Abstract
The oleaginous yeast Yarrowia lipolytica is an industrially important host for production of organic acids, oleochemicals, lipids, and proteins with broad biotechnological applications. Albeit known for decades, the unique native metabolism of Y. lipolytica for using complex fermentable sugars, which are abundant in lignocellulosic biomass, is poorly understood. In this study, we activated and elucidated the native sugar metabolism in Y. lipolytica for cell growth on xylose and cellobiose as well as their mixtures with glucose through comprehensive metabolic and transcriptomic analyses. We identified 7 putative glucose-specific transporters, 16 putative xylose-specific transporters, and 4 putative cellobiose-specific transporters that are transcriptionally upregulated for growth on respective single sugars. Y. lipolytica is capable of using xylose as a carbon source, but xylose dehydrogenase is the key bottleneck of xylose assimilation and is transcriptionally repressed by glucose. Y. lipolytica has a set of 5 extracellular and 6 intracellular β-glucosidases and is capable of assimilating cellobiose via extra- and intracellular mechanisms, the latter being dominant for growth on cellobiose as a sole carbon source. Strikingly, Y. lipolytica exhibited enhanced sugar utilization for growth in mixed sugars, with strong carbon catabolite activation for growth on the mixture of xylose and cellobiose and with mild carbon catabolite repression of glucose on xylose and cellobiose. The results of this study shed light on fundamental understanding of the complex native sugar metabolism of Y. lipolytica and will help guide inverse metabolic engineering of Y. lipolytica for enhanced conversion of biomass-derived fermentable sugars to chemicals and fuels.
INTRODUCTION
Lignocellulosic biomasses, derived from agricultural residues or nonfood crops, are potential renewable feedstocks for sustainable microbial production of biofuels and biochemicals (1). Lignocellulosic biomass is more complex and recalcitrant than corn starch, containing mixed sugars such as C6 sugars (e.g., glucose) and C5 sugars (e.g., xylose) (2). Most microorganisms do not efficiently consume these mixed sugars due to the well-known carbon catabolite repression (CCR) effect (3). The underlying CCR mechanism is governed by complex enzymatic and transcriptional regulation of metabolic processes (e.g., sugar transporters, sugar-degrading enzymes, etc.) that make microbial cell factories preferentially use one sugar (e.g., glucose) instead of other sugars (e.g., xylose and cellobiose) (4). For instance, a higher-level CCR effect causes diauxic growth (5); a milder effect allows simultaneous sugar utilization but often makes the specific uptake rate of one sugar higher than that of others (6). For biotechnological application, it is highly desirable to engineer microorganisms as microbial cell factories that can efficiently convert complex biomass-derived sugars to desirable chemicals with minimal CCR effect (7, 8).
Fig. 1 shows assimilation pathways of glucose, xylose, and cellobiose in native yeasts. Most yeasts such as Saccharomyces cerevisiae can consume only C6 sugars (9), while a few other yeasts such as Pichia stipitis (also known as Scheffersomyces stipitis) can assimilate both C6 and C5 sugars (10). To grow on xylose as a sole carbon source, yeasts need xylose-specific transporters to import xylose into the cytosol followed by conversion of xylose to xylitol mediated by a xylose reductase (XYL1), conversion of xylitol to xylulose mediated by a xylulose dehydrogenase (XYL2), and conversion of xylulose to xylulose-5-phospate mediated by a xylulose kinase (XYL3) and finally to other precursor metabolites of core metabolism required for cell growth (11). While native yeasts use XYL1 and XYL2 to convert xylose to xylulose, genetically engineered yeasts expressing a bacterial isomerase for enhanced xylose utilization have also been successfully demonstrated (12). For growth on cellobiose, yeasts can use two possible routes. In route 1, cellobiose is first extracellularly degraded into glucose by a secreted β-glucosidase (BGL), and glucose is then imported into the cell (13). In route 2, cellobiose is first transported into the cell by a cellodextrin-specific transporter and subsequently converted to glucose by an intracellular BGL (14). Both routes were engineered in S. cerevisiae, and route 2 has been shown to be effective for simultaneous coutilization of xylose and cellobiose (15–17). For effective sugar assimilation, sugar-specific transporters have significant roles and are commonly regulated at both the transcriptional and enzymatic levels (18). Recent characterization of conserved structural motifs and amino acids responsible for xylose- and cellodextrin-specific transporters in S. cerevisiae provides useful insights into complex sugar utilization (19–22).
FIG 1.
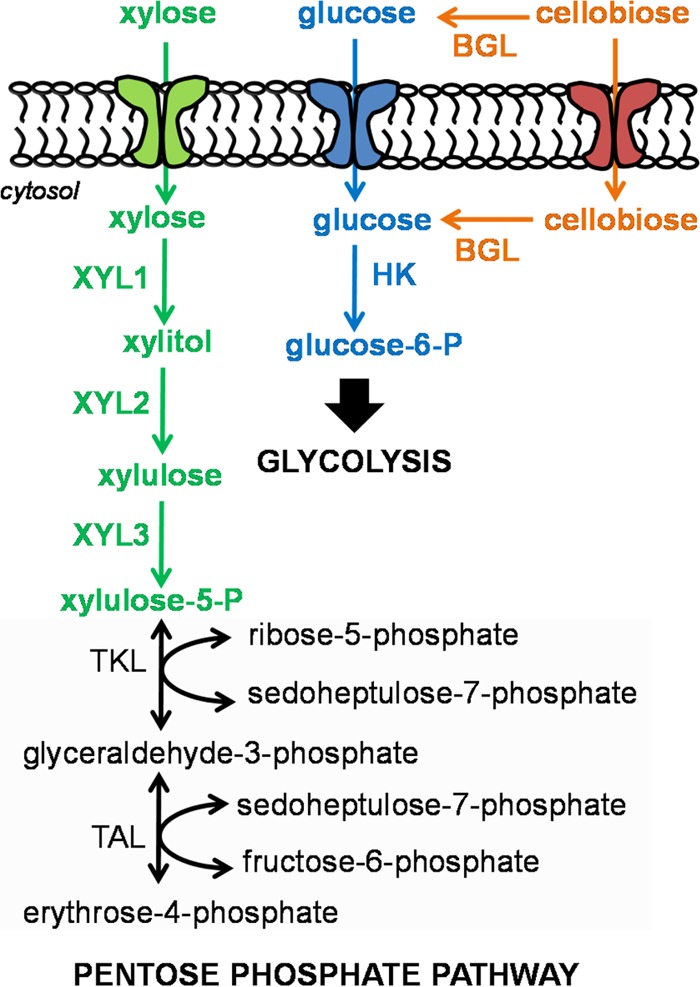
Degradation pathways of glucose (in blue), xylose (in green), and cellobiose (in orange) in yeasts. A simplified pentose phosphate pathway is presented in gray box. Abbreviations: XYL1, xylose reductase; XYL2, xylitol dehydrogenase; XYL3, xylulose kinase; TKL, transketolase; TAL, transaldolase; BGL, β-glucosidase.
Yarrowia lipolytica, classified as an oleaginous yeast that is generally regarded as safe (GRAS), has recently emerged as a potential microbial cell factory with robust phenotypes advantageous for production of biofuels and biochemicals. Y. lipolytica not only can be harnessed to produce large amounts of intracellular neutral lipids (>90% of dry cell weight [DCW]) (23, 24), oleochemicals (25), food supplements (e.g., omega-3 eicosapentaenoic acid) (26), high-value organics (e.g., citric, α-ketoglutaric, succinic, and pyruvic acids), and proteins (e.g., proteases and lipases) (27) but also is capable of assimilating complex substrates (e.g., organic acids, alcohols, triglycerides, and hydrocarbons) (27) as well as of thriving in a wide pH range (pH 2 to 11) (28) and in the presence of inhibitory acid-pretreated biomass hydrolysates (29) or high (>12% NaCl) salt concentrations (30) or even high (10% [vol/vol]) concentrations of ionic liquids (31). While native Y. lipolytica has been known for decades to use only some C6 sugars such as glucose, mannose, and fructose (32), its capability of assimilating other sugars such as xylose and cellobiose and their mixtures with glucose is poorly understood. For instance, the native xylose and cellobiose degradation pathways have not yet been successfully activated (33, 34) even though Y. lipolytica has putative metabolic enzyme and transport genes required for xylose and cellobiose degradation. Recent studies have focused on introducing the native xylose degradation pathway of P. stipitis in Y. lipolytica (34) as well as on developing the heterologous cellobiose degradation pathways in Y. lipolytica. Strategies for the latter include overexpression of (i) the heterologous cellodextrin transporter and intracellular BGL genes of Neurospora crassa for intracellular cellobiose degradation (15) and (ii) native extracellular BGL genes of Y. lipolytica for extracellular cellobiose degradation (33).
In this study, we activated and elucidated the sugar metabolism of Y. lipolytica for cell growth on xylose and cellobiose as well as on their mixtures with glucose. Through comprehensive metabolic and transcriptomic analyses, we identified sugar-specific putative transporters and metabolic degradation enzymes in Y. lipolytica responsible for xylose and cellobiose assimilation. We discovered that Y. lipolytica exhibited enhanced sugar utilization for growth on mixed sugars, with strong carbon catabolite activation (CCA) for growth on the mixture of xylose and cellobiose and with mild CCR of glucose on xylose and cellobiose. The results sheds light on fundamental understanding of the complex native sugar metabolism of Y. lipolytica and will help guide inverse metabolic engineering of this microorganism for enhanced conversion of biomass-derived fermentable sugars to biofuels and biochemicals.
