Abstract
The gut microbiota of termites and cockroaches represents complex metabolic networks of many diverse microbial populations. The distinct microenvironmental conditions within the gut and possible interactions among the microorganisms make it essential to investigate how far the metabolic properties of pure cultures reflect their activities in their natural environment. We established the cockroach Shelfordella lateralis as a gnotobiotic model and inoculated germfree nymphs with two bacterial strains isolated from the guts of conventional cockroaches. Fluorescence microscopy revealed that both strains specifically colonized the germfree hindgut. In diassociated cockroaches, the facultatively anaerobic strain EbSL (a new species of Enterobacteriaceae) always outnumbered the obligately anaerobic strain FuSL (a close relative of Fusobacterium varium), irrespective of the sequence of inoculation, which showed that precolonization by facultatively anaerobic bacteria does not necessarily favor colonization by obligate anaerobes. Comparison of the fermentation products of the cultures formed in vitro with those accumulated in situ indicated that the gut environment strongly affected the metabolic activities of both strains. The pure cultures formed the typical products of mixed-acid or butyrate fermentation, whereas the guts of gnotobiotic cockroaches accumulated mostly lactate and acetate. Similar shifts toward more-oxidized products were observed when the pure cultures were exposed to oxygen, which corroborated the strong effects of oxygen on the metabolic fluxes previously observed in termite guts. Oxygen microsensor profiles of the guts of germfree, gnotobiotic, and conventional cockroaches indicated that both gut tissue and microbiota contribute to oxygen consumption and suggest that the oxygen status influences the colonization success.
INTRODUCTION
Many insects, particularly those feeding on a fiber-rich diet, possess a dense and complex microbiota. The most prominent examples are termites, whose ability to thrive on an entirely lignocellulosic diet depends on the digestive and nutritional contributions of microbial symbionts housed in their intestinal tracts (1, 2). During recent years, the microbial community structure of many termites has been studied in detail, and the evolutionary patterns in the gut microbiota of termites and their closest phylogenetic relatives, the cockroaches, are slowly emerging (3). In particular, the application of high-throughput sequencing techniques provides sufficient resolution and sampling depth to distinguish the phylogenetic and environmental drivers of the community structure (4, 5, 6).
The functional roles of individual community members and their interactions, however, are more difficult to elucidate, mostly due to their formidable resistance to cultivation. Metagenomic and metatranscriptomic approaches have provided the first insights into the functional potentials of the gut community (7, 8, 9), but owing to the lack of reference genomes for many deep-branching lineages of the gut microbiota, it remains difficult to assign functional genes to particular members of the respective communities. Improved binning strategies promise a solution for this problem in the near future (10), but the elucidation of emergent community properties, such as specific interactions or metabolic networks, requires an entirely different approach. Even in cases where representative microorganisms have been brought into culture, our lack of knowledge about the abiotic and biotic factors in the gut microenvironment makes it difficult to predict their metabolic activities in situ.
The intestinal tract of insects comprises unique microenvironmental conditions. It is therefore essential to investigate how far the in vitro metabolic properties of pure cultures reflect their in situ activities. Studies with termites have shown that the influx of oxygen, whose importance increases inversely proportionally with the radius of a gut compartment (11), strongly affects the fermentative processes in the entire hindgut community (12), but this remains to be investigated with pure cultures. Early colonization of the (presumably oxic) gut and the modalities of community succession are also unclear. Based on the observation that the first colonizers of mammalian guts are typically facultative anaerobes, it has been postulated that these bacteria create a reduced environment favorable for the colonization of obligate anaerobes, which constitute the majority of the climax community (13, 14), a tempting hypothesis that awaits experimental testing.
Experiments with germfree animals inoculated with one or more strains of defined gut microorganisms may provide excellent opportunities to approach these questions. Such gnotobiotic animals (15) can be used to characterize the responses of pure cultures of gut bacteria to their natural habitat and their interactions with other strains and to construct complex microbial networks. However, such studies so far have been restricted mostly to gnotobiotic mammals, particularly rats, mice, and piglets (16, 17, 18, 19).
While germfree mammals can be obtained only by Cesarean section, germfree insects are easily generated by chemical surface sterilization of eggs (20, 21, 22, 23). Unfortunately, termites are intractable as gnotobiotic models due to their eusociality and obligate dependence on their gut microbiota for food digestion. In contrast, their closest relatives, the nonsocial omnivorous cockroaches, do not depend on colony members and can be raised under axenic conditions. Moreover, their eggs are contained in egg cases (oothecae) that provide additional protection to the eggs, including that against the potentially detrimental effects of the sterilization process (20, 21). Surface sterilization of eggs does not remove the endosymbiotic Blattabacterium sp., an intracellular symbiont that occurs in all cockroaches and is inherited via the germ line (24). However, blattabacteria do not occur in the gut but colonize special cells of the fat body; they cannot be removed without severely affecting the well-being of the host because of their essential role in both nitrogen recycling and provision of nutrients (25).
We selected the cockroach Shelfordella lateralis, an omnivorous member of the cockroach family Blattidae, the sister group of termites (26), as a gnotobiotic model. The intestinal tract of S. lateralis and other cockroaches is colonized by complex microbial communities, which are dominated by diverse lineages of presumably obligately and facultatively anaerobic bacteria (27, 28) and also comprise methanogenic archaea (29, 30) and ciliate protists (31). As in termites, the bacterial microbiota of cockroaches participates in the breakdown of food, supplies the host with short-chain fatty acids, and contributes to the host's nutrition and normal development (32). The gut environment of cockroaches resembles that encountered in many termites (27, 28), which explains why the gut microbiotas of termites and cockroaches share several common lineages (4, 27) and may allow the use of cockroach guts as a surrogate environment for studying the termite gut microbiota.
With this gnotobiotic cockroach model (Fig. 1), we studied the colonization of the germfree gut by a facultatively and an obligately anaerobic bacterium isolated from the same environment, using fluorescence microscopy and real-time PCR, and tested the effect of precolonization by one strain on the colonization success of the other. Moreover, we compared the effects of environmental factors on the metabolic product profiles of the strains in vitro with their activities in the gut environment in situ, including the effect of colonization on the oxygen status of the gut.
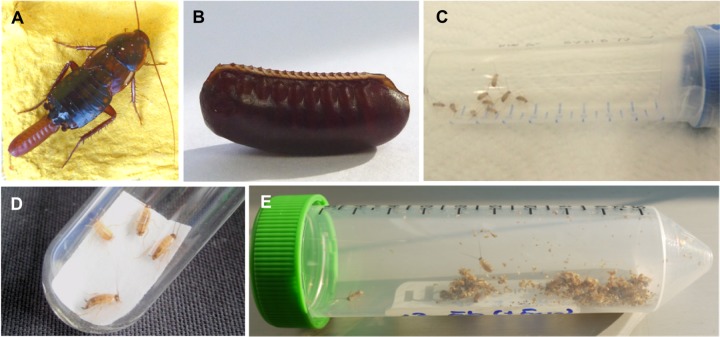
FIG 1.
Shelfordella lateralis as a gnotobiotic model system. (A) Adult female with ootheca; (B) ootheca sterilized with 2% peracetic acid; (C) germfree hatchlings; (D) germfree cockroaches on filter paper strips soaked with bacterial culture; (E) gnotobiotic cockroaches in containers with autoclaved wheat bran.
MATERIALS AND METHODS
Generation of germfree cockroaches.
Shelfordella lateralis was obtained from a commercial breeder and maintained as previously described (27). Oothecae were washed in water to remove dirt particles and to select those that floated at the surface, which is indicative of maturity. Only specimens without indentations or other damage were used for experiments.
Oothecae were surface sterilized in a laminar-flow workbench under aseptic conditions using the protocol described by Doll et al. (20) with several modifications. After a brief immersion in 0.1% sodium dodecylbenzenesulfonate, oothecae were placed in 2% peracetic acid solution for 5 min, rinsed in sterile water, and transferred to sterile 50-ml polypropylene tubes. The tubes were incubated at 25°C. Cockroaches typically hatched from the oothecae within 1 month.
