ABSTRACT
Sarcosine (N-methylglycine) is present in many environments inhabited by pseudomonads and is likely most often encountered as an intermediate in the metabolism of choline, carnitine, creatine, and glyphosate. While the enzymology of sarcosine metabolism has been relatively well studied in bacteria, the regulatory mechanisms governing catabolism have remained largely unknown. We previously determined that the sarcosine-catabolic (sox) operon of Pseudomonas aeruginosa is induced by the AraC family regulator GbdR in response to glycine betaine and dimethylglycine. However, induction of these genes was still observed in response to sarcosine in a gbdR deletion mutant, indicating that an independent sarcosine-responsive transcription factor also acted at this locus. Our goal in this study was to identify and characterize this regulator. Using a transposon-based genetic screen, we identified PA4184, or SouR (sarcosine oxidation and utilization regulator), as the sarcosine-responsive regulator of the sox operon, with tight induction specificity for sarcosine. The souR gene is required for appreciable growth on sarcosine as a carbon and nitrogen source. We also characterized the transcriptome response to sarcosine governed by SouR using microarray analyses and performed electrophoretic mobility shift assays to identify promoters directly regulated by the transcription factor. Finally, we characterized PA3630, or GfnR (glutathione-dependent formaldehyde neutralization regulator), as the regulator of the glutathione-dependent formaldehyde detoxification system in P. aeruginosa that is expressed in response to formaldehyde released during the catabolism of sarcosine. This study expands our understanding of sarcosine metabolic regulation in bacteria through the identification and characterization of the first known sarcosine-responsive transcriptional regulator.
IMPORTANCE The Pseudomonas aeruginosa genome encodes many diverse metabolic pathways, yet the specific transcription regulators controlling their expression remain mostly unknown. Here, we used a genetic screen to identify the sarcosine-specific regulator of the sarcosine oxidase operon, which we have named SouR. SouR is the first bacterial regulator shown to respond to sarcosine, and it is required for growth on sarcosine. Sarcosine is found in its free form and is also an intermediate in the catabolic pathways of glycine betaine, carnitine, creatine, and glyphosate. The similarity of SouR to the regulators of carnitine and glycine betaine catabolism suggests evolutionary diversification within this regulatory family to allow response to structurally similar but physiologically distinct ligands.
INTRODUCTION
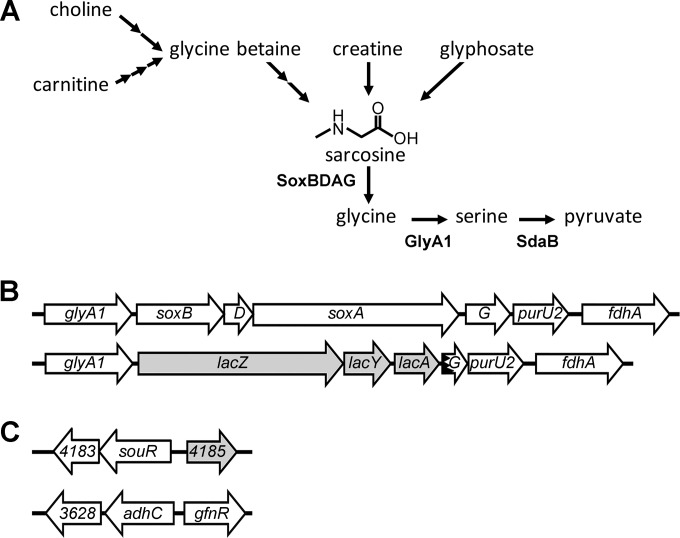
Pseudomonas aeruginosa and other bacteria from similar environments are capable of utilizing sarcosine (N-methylglycine) as a carbon and nitrogen source for growth (1–3). Sarcosine is present in many environments inhabited by pseudomonads, and it is also produced as an intermediate in the metabolism of choline, carnitine, creatine, and glyphosate (Fig. 1A). Choline is abundant in many eukaryote-associated environments, including clinically important sites of opportunistic infection by P. aeruginosa, such as the lung (4), where phosphatidylcholine constitutes an estimated 85% of the dry weight of human pulmonary surfactant (5). Within this environment, P. aeruginosa acquires choline from phosphatidylcholine via the virulence factors phospholipase C (PlcH) and phosphorylcholine phosphatase (PchP) (6, 7). Burns and deep lacerations also expose P. aeruginosa to readily available sarcosine precursors, including carnitine in muscle tissue and choline released from damaged cell membranes (7, 8). Furthermore, Pseudomonas putida and some isolates of P. aeruginosa can metabolize creatine to generate sarcosine (9–11), while other pseudomonads obtain sarcosine through metabolism of the herbicide glyphosate (12–14).
FIG 1.
Sarcosine catabolism in Pseudomonas species. (A) Diagram of sarcosine catabolism in P. aeruginosa and related species. Environmental sources of sarcosine that can be metabolized by Pseudomonas species are shown, along with the structure of sarcosine and the name of each enzyme involved in the conversion of sarcosine into glycine, serine, and pyruvate. (B) Genomic depiction of the sarcosine-catabolic operon in wild-type P. aeruginosa and the altered locus that functions as the transcriptional reporter strain in this study. (C) Genomic arrangement of the souR locus and glutathione-dependent formaldehyde detoxification system genes in P. aeruginosa. For panels B and C, shading denotes genes not involved in sarcosine metabolism. The jagged line in the arrow representing soxG denotes gene disruption during the lacZYA insertion.
Aerobic bacterial sarcosine catabolism proceeds via oxidative demethylation catalyzed by one of two classes of sarcosine oxidase. Monomeric sarcosine oxidases are the simplest form of these enzymes and produce glycine, hydrogen peroxide, and formaldehyde from sarcosine (15). In contrast, heterotetrameric sarcosine oxidases (TsoX) are more complex and assimilate the N-methyl group of sarcosine into the C1 carbon pool through a 5,10-methylenetetrahydrofolate intermediate instead of releasing it as formaldehyde (15, 16). In P. aeruginosa and a variety of soil bacteria, TsoX is encoded in an operon by soxBDAG (Fig. 1B), along with a serine hydroxymethyltransferase, encoded by glyA1, and the 10-formyltetrahydrofolate hydrolase encoded by purU2 (17–19), which together function to transform sarcosine into metabolites used for energy production and biosynthesis. In the absence of sufficient tetrahydrofolate, TsoX demethylation of sarcosine releases formaldehyde (15, 16), and P. aeruginosa and other proteobacteria encode a sarcosine-inducible glutathione-independent formaldehyde dehydrogenase (fdhA) adjacent to the soxBDAG locus that converts formaldehyde to formate and generates reducing potential through NADH synthesis (20).
Although the enzymology of sarcosine catabolism has been relatively well studied in bacteria, the regulatory mechanisms governing this process are largely unknown. We previously determined that expression of the sox operon of P. aeruginosa is induced in response to glycine betaine and dimethylglycine through the AraC family regulator GbdR (21, 22). Consistent with previous reports (1, 2), however, we also observed induction of the operon in response to sarcosine in a gbdR deletion mutant, indicating that an independent sarcosine-responsive transcription factor also acts at the locus (21).
Here, we report the identification and characterization of the first known sarcosine-responsive transcription factor, PA4184, which we have named SouR (sarcosine oxidation and utilization regulator). SouR regulates the soxBDAG operon in P. aeruginosa, and we have determined that it is necessary for appreciable growth when sarcosine is utilized as a sole carbon and nitrogen source. We further determined that transcriptional activation by SouR is specific for sarcosine and characterized the transcriptome response to sarcosine governed by the regulator. During this research, we also characterized PA3630, which we have named GfnR (glutathione-dependent formaldehyde neutralization regulator), as the regulator of the glutathione-dependent formaldehyde detoxification system in P. aeruginosa that is expressed during the catabolism of sarcosine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type (WT) P. aeruginosa PA14 (50), transposon mutants, and deletion strains (see Table S1 in the supplemental material) were maintained on Lennox broth (LB) or Pseudomonas isolation agar (PIA) supplemented with 50 μg/ml gentamicin when appropriate. Escherichia coli strains used in this study (see Table S1 in the supplemental material) were maintained on LB supplemented with gentamicin (7 μg/ml liquid and 10 μg/ml agar) or carbenicillin (100 μg/ml) when necessary. During genetic manipulations, selection for P. aeruginosa over E. coli was performed using PIA supplemented with 50 μg/ml gentamicin. The growth and selection conditions used in the genetic screen are described in detail below. Growth and transcriptional induction assays in P. aeruginosa were performed using MOPS (morpholinepropanesulfonic acid) minimal medium (23) as modified by our group (8, 24, 25).
Construction of deletion strains, complementation constructs, and the sarcosine oxidase operon reporter.
All amplifications and cloning steps were performed using Q5 DNA polymerase and restriction enzymes purchased from New England BioLabs (Ipswich, MA). General nucleic acid procedures were performed using Qiagen kits unless otherwise noted. The gene numbers generally referred to in this study are based on the PAO1 orthologs. The sequences of the primers used to generate each construct are listed in Table S2 in the supplemental material.
