ABSTRACT
Broad-spectrum O-linked protein glycosylation is well characterized in the major Neisseria species of importance to human health and disease. Within strains of Neisseria gonorrhoeae, N. meningitidis, and N. lactamica, protein glycosylation (pgl) gene content and the corresponding oligosaccharide structure are fairly well conserved, although intra- and interstrain variability occurs. The status of such systems in distantly related commensal species, however, remains largely unexplored. Using a strain of deeply branching Neisseria elongata subsp. glycolytica, a heretofore unrecognized tetrasaccharide glycoform consisting of di-N-acetylbacillosamine-glucose-di-N-acetyl hexuronic acid-N-acetylhexosamine (diNAcBac-Glc-diNAcHexA-HexNAc) was identified. Directed mutagenesis, mass spectrometric analysis, and glycan serotyping confirmed that the oligosaccharide is an extended version of the diNAcBac-Glc-based structure seen in N. gonorrhoeae and N. meningitidis generated by the successive actions of PglB, PglC, and PglD and glucosyltransferase PglH orthologues. In addition, a null mutation in the orthologue of the broadly conserved but enigmatic pglG gene precluded expression of the extended glycoform, providing the first evidence that its product is a functional glycosyltransferase. Despite clear evidence for a substantial number of glycoprotein substrates, the major pilin subunit of the endogenous type IV pilus was not glycosylated. The latter finding raises obvious questions as to the relative distribution of pilin glycosylation within the genus, how protein glycosylation substrates are selected, and the overall structure-function relationships of broad-spectrum protein glycosylation. Together, the results of this study provide a foundation upon which to assess neisserial O-linked protein glycosylation diversity at the genus level.
IMPORTANCE Broad-spectrum protein glycosylation systems are well characterized in the pathogenic Neisseria species N. gonorrhoeae and N. meningitidis. A number of lines of evidence indicate that the glycan components in these systems are subject to diversifying selection and suggest that glycan variation may be driven in the context of glycosylation of the abundant and surface-localized pilin protein PilE, the major subunit of type IV pili. Here, we examined protein glycosylation in a distantly related, nonpathogenic neisserial species, Neisseria elongata subsp. glycolytica. This system has clear similarities to the systems found in pathogenic species but makes novel glycoforms utilizing a glycosyltransferase that is widely conserved at the genus level but whose function until now remained unknown. Remarkably, PilE pilin is not glycosylated in this species, a finding that raises important questions about the evolutionary trajectories and overall structure-function relationships of broad-spectrum protein glycosylation systems in bacteria.
INTRODUCTION
Arrays of glycoconjugates varying in structure and function predominate at bacterial cell surfaces and extracytoplasmic milieus. Despite their prevalence and inferred significance, the forces driving glycan diversification and the underlying evolutionary principles are poorly understood. This situation is particularly applicable to protein glycosylation systems, many of which are only beginning to be identified. Delineating selection at the genetic level in such systems is confounded as carbohydrates are secondary gene products and because, in many instances, the biosynthetic pathways and the glycans themselves are incompletely defined. Many systems display robust inter- or intrastrain glycan variability, implying that such diversification is adaptive. For example, flagellin glycosylation systems in Clostridium species and Campylobacter jejuni display remarkable glycoform plasticity (1–3). Here and elsewhere, however, strict correlations between glycosylation genotypes and phenotypes have been difficult to identify. Likewise, attempts at reconciling the glycosylation gene repertoire with particular ecotypes and populations are challenging. Thus, there are significant gaps in our understanding of the long-term evolutionary dynamics and trends in these systems.
Knowledge of broad-spectrum (also known as general) bacterial protein glycosylation systems, in which numerous, diverse proteins are targeted for modification, is rapidly accumulating through the use of phylogenetic, immunological, structural, and glycoproteomic approaches. In particular, recent efforts have strived to address how these systems are evolving at or above the species level. For the N-linked protein glycosylation system of species within the genus Campylobacter, a high degree of conservation of a basic heptasaccharide glycan structure within species representing thermotolerant taxa has been documented (4). However, an unexpected diversity of N-glycan structures differing in length and composition was found in species comprising nonthermotolerant Campylobacter species. In a phylum-wide study of O-linked protein glycosylation within the Bacteroidetes, expression of the core glycan was conserved in all species, while synthesis of the outer glycan was divergent although maintained throughout (5). Using isolates of Acinetobacter baumannii, >10 distinct glycoforms associated with general O-linked protein glycosylation were identified (6). Hence, variation in glycan structures within a genus is well documented.
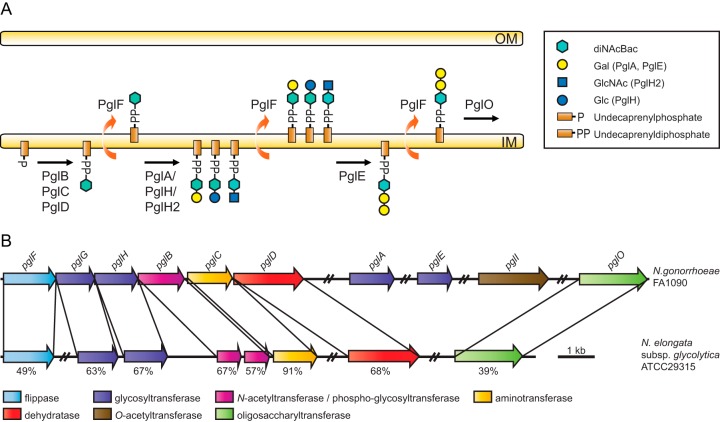
The O-linked protein glycosylation system expressed by highly related pathogenic species within the genus Neisseria, N. gonorrhoeae and N. meningitidis, share many overall features with those noted above and have some that are unique. Based on genetic and biochemical studies, the following model for the neisserial protein glycosylation pathway has emerged (7–11) (Fig. 1A). The PglD and PglC proteins carry out NAD+-dependent dehydratase and aminotransferase reactions, respectively, to convert UDP-GlcNAc to UDP-2-acetamido-4-amino-2,4,6-trideoxy-α-d-glucose (11). PglB acts as a bifunctional enzyme, which catalyzes the amino acetylation of UDP-2-acetamido-4-amino-2,4,6-trideoxy-α-d-glucose to form UDP-di-N-acetylbacillosamine (diNAcBac) and the transfer of the phospho-sugar to undecaprenyl phosphate (Und-P) (11). PglB2, an alternate enzyme to PglB, encoded by pglB2 alleles found in many N. meningitidis strains, contains a domain thought to transfer a glyceroyl moiety instead of an acetyl group to produce 4-glyceramido-2-acetamido-2,4,6-trideoxy-α-d-hexose (GATDH) (12). Further extension of these undecaprenyl diphosphate (Und-PP)-sugars occurs via two exclusive pathways involving distinct glycosyltransferases. One pathway involves PglH or its allelic variant PglH2 that adds a Glc or GlcNAc, respectively, to the Und-PP-sugars described recently (9, 10). A second pathway involves the PglA and PglE glycosyltransferases that add successive Gal units to the Und-PP-sugars (7, 13). The PglH/PglH2-generated Und-PP-disaccharides cannot be further modified by PglE (9, 10). Furthermore, strains expressing both PglA and PglH simultaneously express both glycoforms. Both di- and trisaccharide forms can also be modified via O-acetylation mediated by PglI (7, 10).
FIG 1.
(A) Current model of the broad-spectrum O-linked glycosylation pathway expressed by neisserial species. PglB, -C, and -D are required for the synthesis of UDP-N′-diacetylbacillosamine (UDP-diNAcBac) and transfer of the phospho-diNAcBac to undecaprenyl phosphate. This glycoform can be further modified by the glycosyltransferases PglA and PglH/PglH2. The PglE glycosyltransferase can modify only the disaccharide glycoform generated by PglA. See the key for sugars utilized by the glycosyltransferases. The flippase PglF and the oligosaccharyltransferase PglO have relaxed specificity such that they can utilize the mono-, di-, and trisaccharide undecaprenyl diphosphate glycoforms. In conjunction with phase-variable (on-off) expression of the glycosyltransferases, the latter properties enable the system to support glycan antigenic variation and microheterogeneity. The system can also accommodate the synthesis of undecaprenyl diphosphate-4-glyceramido-2-acetamido-2,4,6-trideoxy-α-d-hexose (Und-PP-GATDH) mediated by pglB2 alleles found in some meningococcal strains. OM, outer membrane; IM, inner membrane. (B) Comparative pgl gene content and synteny between N. elongata subsp. glycolytica strain ATCC 29315 and N. gonorrhoeae strain FA1090. The core pgl locus of N. elongata subsp. glycolytica encompasses pglG, pglH, pglC, and a split (two-ORF) version of pglB, whereas pglF and pglD are localized elsewhere in the genome. The pglO gene is also localized outside the core locus in N. gonorrhoeae and N. meningitidis as well as in N. elongata subsp. glycolytica, while the pglA, pglE, and pglI genes are not found in N. elongata subsp. glycolytica. The numbers indicate the levels of protein sequence identities (maximum identity as determined by BLASTp).