The efficiency of the sterilization protocol was initially evaluated by transferring freshly hatched cockroaches to 500-ml bottles containing Luria-Bertani (LB) agar and sterile food (Corn Flakes; Kellogg's, Hamburg, Germany). The agar surface was checked over several days for the absence of bacterial or fungal growth. To detect potential contaminants that escape cultivation, whole cockroaches and their feces were homogenized and disrupted by bead beating (FastPrep-24; MP Biomedicals, Irvine, CA, USA) for 45 s at 6.5 m s−1. After DNA extraction with the FastDNA spin kit for soil (MP Biomedicals) according to the manufacturer's manual, 16S rRNA genes were amplified using the Bacteria-specific primer pair 27f and 1492r (33); amplicons were purified and sequenced as described earlier (34).
Once the protocol was firmly established, the axenic status of the cockroaches was routinely tested by sacrificing one hatchling of each ootheca; each sacrificed hatchling was crushed with sterile forceps and smeared onto the surface of an LB agar plate. The plates were incubated at 25°C for several weeks and monitored for the absence of microbial growth. In the rare cases when a plate showed growth of bacteria or fungi, all results obtained with the batch of cockroaches from that ootheca were discarded. In addition, the gut contents of individual germfree cockroaches were randomly inspected for the absence of bacteria by phase-contrast microscopy.
Isolation of bacterial strains from cockroach hindguts.
Pure cultures of numerically abundant gut bacteria were isolated from normal adult females of S. lateralis by plating serial dilutions of hindgut homogenates on solid medium. Facultatively anaerobic bacteria were obtained on nutrient agar plates (Difco; Becton, Dickinson, Franklin Lakes, NJ, USA) and incubated under air at 30°C. Obligately anaerobic bacteria were isolated on fastidious-anaerobe agar (35) that had been incubated in an anoxic jar under an atmosphere of N2-CO2 (80:20 [vol/vol]) at 25°C. To identify the strains, DNA was extracted, and 16S rRNA genes were amplified and sequenced as described above. Two strains were selected for the colonization experiments in the gnotobiotic cockroaches: the facultatively anaerobic strain EbSL and the obligately anaerobic strain FuSL. Both strains have been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) culture collection (strain EbSL, DSM 100672; strain FuSL, DSM 100562).
Green fluorescent protein labeling of strain EbSL.
Competent cells of strain EbSL were prepared by following the protocol of Sharma and Schimke (36). Briefly, overnight cultures were grown at 30°C in YENB medium (0.75% yeast extract, 0.8% nutrient broth) and transferred to fresh medium (200 ml). In the exponential phase, the culture was chilled on ice and centrifuged at 4°C for 10 min at 4,000 × g. Cells were washed twice in sterile distilled water and once in 10% glycerol and finally resuspended in 150 μl of 10% glycerol. All solutions were kept on ice. Competent cells were stored at −80°C prior to transformation.
Strain EbSL was transformed with plasmid pGFPuv (Clontech, Palo Alto, CA, USA), which carries genes for ampicillin resistance and green fluorescent protein (GFP) expression under the control of the lacZ promoter; the plasmid has a narrow host range that includes Enterobacteriaceae and contains no mobilizing or conjugation functions. After the plasmid (500 ng) was added to a 50-μl suspension of competent cells of strain EbSL, the cells were transformed by electroporation in 0.2-cm-gap electroporation cuvettes (Sigma-Aldrich) using an Escherichia coli Gene Pulser (Bio-Rad) with settings of 25 μF, 2.5 kV, and 200 Ω. Cells were recovered in LB medium for 1 h at 30°C and were then streaked on LB agar supplemented with ampicillin (100 μg/ml); plates were incubated overnight at 30°C. Transformants were identified by their green fluorescence under UV light.
Plasmid stability was assessed by growing each transformed isolate in antibiotic-free medium overnight at 30°C. Two independent cultures of each isolate were serially diluted and plated in triplicate onto LB agar and LB agar supplemented with ampicillin. The fraction of plasmid-containing cells was calculated as the number of colonies on LB-ampicillin plates divided by the number of colonies on LB plates. The retention of plasmid pGFPuv in strain EbSL was 95%.
Inoculation of germfree cockroaches.
Strain EbSL and strain FuSL were routinely grown in AM5 medium (37) containing 5 mM glucose, 0.2% yeast extract, 0.4% Casamino Acids, 2 mM cysteine, and 1 mM dithiothreitol (DTT) (but no ampicillin) and kept under a headspace of N2-CO2 (80:20 [vol/vol]). The entire inoculation procedure was carried out in a laminar-flow workbench under aseptic conditions. Aliquots (200 μl) of cultures in the exponential growth phase (optical density at 578 nm [OD578] of 0.4 to 0.45) were applied onto sterile filter paper strips, which were immediately placed into sterile 50-ml tubes with five newly hatched germfree cockroaches. Inoculated cockroaches were incubated at 25°C. One day after each inoculation, cockroaches were transferred to a fresh tube containing autoclaved wheat bran (Spielberger, Brackenheim, Germany) soaked with water. Incubations were terminated at different time points during the first instar, and batches were analyzed as described below.
Localization of bacteria within the gut.
GFP-labeled strain EbSL was grown aerobically on LB medium with ampicillin overnight to allow maturation of the GFP fluorophore and inoculated into germfree cockroaches as described above. Cells were localized in the gut by observing the intact foregut, midgut, and hindgut sections under a fluorescence microscope.
In the case of strain FuSL, pooled gut sections (foregut, midgut, and hindgut; 5 each) were homogenized via sonication (ultrasonic processor UP50H; Hielscher Ultrasonics, Teltow, Germany), and the cells were detected using fluorescence in situ hybridization (FISH) with the Bacteria-specific probe EUB338 (38) at 46°C as previously described (39).
Quantitative PCR.
Cockroaches were dissected with sterile forceps, five hindguts from the same batch were pooled and homogenized, and microbial cells were disrupted by two cycles of beat beating (FastPrep-24) for 45 s at 6.5 m s−1. DNA was extracted with the NucleoSpin soil kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's manual. Samples were used for subsequent quantitative real-time PCR (qPCR) analysis.
Standard curves were generated using purified 16S rRNA PCR products of the target strains, which were checked photometrically for purity (NanoDrop; PeqLab, Erlangen, Germany) and quantified fluorimetrically (Qubit; Invitrogen, Eugene, OR, USA). The number of 16S rRNA genes of target strains was determined by qPCR as described by Stubner (40) using primers specific for Enterobacteriaceae (5′-ATGGCTGTCGTCAGCTCGT-3′ and 5′-CCTACTTCTTTTGCAACCCACTC-3′) (41) and for Fusobacteriaceae (5′-GGATTTATTGGGCGTAAAGC-3′ and 5′-GGCATTCCTACAAATATCTACGAA-3′) (42) which matched the 16S rRNA gene sequences of the target strains. The total number of bacterial 16S rRNA genes in conventional cockroach guts was determined using the general Bacteria primer pair 519fc (5′-CAGCMGCCGCGGTAANWC-3′) and 907r (5′-CCGTCAATTCMTTTRAGTT-3′) (33) (primer 519fc as modified by Stubner [40]).
Each sample was analyzed in duplicate with at least three independent determinations, which typically showed a 10 to 30% deviation. All calibration curves were linear over a range of at least 6 orders of magnitude. Cell densities of strains FuSL and EbSL in the hindgut were estimated using the copy number of 16S rRNA genes in the genomes of Enterobacter spp. (seven) and Fusobacterium nucleatum (five) (rRNA database [43]).
Fermentation products of pure cultures in vitro.
