Abstract
Many Gram-negative pathogens express a type III secretion (T3SS) system to enable growth and survival within a host. The three human-pathogenic Yersinia species, Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, encode the Ysc T3SS, whose expression is controlled by an AraC-like master regulator called LcrF. In this review, we discuss LcrF structure and function as well as the environmental cues and pathways known to regulate LcrF expression. Similarities and differences in binding motifs and modes of action between LcrF and the Pseudomonas aeruginosa homolog ExsA are summarized. In addition, we present a new bioinformatics analysis that identifies putative LcrF binding sites within Yersinia target gene promoters.
INTRODUCTION
There are three Yersinia species pathogenic to humans. Y. pestis is the causative agent of bubonic and pneumonic plague and is transmitted through a flea vector or through airborne transmission from one mammalian host to another (1). In contrast, the enteropathogenic Yersinia species Y. enterocolitica and Y. pseudotuberculosis grow in the environment but can be transmitted to mammalian hosts through ingestion of contaminated food or water (1). Y. enterocolitica and Y. pseudotuberculosis cause typically self-limiting mesenteric lymphadenitis or gastroenteritis in otherwise healthy individuals but can cause a serious blood-borne infection in people with iron overload disorders such as hereditary hemochromatosis (2). In addition, sequelae following enteropathogenic Yersinia infection, such as erythema nodosum and reactive arthritis, have also been reported (3–5).
Human-pathogenic Yersinia species share a virulence plasmid, called pCD1 in Y. pestis and pYV in enteropathogenic yersiniae, encoding the Ysc type III secretion system (T3SS) essential for causing disease (6). These 70-kb plasmids carry dozens of T3SS structural genes and encode five or six T3SS effector proteins called Yops and their dedicated chaperones, as well as genes encoding proteins involved in regulating expression and function of the T3SS. One of these regulatory proteins, LcrF, serves as the Yersinia T3SS master regulator, controlling transcription of a large number of plasmid-borne genes. Several environmental cues influence expression of LcrF itself, possibly enabling Yersinia to control T3SS expression during transitions from one niche to another. Recent reviews have highlighted important advances in our understanding of T3SS structure and modulation of the innate immune response by T3SS effector proteins (7). In this review, we focus on factors controlling LcrF expression, the target genes LcrF regulates, and how LcrF activity influences Yersinia pathogenesis.
LcrF HISTORY, STRUCTURE, AND FUNCTION
It has long been appreciated that human-pathogenic Yersinia carrying T3SS genes requires millimolar concentrations of calcium to grow at 37°C, and this phenomenon was termed the low-calcium response, or Lcr (8, 9). The absence of calcium, in combination with a shift to 37°C, triggers secretion of T3SS effector proteins, mimicking the effect of host cell contact. Yersinia undergoes growth arrest during active type III secretion, explaining why calcium is required for growth at 37°C. Mechanistic explanations for how calcium ions regulate Yop secretion and why type III secretion is associated with cessation of growth in vitro remain unclear. However, a number of T3SS genes were originally named for the low-calcium response, as mutations in these genes were shown to alter the Lcr phenotype (10, 11).
lcrF was first identified in Y. pestis by Goguen and colleagues as a gene required for thermal induction of several pCD1 genes (12). Using a similar approach, VirF, the Y. enterocolitica LcrF homolog, was discovered (13). Finally, while sequence analysis of Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis virulence plasmids revealed evidence of several rearrangements, the low-calcium response at 37°C was found to be highly conserved in all three species, suggesting the presence of LcrF in Y. pseudotuberculosis as well (14). In this review, we refer to the regulator as LcrF unless specifically referring to the Y. enterocolitica VirF homolog.
LcrF is a 30-kDa AraC-like protein that shares homology with AraC in its carboxy-terminal DNA binding region (15). AraC is well known for its role in Escherichia coli as a DNA binding transcriptional regulator (reviewed in reference 16). Early on, Y. pestis LcrF and Y. enterocolitica VirF were shown by the use of gel shift assays to bind directly to sequences in the yopE and yopH promoters (17). The amino-terminal domains for AraC-like proteins have been shown to be involved in self-association. Additionally, the amino-terminal domain of several AraC-like proteins binds cofactors that influence the ability of the protein to regulate transcription (16). For example, binding of the arabinose cofactor to E. coli AraC induces a conformational change, allowing AraC to activate transcription (16). It is thought that LcrF exists as a dimer in solution through the self-association of its amino-terminal domain; however, unlike that of E. coli AraC, the amino-terminal domain of LcrF has not been shown to bind additional cofactors.
As in all other AraC family transcriptional regulators, the carboxy-terminal DNA binding region of LcrF contains two helix-turn-helix (HTH) domains (18). Like all HTH domains, the recognition helix binds specific DNA residues within the major groove (19). DNA binding sites of LcrF and its homologs have been experimentally investigated in a number of studies (15–17, 20, 21). The first study, using DNase protection assays performed on promoters of genes activated by LcrF, suggested a common DNA binding motif, TTTTaGYcTgTat (capital letters represent more highly conserved residues, and Y stands for C or T) (17). Wattiau and Cornelis identified this “half-site” upstream of several T3SS genes. However, these proposed LcrF binding sites were highly variable in terms of distance from the transcriptional start site, directionality, and distance of the half-sites from each other (17). In a more recent analysis, King et al. observed that the carboxy-terminal domain of LcrF is nearly identical to the DNA binding region of the homologous AraC-like master regulator ExsA of the Pseudomonas aeruginosa T3SS (18). The authors further showed that Y. pestis LcrF binds and activates ExsA-dependent promoters in P. aeruginosa. Similarly, ExsA was able to induce expression of T3SS genes in Y. pestis in the absence of LcrF (18). LcrF and ExsA were shown to interact with a common nucleotide sequence motif (AaAAAnwnMygrCynnnmYTGyaAk), which is also recognized by activators of T3SS genes from Photorhabdus luminescens, Aeromonas hydrophilus, and Vibrio parahaemolyticus (W stands for A or T, M for A or C, Y for C or T, R for A or G, and K for G or T [with uppercase letters representing more highly conserved residues]) (18, 21, 22).