MATERIALS AND METHODS
Strains and plasmids.
A list of the plasmids and strains used in this study is provided in Table S1 in the supplemental material. Plasmid pSL16-CEN1-1-227, containing the CYC1 terminator (TCYC1) and leucine selection marker, was kindly provided by M. Matsuoka (Sojo University, Japan) (35). Plasmid pSR001 was created by inserting the Y. lipolytica TEF promoter (PTEF) (36) in front of TCYC1 of pSL16-CEN1-1-227 as described elsewhere (31). The list of primers used in this study is shown in Table S2. Plasmid pSR002, carrying the native xylose dehydrogenase (XYL2) gene, was constructed by the use of Gibson gene assembly (37) of two DNA fragments, the XYL2 gene fragment amplified from the genomic DNA (gDNA) of Y. lipolytica using primers Xyl2YL_Fwd/Xyl2YL_Rev and the backbone amplified from pSR001 using primers pSR001_Fwd/pSR001_Rev. Likewise, plasmid pSR003, carrying xylose-specific/cellobiose-specific transporter YALI0D01111g (TRP7YL), was constructed by the assembly of two DNA fragments, the TRP7YL gene amplified from the gDNA of Y. lipolytica using primers TRP7YL_Fwd/TRP7YL_Rev and the backbone amplified from pSR001 using primers pSR001_Fwd/pSR001_Rev.
The Escherichia coli TOP10 strain was primarily used for molecular cloning. Y. lipolytica strain ATCC MYA-2613 (or YlSR001), a thiamine, leucine, and uracil auxotroph, was obtained from the ATCC strain collection. The constructed plasmids, namely, pSR001, pSR002, and pSR003, were transformed into YlSR001 via electroporation (38) to create Y. lipolytica strains YlSR101, YlSR102, and YlSR103, respectively.
Medium and cell culturing. (i) Media.
The complex Luria-Bertani medium, which contained 5 g/liter yeast extract, 10 g/liter tryptone, and 5 g/liter NaCl plus 100 μg/ml ampicillin for selection, was used for molecular cloning in E. coli. For characterization of Y. lipolytica, we used the defined Synthetic Complete Supplement Mixture (SC)-Leu media (pH 5.5), which contained yeast nitrogen base (catalog no. Y0626; Sigma-Aldrich, MO, USA), synthetic dropout amino acid mixture without leucine (Y1376; Sigma-Aldrich), and sugars. Each type of sugar (e.g., glucose, xylose, or cellobiose) was prepared with an initial concentration of 10 g/liter in media containing either single or mixed sugars.
(ii) Cell culturing.
Growth of Y. lipolytica was conducted at 28°C and 190 rpm in 250-ml baffled flasks that contained defined media with working volumes of 25 ml. For detailed strain characterization, single colonies were inoculated in defined media until reaching the exponential phase (optical density at 600 nm [OD600] of ∼2, where an OD of 1 represents ∼0.53 g/liter and ∼3 × 107 viable cells/ml). Next, cells were centrifuged at 4,700 × g for 3 min, washed with 10 mM phosphate-buffered saline (PBS), and resuspended in the fresh medium with an initial OD of 0.1. Cell growth was monitored throughout the cell culturing period, and samples were collected for high-pressure liquid chromatography (HPLC), real-time PCR (RT-PCR), and cell-free enzyme assays. Before the detailed strain characterization was performed in media containing xylose and/or cellobiose, we adapted cells through 5 rounds of culture transfers during the exponential growth to activate xylose and cellobiose degradation pathways. All experiments for the detailed strain characterization were performed in at least 4 biological replicates.
The (average) specific cell growth rate μ (1/h) and sugar uptake rate rs (mmol/gram dry cell weight [gDCW]/h) values were determined as follows:
| (1) |
| (2) |
where Si (g/liter) and Xi (g/liter) are the substrate and biomass concentrations, respectively, at time ti (h).
Analytical methods. (i) HPLC.
To quantify extracellular metabolites (e.g., sugars, xylitol, and xylulose), 1 ml of culture supernatants was filtered through 0.2-μm-pore-size filters prior to the HPLC run. A Shimadzu HPLC system, equipped with a refractive index detector (RID) and a UV light detector (Shimadzu Scientific Instruments, Inc., MD, USA) and an Aminex 87H column (catalog no. 1250140; Bio-Rad, CA, USA), was used to run samples. The running method used 10 mN H2SO4 as a mobile phase and operation at a flow rate of 0.6 ml/min and an oven temperature set at 48°C (39).
(ii) Transcriptomics by RT-PCR.
To extract mRNA samples for RT-PCR, exponentially grown cells were first treated with RNA Protect cell reagent (catalog no. 76526; Qiagen Inc., CA, USA). About 2 × 107 cells were mixed with acid-washed glass beads and were then mechanically disrupted by the use of a Mini-Beadbeater-16 cell disrupter (model 607; BioSpec Product Inc., OK, USA). Disrupted cell samples were centrifuged at 17,000 × g for 2 min, and supernatants were collected for mRNA purification using a Qiagen RNeasy minikit (catalog no. 74104; Qiagen Inc., CA, USA). The mRNA samples were first treated with DNase to remove gDNA contamination by the use of an RNease-free DNase kit (catalog no. 79254; Qiagen Inc., CA, USA) and were subsequently used to synthesize cDNA using a QuantiTect reverse-transcription kit (catalog no. 205311; Qiagen Inc., CA, USA).
The RT-PCR samples were prepared from the synthesized cDNA using a QuantiTect SYBR green PCR kit (catalog no. 204143; Qiagen Inc., CA, USA) and were run on a StepOnePlus real-time PCR system (Applied Biosystems, CA, USA) to quantify mRNA expression levels of targeted genes. Primers used for RT-PCR are listed in Table S2 in the supplemental material. For the analysis, the mRNA expression level of a targeted gene [e.g., 2−ΔCT = 2−(CT, target − CT, actin), where CTi is the critical threshold cycle value for gene i] in each sample was normalized with respect to that of the housekeeping actin gene (e.g., YALI0D08272g). To compare the relative levels of gene expression occurring under two different growth conditions, we used the CT log2 ratio (e.g., log22−ΔΔCT = −ΔΔCT = −(ΔCTcondition1 − ΔCTcondition2) of the normalized mRNA expression levels for the targeted gene for comparisons of condition 1 (e.g., growth on xylose) to condition 2 (e.g., growth on glucose as a reference) (40). The relative mRNA expression level for each gene under a given growth condition was reported as the average result ± 1 standard deviation from a data set of at least 4 biological replicates. Student's t test was performed to evaluate statistical significance.
Enzyme assays.
To prepare samples for cell-free enzyme assays, 1 × 107 cells in late exponential growth were collected and centrifuged at 4,700 × g for 3 min. Cell pellets were resuspended in Y-PER yeast protein extraction reagent (catalog no.78990; Thermo Scientific, IL, USA), which contained EDTA-free pierce protease inhibitors (catalog no. 88266; Thermos Scientific, IL, USA), and were incubated with agitation at room temperature for 20 min. Lysed cell samples were then centrifuged at 17,000 × g for 10 min, and the soluble fractions were collected for enzyme assays. Unless specified, all the enzyme assay reactions were conducted in 384-well plates with a 50-μl working reaction volume at 28°C. Kinetics measurement was conducted with a BioTek Synergy HT microplate reader, and data were processed using Gen5 software (BioTek Instruments, Inc., VT, USA). Protein concentrations were measured by the Bradford assay (41). All enzyme assay experiments were performed with at least 4 biological replicates.
BGL assay.
The extracellular and intracellular β-glucosidase (BGL) activities were measured in the cell supernatants and whole-cell lysates, respectively. The BGL assay reaction was conducted at pH 6 and 28°C in 25 mM sodium phosphate buffer that contained 10 mM p-nitrophenyl-β-d-glucopyranoside and 1.5 μg total proteins from either cell supernatants or whole-cell lysates. The BGL activities were determined by the emission of p-nitrophenol (p-NP) at 400 nm. One unit of the BGL activity was defined as 1 μmol of p-NP reduced per mg protein per min (42).
XYL1 assay.
The xylose reductase (XYL1) assay was conducted at pH 6 and 28°C in 25 mM sodium phosphate buffer that contained 0.5 mM NAD(P)H, 1.5 μg total proteins from whole-cell lysates, and 300 mM xylose. The XYL1 activity was measured by the oxidation of NAD(P)H at 340 nm. One unit of XYL1 activity was defined as 1 μmol of NAD(P)H consumed per mg protein per min (43).
XYL2 assay.
The xylitol dehydrogenase (XYL2) assay was conducted at pH 6 and 28°C in 25 mM sodium phosphate buffer that contained 0.5 mM NAD(P)+, 1.5 μg total proteins from whole-cell lysates, and 300 mM xylitol. The XYL2 activity was measured by the reduction of NAD(P)+ at 340 nm. One unit of XYL2 activity was defined as 1 μmol of NAD(P)H produced per mg protein per min (43).
XYL3 assay.