In-frame chromosomal deletions of souR (PA4184) and gfnR (PA3630) were created using splice overlap extension (SOE) as previously described, using pMQ30-based allelic replacement (26). Briefly, two ∼1-kb regions directly upstream and downstream of the gene to be deleted were amplified from PA14 genomic DNA with primers PA14_9770KO_F1, PA14_9770KO_R1, PA14_9770KO_F2, and PA14_9770KO_R2 and primers PA3630KO_F1, PA3630KO_ R1, PA3630KO_F2, and PA3630KO_R2; ligated into pCR-Blunt (Invitrogen); and transformed into E. coli DH5α cells. After selection on kanamycin and plasmid preparation, the overlap extension products were excised with XbaI and HindIII, gel purified, and ligated into similarly cut pMQ30 before being transformed into DH5α cells. Transformants were selected on LB with 10 μg/ml gentamicin, and plasmid DNA was purified from resistant colonies to generate the pGW008 (ΔsouR) and pGW023 (ΔPA3630) deletion constructs. pGW008 and pGW023 were electroporated into the conjugative E. coli S17 λpir strain. Donor S17 λpir strains were mixed with recipient PA14 strains, and single-crossover mutants were selected for growth on PIA supplemented with 50 μg/ml gentamicin. Recombinants were verified by PCR after selecting for loss of sacB by growth on 5% sucrose LB plates lacking sodium chloride (26, 27) to yield strains GGW034 (PA14 ΔsouR), GGW036 (PA14 ΔgbdR ΔsouR), GGW076 (PA14 ΔgfnR), and GGW078 (PA14 ΔgbdR ΔgfnR).
The souR complementation construct included the souR open reading frame (ORF) and native promoter cloned into pMQ80 using primers with engineered KpnI and HindIII restriction sites (PA14_9770_RescueF and PA14_9770_RescueR). This construct was designated pGW007 (pSouR). Complementation of the ΔgnfR strain was achieved by chromosomal integration of the gfnR ORF and its native promoter at the attTn7 site, as described by Choi and Schweizer (28). Briefly, the PA3630 gene and promoter region were amplified from PA14 genomic DNA using the primers PA3630_RescueF and PA3630_RescueR, which incorporated flanking HindIII and KpnI restriction sites. The amplified product was digested, ligated into similarly cut pUC18-mini-Tn7T-Gm, and transformed into DH5α, and transformants were selected for gentamicin resistance. This construct was designated pGW024. pGW024 and pTNS2 were coelectroporated into the target strains as previously described (28, 29).
Chromosomal soxB′-lacZYA-′soxG operonic reporter strains were engineered through allelic replacement using a pMQ30-based strategy (26). Briefly, regions ∼1 kb upstream of the soxB translational start site and ∼1 kb downstream of the soxG stop codon were amplified from PA14 genomic DNA with SOE-based primers (soxKO_F1, soxKO_R1, soxKO_F2, and soxKO_R2) incorporating an engineered NcoI site into the overlap portion of the construct, ligated, and transformed, and the resultant plasmid was purified as described above. The plasmid was linearized between the soxB and soxG fragments with NcoI and treated with Klenow to generate blunt ends, which allowed ligation of lacZYA (obtained from pMW5 following KpnI and EcoRI digestion and Klenow treatment). Following transformation into DH5α cells, the plasmid DNA was purified and digested with KpnI and HindIII to excise soxB′-lacZYA-′soxG for ligation into similarly cut pMQ30, yielding pGW005. pGW005 was transformed into E. coli S17 λpir (GGW040) and mixed with PA14 recipient strains to create the chromosomal soxB′-lacZYA-′soxG strains, which are effectively ΔsoxBDAG and cannot grow on sarcosine, as lacZYA has replaced most of the operon.
Genetic screen to identify the sarcosine-responsive regulator of soxBDAG expression.
Transposon mutagenesis was performed on PA14 ΔgbdR soxB′-lacZYA-′soxG (GGW039) via conjugation with E. coli SM10 harboring pBT20, a Mariner-based transposon (TnM) (51), using methods modified from Wong and Mekalanos and Kulasekara et al. (30, 31). Briefly, the transposon donor was grown overnight on LB agar supplemented with 100 μg/ml of carbenicillin while GGW039, the recipient, was cultured on PIA. After 24 h, cells of each species were scraped from the plates and resuspended in LB to final concentrations of 40 (donor) and 20 (recipient) optical density at 600 nm (OD600) units. For mating, equal volumes of each strain were mixed, and 50-μl aliquots were spotted onto LB agar and incubated for 2 h at room temperature. To simultaneously select for P. aeruginosa transposon integrants and conduct the screen, cells from the conjugation mixture were resuspended in 2 ml of MOPS, and 400 μl was plated on PIA with 50 μg/ml of gentamicin, 100 μg/ml of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in the presence or absence of 2 mM sarcosine. Colonies exhibiting low or no β-galactosidase activity were tested by Miller assay in liquid media (as described below) before identifying the transposon insertion sites using two rounds of PCR with a TnM-specific forward primer (Rnd1-TnM20) and an arbitrary primer (Rnd1-PA-Arb-2), followed by a second round of amplification using the Rnd2-TnM20 and Rnd2-Arb-primer primer set, as previously described (31, 32). Sequencing was performed using the TnM-specific primer BT20TnMSeq (31), and reads were mapped to their respective loci within PA14 and PAO1 genomes using the BLAST function on the Pseudomonas genome database (33).
Testing activation specificity of SouR.
The small-molecule specificity required for SouR-dependent activation was examined using β-galactosidase assays, as described previously (7, 22). GGW039 (PA14 ΔgbdR soxB′-lacZYA-′soxG) was grown overnight at 37°C on a rotating wheel in MOPS supplemented with 25 mM sodium pyruvate and 5 mM d-glucose. Cells were collected by centrifugation, washed in MOPS, and resuspended in either MOPS–20 mM pyruvate or MOPS–20 mM pyruvate plus 1 mM either glycine betaine, dimethylglycine, sarcosine, ethylglycine, or glycine. Inductions were then carried out at 37°C on a shaker set to 170 rpm for 3 h before β-galactosidase activity was measured according to the method of Miller (34).
Growth assays.
Growth assays were performed as previously described (8). Briefly, strains were grown overnight at 37°C on a roller drum in MOPS medium supplemented with 25 mM sodium pyruvate and 5 mM d-glucose. Cells were collected by centrifugation, washed with MOPS (with no carbon source), resuspended, and added to 48-well tissue culture plates to a final optical density of 0.05 OD600 units in MOPS supplemented with 40 mM sarcosine as the sole carbon and nitrogen source or 40 mM sodium pyruvate in MOPS with ammonium chloride as the nitrogen source. Growth was measured by OD600 using a Synergy 2 Biotek plate reader.
MBP-SouR fusion construct and protein purification.
A maltose binding protein-SouR fusion (MBP-SouR) was engineered into the pMALc2x vector, as previously described for AraC family transcription factors (22, 35). Briefly, souR was amplified from genomic DNA with primers (souR_MBP_F and souR_MBP_R) designed to exclude the start codon and incorporate flanking restriction sites to facilitate ligation in frame with the MBP ORF, generating pGW015. Following cloning in E. coli DH5α, purified pGW015 was transformed into chemically competent E. coli T7 lysY lysIq (New England BioLabs) to generate the MBP-SouR expression strain, GGW47.
GGW47 was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. Cells were collected by centrifugation, rinsed twice in MOPS, and resuspended in 3 ml of cold (150 mM) Tris HCl (pH 7.2) containing Halt 1× protease inhibitor cocktail (Thermo Scientific). The cells were lysed using a French press, DNase I treated, and sheared using a 21-gauge needle. Following centrifugation at 4°C and 13,000 rpm, the soluble fraction was applied to a 3-ml amylose resin column (New England BioLabs). The column was rinsed four times with 6 ml of column buffer (20 mM Tris HCl, 150 mM NaCl, 1 mM EDTA [pH 7.4]) before protein was eluted in amylose elution buffer (20 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 10 mM maltose [pH 7.4]). Elution fractions were evaluated by SDS-PAGE. Fractions containing MBP-SouR were pooled and dialyzed against 20 mM Tris-HCl, pH 7.5, at 4°C in a 10-kDa molecular mass cutoff Slide-A-Lyzer cassette (Pierce). Protein aliquots were stored frozen at −80°C until use.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (22) using DNA probes spanning the promoter of potential SouR regulon members. Probes were constructed by PCR amplification, where one of the primers was 5′ biotinylated (IDT), and were subsequently purified using Qiagen's PCR Clean Up kit. EMSAs were conducted using the Pierce Lightshift kit following the manufacturer's instructions, with changes made as previously described (22) and with salmon sperm DNA (Invitrogen) substituted for poly(dI-dC) at a final concentration of 500 ng/μl. Binding, electrophoresis, and detection were done as previously described (22). The sequences of the primers used in the construction of EMSA probes for adhC (PA3629-prom-5′-biotin and PA3629-prom-3′), glyA1 (glyA1-prom-5′-biotin and glyA1-prom-3′), PA2762 (PA2762-prom-5′-biotin and PA2762-prom-3′), sdaB (cbcX-prom-5′-biotin and cbcX-prom-3′), and the negative-control dhcA (PA1999-prom-3′ and PA1999-prom-5′-biot) are listed in Table S2 in the supplemental material.