Some N. gonorrhoeae and N. meningitidis strains lack pglH due to an extensive deletion that encompasses its 5′ end and the 3′ end of a large open reading frame (ORF), known as orf2 (pglG), found upstream of it (13, 14). Like PglH, the predicted product of pglG is annotated as a glycosyltransferase with domains of CAZy families 1 and 4 (13). However, the function of pglG remains enigmatic, as no connections to any glycosylation phenotype have so far been established.
Finally, high levels of intra- and interstrain glycoform variation are observed, which are attributable to differences in pgl gene expression status (due to phase variation) and gene content, respectively. Thus, individual strains can express anywhere between 1 and 7 glycoforms ranging in size from mono- to trisaccharides, and within meningococcal strains, 18 distinct protein-targeted glycans have been defined (9). Furthermore, relatively high levels of microheterogeneity have been observed, which result from the expression of hypomorphic alleles of glycosyltransferase genes, thus altering the relative abundance of specific glycoforms (10, 15).
O-Linked protein glycosylation does not appear to be a classical virulence determinant in Neisseria as it is not differentially distributed among disease-associated and carriage strains of N. meningitidis and is also found in the nonpathogenic species N. lactamica (8). Nonetheless, the glycoform structure appears to be under positive, diversifying selection, and major shifts in pgl genotypes arising by horizontal gene transfer within otherwise stable clonal complexes have been documented (16, 17). With regard to glycosylation function, most phenotypic traits have been linked to the pilin protein subunit of the type IV pilus colonization factor, which itself often undergoes significant antigenic variation (18–20). That said, the status of such systems in distantly related commensal species remains largely unexplored. Here, the results of examining protein glycosylation in N. elongata subsp. glycolytica, a deeply branching species within the neisserial phylogeny, reveal a previously unseen glycoform, the first evidence that pglG encodes a functional glycosyltransferase, and, for the first time, a neisserial species in which pilin is not modified by its broad-spectrum glycosylation system.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains are described in Table S1 in the supplemental material and were derived from N. elongata subsp. glycolytica strain ATCC 29315. Strains were cultured on conventional GC plates, either by using GC agar Base (Oxoid) or as described previously (21), except that Thiotone E peptone was replaced with Proteose Peptone no. 3 (Difco). Mutants were obtained by transforming N. elongata subsp. glycolytica with crude cell lysates, PCR-amplified DNA, or purified plasmid DNA. The various constructs used are described below. Transformation of N. elongata subsp. glycolytica was performed by using a protocol described previously for N. gonorrhoeae (21), and mutants were selected on plates containing the appropriate antibiotics. The following antibiotics were used: kanamycin (50 μg/ml), tetracycline (20 μg/ml), and chloramphenicol (30 μg/ml).
Genome databases.
Genome analysis was performed by using sequences from the Bacterial Isolate Genome Sequence Database (22) and from the National Center for Biotechnology Information (RefSeq accession no. NZ_ADBF00000000.1).
Plasmid and strain construction.
Information about plasmid and strain construction can be found in Table S1 in the supplemental material.
Affinity purification of glycoproteins.
Purification of His-tagged proteins was performed as previously described (23).
In-gel protein digestion.
In-gel digestion of Coomassie-stained gel slices containing purified proteins with trypsin (Sigma) and AspN (Roche) was performed as previously described (24, 25). In-gel digestions with chymotrypsin (Sigma) were performed as follows: ∼30 μl of chymotrypsin (16 ng/μl) in chymotrypsin solution (100 mM Tris-HCl, 10 mM CaCl2 [pH 7.8]) was added to gel slices, and the slices were incubated on ice for ∼30 min. When gel slices were completely rehydrated, redundant chymotrypsin was removed, and 60 μl of a chymotrypsin solution was added before the gel slices were incubated at 25°C overnight. Extraction of proteolytic peptides was performed as previously described (24, 25).
Reverse-phase LC-MS2 analysis of proteolytic peptides.
Reverse-phase nanoflow liquid chromatography-mass spectrometry (LC-MS) and liquid chromatography-tandem mass spectrometry (LC-MS2) analyses of proteolytically derived peptides were performed by using a system consisting of two Agilent 1200 high-performance liquid chromatography (HPLC) binary pumps (nanoflow and capillary) with an autosampler, a column heater, and an integrated switching valve. The LC system was coupled via a nano-electrospray ionization (nESI) source to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). For analyses, 5 μl of the peptide solution was injected into a 5- by 0.3-mm extraction column filled with Zorbax 300 SB-C18 with a 0.3-mm inner diameter and a 5-μm particle size (Agilent). Samples were washed with a mobile phase (97% H2O–0.1% formic acid–3% acetonitrile). The flow rate provided by the capillary pump was 10 μl/min. After 5 min, the integrated switching valve was activated, and peptides were eluted in the back-flush mode from the extraction column onto a 150- by 0.075-mm, 3-μm C18 resin column (GlycproSIL C18, 80 Å; Glycpromass). The mobile phase consisted of acetonitrile and MS-grade water, both containing 0.1% formic acid. Chromatographic separation was achieved by using a binary gradient from 5 to 55% acetonitrile in water in 60 min. The flow rate provided by the nanoflow pump was 0.2 μl/min. Nanospray ionization was achieved by applying 1.2 kV between an 8-μm-diameter emitter (PicoTip emitter; New Objective) and the capillary entrance of the Orbitrap instrument. Mass spectra were acquired in the positive-ion mode by applying a data-dependent automatic switch between the survey scan and MS2 acquisition on the Thermo Scientific LTQ Orbitrap XL mass spectrometer operated with Xcalibur 2.0 software. Peptide samples were analyzed with collision-induced dissociation (CID) and higher-energy C-cell dissociation (HCD) fragmentation methods, acquiring one Orbitrap survey scan in the mass range of m/z 200 to 2,000, followed by MS2 of the most intense ions in the Orbitrap analysis. The target value for the LTQ Orbitrap XL analysis was 1,000,000 for survey scans at a resolution of 30,000 at m/z 400 by using lock masses for recalibration to improve the mass accuracy of precursor ions. Fragmentation for LTQ analysis was performed by collision-induced dissociation with a target value of 5,000 ions. The ion selection threshold was 500 counts. Selected sequenced ions were dynamically excluded for 180 s.
The normalized collision energy (NCE) for standard HCD and CID experiments was set at 30%. Some experiments utilized a higher (35%) or lower (15%) NCE. Mass spectra generated with such NCEs are marked with an @ preceding the NCE value.
ETD analysis of proteolytic peptides.
Electron transfer dissociation (ETD) experiments with proteolytically derived peptides were performed with an nESI-LC-MS2 LTQ Orbitrap XL system (operated by Xcalibur 2.0) similar to that used for the HCD/CID analysis described above, with some modifications. Chromatographic separation was achieved by using a binary gradient from 7 to 55% acetonitrile in water in 25 min. Mass spectra were acquired in the data-dependent mode, automatically switching between an Orbitrap survey scan at a resolution of 60,000 at m/z 400 and ETD MS2 acquisition with the LTQ instrument for the two most intense ions from a parent mass list generated by including the 12C and 13C m/z values of glycosylated peptides. The isolation width was set at 3.0 m/z units. The activation time was set to 150 ms, with no supplement activation. The ion selection threshold was 500 counts. No dynamic exclusion was set. Before ETD experiments were performed, the reagent gas reached an intensity of 1E6.
Data analysis.