A bioluminescence assay kit (catalog no. K254-200; Biovision, CA, USA) was used to measure xylulokinase (XYL3) activity. The XYL3 assay reaction was conducted at pH 6 and 28°C in 25 mM sodium phosphate buffer that contained 5 μl ATP, 5 μl ATP monitoring enzymes, 1.5 μg total proteins from whole-cell lysates, and 200 mM xylulose. One unit of XYL3 activity was defined as 1 relative light unit (RLU) per μg protein per s.
Bioinformatics.
We applied the BLASTP tool (44) to search for putative sugar transporters and the metabolic enzymes of the xylose- and cellobiose-degrading pathways in Y. lipolytica. To find sugar transporters, we used the glucose transporter of S. cerevisiae (HXT7) (45), the xylose transporter of P. stipitis (XUT4) (46), the cellobiose transporter of N. crassa CDT1 (NCU00801) (14), and the xylobiose transporter of Streptomyces thermoviolaceus (BxlF) (47) as the templates. Using the MEGA5.2 tool, we performed ClustalW alignment for the BLASTP-searched putative sugar transporters and identified conserved structural domains and amino acid residues (48). The alignment results were also edited using the Jalview 2.8 tool (49) for enhanced visual presentation. To find the metabolic enzymes responsible for the xylose and cellobiose degradation, we used the XYL1, XYL2, and XYL3 proteins of P. stipitis (50, 51) as well as BGLs of A. niger (BGL1; Q30BH9) (52), A. fumigatus (BTGE; B0Y9Q9) (53), and Candida wickerhamii (BGLA; Q12602) (54) as the templates. We further applied the PSORTII tool (55) to predict the cellular localization of the BLASTP-searched putative BGL genes of Y. lipolytica.
To evaluate whether the xylose and cellobiose degradation pathways in our characterized Y. lipolytica strain, ATCC MYA-2613 (taxid 4952), are conserved across different isolated Y. lipolytica strains, we used the BLASTN tool (chosen due to intron interference) to search for sequence homology against WSH-Z06 (taxid 1437815) and CLIB122 (taxid 284591), whose genome sequences are available.
RESULTS
Y. lipolytica is capable of using xylose as a sole carbon source.
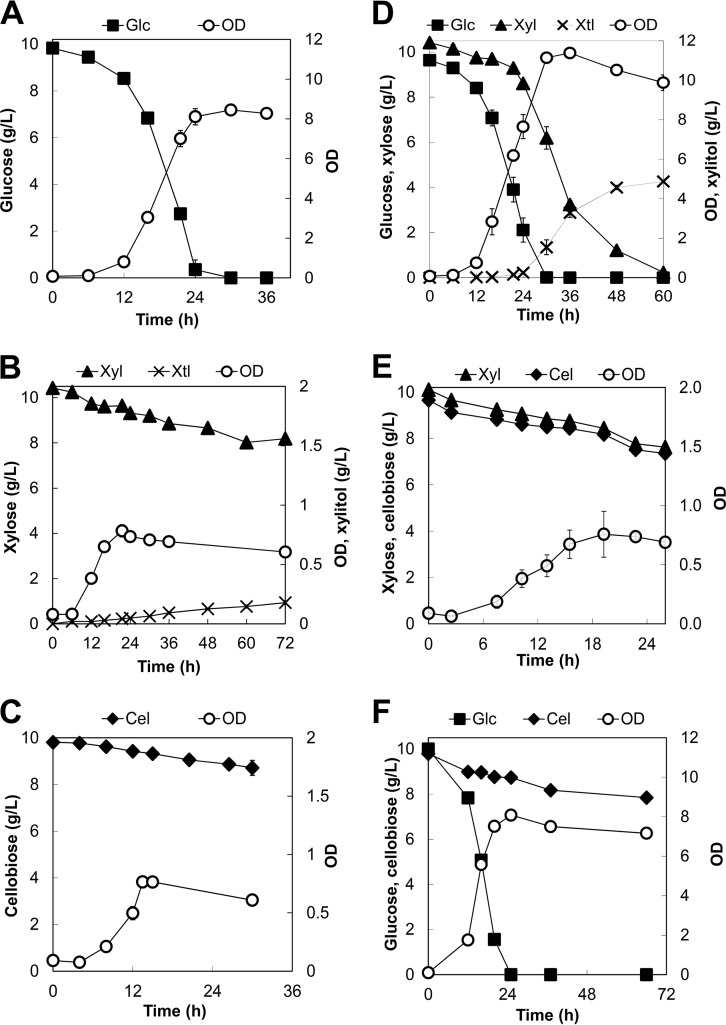
Genome mining confirmed that Y. lipolytica has the putative xylose-degrading metabolic enzymes (XYL1, XYL2, and XYL3) required for xylose assimilation (see Table S3 in the supplemental material). To activate the xylose degradation pathway, we adapted Y. lipolytica to grow on defined medium containing 10 g/liter xylose as a sole carbon source. We controlled the xylose growth adaptation to ensure that it would last no more than 15 generations (see Fig. S1A in the supplemental material) because this short adaptation was sufficient to achieve stable growth. After the adaptation, Y. lipolytica grew on xylose with a specific growth rate of 0.10 ± 0.02 (1/h) and a specific xylose uptake rate of 1.71 ± 0.42 mmol/gDCW/h (Table 1). After 72 h, Y. lipolytica had consumed about 2.23 ± 0.10 g/liter xylose and had produced 0.18 ± 0.02 g/liter xylitol as a major byproduct (Fig. 2B).
TABLE 1.
Specific sugar uptake rates of Y. lipolytica for growth on single and mixed sugarsa
| Parameter | Uptake rate (mmol/gDCW/h) |
|||||
|---|---|---|---|---|---|---|
| Single sugar |
Mixed sugars |
|||||
| Glc | Xyl | Cel | Xyl + Glc | Xyl + Cel | Glc + Cel | |
| rGlc | 2.98 ± 0.78 | 2.13 ± 0.50 | 2.13 ± 0.52 | |||
| rXyl | 1.71 ± 0.42 | 0.94 ± 0.39 | 4.68 ± 0.90 | |||
| rCel | 0.95 ± 0.32 | 1.68 ± 0.25 | 0.16 ± 0.07 | |||
Each data point represents the average ± 1 standard deviation of the results from at least 4 biological replicates. Glc, glucose; Xyl, xylose; Cel, cellobiose.
FIG 2.
Profiles of cell growth and metabolites for growth of the parent Y. lipolytica strain on single sugars, including (A) glucose, (B) xylose, and (C) cellobiose, as well as on mixed sugars, including (D) xylose and glucose, (E) xylose and cellobiose, and (F) glucose and cellobiose. Each data point represents an average value ± 1 standard deviation calculated from the results of 3 of 6 biological replicate experiments that were conducted in parallel. Abbreviations: Glc, glucose; Xyl, xylose; Cel, cellobiose; Xtl, xylitol.
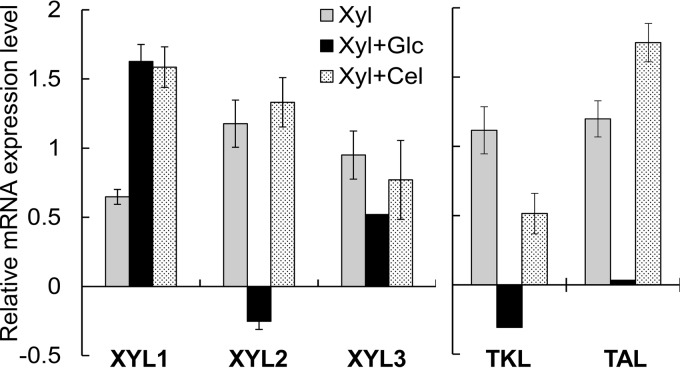
Consistently, we detected the enzymatic activities of XYL1, XYL2, and XYL3 for growth of Y. lipolytica on xylose (Table 2). XYL1 and XYL2 exhibited activities toward NAD(P)H and NAD(P), respectively, as shown by the use of the whole-cell lysates (see Table S4 in the supplemental material). In addition, we confirmed that the XYL1, XYL2, and XYL3 genes were upregulated 1.57 ± 0.06-fold, 2.27 ± 0.27-fold, and 1.94 ± 0.23-fold higher for growth on xylose than on glucose, respectively (Fig. 3). Both the transketolase (TKL) and transaldolase (TAL) genes of the pentose phosphate pathway, immediately downstream of the xylose degradation pathway (Fig. 1), were also upregulated, which further supports the idea of the assimilation of xylose for growth in Y. lipolytica. In comparison with growth on glucose (Fig. 2A), Y. lipolytica reached a much lower plateau OD of 0.78 ± 0.02, produced a high yield of xylitol, and did not completely consume xylose after 24 h (Fig. 2B). This observed phenotype suggests that the xylitol dehydrogenase (XYL2) step might have been limiting and hence might have impeded efficient xylose assimilation.
TABLE 2.
Enzyme activities of xylose reductase, xylitol dehydrogenase, and xylulose kinase of Y. lipolytica growing on xylose and its mixtures with glucose and cellobiosea
| Substrate(s) | Enzyme activity (U) |
||
|---|---|---|---|
| XYL1 | XYL2 | XYL3 | |
| Xyl | 7.63 ± 1.56 | 3.75 ± 1.79 | 15.70 ± 3.51 |
| Xyl + Glc | 12.22 ± 3.16 | 41.42 ± 9.45 | 53.76 ± 13.60 |
| Xyl + Cel | 4.74 ± 0.00 | 40.38 ± 24.0 | 12.01 ± 5.83 |
The measurement of the xylose reductase (XYL1) and xylitol dehydrogenase (XYL2) in vitro activity levels shown in the table used the NADH and NAD cofactors, respectively. Each data point represents the average ± 1 standard deviation of the results from 4 biological replicates. XYL3, xylulose kinase.