To reconcile these two dissimilarly presented LcrF consensus binding sites, we attempted to align the promoter regions of genes known to be controlled by LcrF. First, all previously identified LcrF binding site sequences from the virA, virB, virC, yopE, lcrG, and yopH promoters, as well as known ExsA binding sites from P. aeruginosa, were input into MEME motif discovery to identify a consensus motif (23). The resulting motif was subsequently used to search the virulence plasmids of the three human-pathogenic Yersinia species to identify all putative LcrF binding sites using FIMO (Find Individual Motif Occurrences), where motifs were called with a P value threshold of <0.0001 (24). Alignment of LcrF binding sites of selected genes (Fig. 1A) shows a 5′-AAAA-N6-GNCT-N5-TGANA-3′ motif located 20 to 21 bp upstream of the predicted −10 TATA box for most genes. This consensus motif is similar to that of ExsA described above. Sequences containing these motifs 20 to 21 bp upstream of the −10 TATA box overlap well the regions found experimentally to bind LcrF or ExsA (Fig. 1A, solid underlines) (15–18, 20, 21). In the case of the yopH and sycE genes, a second 5′-AAAA-N6-GNCT-N5-TGANA-3′ motif was found further upstream and coincided with regions previously shown to be weakly bound by LcrF (Fig. 1A, dashed underlines) (17).
FIG 1.
Alignment of verified and putative LcrF and ExsA binding sites within target gene promoters. (A) Promoter regions of Yersinia and Pseudomonas genes controlled by LcrF and ExsA, respectively, were aligned using SeaView (81). The sequences are from Y. enterocolitica 8081 (NC 008791), Y. pestis CO92 (NC 003131), Y. pseudotuberculosis IP 32953 (NC 006153), P. aeruginosa (NC 002516), and P. aeruginosa UCBPP PA14 (NC 008463). Predicted −10 regions are highlighted in blue. Identified 5′-AAAA-N6-GNCT-N5-TGANA-3′ consensus sites containing three conserved regions are highlighted in red, and the conserved nucleotides at each position of the motif are denoted in bold above the alignment, with uppercase letters denoting highly conserved residues and lowercase letters denoting more-degenerate residues. LcrF binding sites 1 and 2 are indicated by arrows. The sequences of the virA, virB, and yopH promoters from all three Yersinia species are identical, and thus a single sequence is shown. virA and virB are proximal but are divergently encoded. Thus, we propose that what appears to be an inverted 5′-AAAA-N6-GNCT-N5-TGANA-3′ motif upstream of the virA and virB promoters (blue solid-line box) actually belongs to the divergent virB and virA promoters, respectively. yopH and sycE have two tandem 5′-AAAA-N6-GNCT-N5-TGANA-3′ motifs (both highlighted in red), which overlap the protected regions identified by Wattiau and Cornelis (17). (B) Sequences upstream of Yersinia yopE were aligned using SeaView. Two regions containing inverted 5′-AAAA-N6-GNCT-N5-TGANA-3′ motifs which may belong to sycE are outlined in blue. The putative translational yopE start site is denoted in bold. (A and B) YtxR binding sites upstream of yopH (A) and yopE (B) are underlined in green (38). Regions experimentally found to be strongly bound by LcrF/VirF or ExsA are underlined with black solid lines, and those found to be weakly bound are underlined with black dashed lines (17). Identified putative LcrF binding sites are highlighted in red and the conserved nucleotides denoted in bold above the alignment, with uppercase letters denoting highly conserved residues and lowercase letters denoting more-degenerate residues.
A closer look at the yopE promoter revealed surprising features that required clarification. A putative LcrF binding site within the yopE promoter was not located 20 to 21 bp upstream of a −10 TATA box (Fig. 1B), as are the majority of putative LcrF sites, but overlapped the annotated translational start site of yopE (Fig. 1A). The −10 region of yopE, however, may be located 30 bp upstream of the translational start site, which is 12 bp upstream of the transcriptional start site (black arrow in Fig. 1B) (25). Interestingly, Wattiau and Cornelis identified two different regions further upstream from Y. enterocolitica yopE that were protected by VirF during DNase I footprinting (Fig. 1B in blue boxes) (17). These regions contain sequences that are the reverse complement of the 5′-AAAA-N6-GNCT-N5-TGANA-3′ motif. Because sycE and yopE are located in close proximity but are transcribed in opposite directions, it is possible that the two protected regions upstream of yopE, one strongly bound by VirF (underlined by a solid line in Fig. 1B) and the other weakly bound (underlined by dashed lines in Fig. 1B) (17), are in fact LcrF binding motifs on the reverse DNA strand, belonging to the sycE promoter. This LcrF binding site phenomenon of two adjacent, coregulated, but divergent genes can also be seen in the case of virA and virB (blue solid-line box in Fig. 1A). It is worth noting that some AraC-like activators require binding sites downstream of the −10 region (26, 27). For example, in the E. coli AraC-like activator Rns, it was found that the inverted Rns motifs upstream of target gene −10 regions and the motifs downstream of −10 regions function in synergy (26, 27). Therefore, it is possible that both the putative LcrF binding sites far upstream and those downstream of the yopE transcriptional start site are required for activation of the yopE promoter. Interestingly, yopE was reported to be transcribed strongly under T3SS-inducing conditions, while sycE was minimally transcribed (28). Because of this departure from the known characteristics of other LcrF binding sites, whether the putative LcrF binding sites within the yopE-sycE promoter regions are functional and, if so, how they function remain to be determined.
Putative LcrF binding sites appear in a number of locations in the virulence plasmids of Yersinia. As shown in Fig. 1A, a 5′-AAAA-N6-GNCT-N5-TGANA-3′ motif was found within the sycE, yopO, yadA, yopK, sycH, yscH, and yscC promoters. The transcriptional dependence of yadA and yopK (homologous to yopQ in Y. enterocolitica) on LcrF and VirF was previously demonstrated in transcriptional reporter assays in Y. pseudotuberculosis (29) and Y. enterocolitica (20), respectively. It is worth mentioning that we could not find LcrF binding sites upstream of two secreted effectors, yopJ and yopM, unless the threshold for motif calling was much lower (data not shown). Note also that yscC and yopK (from Y. enterocolitica and Y. pestis) do not seem to have a consensus TATA box following the TGANA motif, although these two genes are expressed under T3SS-inducing conditions (28). In contrast, the two LcrF binding sites upstream of virA and virB (Fig. 1A) have a TATA box at a proper distance but exhibit a weak dependence on LcrF (20).
Our analysis identified three putative LcrF binding sites within virC (yscA to yscL), upstream of yscA, yscC, and yscH (Fig. 1A). Transcriptional regulation of this region seems to be complex. Michiels et al. proposed the presence of a single operon from yscA to yscL in Y. enterocolitica (30). However, Haddix and Straley demonstrated the presence of at least two operons within the Y. pestis virC region, one starting upstream of yscA and the other starting upstream of yscF (31). The exact operon structure of the virC locus, as well as the LcrF binding sites important for virC gene expression, remains to be clarified.