The generated mass spectrometric data were manually analyzed with an in-house-generated N. elongata subsp. glycolytica protein databases by using the Xcalibur Qual browser (V. 3.0.63). To generate theoretical peptide sequences and peptide masses with trypsin, AspN, and chymotrypsin, we utilized PeptideMass on the ExPASy server (http://web.expasy.org/peptide_mass/) (26). To verify fragment ions (b/c or y/z ions) and to identify glycan-modified amino acids, we utilized ProteinProspector, developed by the UCSF Mass Spectrometry Facility (http://prospector.ucsf.edu/prospector/mshome.htm).
Top-down and deconvolution analyses.
Purified PilE samples were cleaned and prepared as previously described (20). Top-down ESI-MS of intact PilE protein was performed on the LTQ Orbitrap XL instrument as previously described (9). Deconvoluted masses were reported as monoprotonated [M + H]+.
Nucleotide sequence accession number.
The N. elongata subsp. glycolytica pglH sequence is accessible under BankIt identification number 1835047 and GenBank accession number KT206226.
RESULTS
Identification and characterization of N. elongata subsp. glycolytica pgl genes.
A recent comparative genome sequence analysis of members of the genus Neisseria established seven distinct groups whose compositions and relationships were mostly congruent with current species designations and relationships (27). By examining pgl gene content in neisserial species, we noticed, consistent with previously reported findings (28), two major trends within the so-called commensal groups, including the N. cinerea, N. subflava, and N. mucosa groups and congeners. They all carried intact and linked copies of pglG and pglH and lacked recognizable forms of pglA and pglE. We chose here to focus our efforts on the type strain of N. elongata subsp. glycolytica (ATCC 29315) as it (i) is amenable to genetic manipulation (29), (ii) expresses a characterized type IV pilus (comprised of pilin, the most abundant glycoprotein in characterized species) (29), (iii) occupies a deeply branching position in neisserial phylogeny (27), and (iv) showed evidence of protein glycosylation in a previous study (30).
Using N. elongata subsp. glycolytica genome sequences, five contigs carrying ORFs corresponding to canonical neisserial pgl genes were found (Fig. 1B; see also Table S2 in the supplemental material). Analyses of two of these contigs, one bearing an orthologue of pglG and another carrying orthologues of pglB and pglC, revealed the presence of nonoverlapping sequences sharing significant identities to the 5′ and 3′ ends of a pglH orthologue, respectively. The adjacent nature of these contigs and the presence of a single intact pglH gene were confirmed by PCR and sequencing. Interestingly, the N. elongata subsp. glycolytica equivalent of the N. gonorrhoeae pglB gene was found as two separate ORFs (pglBa and pglBb) corresponding to the phosphoglycosyltransferase and acetyltransferase domains. The expression and functionality of this two-ORF system has been confirmed by complementation of a pglB-null mutant in N. gonorrhoeae (our unpublished data). The three other contigs found carry ORFs orthologous to PglD (the NAD+-dependent dehydratase), PglF (a pgl-associated flippase implicated in the translocation of undecaprenyl diphosphate-linked glycan across the periplasmic membrane), and PglO (also known as PglL) (protein-targeting oligosaccharyltransferase). Thus, this strain has the genetic minimal repertoire to express glycoproteins with O-linked N-diacetylbacillosamine (diNAcBac)-based glycoforms. Corresponding findings were made for pgl gene content and organization in the genomes of N. elongata subsp. elongata and N. elongata subsp. nitroreducens (data not shown).
To examine protein glycosylation proficiency in more detail in N. elongata subsp. glycolytica, whole-cell lysates were examined by using immunoblotting with rabbit monoclonal antibody (MAb) npg1 (recognizing a diNAcBac monosaccharide-specific epitope) (Fig. 2, top) and a rabbit polyclonal serum specific for diNAcBac-Glc disaccharide epitopes (Fig. 2, bottom). In the wild-type strain, no reactivity was detected with either of these antibodies, suggesting that the strain either is incapable of synthesizing O-glycans or produces as-yet-uncharacterized oligosaccharide structures. Given the conserved linkage of the pglH-related ORF to other candidate pgl genes along with the established role of PglH in generating diNAcBac-derived disaccharides in N. gonorrhoeae and N. meningitidis, a null mutant with a pglH disruption was tested for reactivity with MAb npg1 (recognizing a diNAcBac monosaccharide glycoform epitope) (Fig. 2, top). The extract from this mutant reacted strongly with MAb npg1 in a manner consistent with the presence of multiple glycoproteins modified with diNAcBac monosaccharide. We then tested whether this phenotype was dependent on other putative N. elongata subsp. glycolytica pgl genes by disrupting each of them in the background of a strain carrying the pglH mutation. With the exception of the pglG and pglF mutants, disruption of each of the other ORFs abolished npg1 reactivity.
FIG 2.
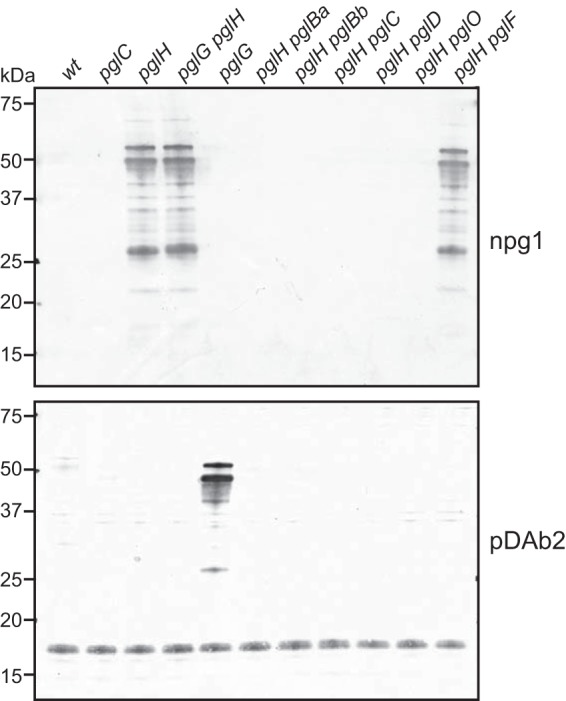
Glycan serotyping analysis of pgl mutants reveals that N. elongata subsp. glycolytica expresses a diNAcBac-Glc-based oligosaccharide. (Top) Immunoblot analysis of whole-cell lysates from wild-type (wt) (KS944), pglC (KS945), pglH (KS1000), pglG (KS1001), pglG pglH (KS1002), pglH pglBa (KS1041), pglH pglBb (KS1043), pglH pglC (KS1050), pglH pglD (KS1051), pglH pglF (KS1040), and pglH pglO (KS1006) strains using diNAcBac epitope-specific monoclonal antibody npg1. (Bottom) Immunoblot analysis of the same samples as those described above with the diNAcBac-Glc epitope-recognizing polyclonal antibody pDAb2 (8).
Given both the linkage to pglH and the predicted structural similarities to PglH and other glycosyltransferases, we hypothesized that a pglG-corresponding protein could modify a PglH-dependent oligosaccharide structure. To test this, whole-cell lysates from all the N. elongata subsp. glycolytica mutants described above were tested for reactivity with a polyclonal rabbit serum recognizing PglH-dependent diNAcBac-Glc disaccharide epitopes (Fig. 2, bottom). In this case, only the pglG mutant showed positive signals, and this reactivity was pglH dependent. Moreover, the pglG pglH double mutant showed reactivity with npg1. Thus, the presence of pglG correlates with the alteration or masking of the PglH-dependent oligosaccharide epitope.
Based on these results, we cannot conclude whether the pglF-encoded flippase orthologue in N. elongata subsp. glycolytica contributes to protein glycosylation. Still, we presume that there are other as-yet-unidentified proteins that function to complement the mutant due to some overlap or redundancy in flippase function, as was seen previously in N. gonorrhoeae (23, 24). However, mutations in the N. elongata subsp. glycolytica ORFs orthologous to pglB, pglC, pglD, pglH, and pglO produced phenotypes in N. elongata subsp. glycolytica consistent with the ORFs having the same functions as in N. gonorrhoeae. We deduce, therefore, that N. elongata subsp. glycolytica expresses a functional pgl system requiring these ORFs and whose glycans are structurally modified by its PglH orthologue. Furthermore, here, we provide the first evidence that pglG encodes a functional glycosyltransferase in N. elongata subsp. glycolytica.
Identification of N. elongata subsp. glycolytica glycoproteins.