FIG 3.
Relative mRNA expression levels (in log2 scale) of the xylose pathway (XYL1, XYL2, and XYL3) and pentose phosphate pathway (TKL and TAL) genes of the parent Y. lipolytica strain for growth on xylose (Xyl), a mixture of xylose and glucose (Xyl+Glc), and a mixture of xylose and cellobiose (Xyl+Cel). The reference condition for normalization is growth on glucose alone. Each data point represents an average value ± 1 standard deviation calculated from the results of 4 biological replicates.
Y. lipolytica is capable of using cellobiose as a sole carbon source.
To determine whether Y. lipolytica is capable of utilizing cellobiose as a sole carbon source, we first performed genome mining to identify putative BGLs. We found that Y. lipolytica has a total of 11 putative BGLs, of which 5 are secreted extracellularly, 3 are located in the cytosol, and 3 are located in the nucleus (see Table S3 in the supplemental material). As in the xylose experiments, to activate the cellobiose degradation pathway, we adapted Y. lipolytica to grow in defined medium containing 10 g/liter cellobiose as a sole carbon source (see Fig. S1B in the supplemental material). After adaptation, Y. lipolytica grew on cellobiose with a specific growth rate of 0.11 ± 0.02 (1/h) and a specific cellobiose uptake rate of 0.95 ± 0.32 (mmol/gDCW/h) (Table 1). However, the cellobiose assimilation was inefficient; Y. lipolytica consumed only 0.99 ± 0.22 g/liter cellobiose and achieved a relatively low plateau OD of 0.76 ± 0.02 after 15 h (Fig. 2C).
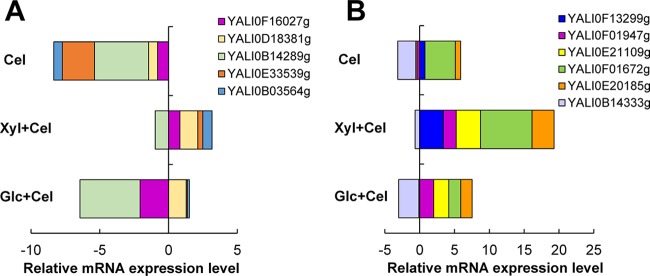
To elucidate how Y. lipolytica assimilated cellobiose, we characterized the mRNA expression levels of the 11 putative BGL genes (Fig. 4; see also Table S5 in the supplemental material). None of the 5 extracellularly localized putative BGL genes were upregulated during growth on cellobiose (Fig. 4A). Similarly, three of six putative intracellular BGL genes—YALI0B14333g (encoding cytosolic BGL), YALI0E21109g (encoding nucleus BGL), and YALI0F01947g (encoding nucleus BGL)—were not induced by cellobiose. In contrast, the three remaining putative intracellular BGL genes—YALI0E20185g (encoding cytosolic BGL), YALI0F01672g (encoding cytosolic BGL), and YALI0F13299g (encoding nucleus BGL)—were significantly upregulated at levels 1.71 ± 0.19-fold, 20.48 ± 0.61-fold, and 1.70 ± 0.11-fold higher for growth on cellobiose than glucose, respectively, and were likely responsible for cellobiose assimilation in Y. lipolytica (Fig. 4B). Consistently, we detected the intracellular BGL activity from the cell lysate as well as the extracellular BGL activity in the culture supernatant for growth on cellobiose; however, the extracellular BGL activity was low, at about 2 orders of magnitude lower than the intracellular activity (Table 3).
FIG 4.
Relative mRNA expression levels (in log2 scale) of (A) putative extracellular BGL genes and (B) putative intracellular BGL genes for growth of the parent Y. lipolytica on cellobiose (Cel) and on a mixture of xylose and cellobiose (Xyl+Cel) as well as on a mixture of glucose and cellobiose (Glc+Cel). The reference condition for normalization is growth on glucose alone. Each data point represents an average value ± 1 standard deviation calculated from the results of 4 biological replicates. Error data were not included in the figure due to the crowding effect but are presented in Table S5 in the supplemental material for reference.
TABLE 3.
Enzyme activity of intracellular and extracellular β-glucosidases of Y. lipolytica growing on single and mixed sugarsa
| Substrate(s) | Enzyme activity (U) |
|
|---|---|---|
| Intra BGL | Extra BGL | |
| Glc | 1.58 ± 0.26 | ND |
| Xyl | ND | 0.04 ± 0.00 |
| Cel | 1.94 ± 0.59 | 0.01 ± 0.00 |
| Xyl + Glc | 2.67 ± 1.46 | 0.03 ± 0.00 |
| Xyl + Cel | 2.71 ± 0.19 | 0.22 ± 0.00 |
| Glc + Cel | 3.98 ± 0.62 | 0.27 ± 0.00 |
Each data point represents the average ± 1 standard deviation of the results from 4 biological replicates. Abbreviations: intra, intracellular; extra, extracellular; BGLs, β-glucosidases; ND, not detected.
Elucidation of functional roles of sugar transporters in Y. lipolytica. (i) Identification of putative sugar transporters for glucose, xylose, and cellobiose.
Sugar transporters play a significant role in cellular metabolism but are poorly understood in Y. lipolytica. To have a complete picture of complex sugar utilization, we first performed genome mining of Y. lipolytica to identify putative sugar transporters responsible for glucose, xylose, and cellobiose assimilation. We identified a total of 23 putative glucose transporter genes, 22 and 19 of which were also putative xylose and cellobiose transporter genes, respectively (Fig. 5A). These transporters, belonging to the major facilitator superfamily (MFS) sugar transporters (18, 56, 57), have 12 transmembrane (TM) domains and the well-known “diffused” motif between TM4 and TM5 that are known to be responsible for catalytic and structural functions (58). Since these transporters have short C-terminal tails, they are unlikely to function similarly to sugar sensors (59, 60). Among these 23 putative glucose transporter genes, YALI0A14212g was predicted to be unique to glucose whereas the other 3 genes, including YALI0B21230g, YALI0F18084g, and YALI0D00132g, were predicted to be unique for both glucose and xylose but not cellobiose.
FIG 5.
(A) Venn diagram of the putative glucose, xylose, and cellobiose transporter genes that were subjected to BLASTP searches in the parent Y. lipolytica strain. (B) The G-G/F-X-X-X-G consensus sequence. (C) The patterns of T213/N370 amino acid residues.
To elucidate the functional roles of these 23 putative transporters for growth of Y. lipolytica on glucose, xylose, and cellobiose, we performed a comprehensive transcriptional analysis (Fig. 6; see also Table S6 in the supplemental material). By examining the relative mRNA expression levels of putative transporter genes that were downregulated or unchanged for growth of Y. lipolytica on xylose or cellobiose alone, we confirmed that 7 of the 23 putative glucose transporters were induced by glucose. We found that 16 of the 22 putative xylose transporter genes were overexpressed up to 3 orders of magnitude (>1,000-fold) higher for growth on xylose than glucose (Fig. 6). We verified that 4 of the 19 putative cellobiose transporter genes, comprised of YALI0D01111g, YALI0C04730g, YALI0D00363g, and YALI0B00396g, were expressed up to 62-fold higher for growth on cellobiose than glucose. These 4 putative cellobiose transporter genes were also highly upregulated by xylose induction.
FIG 6.
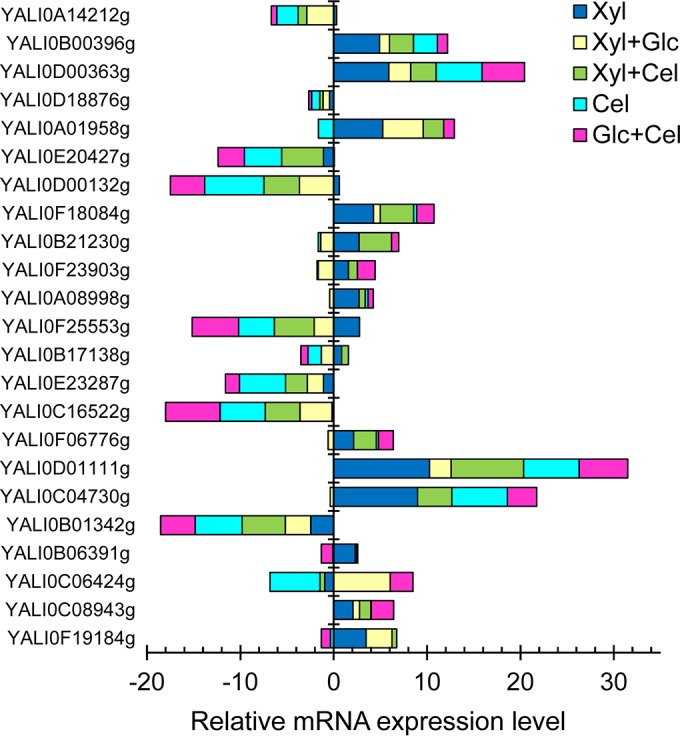
Relative mRNA expression levels (in log2 scale) of putative sugar transporter genes of Y. lipolytica that grow on single sugars, including glucose (Glc), xylose (Xyl), and cellobiose (Cel), as well as on mixtures of xylose and glucose (Xyl+Glc), of xylose and cellobiose (Xyl+Cel), and of glucose and cellobiose (Glc+Cel). The reference condition for normalization is growth on glucose alone. Each data point represents an average value ± 1 standard deviation calculated from the results of 4 biological replicates. Error data were not included in the figure due to the crowding effect but are presented in Table S6 in the supplemental material for reference.
