ABSTRACT
A putative biosynthetic gene cluster of the enterocin NKR-5-3B (Ent53B), a novel circular bacteriocin, was analyzed by sequencing the flanking regions around enkB, the Ent53B structural gene, using a fosmid library. A region approximately 9 kb in length was obtained, and the enkB1, enkB2, enkB3, and enkB4 genes, encoding putative biosynthetic proteins involved in the production, maturation, and secretion of Ent53B, were identified. We also determined the identity of proteins mediating self-immunity against the effects of Ent53B. Heterologous expression systems in various heterologous hosts, such as Enterococcus faecalis and Lactococcus lactis strains, were successfully established. The production and secretion of the mature Ent53B required the cooperative functions of five genes. Ent53B was produced only by those heterologous hosts that expressed protein products of the enkB, enkB1, enkB2, enkB3, and enkB4 genes. Moreover, self-immunity against the antimicrobial action of Ent53B was conferred by at least two independent mechanisms. Heterologous hosts harboring the intact enkB4 gene and/or a combination of intact enkB1 and enkB3 genes were immune to the inhibitory action of Ent53B.
IMPORTANCE In addition to their potential application as food preservatives, circular bacteriocins are now considered possible alternatives to therapeutic antibiotics due to the exceptional stability conferred by their circular structure. The successful practical application of circular bacteriocins will become possible only if the molecular details of their biosynthesis are fully understood. The results of the present study offer a new perspective on the possible mechanism of circular bacteriocin biosynthesis. In addition, since some enterococcal strains are associated with pathogenicity, virulence, and drug resistance, the establishment of the first multigenus host heterologous production of Ent53B has very high practical significance, as it widens the scope of possible Ent53B applications.
INTRODUCTION
Bacteriocins are ribosomally synthesized antimicrobial peptides that generally exert their antagonistic activity toward strains that are closely related to the producer strain (1, 2), although an increasing number of bacteriocins have been reported to have a broad activity range (3, 4). In the last decade, the interest in bacteriocins, especially those from lactic acid bacteria (LAB), has increased considerably, as they potentially can be used as natural food preservatives and therapeutic antibiotics (5–8).
Over the years, various classification schemes of bacteriocins from Gram-positive bacteria have been suggested, which commonly divide bacteriocins into two groups: class I (lantibiotics) and class II (nonlantibiotics) (9–12). Lantibiotics are small heat-stable peptides which contain unusual amino acids, such as lanthionine and/or methyllanthionine, as a result of posttranslational modifications of some common amino acid residues (13, 14). In contrast, class II bacteriocins, which are also heat-stable peptides, do not undergo extensive posttranslational modifications and therefore do not contain unusual amino acid residues. Class II bacteriocins can be further classified into as many as four subclasses (11, 15, 16). These are class IIa (anti-Listeria pediocin-like bacteriocins), class IIb (two-peptide bacteriocins), class IIc (circular bacteriocins), and class IId (linear one-peptide non-pediocin-like bacteriocins) (11). In recent years, circular bacteriocins (class IIc) have attracted considerable attention (10, 17–19). In comparison to their linear counterparts, circular peptides, including circular bacteriocins, have been perceived to be more promising substances for a variety of pharmaceutical applications due to their more favorable structural properties, higher thermal stress resistance, and superior proteolytic stability (18, 20). The remarkable stability of class IIc bacteriocins is conferred by the circular nature of these peptides (21).
Circular bacteriocins are synthesized as linear precursor peptides containing a leader peptide (2 to 35 amino acid residues) attached to a propeptide (58 to 70 amino acid residues). The leader peptide is cleaved off from the propeptide, while the linear propeptide undergoes a dehydration reaction between the N- and C-terminal residues during maturation, thereby resulting in the covalent binding of the terminal ends (10, 17). However, the exact sequence and detailed mechanism of these processes remain a mystery. Moreover, the enzymes responsible for the cleavage of the leader peptide and ligation of the N and C termini have not yet been identified (10, 17). Elucidation of the mechanisms of circular bacteriocin synthesis and processing, which lead to the production of mature circular peptides, will undoubtedly help realize the potential of these substances as biological delivery agents (17). Furthermore, such information will also help develop circular bacteriocins into promising scaffolds for drug design by means of genetic engineering (18, 21).
Enterocin AS-48, the first representative of circular bacteriocins, was first discovered in 1986 (22), although its circular nature was reported in 1994 (23), and it remains the most extensively studied circular bacteriocin to date. Since then, a number of circular bacteriocins from various bacterial species have been isolated: gassericin A (24), subtilosin A (25), circularin A (3), butyrivibriocin AR10 (26), uberolysin (27), carnocyclin A (28), lactocyclicin Q (29), garvicin ML (30), leucocyclicin Q (31), acidocin B (32), and, most recently, enterocin NKR-5-3B (33). Enterocin NKR-5-3B (Ent53B), one of several bacteriocins produced by Enterococcus faecium NKR-5-3 previously isolated from the Thai fermented fish pla-ra (34–36), is a 64-amino-acid novel circular bacteriocin with a molecular mass of 6,316.4 Da. A nuclear magnetic resonance (NMR)-derived three-dimensional solution structure of Ent53B revealed the presence of four helical segments that enclose a tightly packed hydrophobic core (33). It is a strongly amphiphilic peptide with a net positive charge of 5 at neutral pH. Ent53B amphiphilic properties are conferred by a cluster of basic amino acid residues on one surface around the end of helix 4, the initial portion of helix 1, and the central region of helix 2 (33). The spectrum of Ent53B antimicrobial activity was wider than that of other NKR-5-3 enterocins, possibly because Ent53B amphiphilic properties facilitated its binding to negatively charged cell membranes of target cells (33, 34).