Promoter mapping.
To identify the SouR and GbdR binding region within the sarcosine oxidase operon promoter, full-length and truncated PglyA1 fragments were engineered into the pMW5 lacZYA reporter vector. Briefly, the region upstream of glyA1 was amplified from PA14 genomic DNA using primers that incorporated flanking HindIII and KpnI restriction sites (PglyA1_F1, PglyA1_50bp_del_F2, PglyA1_100bp_del_F3, PglyA1_150bp_del_F4, and PglyA1_R). Amplicons were digested and ligated into similarly cut pMW5, creating the plasmids pGW011 through pGW014. Following transformation into DH5α and verification, these plasmids were transformed into the PA14 wild-type, ΔgbdR, ΔsouR, and ΔsouR ΔgbdR strains via electroporation. PglyA1 induction in response to 1 mM pyruvate, sarcosine, or glycine betaine was measured as described above, with the addition of 20 μg/ml of gentamicin, and β-galactosidase activity was quantified according to the method of Miller (34).
Growth conditions and RNA preparation for microarrays and qRT-PCR.
PA14 ΔgbdR and PA14 ΔgbdR ΔsouR (and PA14 ΔgbdR ΔgfnR for quantitative reverse transcription [qRT]-PCR) were grown overnight in 3 ml of MOPS minimal medium supplemented with 20 mM sodium pyruvate and 5 mM d-glucose at 37°C on a rotating wheel. Cells were collected by centrifugation, washed with prewarmed MOPS, and resuspended in MOPS with 20 mM sodium pyruvate at an OD600 of 0.6. Six hundred microliters of each strain was then added to 12-well tissue culture plates containing 600 μl of prewarmed MOPS with 20 mM sodium pyruvate and 2 mM sarcosine or MOPS with 20 mM sodium pyruvate (no-induction control) to achieve a final OD600 of 0.3. The inductions were carried out for 3 h at 37°C with shaking at 170 rpm. Following induction, cells were collected by centrifugation, resuspended in 400 μl of fresh MOPS, and mixed with 800 μl of RNA Protect Bacterial Reagent (Qiagen). The cells were again centrifuged, and the supernatant was decanted before the pellets were frozen at −80°C.
RNA was prepared using a Qiagen RNeasy kit, following the manufacturer's protocol with the following changes. Prior to extraction, cell pellets were resuspended in 200 μl of Tris-EDTA (TE) supplemented with 3 mg/ml lysozyme and incubated at room temperature for 20 min. An on-column DNase I treatment was performed before the RNA was eluted in RNase-free water. Samples were then treated a second time with RNase-free DNase I (NEB) and incubated for 1 h at 37°C before a second round of RNeasy column purification was performed.
Microarray methodology.
Microarray analysis was performed by the Vermont Genetics Network Microarray Facility using Affymetrix P. aeruginosa PAO1 gene chips and DNA probes generated by the NuGen Pico system. Each condition was analyzed in duplicate, and signals from all the probes for a given gene were averaged into one probe intensity using the Expression Console and Transcriptome Analysis Console software package version 2.0 (Affymetrix). Potential SouR regulon members were identified as those exhibiting at least a 2.5-fold change in detection between sarcosine-induced and control cultures using robust multiarray average (RMA) analysis and a P value of <0.05.
Quantitative RT-PCR.
Growth conditions and RNA preparations were as described above (in biological triplicate). cDNA was generated using Superscript IV with the 5′-NSNSNSNSNS-3′ primer previously described (36) and 20 ng of total RNA isolated from each strain under each condition. Quantitative PCR was performed with technical duplicates using Luminaris HiGreen fluorescein qPCR master mix (Thermo Fisher) and primers described previously (22). Due to difficulties in the amplification of sdaB with Taq-based Luminaris mix, quantitative PCR was performed with NEB's Q5 2× master mix supplemented with SYBR green I nucleic acid gel stain (Thermo Fisher) at a final concentration of 0.2×. For each gene, transcript abundance was determined using a five-step standard-curve dilution series with cDNA from the ΔgbdR strain exposed to sarcosine (the highest-responding strain and condition), as described previously (22). Each sample for each transcript was normalized to its cognate rplU abundance before conversion to relative expression based on the average expression level in the noninduced (pyruvate) control sample of each strain.
Formaldehyde susceptibility assay.
PA14 WT, ΔgfnR, ΔgfnR attTn7::gfnR, and ΔgfnR attTn7::EV (empty-vector) strains were grown overnight at 37°C in MOPS medium supplemented with 25 mM sodium pyruvate and 5 mM glucose. Cells were collected by centrifugation, washed with fresh MOPS medium, and resuspended in 48-well tissue culture plates to a final optical density of 0.05 OD600 units in MOPS containing 20 mM sodium pyruvate and 5 mM glucose or MOPS with 20 mM sodium pyruvate, 5 mM d-glucose, and 0.75 mM formaldehyde. Susceptibility to formaldehyde was assessed by growth in the presence of formaldehyde through OD600, using a Synergy 2 Biotek plate reader. The concentration of formaldehyde utilized in this assay was arrived at by titrating the ability of PA14 WT to grow in MOPS medium with 25 mM sodium pyruvate and 5 mM glucose supplemented with 0.25 mM, 0.5 mM, 0.75 mM, or 1.0 mM formaldehyde. The highest formaldehyde concentration that did not impede growth of PA14 WT after 24 h under these conditions was then chosen for assessing the susceptibilities of gfnR deletion and complementation strains.
Microarray data accession number.
The array data are available in the GEO database under accession number GSE72613.
RESULTS
Identification of the sarcosine-responsive regulator of the sarcosine-catabolic operon.
Our previous work demonstrated that while GbdR could control the sarcosine oxidase operon, soxBDAG could still be induced in a gbdR deletion strain in response to sarcosine (21), indicating that an unidentified sarcosine-responsive transcription factor regulated the sarcosine oxidase genes. The sarcosine oxidase operon consists of glyA1-soxBDAG-purU2 (PA14_71460 to PA14_714530 and PA5415 to PA5420), which we refer to as the sox operon, and is controlled from the PglyA1 promoter. To identify the sarcosine-responsive regulator of the sox operon, an operonic lacZYA transcriptional reporter was engineered into the sox locus of a ΔgbdR strain, generating both a reporter and a simultaneous deletion of most of the operon (ΔgbdR soxB′-lacZYA-′soxG). This parent strain was mutagenized with the Mariner transposon from pBT20, and approximately 60,000 transposon insertion mutants were screened for the ability to cleave X-Gal in response to sarcosine. In total, 23 colonies that failed to induce β-galactosidase in the presence of sarcosine were identified. Sixteen of these mutants carried unique insertions in the lacZYA locus, while seven unique insertions mapped within PA14_09770 (PA4184), predicted to encode an AraC family transcription regulator. The unique rate of insertion into lacZYA and PA4184 suggests that the screen was saturated for identification of activators.
An ortholog search of PA4184 against the Pseudomonas genome database (33) revealed the widespread conservation of the gene among sequenced pseudomonads. Unique to P. aeruginosa, however, PA4184 is part of an operon with a gene, PA4183 (PA14_09780) (Fig. 1B), encoding a protein of unknown function exhibiting modest structural homology with the glyoxylase I family of enzymes. A reciprocal BLASTP search of PA4183 against the genome database failed to identify homologs outside P. aeruginosa.
PA4184 is a sarcosine-responsive transcription regulator.