Recent findings have shown that ExsA/LcrF-dependent promoters are σ70 promoters (32). The conserved TGANA sequence within the broader LcrF binding motif was previously mistaken for a −35 box and is thought to bind one of the two monomers of LcrF or ExsA dimers (20). Notably, the distance between the −10 box and the TGANA motif of LcrF-and-ExsA-dependent genes is 21 to 22 nucleotides (nt), while the spacing seen in typical σ70-dependent promoters is 17 nucleotides between the −10 and −35 boxes (32). Decreasing the distance between the −10 box and the TGANA motif from 21 or 22 nucleotides to 17 nucleotides in the ExsA-dependent exoT and exsD genes abolishes transcriptional activation of these genes, suggesting that the 21-to-22-nucleotide spacing ensures that activation of these promoters does not occur without ExsA binding (32).
Despite the resemblance in the consensus binding sequences, the oligomeric states of LcrF and ExsA during DNA binding as well as their binding properties are distinct, leading to differences in the DNA binding specificity and kinetics of transcriptional activation. ExsA is predominantly monomeric in solution, and two molecules are sequentially recruited to target promoters and generate two higher-order DNA-protein complexes to activate T3SS genes, whereas LcrF is dimeric and its presence results in the formation of only one large higher-order DNA-protein complex (18, 33). In addition, LcrF-induced promoter bending is more pronounced and may account for its overall (2.5-fold to 20-fold) higher activator activity relative to that of ExsA (18, 22). Lower basal activity of ExsA could be compensated by its positive autoregulatory feedback loop, which leads to a rapid increase of ExsA expression under inducing conditions (34). While mutations in critical nucleotides within the major groove disrupted both LcrF and ExsA DNA binding, LcrF, but not ExsA, was able to tolerate certain mutations within the consensus binding site, presumably because LcrF binds DNA as a preformed dimer and not in the sequential, ordered manner seen with ExsA monomers (18). Indeed, an LcrF mutant unable to dimerize was shown to bind to each half-site but, similarly to ExsA, was more sensitive to mutations within the binding site (18).
REGULATION OF LcrF TARGET GENES
To date, LcrF/VirF is the only characterized transcriptional activator of T3SS genes in pathogenic Yersinia, and all known LcrF target genes are carried on pCD1/pYV. Several groups have shown transcriptional activation or direct binding of LcrF to the promoter regions of the yopE, yopH, yadA, and ylpA genes as well as of the yopBD-lcrGVH and virC operons, which encode T3SS effector, regulatory, and structural proteins as well as the YadA adhesin and the YlpA lipoprotein (17, 20, 29, 30, 35–37).
Several reports have suggested that regulation of T3SS genes can be mediated by proteins antagonizing LcrF (38, 39). Darwin and colleagues identified YtxR as a global transcriptional regulator that is conserved in all human-pathogenic Yersinia species. Overexpression of YtxR rendered Yersinia defective in secretion of Yops into the bacterial culture supernatant (38). Using a DNase footprinting approach, the authors showed that YtxR protected specific regions in the yopE-sycE and yopH promoters that overlapped known LcrF binding sites (denoted by green lines in Fig. 1A), suggesting that YtxR competes with LcrF for binding to T3SS gene promoters (38). While YtxR may introduce an additional layer of regulation to T3SS gene expression, the environmental conditions under which YtxR is expressed have yet to be elucidated (38).
Recently, Li et al. proposed another model in which the pYV-encoded regulatory protein LcrQ inhibits LcrF activity (39). Y. pseudotuberculosis LcrQ was first discovered as a gene required for calcium dependence at 37°C and was shown to be secreted by the T3SS, relieving repression of Yop expression when the concentration of LcrQ in the bacterial cytoplasm decreased as a result of active secretion (40, 41). The Y. enterocolitica LcrQ orthologs YscM1 and YscM2 were subsequently suggested to control Yop expression in combination with pYV-encoded factors other than VirF (42). Indeed, the T3SS YopD translocon protein and its LcrH/SycD chaperone are thought to cooperate with LcrQ or YscM1/YscM2 to negatively control T3SS gene expression (43–47). LcrQ has no obvious DNA binding domain and does not bind Yop promoter regions, as has been shown for LcrF and YtxR (39). In fact, it has been suggested that Y. enterocolitica YscM1 and YscM2, in cooperation with the YopD-LcrH complex, bind to the 5′ untranslated regions of Yop mRNAs to inhibit translation (46). However, Li et al. proposed that Y. pseudotuberculosis LcrQ inhibits LcrF activity until the T3SS is assembled and LcrQ is secreted out of the cell. The authors showed that overexpression of LcrF mimicked the secretion profiles of a ΔlcrQ mutant. This suggested that the LcrF/LcrQ ratio may be important for activating T3SS gene transcription, but they could not detect a direct interaction of LcrF and LcrQ (39). Moreover, previous data indicated that Y. enterocolitica YscM1 was not capable of inhibiting VirF activity on the yopH promoter in the absence of other pYV-encoded factors (42). Therefore, how LcrQ and YscM1/YscM2 repress T3SS gene expression in pathogenic Yersinia remains to be clarified.
TRANSCRIPTIONAL CONTROL OF LcrF
Thermoregulation of lcrF transcription and ymoA.
There are several lines of evidence showing that transcription of lcrF is regulated by temperature. The first observations were made with Y. enterocolitica, in which induction of a virF-cat fusion, and a substantial increase in the abundance of the virF transcript, was detected upon temperature upshift (15, 48). A comprehensive expression analysis revealed that the lcrF gene of Y. pseudotuberculosis is transcribed from a σ70-dependent promoter located upstream of the yscW gene (named virG in Y. enterocolitica), which in turn is located 124 bp upstream of the lcrF coding sequence (Fig. 2). Temperature-dependent yscW-lcrF transcription from this promoter is controlled by the nucleoid-associated YmoA protein, which shows homology to the Hha protein of E. coli (49). YmoA (for “Yersinia modulator”) was identified in a search for chromosomal insertion mutants of Y. enterocolitica transcribing virF and, hence, VirF-dependent yop and yadA genes at low temperatures (48). Mutations in ymoA led to increased virF and yop expression at 25°C and decreased expression at 37°C compared to that in the control strain, although ymoA mutants still induced expression of virF and yop upon temperature shift (48). Elevated expression of a yscW-lacZ fusion in a ymoA-deficient Y. pseudotuberculosis strain further suggested that YmoA represses yscW-lcrF transcription from a promoter located 264 nt upstream of the start codon of yscW (49) (Fig. 2). As YmoA dependency was lost when sequences downstream of the yscW transcriptional start site were deleted but was maintained when the regulatory region upstream of the yscW promoter was removed, it is assumed that YmoA influences lcrF expression via sequences located downstream of the yscW promoter (49). However, even high concentrations of purified YmoA homodimers were unable to interact specifically with the yscW regulatory region, indicating that an additional factor contributes to YmoA-mediated repression of yscW-lcrF transcription at moderate temperatures (15°C to 30°C) (49).
