Abstract
N-Glycosylation can modulate enzyme structure and function. In this study, we identified two pepsin-resistant histidine acid phosphatase (HAP) phytases from Yersinia kristensenii (YkAPPA) and Yersinia rohdei (YrAPPA), each having an N-glycosylation motif, and one pepsin-sensitive HAP phytase from Yersinia enterocolitica (YeAPPA) that lacked an N-glycosylation site. Site-directed mutagenesis was employed to construct mutants by altering the N-glycosylation status of each enzyme, and the mutant and wild-type enzymes were expressed in Pichia pastoris for biochemical characterization. Compared with those of the N-glycosylation site deletion mutants and N-deglycosylated enzymes, all N-glycosylated counterparts exhibited enhanced pepsin resistance. Introduction of the N-glycosylation site into YeAPPA as YkAPPA and YrAPPA conferred pepsin resistance, shifted the pH optimum (0.5 and 1.5 pH units downward, respectively) and improved stability at acidic pH (83.2 and 98.8% residual activities at pH 2.0 for 1 h). Replacing the pepsin cleavage sites L197 and L396 in the immediate vicinity of the N-glycosylation motifs of YkAPPA and YrAPPA with V promoted their resistance to pepsin digestion when produced in Escherichia coli but had no effect on the pepsin resistance of N-glycosylated enzymes produced in P. pastoris. Thus, N-glycosylation may improve pepsin resistance by enhancing the stability at acidic pH and reducing pepsin's accessibility to peptic cleavage sites. This study provides a strategy, namely, the manipulation of N-glycosylation, for improvement of phytase properties for use in animal feed.
INTRODUCTION
As much as 80% of the total phosphorus in cereal grains, oilseeds, and legumes exists in the form of phytate (myo-inositol hexakisphosphate), one of the most important functional ingredients in animal feed (1). However, phytate usually forms indigestible complexes with mineral cations and proteins, thereby causing reductions in nutrient utilization (2, 3). The dietary phytate undigested by monogastric animals is also released into the environment, where it causes phosphorus pollution (4, 5).
The enzyme phytase catalyzes the sequential release of inorganic phosphate and lower myo-inositol phosphates from phytate (6); thus, inclusion of exogenous phytases in animal feeds as enzyme additives can enhance the nutrient uptake, lower the feedstuff production costs, and protect the environment in regions where intensive animal farming is conducted (7, 8). Numerous phytases have been identified from microorganisms, animals, and plants (9–11), but only a few of them are commercially produced (12). The extensive application of phytases is limited by enzyme lability and protease sensitivity under high processing temperatures and low physiological pHs. Therefore, improving phytase stability at low pHs and high protease concentrations is desirable.
Pepsin, a monomeric protein composed of two β-barrel-like domains, represents the main proteolytic enzyme in gastric juice (13). The acidic residues D32 and D215 are mainly responsible for proteolysis of substrates, which occurs by a base catalysis mechanism (14, 15). Pepsin is an aspartic protease that catalyzes the hydrolytic cleavage of peptide bonds of a broad range of enzymes, with preference for F and L residues (16). The enzyme exhibits the highest catalytic activity at pH 2.0 and a temperature of 37 to 42°C (17, 18). Clearly, any feed enzyme should be stable under acidic gastric conditions and be highly resistant to pepsin digestion.
Some efforts have been made to study the effect of protein structure on proteolysis by pepsin. In the stomach, the stability of protein polymers depends on their particular chemical structures (19). For example, enzymatic cross-linking modification of β-casein results in the formation of high-molecular-mass polymers with a compact structure and enhanced resistance to in vitro pepsin digestion (20).
N-Glycosylation modification at the N in the N-X-S/T sequon commonly occurs in eukaryotic expression systems (21). Previous studies indicate that N-glycosylation confers resistance to proteolysis. For example, the N-deglycosylated form of glucoamylase from Aspergillus niger is more sensitive to proteolysis by subtilisin than is the native N-glycosylated enzyme (22). Similarly, bovine pancreatic RNase B occurs naturally as an N-glycoenzyme, and the glycoforms show increased resistance to pronase compared with those of their unglycosylated counterparts (23). The N-glycosylated form of β-glucuronidase also exhibits higher resistance to proteolytic degradation by pepsin than is seen with its N-deglycosylated form (24). Site-directed mutagenesis has confirmed that the N-glycosylation of scavenger receptor SREC-I at N289 confers resistance against trypsin digestion (25). Some N-glycosylated microbial phytases also exhibit resistance to degradation by pepsin (26–28). Thus, N-glycosylation represents a stabilizing factor against proteolytic cleavage by proteases (29). Apart from conferring an increased resistance to proteolysis, N-glycosylation can also enhance enzyme thermostability and alter catalytic activity (30–33). The improved conformational stability of enzymes derived from the steric interactions between N-linked glycan and protein can decrease the enzyme flexibility or increase the rigidity of the enzyme structure (34, 35).
Our previous studies have reported three Yersinia phytases of the histidine acid phosphatase (HAP) type: YkAPPA from Yersinia kristensenii, YrAPPA from Y. rohdei, and YeAPPA from Y. enterocolitica (9, 36, 37). In the present study, sequence analysis indicated that YkAPPA and YrAPPA each have an N-glycosylation motif, whereas YeAPPA does not. Site-directed mutagenesis and comparison of the enzyme properties of wild-type and mutant proteins showed that N-glycosylated phytases are more stable at the physiological pH of the stomach and are more resistant to pepsin digestion and acidity. In contrast, N-glycosylated phytases were equally susceptible to degradation by trypsin. Further analysis of the N-glycosylation effects on pepsin cleavage indicated that the N-linked glycan may enhance enzyme pepsin resistance by improving enzyme stability at acidic pH and reducing accessibility of pepsin to peptic cleavage sites.