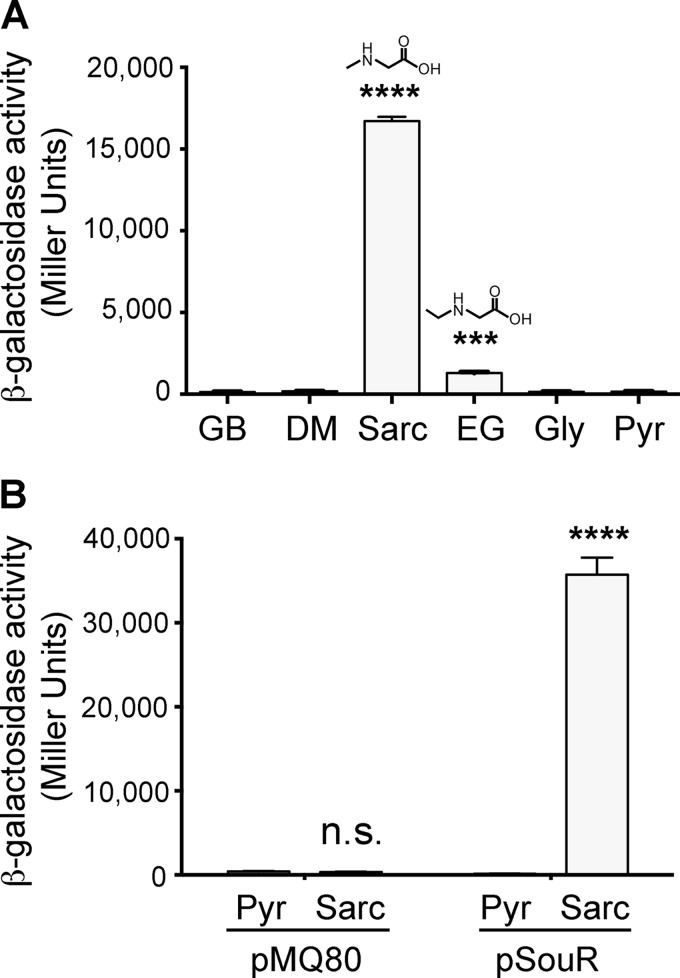
The induction specificity of PA4184 was examined through β-galactosidase assays performed using the same reporter strain described above (ΔgbdR soxB′-lacZYA-′soxG) with sarcosine or structurally related compounds. Glycine betaine, dimethylglycine, glycine, and pyruvate failed to induce transcription of the sox operon, while incubation with sarcosine, and to a lesser extent the synthetic compound ethylglycine, resulted in induction of β-galactosidase activity (Fig. 2A). This induction was dependent on PA4184, as the same assay conducted in the PA4184 deletion strains yielded no β-galactosidase activity (Fig. 2B and data not shown). Moreover, transcription from the soxBDAG operon in response to sarcosine was restored in a ΔgbdR ΔPA4184 soxB′-lacZYA-′soxG strain carrying PA4184 on a plasmid under the control of its native promoter (Fig. 2B). These results confirm that PA4184 is required for transcriptional induction of the sarcosine oxidase operon in response to sarcosine. Furthermore, the ability of ethylglycine to stimulate transcription from the promoter implies the necessity for the secondary amine moiety in the recognition of the inducing compound by PA4184. Based on these data and the growth data reported below, we renamed PA4184 as souR (sarcosine oxidation and utilization regulator), which encodes an AraC family transcription regulator.
FIG 2.
Activating ligand specificity and necessity for SouR in sarcosine-dependent induction of the sox operon. (A) Results from a β-galactosidase assay of a ΔgbdR soxB′-lacZYA-′soxG strain exposed in MOPS-pyruvate (Pyr) to 1 mM either glycine betaine (GB), dimethylglycine (DM), sarcosine (Sarc), ethylglycine (EG), or glycine (Gly) or no compound (Pyr) as a control. For convenience, the structures for sarcosine and ethylglycine are shown over their respective bars. (B) Results of a β-galactosidase assay of a ΔgbdR ΔsouR soxB′-lacZYA-′soxG strain exposed in MOPS-pyruvate (Pyr) with or without 1 mM sarcosine with the addition of the empty vector (pMQ80) or the plasmid carrying souR and its native promoter (pSouR). Statistical significance was determined using one-way analysis of variance (ANOVA) with Dunnett's posttest, with the uninduced (Pyr) condition as the comparator for all other data. n.s., not significant; ***, P < 0.001; ****, P < 0.0001. The data shown are representative of the results of three independent experiments, and the error bars represent standard deviations.
souR is essential for growth on sarcosine as a sole carbon and nitrogen source.
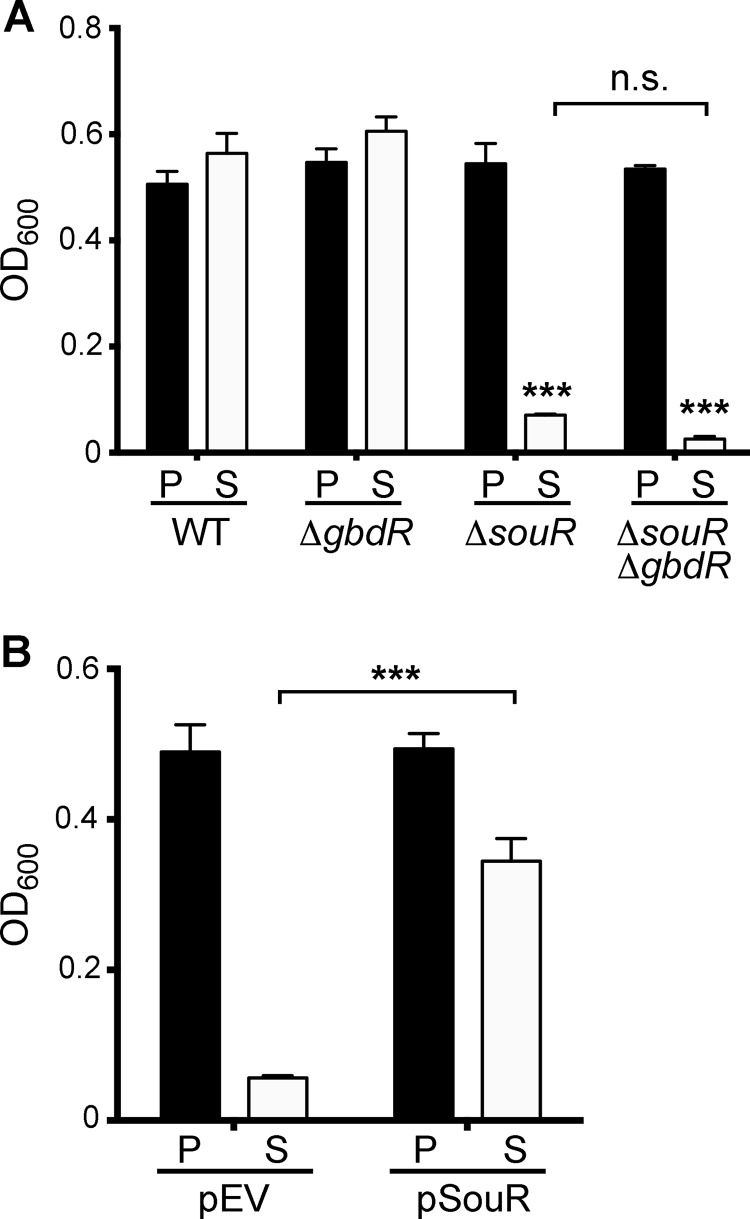
P. aeruginosa can use sarcosine as a sole carbon and nitrogen source for growth (1). To assess the requirement for souR in the metabolism of sarcosine by P. aeruginosa, growth assays were performed with WT, ΔgbdR, ΔsouR, and ΔgbdR ΔsouR strains cultured in MOPS minimal medium with sarcosine as the sole carbon and nitrogen source. Deletion of souR resulted in substantial growth defects compared to the WT, and there was no detectable growth in the ΔgbdR ΔsouR double-deletion mutant (Fig. 3A). The necessity for souR for this activity was confirmed by transcomplementation with a plasmid carrying souR with its native promoter, which restored growth (Fig. 3B). All the deletion strains grew similarly to the WT when cultured in MOPS medium supplemented with pyruvate and ammonium chloride as carbon and nitrogen sources, respectively, indicating that the observed growth defects are sarcosine specific (Fig. 3).
FIG 3.
Role of souR during growth on sarcosine. (A) Culture density (OD600) after 24 h of growth in wild-type, ΔgbdR, ΔsouR, and ΔgbdR ΔsouR cells in MOPS minimal medium without nitrogen supplemented with either 20 mM pyruvate and 10 mM ammonium chloride (P) or 40 mM sarcosine (S). (B) Culture density (OD600) after 24 h of growth in ΔgbdR ΔsouR transformed with the empty vector (pMQ80) (pEV) or souR with its native promoter (pSouR). Statistical significance was determined using two-way ANOVA with Tukey's posttest (A) and with Sidak's posttest (B). n.s., not significant; ***, P < 0.001. The data shown are summaries of the results of three independent experiments, each with three biological replicates; the error bars represent standard errors of the means.
