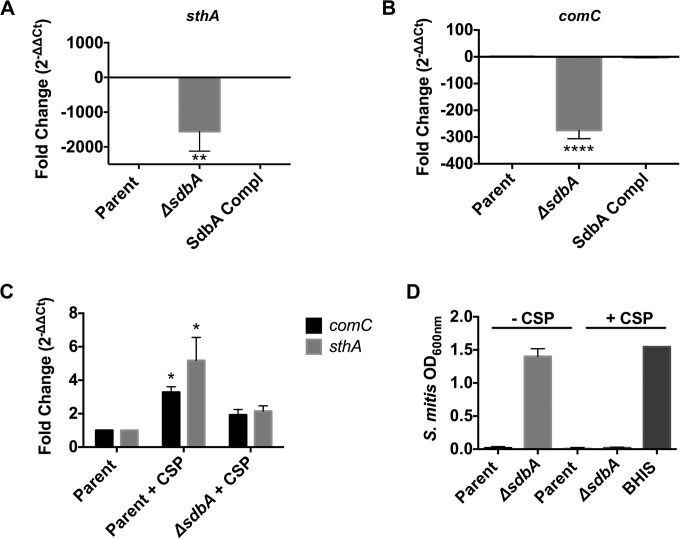
FIG 3.
The ΔsdbA mutant requires exogenous CSP to activate the Com two-component system. (A and B) Expression of sthA (A) and comC (B) in the parent strain, the ΔsdbA mutant, and the sdbA-complemented mutant (SdbA Compl) grown in BHI medium with 5% serum (BHIS). (C) Expression of comC and sthA following induction with exogenous CSP. Synthetic CSP was added to cultures at an OD600 of 0.150, and the cultures were grown for an additional 30 min prior to RNA isolation. Bars, expression levels relative to the parent strain grown in BHIS medium without exogenous CSP. (D) Bacteriocin activity assay with S. mitis as the target strain. CSP was added to S. gordonii cultures to induce bacteriocin production 30 min before the culture supernatants were filter sterilized and inoculated with the target strain. BHIS medium with synthetic CSP was used as a negative control, and results confirm that CSP does not affect the growth of S. mitis. Results are means ± SDs from three experiments. ****, P < 0.0001, compared with the parent strain; **, P < 0.01, compared with the parent strain; *, P < 0.05, compared with the parent strain.
The Com pathway is not activated in the ΔsdbA mutant.
Bacteriocin expression in S. gordonii is regulated by the Com two-component system (10). The process is initiated by an autoinducer encoded by comC, which is processed during secretion to produce the mature competence-stimulating peptide (CSP). When CSP levels reach a threshold concentration, the ComDE two-component system becomes activated, ultimately inducing expression of the bacteriocin genes sthA and sthB (10, 13). The lack of sthA expression in the ΔsdbA mutant suggested that the ComDE pathway had not been activated, which is consistent with our previous observation that the ΔsdbA mutant is also defective in genetic competence (26) (see Fig. S1 in the supplemental material), a phenotype that is regulated by the same system (13).
To assess the activity of the Com system in the ΔsdbA mutant, we tested the expression of comC, which encodes the CSP that activates the system. CSP is required for expression of the alternative sigma factors ComR1 and ComR2, which are needed for bacteriocin expression. Consistent with the lack of bacteriocin activity and competence, comC expression in the ΔsdbA mutant was downregulated an average of 247-fold, relative to the parent strain (Fig. 3B). Complementation of sdbA restored comC expression.
Two key elements required for activity of the Com pathway are the ComDE two-component system, which senses extracellular CSP, and the ComAB transporter, which secretes both CSP and bacteriocins. Inactivation of any of these components has been shown to abolish bacteriocin production (10) and could explain the ΔsdbA mutant phenotype. To determine whether the ComDE system was functional in the ΔsdbA mutant, we tested the ability of exogenous synthetic CSP to activate the pathway artificially. When exogenous CSP was added to the culture medium, the expression of comC and sthA by the ΔsdbA mutant was restored to levels similar to those in the parent strain, with 1.9- and 2.2-fold increases, respectively, over the parent strain grown in BHIS medium (Fig. 3C). Induction was stronger for the parent strain stimulated with CSP, with a 3.3-fold increase in comC expression and a 5.2-fold increase in sthA expression, compared with growth with no exogenous CSP. The ability of the ΔsdbA mutant to respond to exogenous CSP suggested that the ComDE system was functional.