FIG 2.
Regulatory elements encoded within the yscW-lcrF sequence. Nucleotide sequences of the yscW-lcrF promoter regions for Y. pestis CO92, Y. enterocolitica 8081, and Y. pseudotuberculosis YPIII were aligned using ClustalW2. Nucleotides whose sequences are not identical are marked in red, while the conserved nucleotides are indicated by asterisks. The identified binding sequence for IscR is marked in purple, and critical residues required for IscR binding within this motif are in bold (28, 64). The binding site for RcsB is marked in green (71). The region experimentally determined to be involved in YmoA/H-NS binding, marked in brown, is downstream of the transcriptional start site (49). The sequence encoding the RNA thermometer is marked in yellow and is encoded within the intergenic region between yscW and lcrF (49). The DNA bending region identified within the yscW-lcrF operon is denoted with a dotted line and is within the intergenic region between yscW and lcrF (59). The −10 and −35 boxes, the transcriptional start site, the ribosome binding sites (rbs) upstream of the yscW and lcrF coding sequence, and the fourU element in the intergenic region between the yscW and lcrF coding regions are marked in bold (49). All coding sequences are marked in blue.
The small size of YmoA, its unusual high number of charged amino acid residues, and its influence on the fragility of the chromosomal DNA suggested that YmoA is a histone-like protein involved in chromosome structure similarly to histone-like nucleoid-structuring (H-NS) protein and controls lcrF expression through temperature-induced changes in DNA topology (48, 50, 51). Domains of members of the Hha/YmoA protein family have a striking similarity to the oligomerization domain of the H-NS nucleoid-structuring protein and its paralogs and were shown to interact specifically with different enterobacterial H-NS proteins (52, 53). Band shift analyses performed with yscW promoter fragments demonstrated that YmoA copurified with H-NS was able to interact specifically with sequences located downstream of the yscW promoter (Fig. 2) (49). Therefore, heterocomplex formation of YmoA with H-NS seems responsible for the thermoregulation of the yscW-lcrF operon. Interestingly, H-NS alone is also able to interact with yscW promoter fragments (49). In this context, it would be important to know how H-NS homodimers and H-NS/YmoA heterodimers differ in their abilities to interact with the yscW promoter region and how this influences expression of the yscW-lcrF operon in response to temperature. Several independent attempts to construct an hns-deficient mutant in Yersinia to address this issue failed, indicating that H-NS is essential for the biological fitness of members of this genus (54, 55).
It is also quite reasonable that the DNA binding abilities of H-NS/H-NS and H-NS/YmoA complexes are differentially influenced by temperature-induced topological changes of the promoter and/or intrinsic conformational alterations of the regulatory proteins (56, 57). In fact, it has been reported that the DNA topology of the virulence plasmid undergoes a conformational change upon upshift to 37°C, leading to significant derepression of virF-lcrF expression (15, 30, 58, 59). Rohde et al. observed that Y. enterocolitica mutants resistant to the DNA gyrase inhibitor novobiocin constitutively expressed Yops, similarly to a ΔymoA mutant (59). Using a two-dimensional (2-D) gel-based assay, the authors identified several DNA-intrinsic bends in the pYV plasmid. Interestingly, the presence of a DNA bend within the intergenic region between the yscW and virF genes was identified (Fig. 2). This bend was shown to melt at 37°C, suggesting that this intrinsic bend could potentially inhibit transcription of virF at noninducing temperatures (59).
Lastly, YmoA of Y. pestis and Y. pseudotuberculosis is subject to proteolysis by the Lon and ClpP proteases at 37°C but not at common environmental temperatures between 15°C and 30°C (49, 57). As a result, YmoA-mediated repression of the yscW-lcrF operon is rapidly eliminated at 37°C to induce the T3SS, but the repression effect remains present at all temperatures in a clpP-lon deletion mutant (57). Nevertheless, LcrF synthesis is still significantly enhanced in a ymoA-deficient strain at host body temperature compared to 25°C, indicating the importance of thermally induced DNA bending, the RNA thermometer (see below), and, possibly, H-NS homodimers.
Cross-regulation with flagellar system.
Many Yersinia flagellar genes, including the alternative flagellum-specific σ28 sigma factor encoded by fliA, are strictly controlled by temperature. In contrast to T3SS genes, they are upregulated only at moderate temperatures and repressed at body temperature (60). Yersinia likely utilizes flagellar motility in the environment at temperatures under 37°C and does not require the T3SS prior to transmission into a mammalian host. This suggests an inverse regulation of flagellar and T3SS genes, and some evidence exists that σ28/FliA is crucial for this process (61). Through microarray analysis comparing the Y. enterocolitica wild-type strain and a ΔfliA mutant, several pYV-borne genes such as virF were found to be upregulated in the ΔfliA mutant at 25°C (61). Furthermore, a putative binding site for FliA was identified in the virF promoter (61), suggesting that FliA binds to the virF promoter to repress transcription under temperature conditions that induce flagellar expression and assembly (61). However, we could not identify the putative FliA site described by Horne and Prüss (61) within the virF upstream region either by scanning for the exact FliA binding site sequence suggested by the authors or by using known FliA binding sites to make a motif model to search against the Y. enterocolitica genome (unpublished observations). While a discernible FliA motif may not be present in the virF promoter region in the current Y. enterocolitica genome assembly, the discrepancy might be due to differences between the current genome assembly and the one used in Horne and Prüss. Interestingly, Y. pseudotuberculosis bacteria lacking the RNA chaperone Hfq are hypermotile but defective in type III secretion (62). Thus, further studies are needed to identify the mechanism(s) involved in maintaining this inverse relationship between flagellar motility and T3SS expression.
IscR.
It has been recently suggested that LcrF might be affected by environmental signals other than temperature. Through a forward genetic screen for modulators of the Y. pseudotuberculosis T3SS, the iron-sulfur cluster coordinating transcription regulator IscR was identified as important for type III secretion and Yersinia virulence (28). IscR is a global transcriptional regulator that has been extensively characterized in E. coli (63). The ability of IscR to modulate transcription of target genes depends on coordination of a [2Fe-2S] cluster, and IscR is an active transcription factor in both the apo-IscR and holo-IscR forms (63–65). This is due to the ability of IscR to recognize two separate DNA binding motifs: type I motifs are bound by holo-IscR, while type II motifs are recognized by both apo-IscR and holo-IscR (63, 64, 66). In Y. pseudotuberculosis, it was shown that IscR binds to a type II motif within the lcrF promoter (Fig. 2) (28), suggesting that IscR controls transcription of Yersinia type III secretion directly. Furthermore, this motif within the lcrF promoter is 100% conserved between Y. pseudotuberculosis and Y. pestis species and contains all nine residues found to be critical for IscR binding in E. coli (64). While the IscR binding site is not 100% conserved in Y. enterocolitica, the nine residues that were found to be critical for IscR binding are conserved (Fig. 2, marked in bold). This indicates a possible mechanism of T3SS gene regulation that is conserved among the three pathogens. It has been suggested that oxidative stress and oxygen limitation (as a result of Fe-S cluster damage) as well as iron availability influence the apo-IscR/holo-IscR ratio and that these environmental signals may affect IscR expression and activity (67–70). Therefore, while it has not yet been demonstrated, oxidative stress, oxygen limitation, or iron availability may influence expression of LcrF and the T3SS.