MATERIALS AND METHODS
Materials.
Escherichia coli Trans1-T1 from TransGen (China) was used as the host strain for plasmid amplification. Pichia pastoris GS115 and the pPIC9γ vector from Invitrogen (USA) were used as the host and vector for the eukaryotic expression system. The prokaryotic expression vector pET-22b and E. coli BL21(DE3) cells were supplied by Novagen (USA) and TransGen, respectively. Pfu DNA polymerase, restriction endonucleases, T4 DNA ligase, endo-β-N-acetylglucosaminidase H (endo H), and peptide-N4-(N-acetyl-β-d-glucosaminyl) asparagine amidase F (PNGase F) from Flavobacterium meningosepticum were obtained from Tiangen (China) and New England BioLabs (United Kingdom), respectively. Phytate (sodium salt), pepsin (P0685), and trypsin (T0458) were purchased from Sigma (USA). All other chemicals used in this study were of the best grade available.
Identification of key N-glycosylation motifs.
Putative N-glycosylation motifs of YkAPPA, YrAPPA, and YeAPPA were identified according to a multiple-sequence alignment using the ClustalW program. The tertiary structures of the Yersinia phytases were modeled by Discovery studio 2.5.5 software (Accelrys, USA) with the E. coli phytase (PDB code: 1DKP) as the template and were superimposed for comparison of the N-glycosylation motifs.
Site-directed mutagenesis of wild-type phytases.
The recombinant pPIC9γ plasmids harboring phytase genes YkAPPA, YrAPPA, and YeAPPA (GenBank accession no. EU203664, EF608455, and GU936684, respectively) were maintained in our laboratory and used as PCR templates for site-directed mutagenesis (9, 36, 37). Site-directed mutagenesis was performed using overlap extension PCR with the specific primers shown in Table S1 in the supplemental material, as previously described (38). S200 in YkAPPA and NLT397-399 in YrAPPA were replaced with A and DMK, respectively, to delete their N-glycosylation sites. A200S and DMK397-399NLT were introduced into YeAPPA to yield single N-glycosylated mutants. D397 and K399 of YkAPPA were replaced with N and T to produce the N-diglycosylated mutant YkAPPA-D397N/K399T. To determine whether the N-glycans can sterically protect the adjacent pepsin cleavage sites, L197 in YkAPPA and L396 in YrAPPA, which are in close proximity to the N-glycosylation sequons, were replaced with V. The desired PCR products were introduced into the pEASY-T3 vector, and the positive clones were sequenced for verification.
Enzyme expression and purification.
The gene fragments encoding the wild-type and mutant enzymes were inserted into the EcoRI and NotI sites of pPIC9γ, respectively. After linearization with BglII, the recombinant phytases were individually expressed in P. pastoris GS115 competent cells using a Pichia expression kit (Invitrogen) according to the manufacturer's instructions. The positive transformants were screened for phytase activity, and those with the highest activity were selected for high-cell-density fermentation in 1-liter conical flasks. All crude enzyme solutions were purified to homogeneity using Vivaflow 50 ultrafiltration membranes with a 10-kDa molecular mass cutoff (Vivascience, Germany) and HiTrap Q Sepharose XL fast protein liquid chromatography (Amersham Pharmacia Biotech, Sweden) (37).
For expression in E. coli, the gene fragments encoding the wild-type and mutant enzymes were fused to a C-terminal 6-His tag and cloned into the pET-22b vector at the EcoRI and NotI sites, respectively. His tag fusion proteins were then expressed in E. coli BL21(DE3). The crude enzyme solutions were purified using a fast protein liquid chromatography system consisting of nickel-nitrilotriacetic acid (Ni-NTA) and DEAE columns (33).
Total enzyme concentration was determined using the Bio-Rad protein assay kit (Thermo Fisher Scientific, USA). Purified glycoenzymes were N-deglycosylated by endo H according to the manufacturer's instructions. The N-deglycosylated and untreated enzymes were analyzed in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and stained with Coomassie brilliant blue R-250 (39). The N-glycosylation level of each phytase was determined by the proportion of the N-glycans being estimated to account for the molecular mass, i.e., (actual molecular weight − theoretical molecular weight) × 100/actual molecular weight, as described previously (27). The protein band intensity was estimated by using ImageJ software (http://rsbweb.nih.gov/ij/).
Phytase activity assay.
Standard phytase activity was assayed with the ferrous sulfate-molybdenum blue method (40). A 50-μl volume of each enzyme was incubated with 950 μl of substrate solution (1.5 mM sodium phytate, 0.25 M sodium acetate [pH 4.5]) at 37°C for 30 min. The reaction was terminated by addition of 1 ml of 10% (wt/vol) trichloroacetic acid, followed by the addition of 2 ml of color reagent (1% [wt/vol] ammonium molybdate, 3.2% [vol/vol] sulfuric acid, 7.2% [wt/vol] ferrous sulfate). The released inorganic phosphate was monitored by measuring the absorbance at 700 nm. One unit of phytase activity was defined as the amount of enzyme required to liberate 1 μmol of phosphate per minute under standard conditions. All reactions were performed in triplicate.