Heng et al. demonstrated previously that the ComAB transporter is essential for bacteriocin secretion in S. gordonii and mutation of either ComA or ComB eliminated bacteriocin activity, even when expression of the bacteriocin genes was induced with exogenous CSP (10). Thus, we hypothesized that the ComAB transporter might be inactive in the ΔsdbA mutant. If the ComAB transporter was inactive, then we would expect to see strong induction of the bacteriocin genes without a corresponding increase in inhibitory activity against Sth-sensitive strains. To determine whether sthA expression induced by exogenous CSP led to bacteriocin secretion in the ΔsdbA mutant, we tested for the presence of bacteriocins in culture supernatants by using immunoaffinity chromatography. Secreted bacteriocins were successfully purified from culture supernatants obtained from the ΔsdbA mutant following induction with CSP, indicating that the transporter was functional or that the ΔsdbA mutant was secreting the bacteriocins using an unknown mechanism (Fig. 2B). The bacteriocins were biologically active, and supernatant from the ΔsdbA mutant efficiently inhibited growth of the target strain S. mitis (Fig. 3D). Thus, the ΔsdbA mutant was capable of bacteriocin production, and the lack of bacteriocin activity appeared to stem from the initial activation of the Com signaling pathway that regulates bacteriocin expression.
Expression of the CiaRH two-component system is upregulated in the ΔsdbA mutant.
Given that the components of the Com signaling system appeared to be functional, we reasoned that the lack of bacteriocin activity in the ΔsdbA mutant might involve an indirect mechanism. In S. pneumoniae, the CiaRH two-component system represses both bacteriocin production and the Com signaling system (17, 46), which was strikingly similar to the phenotype we observed in the ΔsdbA mutant (26). Although the signals detected by the sensor protein CiaH are not known, certain conditions can increase the activity of the system (47–50), and we hypothesized that inactivation of sdbA might create a signal that increases CiaRH activity, leading to loss of bacteriocin production.
To determine whether CiaRH was activated in the ΔsdbA mutant, we used qPCR to assess the expression of known cia-induced genes. We tested ciaR and degP because they have been identified as part of the cia regulons in S. pneumoniae and S. mutans (47, 50), and the S. gordonii degP gene contained a CiaR binding motif that matched the sequence and location reported for S. pneumoniae (51).
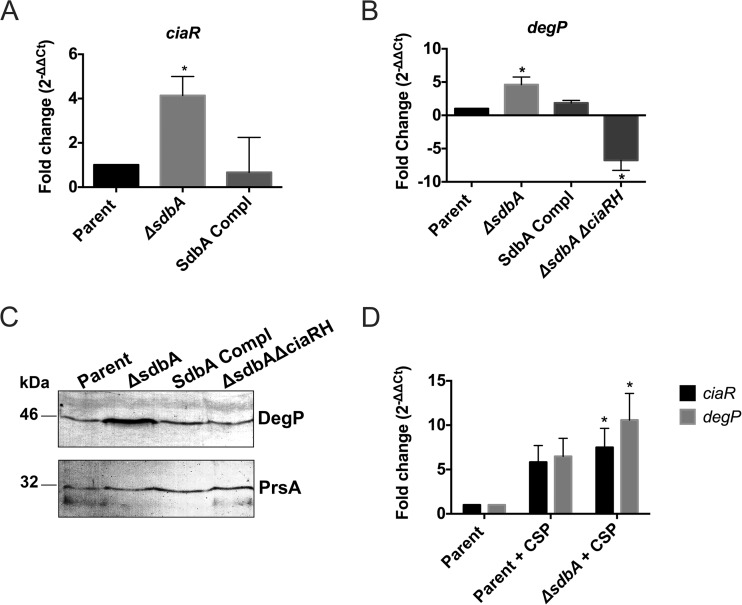
The qPCR results showed that ciaR expression in the ΔsdbA mutant was increased 4-fold over that in the parent strain (P < 0.05) (Fig. 4A). Similarly, degP expression in the ΔsdbA mutant was upregulated 4.5-fold, compared to the parent strain (P < 0.05) (Fig. 4B). This increase was CiaRH dependent, and degP expression was reduced 6.8-fold in the ΔsdbA ΔciaRH strain. The expression data were confirmed by Western blotting, which showed notably higher levels of DegP protein in the ΔsdbA mutant than in the parent strain (Fig. 4C). Complementation of sdbA returned the levels of ciaR and degP expression to those of the parent strain.