In the present study, we attempted to obtain at least a partial solution to the enigma of the circular bacteriocin biosynthesis mechanism. We have identified five genes required for the production and maturation of Ent53B in various heterologous hosts. We also determined the proteins involved in two independent mechanisms of self-immunity developed by the producer strain to protect against the inhibitory action of Ent53B.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
The strains and plasmids used in this study are summarized in Table 1. E. faecium NKR-5-3 was cultivated in M17 medium (Merck, Darmstadt, Germany), while the indicator strain Lactococcus lactis ATCC 19435T was cultivated in MRS medium (Oxoid, Hampshire, England) and in Lactobacilli Agar AOAC medium (Becton Dickinson, Sparks, MD), both at 30°C, in liquid and solid culture, respectively. The circularin A (CirA) producer strain Clostridium beijerinckii ATCC 25752 was cultivated anaerobically in reinforced clostridial medium (RCM) (Becton Dickinson) at 37°C. The cloning strain Escherichia coli DH5α was cultivated in the Luria-Bertani (LB) medium (Becton Dickinson) under constant agitation of 180 strokes/min at 37°C. The expression host strains Enterococcus faecalis JH2-2, L. lactis IL1403, L. lactis NZ9000, and L. lactis ATCC 19435T were cultivated in the M17 medium supplemented with 0.5% glucose (GM17) at 30°C. Chloramphenicol and erythromycin were used as antibiotic markers in selective medium at a final concentration of 10 μg/ml. All microorganisms were stored at −80°C in their respective media supplemented with 30% glycerol and cultivated twice before use.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. faecium NKR-5-3 | Enterocin NKR-5-3B producer strain | 34 |
| Clostridium beijerinckii ATCC 25752 | Circularin A producer strain | 3 |
| E. faecalis JH2-2 | Plasmid-free derivative of E. faecalis JH-2 | 40 |
| L. lactis ATCC 19435T | Bacteriocin indicator strain, heterologous host strain | ATCCb |
| L. lactis IL1403 | Heterologous host strain, plasmid free | 61 |
| L. lactis NZ9000 | Heterologous host strain, nisRK | 62 |
| E. coli DH5α | Plasmid storage strain | Novagen |
| Plasmids | ||
| pMG36c | Cmr, pWV01-based cloning vector carrying a strong Lactococcus-based promoter, P32 | 4 |
| pIL253 | Emr, theta-replicating vector | 38 |
| pIL-Pnis | Emr, pIL253 derivative with a nisin-inducible promoter, Pnis | This study |
| pNK-B1234 | Cmr, pMG36c derivative containing enkB, enkB1, enkB2, enkB3, and enkB4 | This study |
| pNK-B | Cmr, pMG36c derivative containing enkB | This study |
| pNK-B234 | Cmr, pNK-B1234 ΔenkB1 | This study |
| pNK-B134 | Cmr, pNK-B1234 ΔenkB2 | This study |
| pNK-B124 | Cmr, pNK-B1234 ΔenkB3 | This study |
| pNK-B123 | Cmr, pNK-B1234 ΔenkB4 | This study |
| pNK-B34 | Cmr, pNK-B1234 ΔenkB1 ΔenkB2 | This study |
| pNK-B24 | Cmr, pNK-B1234 ΔenkB1 ΔenkB3 | This study |
| pNK-B23 | Cmr, pNK-B1234 ΔenkB1 ΔenkB4 | This study |
| pNK-B14 | Cmr, pNK-B1234 ΔenkB2 ΔenkB3 | This study |
| pNK-B13 | Cmr, pNK-B1234 ΔenkB2 ΔenkB4 | This study |
| pNK-B12 | Cmr, pNK-B1234 ΔenkB3 ΔenkB4 | This study |
| pNK-B4 | Cmr, pNK-B1234 ΔenkB1 ΔenkB2 ΔenkB3 | This study |
| pNK-B3 | Cmr, pNK-B1234 ΔenkB1 ΔenkB2 ΔenkB4 | This study |
| pNK-B2 | Cmr, pNK-B1234 ΔenkB1 ΔenkB3 ΔenkB4 | This study |
| pNK-B1 | Cmr, pNK-B1234 ΔenkB2 ΔenkB3 ΔenkB4 | This study |
| pIL-1 | Emr, pIL-Pnis derivative containing enkB1 | This study |
| pIL-2 | Emr, pIL-Pnis derivative containing enkB2 | This study |
| pIL-3 | Emr, pIL-Pnis derivative containing enkB2 | This study |
| pIL-4 | Emr, pIL-Pnis derivative containing enkB4 | This study |
Cmr, chloramphenicol resistant; Emr, erythromycin resistant.
ATCC, American Type Culture Collection, Rockville, MD.
Determination of the Ent53B gene locus.
To find the enkB gene locus, we utilized the fosmid library of the E. faecium NKR-5-3 genome, which was used previously to obtain the loci of genes encoding NKR-5-3 enterocins (36). Briefly, the total genome DNA of E. faecium NKR-5-3 was extracted, physically digested by sonication into fragments of approximately 40 kb in length, and cloned into the CopyControl pCC1FOS vector (Epicentre, WI, USA) using T4 DNA ligase (TaKaRa, Osaka, Japan). Subsequent plasmids were packed into phage particles using an in vitro packaging system (MaxPlax Lambda packaging extract; Epicentre) to prepare the NKR-5-3 phage library solution. A 1-μl aliquot of the phage solution was then added to 100 μl of an E. coli EPI-300 cell suspension previously cultivated in the LB broth (optical density at 600 nm [OD600], 0.7), supplemented with 10 mM MgSO4 and 0.2% (wt/vol) maltose, and incubated at 37°C for 1 h. Following the incubation, the cell suspension was plated on the LB agar medium containing 12.5 μg/ml chloramphenicol. Positive colonies carrying the NKR-5-3 genome library were screened and identified using colony PCR with appropriate primers (36). To obtain the enkB gene locus, consecutive DNA sequencing by primer walking was performed using the NKR-5-3 fosmid library as the template.
Cloning of Ent53B genetic determinants.
Basic molecular cloning was performed using standard methods described by Sambrook and Russell (37). Ent53B genetic determinants were consecutively cloned from two fragments into the wide-host-range pMG36c vector, which carries the strong lactococcal promoter P32. The PCR primers used are summarized in Table 2. The first fragment containing the enkB and enkB1 genes was amplified by PCR using the SacI-ent53B-f and XbaI-enkB1-r primers. Next, the PCR product was digested with the SacI and XbaI restriction enzymes and subsequently ligated to the SacI- and XbaI-digested pMG36c plasmid to generate the pNK-B1 construct. The second fragment, which contained the enkB2, enkB3, and enkB4 genes, was then amplified using the PstI-enkB2-f and SphI-enkB4-r primers. The resulting PCR product was then digested with the PstI and SphI restriction enzymes and subsequently ligated to the PstI- and SphI-digested pNK-B1 plasmid to generate the pNK-B1234 plasmid construct, which contained the whole region encompassing all five genes, from enkB to enkB4.
TABLE 2.
Primers used in the study
| Primer name | Sequence (5′ → 3′)a | Tm (°C)b |
|---|---|---|
| SacI-enkB-f | AAAGAGCTCATTCAGTGGTGGTAGTAACAGT | 51 |
| XbaI-enkB-r | AAATCTAGAGACCATGGCCAATTTATTCG | 58 |
| XbaI-enkB1-r | AAATCTAGATATAAGAATTCCTCCCATCC | 52 |
| Pst-enkB1-f | TGCTGCAGCGAAATCATTGATTA | 63 |
| Sph-enkB1-r | TCTATAAGCATGCCTCCCATCC | 61 |
| PstI-enkB2-f | TCCCTGCAGTATTTGTTCTTTTTGG | 63 |
| SphI-enkB2-r | TGCATGCTTTAAAGTATAATTCAGC | 57 |
| PstI-enkB3-f | TTGCTGCAGTAGTAGAAGGGGTATTC | 61 |
| Sph-I-enkB3-r | AGCATGCAATTCCACAAATTATTAG | 59 |
| PstI-enkB4-f | ACTGCAGACAAAAAAATTGAGCTAAC | 60 |
| SphI-enkB4-r | AGCATGCAGAATATTCACAGCC | 60 |
| Inv-enkB3-f | ACAGGAGGGGTTGAGACTAACT | 57 |
| Inv-enkB3-r | TCGATGAGTATTGTTGGAGTG | 55 |
Restriction sites are underlined.