Pepsin and trypsin digestion assays.
The protease resistance of the wild-type and mutant phytases was determined by incubating 30 μg of each purified recombinant phytase or phytase mixtures of 1/1 with pepsin (0.25 M glycine-HCl, pH 2.0) or trypsin (0.25 M Tris-HCl, pH 7.0) at 37°C for 2 h with various protease/phytase mass ratios ranging from 1/1,000 to 1/1. The time course of proteolysis was determined in reaction mixtures containing 30 μg of enzyme and pepsin or trypsin at a ratio of 1/40 (wt/wt) at 37°C for various durations (0 to 2 h). The reaction mixtures were aliquoted into two parts. One part of the reaction mixture was used to measure residual phytase activity, as described above; the other was combined with 1.0 mM phenylmethylsulfonyl fluoride (PMSF) to stop the proteolytic reaction. After denaturation by boiling in SDS-β-mercaptoethanol, these mixtures were run on an SDS-PAGE gel and stained with CBR-250.
Stabilities of wild-type and mutant enzymes at acidic pH.
The stabilities of the wild-type and mutant phytases at acidic pH were determined by incubating each purified recombinant phytase in 0.25 M glycine-HCl (pH 2.0) at 37°C over 2 h. The stability at acidic pH was observed by SDS-PAGE, and the residual activity was determined as described above.
Biochemical characterization of wild-type and mutant enzymes.
The effects of pH and temperature on the enzyme activity were investigated at a temperature range of 35 to 70°C in a 0.25 M buffer system: glycine-HCl, pH 1.0 to 3.5; sodium acetate-acetic acid, pH 3.5 to 6.0; Tris-HCl, pH 6.0 to 8.5; and glycine-NaOH, pH 8.5 to 12.0. The pH optima were evaluated at 37°C for 30 min over a pH range of 1.0 to 9.0. The temperature optima were determined at each optimal pH by running assays for 30 min at 35 to 70°C. The pH stability was assessed by measuring the residual activities after incubation at 37°C for 1 h at pH 1.0 to 12.0 without substrate. The thermal stability was determined by incubating the enzymes at optimal pH and specific temperatures (60°C for YkAPPA and YrAPPA or 42°C for YeAPPA) for the desired times in the absence of substrate, and the residual phytase activity was measured by following the standard procedure. The nontreated enzyme was considered the control (100%).
RESULTS
Identification of key N-glycosylation motifs.
Each of the three Yersinia phytases contained 441 amino acids and had a theoretical molecular mass of 45.9 kDa. YeAPPA had 83.9 and 88.0% sequence identity to YrAPPA and YkAPPA, respectively. Multiple-sequence alignment and homology modeling of the three Yersinia phytases revealed a single N-linked glycosylation motif, NFS198-200 for YkAPPA and NLT397-399 for YrAPPA, on the protein surface, but no putative N-glycosylation site was identified in YeAPPA (Fig. 1).
FIG 1.
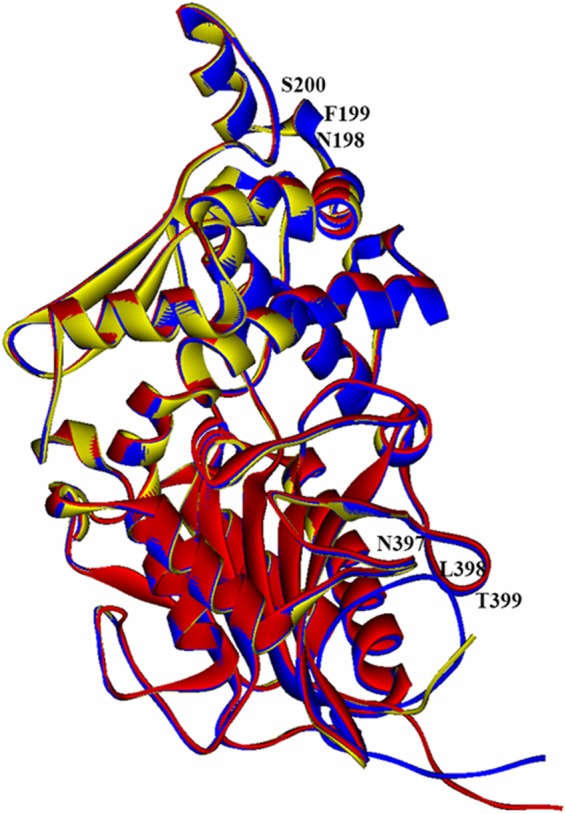
Superimposition of the tertiary structures of YeAPPA (red), YkAPPA (yellow), and YrAPPA (blue) using E. coli phytase (1DKP) as the template with N-glycosylation sites indicated.
Phytase expression, purification, and SDS analysis.