The cultures of strains FuSL and EbSL were routinely grown in AM5 medium amended with 5 mM glucose, 0.1% yeast extract, 0.1% Casamino Acids, 1 mM DTT as the reducing agent, and 0.8 mg/liter resazurin as the redox indicator. Tubes were inoculated with 4% preculture and incubated at 30°C. The influence of oxygen on the growth and fermentation products was tested in nonreduced medium by the addition of different amounts of sterile oxygen to the headspace of cultures incubated on a rotary shaker.
Growth was determined photometrically by following the increase in OD578 using a culture tube photometer (Spectronic 20+, path length of ca. 1.3 cm; Milton Roy); optical densities of >0.8 were calculated after appropriate dilution. After centrifugation of the fully grown cultures, the cell-free supernatants were acidified with H2SO4 (50 mM final concentration) and analyzed by high-performance liquid chromatography (HPLC) with an ion-exclusion column (Resin H+ IEX column, 8 μm; Grom, Rottenburg, Germany) and a refractive index detector (27). The hydrogen concentration in the culture headspaces was analyzed by gas chromatography with a thermal conductivity detector (44).
For computation of electron recoveries, all metabolites were formally oxidized to CO2, and the number of electrons theoretically released from the respective amounts of products was compared with that from the amount of substrate consumed (45). The amount of glucose assimilated into the biomass was estimated using the turbidity of the culture (OD578 of 0.1 corresponds to a dry weight of 30 mg liter−1) and an elemental composition of C4H8O2N for bacterial cells (46), which corresponds to 6.9 mmol glucose per g cell mass.
Detection of metabolites in situ.
For the detection of metabolites in the cockroach hindgut, cockroaches (first-instar nymphs, 9 days after inoculation) were dissected under a stereomicroscope using sterile forceps. The fat body surrounding each hindgut was carefully removed, and 10 hindguts were pooled and homogenized in 150 μl water by sonication (ultrasonic processor UP50H). Samples were prepared and analyzed by HPLC as previously described in Schauer et al. (27).
Hydrogen emission by living cockroaches (first-instar nymphs, 7 days after inoculation) was measured by gas chromatography with a packed Mol Sieve 5A column (80/100 mesh; 70 cm by 6.35 mm) and a reduction gas detector (RGD2; Trace Analytical, Menlo Park, CA, USA). For the measurement, pools of ca. 10 cockroaches were placed in 15-ml glass vials closed with a rubber stopper. With the respiratory activity of S. lateralis (47), it was estimated that the oxygen concentration in the vials decreased by 0.3 to 1.1% per hour of incubation. Hydrogen production rates were determined from the linear increase of the hydrogen concentration; at least three time points were taken over a period of 5 to 6 h. In the rare cases when the slope slightly decreased at the end of the incubation, the initial rates were used.
Oxygen microsensor measurements.
Guts from first-instar nymphs were dissected, placed in a chamber with a bottom layer of 2% agarose, and immediately embedded at a depth of approximately 2 mm in Ringer's solution solidified with 0.5% agarose. Axial profiles of intestinal oxygen concentrations at the gut center were measured with microsensors (10- or 25-μm tip diameter; Unisense, Aarhus, Denmark) as described previously (48).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of strain EbSL and strain FuSL have been submitted to GenBank under accession numbers KU043525 and KU043524, respectively.
RESULTS
Establishment of the sterilization protocol.
Based on previous protocols for the axenic rearing of insects (20, 21, 22), we tested several biocides for their applicability in the surface sterilization of S. lateralis oothecae. The highest hatching rates were obtained when oothecae were exposed to 2% peracetic acid for 5 min; since these conditions yielded sterile hatchlings, they were subsequently used as the standard protocol. Sterilization was not reliable at lower concentrations of peracetic acid (0.5%), and longer exposure times (10 or 20 min) severely reduced the hatching rate. Preliminary experiments showed that oothecae treated with benzalkonium chloride (10%, 10 min) often showed microbial growth, whereas no living cockroaches hatched from oothecae treated with sodium hypochlorite (0.25%, 5 min).
The ootheca subjected to the standard protocol yielded an average of 10 ± 3.6 hatchlings (n = 22), which was not significantly different from the number of hatchlings obtained from the untreated controls (11 ± 3.1; n = 20). However, surface sterilization significantly influenced the number of oothecae that yielded healthy hatchlings (36% of treated oothecae versus 92% of untreated controls; n = 50 each). No bacterial 16S rRNA genes were amplified from the hatchling feces. The only 16S rRNA gene amplified from DNA extracts of whole cockroaches was that of a Blattabacterium sp. (99% sequence similarity to the Blattabacterium sp. from Periplaneta americana, a close relative of S. lateralis), which was expected because of the omnipresence of this essential, maternally transmitted endosymbiont in the fat body of cockroaches. This was in agreement with the results of phase-contrast microscopy of gut homogenates, which confirmed the general absence of bacteria from the gut but always showed a small number of large, rod-shaped cells (4 to 6 μm long and 1 to 1.5 μm wide) with the same morphology as the blattabacteria located in the fat body tissue surrounding the gut.
The routinely conducted controls, in which one hatchling from each clutch was crushed with sterile forceps and smeared onto the surface of an LB agar plate, showed bacterial or fungal growth in only 2% of all oothecae. In such cases, the results obtained with that batch were discarded.
Isolation of bacterial strains from the cockroach gut.
Representative isolates of numerically abundant hindgut bacteria of S. lateralis were obtained by serial dilution of hindgut homogenates on solid medium. We selected one facultatively anaerobic strain and one strictly anaerobic strain for further investigations.
The facultatively anaerobic strain EbSL is a hitherto uncultured representative in the family Enterobacteriaceae. 16S rRNA gene sequencing showed 95 to 97% sequence similarity to that of the species described in the genera Pantoea and Cronobacter (formerly Enterobacter) and Shimwellia; strain EbSL represents a new genus of Enterobacteriaceae and will be described in a separate study.
The obligately anaerobic strain FuSL shares more than 99% sequence similarity with Fusobacterium varium and showed the phenotypic properties of Fusobacterium species previously isolated from cockroach guts, including a pleomorphic cell shape during growth in rich medium (49).
Colonization of germfree cockroaches.
Germfree cockroaches were inoculated with pure cultures of strains EbSL and FuSL, either in monoassociation or diassociation. Phase-contrast microscopy already indicated dense colonization of the hindgut compartment, but gut particles interfered with an accurate localization.
Fluorescence microscopy of monocolonized guts showed that both strains preferentially colonized the hindgut (Fig. 2A to C); very few cells were observed in the foregut or midgut section. In hindgut homogenates of diassociated cockroaches, the majority of the cells hybridizing with the Bacteria-specific probe also showed GFP fluorescence (Fig. 2D), which indicated that strain EbSL was more abundant than strain FuSL. Again, almost no cells were detected in the foregut and midgut.
FIG 2.