Tm, melting temperature.
In-frame gene deletion plasmids were constructed by either an inverse PCR strategy, using pNK-B1234 as a DNA template, or by direct cloning of the target region into the pNK-B1 or pNK-B plasmid constructs digested with the PstI and SphI restriction enzymes. The pNK-B134, pNK-B123, pNK-B14, pNK-B13, and pNK-B12 plasmids were constructed using pNK-B1 as the DNA template. The primers used were PstI-enkB3-f and SphI-enkB4-r for pNK-B134, PstI-enkB2-f and SphI-enkB3-r for pNK123, PstI-enkB4-f and SphI-enkB4-r for pNK-B14, PstI-enkB3-f and SphI-enkB3-r for pNK-B13, and PstI-enkB2-f and SphI-enkB2-r for pNK-B12. The resulting PCR products were individually digested with the PstI and SphI restriction enzymes and ligated to the PstI- and SphI-digested pNK-B1 plasmid. The pNK-B234, pNK-B34, pNK-B23, pNK-B4, pNK-B3, and pNK-B2 plasmids were constructed by cloning the target regions amplified by PCR into the pNK-B plasmid construct. The primers used to amplify the target regions were PstI-enkB2-f and SphI-enkB4-r for pNK-B234, PstI-enkB3-f and SphI-enkB4-r for pNK-B34, PstI-enkB2-f and SphI-enkB3-r for pNK-B23, PstI-enkB4-f and SphI-enkB4-r for pNK-B4, PstI-enkB3-f and SphI-enkB3-r for pNK-B3, and PstI-enkB2-f and SphI-enkB2-r for pNK-B2. The resulting PCR products were individually digested with the PstI and SphI restriction enzymes and subsequently ligated to the PstI- and SphI-digested pNK-B plasmid.
An inverse PCR strategy was employed to obtain the pNK-B124 plasmid. The outward-facing primers Inv-enkB3-f and Inv-enkB3-r were designed to generate a PCR product of the entire plasmid without enkB3 using pNK-B1234 as the DNA template. On the other hand, in order to obtain the pNK-B24 plasmid, another inverse PCR was performed with the pNK-B124 plasmid construct as the DNA template using the same set of primers. The PCR products were then treated with a kinase (T4 polynucleotide kinase; Toyobo, Osaka, Japan) in the kinase buffer (Toyobo) and subsequently self-ligated using Ligation high reagent (Toyobo). All plasmid constructs were transformed in E. coli DH5α for plasmid storage and in E. faecalis JH2-2, L. lactis IL1403, L. lactis NZ9000, and L. lactis ATCC 19435T for expression analysis.
Complementation assay.
Complementation of the lacking gene in the enkB gene cluster was done with a plasmid containing the nisin-inducible promoter (Pnis), pIL-Pnis, originally modified from pIL253 (38). Each lacking gene was cloned behind the Pnis promoter into the pIL-Pnis plasmid and introduced into L. lactis NZ9000 that harbored the single-gene-deletion plasmid pNK-BΔX. NZ9000 was used as the host in the complementation assay because it has a native nisRK gene in its genome, which is essential for the nisin induction system. Overnight cultures of complemented strains were inoculated into fresh GM17 medium at a dilution of 1:100, grown to an OD600 of 0.4, and induced with nisin at a final concentration of 5 ng/ml. After an overnight incubation, cell-free supernatants were subsequently tested for Ent53B production using the spot-on-lawn assay described previously (39).
Bacteriocin production and immunity assay.
The ability of recombinant E. faecalis JH2-2 strains expressing different enkB gene cluster combinations to produce mature Ent53B was evaluated using the colony overlay assay, as described previously (3), but with a slight modification. The bacteriocin activity of the cell-free supernatant of recombinant strains was quantified by a spot-on-lawn assay described previously (39), using L. lactis ATCC 19435T as an indicator strain. Briefly, 10 μl of the cell-free supernatant was spotted onto a double-layered agar plate, which contained 5 ml of Lactobacilli Agar AOAC medium inoculated with an overnight culture of an indicator strain as the upper layer and 10 ml of the MRS broth supplemented with 1.5% agar as the bottom layer. After an overnight incubation at 30°C, the bacterial lawns were checked and inhibition zones measured.
The self-immunity assay was performed by determining the MIC of Ent53B against recombinant E. faecalis JH2-2 strains expressing different enkB gene cluster combinations. The MICs were determined by the spot-on-lawn assay, as described previously (39), using serial dilutions of purified Ent53B at known concentrations and utilizing recombinant strains as the indicator bacteria. Immunity was recorded as a strain-specific change in the MIC relative to the MIC observed in the strain expressing the control vector.
Computer analysis of DNA sequence.
Putative open reading frames, isoelectric points, molecular weights, and sequence alignments were identified and analyzed using the GENETYX-WIN software, version 8.0.1 (Genetyx, Tokyo, Japan). Homology comparisons were done using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/). Putative transmembrane helices were identified using the SOSUI prediction system (http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html).
Nucleotide sequence accession number.
The nucleotide sequence of the enkB gene cluster has been deposited in the DDBJ database under the accession number LC068607.
RESULTS
Sequence analysis of the region encoding the Ent53B locus.
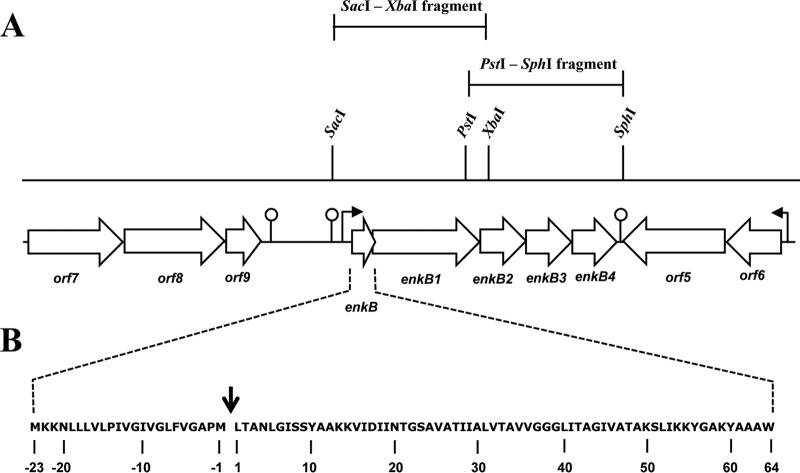
Ent53B is one of the five bacteriocin peptides produced by the NKR-5-3 strain. However, its structural gene, enkB, is located at a locus which is different from the rest of the NKR-5-3 enterocin genes (36). We sequenced the 9-kb region surrounding enkB and found that it contains at least 10 open reading frames, including enkB (Fig. 1A). A putative promoter was identified upstream of the enkB gene, with its putative terminator downstream of the enkB4 gene. This finding suggested that the five genes, enkB, enkB1, enkB2, enkB3, and enkB4, comprise one polycistronic message. This was confirmed when we carried out reverse transcription-PCR (RT-PCR) with different sets of primers to detect PCR products that cover intergenic regions within the enkB through enkB4 genes (data not shown). The enkB gene cluster is tightly organized. The enkB and enkB1 genes have a 26-bp overlap, which indicates that their expression is regulated by translational coupling. The significance of this phenomenon is discussed below.