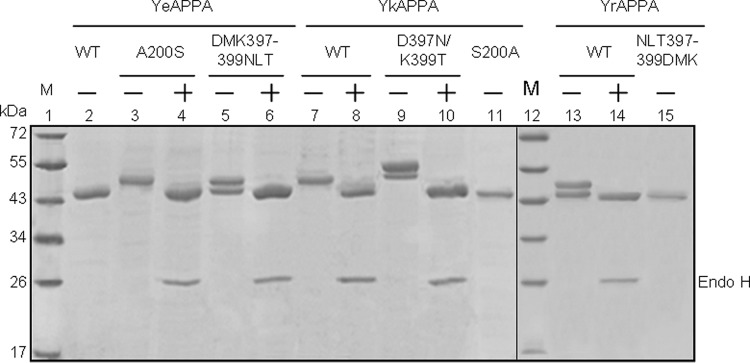
The wild-type and mutant phytases were individually expressed in P. pastoris and purified to electrophoretic homogeneity, as determined by SDS-PAGE (Fig. 2). YeAPPA showed a molecular mass of ∼46 kDa, similar to its expected theoretical mass of 45.9 kDa, while YkAPPA and YrAPPA showed a single band of ∼52 kDa and two bands of ∼46.5 and ∼52 kDa, respectively (Fig. 2, lanes 2, 7, and 13). After treatment with endo H, which removed N-linked glycosylation, both YkAPPA and YrAPPA showed a single band of ∼46 kDa (Fig. 2, lanes 8 and 14). These results indicated that YkAPPA produced in P. pastoris is a homogeneous N-glycoenzyme, with the N-glycan being estimated to account for 11.5% of the molecular mass, while the yeast-produced YrAPPA is presumed to be heterogeneously N-glycosylated and the total carbohydrate contents of the 46.5- and 52-kDa forms accounted for 1.1 and 11.5% of the molecular mass, respectively.
FIG 2.
SDS-PAGE analysis of the apparent molecular masses of wild-type and mutant phytases produced in P. pastoris. Lanes: 1 and 12, protein markers (M); 2, 11 and 15, the nonglycosylated wild-type (WT) YeAPPA and mutants YkAPPA-S200A and YrAPPANLT397-399DMK; 3 to 10, 12 and 13, the N-glycosylated YeAPPA-A200S, YeAPPA-DMK397-399NLT, YkAPPA, YkAPPA-D397N/K399T, and YrAPPA before (−) and after (+) N-deglycosylation with endo H, respectively.
The mutants YkAPPA-S200A and YrAPPA-NLT397-399DMK, containing no N-glycosylation motifs, were produced in P. pastoris and exhibited a single band with expected theoretical mass of ∼46 kDa (Fig. 2, lanes 11 and 15). The P. pastoris-produced YeAPPA-A200S has a single N-glycosylation motif, NFS, and showed a single band of ∼52 kDa (Fig. 2, lane 3), while the yeast-produced YeAPPA-DMK397-399NLT, with a single N-glycosylation motif, NLT, showed two bands of ∼46.5 and ∼52 kDa (Fig. 2, lane 5). YkAPPA-D397N/K399T, with double N-glycosylation motifs, was also produced in P. pastoris and exhibited two bands of ∼53 and ∼55 kDa (Fig. 2, lane 9). After removal of the N-glycans with endo H, the YeAPPA-A200S, YeAPPA-DMK397-399NLT, and YkAPPA-D397N/K399T mutants showed a single band of the expected theoretical mass (Fig. 2, lanes 4, 6, and 10). Thus, the N-glycans of yeast-produced YeAPPA-A200S, YeAPPA-DMK397-399NLT, and YkAPPA-D397N/K399T accounted for about 11.5, 1.1 to 11.5, and 13.21 to 16.36% of the molecular masses, respectively. The results indicated the successful deletion or introduction of N-glycosylation sites into Yersinia phytases and differential N-glycosylation levels varied with the number of the glycosylation sites and the sizes of the N-glycans.
Pepsin and trypsin resistances of wild-type and mutant phytases.
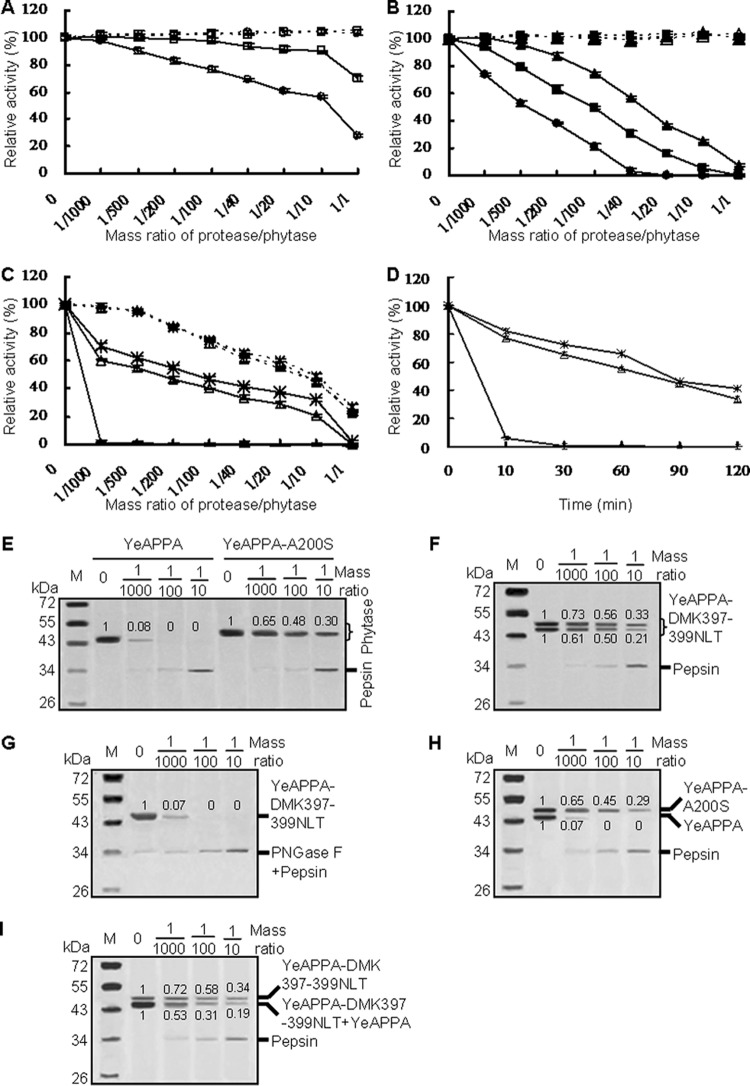
The three wild-type phytases varied in protease resistance over a broad range of protease/phytase mass ratios following incubation at 37°C for 2 h (Fig. 3A to C). After treatment with pepsin in 0.25 M glycine-HCl (pH 2.0), the YrAPPA activity remained unchanged or was slightly decreased at protease/phytase ratios of 1/1,000 to 1/100 but decreased to 70.2% of the initial value at the ratios from 1/40 to 1/1 (Fig. 3A). The pepsin resistance of YkAPPA decreased with increased pepsin/phytase ratios, retaining 93.2%, 49.4%, and 5.4% activity after pepsin treatment at ratios of 1/1,000, 1/100, and 1/10, respectively (Fig. 3B). In contrast, the nonglycosylated YeAPPA retained less than 1.3% activity after pepsin treatment at all ratios (Fig. 3C). When treated with trypsin in 0.25 M Tris-HCl (pH 7.0) at various ratios from 1/1,000 to 1/1, YkAPPA and YrAPPA retained almost all of their activities, while YeAPPA lost 2.5 to 78.0% activity (Fig. 3A to C). These results indicated that N-glycosylated phytases are highly resistant to trypsin and moderately resistant to pepsin degradation, while the nonglycosylated counterparts are highly sensitive to pepsin and moderately resistant to trypsin. Thus, N-glycosylation may play a key role in the stability of phytases against pepsin.