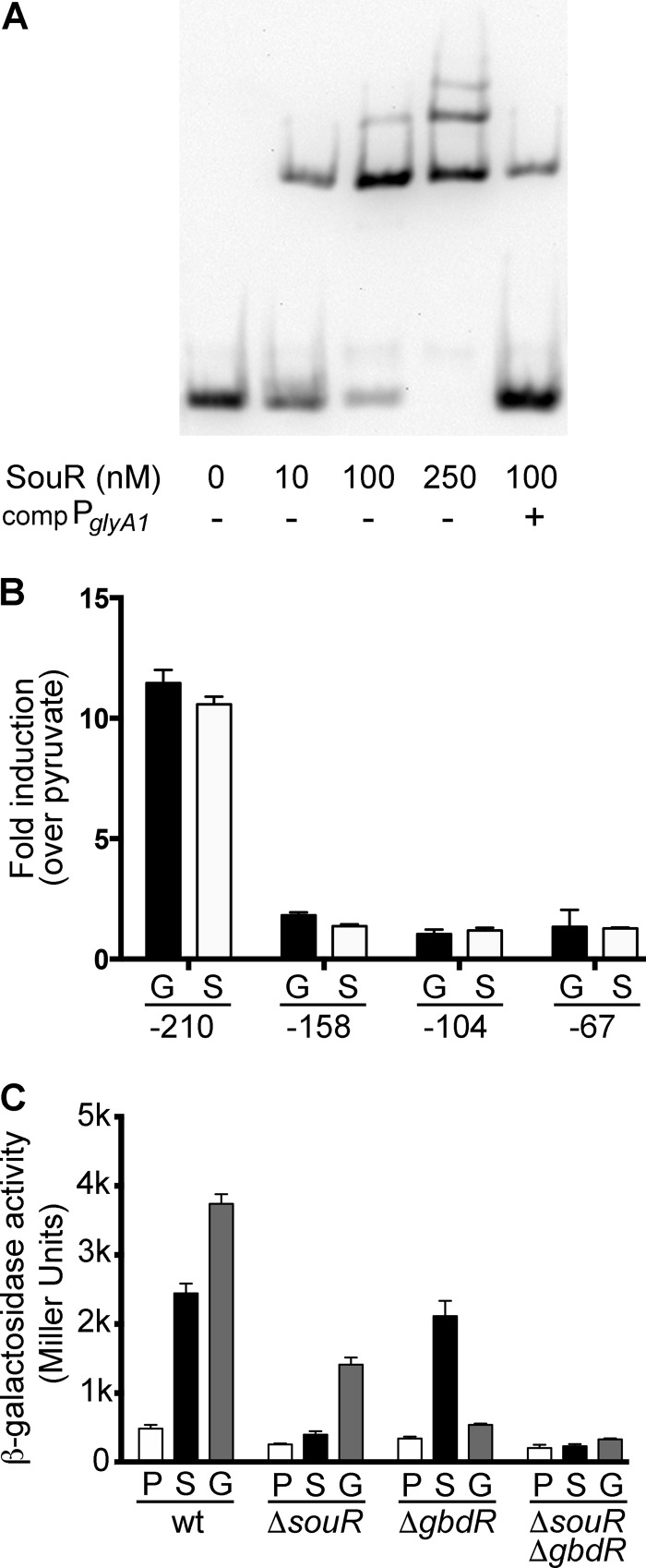
SouR and GbdR bind within the same region of PglyA1.
We previously determined that GbdR recognizes a binding site within the promoter of glyA1 using an EMSA with a maltose binding protein-GbdR fusion (22). Here, we show that a maltose binding protein-SouR fusion also binds the promoter of glyA1 and that this binding was sensitive to competition with unlabeled PglyA1 DNA (Fig. 4A). As previously reported for MBP-GbdR (17), the MBP-SouR DNA interaction was not affected by the presence of sarcosine (data not shown). Promoter mapping was used to determine where the SouR and GbdR binding sites were within PglyA1. Serial truncations of PglyA1 were engineered into the pMW5 promoterless lacZ reporter plasmid and transformed into ΔgbdR and ΔsouR cells. In both scenarios, deletion of the region between bp −210 and −158 upstream from the glyA1 translational start site resulted in loss of induction of β-galactosidase activity in response to sarcosine (in the ΔgbdR strain) and glycine betaine (in the ΔsouR strain), indicating that SouR and GbdR require the same region of the promoter (Fig. 4B). SouR and GbdR appear to function independently at the promoter, and either one can support induction in response to their cognate inducing molecules (Fig. 4C). The full promoter deletion series in each of the four strains shown in Fig. 4C are presented in Fig. S1 in the supplemental material. These data demonstrate that the minimal requirements for induction by GbdR or SouR are present between −210 and −158.
FIG 4.
SouR interaction with the PglyA1 promoter. (A) EMSA performed with MBP-SouR (SouR) and biotinylated PglyA1 probe. The data are representative of four independent experiments performed with two separately purified batches of MBP-SouR. The presence (+) or absence (−) of unlabeled competitor (comp) PglyA1 probe is noted below each lane. (B) Results from a β-galactosidase assay for promoter mapping to identify regions within PglyA1 required for souR- and gbdR-dependent induction. The ΔgbdR cells were exposed to 1 mM sarcosine (S), and ΔsouR cells were exposed to glycine betaine (G) and compared to controls with pyruvate. The size of each PglyA1 promoter construct is noted as the beginning position relative to the glyA1 translational start site. Fold induction was calculated as a multiple of the pyruvate condition for each strain. (C) Results from a β-galactosidase assay for the −210 PglyA1 promoter in wild-type, ΔsouR, ΔgbdR, and ΔsouR ΔgbdR strains. Cells were induced as for panel B. The data shown are representative of three biological replicates, and the error bars represent standard deviations.
Characterization of sarcosine-induced transcripts and determination of the SouR regulon.
Using Affymetrix P. aeruginosa microarrays, we characterized the transcriptional response of P. aeruginosa ΔgbdR and ΔgbdR ΔsouR in the presence and absence of sarcosine, which allowed us to distinguish SouR-dependent transcriptional changes from the total cellular response to sarcosine. Potential SouR regulon members were those transcripts exhibiting at least a 2.5-fold induction in the ΔgbdR strain (with souR intact) and no induction in the ΔgbdR ΔsouR strain in response to sarcosine.
The ΔgbdR and ΔgbdR ΔsouR strains revealed no statistically significant differences in their expression profiles during exposure to MOPS-pyruvate medium (data not shown) (see GEO database accession number GSE72613). In contrast, the transcriptional responses of the two strains to sarcosine were markedly different. As expected from the results of the genetic screen, transcription of the sox operon (PA5415 to PA5420) and the glutathione-independent formaldehyde dehydrogenase gene (fdhA) were induced in the strain expressing SouR (ΔgbdR) compared to the MOPS-pyruvate control (Table 1). Sarcosine also induced expression of the glutathione-dependent formaldehyde detoxification system encoded by PA14_17410 and adhC (PA3628 and adhC) in a SouR-dependent manner (Table 1). Since sarcosine catabolism by Pseudomonas species is known to generate formaldehyde (15), the expression of a second detoxification system was not completely unanticipated. In the souR deletion (ΔgbdR ΔsouR) strain, sarcosine failed to induce transcription of the sox operon, fdhA, or the glutathione-dependent formaldehyde detoxification operon. Surprisingly, the dipeptide transport operon including the opdP porin and associated ABC transporter genes exhibited a roughly 4-fold increase in expression over the pyruvate control in the absence of SouR and the presence of sarcosine (Table 1).
TABLE 1.
Transcript changes (fold abundance) related to sarcosine and SouR
| Gene no. | Gene name | Fold changea in: |
||
|---|---|---|---|---|
| Δgbd strain (Sarc vs Pyr) | ΔgbdR ΔsouR strain (Sarc vs Pyr) | ΔgbdR strain vs ΔgbdR ΔsouR strain (Sarc) | ||
| PA1168 | −1.8 | −2.5 | −1.8 | |
| PA1247 | aprE | 1.6 | 3.1 | −2.4 |
| PA1250 | aprI | 2.4 | 2.8 | −2.4 |
| PA2513 | antB | −1.9 | −12.2 | 2.4 |
| PA3628 | 4.2 | −1.3 | 5.2 | |
| PA3629 | adhC | 3.4 | −1.1 | 3.6 |
| PA4385 | groEL | 1.1 | 3.3 | −2.3 |
| PA4386 | groES | 1.3 | 3.5 | −2.2 |
| PA4498 | mdpA | 1.2 | 2.8 | −2.6 |
| PA4501 | odpD | 1.4 | 3.6 | −3.4 |
| PA4502 | 1.1 | 4.2 | −4.0 | |
| PA4504 | 1.4 | 5.0 | −4.3 | |
| PA4505 | 1.1 | 4.1 | −3.9 | |
| PA4506 | 1.1 | 4.6 | −4.6 | |
| PA4761 | dnaK | −1.1 | 3.1 | −2.7 |
| PA5415 | glyA1 | 2.7 | 1.1 | 2.7 |
| PA5416 | soxB | 3.5 | 1.1 | 4.0 |
| PA5417 | soxD | 12.3 | −1.1 | 11.6 |
| PA5418 | soxAb | 3.1 | 1.0 | 2.1 |
| PA5419 | soxG | 8.0 | 1.1 | 8.2 |
| PA5420 | purU2 | 8.3 | 1.1 | 10.2 |
| PA5421 | fdhA | 5.5 | 1.1 | 5.4 |
Increase in transcript abundance in the presence of sarcosine compared to the pyruvate control (Sarc vs Pyr) or in the ΔgbdR strain compared to the ΔgbdR ΔsouR strain in the presence of sarcosine (Sarc). All changes in boldface are >2.5-fold different and significant, with P values of <0.05.
Signal from the soxA probe was low due to poor hybridization.
Testing SouR binding to the promoters of potential regulon members.
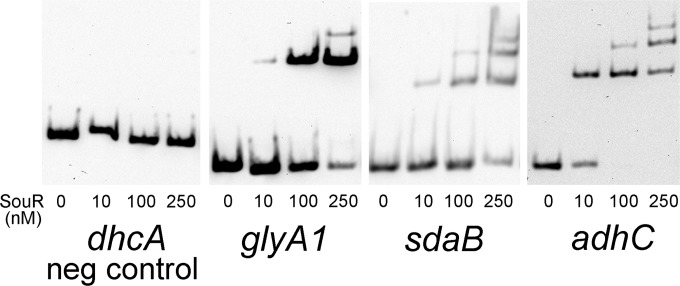
A short induction period (3 h) was used in our microarray studies to limit the expression of genes involved in secondary processes downstream of sarcosine metabolism. Nevertheless, alterations within the transcriptome could reflect the response to metabolic intermediates generated during sarcosine catabolism, including formaldehyde, glycine, serine, and pyruvate (Fig. 1A). Therefore, EMSAs were performed with MBP-SouR and biotinylated probes from the adhC and sdaB promoter regions to determine if they were directly bound by SouR. Although expression of the serine dehydratase gene transcript, sdaB, failed to surpass our 2.5-fold cutoff (2.32-fold change), we included the promoter of the gene in our EMSAs because sdaB has previously been identified as a member of the GbdR regulon and plays a critical role in the conversion of serine to pyruvate during sarcosine metabolism (22). As shown in Fig. 5, MBP-SouR specifically bound to the promoters of glyA1, adhC, and sdaB but not to the promoter region of dhcA (negative control) (8, 22).