FIG 1.
Organization of the enkB biosynthetic locus and surrounding flanking regions in E. faecium NKR-5-3. The open arrows indicate the genes and their orientations. The bent arrows show the putative promoter. The circles represent putative terminators. (A) Positions of the SacI-XbaI and PstI-SphI cloning fragment units. (B) The deduced enkB translation product is shown with the cleavage site of the leader peptide, indicated by the arrow.
Possible functions and identities of the deduced protein sequences from these putative genes were obtained by BLAST database search and directly compared to previously described proteins involved in circular bacteriocin biosynthesis (Table 3). The orientation of the enkB gene locus resembled that of similar loci of the circular bacteriocins circularin A (4) and uberolysin (27). Moreover, a direct comparison shows that protein products of the enkB genes, EnkB1 to EnkB4, appear to be equivalent to CirBCDE of circularin A and UblBCDE of uberolysin, as they showed some degree of similarity (Table 3). Furthermore, EnkB3, like its circularin A (CirD) and uberolysin (UblD) counterparts, contains an ATP-binding domain (Fig. 2). Open reading frame 5 (Orf5) and Orf6 are homologues to the two-component regulatory system constituents histidine kinases (HKs) and response regulators (RRs), respectively. Orf5 contains a conserved domain that belongs to the HATPase_c superfamily, with up to 86% sequence identity with putative HKs of various E. faecalis and E. faecium strains, while Orf6 shares up to 31% sequence identity with RR proteins from many E. faecalis strains.
TABLE 3.
Characteristics of the putative genes in the enterocin NKR-5-3B gene cluster
| ORFa | Size (bp) | Deduced protein characteristic |
||||
|---|---|---|---|---|---|---|
| Size (kDa) | pI | TMb | Homologue(s) (% identity)c | Putative function | ||
| orf7 | 711 | 26.0 | 8.97 | 6 | Arsenic resistance protein (83) | |
| orf8 | 1,083 | 39.6 | 9.53 | 10 | Arsenic resistance protein complex (84) | |
| orf9 | 408 | 15.0 | 4.59 | 0 | Arsenate reductase (98) | |
| enkB | 261 | 8.7 | 10.49 | 0 | CirA (20), UblA (30) | Ent53B prepeptide |
| enkB1 | 1,185 | 45.9 | 10.31 | 10 | CirB (17), UblB (17) | Secretion/immunity |
| enkB2 | 519 | 21.6 | 9.40 | 6 | CirC (12), UblC (21) | Maturation |
| enkB3 | 591 | 22.1 | 5.08 | 0 | CirD (26), UblD (29) | Secretion/immunity |
| enkB4 | 498 | 18.6 | 9.79 | 5 | CirE (41), UblE (18) | Immunity |
| orf5 | 1,311 | 50.8 | 8.55 | 6 | Histidine kinases (up to 86) | |
| orf6 | 573 | 21.6 | 9.55 | 5 | Response regulators (up to 31) | |
ORF, open reading frame.
TM, number of putative transmembrane helices, as predicted using the SOSUI prediction program.
Homologues were identified using a BLAST search against the NCBI protein database or by direct comparison to proteins involved in known circular bacteriocin production.
FIG 2.
Comparative alignment of EnkB3 and other ATP-binding proteins involved in the synthesis of other circular bacteriocins: enterocin AS-48 (AS-48D), circularin A (CirD), and uberolysin (UblD). Identical residues and conserved regions are indicated by asterisks and periods, respectively. Gaps in residues are indicated by dashes to improve alignment visualization. Walker A, Walker B, and ABC-transporter (ABC-trans.) signature conserved motifs are indicated accordingly.
Based on the result of the homology analysis, Orf7, Orf8, and Orf9 most likely do not participate in the biosynthesis of EnkB. Both Orf7 and Orf8 demonstrated up to 83% sequence identity with the arsenic resistance protein complex in various bacteria, such as E. faecalis, Listeria innocua, L. lactis, and Bacillus subtilis. These proteins belong to the arsenical resistance-3 (ACR3) family of arsenite efflux pumps that play a role in inorganic ion transport and metabolism, whereas Orf9 showed 98% identity to arsenate reductase proteins of some E. faecalis strains.
The Ent53B precursor peptide is an 87-amino-acid polypeptide encoded by enkB (Fig. 1B). It contains a long leader peptide consisting of 23 amino acid residues, which is longer than a similar leader peptide sequence in its circular bacteriocin homologues circularin A (3 amino acids) and uberolysin (6 amino acids).
Genes responsible for the production and maturation of Ent53B.
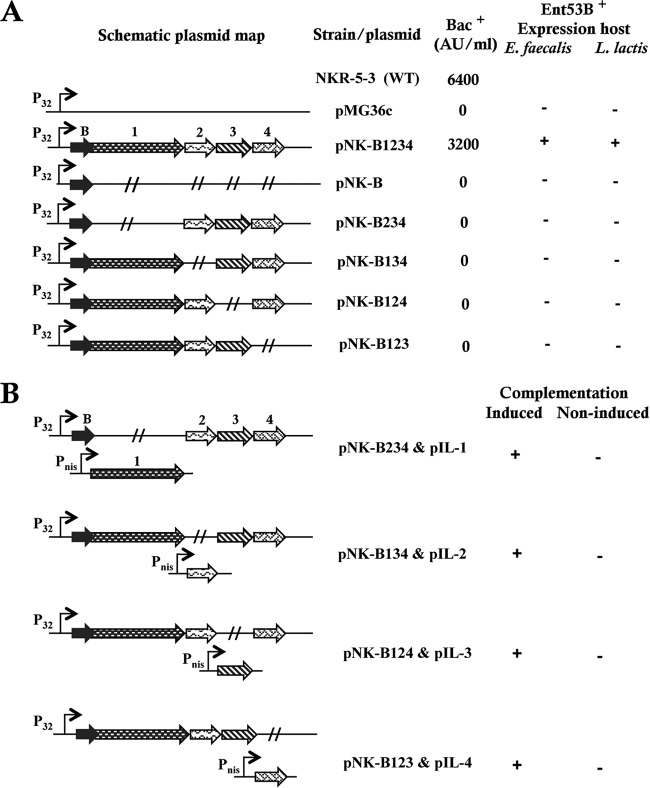
In an effort to determine which genes in the enkB gene cluster are involved in the production and maturation of Ent53B, plasmids containing deletions of the individual genes enkB1, enkB2, enkB3, and enkB4 were constructed using the wide-host-range cloning vector pMG36c, which carries a powerful constitutive lactococcal promoter, P32 (40). The production of the mature circular bacteriocin Ent53B was observed only in E. faecalis and L. lactis mutants harboring the pNK-B1234 plasmid, which expressed all five genes, enkB, enkB1, enkB2, enkB3, and enkB4, as determined by the spot-on-lawn assay (Fig. 3A). This result suggests that the production and secretion of the NKR-5-3B enterocin require the cooperative function of the products of all these genes. It is noteworthy that the antimicrobial activity of E. faecium NKR-5-3 (wild type [WT]) is higher than that of the heterologous expression strain, since the WT strain produces other bacteriocins in addition to the enterocin NKR-5-3B (34).