RcsB and CpxR in response to extracytoplasmic stress.
A recent study reported that T3SS/yop expression in Yersinia is regulated by the Rcs phosphorelay system—a complex signaling pathway used by members of the Enterobacteriaceae to adapt their cellular physiology, biofilm and capsule formation, and motility in response to perturbations in external or surface-associated processes, e.g., overproduction of envelope components, osmotic shock, or desiccation (71). Overexpression of the wild type or a constitutive active variant of the response regulator RcsB enhanced mRNA levels of LcrF as well as Yop protein expression and secretion, suggesting that RcsB influences T3SS/Yop through LcrF (71). This was confirmed by the fact that activated/phosphorylated RcsB has the capacity to bind directly to a conserved RcsB box just upstream of the −35 promoter element of the yscW-lcrF operon (Fig. 2). RcsB binding most likely enhances RNA polymerase binding and/or function (71).
In addition to RcsB, it has been established that a second response regulator, CpxR, of another prominent phosphorelay system that responds to extracytoplasmic stress conditions targets the yscW-lcrF promoter region. However, in contrast to RcsB, CpxR represses transcription of the operon, suggesting that the two regulatory components either are part of a joint regulatory cascade or are separately induced during different infection stages or in different niches (71, 72).
Effects on lcrF revealed by global expression analyses.
To gain insight into genes expressed by Yersinia during septicemia, global transcription patterns of bacteria grown in human plasma were compared with those of bacteria grown in Luria-Bertani broth, a standard laboratory medium (73, 74). Y. pestis virG and yscW and Y. pseudotuberculosis lcrF were specifically upregulated in human plasma at 37°C, indicating that induction of the yscW-lcrF operon is important for virulence during the septicemic phase of the infection. A strong upregulation of lcrF gene expression was also observed at various time points after nasal infection of mice with Y. pestis in the lungs, spleen, and liver, supporting previous studies indicating that LcrF-controlled T3SS/yop genes are crucial in resisting immune and inflammatory defensive responses during the development of pneumonic plague (75).
A transcriptomic study designed to identify genes under the control of the recently recognized YbeY endonuclease demonstrated that deletion of the ybeY gene led to an upregulation of lcrF and the T3SS/yop genes in Y. enterocolitica serotype O:3 under conditions in which these genes are usually repressed (i.e., at 22°C) (76). This derepression was not caused by lower YmoA levels, since the amount of ymoA transcript was increased in the ybeY mutant. Instead, the authors suggested that lcrF upregulation in the ybeY mutant was due to increased copy numbers of the virulence plasmid or to altered regulation of global regulators (e.g., Hfq, nucleoid-structuring proteins) affecting the noncoding RNA network and/or DNA supercoiling (76).
Transcriptional profiling further revealed that bacterial membrane permeability may affect expression of LcrF and the T3SS/Yops. Microarray analysis showed that LcrF was upregulated by ∼2-fold in a ΔrovA Y. pestis mutant grown under T3SS-inducing conditions (77). This result seems surprising, as RovA is a transcriptional regulator that is upregulated at 25°C and best known for inducing expression of invasin, an important virulence factor in enteropathogenic Yersinia but absent from Y. pestis. Through gel shift analysis, the possibility that RovA binds to the lcrF promoter to modulate transcription was ruled out. However, small electron-dense particles were found surrounding the ΔrovA mutant membrane by the use of transmission electron microscopy and the membrane permeability of the mutant was decreased compared to that of the wild type (77). The authors suggested that membrane construction is altered in the absence of RovA and that this might impact assembly and regulation of the T3SS. However, the connection between membrane integrity and LcrF has not been further explored.
TRANSLATIONAL CONTROL OF lcrF IN RESPONSE TO TEMPERATURE
Several early studies on temperature sensing in Yersinia reported that levels of protein encoded by LcrF-regulated genes such as yopE still change in response to temperature even under conditions in which lcrF transcription remains constant (37). Moreover, forced transcription of lcrF at low temperatures did not cause induction of LcrF-dependent genes (20). This implied that posttranscriptional mechanisms modulate LcrF synthesis and/or its specific activity in response to temperature. Hoe et al. (78) reported that Y. pestis LcrF is controlled at the translational level in response to temperature. This thermal control was maintained when lcrF, containing only 208 bp of the upstream 5′ untranslated region, was transcribed by the T7 polymerase in E. coli (78). Based on these data, a simple model of thermal regulation of lcrF translation was proposed in which the presence of a short predicted thermolabile stem-loop structure, or RNA thermometer, sequesters the lcrF ribosome binding site (rbs) sequence and blocks translation initiation at moderate temperatures. The decreased stability or melting of this structure at higher temperatures liberates the rbs sequence and allows formation of a productive mRNA complex and efficient translation (78).
A comprehensive secondary-structure prediction of the 124-bp intergenic region of yscW and lcrF performed using algorithms such as mfold and RNAfold suggested that this untranslated region of the bicistronic operon folds into two hairpin structures with a free energy of −19.67 kcal/mol (Fig. 2) (49). The second temperature-sensitive stem-loop includes a stretch of four uridines base-paired with the AGGA sequence of the rbs (49). This short motif with its simple design is referred to as a fourU element. It was first discovered in the agsA (aggregation suppression A) heat shock gene in Salmonella enterica serovar Typhimurium and bears resemblance to other potential fourU elements identified in the 5′ untranslated region of the groES and dnaJ heat shock genes of Staphylococcus aureus and Brucella melitensis (79). The presence of small loops and several noncanonical base pairs coupled with a network of weak hydrogen bonds, which facilitate liberation of the lcrF rbs, argued for a thermolabile RNA structure prone to melting within a physiological temperature range (49).