FIG 5.
SouR binding to promoters of potential regulon members. Shown are EMSAs with purified MBP-SouR and biotinylated probes of promoter regions from operons induced by sarcosine. Each blot represents a separate biotinylated probe, with dhcA included as a negative control. The MBP-SouR concentrations are shown below the lanes. The data are representative of the results of at least three independent experiments with two separate batches of purified MBP-SouR.
Effects of SouR regulon members and sarcosine-induced genes on sarcosine catabolism.
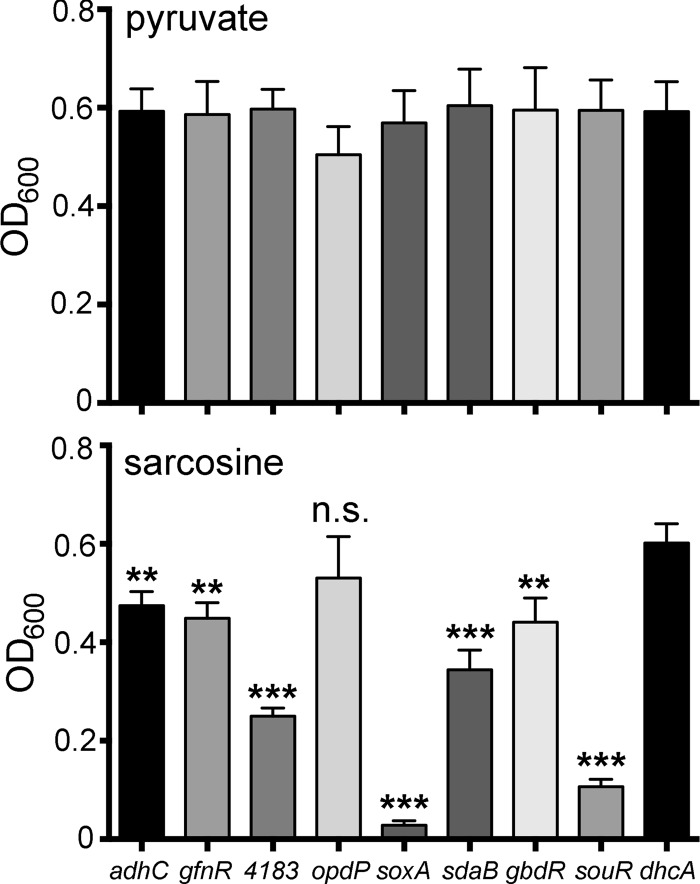
The genes within the sox operon and their respective roles in the metabolism of sarcosine have been well characterized (3, 15, 17, 18). However, the contributions of the other genes in the sarcosine regulon—PA4183, the glutathione-dependent formaldehyde detoxification system (PA3628 and adhC), and the sarcosine-induced dipeptide porin and transport system (PA4501 to PA4506)—in the metabolism of sarcosine were unknown. To determine the requirement for these genes in this process, transposon mutants were selected from the PA14 transposon mutant library (37) and screened for the ability to grow in MOPS minimal medium using 40 mM sarcosine as the sole carbon and nitrogen sources. With the exception of soxA::TnM, all of the TnM disruption mutants from the sarcosine regulon tested were capable of utilizing sarcosine as a carbon and nitrogen source to some extent (Fig. 6). However, growth was significantly slower than that of the positive growth control (dhcA::TnM) strain for all strains except opdP::TnM (Fig. 6). As a whole, these data indicate that the glutathione-dependent formaldehyde detoxification genes, PA4183, sdaB, souR, and gbdR, are not absolutely necessary for the metabolism of sarcosine but are important for achieving optimal growth under these conditions.
FIG 6.
Roles of SouR- and sarcosine-regulated genes during growth on sarcosine. Shown are the culture densities (OD600) after 24 h of growth in MOPS minimal medium without nitrogen supplemented with either 20 mM pyruvate and 10 mM ammonium chloride (top) or 40 mM sarcosine (bottom) for the transposon insertion mutants labeled on the x axis. The dhcA insertion mutant is known not to have a role in this pathway and served as the positive growth control (no growth defect). Statistical significance was determined using one-way ANOVA with Dunnett's posttest, with growth in the dhcA mutant as the comparator for all other data. n.s., not significant; **, P < 0.01; ***, P < 0.001. The data shown are summaries of the results of three independent experiments, each with three biological replicates; the error bars represent standard errors of the means.
PA3630 encodes the transcription regulator of the glutathione-dependent formaldehyde detoxification genes.
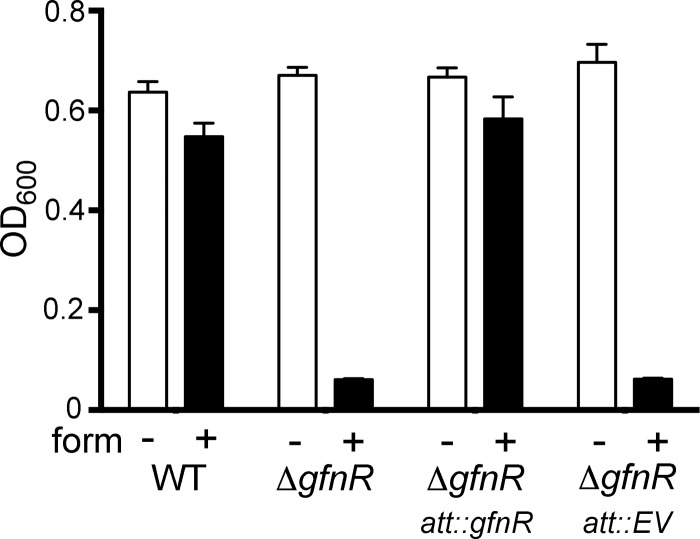
Although EMSAs demonstrated a clear interaction between MBP-SouR and the promoter region of the glutathione-dependent formaldehyde detoxification operon (PA3628 and adhC), we suspected that the divergently transcribed LysR family transcription factor encoded by PA3630 (Fig. 1B) might also influence the expression of these genes in response to formaldehyde generated endogenously through the metabolism of sarcosine. Evidence for this function is supported by a search of PA3628, adhC, and PA3630 using the String database (38), which revealed that the synteny of these genes is conserved among hundreds of proteobacterial taxa. To test the role of PA3630 in the cellular response to formaldehyde, an unmarked deletion of PA3630 was generated. Growth of the strain was severely attenuated compared to the WT when cultured in minimal medium containing 0.75 mM formaldehyde. Moreover, integration of PA3630 at the attTn7 site restored growth of the deletion strain to wild-type levels (Fig. 7). These data suggest that PA3630 encodes a formaldehyde-responsive regulator of the glutathione-dependent formaldehyde detoxification genes, and we propose the name GfnR (glutathione-dependent formaldehyde neutralization regulator) to reflect this function.
FIG 7.
Role of gfnR during growth in the presence of formaldehyde (form). Shown are the culture densities (OD600) after 24 h of growth in MOPS minimal medium, 20 mM sodium pyruvate, and 5 mM d-glucose in the presence (+) and absence (−) of 0.75 mM formaldehyde for wild-type, ΔgfnR, ΔgfnR attTn7::gfnR, and ΔgfnR attTn7::EV (empty-vector) strains. The data are representative of the results of three separate experiments, and the error bars represent standard deviations.
Confirmation of SouR and GfnR regulon members.
Quantitative RT-PCR was performed to confirm the expression of SouR regulon members identified through microarray analysis, as well as to distinguish the regulatory contribution of GfnR from that of SouR in the expression of the glutathione-dependent formaldehyde detoxification system in response to sarcosine. While the expression of sdaB and the sox operon was induced by sarcosine in a SouR-dependent manner, induction of adhC was more stochastic, as a greater-than-2-fold increase in expression was observed in only half of the ΔgbdR replicates exposed to sarcosine (3 out of 6 biological replicates) (Table 2). However, induction of adhC and PA3628 was not observed in the ΔgbdR ΔgfnR strain in response to sarcosine (Table 2), indicating that the expression of the glutathione-dependent formaldehyde detoxification system is likely induced by GfnR in response to formaldehyde generated through sarcosine catabolism.
TABLE 2.