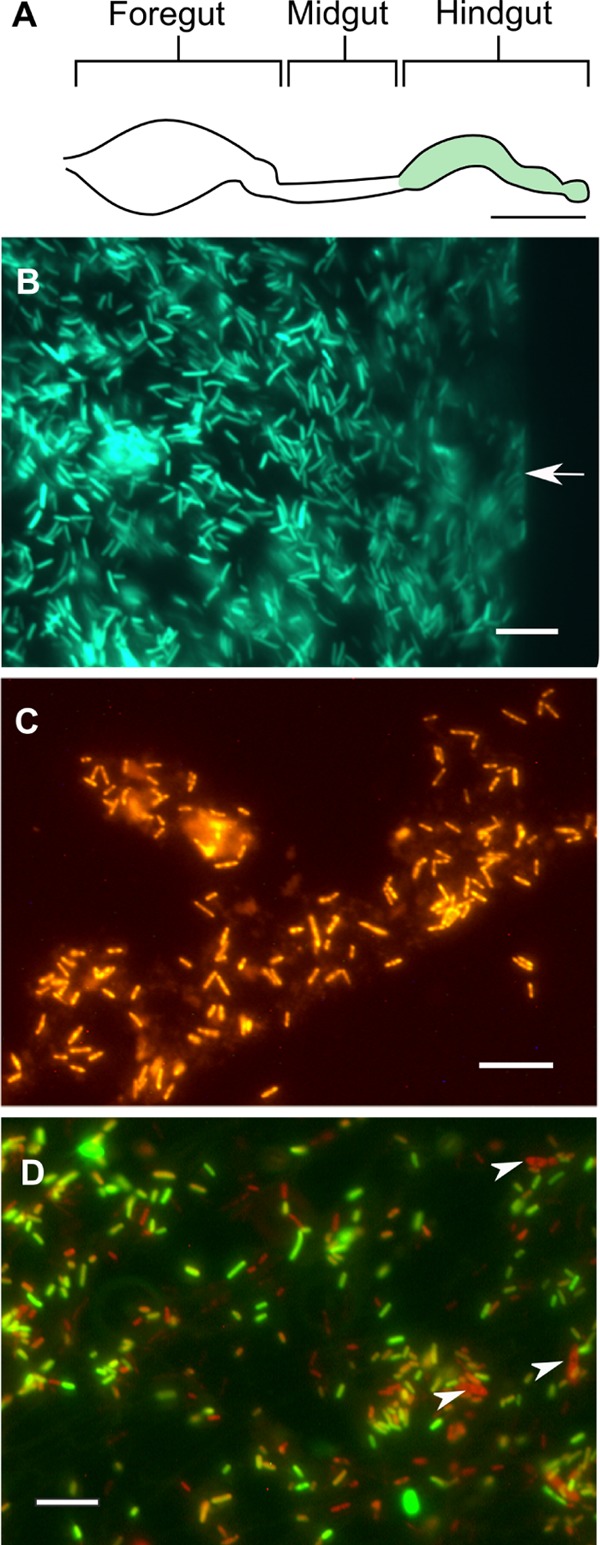
Epifluorescence micrographs of hindguts colonized with strains EbSL and FuSL. (A) Scheme illustrating the structure of the intestinal tract of Shelfordella lateralis and the exclusive location of fluorescent cells throughout the hindgut. (B) Periphery of an intact hindgut colonized by GFP-labeled strain EbSL (green). The gut wall is indicated by an arrow. (C) Hindgut homogenate of cockroaches colonized by strain FuSL, hybridized with a Cy3-labeled Bacteria-specific oligonucleotide probe (orange). (D) Hindgut homogenate of cockroaches diassociated with both strains; the image is an overlay of the GFP fluorescence and FISH signal. Cells of Blattabacterium sp. are indicated by arrowheads. Bars, ca. 2 mm (A) and 10 μm (B to D).
These observations were in agreement with the quantitative assessment of 16S rRNA genes by qPCR, which indicated that the estimated cell density of strain EbSL in diassociated cockroaches was always an order of magnitude higher than that of strain FuSL, irrespective of the sequence of inoculation and the time of incubation (Table 1). While the cell density of strain FuSL was about five times higher in monoassociation than in diassociation with strain EbSL, the colonization success of strain EbSL was not significantly affected by the presence of strain FuSL. The number of 16S rRNA genes in monoassociated cockroaches was in the same range as the total number of bacterial 16S rRNA genes in conventional cockroaches, where Enterobacteriaceae and Fusobacteriaceae formed only a small fraction of the entire community (Table 1).
TABLE 1.
Quantification of strains EbSL and FuSL in the guts of gnotobiotic cockroaches via qPCR with family-specific primers
| Inoculum | Incubation time (no. of days) | No. of 16S rRNA genes (107 copies mg−1 gut)a |
Cell density (107 cells mg−1 gut)a,b |
||
|---|---|---|---|---|---|
| EbSL | FuSL | EbSL | FuSL | ||
| Strain EbSLc | 5 | 29.9 ± 15.7 | 4.3 ± 2.2 | ||
| 7 | 39.8 ± 6.4 | 5.7 ± 0.9 | |||
| 10 | 40.4 ± 1.2 | 5.8 ± 0.2 | |||
| Strain FuSLc | 7 | 8.4 ± 3.8 | 1.7 ± 0.8 | ||
| 10 | 8.7 ± 2.5 | 1.7 ± 0.5 | |||
| EbSL + FuSLd | 9 | 32.9 ± 14.0 | 2.08 ± 0.4 | 4.7 ± 2.0 | 0.4 ± 0.1 |
| FuSL + EbSLd | 9 | 36.3 ± 12.2 | 1.43 ± 0.4 | 5.1 ± 1.7 | 0.3 ± 0.1 |
| Conventionale | 9 | 0.1 ± 0.1 | 0.3 ± 0.4 | 0.01 ± 0.01 | 0.06 ± 0.08 |
Values are means (± standard deviations [SD]) from three replicate experiments with five hindguts each. Results from conventional cockroaches are shown for comparison.
Estimated using the rRNA gene copy number of seven for Enterobacter spp. and five for Fusobacterium nucleatum.
Cockroaches monoassociated with either strain EbSL or strain FuSL.
Cockroaches diassociated with strains EbSL and FuSL, inoculated in the given order (the second strain 2 days after the first).
Conventional cockroaches (first instar) from two replicate experiments. The total number of bacterial 16S rRNA genes was between 8.2 and 24.8 × 107 per mg gut.
In situ activities in gnotobiotic cockroaches.
The metabolic activities of strains EbSL and FuSL within the cockroach gut were assessed by comparing the metabolites in gut homogenates of gnotobiotic cockroaches with those of germfree and conventional cockroaches. In all cases, the homogenates contained high concentrations of glucose, which indicated that dietary starch is depolymerized by host enzymes. Glucose levels were similar in the gut homogenates of germfree and gnotobiotic cockroaches but lower in those of conventional animals. In monoassociated cockroaches, the prevailing fermentation products in gut homogenates were acetate and lactate; in some batches of cockroaches associated with strain EbSL, the gut homogenates also contained small amounts of ethanol and succinate. Gut homogenates of conventional cockroaches contained acetate and lactate, but lactate accumulated in smaller amounts. In gut homogenates of germfree cockroaches, only small amounts of lactate were detected (Table 2).
TABLE 2.
Gut metabolites and hydrogen emission rates of gnotobiotic S. lateralis monoassociated with strain EbSL or strain FuSL and germfree and conventional cockroaches of the same age group
| Cockroach | Gut metabolite concn (nmol mg−1)a |
Hydrogen emission rate (pmol mg−1 h−1)b | ||||
|---|---|---|---|---|---|---|
| Glucose | Acetate | Lactate | Ethanol | Succinate | ||
| Inoculated with strain EbSL | 28.8 ± 3.8 | 13.3 ± 6.1 | 15.8 ± 4.2 | 4.4 ± 7.7 | 1.5 ± 2.6 | 76 ± 78 |
| Inoculated with strain FuSL | 31.2 ± 10.6 | 20.8 ± 5.5 | 14.0 ± 2.6 | — | — | 58 ± 39 |
| Conventional | 7.2 ± 1.9 | 22.9 ± 2.1 | 6.6 ± 3.5 | — | — | 235 ± 145 |
| Germfree | 36.2 ± 12.3 | — | 3.6 ± 2.9 | — | — | —c |
Values are means (± SD) from four replicate experiments, using homogenates of 10 hindguts with an average fresh weight of 0.2 ± 0.06 mg per gut. —, below the detection limit (<1 nmol mg−1).
Values are means (± SD) from three to four replicate experiments with pools of ca. 10 cockroaches, each with a fresh weight of 8.0 ± 2.3 mg per cockroach.
—, below the detection limit (<1 pmol mg−1 h−1).