FIG 3.
Identification of genes responsible for the production and maturation of Ent53B. (A) Schematic representation of constructed plasmids and their productivity of Ent53B. Each plasmid was cloned and expressed in E. faecalis (JH2-2) and L. lactis (ATCC 19435T, IL1403, and NZ9000) host strains. Bacteriocin (Bac) production by E. faecium NKR-5-3 (WT) and E. faecalis JH2-2 mutants was also quantified in arbitrary units (AU) per milliliter. Ent53B production by each mutant was visualized by the colony overlay assay using L. lactis ATCC 19435T as an indicator strain, and plus and minus signs indicate positive and negative production, respectively (see also Fig. 4). (B) Schematic representation of complementation of deleted genes using the inducible pIL-Pnis plasmid. Ent53B production (complementation) was visualized and denoted as described above.
Complementation of the lacking gene using the nisin-inducible plasmid pIL-Pnis enabled all the nonproducing recombinants to produce the mature Ent53B bacteriocin in the presence but not in the absence of the inducing factor nisin A (Fig. 3B). This result also confirms that the production of the mature Ent53B bacteriocin requires the cooperative function of the proteins encoded by these genes.
Self-immunity to Ent53B.
In order to examine the self-immunity mechanisms against the inhibitory action of Ent53B, various combinations of the enkB gene cluster were cloned using pMG36c as a cloning vector. The resulting recombinants were assayed for their immunity to Ent53B. Only the clones expressing enkB4 (pNK-B1234, pNK-B4, pNK-B34, pNK-B24, pNK-B14, pNK-B234, pNK-B134, and pNK-B124) and/or a combination of enkB1 and enkB3 plasmids (pNK-B13 and pNK-B123) showed immunity to the inhibitory action of Ent53B. E. faecalis JH2-2 is generally sensitive to Ent53B, with an MIC of 0.826 μM. The vector control pMG36c strain showed no change in MIC relative to the WT JH2-2 strain. However, the presence of enkB4 and/or a combination of enkB1- and enkB3-expressing plasmids conferred strong immunity to Ent53B, as was deduced from the 512-fold and 256-fold increases in the corresponding MICs (Table 4). Recombinant strains, which expressed neither enkB4 nor a combination of enkB1 and enkB3, exhibited MICs comparable to those seen in the control strains. The strains, which expressed only enkB1 or enkB3, did not possess any immunity to Ent53B. This suggests that the respective gene products, EnkB1 and EnkB3, form a protein complex that can confer immunity to the host.
TABLE 4.
Self-immunity of the recombinant strains expressing different gene combinations to enterocin NKR-5-3B
| Strain or relevant plasmida | MIC (μM)b | Immunityc |
|---|---|---|
| E. faecalis JH2-2 | 0.826 | − |
| pMG36c (vector control) | 0.826 | − |
| pNK-B | 0.826 | − |
| pNK-B4 | >423 | ++ |
| pNK-B3 | 0.826 | − |
| pNK-B2 | 1.652 | − |
| pNK-B1 | 0.826 | − |
| pNK-B34 | >423 | ++ |
| pNK-B24 | >423 | ++ |
| pNK-B23 | 0.826 | − |
| pNK-B14 | >423 | ++ |
| pNK-B13 | 211.5 | + |
| pNK-B12 | 0.826 | − |
| pNK-B234 | >423 | ++ |
| pNK-B134 | >423 | ++ |
| pNK-B124 | >423 | ++ |
| pNK-B123 | 211.5 | + |
| pNK-B1234 | >423 | ++ |
E. faecalis JH2-2 was used as a host strain. Strains carrying the indicated plasmids were assayed as described in Materials and Methods.
MIC for the indicated strain or the strain expressing the indicated plasmid. The highest concentration of Ent53B used was 423 μM.
++, +, and − represent strong immunity, moderate immunity, and no immunity, respectively.
DISCUSSION
Ent53B is a novel circular bacteriocin recently isolated from E. faecium NKR-5-3, a strain also known to produce several other bacteriocins (33–35). Structural characterization and NMR three-dimensional (3D) solution structure analysis of Ent53B revealed the presence of a stable circular backbone with four helical segments, which enclose a tightly packed hydrophobic core (33). In this study, we performed a genetic characterization of the E. faecium NKR-5-3 bacteriocin cluster, which is responsible for the production, secretion, maturation, and self-immunity of Ent53B. Production of the mature circular Ent53B appears to require the cooperative function of five genes, namely, enkB, enkB1, enkB2, enkB3, and enkB4. When any of these five genes were absent, heterologous hosts did not produce the mature circular Ent53B, whereas complementation of the missing gene restored their ability to synthesize this bacteriocin. Similar observations were made in the case of the CirA bacteriocin, which coincidentally has a genetic organization similar to that of Ent53B. Heterologous production of the mature CirA was possible only when the host expressed all five genes, cirA to cirE (4). However, in the absence of CirE, the immunity protein, CirA mutants failed to exhibit a phenotype (4), whereas it was relatively easy to observe a phenotype in the recombinant pNK-B123 strain, which lacked Ent53B4 (Fig. 3A). Furthermore, the growth of pNK-B123 was comparable to that of other recombinant strains (data not shown), which indicated that the growth was not affected by stress and, therefore, mature Ent53B was obviously not produced by this strain. Taken together, these results suggest that protein products of genes enkB, enkB1, enkB2, enkB3, and enkB4 form a protein complex that cooperatively mediates Ent53B processing and its transport into the extracellular space. It is still unclear, however, whether the cleavage of the leader peptide, the N- and C-terminal head-to-tail circularization, and secretion to the extracellular space are interlinked phenomena. In this regard, it was suggested recently that the head-to-tail circularization and the secretion of leucocyclicin Q are separate processes (41). In addition, it was reported recently that the cleavage of the garvicin ML leader peptide and bacteriocin circularization are separate processes, in which the bacteriocin circularization precedes cleavage of the garvicin ML leader peptide (42). It should be noted, however, that both leucocyclicin Q and garvicin ML have short leader peptide sequences (only 2 and 3 amino acids long, respectively), whereas Ent53B has a longer (23-amino-acid) leader peptide sequence. The role of the leader peptide in the maturation (i.e., cleavage and posttranslational modification) and transport of bacteriocins has been well established (43–46).