Studies of protein glycosylation in N. gonorrhoeae and N. meningitidis have identified numerous targets of the pgl system (23, 25, 31). We turned our attention to candidate glycoproteins in N. elongata subsp. glycolytica based on their possession of domains of low structural complexity (rich in alanine, proline, and serine) implicated in substrate targeting in gonococcal and meningococcal glycoproteins (23, 31). Specifically, these proteins were NirK (a nitrite reductase encoded by nirK), cytochrome c5 (a c-type diheme cytochrome encoded by cycB), and CcoP (a c-type triheme cytochrome encoded by ccoP). Derivatives of N. elongata subsp. glycolytica in which the ORFs of these candidate glycoproteins were tagged by virtue of C-terminal 6-His extensions were constructed. Following their affinity purification, immunoblotting using anti-His antibodies revealed a clear increase in mobility for each protein when isolated from a pglC mutant background versus that observed in the wild-type background (Fig. 3). The same behavior was observed previously for the N. elongata subsp. glycolytica cN protein (a c-type monoheme cytochrome) (30). Thus, these four proteins display behavior consistent with being substrates for glycosylation.
FIG 3.
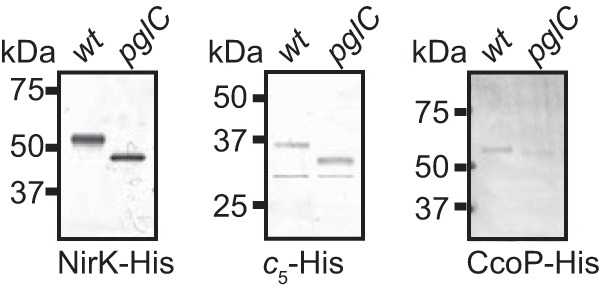
Identification of NirK, cytochrome c5, and CcoP as N. elongata subsp. glycolytica glycoproteins. C-terminally 6-His-tagged proteins were affinity purified from wild-type (wt) and pglC backgrounds and subjected to immunoblotting with a tetra-His epitope-recognizing antibody. Strains used were nirK-His (KS992), nirK-His pglC (KS994), cycB-His (KS997), cycB-His pglC (KS1005), ccoP-His (KS996), and ccoP-His pglC (KS1003) strains.
N. elongata subsp. glycolytica glycan structure determined by mass spectrometry.
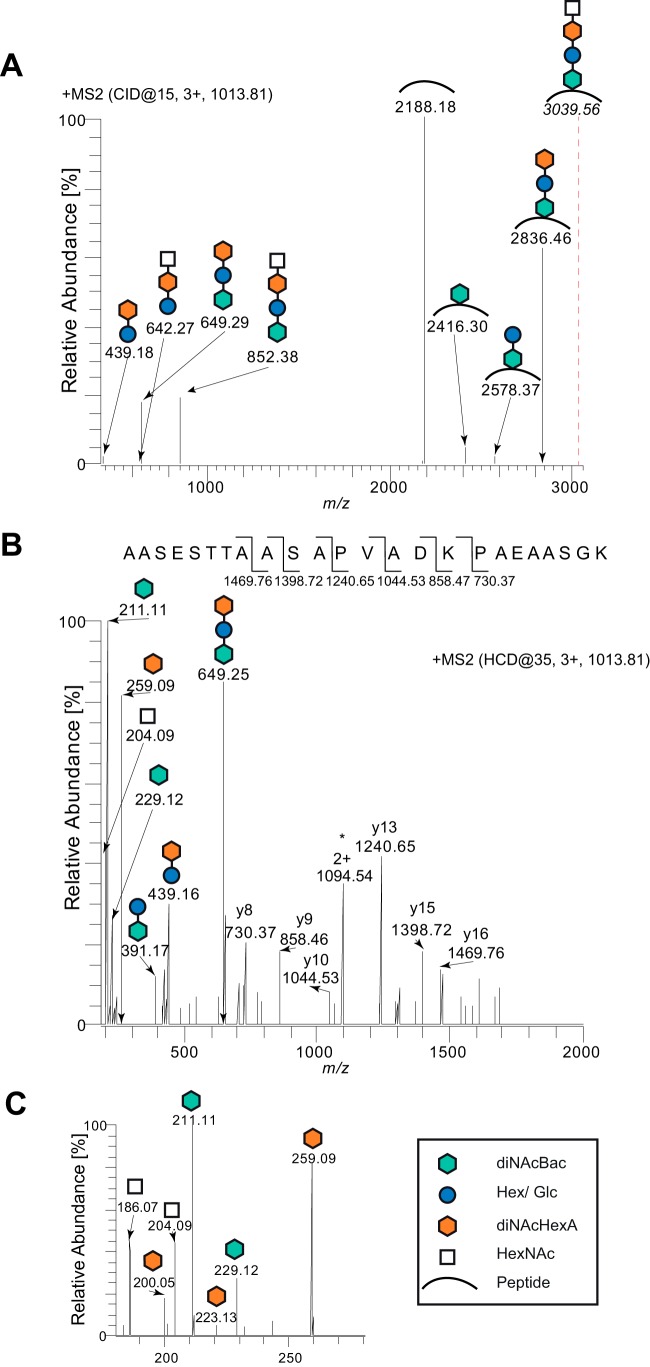
To determine the structure and composition of the endogenous glycan, trypsin-derived peptides from affinity-purified CcoP-His were subjected to consecutive mass spectrometry (MS) fragmentation utilizing both low and high fragmentation energies. The deconvoluted MS2 spectrum of the triply charged peptide at m/z 1,013.81 generated with low fragmentation energy showed a fragmentation pattern consistent with a glycan modification (Fig. 4). The ions in the high-mass area point to a linear tetrasaccharide comprised of a terminal HexNAc followed by an unknown moiety with a mass of 258.1 Da, a hexose, and diNAcBac at the reducing end of the oligosaccharide.
FIG 4.
N. elongata subsp. glycolytica glycan structure determined by mass spectrometry. (A) Deconvoluted MS2 spectrum of the glycopeptide at m/z 1,013.81 generated by low collision energy. The generated ions reveal a linear glycan consisting of a terminal HexNAc followed by an unknown moiety of 258.1 Da, a hexose, and, finally, a diNAcBac. The theoretical mass of the intact glycopeptide is indicated with a dotted line. (B) The MS2 spectrum of the glycopeptide at m/z 1,013.81 generated by higher collision energy. b and y ions identifying the peptide sequence 350AASESTTAASAPVADKPAEAASGK373 are marked. Reporter ions used for the identification of individual glycans are marked with the same symbols as those shown in the key in panel C. The charge state is marked with the number of protons followed by a plus. An asterisk denotes the unmodified full-length peptide. (C) Enlarged window of data from panel B between m/z 180 and m/z 280 of the MS2 spectrum. The strain used was the ccoP-His strain (KS996).
To verify the glycan composition, we investigated the MS2 spectrum of a triply charged glycopeptide generated with a higher fragmentation energy for primary glycan reporter ions (Fig. 4B) and internal secondary glycan fragment reporter ions (Fig. 4C). Sequencing of the y ions identified the glycopeptide sequence as 350AASESTTAASAPVADKPAEAASGK373, with a theoretical mass of 2,188.073 Da ([M + H]+), and thereby verified that the mass of the full-length glycan was 851.37 Da (Fig. 4B). Multiple reporter ions were also detected in the low-mass area, confirming the glycan composition. The ions at m/z 229.12 and m/z 211.11 confirm the presence of diNAcBac (24, 25), whereas the presence of hexose was verified by the ion at m/z 391.17, previously shown to correlate with the disaccharide diNAcBac-Hex (23, 24). In addition, an abundant reporter ion at m/z 259.09 (corresponding to the protonated version of the unknown moiety of 258.1 Da) was detected. Both the accurate mass and abundance of the m/z 259.09 reporter ion are consistent with the presence of di-N-acetyl-hexuronic acid (diNAcHexA) (32). Moreover, two secondary fragmentation glycan ions detected at m/z 223.12 and at m/z 200.15 (Fig. 4C) are also consistent with the presence of diNAcHexA (32). Finally, the ions at m/z 204.09 and 186.07 (Fig. 4C) confirmed the presence of a HexNAc (9, 33). Corroborative evidence for the tetrasaccharide was also found by using higher-energy fragmentation of glycopeptides derived from the cytochrome c-type heme proteins c5 and cN (see Fig. S1 in the supplemental material). Taken together, these MS2 spectra indicate that the N. elongata subsp. glycolytica protein glycan is a linear tetrasaccharide consisting of diNAcBac-Hex-unknown m/z 258.1 moiety-HexNAc, where it is likely that the third sugar corresponds to diNAcHexA.