FIG 3.
Proteolytic resistance of N-deglycosylated and N-glycosylated phytases produced in P. pastoris. (A to C) Evaluation of resistance to pepsin at pH 2.0 (full lines) and trypsin at pH 7.0 (dotted lines) at various protease/phytase mass ratios and 37°C for 2 h. (D) Time course of resistance to pepsin at the pepsin/phytase mass ratio of 1/40 over 2 h. For panels A to D, the phytase activity toward sodium phytate (1.5 mM) at 37°C for 30 min was regarded as 100%, and the residual activity is indicated as a percentage of activity of untreated enzymes, with means ± SDs from triplicate determinations. Symbols: □, YrAPPA; ○, YrAPPA-NLT397-399DMK; ■, YkAPPA; ▲, YkAPPA-D397N/K399T; ●, YkAPPA-S200A;  , YeAPPA-DMK397-399NLT; △, YeAPPA-A200S;
, YeAPPA-DMK397-399NLT; △, YeAPPA-A200S;  , YeAPPA. (E to G) SDS-PAGE analysis of the proteolytic products of the wild-type and mutant YeAPPA and enzymatically N-deglycosylated YeAPPA-DMK397-399NLT by pepsin. (H and I) SDS-PAGE analysis of the proteolytic products of the phytase mixture of YeAPPA and YeAPPA-A200S or YeAPPA-DMK397-399NLT. The phytase band intensity was estimated by using ImageJ software.
, YeAPPA. (E to G) SDS-PAGE analysis of the proteolytic products of the wild-type and mutant YeAPPA and enzymatically N-deglycosylated YeAPPA-DMK397-399NLT by pepsin. (H and I) SDS-PAGE analysis of the proteolytic products of the phytase mixture of YeAPPA and YeAPPA-A200S or YeAPPA-DMK397-399NLT. The phytase band intensity was estimated by using ImageJ software.
The protease resistances of the N-glycosylation site deletion mutants YrAPPA-NLT397-399DMK and YkAPPA-S200A were also determined after pepsin or trypsin treatment at 37°C for 2 h and compared with those of their N-glycosylated counterparts. When treated with pepsin at pH 2.0 for 2 h, YrAPPA-NLT397-399DMK and YkAPPA-S200A retained less activity than the wild types at the pepsin/phytase mass ratios of 1/1,000 to 1/1 and 1/1,000 to 1/10, respectively (Fig. 3A and B). SDS-PAGE analysis further demonstrated a greater susceptibility to pepsin cleavage for the N-deglycosylated mutants and enzymatically N-deglycosylated phytases than for the wild types (see Fig. S1A to C and E in the supplemental material). After incubation with pepsin, the 46.5- and 52-kDa phytase forms of YrAPPA were mildly degraded, retaining more than 61% and 79% of the proteins, respectively (see Fig. S1A), but the N-deglycosylated variant YrAPPA-DMK397-399NLT and enzymatically N-deglycosylated YrAPPA were much more susceptible to the pepsin cleavage and retained only about 30% of the proteins at the pepsin/phytase mass ratio of 1/1 (see Fig. S1B and C). The phytase mixture of the 46.5-kDa YrAPPA form and YrAPPA-NLT397-399DMK were degraded by pepsin at a higher rate than the 46.5-kDa YrAPPA form alone at ratios of 1/100 to 1/1 (see Fig. S1D). YkAPPA was degraded by pepsin at a lower rate than YkAPPA-S200A at ratios of 1/100 to 1/10, as determined by SDS-PAGE (see Fig. S1E). After trypsin treatment at pH 7.0 for 2 h, each N-deglycosylated mutant had an activity similar to that of the wild type at various trypsin/phytase mass ratios (Fig. 3A and B). These results indicated that N-glycosylation may account for the resistance of YkAPPA and YrAPPA against pepsin but not trypsin degradation.