FIG 4.
The CiaRH system is activated in the ΔsdbA mutant. (A) Expression of the cia-induced gene ciaR in the parent strain, the ΔsdbA mutant, and the sdbA-complemented mutant (SdbA Compl). (B) Expression of the cia-induced gene degP in the parent strain, the ΔsdbA mutant, the sdbA-complemented mutant, and the ΔsdbA ΔciaRH mutant. (C) Western blot showing DegP detected in cell extracts from the parent strain, the ΔsdbA mutant, the sdbA-complemented mutant, and the ΔsdbA ΔciaRH mutant. The same samples were electrophoresed on duplicate gels and reacted with either anti-HtrA (DegP) antiserum or anti-PrsA antiserum (as a loading control). (D) Expression of ciaR and degP in cultures induced with exogenous CSP. Results are means ± SDs from three experiments. *, P < 0.05, compared with the parent strain.
Interestingly, cultures induced with exogenous CSP showed even higher levels of ciaR and degP expression, i.e., levels increased 7.5- and 10.6-fold, respectively, in the ΔsdbA mutant (Fig. 4D). The parent strain also showed increased ciaR and degP expression in the presence of exogenous CSP, but the increase was not statistically significant, compared with growth with no CSP (P = 0.062 and P = 0.056, respectively) (Fig. 4D). This is consistent with a previous investigation of CSP-induced genes in S. gordonii, which identified a 2.25-fold increase in degP expression but did not identify ciaR among the CSP-upregulated genes (13). Thus, there might be some cross-regulation between the ComDE and CiaRH systems, or the stress of competence induction might affect CiaRH activity. Taken together, the results indicate that mutation of sdbA generates a signal that results in increased CiaRH activity, which is further upregulated with CSP induction.
CiaRH shuts down bacteriocin production in the ΔsdbA mutant.
Upregulation of CiaRH inhibits the Com pathway in S. pneumoniae by inhibiting CSP production (46). The mechanism involves small noncoding RNAs that are regulated by CiaRH, which are thought to bind and to prevent translation of comC mRNA, thereby inhibiting CSP production (46). The importance of CSP to bacteriocin production in S. gordonii was demonstrated previously, and ΔcomC mutants lack bacteriocins (10). Thus, we hypothesized that the enhanced CiaRH activity observed in the ΔsdbA mutant could lead to repression of the Com pathway. This scenario is consistent with our finding that synthetic CSP activates bacteriocin production in the ΔsdbA mutant despite upregulating ciaR expression, since it would circumvent the need for CSP production to activate the pathway.