The hydrogen production by the microbiota was assessed by measuring hydrogen emission of cockroaches in vivo. Both conventional and gnotobiotic cockroaches emitted hydrogen, albeit at different rates (Table 2). The hydrogen emission rates of monoassociated cockroaches were in the same range as the rates of those in diassociation (70 ± 23 nmol g−1 h−1), with strong variations among replicates. The hydrogen emission of conventional cockroaches, however, was significantly higher. As expected, germfree cockroaches did not emit any hydrogen.
Influence of cultivation conditions on metabolic activities in vitro.
When pure cultures of strains EbSL and FuSL were grown in vitro on glucose under anoxic conditions (Table 3), their fermentation products differed substantially from those produced in situ in the guts of monoassociated cockroaches.
TABLE 3.
Growth and fermentation products of strains EbSL and FuSL cultivated on basal medium with 5 mM glucose at different oxygen concentrations in the headspacea
| Strain and oxygen concn (%)b | Turbidity (OD578)c | Dissimilated glucose (mM)d | Concn (mM) of indicated producte |
Electron recovery (%)f | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Formate | Ethanol | Succinate | Acetate | Hydrogen | Butyrate | Lactate | ||||
| FuSL | ||||||||||
| 0 | 0.65 | 3.7 | — | — | — | 5.3 | 6.4 | 4.4 | 0.2 | 166 |
| 1 | 0.32 | 4.3 | — | — | — | 6.1 | 6.4 | 2.1 | 0.2 | 102 |
| 2 | 0.01 | 0.8 | — | — | — | 1.0 | 0.3 | — | — | 46 |
| EbSL | ||||||||||
| 0 | 0.56 | 3.9 | 5.4 | 3.6 | 1.1 | 3.7 | 0.6 | — | — | 108 |
| 1 | 0.61 | 3.7 | 2.1 | 4.0 | 1.2 | 4.5 | 2.4 | — | — | 122 |
| 2 | 0.62 | 3.7 | 1.5 | 3.3 | 1.0 | 4.3 | 2.4 | — | — | 108 |
| 4 | 0.66 | 3.6 | 1.1 | 2.8 | 0.9 | 4.7 | 2.1 | — | — | 101 |
| 8 | 0.78 | 3.4 | 0.4 | 1.5 | 0.6 | 4.7 | 1.0 | — | — | 83 |
| 21 | 2.38 | 2.0g | — | — | 0.4 | 0.9 | — | — | — | 26 |
Values are means of results from duplicate cultures (typically <10% deviation).
Initial values.
Values include cell mass formed on basal medium (0.1% Casamino Acids and 0.1% yeast extract).
Dissimilated glucose accounts for the amount of glucose assimilated into the biomass.
Values include the products formed on basal medium (0.1% Casamino Acids and 0.1% yeast extract). —, below the detection limit (<0.02 mM).
Electron recoveries in fermentation products, based on dissimilated glucose.
Corrected for the large amount of cell mass formed on basal medium under oxic conditions (OD578 of 0.93).
The major fermentation products of the strictly anaerobic strain FuSL in pure culture were butyrate, acetate, and hydrogen. With only 1% oxygen in the headspace, the growth rate, cell yield, and butyrate production decreased significantly, whereas acetate production slightly increased (Table 3). Oxygen was completely consumed in the fully grown cultures, as indicated by the reduced status of the redox indicator resazurin at the end of the incubation. With 2% oxygen in the headspace, however, growth, glucose consumption, and product formation ceased almost completely; the medium was still oxidized at the end of the incubation. The high electron recovery under anoxic conditions (Table 3) can be explained by the substantial amounts of products (3 mM acetate and 1.5 mM butyrate) produced by the amino-acid-fermenting strain FuSL on basal medium (0.1% Casamino Acids and 0.1% yeast extract). After subtraction of the products formed in the absence of glucose, the electron recovery decreased to 110%, yielding a reaction stoichiometry of 0.60 mol butyrate, 0.47 mol acetate, and 1.12 mol H2 per mol glucose.
The major fermentation products of the facultatively anaerobic strain EbSL in pure culture were formate, ethanol, acetate, succinate, and hydrogen, which are typical of mixed-acid fermentations. In contrast to the yield of the strictly anaerobic strain FuSL, the cell yield of strain EbSL increased with the oxygen concentration in the headspace, and less formate and ethanol were produced (Table 3). Glucose was consumed completely at all oxygen concentrations. The electron recoveries under anoxic conditions were much lower than in the case of strain FuSL and decreased to 90% when the amounts of fermentation products formed on basal medium were subtracted (1 mM acetate and minor amounts of formate, ethanol, and succinate), resulting in corrected reaction stoichiometries of 0.95 mol formate, 0.65 mol ethanol, 0.53 mol acetate, 0.11 mol succinate, and 0.12 mol H2 per mol glucose.
The fermentation products were also influenced by the glucose concentration in the medium (Table 4). In the case of strain EbSL, lactate was entirely absent in cultures grown on 5 mM glucose but increased at higher glucose concentrations. Strain FuSL also produced more lactate at higher glucose concentrations, but the effect was less pronounced.
TABLE 4.
Metabolic products of strains EbSL and FuSL cultivated with different glucose concentrations under anoxic conditionsa
| Strain and glucose concn (mM) | Concn (mol per mol glucose) of indicated productb |
|||||
|---|---|---|---|---|---|---|
| Formate | Ethanol | Succinate | Acetate | Butyrate | Lactate | |
| FuSL | ||||||
| 5 | — | — | — | 0.47 | 0.60 | 0.03 |
| 10 | — | — | — | 0.22 | 0.56 | 0.10 |
| 15c | — | — | — | 0.20 | 0.53 | 0.11 |
| EbSL | ||||||
| 5 | 0.95 | 0.65 | 0.11 | 0.53 | — | — |
| 10 | 0.68 | 0.76 | 0.19 | 0.59 | — | 0.36 |
| 15 | 0.39 | 0.62 | 0.20 | 0.39 | — | 0.70 |
The hydrogen concentration was not determined. Values are means of results from duplicate cultures (<10% deviation).
Calculated from consumed glucose, after subtraction of products formed on basal medium (0.1% Casamino Acids and 0.1% yeast extract). —, below the detection limit (<0.02 mM).
Only 12.3 mM glucose was consumed.
Oxygen status of the gut.
Microsensor measurements revealed strong differences in the oxygen status in the different regions of the agarose-embedded guts of conventional, germfree, and gnotobiotic cockroaches (Fig. 3). In all cockroaches, oxygen partial pressures were generally low in the foregut and midgut compartments, often under the detection limit of the sensor (ca. 0.15 kPa). In germfree and conventional guts, the axial oxygen profiles were more varied than in gnotobiotic guts. The highest partial pressures were encountered in the posterior hindgut, where the oxygen levels in the germfree cockroaches surpassed those in the gnotobiotic and conventional cockroaches. Guts colonized with strain EbSL consistently exhibited only very low oxygen concentrations in all compartments.
FIG 3.
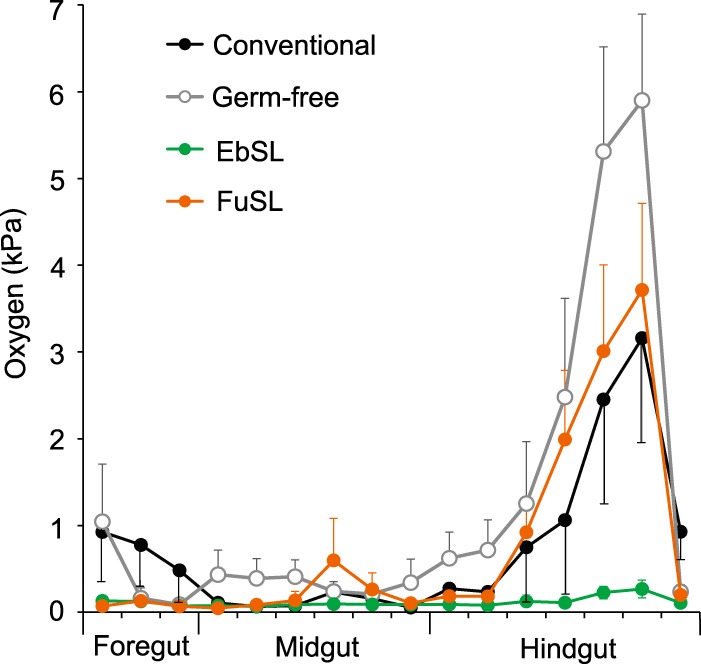
Oxygen partial pressure in the guts of gnotobiotic, germfree, and conventional first-instar cockroaches. The axial profiles were measured with microsensors at the gut center. The symbols are means (± standard errors of the mean [SEM]) from replicate measurements with eight different guts. Error bars are shown in one direction only for clarity.