Effects of souR and gfnR mutations on sarcosine regulation of regulon members
| Transcript | Relative expressiona in: |
||
|---|---|---|---|
| ΔgbdR strain | ΔgbdR ΔsouR strain | ΔgbdR ΔgfnR strain | |
| soxA | 32.9 (8.0)b | 1.11 (0.2) | 31.0 (11.0)b |
| adhC | 3.60 (2.8)d | 1.23 (0.2) | 1.21 (0.3) |
| sdaB | 5.40 (2.4)c | 0.73 (0.1) | Not determined |
Relative expression was calculated based on the expression in the WT in the pyruvate control normalized to the rplU transcript; standard deviations are in parentheses. The data were analyzed using one-way ANOVA within each transcript using a Dunnett's corrected posttest with the pyruvate condition as the comparator; unmarked relative expression numbers are not statistically significant.
P < 0.01.
P < 0.05.
The adhC transcript is stochastically induced under these conditions, and while not different using the above-mentioned parametric analysis, the data are not normally distributed. Analysis with the nonparametric Mann-Whitney test showed significance (P = 0.026).
DISCUSSION
P. aeruginosa is ubiquitous in nature and is often described as an optimal exploiter of nutrient pulses, largely as a result of the diverse metabolic potential encoded within its genome. Related to this metabolic flexibility, close to 10% of P. aeruginosa genes are predicted to encode transcription factors (39), many of which likely allow the organism to sense potential nutrient sources and regulate enzymatic pathways to exploit a variety of metabolic niches. Sarcosine is present in a range of environments inhabited by P. aeruginosa, although it is likely encountered most often as an intermediate metabolite of glycine betaine, carnitine, glyphosate, or creatine catabolism (6–14, 22) (Fig. 1A). We propose that the capacity to sense and metabolize sarcosine provides Pseudomonas with a fitness advantage in certain environments through the ability to fully catabolize a carbon and nitrogen source that competitors cannot.
In this study, we utilized a genetic screen to identify an AraC family transcription factor, SouR (PA4184), as the sarcosine-responsive regulator of sarcosine catabolism in P. aeruginosa. SouR is required for appreciable growth when sarcosine is utilized as a sole carbon and nitrogen source, and transcriptional induction is limited to sarcosine, a natural metabolite, and ethylglycine, a nonnatural sarcosine analog. Together, the data support SouR as the first known sarcosine-responsive transcription factor. While previous work by Nishiya and Imanaka reported SoxR as a repressor of monomeric sarcosine oxidase in Arthrobacter sp. spe4, the authors noted that sarcosine failed to relieve repression in vitro (40). Moreover, a follow-up study determined that soxR and the monomeric sarcosine oxidase genes clustered with genes involved in the degradation of creatinine and creatine (41). Since sarcosine is generated during creatine metabolism, it is likely that either creatinine or creatine acts as the inducing ligand of SoxR in Arthrobacter.
While all pseudomonads sequenced to date encode clear orthologs of SouR, only P. aeruginosa isolates carry the gene as part of a two-gene operon with PA4183. PA4183 encodes a protein of unknown function that shares modest structural similarity with members of the glyoxylase I family of enzymes (PF00903). The lack of genus-wide conservation of PA4183 outside P. aeruginosa suggests that the gene is likely to play an accessory role in the metabolism of sarcosine, and growth assays performed with a PA4183 transposon mutant support this theory (Fig. 6). However, we have no current hypothesis as to the role of PA4183 in P. aeruginosa sarcosine catabolism.
SouR is a member of the glutamine amidotransferase I-like transcription regulator (GATR) subfamily of the AraC regulator family (CD03137). Little is known about this group aside from their widespread distribution among Gram-negative taxa. Like other members of the AraC family, GATRs exhibit a two-domain layout with a C-terminal AraC-like helix-turn-helix DNA binding domain. Unlike those of other members of the AraC family, the amino-terminal domain is a glutamine amidotransferase I-like domain (42), likely involved in the recognition of the inducing molecules. Pseudomonas species encode a number of GATRs, with seven members conserved among the core genomes of sequenced and annotated P. aeruginosa isolates. Interestingly, multiple GATRs regulate glycine betaine acquisition and catabolism in P. aeruginosa, with the GATR member GbdR controlling glycine betaine-catabolic genes in response to glycine betaine and dimethylglycine (21, 22) and the GATR member CdhR regulating the carnitine catabolic pathway in response to carnitine (8).
Evidence suggests that SouR and CdhR may be paralogs of GbdR that arose through gene duplication. SouR and CdhR display close homology to GbdR (58% and 62% similarity, respectively), and their phylogenetic distribution hints at common ancestry, as orthologs of SouR and CdhR are present only in taxa that also encode the glycine betaine-catabolic pathway regulated by GbdR. In contrast, GbdR orthologs are widespread in taxa that lack clear SouR and CdhR orthologs. Here, we have shown that SouR and GbdR both regulate the expression of the glyA1 promoter (the promoter of the sox operon), and we determined that they likely recognize the same binding region (Fig. 4B and C; see Fig. S1 in the supplemental material). We are currently investigating whether CdhR and GbdR also regulate genes from the same binding region in one or more promoters. Such coregulation may indicate a hierarchy of binding priority contributing to regulation as a means to control flux through the intermediate metabolite pools in the glycine betaine-catabolic pathway.
Additional transcripts regulated by SouR were identified through microarrays, EMSAs with MBP-tagged SouR, and quantitative RT-PCR, which point to additional overlap between the GbdR and SouR regulons. The serine dehydratase gene, sdaB, is a member of the GbdR regulon (22), and we were initially surprised that sarcosine failed to induce transcription of the gene above the expression fold change cutoff in our microarrays, as the activity of the enzyme links sarcosine catabolism to central metabolism by converting serine generated from glycine and 5,10-methyltetrahydrofolate via GlyA1 (Fig. 1A) to pyruvate and ammonium. However, the expression of SdaB did increase 2.3-fold in response to sarcosine; hence, we included the promoter of the gene in our EMSAs with MBP-tagged SouR. In doing so, we determined that SouR, like GbdR, could bind the promoter region of sdaB (Fig. 5). Furthermore, quantitative RT-PCR revealed that the expression of sdaB is induced by SouR in response to sarcosine (Table 2). Thus, the expression cutoff used in our microarrays (2.5-fold change) was likely conservative, and additional, less dramatically induced SouR regulon members, like sdaB, might exist.
Our microarrays also revealed that the dipeptide transport system (PA4501 to PA4506) was induced by sarcosine in the ΔgbdR ΔsouR strain but not in the ΔgbdR strain. Regulation of this system is complex, and expression has been shown to be influenced by numerous dipeptides, as well as the amino acid arginine (43–45). Similarly, the substrate specificities of the OpdP porin (PA4501) and associated transporter proteins have recently been examined, and the system was found to be implicated in the uptake and metabolism of over 100 unique dipeptides (46). Growth assays performed with an opdP transposon disruption mutant revealed a wild-type growth phenotype when utilizing sarcosine as a sole carbon and nitrogen source, indicating that the system does not contribute significantly to the catabolism of the molecule (Fig. 6). We instead hypothesize that the opdP operon might be induced in the ΔgbdR ΔsouR genotype as a consequence of perceived nutrient deprivation and/or the strain's inability to metabolize sarcosine. In the latter scenario, we speculate that the accumulation of sarcosine within the cytosol might promote detection by the low-specificity regulator governing expression of the dipeptide transport operon.
Glycine betaine and sarcosine catabolism in proteobacterial species generates formaldehyde in the absence of tetrahydrofolate (15, 17). However, these bacteria also encode (fdhA) a sarcosine-inducible glutathione-independent formaldehyde dehydrogenase that functions in converting formaldehyde to formate (20). Our microarrays revealed that a second formaldehyde detoxification system is expressed during sarcosine catabolism in P. aeruginosa. adhC and PA3628 encode a glutathione-dependent formaldehyde dehydrogenase and a formate esterase that are nearly universally conserved among Gram-negative bacteria. This system has been well characterized in proteobacteria and has been demonstrated to function in protecting cells against the effects of intracellular formaldehyde (20, 47). Interestingly, while EMSAs revealed that MBP-tagged SouR is capable of binding to the promoter region of the adhC and PA3628 operon, qRT-PCR data suggest that expression of these genes is likely influenced by a second regulator in response to formaldehyde production (Fig. 5A and Table 2). Searching the Pseudomonas genome database, we identified an uncharacterized LysR family regulator (PA3630) that is divergently transcribed from the PA3628 and adhC operon in all Pseudomonas genomes annotated to date (33). Moreover, a study in P. putida revealed that the expression of this transcription factor is upregulated, along with the (then uncharacterized) glutathione-dependent formaldehyde detoxification operon, following exposure to formaldehyde (48).
Here, using formaldehyde susceptibility challenge with chromosomal deletion and complementation strains, we have shown that GfnR (PA3630) is required for optimal growth of P. aeruginosa in the presence of formaldehyde (Fig. 7). Alternative regulatory mechanisms have been described for the glutathione-dependent formaldehyde detoxification system in proteobacteria, including the frmR repressor of E. coli (47) and the fhlRS two-component sensor system of Paracoccus denitrificans (49). Nevertheless, a synteny search of the String database (38) revealed widespread conservation of gfnR orthologs in association with the detoxification genes among hundreds of taxa, indicating that the LysR family regulatory mechanism is likely prevalent among proteobacteria.