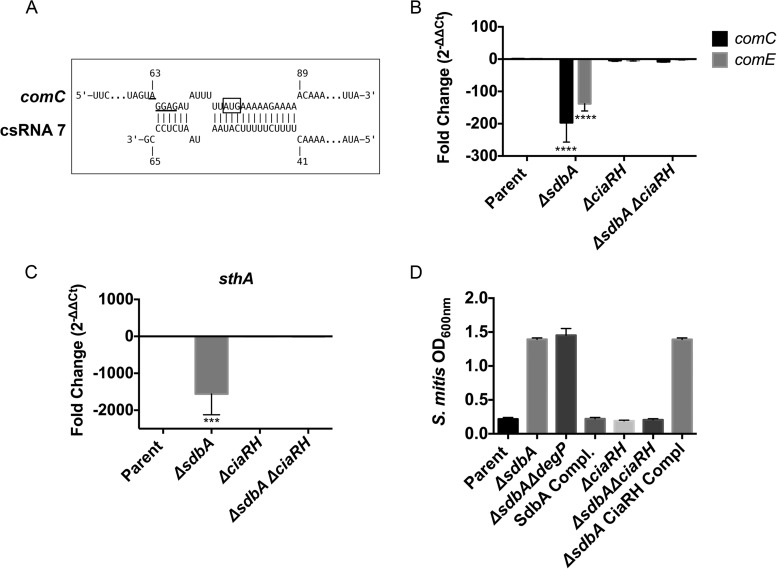
Using published sequences for predicted S. gordonii cia-dependent small RNAs (csRNAs) identified by Marx et al. (52), we searched the genome of S. gordonii for potential targets by using the program IntaRNA (53). Although the bacteriocin genes sthA and sthB were not among the predicted targets, comC was identified as a statistically significant (P = 0.017 to 0.03) target for multiple csRNAs (csRNA2-1, csRNA2-2, and csRNA7) (Fig. 5A). The predicted csRNA binding sites were centered on the start codon and the ribosomal binding site and were almost identical to the experimentally verified csRNA binding sites on S. pneumoniae comC (46).
FIG 5.
CiaRH represses comC and sthA in the ΔsdbA mutant. (A) Interaction between csRNA7 (52) and comC predicted by IntaRNA (53). The csRNA sequence was searched against the RefSeq sequence for S. gordonii Challis (GenBank accession no. NC_009785). The ribosomal binding site is underlined, and the box indicates the start codon. (B and C) Expression of comC and comE (B) and sthA (C) in the parent strain, the ΔsdbA mutant, the ΔciaRH mutant, and the ΔsdbA ΔciaRH mutant. (D) Bacteriocin activity of the parent strain, the ΔsdbA mutant, the ΔsdbA ΔdegP mutant, the sdbA-complemented mutant (SdbA Compl), the ΔciaRH mutant, the ΔsdbA ΔciaRH mutant, and the ciaRH-complemented mutant (ΔsdbA CiaRH Compl). Supernatants were filtered sterilized and inoculated with S. mitis as the target strain. Results are means ± SDs from three experiments. ****, P < 0.0001, compared with the parent strain; ***, P < 0.001, compared with the parent strain.
To determine whether CiaRH affected comC and sthA expression in the ΔsdbA mutant, we assessed expression in a ΔsdbA ΔciaRH mutant. Consistent with a role in inhibiting the Com pathway, inactivation of ciaRH in the ΔsdbA mutant returned expression of comC and comE to levels that were not significantly different from those in the parent strain (Fig. 5A). Similarly, sthA showed derepression in the ΔsdbA ΔciaRH mutant (Fig. 5B). Mutation of ciaRH in the parent strain did not significantly affect the expression of comC or sthA (Fig. 5A and B). Thus, CiaRH influences comC and sthA levels in the ΔsdbA mutant.
Inactivation of CiaRH restores bacteriocin activity to the ΔsdbA mutant.
Finally, we used an activity assay to determine whether mutation of ciaRH restored bacteriocin production to the ΔsdbA mutant. Consistent with the qPCR data, the ΔsdbA ΔciaRH mutant produced biologically active bacteriocins, sufficient to inhibit the growth of S. mitis (Fig. 5C). Mutation of ciaRH in the parent strain had no effect on activity, whereas complementation with a single copy of ciaRH on the chromosome of the ΔsdbA ΔciaRH mutant eliminated bacteriocin activity. This confirmed that the CiaRH signaling system mediated the lack of bacteriocin activity in the ΔsdbA mutant.