Effect of pgl mutations on glycan structure.
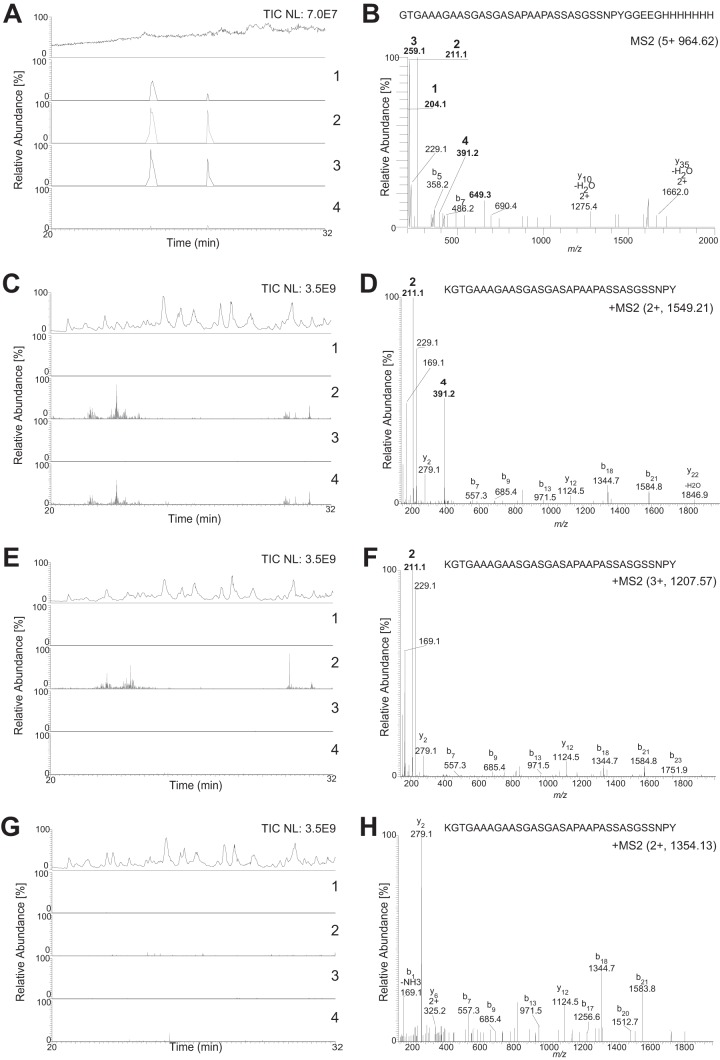
To further investigate the role of pgl glycosyltransferases in protein glycosylation, affinity-tagged NirK was purified from selected pgl-null mutants and subjected to proteolytic digestion and MS2 analyses. As described above, the tetrasaccharide-generated MS2 spectra contained glycan reporter ions for HexNAc (m/z 204.086), diNAcBac (m/z 211.108), diNAcHexA (m/z 259.093), and diNAcBac-Hex (m/z 391.170), and here, these ions were used as a readout of glycan composition. In the LC-MS2 chromatogram of trypsinated wild-type NirK-His, peaks representing the presence of all four glycan reporter ions were detected at the same position (Fig. 5A), meaning that glycopeptides in the wild-type background were modified with full-length glycans. To demonstrate that the peaks representing the glycan reporter ions occur on glycopeptides, a representative MS2 spectrum was selected from one of the spectra with overlapping reporter ion peaks in the LC-MS2 chromatogram. As shown in Fig. 5B, the presence of the four glycan reporter ions in the low-mass area verify the trypsin-generated peptide 372GTGAAAGAASGASGASAPAAPASSASGSSNPYGGEEGHHHHHHH415 from NirK-His as a bona fide glycopeptide. The LC-MS2 chromatogram and the representative spectrum from a glycopeptide from chymotrypsin-digested NirK-His purified from a pglG background displayed only the reporter ions for diNAcBac (m/z 211.1) and diNAcBac-Hex (m/z 391.2) (Fig. 5C and D), while those derived from the analogous glycopeptide from a pglH background displayed only the diNAcBac reporter ion (Fig. 5D and E). Finally, no glycan-related reporter ions were detected in the chromatogram or spectrum from the equivalent peptide isolated from a pglC background (Fig. 5F and G). Together, these results validated the findings made by glycan serotyping of the mutant strains (Fig. 2) and showed that N. elongata subsp. glycolytica PglH participates in the synthesis of Und-PP-diNAcBac-Glc similarly to its gonococcal and meningococcal orthologues. Furthermore, the data confirm the inference that PglG is required for the extension of the Und-PP-diNAcBac-Glc precursor.
FIG 5.
Effect of pgl gene mutations on glycan structure. Shown are LC-MS2 chromatograms of proteolytically derived peptides from affinity-purified NirK from pgl mutants at between 20 and 32 min. Total ion chromatogram (TIC) intensity values represent the amount of peptides entering the mass spectrometer. The selected ion chromatograms are for the four glycan reporter ions characteristic for a tetrasaccharide, HexNAc at m/z 204.086 (1), diNAcBac at m/z 211.108 (2), diNAcHexA at m/z 259.093 (3), and diNAcBac-Hex at m/z 391.170 (4). All glycan reporter ions were searched with an 8-ppm window. All selected ion chromatogram values were normalized to the most abundant reporter ion in each sample. The MS2 spectrum demonstrates the presence of glycan reporter ions (marked in boldface type and numbered as described above). (A) LC-MS2 chromatogram of trypsin-derived peptides from affinity-purified NirK-His from a wild-type background (KS992). The total ion chromatogram intensity value was set at 7.0E7. All selected ion chromatogram values were normalized to the most abundant glycan reporter ion, diNAcHexA at 1.5E4. (B) MS2 spectrum of the singly modified trypsin-generated peptide 372GTGAAAGAASGASGASAPAAPASSASGSSNPYGGEEGHHHHHHH415 from NirK-His from panel A. (C) LC-MS2 chromatogram of chymotrypsin-derived peptides from affinity-purified NirK-His from a pglG background (KS1032). The total ion chromatogram intensity value was set at 3.5E9. All selected ion chromatogram values were normalized to the most abundant glycan reporter ion, diNAcBac at 1.0E6. (D) MS2 spectrum of the singly modified chymotrypsin-generated peptide 371KGTGAAAGAASGASGASAPAAPASSASGSSNPY403 from NirK-His from panel C. (E) LC-MS2 chromatogram of chymotrypsin-derived peptides from affinity-purified NirK-His from a pglH background (KS1052). The total ion chromatogram intensity value was set at 3.5E9. All selected ion chromatogram values were normalized to the most abundant glycan reporter ion, diNAcBac at 1.0E6. (F) MS2 spectrum of the quadruple-modified chymotrypsin-generated peptide 371KGTGAAAGAASGASGASAPAAPASSASGSSNPY403 from NirK-His from panel E. (G) LC-MS2 chromatogram of chymotrypsin-derived peptides from affinity-purified NirK-His from a pglC background (KS994). The total ion chromatogram intensity value was set at 3.5E9. All selected ion chromatogram values were set at 5.0E5. (H) MS2 spectrum of the unmodified chymotrypsin-generated peptide 371KGTGAAAGAASGASGASAPAAPASSASGSSNPY403 from NirK-His from panel G.
Determination of the glycan linkage site.
To determine the nature of glycan attachment, the sites of modification were characterized by using ETD fragmentation. By using the cN protein affinity purified from a pglH background (and thus bearing the diNAcBac monosaccharide), three attachment sites were localized (Fig. 6). Within the glycopeptide 38NNTASAVDAAASSVQEAASK57, two diNAcBac modification sites were identified at serine 42 and serine 49 (Fig. 6A), while within the glycopeptide peptide 27EAAQAIASDVK37, a single diNAcBac modification was identified at serine 34 (Fig. 6B). Given the presence of serine in all other glycopeptides we identified in N. elongata subsp. glycolytica (and cases where serine is the sole hydroxyl-containing amino acid) (see Fig. S1 in the supplemental material), glycan attachment is clearly O linked and likely occurs exclusively at this residue. This is in line with attachment site specificities seen in N. gonorrhoeae and N. meningitidis. Likewise, all N. elongata subsp. glycolytica attachment sites (determined and inferred) are localized to low-complexity regions rich in proline and alanine, with the latter often being found in the +1 position relative to serine. Furthermore, we found evidence that N. elongata subsp. glycolytica glycoproteins undergo multisite modification (see Fig. S2 in the supplemental material), similarly to what is seen in the gonococcal and meningococcal systems. Thus, the key features governing glycan attachment site selection are conserved within these neisserial species.