DISCUSSION
Our gnotobiotic cockroach model offered the unique opportunity to study the effect of the gut environment on a defined microbiota of autochthonous gut bacteria. The fermentation product patterns of the model strains under in situ conditions, which differed from those observed in anoxic cultures, were elicited in vitro by the influence of oxygen and glucose. Microsensor measurements confirmed the assumption that the colonization of the gut with a facultative anaerobe creates an anoxic environment. However, precolonization with the facultatively anaerobic strain EbSL did not favor colonization by the obligately anaerobic strain FuSL, which suggested that the differences in their levels of colonization success are most likely due to their different responses to oxygen.
Model organisms specifically colonize the germfree hindgut.
GFP labeling is a useful tool to localize bacterial strains in the gut. Husseneder and Grace (50) isolated an indigenous strain of Enterobacter cloacae from termite guts and introduced a GFP label to monitor its fate after inoculation into the conventional gut microbiota of termites, where it persisted up to 11 weeks after inoculation. In our study, the GFP label allowed us to accurately localize strain EbSL in the gnotobiotic cockroach gut without any interference from gut tissue or food particles (Fig. 2B).
The exclusive colonization of the germfree cockroach hindgut by strains EbSL and FuSL suggests that only this gut compartment provides a favorable environment for microbial colonization. This is in agreement with observations of conventional cockroaches, where the hindgut shows the highest density and diversity of bacteria among all gut compartments (27, 51). It is likely that the colonization of foregut and midgut is suppressed by the high activities of digestive enzymes in these compartments (52, 53, 54).
The colonization densities of strains EbSL and FuSL in monoassociation and diassociation are much higher than those of all Enterobacteriaceae and Fusobacteriaceae in the hindguts of conventional cockroaches and, in the case of strain EbSL, even surpassed the total cell counts in the hindguts of conventional adults (2.2 [±1.6] × 107 cells mg−1 gut) (27). The high colonization densities in gnotobiotic cockroaches are most likely explained by the absence of other microorganisms, particularly the ciliate protists, which occupy a substantial portion of the hindgut volume in conventional cockroaches (31).
Colonization sequence does not explain colonization success.
Although colonization by strain EbSL created a mostly anoxic environment in the hindgut, precolonization with this strain did not enhance the colonization success of strain FuSL. This suggests that early colonization with a facultative anaerobe does not necessarily favor colonization by obligate anaerobes. On the contrary, the colonization densities of strain FuSL in the presence of strain EbSL were even lower than in monoassociation, which suggested that the obligate anaerobe is outcompeted by the facultative anaerobe, irrespective of the sequence of colonization. The basis for this phenomenon is not entirely clear, and it is possible that the situation in this simple gnotobiotic model system differs from that in a complex community. Since the fermentation product concentrations in the two strains are similar, the higher growth yields of strain EbSL might be explained by its capacity for respiration. However, glucose does not appear to be a limiting factor in the gnotobiotic gut, so that antagonistic effects resulting in the suppression of strain FuSL also cannot be excluded. Similarly, in disassociated gnotobiotic rats, the relative abundance of Fusobacterium varium was almost an order of magnitude lower than that of Bacteroides thetaiotaomicron (16).
In view of the complete growth inhibition of the obligately anaerobic strain FuSL at only 2% oxygen in the headspace, its capacity to robustly colonize the hindguts of germfree cockroaches in monoassociation is remarkable. The decreased oxygen partial pressure in hindguts colonized with strain FuSL in comparison to that in germfree guts indicated that strain FuSL is able to remove at least some of the oxygen diffusing into its habitat. However, its oxygen tolerance is lower than that of Fusobacterium nucleatum, which survives prolonged exposure to air (55) and can grow in dense cultures in a chemostat even under atmospheric oxygen partial pressure (56).
The colonization of the germfree hindgut with strain FuSL is probably facilitated by the low oxygen partial pressure in the anterior hindgut, which is <1 kPa even in germfree cockroaches, most likely due to the respiratory activity of the gut epithelium. Oxygen consumption by the gut tissue appears to be significant and is probably responsible for the low oxygen partial pressures in the midgut and the production of small amounts of lactate in the hindguts of germfree nymphs, which is likely caused by a switch to anaerobic metabolism in the gut tissue due to the limit in the oxygen supply.
The further reduction of the oxygen partial pressure in the hindgut after successful colonization by strain FuSL indicated that the strain itself is able to reduce oxygen, which is confirmed by the results obtained in vitro. The removal of oxygen by nonrespiratory activities is a common phenomenon in anaerobes (57, 58, 59, 60) and has been documented for lactic acid bacteria, homoacetogens, and methanogens isolated from the intestinal tracts of termites (37, 45, 61, 62, 63); many obligate anaerobes possess enzymes that detoxify oxygen or oxygen radicals (64, 65).
Metabolic activities in situ are controlled by oxygen.
The strong differences between the fermentation products in anoxic cultures and in association with cockroaches cannot be explained by the selective resorption of metabolites by the gut wall. Fermentation products such as acetate that are formed at high rates will inevitably accumulate at the gut center, no matter how efficiently they are resorbed at the gut epithelium (12). Conversely, metabolites such as butyrate and formate, which do not accumulate in gnotobiotic cockroaches, lack the concentration gradients required for efficient diffusive transport toward the epithelium.
Therefore, the differences between the product patterns observed under in situ conditions and in anoxic cultures must be caused by different rates of formation, which indicates that the metabolism of both strains is strongly affected by microenvironmental factors in the gut habitat. The most obvious factor is the presence of oxygen, whose influx rates into the small guts of first-instar nymphs must be substantial (11, 66). A strong effect of inflowing oxygen on the fermentative processes has been documented for termites, where hindgut fermentations shift to more-oxidized products when intact guts are incubated under oxic conditions (12).
This effect explains the absence of butyrate and the strong accumulation of acetate in cockroaches colonized by strain FuSL, which is confirmed by the effect of oxygen on butyrate production in vitro. A shift from butyrate to acetate has also been described for the more oxygen-tolerant Fusobacterium nucleatum when incubated under oxic conditions (56). The strong influence of oxygen on hindgut metabolism is corroborated by the absence of formate and the low concentrations of ethanol observed in cockroaches colonized with strain EbSL and the corresponding effects of oxygen in vitro. In this case, the decreased formate production may result from the inhibitory effects of oxygen on pyruvate-formate lyase, as has been reported for Streptococcus mutans and Streptococcus sanguis (67, 68, 69). However, the increase in hydrogen production by strain EbSL in the presence of low oxygen concentrations in vitro also suggests an enhanced turnover of formate owing to increased formate-hydrogen lyase activity in situ.