DISCUSSION
In this study, we identified the CiaRH two-component system as an important regulator of S. gordonii bacteriocin production, and we found that CiaRH was upregulated in mutants lacking the disulfide bond-forming enzyme SdbA. Activity assays and immunoaffinity chromatography confirmed that mutation of sdbA abolished bacteriocin production. However, production could be restored by exogenous CSP, suggesting that the individual components required for bacteriocin processing and secretion remained functional and that these proteins might not require disulfide bonds formed by SdbA. Instead, mutation of sdbA generated a signal that caused increased CiaRH activity, which in turn inhibited bacteriocin production (Fig. 6).
The CiaRH system has been found to affect genetic competence, stress resistance, colonization, and bacteriocin production in both S. pneumoniae and S. mutans (17, 22, 47, 50, 54); however, the activity of the CiaRH system in S. gordonii has not been investigated as thoroughly. In one of the few studies looking at the S. gordonii CiaRH system, Liu and Burne (54) determined that the CiaRH system is important for survival at low pH and efficient induction of the arginine deiminase system, which increases acid tolerance. Our data add to the roles of the CiaRH system in S. gordonii, showing that it is involved in regulation of the protease DegP and bacteriocin production.
The CiaRH system regulates htrA (degP) expression in S. pneumoniae and S. mutans (35, 50, 55, 56), and our results suggest that degP is part of the ciaRH regulon in S. gordonii as well. The CiaRH system was required for upregulation of degP in the ΔsdbA mutant, and mutation of ciaRH decreased degP expression, suggesting that the system might also affect basal levels of DegP (Fig. 4B). Sequence analysis of the degP promoter region revealed a possible CiaR binding site matching that described by Halfmann et al., consisting of a consensus sequence of TTTAAG-5 bp-(T/A)TTAAG located approximately 10 bp upstream from the −10 position (57).
While regulation of DegP by the CiaRH system appears to be consistent across various streptococci, the role for CiaRH in bacteriocin production is surprisingly species specific. For example, CiaH positively regulates bacteriocin production in S. mutans, and ΔciaH mutants lack mutacin activity (22). In contrast, similar to our findings in S. gordonii, the CiaRH system in S. pneumoniae represses bacteriocin activity (17, 18). In S. pneumoniae, the CiaRH system inhibits bacteriocin production through HtrA (DegP), which has been shown to disrupt the BlpAB transporter required for bacteriocin processing and secretion. Thus, inactivation of HtrA can alleviate CiaRH-mediated repression and increase the amounts of secreted bacteriocins (18).
Although degP was upregulated in the S. gordonii ΔsdbA mutant, the protease was not required for inhibition of bacteriocin production, and mutation of degP failed to restore bacteriocin activity to the ΔsdbA mutant (Fig. 5C). Additional investigation will be required to determine whether DegP affects bacteriocin activity or CSP levels in S. gordonii. Despite multiple attempts to detect and to quantify CSP in the culture medium using anti-CSP antibodies, we were unable to detect the peptide due to a strong cross-reaction with the BHIS medium required for activation of the Com pathway.
A key difference in bacteriocin production in S. pneumoniae versus S. gordonii is in the systems that regulate production; unlike in S. pneumoniae, the Com pathway controls the only bacteriocins produced by S. gordonii (10). The CiaRH system has long been known to influence the Com pathway in S. pneumoniae (58). Mutations that upregulate CiaR activity in S. pneumoniae result in loss of competence, while ΔciaR mutants grow poorly and are susceptible to lysis upon exposure to CSP (55). Unlike in S. pneumoniae, we did not observe any lysis or growth defects in the S. gordonii ΔciaRH or ΔsdbA ΔciaRH mutant when CSP was added to the culture. Our finding that mutation of ciaRH resulted in derepression of comC in the ΔsdbA mutant but not overexpression, compared to the parent strain, is consistent with previously reported results for S. pneumoniae, in which comC expression in a ΔciaR mutant was similar to that in the parent strain (55). This might be because CiaR is not inhibiting competence by binding to comC to downregulate transcription but is working via a different mechanism that involves posttranscriptional regulation.