(ii) Analysis of highly conserved structural motif and amino acid residues identified novel xylose-specific transporters of Y. lipolytica.
While the structural topology and functional role of sugar transporters of eukaryotes can be classified with high confidence by bioinformatics (18, 56, 57), fundamental knowledge regarding prediction of sugar specificity and transport efficiency is still lacking. Recent studies showed that the highly conserved structural motif G-G/F-X-X-X-G in the TM1 as well as the conserved T213 or N370 amino acid residue of the S. cerevisiae HXT7 in TM5 or TM8, respectively, can govern the specificity and efficiency of the xylose uptake over glucose in S. cerevisiae (20, 22). Motivated by these studies, we first investigated whether the G-G/F-X-X-X-G motif is conserved for the putative xylose transporters of Y. lipolytica. Among the 16 putative xylose transporters of Y. lipolytica that were upregulated for growth on xylose, we found that 8 of these transporters have the G-G/F-X-X-X-G motif containing the highly conserved L/F-L-F amino acids in place of X-X-X regions (Fig. 5B and Table 4), responsible for high-level xylose-specific uptake in S. cerevisiae (22). We also determined that 3 of the remaining 8 putative xylose transporters of Y. lipolytica have a similar but not identical G-G/F-X-X-X-G motif where variables occur at the first two amino acid residues, including G-V for YALI0F06776p, S-G for YALI0F23903p, and G-T for YALI0B21230p. These similar motifs were also found to be responsible for xylose-specific uptake in S. cerevisiae (22).
TABLE 4.
Classification of putative glucose transporters, putative xylose transporters, and putative cellobiose transporters of Y. lipolytica that are transcriptionally induced by their respective single sugarsa
| Transporter category and gene | G-G/F-X-X-X-G motif | Amino acid |
Relative mRNA expression in indicated sugar mixture | ||
|---|---|---|---|---|---|
| T213 | N370 | ||||
| Putative glucose transporters | Xyl+Glc | Glc+Cel | |||
| YALI0C06424g | GGILFG | T | N | +++ | + |
| YALI0A14212g | ND | V | F | - | n.c. |
| YALI0B01342g | GGMLFG | T | N | - | – |
| YALI0C16522g | GFLLFG | T | Y | – | — |
| YALI0E23287g | GGMLFG | T | N | - | - |
| YALI0E20427g | ND | Y | K | n.c. | - |
| YALI0D18876g | ND | Y | K | n.c. | n.c. |
| Putative xylose transporters | Xyl+Glc | Xyl+Cel | |||
| YALI0B21230g | GTLQFG | N | V | – | + |
| YALI0F19184g | GGFVFG | T | N | n.c. | - |
| YALI0A01958g | ND | Y | K | n.c. | - |
| YALI0A08998g | GFLLFG | T | Y | - | - |
| YALI0F23903g | SGFLFG | T | N | - | n.c. |
| YALI0C08943g | GGLLYG | T | N | - | n.c. |
| YALI0F18084g | AAAVQG | A | N | – | n.c. |
| YALI0B06391g | GGLLFG | T | N | - | - |
| YALI0D00132g | AAAVQG | A | N | – | — |
| YALI0F06776g | GVFLFG | I | Y | - | n.c. |
| YALI0B17138g | GFLLFG | T | Y | - | n.c. |
| YALI0F25553g | GFFLFG | S | Y | — | — |
| YALI0D01111g | GFLLFG | T | Y | — | - |
| YALI0C04730g | GFLLFG | T | Y | — | — |
| YALI0D00363g | ND | Y | K | – | - |
| YALI0B00396g | ND | V | V | – | - |
| Putative cellobiose transporters | Xyl+Cel | Glc+Cel | |||
| YALI0D01111g | GFLLFG | T | Y | + | n.c. |
| YALI0C04730g | GFLLFG | T | Y | - | - |
| YALI0D00363g | ND | Y | K | - | n.c. |
| YALI0B00396g | ND | V | V | n.c. | - |
For the relative mRNA expression levels of these transporters in sugar mixtures, qualitative scale designations corresponding to the effect of the presence of one sugar on the other in the mixture are defined as follows: +++, R (ratio or fold change) ≥ 20.0; ++, 10 ≤ R < 20, +, 1.5 ≤ R < 10; n.c. (no significant change), 0.5 ≤ R < 1.5, -, 0.1 ≤ R < 0.5; –, 0.05 ≤ R < 0.1; —, R < 0.05. The relative mRNA expression levels of putative transporter genes in each sugar transporter class were determined by the ratios of the normalized mRNA expression levels between growth in the mixtures and in the respective single-sugar experiments.
Since the replacement of T213 and N370 in S. cerevisiae HXT7 by a hydrophobic or small hydrophilic amino acid could change the substrate binding pocket and improve the xylose specificity (20, 22), we examined these conserved amino acid residues in the putative xylose transporters of Y. lipolytica (Fig. 5C). The results show that 8 of the 16 putative xylose-specific transporters induced by growth on xylose have T213 replaced by N, Y, A, I, S, or V whereas 10 of the 16 putative xylose transporters of Y. lipolytica have N370 replaced by V, K, or Y (Table 4). Among the 8 putative xylose transporters of Y. lipolytica that contain the G-G/F-X-X-X-G motif, 1 transporter has both T213 and N370 replacement whereas the remaining 4 transporters have either T213 or N370 replacement. Altogether, these results suggest that Y. lipolytica likely has at least 5 putative xylose-specific transporters induced by xylose.
Synergistic effect of mixed-sugar utilization in Y. lipolytica.
The CCR effect often causes inefficient utilization of mixed, complex fermentable sugars derived from lignocellulosic biomass for production of desirable biochemicals and biofuels (15). To investigate whether the CCR effect exists in Y. lipolytica, we characterized growth of Y. lipolytica on various mixtures of glucose, xylose, and cellobiose.
(i) Elucidation of growth characteristics of Y. lipolytica in a mixture of glucose and xylose.
In a mixture of 10 g/liter glucose and 10 g/liter xylose, Y. lipolytica grew with a specific growth rate of 0.15 ± 0.01 (1/h). It consumed all glucose after 30 h (Fig. 2D) with a specific glucose uptake rate of 2.13 ± 0.50 mmol/gDCW/h (Table 1), which is similar to the growth phenotype seen with glucose alone without any observed inhibitory effect by xylose. Unlike growth on xylose alone, Y. lipolytica was able to consume all of the xylose after 60 h with a specific xylose uptake rate of 0.94 ± 0.39 mmol/gDCW/h. Even though xylose assimilation was significantly enhanced, it did not entirely contribute to biomass production because a relatively large amount of 4.87 g/liter xylitol was produced as a major product (Fig. 2D) together with other minor byproducts (e.g., pyruvic, succinic, and a-ketoglutaric acids; data not shown). This observed phenotype further supports the hypothesis that XYL2 is the rate-limiting step of the xylose degradation pathway in Y. lipolytica. Even though the xylose consumption rate was lower than that of glucose, Y. lipolytica did not exhibit the diauxic growth commonly caused by the severe CCR effect and was able to consume glucose and xylose simultaneously (Fig. 2D; see also Fig. S2A in the supplemental material).
To further elucidate the driving force for enhanced xylose assimilation for growth on the mixture of glucose and xylose, we performed transcriptomic and metabolic analyses of the xylose degradation pathway. Like growth on xylose alone, both XYL1 and XYL3 genes were upregulated by 3.09 ± 0.26 and 1.43 ± 0.00-fold, respectively, higher for growth on mixed sugars than on glucose alone (Fig. 3). The observed gene upregulation correlated with relatively high enzyme activities of XYL1 and XYL3 (Table 2). In contrast to growth on xylose alone, the XYL2 gene was slightly repressed for growth on the mixture of glucose and xylose, which strongly correlated with the relatively high accumulation of xylitol. Like the XYL2 gene, the TKL gene, but not the TAL gene, was transcriptionally repressed (Fig. 3).
Consistent with the observed simultaneous assimilation of glucose and xylose, we found that some of the putative glucose and xylose transporters were not transcriptionally repressed by the CCR effect of glucose on xylose and vice versa. For instance, among the 7 putative glucose transporter genes induced by growth on glucose alone, the expression of YALI0C06424g was synergistically enhanced in the presence of xylose whereas both YALI0E20427g and YALI0D18876g remained unaffected (Table 4). Likewise, among the 16 putative xylose transporter genes, the expression levels of YALI0F19184g and YALI0A01958g were not affected by glucose (Table 4).
(ii) Elucidation of growth characteristics of Y. lipolytica in a mixture of xylose and cellobiose.