The effect of introducing N-glycosylation into YeAPPA on protease resistance was also assessed under similar conditions. After pepsin treatment at pH 2.0 and 37°C for 2 h, YeAPPA-A200S and YeAPPA-DMK397-399NLT retained higher activity (≥21.1 and ≥32.1%, respectively, versus ≤1.3% activity of YeAPPA) at pepsin/phytase ratios of 1/1,000 to 1/10 (Fig. 3C). SDS-PAGE revealed a greater stability of the N-glycosylation variants of YeAPPA than those of the wild type and enzymatically N-deglycosylated variant (Fig. 3E to G). When treated with pepsin for 2 h, the N-glycosylated variants of YeAPPA were degraded at a lower rate than YeAPPA and YeAPPA-DMK397-399NLT with increasing ratios from 1/1,000 to 1/10. After YeAPPA and YeAPPA-A200S were mixed at a ratio of 1/1, the N-glycan of YeAPPA-A200S failed to protect YeAPPA against proteolysis by pepsin (Fig. 3H). The phytase mixture of YeAPPA and the 46.5-kDa YeAPPA-DMK397-399NLT form was less stable against pepsin than the 46.5-kDa YeAPPA-A200S form alone (Fig. 3I). In the presence of trypsin, the N-glycosylated variants showed an activity similar to that of the wild type (Fig. 3C). Thus, introduction of N-glycosylation sites into YeAPPA conferred resistance for pepsin but not to trypsin.
One more N-glycosylation site (NLT397-399) was introduced into YkAPPA to confirm the additive effect of N-glycosylation on the resistance to protease degradation. The residual activity after 2 h of pepsin treatment was greater for YkAPPA-D397N/K399T than for YkAPPA at all tested ratios (Fig. 3B). SDS-PAGE analysis showed a higher resistance to pepsin degradation for YkAPPA-D397N/K399T than for YkAPPA (see Fig. S1E and F). Pepsin treatment for 2 h resulted in a slower degradation of YkAPPA-D397N/K399T than of YkAPPA at ratios of 1/100 to 1/1. Residual YkAPPA-D397N/K399T was apparent but YkAPPA completely disappeared at the highest ratio, 1/1, but no difference in resistance to trypsin was detected in YkAPPA-D397N/K399T and YkAPPA (Fig. 3B). The results confirmed the additive effects of N-glycosylation of phytase on its resistance to pepsin degradation.
Pepsin resistance was also determined for all tested phytases over time at a pepsin/phytase ratio of 1/40. All phytases showed a time-dependent decrease in activity after pepsin treatment. The single N-glycosylated phytases YrAPPA, YkAPPA, YeAPPA-A200S, and YeAPPA-DMK397-399NLT lost activity over time but at a lower rate than the N-deglycosylated forms YrAPPA-NLT397-399DMK and YkAPPA-S200A and the nonglycosylated YeAPPA. The doubly N-glycosylated variant YkAPPA-D397N/K399T lost activity at the lowest rate among the wild type and variants of YkAPPA (Fig. 3D and data not shown). The time course results further confirmed the role and additive effect of N-glycosylation in the resistance to pepsin.
Stabilities of wild-type and mutant phytases at acidic pH.
The effects of N-glycosylation on phytase stability at acidic pH were determined at pH 2.0 and 37°C for various times, and N-glycosylated phytases were compared with their N-deglycosylated or nonglycosylation counterparts (Fig. 4). Under the acid condition of pH 2.0 at 37°C for 2 h, YrAPPA and N-glycosylation-deleted variant YrAPPA-NLT397-399DMK were highly stable, while YkAPPA and the N-glycosylation site deletion variant YkAPPA-S200A lost 6.7 and 13.2% activity, respectively (Fig. 4A). The results indicated that N-glycosylation may account for the appreciable stability of YkAPPA at acidic pH (2.0). In comparison with the low residual activity (1.3%) of YeAPPA at pH 2.0 after 2 h, the N-glycosylated mutants YeAPPA-A200S and YeAPPA-DMK397-399NLT retained 61.2 and 72.1% of the original activity (Fig. 4B), respectively. The SDS-PAGE analysis showed higher stability at acidic pH for the N-glycosylation variants of YeAPPA than for the wild type and enzymatically N-deglycosylated variant by PNGase F (Fig. 4C and D). Under treatment conditions, the yeast-produced YeAPPA-DMK397-399NLT has 46.5- and 52-kDa forms of nearly equal intensities. Under the acid condition of pH 2.0 at 37°C for 2 h, the 52-kDa form was a little more stable than the 46.5-kDa form. The 46.5- and 52-kDa YeAPPA-DMK397-399NLT forms and YeAPPA-A200S were significantly more stable than nonglycosylated YeAPPA and N-deglycosylated YeAPPA-DMK397-399NLT with PNGase F. These results suggested that introduction of N-glycosylation sites into YeAPPA increased the stability at acidic pH. The doubly N-glycosylated YkAPPA-N397D/T399K was more stable at acidic pH than the singly N-glycosylated YkAPPA, which retained 102.8 and 93.3% activity at pH 2.0 and 37°C for 2 h, respectively (Fig. 4A). These results confirmed the additive effects of N-glycosylation of phytase on the stability at acidic pH.