Several studies have analyzed the S. pneumoniae CiaRH regulon through microarray analysis (50, 55, 59), and CiaR controls the expression of 25 genes, including five small noncoding RNAs called csRNAs (51, 57). Analysis of the csRNAs provided the first direct link between CiaRH signaling and genetic competence; comC was identified as a target of multiple csRNAs (46). The csRNAs are thought to work by binding to the Shine-Dalgarno sequence of complementary transcripts to prevent translation (46, 60, 61). S. gordonii is predicted to encode several csRNAs (60), and this could be a potential mechanism for CiaRH to influence bacteriocin production. However, additional analysis that is beyond the scope of this study will be required to determine the biological roles of S. gordonii csRNAs.
The signals that activate CiaRH are unknown (51), and it is not clear how mutation of sdbA induces CiaRH in S. gordonii. Because SdbA is required for disulfide bond formation (26), CiaRH might respond to general stress created by misfolding of SdbA substrates or by the loss of function of a specific SdbA substrate. Bacteria sense envelope stress by using two-component systems, and thiol-disulfide oxidoreductases have been linked to these stress responses in both Gram-negative and Gram-positive species (61–64). For example, Bacillus subtilis senses envelope stress by using a system called CssRS, which, like CiaRH in streptococci, regulates expression of the DegP-family proteases HtrA and HtrB (65, 66). Notably, expression of a misfolded disulfide-bonded protein, alkaline phosphatase, resulted in strong induction of the CssRS system and a 4-fold increase in htrB expression (66). A similar scenario might occur in S. gordonii, where mutation of sdbA causes protein misfolding that either directly or indirectly triggers increased CiaRH activity. We showed recently that the major autolysin AtlS was misfolded in the S. gordonii ΔsdbA mutant (26).
Interestingly, protein misfolding has been shown to enhance, rather than repress, ComDE activity in S. pneumoniae (67). This increase is mediated by HtrA (DegP), which degrades CSP. Under conditions with high levels of ribosomal coding errors, the increased amounts of misfolded protein are thought to inhibit HtrA competitively and to prevent degradation of CSP (67, 68). Thus, protein quality control, CiaRH, and ComDE appear to be related in various streptococci, although there are clear species-specific differences in how these systems affect one another.
In conclusion, we have demonstrated that mutation of the gene for the disulfide oxidoreductase SdbA activates the CiaRH two-component system, which in turn shut downs bacteriocin expression. Our data reveal a link in S. gordonii between CiaRH and the Com system, both of which play important roles in bacteriocin production, genetic competence, stress resistance, and biofilm formation.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Natural Sciences and Engineering Research Council of Canada. L.D. was the recipient of an NSERC postgraduate scholarship, an IWK graduate studentship, and a Nova Scotia Health Research Foundation Scotia Scholars Award.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00800-15.