During simultaneous saccharification and fermentation of lignocellulose, microbial cell factories, which can efficiently coutilize xylose and cellobiose, are desirable because they help eliminate external supply of BGLs, alleviate feedback inhibition of extracellular cellulases, and avoid the CCR effect exerted by glucose intracellularly. To investigate whether Y. lipolytica is capable of assimilating xylose and cellobiose simultaneously, we cultured Y. lipolytica in defined medium containing 10 g/liter xylose and 10 g/liter cellobiose. The results show that Y. lipolytica was able to simultaneously utilize xylose and cellobiose with a specific growth rate of 0.10 ± 0.01 (1/h) (Fig. 2E; see also Fig. S2B in the supplemental material). We observed that the assimilation of both xylose and cellobiose was synergistically enhanced, with the specific sugar uptake rates increased by 2.78 ± 0.22-fold and 1.90 ± 0.54-fold, respectively (Table 1).
We further performed comprehensive metabolic and transcriptomic analyses to elucidate the driving force for synergistically enhanced coutilization of xylose and cellobiose. For the xylose degradation pathway, the expression levels of all XYL1, XYL2, and XYL3 genes were upregulated to 3.01 ± 0.31-fold, 2.53 ± 0.32-fold, and 1.73 ± 0.35-fold (P value < 0.01) higher for growth on the mixture than on glucose alone (Fig. 3). The upregulation of TKL and TAL genes further supports the enhanced xylose assimilation in Y. lipolytica. The enhanced xylose utilization in the mixture also correlated with not only the upregulation of the xylose-degrading genes (Fig. 3) but also their enzyme activities (Table 2). Unlike the results seen with respect to growth on the mixture of xylose and glucose, the XYL2 gene was not transcriptionally repressed by cellobiose. Notably, in comparison with the growth seen on xylose alone, XYL1 and TAL genes were upregulated (Fig. 3) by 1.92 ± 0.20-fold and 1.47 ± 0.14-fold (P value < 0.01), respectively, which highly correlated with the enhanced xylose assimilation. For the cellobiose degradation pathway, the enhanced cellobiose utilization in the mixture correlated well with significant increases in the transcriptional expression of BGL genes and their enzyme activities of 166.04 ± 16.05-fold and 1.74 ± 0.24-fold, respectively, in comparison with growth on glucose alone and to 12.66 ± 1.30-fold and 1.51 ± 0.51-fold (P value < 0.01), respectively, in comparison with growth on cellobiose alone (Fig. 4 and Table 3). Strikingly, most of the extracellular BGL genes were also overexpressed, which was not observed for growth on cellobiose alone.
Transcriptional analysis of putative xylose and cellobiose transporters further supports the idea of improved simultaneous coutilization of xylose and cellobiose (Fig. 6 and Table 4). Among the 16 putative xylose transporter genes, the expression of YALI0B21230g was synergistically enhanced in the presence of cellobiose whereas the expression of YALI0F23903g, YALI0C08943g, YALI0F18084g, YALI0F06776g, and YALI0B17138g remained unaffected. Likewise, among the 4 putative cellobiose transporters, YALI0D01111g expression was synergistically enhanced whereas YALI0B00396g expression remained unaffected in the presence of xylose.
(iii) Elucidation of growth characteristics of Y. lipolytica in a mixture of glucose and cellobiose.
To investigate whether Y. lipolytica is capable of coutilizing glucose and cellobiose and whether it exhibits any CCR effect of glucose on cellobiose assimilation or vice versa, we characterized growth of Y. lipolytica in the mixture of 10 g/liter glucose and 10 g/liter cellobiose. Y. lipolytica grew with a specific growth rate of 0.19 ± 0.02 (1/h) and a specific glucose uptake rate of 2.13 ± 0.52 mmol/gDCW/h and was able to consume all of the glucose after 30 h (Fig. 2F). However, only 2.00 ± 0.12 g/liter of cellobiose was consumed after 96 h, with a specific cellobiose uptake rate of 0.16 ± 0.07 mmol/gDCW/h. The results also show that Y. lipolytica was able to simultaneously use glucose and cellobiose and did not exhibit a severe CCR effect of glucose on cellobiose assimilation (Fig. 2F; see also Fig. S2C in the supplemental material).
Transcriptional analysis shows that the expression of the BGL genes was slightly greater for growth on the mixture of glucose and cellobiose than on cellobiose alone at up to 5.71 ± 2.71-fold (Fig. 4), which correlated well with the increase in the total BGL activity by 2.15 ± 0.47-fold (Table 3). However, cellobiose consumption was not significantly enhanced as expected (Fig. 2F and Table 1), suggesting that the cellobiose transporters represent the rate-limiting step and likely that there is a CCR effect of glucose on cellobiose transporters. In fact, transcriptional analysis of the 4 putative cellobiose transporters shows that they were either downregulated or not affected in growth on the mixed sugars in comparison to growth on cellobiose alone (Table 4).
Alleviation of bottlenecks of xylose and cellobiose assimilation in Y. lipolytica. (i) Effect of overexpression of the native XYL2 on xylose assimilation.
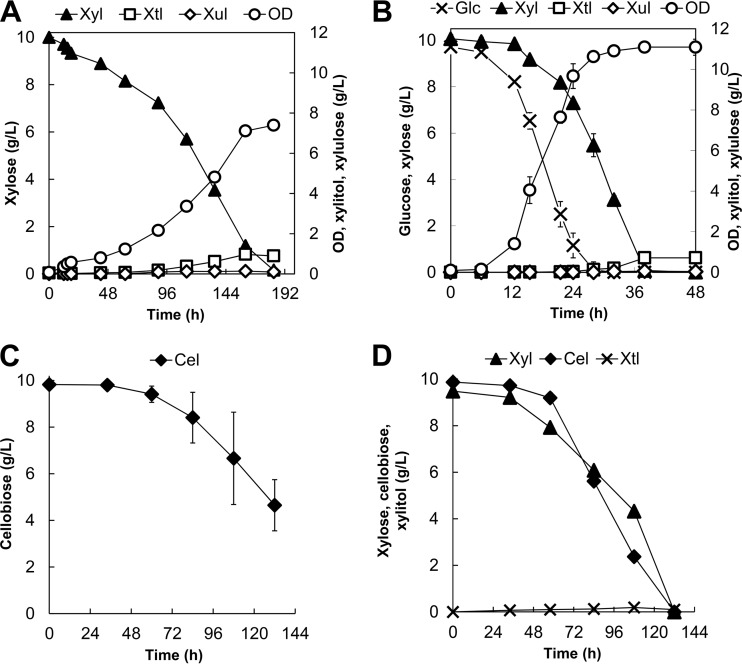
Characterization of growth of the parent Y. lipolytica YlSR101 strain on xylose alone and on its mixture with glucose clearly suggests that the XYL2 step represents the most dominant bottleneck. To alleviate this bottleneck, we constructed strain YlSR102, which overexpressed the native XYL2 gene under the control of the constitutive TEF promoter. The strain characterization for growth on xylose alone clearly showed that YlSR102 has significantly improved growth on xylose, with an enhanced plateau OD of 7.39 ± 0.20, about 10-fold higher than that seen with the parent strain, and complete consumption of xylose as well as significantly lower accumulation of xylitol (Fig. 7A).
FIG 7.
Profiles of cell growth and metabolites for (A) growth of YlSR102 on xylose, (B) growth of YlSR102 on the mixture of glucose and xylose, (C) growth of YlSR103 on cellobiose, and (D) growth of YlSR103 on the mixture of xylose and cellobiose. Each data point represents an average value ± 1 standard deviation calculated from the results of 3 of 6 biological replicate experiments that were conducted in parallel. Abbreviations: Xyl, xylose; Cel, cellobiose; Xtl, xylitol; Xul, xylulose.
Likewise, for growth on the mixture of glucose and xylose, YlSR102 could consume all 10 g/liter xylose without xylitol accumulation within 36 h, about 1.7 times faster than YlSR101 (Fig. 7B). As expected, YlSR102 also exhibited the simultaneous coutilization of glucose and xylose (see Fig. S2A in the supplemental material).
Taken altogether, these results provide evidence that the XYL2 step is the most critical bottleneck affecting the efficient xylose assimilation in the native Y. lipolytica strain.
(ii) Effect of overexpression of the native cellobiose transporter on cellobiose assimilation.
Growth characterization of the parent Y. lipolytica YlSR101 strain on cellobiose alone and on its mixture with either xylose or glucose clearly suggested that the cellobiose assimilation is inefficient, with less than 20% of cellobiose in the feed being consumed. We hypothesized that cellobiose transporters might have been the dominant rate-limiting step in efficient cellobiose assimilation in the parent Y. lipolytica. To test this hypothesis, we constructed the Y. lipolytica YlSR103 mutant, which overexpressed the YALI0D01111g putative cellobiose transporter gene. This gene was chosen because it was induced at the highest levels by both cellobiose and xylose (Fig. 6).
For growth on cellobiose alone, YlSR103 improved both cell growth and cellobiose assimilation in comparison with the YlSR101 parent strain. YlSR103 reached a final biomass titer of 2.80 ± 0.70 gDCW/liter (or OD = ∼5.25 ± 1.30) and was able to consume 5.18 ± 1.10 g/liter cellobiose (Fig. 7C). Unlike the parent strain, interestingly, YlSR103 exhibited a morphological change where cells formed pellets in the aqueous medium (61). For this reason, cell mass was analyzed only at the end of the experiments. In growth on the mixture of cellobiose and xylose, YlSR103 was capable of completely and simultaneously assimilating both cellobiose and xylose and outperformed the parent YlSR101 strain (Fig. 7D).