FIG 4.
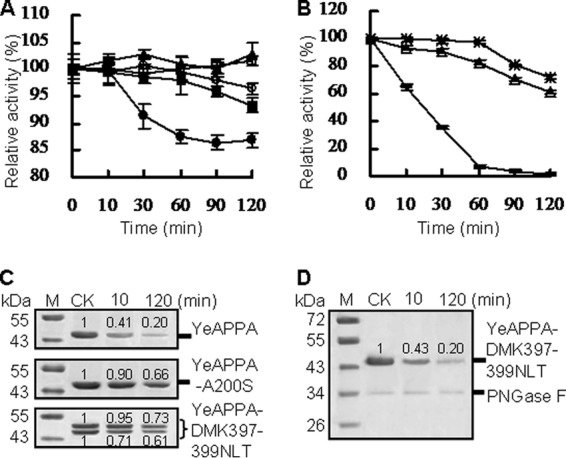
Time course stability of N-deglycosylated and N-glycosylated phytases produced in P. pastoris at pH 2.0 and 37°C over 2 h. (A and B) Residual activities of phytases after acid treatment. For panels A and B, the 100% activity and residual activity were calculated as for Fig. 3; data are represented as means ± SDs. Symbols: □, YrAPPA; ○, YrAPPA-NLT397-399DMK; ■, YkAPPA; ▲, YkAPPA-D397N/K399T; ●, YkAPPA-S200A;  , YeAPPA-DMK397-399NLT; △, YeAPPA-A200S;
, YeAPPA-DMK397-399NLT; △, YeAPPA-A200S;  , YeAPPA. (C and D) SDS-PAGE analysis of phytase stability after acid treatment. The phytase band intensity was estimated by using ImageJ software. The untreated phytases were used as the control.
, YeAPPA. (C and D) SDS-PAGE analysis of phytase stability after acid treatment. The phytase band intensity was estimated by using ImageJ software. The untreated phytases were used as the control.
Characterization of N-deglycosylated and N-glycosylated mutant enzymes.
The pH and temperature activity profiles of N-glycosylated mutants YeAPPA-A200S and YeAPPA-NLT397-399DMK were determined and compared with those of wild-type YeAPPA (Table 1). Each mutant had a downward shift of the pH optimum, 0.5 and 1.5 pH units for YeAPPA-A200S and YeAPPA-NLT397-399DMK, respectively, and remained stable over a broad pH range. The mutant enzymes had a temperature optimum (45°C) similar to that of the wild type but showed increased thermostability at 42°C. In contrast, the nonglycosylated and doubly N-glycosylated mutants YkAPPA-S200A and YkAPPA-D397N/K399T had the same pH optimum (pH 4.5) as YkAPPA and remained stable over the same pH range, 2.0 to 10.0 (Table 1). After incubation at pH 1.0 for 1 h, YkAPPA-D397N/K399T and YkAPPA retained 80.9 and 65.2% residual activity but YkAPPA-S200A activity dropped to 42.6% (Table 1). The results indicated that the N-glycosylated YkAPPA and YkAPPA-D397N/K399T were more acid tolerant than the N-deglycosylated variant, as shown above.
TABLE 1.
Glycosylation motifs and pH and temperature properties of wild-type phytases and their mutants
| Enzyme | Glycosylation sequon | pH optimum | Temp optimum (°C) | pH stabilitya | Temp stabilitya |
|---|---|---|---|---|---|
| YeAPPA (wild type) | —b | 5 | 45 | pH 1.5–2.5, <23.7%; pH 3–9, >83.8% | 42°C, 2 h, 17.6% |
| YeAPPA-A200S | NFS | 4.5 | 45 | pH 1.5–2.5, >30.8%; pH 3–9, >84.7% | 42°C, 2 h, 19.6% |
| YeAPPA-D397N/M398L/K399T | NLT | 3.5 | 45 | pH 1.5–2.5, >80.5%; pH 3–9, >99.4% | 42°C, 2 h, 36.5% |
| YkAPPA (wild type) | NFS | 4.5 | 55 | pH 1–1.5, 65.2%-80.7%; pH 2–10, >93.5% | 60°C, 2 h, 25.4% |
| YkAPPA-S200A | — | 4.5 | 55 | pH 1–1.5, <59.3%; pH 2–10, >86.1% | 60°C, 2 h, 5.6% |
| YkAPPA-D397N/K399T | NFS and NLT | 4.5 | 60 | pH 1–1.5, >80.9%; pH 2–10, >96.4% | 60°C, 2 h, 37.5% |
| YrAPPA (wild type) | NLT | 4.5 | 60 | pH 1–12, >85.8% | 60°C, 2 h, 28.5% |
| YrAPPA-N397D/L398 M/T399K | — | 4.5 | 60 | pH 2–11, >97.7%; pH 1, 12, <75.3% | 60°C, 2 h, 15.2% |
The phytase activity toward sodium phytate (1.5 mM) at 37°C for 30 min was regarded as 100%. The residual activity was expressed as a percentage of the activity of untreated enzyme.
—, no typical N-glycosylation site is found in wild-type or mutant phytases.