FIG 6.
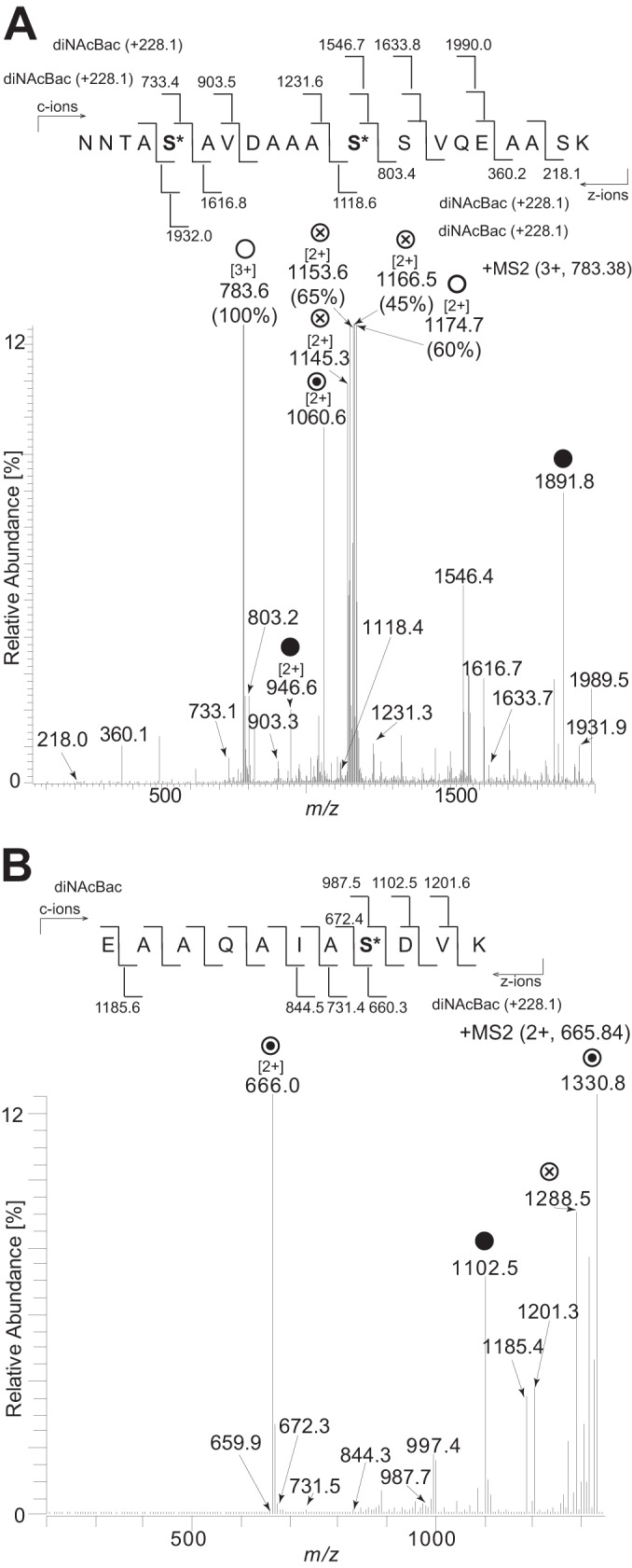
Identification of serines as glycan occupancy sites by ETD analysis. (A) Fragmentation of the triply charged peptide at m/z 783.38 corresponding to the peptide 38NNTASAVDAAASSVQEAASK57 carrying two diNAcBac modifications at serine 42 and serine 49. (B) Fragmentation of the doubly charged peptide at m/z 665.84 corresponding to the peptide 27EAAQAIASDVK37 carrying a diNAcBac modification at serine 34. ○ denotes ions corresponding to the full-length peptide with two diNAcBac modifications. ⊗ denotes ions generated by internal glycan fragmentation. ⊙ denotes ions corresponding to the full-length peptide with a single diNAcBac modification. ● denotes the mass of the full-length unmodified peptide. Bracketed numbers denote the charge state of the ion if different from 1. Numbers in parentheses denote the full relative intensity of the ion. The strain used was the cycC-His pglH strain (KS951).
The major pilin protein of N. elongata subsp. glycolytica is not glycosylated.
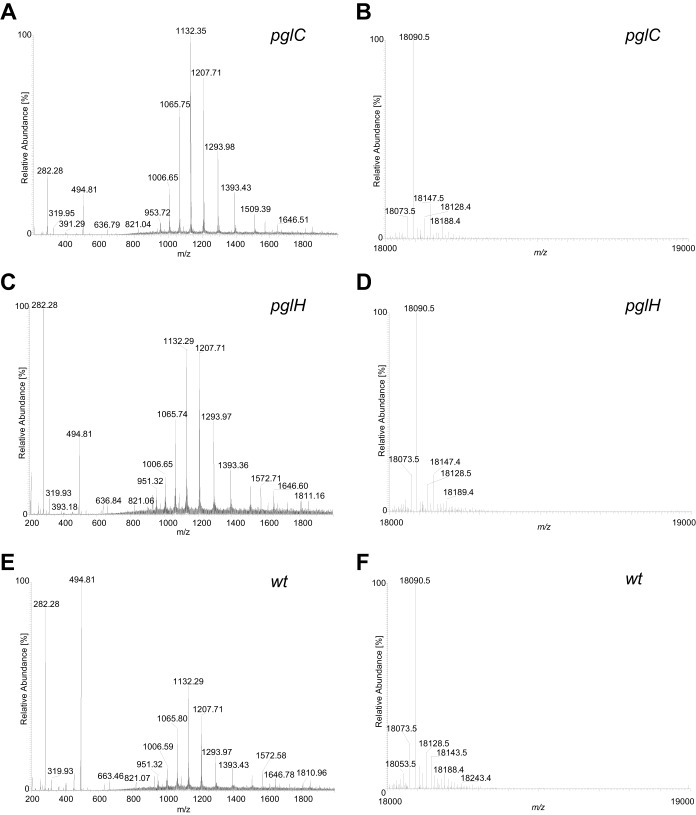
The major pilin subunit of type IV pili is the most abundant glycoprotein in all Neisseria species studied to date. To address the status of pilin subunit posttranslational modification, type IV pili were purified and analyzed. The predominant protein in these preparations had an electrophoretic mass of 17 kDa, and this protein was excised following SDS-PAGE and processed for MS analyses. Tryptic digestion and MS processing revealed the major band to correspond to PilE, as identified previously (UniProt accession number D4DRI6; locus tag NEIELOOT_01679) (29), and although nearly complete peptide coverage was achieved, no evidence for glycosylation was obtained (Fig. 7). In line with this, no detectable alteration in migration was seen for PilE derived from the pglC versus the wild-type background. Samples from these backgrounds as well as from the pglH mutant were examined by immunoblotting with npg1 (which reacts with all glycoproteins in the latter background). Again, no signal for PilE glycosylation was detected in the correct size range for pilin (Fig. 7). It is worth noting that the predominant 17-kDa protein species detected by the pDAb2 antiserum (Fig. 2, bottom) in all samples is PilE. There, the ability of the pDAb2 antiserum to react with PilE reflected the fact that the immunogen was glycosylated gonococcal pilin protein and that N. elongata subsp. glycolytica PilE likely shares common epitopes with those of gonococcal pilin. Thus, it was apparent that PilE migration patterns were indistinguishable regardless of the glycosylation status of the host strain. Finally, in order to definitively address potential PilE glycan modification, intact pilins derived from wild-type and pgl backgrounds were examined by using a top-down MS approach. Not only does this technique allow accurate PilE molecular mass determination, it also facilitates the detection of potential oxonium ions arising from the cleavage of glycosidic bonds during in-source fragmentation. However, the profiles of all samples seen in the deconvoluted molecular mass spectra from intact pilin were identical to one another, with dominant signals at 18,090.5 Da corresponding to the processed PilE protein (cleaved at position G−1-F+1, N-methylated, and with a single reduced disulfide bridge) (Fig. 8). Although background ions were detected in the low-mass area of the ESI-MS spectra, these ions were the same in all samples and unrelated to potential covalently attached glycans.