Taken altogether, these results clearly support the idea that the cellobiose transporter represents the rate-limiting step of cellobiose assimilation in the native Y. lipolytica strain.
DISCUSSION
In this study, we activated and elucidated the native sugar metabolism of Y. lipolytica in growth on xylose and cellobiose as well as on their mixtures with glucose through metabolic and transcriptomic analyses. We discovered that Y. lipolytica has putative sugar-specific transporters as well as metabolic enzymes to assimilate these sugars. We found that Y. lipolytica exhibited a mild CCR effect of glucose on xylose and cellobiose by exerting transcriptional repression of the xylose-degrading metabolic enzymes XYL2 and TKL and of the putative cellobiose transporters. In contrast, the CCA effect enhanced the coutilization of xylose and cellobiose. Regardless of the sugar mixture used, Y. lipolytica was able to simultaneously consume the sugars in the mixtures, likely due to mild CCR and beneficial CCA effects exerted on the sugar degradation pathways.
One important factor in activation of the xylose and cellobiose degradation pathways of the native Y. lipolytica strain is that of adapting it to grow on these sugars through serial culture transfers. During the first transfer, cell growth was very slow and was observed only after 3 days (see Fig. S1 in the supplemental material). Since studies by other groups did not investigate cell growth of the native Y. lipolytica after 2 days (34, 62), the xylose and cellobiose degradation pathways were not activated in those studies. In our study, since the adaptation period was conducted for less than 15 generations, which was sufficient to achieve relatively stable cell growth, the adapted phenotypes of the native Y. lipolytica were reversible (results not shown).
By demonstrating that native Y. lipolytica has 16 putative xylose transporters and xylose-degrading metabolic enzymes (e.g., XYL1, XYL2, XYL3, TKL, and TAL) induced by xylose, we have provided metabolic and genetic evidence of how the native Y. lipolytica could grow on xylose as a sole carbon source. However, the xylose consumption was inefficient because the XYL2 step was limiting and was also transcriptionally repressed by glucose. While the engineered XYL2-overexpressing YlSR102 strain proved very effective for overcoming this rate-limiting step, additional optimization is required to engineer the xylose degradation pathway of Y. lipolytica to operate as efficiently as those of the native yeast P. stipitis strain (63) and the recombinant S. cerevisiae strain (43).
Likewise, the native Y. lipolytica strain was able to grow on cellobiose as a sole carbon source because it has 4 putative cellobiose transporters as well as 11 BGLs induced by cellobiose. Six of these 11 BGLs, including YALI0D18381, YALI0B14333, YALI0B14289, YALI0E20185, YALI0F01672, and YALI0F16027, were the same as those reported very recently (during the preparation of our manuscript) (33). Furthermore, we identified 5 additional BGLs, YALI0B03564p, YALI0E33539p, YALI0E21109p, YALI0F01947p, and YALI0F13299p, which also have functional roles in the presence of cellobiose. The transcriptional and enzymatic activities of BGL genes strongly depend on the types of sugars used for cell growth. For instance, most of intracellular BGL genes were activated by growth on cellobiose alone or on its mixture with glucose, suggesting that the native Y. lipolytica assimilated cellobiose intracellularly. In contrast, growth on the mixture of cellobiose and xylose resulted in overexpression of most of the extracellular and intracellular BGL genes, suggesting that the native Y. lipolytica strain could degrade cellobiose both intracellularly and extracellularly.
In addition, the native Y. lipolytica strain exhibited leaky expression of some BGL genes, as the BGL activity was detected regardless of whether the growth was on glucose or xylose or both (Table 3). This phenotype is often found in the many biomass-degrading fungi that tend to produce base-level cellulases regardless of growth conditions (64, 65). While native Y. lipolytica could grow on cellobiose as a sole carbon source, the cellobiose assimilation was inefficient compared to that seen with native N. crassa (14), native A. niger (66), recombinant S. cerevisiae (67), or recombinant Y. lipolytica (62), likely due to the limitation of the cellobiose transporter. With the knowledge of putative cellobiose transporters and BGL genes identified in this study, further optimization of the cellobiose degradation pathway is highly feasible.
Even though metabolic and transcriptomic analyses clearly demonstrate the activation of xylose and cellobiose degradation pathways, the catalytic activity and specificity for many metabolic enzymes/transporters need to be characterized in detail due to their potential novelty, which might be masked by transcriptional regulation of the native metabolism. For instance, complementary to the very recent studies that have successfully engineered xylose-specific transporters in S. cerevisiae (22), we discovered at least 5 novel, putative native xylose-specific transporters in Y. lipolytica that have both the G-G/F-X-X-X-G motif and T213/N370 replacement. Even though our study focused on activation and elucidation of the sugar metabolism in the native Y. lipolytica ATCC MYA-2613 strain (taxid 4952), we found that these sugar assimilation pathways are also conserved in other isolated Y. lipolytica strains, WSH-Z06 (taxid 1437815) and CLIB122 (taxid 284591) (see Table S7 in the supplemental material). This result suggests that, once activated, Y. lipolytica WSH-Z06 and CLIB122 can assimilate xylose and cellobiose.
In summary, this report sheds light on fundamental understanding of the complex sugar metabolism of Y. lipolytica and will help guide inverse metabolic engineering of this industrially important microorganism for enhanced conversion of biomass-derived fermentable sugars to chemicals and fuels with broad biotechnological applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Matsuoka (Sojo University, Japan) for kindly providing the plasmid pSL16-CEN1-1-227. We also thank members of the Trinh lab for proofreading and commenting on the manuscript.
Funding Statement
This research was also funded by the Sustainable Energy and Education Research Center (SEERC) at The University of Tennessee, Knoxville, TN.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03582-15.
REFERENCES
- 1.Naik SN, Goud VV, Rout PK, Dalai AK. 2010. Production of first and second generation biofuels: a comprehensive review. Renew Sust Energ Rev 14:578–597. doi: 10.1016/j.rser.2009.10.003. [DOI] [Google Scholar]
- 2.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Magasanik B. 1961. Catabolite repression. Cold Spring Harbor Symp Quant Biol 26:249–256. doi: 10.1101/SQB.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- 4.Gancedo JM. 1998. Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62:334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monod J. 1949. The growth of bacterial cultures. Annu Rev Microbiol 3:371–394. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 6.Kim J-H, Block DE, Mills DA. 2010. Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl Microbiol Biotechnol 88:1077–1085. doi: 10.1007/s00253-010-2839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong Z, Wang Q, Shen H, Hu C, Jin G, Zhao ZK. 2012. Co-fermentation of cellobiose and xylose by Lipomyces starkeyi for lipid production. Bioresour Technol 117:20–24. doi: 10.1016/j.biortech.2012.04.063. [DOI] [PubMed] [Google Scholar]
- 8.Long TM, Su Y-K, Headman J, Higbee A, Willis LB, Jeffries TW. 2012. Cofermentation of glucose, xylose, and cellobiose by the beetle-associated yeast Spathaspora passalidarum. Appl Environ Microbiol 78:5492–5500. doi: 10.1128/AEM.00374-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnett JA. 1976. The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem 32:125–234. doi: 10.1016/S0065-2318(08)60337-6. [DOI] [PubMed] [Google Scholar]
- 10.Du Preez JC, Bosch M, Prior BA. 1986. The fermentation of hexose and pentose sugars by Candida shehatae and Pichia stipitis. Appl Microbiol Biotechnol 23:228–233. doi: 10.1007/BF00261920. [DOI] [Google Scholar]
- 11.Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. 2007. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 12.Kuyper M, Hartog MMP, Toirkens MJ, Almering MJH, Winkler AA, van Dijken JP, Pronk JT. 2005. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 13.van Rooyen R, Hahn-Hagerdal B, La Grange DC, van Zyl WH. 2005. Construction of cellobiose-growing and fermenting Saccharomyces cerevisiae strains. J Biotechnol 120:284–295. doi: 10.1016/j.jbiotec.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 15.Ha S-J, Galazka JM, Kim SR, Choi J-H, Yang X, Seo J-H, Glass NL, Cate JHD, Jin Y-S. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci U S A 108:504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katahira S, Mizuike A, Fukuda H, Kondo A. 2006. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol 72:1136–1143. doi: 10.1007/s00253-006-0402-x. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura N, Yamada R, Katahira S, Tanaka T, Fukuda H, Kondo A. 2008. Effective xylose/cellobiose co-fermentation and ethanol production by xylose-assimilating S. cerevisiae via expression of β-glucosidase on its cell surface. Enzyme Microb Technol 43:233–236. doi: 10.1016/j.enzmictec.2008.04.003. [DOI] [Google Scholar]