The structure of the enkB gene cluster is tightly organized: the enkB and enkB1 genes have a 26-bp overlap. The control of translation of the majority of overlapped genes is usually coupled to the translation of neighboring genes. During this process, known as translation coupling, the terminating ribosomes may be able to reinitiate at the start codon in the overlapping stop codon of an upstream cistron (47, 48). It has been suggested that translation coupling also mediates biosynthesis of CirA (4) and garvicin ML (42).
A distinct feature of bacteriocin-producing strains is their ability to protect themselves from the antimicrobial action of their own bacteriocins (5). In this study, we showed that at least two independent mechanisms confer self-immunity against the antimicrobial action of Ent53B. The first mechanism is the expression of the EnkB4 protein. It contains at least 5 putative transmembrane helices and shares relatively high identity with CirE (41%) and UblE (18%), which confer immunity to the circular bacteriocins circularin A and uberolysin, respectively. Similar to other immunity-conferring proteins, EnkB4 is relatively small and has a high isoelectric point. The second mechanism of self-immunity is mediated by the coexpression of the protein products of the enkB1 and enkB3 genes. EnkB1 and EnkB3 most likely form an energy-requiring ABC transporter protein complex. It is plausible that within this complex, EnkB1 functions as a membrane-bound transporter, while soluble EnkB3 provides the ATP-binding domain. We believe that the self-protection provided by these two proteins is achieved by active pumping of bacteriocin molecules out from the producing cells, although final confirmation of this hypothesis needs further experimental verification. This kind of mechanism has been described for the majority of bacteriocin immunity ABC transporters (49), such as NisFEG of nisin (50) and NukFEG of nukacin ISK-1 (51).
The cooperative nature of EnkB1 and EnkB3 in providing immunity supports the above-mentioned suggestion that these biosynthetic enzymes form a protein complex. Furthermore, proteins conferring immunity to most bacteriocins usually do not play a role in their biosynthesis (1, 9, 11). However, as we discuss above, EnkB4 affects Ent53B maturation, which is a relatively quick process because the precursor peptide is highly susceptible to protease digestion. Notably, we did not observe any intracellular accumulation of the Ent53B precursor in recombinant strains, which failed to produce Ent53B. In our opinion, this observation supports the notion that biosynthetic enzymes form a protein complex. Otherwise, the precursor peptide would be digested before it could be processed to yield the mature bacteriocin molecule.
These two mechanisms of self-protection were also observed in the case of the CirA bacteriocin, in which immunity was conferred by both a dedicated immunity protein (CirE) and an ABC transporter protein complex (CirBD) (4). It should be noted that the Ent53B and CirA proteins share 20% sequence identity, so it is not surprising that the organization of genetic determinants also shows high similarity. This led us to think that CirA genetic determinants might also mediate the processing of Ent53B and vice versa. After all, immunity determinants of class IIa bacteriocins confer cross-immunity to their cognate bacteriocins (52, 53). Unfortunately, our experimental results did not support this hypothesis. The immunity-conferring components of Ent53B did not protect respective Ent53B-producing recombinant strains against the antimicrobial action of CirA after treatment with the cell-free supernatant from a CirA producer strain (data not shown). Furthermore, the genetic determinants of Ent53B did not produce mature CirA when enkB was replaced with the CirA structural gene (data not shown). These results highlight the fact that our understanding of the regulation and functions of antibiotic proteins from the circular bacteriocin family is still very limited.
Successful heterologous expression of the circular bacteriocins AS-48 (54), CirA (4), gassericin A (55), and carnocyclin A (56) has already been reported. However, heterologous expression of these circular bacteriocins was successful only in the same host or when limited to host species of same genus. The full expression of AS-48 was achieved only in hosts belonging to the genus Enterococcus (54), whereas the heterologous expression of CirA was possible only in E. faecalis, not in L. lactis, despite many attempts (4). Here, we report the successful establishment of a heterologous expression system of Ent53B in host strains belonging to different genera (Enterococcus and Lactococcus) (Fig. 4). This result rules out the possibility of some machinery specific only to the Enterococcus genus for the maturation and secretion of Ent53B. Furthermore, this result underlines the cooperative function of protein products of the enkB, enkB1, enkB2, enkB3, and enkB4 genes to produce the mature circular Ent53B. This result has a very important practical implication, since some enterococcal strains are opportunistic pathogens, whereas other strains carry multiple antibiotic resistance genes and exhibit some virulent traits (57–59). Our data will help widen the future applicability of Ent53B in the food fermentation process. This can be achieved by cloning the enkB biosynthetic gene cluster into host strains devoid of possible potential virulence factors or pathogenic traits. Such strains could subsequently be used as defined starter, adjunct, or protective culture in food fermentation processes.
FIG 4.
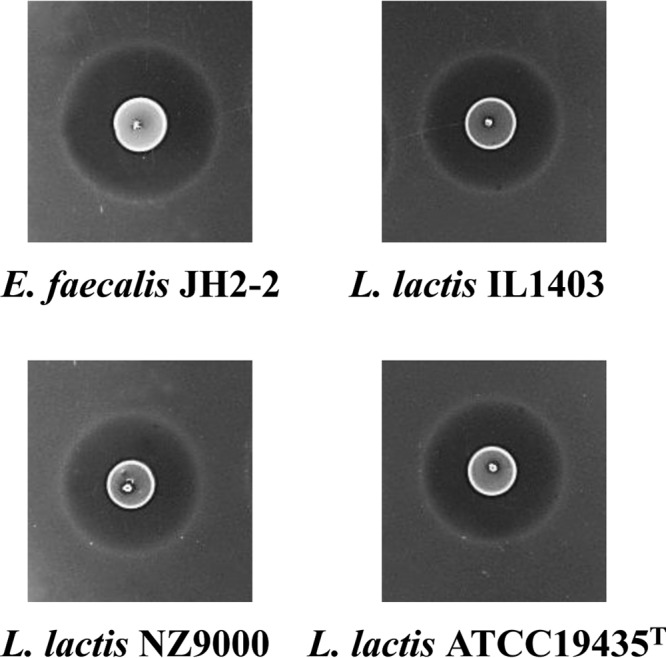
Heterologous production of enterocin NKR-5-3B through the pNK-B plasmid expressed in E. faecalis JH2-2 and L. lactis ATCC 19435T, IL1403, and NZ9000 host strains. Production was visualized using a colony overlay assay using L. lactis ATCC 19435T as an indicator strain.
In conclusion, in the present study, we identified genes required for the production of the Ent53B bacteriocin and genes involved in two mechanisms of bacterial self-protection against Ent53B. We also successfully expressed Ent53B in different heterologous host strains. Our results open new possibilities for the use of Ent53B in the food industry.