REFERENCES
- 1.Nobbs AH, Jenkinson HF. 2015. Interkingdom networking within the oral microbiome. Microbes Infect 17:484–492. doi: 10.1016/j.micinf.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 3.Dû LD, Kolenbrander PE. 2000. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect Immun 68:4834–4837. doi: 10.1128/IAI.68.8.4834-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. 2001. The predominant microflora of nursing caries lesions. Caries Res 35:397–406. doi: 10.1159/000047482. [DOI] [PubMed] [Google Scholar]
- 5.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuramitsu HK, Wang BY. 2006. Virulence properties of cariogenic bacteria. BMC Oral Health 6(Suppl 1):S11. doi: 10.1186/1472-6831-6-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B, Deutch A, Hong J, Kuramitsu H. 2011. Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J Dent Res 90:501–505. doi: 10.1177/0022034510388808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. 2008. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol 66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heng NCK, Tagg JR, Tompkins GR. 2007. Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (Challis). J Bacteriol 189:1468–1472. doi: 10.1128/JB.01174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang BY, Kuramitsu HK. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71:354–362. doi: 10.1128/AEM.71.1.354-362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tompkins GR, Peavey MA, Birchmeier KR, Tagg JR. 1997. Bacteriocin production and sensitivity among coaggregating and noncoaggregating oral streptococci. Oral Microbiol Immunol 12:98–105. doi: 10.1111/j.1399-302X.1997.tb00624.x. [DOI] [PubMed] [Google Scholar]
- 13.Vickerman MM, Iobst S, Jesionowski AM, Gill SR. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J Bacteriol 189:7799–7807. doi: 10.1128/JB.01023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 15.Heng NCK, Tagg JR, Tompkins GR. 2006. Identification and characterization of the loci encoding the competence-associated alternative σ factor of Streptococcus gordonii. FEMS Microbiol Lett 259:27–34. doi: 10.1111/j.1574-6968.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 16.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 17.Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J Bacteriol 191:1509–1518. doi: 10.1128/JB.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kochan TJ, Dawid S. 2013. The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol 195:1561–1572. doi: 10.1128/JB.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol 187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreth J, Merritt J, Zhu L, Shi W, Qi F. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett 265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 22.Qi F, Merritt J, Lux R, Shi W. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect Immun 72:4895–4899. doi: 10.1128/IAI.72.8.4895-4899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senadheera DB, Cordova M, Ayala EA, Chávez de Paz LE, Singh K, Downey JS, Svensäter G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okinaga T, Niu G, Xie Z, Qi F, Merritt J. 2010. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J Bacteriol 192:1844–1852. doi: 10.1128/JB.01667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Okinaga T, Niu G, Qi F, Merritt J. 2010. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol Microbiol 78:1431–1447. doi: 10.1111/j.1365-2958.2010.07417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davey L, Ng CKW, Halperin SA, Lee SF. 2013. Functional analysis of paralogous thiol-disulfide oxidoreductases in Streptococcus gordonii. J Biol Chem 288:16416–16429. doi: 10.1074/jbc.M113.464578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heng NCK, Burtenshaw GA, Jack RW, Tagg JR. 2007. Ubericin A, a class IIa bacteriocin produced by Streptococcus uberis. Appl Environ Microbiol 73:7763–7766. doi: 10.1128/AEM.01818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontaine L, Hols P. 2008. The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpGSt, a thiol-disulfide oxidase. Appl Environ Microbiol 74:1102–1110. doi: 10.1128/AEM.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, Zhong J, Ni J, Chen M, Xiao H, Huan L. 2009. Characteristics of the bovicin HJ50 gene cluster in Streptococcus bovis HJ50. Microbiology 155:584–593. doi: 10.1099/mic.0.022707-0. [DOI] [PubMed] [Google Scholar]
- 30.Knight JB, Halperin SA, West KA, Lee SF. 2008. Expression of a functional single-chain variable-fragment antibody against complement receptor 1 in Streptococcus gordonii. Clin Vaccine Immunol 15:925–931. doi: 10.1128/CVI.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunny GM, Lee LN, LeBlanc DJ. 1991. Improved electroporation and cloning vector system for Gram-positive bacteria. Appl Environ Microbiol 57:1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vats N, Lee SF. 2001. Characterization of a copper transport operon, copYAZ, from Streptococcus mutans. Microbiology 147:653–662. doi: 10.1099/00221287-147-3-653. [DOI] [PubMed] [Google Scholar]