- 18.Bisson LF, Coons DM, Kruckeberg AL, Lewis DA. 1993. Yeast sugar transporters. Crit Rev Biochem Mol Biol 28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- 19.Du J, Li S, Zhao H. 2010. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst 6:2150–2156. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 20.Farwick A, Bruder S, Schadeweg V, Oreb M, Boles E. 2014. Engineering of yeast hexose transporters to transport d-xylose without inhibition by d-glucose. Proc Natl Acad Sci U S A 111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijland JG, Shin HY, de Jong RM, De Waal PP, Klaassen P, Driessen AJM. 29 November 2014, posting date Engineering of an endogenous hexose transporter into a specific d-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels doi: 10.1186/s13068-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young EM, Tong A, Bui H, Spofford C, Alper HS. 2014. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci U S A 111:131–136. doi: 10.1073/pnas.1311970111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, Otoupal P, Alper HS. 20 January 2014, posting date Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 24.Dulermo T, Nicaud JM. 2011. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng 13:482–491. doi: 10.1016/j.ymben.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Abghari A, Chen S. 2014. Yarrowia lipolytica as an oleaginous cell factory platform for the production of fatty acid-based biofuel and bioproducts. Front Energy Res 2:1–21. doi: 10.3389/fenrg.2014.00021. [DOI] [Google Scholar]
- 26.Xue Z, Sharpe PL, Hong S-P, Yadav NS, Xie D, Short DR, Damude HG, Rupert RA, Seip JE, Wang J, Pollak DW, Bostick MW, Bosak MD, Macool DJ, Hollerbach DH, Zhang H, Arcilla DM, Bledsoe SA, Croker K, McCord EF, Tyreus BD, Jackson EN, Zhu Q. 2013. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechol 31:734–740. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- 27.Nicaud JM. 2012. Yarrowia lipolytica. Yeast 29:409–418. doi: 10.1002/yea.2921. [DOI] [PubMed] [Google Scholar]
- 28.Epova E, Guseva M, Kovalyov L, Isakova E, Deryabina Y, Belyakova A, Zylkova M, Shevelev A. 2012. Identification of proteins involved in pH adaptation in extremophile yeast Yarrowia lipolytica, p 209–224. In Heazlewood JL, Petzold CJ (ed), Proteomic applications in biology. InTech, Rijeka, Croatia. doi: 10.5772/28791. [DOI] [Google Scholar]
- 29.Tsigie YA, Wang CY, Truong CT, Ju YH. 2011. Lipid production from Yarrowia lipolytica Po1g grown in sugarcane bagasse hydrolysate. Bioresourc Technol 102:9216–9222. doi: 10.1016/j.biortech.2011.06.047. [DOI] [PubMed] [Google Scholar]
- 30.Andreishcheva EN, Isakova EP, Sidorov NN, Abramova NB, Ushakova NA, Shaposhnikov GL, Soares MIM, Zvyagilskaya RA. 1999. Adaptation to salt stress in a salt-tolerant strain of the yeast Yarrowia lipolytica. Biochemistry (Mosc) 64:1061–1067. [PubMed] [Google Scholar]
- 31.Ryu S, Labbe N, Trinh CT. 2015. Simultaneous saccharification and fermentation of cellulose in ionic liquid for efficient production of alpha-ketoglutaric acid by Yarrowia lipolytica. Appl Microbiol Biotechnol 99:4237–4244. doi: 10.1007/s00253-015-6521-5. [DOI] [PubMed] [Google Scholar]
- 32.Coelho MAZ, Amaral PFF, Belo I. 2010. Yarrowia lipolytica: an industrial workhorse, p 930–944. In Méndez-Villas A. (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology, vol 2 Formatex, Badajoz, Spain. [Google Scholar]
- 33.Guo Z, Duquesne S, Bozonnet S, Cioci G, Nicaud JM, Marty A, O'Donohue MJ. 2015. Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol Biofuels 8:109. doi: 10.1186/s13068-015-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tai M. 2012. Metabolic engineering of oleaginous yeast for the production of biofuels. Ph.D. dissertation. Massachusetts Institute of Technology, Cambridge, MA. [Google Scholar]
- 35.Yamane T, Sakai H, Nagahama K, Ogawa T, Matsuoka M. 2008. Dissection of centromeric DNA from yeast Yarrowia lipolytica and identification of protein-binding site required for plasmid transmission. J Biosci Bioeng 105:571–578. doi: 10.1263/jbb.105.571. [DOI] [PubMed] [Google Scholar]
- 36.Blazeck J, Liu L, Redden H, Alper H. 2011. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol 77:7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–341. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 38.Wang JH, Hung WP, Tsai SH. 2011. High efficiency transformation by electroporation of Yarrowia lipolytica. J Microbiol 49:469–472. doi: 10.1007/s12275-011-0433-6. [DOI] [PubMed] [Google Scholar]
- 39.Trinh CT, Unrean P, Srienc F. 2008. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentose. Appl Environ Microbiol 74:3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Dekker RFH. 1986. Kinetic, inhibition, and stability properties of a commercial β-d-glucosidase (cellobiase) preparation from Aspergillus niger and its suitability in the hydrolysis of lignocellulose. Biotechnol Bioeng 28:1438–1442. doi: 10.1002/bit.260280918. [DOI] [PubMed] [Google Scholar]
- 43.Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin Y-S. 2013. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi: 10.1371/journal.pone.0057048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye L, Berden JA, van Dam K, Kruckeberg AL. 2001. Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiae. Yeast 18:1257–1267. doi: 10.1002/yea.77. [DOI] [PubMed] [Google Scholar]
- 46.Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, Salamov A, Schmutz J, Lindquist E, Dehal P, Shapiro H, Jin Y-S, Passoth V, Richardson PM. 2007. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechol 25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- 47.Tsujibo H, Kosaka M, Ikenishi S, Sato T, Miyamoto K, Inamori Y. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J Bacteriol 186:1029–1037. doi: 10.1128/JB.186.4.1029-1037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho NWY, Chen ZD, Brainard AP. 1998. Genetically engineered Sacccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin YS, Jones S, Shi NQ, Jeffries TW. 2002. Molecular cloning of XYL3 (D-xylulokinase) from Pichia stipitis and characterization of its physiological function. Appl Environ Microbiol 68:1232–1239. doi: 10.1128/AEM.68.3.1232-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dan S, Marton I, Dekel M, Bravdo BA, He SM, Withers SG, Shoseyov O. 2000. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger β-glucosidase. J Biol Chem 275:4973–4980. doi: 10.1074/jbc.275.7.4973. [DOI] [PubMed] [Google Scholar]
- 53.Suh M-J, Fedorova ND, Cagas SE, Hastings S, Fleischmann RD, Peterson SN, Perlin DS, Nierman WC, Pieper R, Momany M. 30 April 2012, posting date Development stage-specific proteomic profiling uncovers small, lineage specific proteins most abundant in the Aspergillus Fumigatus conidial proteome. Proteome Sci doi: 10.1186/1477-5956-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skory CD, Freer SN, Bothast RJ. 1996. Expression and secretion of the Candida wickerhamii extracellular β-glucosidase gene, bglB, in Saccharomyces cerevisiae. Curr Genet 30:417–422. doi: 10.1007/s002940050151. [DOI] [PubMed] [Google Scholar]
- 55.Horton P, Nakai K. 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier, p 147–152. In Gaasterland T, Karp P, Karplus K, Ouzounis G, Sander S, Valencia A (ed), Proceedings of the 5th International Conference on Intelligent Systems for Molecular Biology. The AAAI Press, Menlo Park, CA. [PubMed] [Google Scholar]
- 56.Barrett MP, Walmsley AR, Gould GW. 1999. Structure and function of facilitative sugar transporters. Curr Opin Cell Biol 11:496–502. doi: 10.1016/S0955-0674(99)80072-6. [DOI] [PubMed] [Google Scholar]
- 57.Leandro MJ, Fonseca C, Goncalves P. 2009. Hexose and pentose transport in ascomycetous yeasts: an overview. FEMS Yeast Res 9:511–525. doi: 10.1111/j.1567-1364.2009.00509.x. [DOI] [PubMed] [Google Scholar]
- 58.Pao SS, Paulsen IT, Saier MH. 1998. Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci U S A 93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozcan S, Dover J, Johnston M. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J 17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahearn RJB, Fell DGJW, Johannsen E, Kreger NJW, Kurtzman CP, Kwon-Chung KJ, Meyer SA, Miller MW, Phaff HJ, De Miranda LR, Schlitzer RL, Smith MTH, Tallman AS, Van Der Walt JP, Yarrow D. 1984. General classification of the yeasts, p 1–44. In Kreger-van Rij NJW. (ed), The yeasts, 3rd ed Elsevier, Amsterdam, The Netherlands. doi: 10.1016/B978-0-444-80421-1.50008-5. [DOI] [Google Scholar]
- 62.Lane S, Zhang S, Wei N, Rao C, Jin YS. 2015. Development and physiological characterization of cellobiose-consuming Yarrowia lipolytica. Biotechnol Bioeng 112:1012–1022. doi: 10.1002/bit.25499. [DOI] [PubMed] [Google Scholar]
- 63.Shi J, Zhang M, Zhang L, Wang P, Jiang L, Deng H. 2014. Xylose-fermenting Pichia stipitis by genome shuffling for improved ethanol production. Microb Biotechnol 7:90–99. doi: 10.1111/1751-7915.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. 2012. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci U S A 109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun X, Liu Z, Zheng K, Song X, Qu Y. 2008. The composition of basal and induced cellulase systems in Penicillium decumbens under induction or repression conditions. Enzyme Microb Tech 42:560–567. doi: 10.1016/j.enzmictec.2008.01.020. [DOI] [Google Scholar]
- 66.Sternberg D, Vijayakumar P, Reese ET. 1977. Beta-glucosidase microbial production and effect on enzymatic hydrolysis of cellulose. Can J Microbiol 23:139–147. doi: 10.1139/m77-020. [DOI] [PubMed] [Google Scholar]
- 67.Murai T, Ueda M, Kawaguchi T, Arai M, Tanaka A. 1998. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl Environ Microbiol 64:4857–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.