Enzymatic structural probing experiments using RNases T1 and V1 confirmed in silico predictions and demonstrated temperature-induced partial, but not complete, opening of the second hairpin loop (49). Existence of a thermosensing RNA element (RNA thermometer) was further confirmed by (i) lcrF-lacZ translational fusions, which were thermally induced when the fusion was transcribed from a temperature-independent PBAD promoter, (ii) toe printing assays demonstrating that binding of ribosomes to the lcrF translational start site is restricted to 37°C and does not occur at 25°C, and (iii) base substitutions within the second hairpin of the thermosensing RNA element designed to stabilize or destabilize the second stem-loop (49). These stabilizing point mutations led to a “closed” conformation of the RNA element, resulting in full repression of LcrF synthesis at 37°C. In contrast, the destabilizing mutations allowed an opening of RNA structure (“open” conformation) and enhanced LcrF production at moderate and higher temperatures. Posttranscriptional control in an RNA thermometer-like fashion was further confirmed with a deletion of a stretch of nucleotides implicated in the formation of hairpin II, which completely abolished the thermosensing function of the intergenic RNA element and provoked constitutive increased synthesis of LcrF in a temperature range from 25°C to 37°C. In contrast, an RNA thermometer variant, which consisted only of the second hairpin, was more open than the full RNA thermometer and was characterized by higher LcrF levels. Nevertheless, this shortened version was still temperature inducible, indicating that the first stem-loop is not essential for thermosensing but seems to promote proper folding and/or supports the stability of the second hairpin (49).
In order to test the physiological relevance of the lcrF RNA thermometer and its role in Yersinia virulence, the pathogenicities of a closed RNA thermometer variant and an open variant of Y. pseudotuberculosis were compared with that of the isogenic wild-type strain in an oral mouse infection model. Despite the fact that all mice were challenged with a normally lethal dose of Y. pseudotuberculosis, all animals infected with Yersinia mutants encoding the closed RNA thermometer variant survived and showed no visible signs of infection, similarly to an lcrF-deficient strain, and displayed decreased colonization of the Peyer's patches, mesenteric lymph nodes, liver, and spleen (49). Intriguingly, overexpression of the T3SS/yop virulence program, as displayed by the open variant, was not beneficial and did not cause greater host mortality. In contrast, colonization of some host tissues was slightly reduced, and the average time to death remained unchanged or was even increased by several days, most likely due to biological fitness impediments of the pathogen or increased inflammation in the host (49). This clearly illustrated that the lcrF RNA thermometer, all examples of which are 100% identical in all human-pathogenic Yersinia species, is a decisive posttranscriptional control element evolved to produce just the right amount of LcrF to promote the most ideal infection efficiency.
Apparently, both the YmoA- and RNA thermometer-mediated thermosensing mechanisms achieve a very rapid and efficient response. However, recent reports indicate that this control strategy seems to be complemented by additional regulatory modules adjusting LcrF synthesis according to host cell contact and T3SS-mediated effector translocation. In fact, the YopD translocator protein was recently found to bind to the 5′ untranslated sequences of multiple T3SS/yop genes, including lcrF, which facilitates their degradation (47). The molecular mechanism of YopD-mediated repression of LcrF synthesis is still unknown, but it is possible that YopD binding to the lcrF transcript in the absence of host cells (i) promotes a more closed conformation of the RNA thermometer and/or (ii) accelerates the degradation of the lcrF transcript as a consequence of the blockage of ribosome binding and translation. Alternatively, it is possible that YopD controls expression of additional factors influencing lcrF transcript stability.
CONCLUSIONS
Yersinia bacteria and other T3SS-expressing pathogens have complex regulatory networks in place to control expression of T3SS genes. While the Yersinia T3SS master regulator LcrF was identified almost three decades ago, recent work has greatly expanded our understanding of how expression of LcrF is regulated and what environmental signals might contribute to its regulation. YmoA, RcsB, and IscR all enhance transcription of lcrF (Fig. 3). As Yersinia experiences changes in temperature during the transition from the environment or the flea vector to the mammalian host, as well as various stresses, including iron limitation and reactive oxygen species (ROS) production during infection, sensing these environmental cues to control LcrF and T3SS expression may enable Yersinia to optimize T3SS deployment and virulence. While temperature has indeed been shown to contribute to lcrF transcription, future work focusing on expression of LcrF and T3SS genes in different host niches will enable a more complete understanding of how this T3SS master regulator facilitates optimization of T3SS expression to promote virulence.
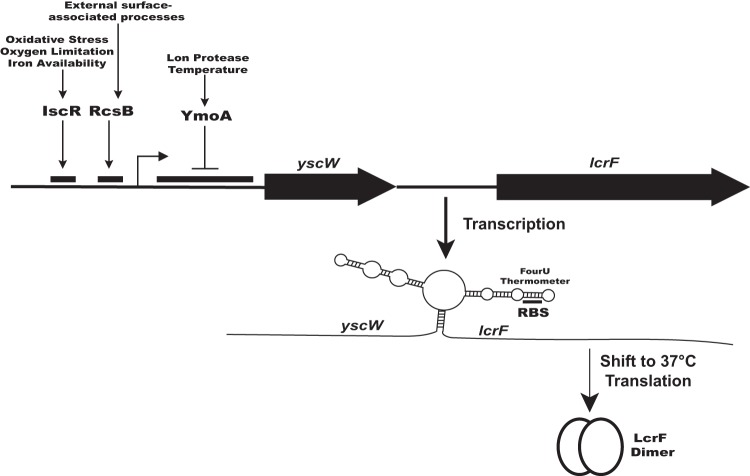
FIG 3.
Multiple environmental signals control lcrF expression and, subsequently, T3SS expression through several distinct transcriptional and translational regulatory mechanisms. The data summarized in this review suggest that the YmoA, RcsB, and IscR regulators control transcription of lcrF in response to temperature, extracytoplasmic stress, iron bioavailability, oxygen tension, and reactive oxygen species. In addition, the RNA thermometer found upstream of lcrF allows LcrF translation only at the mammalian host body temperature, 37°C. As Yersinia transits from the environment or the flea vector to the mammalian host and then from localized to disseminated sites of infection, changes in temperature, iron availability, and stresses such as ROS may direct the regulatory network controlling LcrF, optimizing T3SS deployment and virulence.
Biographies

Leah Schwiesow received her B.S. in chemistry from Gonzaga University in Spokane, WA, in 2009. While an undergraduate, she became interested in molecular biology and worked to characterize the role of calcium in the regulation of photosynthesis in Arabidopsis thaliana. After graduating, she took a position at Dow Agrosciences, where she worked on a team optimizing maize transformation protocols. She started the molecular, cellular, and developmental biology Ph.D. program at the University of California, Santa Cruz, in 2011 and joined Dr. Auerbuch's research group, where she discovered her love for microbes. Her current research is focused on how IscR in Yersinia pseudotuberculosis responds to changing environmental signals within the host to optimize expression of factors important for virulence.