FIG 7.
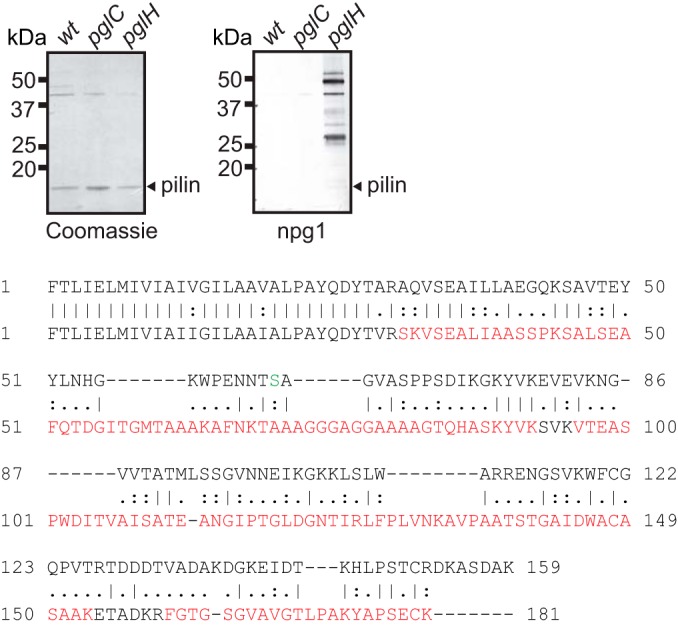
The major pilin protein PilE of N. elongata subsp. glycolytica is not glycosylated. (Top) Coomassie-stained (left) and immunoblotted (right) SDS-PAGE gels of purified pili from wild-type (wt) (KS944), pglC (KS945), and pglH (KS1000) strains. The antibody used was the diNAcBac-reactive MAb npg1. The blot was overdeveloped so as to be able to see reactivity from impurities in the pilin preparation to ascertain sufficient development to see any trace of potential pilin glycosylation. (Bottom) Pairwise alignment of predicted protein sequences of mature PilE from N. gonorrhoeae strain MS11 (top sequence) and N. elongata subsp. glycolytica (bottom sequence). The N. elongata subsp. glycolytica PilE sequence covered by peptides detected by MS is shown in red overlay. The glycosylated residue (serine 63) found in N. gonorrhoeae strain MS11 is highlighted in green (35, 36).
FIG 8.
Intact-protein MS confirms the lack of N. elongata subsp. glycolytica PilE pilin glycosylation. Shown are the in-source fragmentation ESI-MS spectra over a range of 200 to 2,000 m/z and the corresponding deconvoluted molecular mass spectra of intact pilin proteins. The mass at 18,090.5 Da [M + H]+ corresponds to the unmodified mature pilin. Na adducts (+22 Da) and K adducts (+38 Da) were detected but are not marked in the spectra. (A) ESI-MS spectrum of purified pilin from a pglC strain (KS945). (B) Deconvoluted molecular mass spectra of intact pilin from the pglC strain. (C) ESI-MS spectrum of purified pilin from a pglH strain (KS1000). (D) Deconvoluted molecular mass spectra of intact pilin from the pglH strain. (E) ESI-MS spectrum of purified pilin from the wild-type strain (KS945). (F) Deconvoluted molecular mass spectra of intact pilin from the wild-type strain.
DISCUSSION
It is now well established that there is broad-spectrum protein O-glycosylation in bacterial species. For the genus Neisseria, studies of these systems have been focused on isolates of N. gonorrhoeae, N. meningitidis, and the closely related species N. lactamica. However, the prevalence and status of such systems within other neisserial species remain unclear. Here, salient findings relating to the potential for glycan diversity, glycoprotein target selection, and, ultimately, protein glycosylation macroevolution were established.
First and perhaps foremost, we found that N. elongata subsp. glycolytica expresses a previously unknown tetrasaccharide glycoform (summarized in Fig. S3 in the supplemental material). Strains of N. gonorrhoeae, N. meningitidis, and N. lactamica can express trisaccharides by virtue of the PglE galactosyltransferase that further elaborates a PglA-based Und-PP-disaccharide. However, there were no equivalents of either pglA or pglE present in the genomes of N. elongata subsp. glycolytica or of other related commensal strains. In the strain studied here, a null mutation in pglG resulted in the loss of the tetrasaccharide glycoform and the appearance of a disaccharide glycoform. Based on both MS and serotyping data, the altered glycan in this mutant background is a diNAcBac-Hex-disaccharide. Due to the strong reactivity of glycoproteins with the pDAb2 antibodies generated against a diNAcBac-Glc-disaccharide, it seems most likely that Glc resides at the second position of this oligosaccharide. Furthermore, a null mutation in pglH, encoding a glucosyltransferase, led to the expression of a diNAcBac monosaccharide glycoform. Thus, the product of pglG is essential for either the extension of the Und-PP-diNAcBac-Glc-disaccharide or the biosynthesis of the nucleotide-sugar precursor to the third sugar (moiety of 258.1 Da). Given the established presence of glycosyltransferase domains in PglG (13), we favor the former scenario, although this needs to be biochemically confirmed. Whatever the case, this is, to our knowledge, the first phenotype associated with pglG gene status. It is important to note that this result is not attributable to a polar effect of the pglG mutation, as the downstream gene is pglH, and the pglG mutant shows a clear, distinct phenotype from that of the pglG pglH double mutant (Fig. 2). These findings obviously raise the question as to what role pglG and/or PglG might play in other neisserial species and in particular in N. gonorrhoeae and N. meningitidis. The expression of the tetrasaccharide glycoform in the wild-type background strongly suggests that there is another as-yet-unidentified glycosyltransferase that carries out further elaboration of the Und-PP-trisaccharide. A number of putative glycosyltransferase-encoding genes are identifiable in the N. elongata subsp. glycolytica genome sequences, but none of these genes map to the contigs containing pglG and pglH.
A peculiar finding here is that the major type IV pilin subunit PilE of N. elongata subsp. glycolytica is not glycosylated despite the presence of a fully functioning general pgl system (as manifest by the expression of other glycoprotein orthologues). The simplest explanation for this is that the pilin is intrinsically incapable of being O-glycosylated as it lacks the structural determinants required to act as a substrate for the PglO oligosaccharyltransferase. Instead, we observe concurrence between the identities and characteristics of the other glycoproteins in N. gonorrhoeae and those in N. elongata subsp. glycolytica. This situation is remarkable, as most if not all of the phenotypic alterations observed in neisserial pgl mutants or variants are related to pilus-associated properties (18–20). The findings here point to the likely importance of broad-spectrum glycosylation per se. It has also been speculated that the neisserial glycosylation system evolved from an ancestral targeted system modifying solely the abundant PilE substrate to a general system that encompasses other protein substrates (34). The findings made in our work suggest that caution may be warranted in accepting such a model and that more information on the relative distribution of glycosylated pilins and general protein glycosylation is needed to resolve this issue.
While confirming and extending previously reported findings, the data here provide further strong evidence that the pgl gene families have a dynamic evolutionary history. The results reveal not only a surprising degree of conservation, as seen in the use of a basal diNAcBac-Glc disaccharide core, but also the potential for diversification in novel extended sugar residues. Establishing a role for N. elongata subsp. glycolytica PglG in extending diNAcBac-Glc-based glycoforms is important, as it provides a directed approach to address its potential function in gonococci and meningococci, which remains enigmatic. Also, as N. elongata subsp. glycolytica lacks type IV pilus glycosylation, studies of general O-linked glycosylation and associated phenotypes in this species may also help elucidate novel glycosylation functions of broader significance.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by Research Council of Norway grants 166931, 183613, and 183814 and the Centre for Integrative Microbial Evolution at the Department of Biosciences and by funds from the University of Oslo Faculty of Mathematics and Natural Sciences. This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust and European Union. This publication also made use of the Meningitis Research Foundation Meningococcus Genome Library (http://www.meningitis.org/research/genome) developed by Public Health England, the Wellcome Trust Sanger Institute, and the University of Oxford as a collaboration. That project is funded by the Meningitis Research Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00620-15.