The high proportions of lactate among the fermentation products formed in situ in the hindguts of monoassociated cockroaches may also be caused by the high concentrations of free glucose. When grown on 5 mM glucose, strain EbSL formed no lactate, and strain FuSL formed only small amounts. However, at higher glucose concentrations of 10 or 15 mM, which resemble the in situ conditions in the guts of conventional cockroaches, strain EbSL formed increasing amounts of lactate; in the case of strain FuSL, increasing glucose concentrations had the same but less pronounced effects. Such shifts toward increased lactate formation in the presence of nonlimiting concentrations of glucose have also been described for chemostat cultures of Klebsiella aerogenes (70) and several Streptococcus spp. (71, 72) and are in agreement with the production of lactate by Fusobacterium varium grown at high glucose concentrations (73). In conventional cockroaches, the accumulation of lactate was less pronounced, which suggested that the normal gut microbiota, like that of termites, comprises active lactate-consuming populations (12).
Despite the obvious effects of oxygen on the metabolic processes of both strains, the hydrogen emissions of the gnotobiotic cockroaches underscore the prevalence of anaerobic processes in the hindgut. The in vivo emission of hydrogen is in agreement with the increase in hydrogen production in the presence of low oxygen concentrations observed in vitro. This also opens interesting perspectives for future studies, because production of hydrogen allows coupling of fermentative processes with hydrogenotrophic processes and the creation of synthetic methanogenic microbial communities, which may provide new insights into methanogenesis in insects and the factors limiting the colonization by methanogenic archaea in the intestinal tracts of both invertebrates and mammals (30, 74).
Last, the gnotobiotic cockroach model might provide a valuable tool in “synthetic microbial ecology” by helping to identify factors governing community assembly in cockroaches. It will likely also improve our understanding of community ecology and metabolic interactions in the intestinal tract of the closely related termites, where the eusociality of the host and an obligate dependence on its gut microbiota prohibit gnotobiotic studies.
ACKNOWLEDGMENTS
This work was supported within the LOEWE program of the State of Hessen (Center for Synthetic Microbiology) and by the Max Planck Society. C.L.T. received a postdoctoral fellowship from the Alexander von Humboldt Foundation.
We thank Katja Meuser for excellent technical assistance.
REFERENCES
- 1.Brune A, Ohkuma M. 2011. Role of the termite gut microbiota in symbiotic digestion, p 439–475. In Bignell DE, Roisin Y, Lo N (ed), Biology of termites: a modern synthesis. Springer, Dordrecht, Netherlands. [Google Scholar]
- 2.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 3.Brune A, Dietrich C. 2015. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich C, Köhler T, Brune A. 2014. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol 80:2261–2269. doi: 10.1128/AEM.04206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikaelyan A, Dietrich C, Köhler T, Poulsen M, Sillam-Dussès D, Brune A. 2015. Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol Ecol 24:5284–5295. doi: 10.1111/mec.13376. [DOI] [PubMed] [Google Scholar]
- 6.Mikaelyan A, Köhler T, Lampert N, Rohland J, Boga H, Meuser K, Brune A. 2015. Classifying the bacterial gut microbiota of termites and cockroaches: a curated phylogenetic reference database (DictDb). Syst Appl Microbiol 38:472–482. doi: 10.1016/j.syapm.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernández M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 8.He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, Kyrpides NC, Warnecke F, Tringe SG, Hugenholtz P. 2013. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. doi: 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu N, Zhang L, Zhou H, Zhang M, Yan X, Wang Q, Long Y, Xie L, Wang S, Huang Y, Zhou Z. 2013. Metagenomic insights into metabolic capacities of the gut microbiota in a fungus-cultivating termite (Odontotermes yunnanensis). PLoS One 8:e69184. doi: 10.1371/journal.pone.0069184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laczny CC, Sternal T, Plugaru V, Gawron P, Atashpendar A, Margossian HH, Coronado S, van der Maaten L, Vlassis N, Wilmes P. 2015. VizBin—an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 3:1. doi: 10.1186/s40168-014-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brune A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol 16:16–21. doi: 10.1016/S0167-7799(97)01151-7. [DOI] [Google Scholar]
- 12.Tholen A, Brune A. 2000. Impact of oxygen on metabolic fluxes and in situ rates of reductive acetogenesis in the hindgut of the wood-feeding termite Reticulitermes flavipes. Environ Microbiol 2:436–449. doi: 10.1046/j.1462-2920.2000.00127.x. [DOI] [PubMed] [Google Scholar]
- 13.Favier CF, Vaughan EE, De Vos WM, Akkermans ADL. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillilland MG, Erb-Downward JR, Bassis CM, Shen MC, Toews GB, Young VB, Huffnagle GB. 2012. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl Environ Microbiol 78:2359–2366. doi: 10.1128/AEM.05239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon HA, Pesti L. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev 35:390–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noack J, Dongowski G, Hartmann L, Blaut M. 2000. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J Nutr 130:1225–1231. [DOI] [PubMed] [Google Scholar]
- 17.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuel BS, Gordon BI. 2006. A humanized gnotobiotic mouse model of host archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozakova H, Kolinska J, Lojda Z, Rehakova Z, Sinkora J, Zakostelecka M, Splichal I, Tlaskalova-Hogenova H. 2006. Effect of bacterial monoassociation on brush-border enzyme activities in ex-germ-free piglets: comparison of commensal and pathogenic Escherichia coli strains. Microbes Infect 8:2629–2639. doi: 10.1016/j.micinf.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Doll JP, Trexler PC, Reynolds LI, Bernard GR. 1963. The use of peracetic acid to obtain germfree invertebrate eggs for gnotobiotic studies. Am Midl Nat 69:231–239. doi: 10.2307/2422857. [DOI] [Google Scholar]
- 21.Benschoter CA, Wrenn RT. 1972. Germfree techniques for establishment and maintenance of a colony of aseptic german cockroaches. Ann Entomol Soc Am 65:641–644. doi: 10.1093/aesa/65.3.641. [DOI] [Google Scholar]
- 22.Hamilton DR, Bradley RE. 1977. An integrated system for the production of gnotobiotic Anopheles quadrimaculatus. J Invertebr Pathol 30:318–324. doi: 10.1016/0022-2011(77)90140-9. [DOI] [PubMed] [Google Scholar]
- 23.Dillon RJ, Charnley AK. 2002. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res Microbiol 153:503–509. doi: 10.1016/S0923-2508(02)01361-X. [DOI] [PubMed] [Google Scholar]
- 24.Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. 2003. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol 20:907–913. doi: 10.1093/molbev/msg097. [DOI] [PubMed] [Google Scholar]
- 25.Sabree ZL, Kambhampati S, Moran NA. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A 106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inward D, Beccaloni G, Eggleton P. 2007. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol Lett 3:331–335. doi: 10.1098/rsbl.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78:2758–2767. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer E, Lampert N, Mikaelyan A, Köhler T, Maekawa K, Brune A. 2015. Physicochemical conditions, metabolites, and community structure of the bacterial microbiota in the gut of wood-feeding cockroaches (Blaberidae: Panesthiinae). FEMS Microbiol Ecol 91:1–14. doi: 10.1093/femsec/fiu028. [DOI] [PubMed] [Google Scholar]
- 29.Kane MD, Breznak JA. 1991. Effect of host diet on production of organic acids and methane by cockroach gut bacteria. Appl Environ Microbiol 57:2628–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune A. 2010. Methanogenesis in the digestive tracts of insects, p 707–728. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Heidelberg, Germany. [Google Scholar]