In summary, this study has expanded our understanding of how sarcosine metabolism is transcriptionally regulated in P. aeruginosa. SouR is the first sarcosine-responsive transcription factor to be described, and we speculate that the regulator arose from GbdR as a means for Pseudomonas species to independently detect this intermediate of glycine betaine and creatine degradation in the environment. Finally, we identified GfnR as the regulator of the glutathione-dependent formaldehyde detoxification system in P. aeruginosa and determined that homologs are widespread among proteobacterial taxa.
Supplementary Material
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00739-15.
REFERENCES
- 1.Bernheim F. 1950. The sarcosine oxidase in adapted and unadapted cultures of a strain of Pseudomonas aeruginosa. J Bacteriol 60:767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopper PH. 1950. Studies on a sarcosine oxidase of bacterial origin. J Gen Physiol 34:9–17. doi: 10.1085/jgp.34.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan-Abdallah A, Zhao G, Eschenbrenner M, Chen ZW, Mathews FS, Jorns MS. 2005. Cloning, expression and crystallization of heterotetrameric sarcosine oxidase from Pseudomonas maltophilia. Protein Expr Purif 43:33–43. doi: 10.1016/j.pep.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wargo MJ. 2013. Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa. Appl Environ Microbiol 79:2112–2120. doi: 10.1128/AEM.03565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son MS, Matthews WJ Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wargo MJ. 2013. Choline catabolism to glycine betaine contributes to Pseudomonas aeruginosa survival during murine lung infection. PLoS One 8:e56850. doi: 10.1371/journal.pone.0056850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wargo MJ, Ho TC, Gross MJ, Whittaker LA, Hogan DA. 2009. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect Immun 77:1103–1111. doi: 10.1128/IAI.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wargo MJ, Hogan DA. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155:2411–2419. doi: 10.1099/mic.0.028787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nimmo-Smith RH, Appleyard G. 1956. Studies with a pseudomonad able to grow with creatine as main source of carbon and nitrogen. J Gen Microbiol 14:336–350. doi: 10.1099/00221287-14-2-336. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimoto T, Oka I, Tsuru D. 1976. Purification, crystallization, and some properties of creatine amidinohydrolase from Pseudomonas putida. J Biochem 79:1381–1383. [DOI] [PubMed] [Google Scholar]
- 11.Drosinos EH, Board RG. 1994. Metabolic activities of pseudomonads in batch cultures in extract of minced lamb. J Appl Bacteriol 77:613–620. doi: 10.1111/j.1365-2672.1994.tb02809.x. [DOI] [PubMed] [Google Scholar]
- 12.Kishore GM, Jacob GS. 1987. Degradation of glyphosate by Pseudomonas sp. PG2982 via a sarcosine intermediate. J Biol Chem 262:12164–12168. [PubMed] [Google Scholar]
- 13.Fitzgibbon JE, Braymer HD. 1990. Cloning of a gene from Pseudomonas sp. strain PG2982 conferring increased glyphosate resistance. Appl Environ Microbiol 56:3382–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JK, Braymer HD, Larson AD. 1983. Isolation of a Pseudomonas sp. which utilizes the phosphonate herbicide glyphosate. Appl Environ Microbiol 46:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner MA, Schuman Jorns M. 1997. Folate utilization by monomeric versus heterotetrameric sarcosine oxidases. Arch Biochem Biophys 342:176–181. doi: 10.1006/abbi.1997.0106. [DOI] [PubMed] [Google Scholar]
- 16.Zeller HD, Hille R, Jorns MS. 1989. Bacterial sarcosine oxidase: identification of novel substrates and a biradical reaction intermediate. Biochemistry 28:5145–5154. doi: 10.1021/bi00438a035. [DOI] [PubMed] [Google Scholar]
- 17.Chlumsky LJ, Zhang L, Jorns MS. 1995. Sequence analysis of sarcosine oxidase and nearby genes reveals homologies with key enzymes of folate one-carbon metabolism. J Biol Chem 270:18252–18259. doi: 10.1074/jbc.270.31.18252. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Tamamura R, Yajima S, Kanno M, Suguro M. 2005. Corynebacterium sp. U-96 contains a cluster of genes of enzymes for the catabolism of sarcosine to pyruvate. Biosci Biotechnol Biochem 69:952–956. [DOI] [PubMed] [Google Scholar]
- 19.Meskys R, Harris RJ, Casaite V, Basran J, Scrutton NS. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J Biochem 268:3390–3398. doi: 10.1046/j.1432-1327.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 20.Marx CJ, Miller JA, Chistoserdova L, Lidstrom ME. 2004. Multiple formaldehyde oxidation/detoxification pathways in Burkholderia fungorum LB400. J Bacteriol 186:2173–2178. doi: 10.1128/JB.186.7.2173-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wargo MJ, Szwergold BS, Hogan DA. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190:2690–2699. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampel KJ, LaBauve AE, Meadows JA, Fitzsimmons LF, Nock AM, Wargo MJ. 2014. Characterization of the GbdR regulon in Pseudomonas aeruginosa. J Bacteriol 196:7–15. doi: 10.1128/JB.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaBauve AE, Wargo MJ. 2012. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter 6:Unit 6E 1. doi: 10.1002/9780471729259.mc06e01s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meadows JA, Wargo MJ. 2013. Characterization of Pseudomonas aeruginosa growth on O-acylcarnitines and identification of a short-chain acylcarnitine hydrolase. Appl Environ Microbiol 79:3355–3363. doi: 10.1128/AEM.03943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer HD. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834. [PubMed] [Google Scholar]
- 28.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 29.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 30.Wong SM, Mekalanos JJ. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:10191–10196. doi: 10.1073/pnas.97.18.10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol 55:368–380. [DOI] [PubMed] [Google Scholar]
- 32.Chun KT, Edenberg HJ, Kelley MR, Goebl MG. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233–240. [DOI] [PubMed] [Google Scholar]
- 33.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller J. 1972. Experiments in molecular genetics, p 352–355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 35.LaBauve AE, Wargo MJ. 2014. Detection of host-derived sphingosine by Pseudomonas aeruginosa is important for survival in the murine lung. PLoS Pathog 10:e1003889. doi: 10.1371/journal.ppat.1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wargo MJ, Gross MJ, Rajamani S, Allard JL, Lundblad LK, Allen GB, Vasil ML, Leclair LW, Hogan DA. 2011. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am J Respir Crit Care Med 184:345–354. doi: 10.1164/rccm.201103-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balasubramanian D, Schneper L, Merighi M, Smith R, Narasimhan G, Lory S, Mathee K. 2012. The regulatory repertoire of Pseudomonas aeruginosa AmpC ss-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiya Y, Imanaka T. 1996. Analysis of a negative regulator, SoxR, for the Arthrobacter sarcosine oxidase gene. J Ferment Bioeng 81:64–67. doi: 10.1016/0922-338X(96)83122-9. [DOI] [Google Scholar]
- 41.Nishiya Y, Toda A, Imanaka T. 1998. Gene cluster for creatinine degradation in Arthrobacter sp. TE1826. Mol Gen Genet 257:581–586. doi: 10.1007/s004380050685. [DOI] [PubMed] [Google Scholar]
- 42.Bandyopadhyay S, Cookson MR. 2004. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol Biol 4:6. doi: 10.1186/1471-2148-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamber S, Hancock RE. 2006. Involvement of two related porins, OprD and OpdP, in the uptake of arginine by Pseudomonas aeruginosa. FEMS Microbiol Lett 260:23–29. doi: 10.1111/j.1574-6968.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- 44.Tamber S, Ochs MM, Hancock RE. 2006. Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa. J Bacteriol 188:45–54. doi: 10.1128/JB.188.1.45-54.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiely PD, O'Callaghan J, Abbas A, O'Gara F. 2008. Genetic analysis of genes involved in dipeptide metabolism and cytotoxicity in Pseudomonas aeruginosa PAO1. Microbiology 154:2209–2218. doi: 10.1099/mic.0.2007/015032-0. [DOI] [PubMed] [Google Scholar]
- 46.Pletzer D, Lafon C, Braun Y, Kohler T, Page MG, Mourez M, Weingart H. 2014. High-throughput screening of dipeptide utilization mediated by the ABC transporter DppBCDF and its substrate-binding proteins DppA1-A5 in Pseudomonas aeruginosa. PLoS One 9:e111311. doi: 10.1371/journal.pone.0111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herring CD, Blattner FR. 2004. Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR. J Bacteriol 186:6714–6720. doi: 10.1128/JB.186.20.6714-6720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roca A, Rodriguez-Herva JJ, Duque E, Ramos JL. 2008. Physiological responses of Pseudomonas putida to formaldehyde during detoxification. Microb Biotechnol 1:158–169. doi: 10.1111/j.1751-7915.2007.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harms N, Reijnders WN, Koning S, van Spanning RJ. 2001. Two-component system that regulates methanol and formaldehyde oxidation in Paracoccus denitrificans. J Bacteriol 183:664–670. doi: 10.1128/JB.183.2.664-670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic-engineering—transposon mutagenesis in gram-negative bacteria. Bio-Technology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.