ACKNOWLEDGMENTS
We thank the JSPS-National Research Council of Thailand (NRCT) Core University Program on the Development of Thermotolerant Microbial Resources and Their Applications. We also thank the Ministry of Education, Culture, Sports, Science and Technology of Japan for providing a scholarship grant to Rodney H. Perez.
We acknowledge the generosity of Oscar Kuipers (Molecular Genetics Laboratory, Groningen University, the Netherlands), who provided the pIL-Pnis plasmid (a plasmid derivative of Zirex [60]).
Funding Statement
These funders supported this study.
REFERENCES
- 1.Nes IF, Yoon SS, Diep DB. 2007. Ribosomally synthesized antimicrobial peptides (bacteriocins) in lactic acid bacteria: a review. Food Sci Biotechnol 16:675–690. [Google Scholar]
- 2.De Vuyst L, Leroy F. 2007. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol 13:194–199. doi: 10.1159/000104752. [DOI] [PubMed] [Google Scholar]
- 3.Kemperman R, Kuipers A, Karsens H, Nauta A, Kuipers O, Kok J. 2003. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl Environ Microbiol 69:1589–1597. doi: 10.1128/AEM.69.3.1589-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemperman R, Jonker M, Nauta A, Kuipers OP, Kok J. 2003. Functional analysis of the gene cluster involved in production of the bacteriocin circularin A by Clostridium beijerinckii ATCC 25752. Appl Environ Microbiol 69:5839–5848. doi: 10.1128/AEM.69.10.5839-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riley MA, Wertz JE. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357–364. doi: 10.1016/S0300-9084(02)01421-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Hoover DG. 2003. Bacteriocins and their food applications. Compr Rev Food Sci Food Safety 2:82–100. doi: 10.1111/j.1541-4337.2003.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 7.Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. 2006. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez RH, Zendo T, Sonomoto K. 2014. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Fact 13:S3. doi: 10.1186/1475-2859-13-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klaenhammer TR. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12:39–85. doi: 10.1016/0168-6445(93)90057-G. [DOI] [PubMed] [Google Scholar]
- 10.Maqueda M, Sanchez-Hidalgo M, Fernandez M, Montalban-Lopez M, Valdivia E, Martinez-Bueno M. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol Rev 32:2–22. doi: 10.1111/j.1574-6976.2007.00087.x. [DOI] [PubMed] [Google Scholar]
- 11.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 12.Nissen-Meyer J, Nes IF. 1997. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol 167:67–77. doi: 10.1007/s002030050418. [DOI] [PubMed] [Google Scholar]
- 13.McAuliffe O, Ross RP, Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev 25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 14.Dufour A, Hindre T, Haras D, Le Pennec JP. 2007. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol Rev 31:134–167. doi: 10.1111/j.1574-6976.2006.00045.x. [DOI] [PubMed] [Google Scholar]
- 15.van Belkum MJ, Stiles ME. 2000. Nonlantibiotic antibacterial peptides from lactic acid bacteria. Nat Prod Rep 17:323–335. doi: 10.1039/a801347k. [DOI] [PubMed] [Google Scholar]
- 16.Eijsink VG, Axelsson L, Diep DB, Håvarstein LS, Holo H, Nes IF. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Van Leeuwenhoek 81:639–654. doi: 10.1023/A:1020582211262. [DOI] [PubMed] [Google Scholar]
- 17.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol 19:411–418. doi: 10.1016/j.tim.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Conlan BF, Gillon AD, Craik DJ, Anderson MA. 2010. Circular proteins and mechanisms of cyclization. Biopolymers 94:573–583. doi: 10.1002/bip.21422. [DOI] [PubMed] [Google Scholar]
- 19.Masuda Y, Zendo T, Sonomoto K. 2012. New type of non-lantibiotic bacteriocins: circular and leaderless bacteriocins. Benef Microbes 3:3–12. doi: 10.3920/BM2011.0047. [DOI] [PubMed] [Google Scholar]
- 20.Craik DJ, Cemazar M, Daly NL. 2006. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Devel 9:251–260. [PubMed] [Google Scholar]
- 21.Craik DJ, Simonsen S, Daly NL. 2002. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Devel 5:251–260. [PubMed] [Google Scholar]
- 22.Gálvez A, Maqueda M, Valdivia E, Quesada A, Montoya E. 1986. Characterization and partial purification of a broad spectrum antibiotic AS-48 produced by Streptococcus faecalis. Can J Microbiol 32:765–771. doi: 10.1139/m86-141. [DOI] [PubMed] [Google Scholar]
- 23.Samyn B, Martinez-Bueno M, Devreese B, Marqueda M, Galvez A, Valvidia E, Coyette J, Van Beeumen J. 1994. The cyclic structure of the enterococcal peptide AS-48. FEBS Lett 352:87–90. [DOI] [PubMed] [Google Scholar]
- 24.Kawai Y, Saito T, Kitazawa H, Itoh T. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci Biotechnol Biochem 62:2438–2440. doi: 10.1271/bbb.62.2438. [DOI] [PubMed] [Google Scholar]
- 25.Kawulka K, Sprules T, McKay RT, Mercier P, Diaper CM, Zuber P, Vederas JC. 2003. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J Am Chem Soc 125:4726–4727. doi: 10.1021/ja029654t. [DOI] [PubMed] [Google Scholar]
- 26.Kalmokoff ML, Cyr TD, Hefford MA, Whitford MF, Teather RM. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can J Microbiol 49:763–773. doi: 10.1139/w03-101. [DOI] [PubMed] [Google Scholar]
- 27.Wirawan RE, Swanson KM, Kleffmann T, Jack RW, Tagg JR. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630. doi: 10.1099/mic.0.2006/005967-0. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl Environ Microbiol 74:4756–4763. doi: 10.1128/AEM.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawa N, Zendo T, Kiyofuji J, Fujita K, Himeno K, Nakayama J, Sonomoto K. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl Environ Microbiol 75:1552–1558. doi: 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrero J, Brede DA, Skaugen M, Diep DB, Herranz C, Nes IF, Cintas LM, Hernandez PE. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl Environ Microbiol 77:369–373. doi: 10.1128/AEM.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda Y, Ono H, Kitagawa H, Ito H, Mu F, Sawa N, Zendo T, Sonomoto K. 2011. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl Environ Microbiol 77:8164–8170. doi: 10.1128/AEM.06348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acedo JZ, van Belkum MJ, Lohans CT, McKay RT, Miskolzie M, Vederas JC. 2015. Solution structure of acidocin B, a circular bacteriocin produced by Lactobacillus acidophilus M46. Appl Environ Microbiol 81:2910–2918. doi: 10.1128/AEM.04265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himeno K, Rosengren KJ, Inoue T, Perez RH, Colgrave ML, Lee HS, Chan LY, Henriques ST, Fujita K, Ishibashi N, Zendo T, Wilaipun P, Nakayama J, Leelawatcharamas V, Jikuya H, Craik DJ, Sonomoto K. 2015. Identification, characterization, and the three-dimensional structure of the novel circular bacteriocin, enterocin NKR-5-3B, from Enterococcus faecium. Biochemistry 54:4863–4876. [DOI] [PubMed] [Google Scholar]