- 31.Gijzen HJ, Broers CA, Barugahare M, Stumm CK. 1991. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl Environ Microbiol 57:1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zurek L, Keddie BA. 1996. Contribution of the colon and colonic bacterial flora to metabolism and development of the American cockroach Periplaneta americana L. J Insect Physiol 42:743–748. doi: 10.1016/0022-1910(96)00028-5. [DOI] [PubMed] [Google Scholar]
- 33.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., New York, NY. [Google Scholar]
- 34.Strassert JF, Desai MS, Radek R, Brune A. 2010. Identification and localization of the multiple bacterial symbionts of the termite gut flagellate Joenia annectens. Microbiology 156:2068–2079. doi: 10.1099/mic.0.037267-0. [DOI] [PubMed] [Google Scholar]
- 35.Atlas RM. 2010. Handbook of microbiological media, 4th ed, p 2036 Taylor and Francis, Boca Raton, FL. [Google Scholar]
- 36.Sharma RC, Schimke RT. 1996. Preparation of electrocompetent E. coli using salt-free growth medium. Biotechniques 20:42–44. [DOI] [PubMed] [Google Scholar]
- 37.Boga HI, Brune A. 2003. Hydrogen-dependent oxygen reduction by homoacetogenic bacteria isolated from termite guts. Appl Environ Microbiol 69:779–786. doi: 10.1128/AEM.69.2.779-786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amann R, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stingl U, Brune A. 2003. Phylogenetic diversity and whole-cell hybridization of oxymonad flagellates from the hindgut of the wood-feeding lower termite Reticulitermes flavipes. Protist 154:147–155. doi: 10.1078/143446103764928530. [DOI] [PubMed] [Google Scholar]
- 40.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage in ricefield soil by real-time PCR with SYBR green detection. J Microbiol Methods 50:155–164. doi: 10.1016/S0167-7012(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 41.Castillo M, Martín-Orú SM, Manzanilla EG, Badiola I, Martín M, Gasa J. 2006. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet Microbiol 114:165–170. doi: 10.1016/j.vetmic.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 42.Boutaga K, van Winkelho AJ, Vandenbroucke-Grauls CMJE, Savelkoul PHM. 2005. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol Med Microbiol 45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. 2015. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuler S, Conrad R. 1990. Soils contain two different activities for oxidation of hydrogen. FEMS Microbiol Ecol 73:77–84. doi: 10.1111/j.1574-6968.1990.tb03927.x. [DOI] [Google Scholar]
- 45.Tholen A, Schink B, Brune A. 1997. The gut microflora of Reticulitermes flavipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol Ecol 24:137–149. doi: 10.1111/j.1574-6941.1997.tb00430.x. [DOI] [Google Scholar]
- 46.Mayberry WR, Prochazka GJ, Payne WJ. 1968. Factors derived from studies of aerobic growth in minimal media. J Bacteriol 96:1424–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schauer C, Thompson C, Brune A. 2014. Pyrotag sequencing of the gut microbiota of the cockroach Shelfordella lateralis reveals a highly dynamic core but only limited effects of diet on community structure. PLoS One 9:e85861. doi: 10.1371/journal.pone.0085861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brune A, Emerson D, Breznak JA. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl Environ Microbiol 61:2681–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foglesong MA, Cruden DL, Markovetz AJ. 1984. Pleomorphism of Fusobacteria isolated from the cockroach hindgut. J Bacteriol 158:474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Husseneder C, Grace JK. 2005. Genetically engineered termite gut bacteria (Enterobacter cloacae) deliver and spread foreign genes in termite colonies. Appl Microbiol Biotechnol 68:360–367. doi: 10.1007/s00253-005-1914-5. [DOI] [PubMed] [Google Scholar]
- 51.Cruden DL, Markovetz AJ. 1984. Microbial aspects of the cockroach hindgut. Arch Microbiol 138:131–139. doi: 10.1007/BF00413013. [DOI] [PubMed] [Google Scholar]
- 52.Bignell DE. 1981. Nutrition and digestion, p 57–86. In Bell WJ, Adiyodi KG (ed), The American cockroach. Chapman & Hall, London, United Kingdom. [Google Scholar]
- 53.Terra WR, Ferreira C. 1994. Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol B 109:1–62. doi: 10.1016/0300-9629(94)90307-7. [DOI] [Google Scholar]
- 54.Chapman RF. 2013. The alimentary canal, digestion and absorption, p 46–79. In Simpson SJ, Douglas AE (ed), The insects: structure and function, 5th ed Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 55.Loesche WJ. 1969. Oxygen sensitivity of various anaerobic bacteria. Appl Microbiol 18:723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz PI, Zilm PS, Rogers AH. 2000. The response to oxidative stress of Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol Lett 187:31–34. doi: 10.1111/j.1574-6968.2000.tb09132.x. [DOI] [PubMed] [Google Scholar]
- 57.de Vries W, Donkers C, Boellaard M, Stouthamer AH. 1978. Oxygen metabolism by the anaerobic bacterium Veillonella alcalescens. Arch Microbiol 119:167–174. doi: 10.1007/BF00964269. [DOI] [PubMed] [Google Scholar]
- 58.Chen L, Liu MY, Legall J, Fareleira P, Santos H, Xavier AV. 1993. Purification and characterization of an NADH-rubredoxin oxidoreductase involved in the utilization of oxygen by Desulfovibrio gigas. Eur J Biochem 216:443–448. doi: 10.1111/j.1432-1033.1993.tb18162.x. [DOI] [PubMed] [Google Scholar]
- 59.Karnholz A, Kusel K, Gossner A, Schramm A, Drake HL. 2002. Tolerance and metabolic response of acetogenic bacteria toward oxygen. Appl Environ Microbiol 68:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riebe O, Fischer RJ, Wampler DA, Kurtz DM Jr, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 155:16–24. doi: 10.1099/mic.0.022756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tholen A, Pester M, Brune A. 2007. Simultaneous methanogenesis and oxygen reduction by Methanobrevibacter cuticularis at low oxygen fluxes. FEMS Microbiol Ecol 62:303–312. doi: 10.1111/j.1574-6941.2007.00390.x. [DOI] [PubMed] [Google Scholar]
- 62.Leadbetter JR, Breznak JA. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol 62:3620–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graber JR, Breznak JA. 2004. Physiology and nutrition of Treponema primitia, an H2-CO2-acetogenic spirochete from termite hindguts. Appl Environ Microbiol 70:1307–1314. doi: 10.1128/AEM.70.3.1307-1314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jenney FE Jr, Verhagen MFJM, Cui X, Adams MWW. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 65.Brioukhanov AL, Netrusov AI. 2007. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl Biochem Microbiol 43:567–582. doi: 10.1134/S0003683807060014. [DOI] [PubMed] [Google Scholar]
- 66.Ebert A, Brune A. 1997. Hydrogen concentration profiles at the oxic-anoxic interface: a microsensor study of the hindgut of the wood-feeding lower termite Reticulitermes flavipes (Kollar). Appl Environ Microbiol 63:4039–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abbe K, Takahashi S, Yamada T. 1982. Involvement of oxygen sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J Bacteriol 152:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada T, Takahashi-Abbe S, Abbe K. 1985. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect Immun 47:129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahashi N, Abbe K, Takahashi-Abbe S, Yamada T. 1987. Oxygen sensitivity of sugar metabolism and interconversion of pyruvate formate lyase in intact cells of Streptococcus mutans and Streptococcus sanguis. Infect Immun 55:652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teixeira de Mattos MJ, Tempest DW. 1983. Metabolic and energetic aspects of the growth of Klebsiella aerogenes NCTC 418 on glucose in anaerobic chemostat culture. Arch Microbiol 134:80–85. doi: 10.1007/BF00429412. [DOI] [PubMed] [Google Scholar]
- 71.Thomas TD, Ellwood DC, Longyear MV. 1979. Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J Bacteriol 138:109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Condon S. 1987. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev 46:269–280. doi: 10.1111/j.1574-6968.1987.tb02465.x. [DOI] [Google Scholar]
- 73.Resmer KL, White RL. 2011. Metabolic footprinting of the anaerobic bacterium Fusobacterium varium using 1H NMR spectroscopy. Mol Biosyst 7:2220–2227. doi: 10.1039/c1mb05105a. [DOI] [PubMed] [Google Scholar]
- 74.St-Pierre B, Cersosimo LM, Ishaq SL, Wright AD. 2015. Toward the identification of methanogenic archaeal groups as targets of methane mitigation in livestock animals. Front Microbiol 6:776. doi: 10.3389/fmicb.2015.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]