Hanh Lam completed her Ph.D. in plant pathology and plant-microbe biology at Cornell University. Her thesis research was on the plant pathogen Pseudomonas syringae pv. DC3000, focusing on regulation of that organism's type III secretion system. At Cornell, Hanh developed interests in and was trained in bacterial gene regulation, genomics, and computational biology. In 2014, she joined Dr. Auerbuch's research group at the University of California, Santa Cruz, as a postdoctoral scientist. Dr. Hanh's main research focus is on developing methods for high-throughput screening to identify antimicrobial compounds targeting the T3SS. The resulting compounds could be used for therapeutics and as biochemical tools to study T3SS regulation.

Petra Dersch performed the work for her Ph.D. thesis at University Konstanz and the Max Planck Institute for Terrestrial Microbiology in Marburg on the nucleoid-associated H-NS protein of E. coli. She was a postdoctoral fellow with Ralph Isberg at the Tufts Medical School/Howard Hughes Institute in Boston, MA, where she became interested in the function and regulation of crucial virulence-related factors pertaining to enteropathogenic Yersinia species. She continued her work as independent group leader at the Free University in Berlin and as a junior research group leader at the Robert Koch Institute in Berlin. She joined the faculty of microbiology at the Technical University Braunschweig in 2005 and since 2008 has been head of the Department of Molecular Infection Biology at the Helmholtz Center for Infection Research in Braunschweig. Her current main research interest is the global regulation of Yersinia virulence factors by posttranscriptional control mechanisms, with an emphasis on sensory and regulatory RNAs.

Victoria Auerbuch received her diploma from Cornell University in 1997 and her Ph.D. from the University of California, Berkeley (U.C. Berkeley), in 2002, where she worked with Daniel Portnoy. She was a postdoctoral researcher at U.C. Berkeley from 2003 to 2004 and then with Ralph Isberg at Tufts University School of Medicine, where she began working on the immune response of mammalian cells to the Yersinia type III secretion system (T3SS). In 2009, she joined the faculty in the Department of Microbiology and Environmental Toxicology at U.C. Santa Cruz, where she is now an associate professor. Her current main research interests include IscR control of virulence factor utilization in Yersinia and development of T3SS inhibitors.
Funding Statement
We also acknowledge the Vietnam Education Foundation for support (to H.L.).
REFERENCES
- 1.Putzker M, Sauer H, Sobe D. 2001. Plague and other human infections caused by Yersinia species. Clin Lab 47:453–466. [PubMed] [Google Scholar]
- 2.Khan FA, Fisher MA, Khakoo RA. 2007. Association of hemochromatosis with infectious diseases: expanding spectrum. Int J Infect Dis 11:482–487. doi: 10.1016/j.ijid.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Bottone EJ. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect 1:323–333. doi: 10.1016/S1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 4.Smego RA, Frean J, Koornhof HJ. 1999. Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur J Clin Microbiol Infect Dis 18:1–15. doi: 10.1007/s100960050219. [DOI] [PubMed] [Google Scholar]
- 5.Koornhof HJ, Smego RA Jr, Nicol M. 1999. Yersiniosis. II: the pathogenesis of Yersinia infections. Eur J Clin Microbiol Infect Dis 18:87–112. doi: 10.1007/s100960050237. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev 62:1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bliska JB, Wang X, Viboud GI, Brodsky IE. 2013. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol 15:1622–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kupferberg LL, Higuchi K. 1958. Role of calcium ions in the stimulation of growth of virulent strains of Pasteurella pestis. J Bacteriol 76:120–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi K, Kupferberg LL, Smith JL. 1959. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J Bacteriol 77:317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yother J, Goguen JD. 1985. Isolation and characterization of Ca2+-blind mutants of Yersinia pestis. J Bacteriol 164:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry RD, Harmon PA, Bowmer WS, Straley SC. 1986. A low-Ca2+ response operon encodes the V antigen of Yersinia pestis. Infect Immun 54:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yother J, Chamness TW, Goguen JD. 1986. Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J Bacteriol 165:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelis G, Vanootegem JC, Sluiters C. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb Pathog 2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 14.Bölin I, Forsberg A, Norlander L, Skurnik M, Wolf-Watz H. 1988. Identification and mapping of the temperature-inducible, plasmid-encoded proteins of Yersinia spp. Infect Immun 56:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis G, Sluiters C, de Rouvroit CL, Michiels T. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol 171:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleif R. 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 17.Wattiau P, Cornelis GR. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol 176:3878–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King JM, Schesser Bartra S, Plano G, Yahr TL. 2013. ExsA and LcrF recognize similar consensus binding sites, but differences in their oligomeric state influence interactions with promoter DNA. J Bacteriol 195:5639–5650. doi: 10.1128/JB.00990-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee S, Martin RG, Rosner JL, Davies DR. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci U S A 95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert de Rouvroit C, Sluiters C, Cornelis GR. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol Microbiol 6:395–409. doi: 10.1111/j.1365-2958.1992.tb01483.x. [DOI] [PubMed] [Google Scholar]
- 21.Diaz MR, King JM, Yahr TL. 2011. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front Microbiol 2:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. 2008. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 68:657–671. doi: 10.1111/j.1365-2958.2008.06179.x. [DOI] [PubMed] [Google Scholar]
- 23.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuss AM, Heroven AK, Waldmann B, Reinkensmeier J, Jarek M, Beckstette M, Dersch P. 2015. Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet 11:e1005087. doi: 10.1371/journal.pgen.1005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munson GP, Scott JR. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol Microbiol 36:1391–1402. [DOI] [PubMed] [Google Scholar]
- 27.Munson GP, Holcomb LG, Scott JR. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect Immun 69:186–193. doi: 10.1128/IAI.69.1.186-193.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller HK, Kwuan L, Schwiesow L, Bernick DL, Mettert E, Ramirez HA, Ragle JM, Chan PP, Kiley PJ, Lowe TM, Auerbuch V. 2014. IscR is essential for Yersinia pseudotuberculosis type III secretion and virulence. PLoS Pathog 10:e1004194. doi: 10.1371/journal.ppat.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.China B, Michiels T, Cornelis GR. 1990. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol Microbiol 4:1585–1593. doi: 10.1111/j.1365-2958.1990.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 30.Michiels T, Vanooteghem JC, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis GR. 1991. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol 173:4994–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haddix PL, Straley SC. 1992. Structure and regulation of the Yersinia pestis yscBCDEF operon. J Bacteriol 174:4820–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vakulskas CA, Brady KM, Yahr TL. 2009. Mechanism of transcriptional activation by Pseudomonas aeruginosa ExsA. J Bacteriol 191:6654–6664. doi: 10.1128/JB.00902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brutinel ED, Vakulskas CA, Yahr TL. 2009. Functional domains of ExsA, the transcriptional activator of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 191:3811–3821. doi: 10.1128/JB.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahr TL, Frank DW. 1994. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol 176:3832–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michiels T, Cornelis G. 1988. Nucleotide sequence and transcription analysis of yop51 from Yersinia enterocolitica W22703. Microb Pathog 5:449–459. doi: 10.1016/0882-4010(88)90006-X. [DOI] [PubMed] [Google Scholar]