REFERENCES
- 1.Carter AT, Paul CJ, Mason DR, Twine SM, Alston MJ, Logan SM, Austin JW, Peck MW. 2009. Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics 10:115. doi: 10.1186/1471-2164-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champion OL, Gaunt MW, Gundogdu O, Elmi A, Witney AA, Hinds J, Dorrell N, Wren BW. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci U S A 102:16043–16048. doi: 10.1073/pnas.0503252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twine SM, Paul CJ, Vinogradov E, McNally DJ, Brisson JR, Mullen JA, McMullin DR, Jarrell HC, Austin JW, Kelly JF, Logan SM. 2008. Flagellar glycosylation in Clostridium botulinum. FEBS J 275:4428–4444. doi: 10.1111/j.1742-4658.2008.06589.x. [DOI] [PubMed] [Google Scholar]
- 4.Nothaft H, Scott NE, Vinogradov E, Liu X, Hu R, Beadle B, Fodor C, Miller WG, Li J, Cordwell SJ, Szymanski CM. 2012. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics 11:1203–1219. doi: 10.1074/mcp.M112.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coyne MJ, Fletcher CM, Chatzidaki-Livanis M, Posch G, Schaffer C, Comstock LE. 2013. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol Microbiol 88:772–783. doi: 10.1111/mmi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. 2014. Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol Cell Proteomics 13:2354–2370. doi: 10.1074/mcp.M114.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aas FE, Vik A, Vedde J, Koomey M, Egge-Jacobsen W. 2007. Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol 65:607–624. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Børud B, Aas FE, Vik A, Winther-Larsen HC, Egge-Jacobsen W, Koomey M. 2010. Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J Bacteriol 192:2816–2829. doi: 10.1128/JB.00101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Børud B, Anonsen JH, Viburiene R, Cohen EH, Samuelsen AB, Koomey M. 2014. Extended glycan diversity in a bacterial protein glycosylation system linked to allelic polymorphisms and minimal genetic alterations in a glycosyltransferase gene. Mol Microbiol 94:688–699. doi: 10.1111/mmi.12789. [DOI] [PubMed] [Google Scholar]
- 10.Børud B, Viburiene R, Hartley MD, Paulsen BS, Egge-Jacobsen W, Imperiali B, Koomey M. 2011. Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. Proc Natl Acad Sci U S A 108:9643–9648. doi: 10.1073/pnas.1103321108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartley MD, Morrison MJ, Aas FE, Børud B, Koomey M, Imperiali B. 2011. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50:4936–4948. doi: 10.1021/bi2003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamot-Rooke J, Rousseau B, Lanternier F, Mikaty G, Mairey E, Malosse C, Bouchoux G, Pelicic V, Camoin L, Nassif X, Dumenil G. 2007. Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc Natl Acad Sci U S A 104:14783–14788. doi: 10.1073/pnas.0705335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Power PM, Roddam LF, Rutter K, Fitzpatrick SZ, Srikhanta YN, Jennings MP. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol 49:833–847. [DOI] [PubMed] [Google Scholar]
- 14.Kahler CM, Martin LE, Tzeng YL, Miller YK, Sharkey K, Stephens DS, Davies JK. 2001. Polymorphisms in pilin glycosylation locus of Neisseria meningitidis expressing class II pili. Infect Immun 69:3597–3604. doi: 10.1128/IAI.69.6.3597-3604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannessen C, Koomey M, Børud B. 2012. Hypomorphic glycosyltransferase alleles and recoding at contingency loci influence glycan microheterogeneity in the protein glycosylation system of Neisseria species. J Bacteriol 194:5034–5043. doi: 10.1128/JB.00950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krauland MG, Dunning Hotopp JC, Riley DR, Daugherty SC, Marsh JW, Messonnier NE, Mayer LW, Tettelin H, Harrison LH. 2012. Whole genome sequencing to investigate the emergence of clonal complex 23 Neisseria meningitidis serogroup Y disease in the United States. PLoS One 7:e35699. doi: 10.1371/journal.pone.0035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamelas A, Harris SR, Roltgen K, Dangy JP, Hauser J, Kingsley RA, Connor TR, Sie A, Hodgson A, Dougan G, Parkhill J, Bentley SD, Pluschke G. 2014. Emergence of a new epidemic Neisseria meningitidis serogroup A clone in the African meningitis belt: high-resolution picture of genomic changes that mediate immune evasion. mBio 5:e01974-14. doi: 10.1128/mBio.01974-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jen FE, Warren MJ, Schulz BL, Power PM, Swords WE, Weiser JN, Apicella MA, Edwards JL, Jennings MP. 2013. Dual pili post-translational modifications synergize to mediate meningococcal adherence to platelet activating factor receptor on human airway cells. PLoS Pathog 9:e1003377. doi: 10.1371/journal.ppat.1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings MP, Jen FE, Roddam LF, Apicella MA, Edwards JL. 2011. Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol 13:885–896. doi: 10.1111/j.1462-5822.2011.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vik A, Aspholm M, Anonsen JH, Børud B, Roos N, Koomey M. 2012. Insights into type IV pilus biogenesis and dynamics from genetic analysis of a C-terminally tagged pilin: a role for O-linked glycosylation. Mol Microbiol 85:1166–1178. doi: 10.1111/j.1365-2958.2012.08166.x. [DOI] [PubMed] [Google Scholar]
- 21.Tønjum T, Freitag NE, Namork E, Koomey M. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol 16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 22.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aas FE, Egge-Jacobsen W, Winther-Larsen HC, Løvold C, Hitchen PG, Dell A, Koomey M. 2006. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J Biol Chem 281:27712–27723. doi: 10.1074/jbc.M604324200. [DOI] [PubMed] [Google Scholar]
- 25.Anonsen JH, Vik A, Egge-Jacobsen W, Koomey M. 2012. An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J Proteome Res 11:5781–5793. doi: 10.1021/pr300584x. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins MR, Lindskog I, Gasteiger E, Bairoch A, Sanchez JC, Hochstrasser DF, Appel RD. 1997. Detailed peptide characterization using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis 18:403–408. doi: 10.1002/elps.1150180314. [DOI] [PubMed] [Google Scholar]
- 27.Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, Maiden MC. 2012. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158:1570–1580. doi: 10.1099/mic.0.056077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marri PR, Paniscus M, Weyand NJ, Rendon MA, Calton CM, Hernandez DR, Higashi DL, Sodergren E, Weinstock GM, Rounsley SD, So M. 2010. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higashi DL, Biais N, Weyand NJ, Agellon A, Sisko JL, Brown LM, So M. 2011. N. elongata produces type IV pili that mediate interspecies gene transfer with N. gonorrhoeae. PLoS One 6:e21373. doi: 10.1371/journal.pone.0021373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aas FE, Li X, Edwards J, Hongro Solbakken M, Deeudom M, Vik A, Moir J, Koomey M, Aspholm M. 2015. Cytochrome c-based domain modularity governs genus-level diversification of electron transfer to dissimilatory nitrite reduction. Environ Microbiol 17:2114–2132. doi: 10.1111/1462-2920.12661. [DOI] [PubMed] [Google Scholar]
- 31.Ku SC, Schulz BL, Power PM, Jennings MP. 2009. The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem Biophys Res Commun 378:84–89. doi: 10.1016/j.bbrc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 32.Voisin S, Houliston RS, Kelly J, Brisson JR, Watson D, Bardy SL, Jarrell KF, Logan SM. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem 280:16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- 33.Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AV, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P, Packer NH, Larsen MR, Cordwell SJ. 2011. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics 10:M000031–MCP201. doi: 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulz BL, Jen FE, Power PM, Jones CE, Fox KL, Ku SC, Blanchfield JT, Jennings MP. 2013. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS One 8:e62768. doi: 10.1371/journal.pone.0062768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. 1995. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature 378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 36.Hegge FT, Hitchen PG, Aas FE, Kristiansen H, Løvold C, Egge-Jacobsen W, Panico M, Leong WY, Bull V, Virji M, Morris HR, Dell A, Koomey M. 2004. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc Natl Acad Sci U S A 101:10798–10803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.