- 33.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebert ME, Patel KP, Plotnick M, Weiser JN. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J Bacteriol 187:3969–3979. doi: 10.1128/JB.187.12.3969-3979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis E, Kennedy D, Halperin SA, Lee SF. 2011. Role of the cell wall microenvironment in expression of a heterologous SpaP-S1 fusion protein by Streptococcus gordonii. Appl Environ Microbiol 77:1660–1666. doi: 10.1128/AEM.02178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davey L, Halperin SA, Lee SF. 2015. Immunoblotting conditions for small peptides from streptococci. J Microbiol Methods 114:40–42. doi: 10.1016/j.mimet.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 39.Tremblay YDN, Lo H, Li YH, Halperin SA, Lee SF. 2009. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology 155:2856–2865. doi: 10.1099/mic.0.028456-0. [DOI] [PubMed] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Kadokura H, Beckwith J. 2010. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal 13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlegel R, Slade HD. 1972. Bacteriocin production by transformable group H streptococci. J Bacteriol 112:824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishii S, Yano T, Hayashi H. 2006. Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J Biol Chem 281:4726–4731. doi: 10.1074/jbc.M512516200. [DOI] [PubMed] [Google Scholar]
- 45.Claverys JP, Martin B, Håvarstein LS. 2007. Competence-induced fratricide in streptococci. Mol Microbiol 64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 46.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. 2013. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 89:334–349. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 47.Mascher T, Heintz M, Zähner D, Merai M, Hakenbeck R. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in β-lactam resistance. J Bacteriol 188:1959–1968. doi: 10.1128/JB.188.5.1959-1968.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas W, Kaushal D, Sublett J, Obert C, Tuomanen EI. 2005. Vancomycin stress response in a sensitive and a tolerant strain of Streptococcus pneumoniae. J Bacteriol 187:8205–8210. doi: 10.1128/JB.187.23.8205-8210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers PD, Liu TT, Barker KS, Hilliard GM, English BK, Thornton J, Swiatlo E, McDaniel LS. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J Antimicrob Chemother 59:616–626. doi: 10.1093/jac/dkl560. [DOI] [PubMed] [Google Scholar]
- 50.Sebert ME, Palmer LM, Rosenberg M, Weiser JN. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect Immun 70:4059–4067. doi: 10.1128/IAI.70.8.4059-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halfmann A, Schnorpfeil A, Müller M, Marx P, Günzler U, Hakenbeck R, Brückner R. 2011. Activity of the two-component regulatory system CiaRH in Streptococcus pneumoniae R6. J Mol Microbiol Biotechnol 20:96–104. doi: 10.1159/000324893. [DOI] [PubMed] [Google Scholar]
- 52.Marx P, Nuhn M, Kovács M, Hakenbeck R, Brückner R. 2010. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 11:661. doi: 10.1186/1471-2164-11-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Busch A, Richter AS, Backofen R. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Burne RA. 2009. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J Bacteriol 191:7353–7362. doi: 10.1128/JB.01053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dagkessamanskaia A, Moscoso M, Overweg K, Reuter M, Martin B, Wells J, Claverys J. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol 51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 56.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 66:110–126. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 58.Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol 12:505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 59.Mascher T, Zähner D, Merai M, Balmelle N, De Saizieu AB, Hakenbeck R. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J Bacteriol 185:60–70. doi: 10.1128/JB.185.1.60-70.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brantl S, Brückner R. 2014. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol 11:443–456. doi: 10.4161/rna.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev 32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- 62.Wilton J, Acebo P, Herranz C, Gómez A, Amblar M. 2015. Small regulatory RNAs in Streptococcus pneumoniae: discovery and biological functions. Front Genet 6:126. doi: 10.3389/fgene.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarvas M, Harwood CR, Bron S, van Dijl JM. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta 1694:311–327. [DOI] [PubMed] [Google Scholar]
- 64.Raivio TL. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 65.Westers H, Westers L, Darmon E, Van Dijl JM, Quax WJ, Zanen G. 2006. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J 273:3816–3827. doi: 10.1111/j.1742-4658.2006.05389.x. [DOI] [PubMed] [Google Scholar]
- 66.Darmon E, Dorenbos R, Meens J, Freudl R, Antelmann H, Hecker M, Kuipers OP, Bron S, Quax WJ, Dubois JYF, Van Dijl JM. 2006. A disulfide bond-containing alkaline phosphatase triggers a BdbC-dependent secretion stress response in Bacillus subtilis. Appl Environ Microbiol 72:6876–6885. doi: 10.1128/AEM.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2:e00071-11. doi: 10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME. 2012. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem 287:38449–38459. doi: 10.1074/jbc.M112.391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.