- 34.Ishibashi N, Himeno K, Fujita K, Masuda Y, Perez RH, Zendo T, Wilaipun P, Leelawatcharamas V, Nakayama J, Sonomoto K. 2012. Purification and characterization of multiple bacteriocins and an inducing peptide produced by Enterococcus faecium NKR-5-3 from Thai fermented fish. Biosci Biotechnol Biochem 76:947–953. doi: 10.1271/bbb.110972. [DOI] [PubMed] [Google Scholar]
- 35.Perez RH, Himeno K, Ishibashi N, Masuda Y, Zendo T, Fujita K, Wilaipun P, Leelawatcharamas V, Nakayama J, Sonomoto K. 2012. Monitoring of the multiple bacteriocin production by Enterococcus faecium NKR-5-3 through a developed liquid chromatography and mass spectrometry-based quantification system. J Biosci Bioeng 114:490–496. doi: 10.1016/j.jbiosc.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Ishibashi N, Himeno K, Masuda Y, Perez RH, Iwatani S, Zendo T, Wilaipun P, Leelawatcharamas V, Nakayama J, Sonomoto K. 2014. Gene cluster responsible for secretion of and immunity to multiple bacteriocins, the NKR-5-3 enterocins. Appl Environ Microbiol 80:6647–6655. doi: 10.1128/AEM.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Simon D, Chopin A. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 39.Ennahar S, Asou Y, Zendo T, Sonomoto K, Ishizaki A. 2001. Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. Int J Food Microbiol 70:291–301. doi: 10.1016/S0168-1605(01)00565-7. [DOI] [PubMed] [Google Scholar]
- 40.Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol 117:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu F, Masuda Y, Zendo T, Ono H, Kitagawa H, Ito H, Nakayama J, Sonomoto K. 2014. Biological function of a DUF95 superfamily protein involved in the biosynthesis of a circular bacteriocin, leucocyclicin Q. J Biosci Bioeng 117:158–164. [DOI] [PubMed] [Google Scholar]
- 42.Gabrielsen C, Brede DA, Salehian Z, Nes IF, Diep DB. 2014. Functional genetic analysis of the GarML gene cluster in Lactococcus garvieae DCC43 gives new insights into circular bacteriocin biosynthesis. J Bacteriol 196:911–919. doi: 10.1128/JB.01115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oman TJ, van der Donk WA. 2010. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol 6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abts A, Montalban-Lopez M, Kuipers OP, Smits SH, Schmitt L. 2013. NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52:5387–5395. doi: 10.1021/bi4008116. [DOI] [PubMed] [Google Scholar]
- 45.Kuipers A, de Boef E, Rink R, Fekken S, Kluskens LD, Driessen AJ, Leenhouts K, Kuipers OP, Moll GN. 2004. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J Biol Chem 279:22176–22182. doi: 10.1074/jbc.M312789200. [DOI] [PubMed] [Google Scholar]
- 46.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJ, Kuipers OP, Moll GN. 2007. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl Environ Microbiol 73:1792–1796. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstel B, McCarthy JE. 1989. Independent and coupled translational initiation of atp genes in Escherichia coli: experiments using chromosomal and plasmid-borne lacZ fusions. Mol Microbiol 3:851–859. doi: 10.1111/j.1365-2958.1989.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 48.McCarthy JE, Gualerzi C. 1990. Translational control of prokaryotic gene expression. Trends Genet 6:78–85. doi: 10.1016/0168-9525(90)90098-Q. [DOI] [PubMed] [Google Scholar]
- 49.Peschel A, Gotz F. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J Bacteriol 178:531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ra R, Beerthuyzen MM, de Vos WM, Saris PE, Kuipers OP. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227–1233. doi: 10.1099/13500872-145-5-1227. [DOI] [PubMed] [Google Scholar]
- 51.Aso Y, Okuda K, Nagao J, Kanemasa Y, Thi Bich Phuong N, Koga H, Shioya K, Sashihara T, Nakayama J, Sonomoto K. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci Biotechnol Biochem 69:1403–1410. doi: 10.1271/bbb.69.1403. [DOI] [PubMed] [Google Scholar]
- 52.Fimland G, Eijsink VG, Nissen-Meyer J. 2002. Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology 148:3661–3670. doi: 10.1099/00221287-148-11-3661. [DOI] [PubMed] [Google Scholar]
- 53.Johnsen L, Fimland G, Nissen-Meyer J. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 280:9243–9250. doi: 10.1074/jbc.M412712200. [DOI] [PubMed] [Google Scholar]
- 54.Fernández M, Martinez-Bueno M, Martin MC, Valdivia E, Maqueda M. 2007. Heterologous expression of enterocin AS-48 in several strains of lactic acid bacteria. J Appl Microbiol 102:1350–1361. doi: 10.1111/j.1365-2672.2006.03194.x. [DOI] [PubMed] [Google Scholar]
- 55.Ito Y, Kawai Y, Arakawa K, Honme Y, Sasaki T, Saito T. 2009. Conjugative plasmid from Lactobacillus gasseri LA39 that carries genes for production of and immunity to the circular bacteriocin gassericin A. Appl Environ Microbiol 75:6340–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2010. Cloning and characterization of the gene cluster involved in the production of the circular bacteriocin carnocyclin A. Probiotics Antimicrob Proteins 2:218–225. [DOI] [PubMed] [Google Scholar]
- 57.Ben Omar N, Castro A, Lucas R, Abriouel H, Yousif NMK, Franz CMAP, Holzapfel WH, Ruben P-P, Martinez-Canãmero M, Galvez A. 2004. Functional and safety aspects of enterococci isolated from different Spanish foods. Syst Appl Microbiol 27:118–130. doi: 10.1078/0723-2020-00248. [DOI] [PubMed] [Google Scholar]
- 58.Franz CM, Stiles ME, Schleifer KH, Holzapfel WH. 2003. Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122. doi: 10.1016/S0168-1605(03)00174-0. [DOI] [PubMed] [Google Scholar]
- 59.Franz CMAP, Muscholl-Silberhorn AB, Yousif NMK, Vancanneyt M, Swings J, Holzapfel WH. 2001. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl Environ Microbiol 67:4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mu D, Montalbán-López M, Masuda Y, Kuipers OP. 2013. Zirex: a novel zinc-regulated expression system for Lactococcus lactis. Appl Environ Microbiol 79:4503–4508. doi: 10.1128/AEM.00866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolotin A, Mauger S, Malarme K, Ehrlich SD, Sorokin A. 1999. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Van Leeuwenhoek 76:27–76. doi: 10.1023/A:1002048720611. [DOI] [PubMed] [Google Scholar]
- 62.de Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol 178:3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]