- 36.Skurnik M, Toivanen P. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J Bacteriol 174:2047–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoe NP, Minion FC, Goguen JD. 1992. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol 174:4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Axler-DiPerte GL, Hinchliffe SJ, Wren BW, Darwin AJ. 2009. YtxR acts as an overriding transcriptional off switch for the Yersinia enterocolitica Ysc-Yop type 3 secretion system. J Bacteriol 191:514–524. doi: 10.1128/JB.01305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Yan H, Feng L, Li Y, Lu P, Hu Y, Chen S. 2014. LcrQ blocks the role of LcrF in regulating the Ysc-Yop type III secretion genes in Yersinia pseudotuberculosis. PLoS One 9:e92243. doi: 10.1371/journal.pone.0092243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 41.Rimpiläinen M, Forsberg A, Wolf-Watz H. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J Bacteriol 174:3355–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stainier I, Iriarte M, Cornelis GR. 1997. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol 26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 43.Kopaskie KS, Ligtenberg KG, Schneewind O. 2013. Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J Biol Chem 288:35478–35488. doi: 10.1074/jbc.M113.504811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams AW, Straley SC. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J Bacteriol 180:350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson DM, Ramamurthi KS, Tam C, Schneewind O. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol 184:1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cambronne ED, Schneewind O. 2002. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J Bacteriol 184:5880–5893. doi: 10.1128/JB.184.21.5880-5893.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Anderson DM. 2011. Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol Microbiol 80:966–980. doi: 10.1111/j.1365-2958.2011.07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornelis G, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rouvroit C, Sory MP, Vanooteghem JC, Michiels T. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol 5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 49.Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cornelis GR. 1993. Role of the transcription activator virF and the histone-like protein YmoA in the thermoregulation of virulence functions in yersiniae. Zentralbl Bakteriol 278:149–164. doi: 10.1016/S0934-8840(11)80833-9. [DOI] [PubMed] [Google Scholar]
- 51.Mikulskis AV, Cornelis GR. 1994. A new class of proteins regulating gene expression in enterobacteria. Mol Microbiol 11:77–86. doi: 10.1111/j.1365-2958.1994.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 52.Madrid C, Nieto JM, Juarez A. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int J Med Microbiol 291:425–432. [DOI] [PubMed] [Google Scholar]
- 53.Nieto J, Madrid C, Miquelay E, Parra JL, Rodriguez S, Juarez A. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J Bacteriol 184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heroven A, Nagel G, Tran HJ, Parr S, Dersch P. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol 53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 55.Ellison DW, Miller VL. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J Bacteriol 188:5101–5112. doi: 10.1128/JB.00862-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J 391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson M, Silva-Herzog E, Plano GV. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. sMol Microbiol 54:1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x. [DOI] [PubMed] [Google Scholar]
- 58.Rohde JR, Fox JM, Minnich SA. 1994. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol 12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 59.Rohde JR, Luan XS, Rohde H, Fox JM, Minnich SA. 1999. The Yersinia enterocolitica pYV virulence plasmid contains multiple intrinsic DNA bends which melt at 37 degrees C. J Bacteriol 181:4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapatral V, Olson JW, Pepe JC, Miller VL, Minnich SA. 1996. Temperature-dependent regulation of Yersinia enterocolitica Class III flagellar genes. Mol Microbiol 19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 61.Horne SM, Prüss BM. 2006. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch Microbiol 185:115–126. doi: 10.1007/s00203-005-0077-1. [DOI] [PubMed] [Google Scholar]
- 62.Schiano CA, Bellows LE, Lathem WW. 2010. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect Immun 78:2034–2044. doi: 10.1128/IAI.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A 98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nesbit AD, Giel JL, Rose JC, Kiley PJ. 2009. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol 387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Munck E, Kiley PJ. 2012. Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51:4453–4462. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ, Kiley PJ. 2013. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat Struct Mol Biol 20:740–747. doi: 10.1038/nsmb.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mettert EL, Kiley PJ. 2014. Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J Bacteriol 196:4315–4323. doi: 10.1128/JB.01975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol 60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 69.Yeo WS, Lee JH, Lee KC, Roe JH. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol 61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 70.Wu Y, Outten FW. 2009. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol 191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Hu Y, Francis MS, Chen S. 2015. RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ Microbiol 17:1219–1233. doi: 10.1111/1462-2920.12556. [DOI] [PubMed] [Google Scholar]
- 72.Liu J, Thanikkal EJ, Obi IR, Francis MS. 2012. Elevated CpxR∼P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res Microbiol 163:518–530. doi: 10.1016/j.resmic.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 73.Rosso ML, Chauvaux S, Dessein R, Laurans C, Frangeul L, Lacroix C, Schiavo A, Dillies MA, Foulon J, Coppee JY, Medigue C, Carniel E, Simonet M, Marceau M. 2008. Growth of Yersinia pseudotuberculosis in human plasma: impacts on virulence and metabolic gene expression. BMC Microbiol 8:211. doi: 10.1186/1471-2180-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chauvaux S, Rosso ML, Frangeul L, Lacroix C, Labarre L, Schiavo A, Marceau M, Dillies MA, Foulon J, Coppee JY, Medigue C, Simonet M, Carniel E. 2007. Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology 153:3112–3124. doi: 10.1099/mic.0.2007/006213-0. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Wang H, Qiu J, Wang X, Guo Z, Qiu Y, Zhou D, Han Y, Du Z, Li C, Song Y, Yang R. 2009. Transcriptional profiling of a mice plague model: insights into interaction between Yersinia pestis and its host. J Basic Microbiol 49:92–99. doi: 10.1002/jobm.200800027. [DOI] [PubMed] [Google Scholar]
- 76.Leskinen K, Varjosalo M, Skurnik M. 2015. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology 161:285–299. doi: 10.1099/mic.0.083097-0. [DOI] [PubMed] [Google Scholar]
- 77.Yang F, Ke Y, Tan Y, Bi Y, Shi Q, Yang H, Qiu J, Wang X, Guo Z, Ling H, Yang R, Du Z. 2010. Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS One 5:e12840. doi: 10.1371/journal.pone.0012840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoe NP, Goguen JD. 1993. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol 175:7901–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol 65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 80.Venecia K, Young GM. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect Immun 73:5961–5977. doi: 10.1128/IAI.73.9.5961-5977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]