The optimal temperature of YkAPPA and YkAPPA-S200A was about 55°C at pH 4.5, which was 5°C lower than that of YkAPPA-D397N/K399T (Table 1). The thermostability of YkAPPA and its mutants followed the order YkAPPA-D397N/K399T (37.5%) > YkAPPA (25.4%) > YkAPPA-S200A (0%) after 2 h of incubation at 60°C (Table 1). The results indicated that N-glycosylation not only plays a role in the stability at acidic pH and pepsin resistance of phytases but also improves their pH stability and thermostability. This conclusion was further verified by YrAPPA and its N-deglycosylated variant, which exhibited similar pH and temperature optima, remained stable over a narrower pH range, and had reduced thermostability at 60°C (Table 1).
Pepsin resistances of mutants YkAPPA-L197V and YrAPPA-L396V.
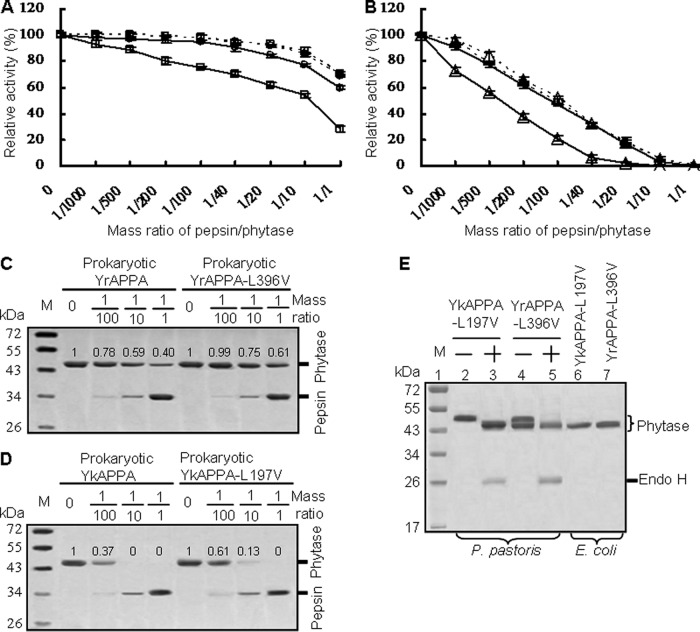
The pepsin cleavage sites in close proximity to the N-glycosylation motifs of Yersinia phytases—L197 in YkAPPA and L396 in YrAPPA—were successfully replaced by residue V. All the wild-type and mutant enzymes were both produced in P. pastoris and E. coli. As shown in Fig. 5E, YkAPPA-L197V and YrAPPA-L396V produced in P. pastoris migrated as a single band of ∼52 kDa and double bands of ∼52 and ∼46.5 kDa on SDS-PAGE (Fig. 5E, lanes 2 and 4, respectively). After N-deglycosylation with endo H, both phytases showed a single band of ∼46 kDa (Fig. 5E, lanes 3 and 5), similar to the predicted size of 45.9 kDa and the apparent size of recombinant phytases produced in E. coli (Fig. 5E, lanes 6 and 7). Moreover, the stability of these mutants at acidic pH was unchanged. After a 2-h incubation at pH 2.0 and 37°C, all YkAPPA-L197V, YkAPPA, YrAPPA-L396V, and YrAPPA produced in P. pastoris or E. coli retained more than 90% activity (data not shown). The results indicated that L replacement with V had no effect on phytase N-glycosylation in yeast and stability at acidic pH, and phytases produced in E. coli were not N-glycosylated.
FIG 5.
Comparison of the pepsin resistance of YrAPPA and YkAPPA and their mutants produced in P. pastoris and E. coli at various pepsin/phytase mass ratios at 37°C and pH 2.0 for 2 h. (A and B) Pepsin resistance evaluation of phytases produced in P. pastoris (dotted lines) and E. coli (full lines). The 100% activity and residual activity were calculated as for Fig. 3; each point data is the mean from three replicates ± SD. Symbols: □, YrAPPA; ○, YrAPPA-L396V; △, YkAPPA;  , YkAPPA-L197V. (C and D) SDS-PAGE analysis of the proteolytic products of the E. coli-produced phytases degraded by pepsin at different ratios. The phytase band intensity was estimated by using ImageJ software. (E) SDS-PAGE analysis of the phytase variants YkAPPA-L197V and YrAPPA-L396V. Lanes: 1, protein marker (M); 2 to 4, the N-glycosylated YkAPPA-L197V and YrAPPA-L396V produced in P. pastoris before (−) and after (+) N-deglycosylation with endo H; 6 and 7, E. coli-expressed variants.
, YkAPPA-L197V. (C and D) SDS-PAGE analysis of the proteolytic products of the E. coli-produced phytases degraded by pepsin at different ratios. The phytase band intensity was estimated by using ImageJ software. (E) SDS-PAGE analysis of the phytase variants YkAPPA-L197V and YrAPPA-L396V. Lanes: 1, protein marker (M); 2 to 4, the N-glycosylated YkAPPA-L197V and YrAPPA-L396V produced in P. pastoris before (−) and after (+) N-deglycosylation with endo H; 6 and 7, E. coli-expressed variants.
When produced in E. coli, the nonglycosylated YrAPPA-L396V and YkAPPA-197V showed higher resistance to pepsin than the nonglycosylated YrAPPA and YrAPPA, retaining higher activity than the nonglycosylated wild types after pepsin treatment at pH 2.0 and 37°C for 2 h at pepsin/phytase ratios from 1/1,000 to 1/1 and from 1/1,000 to 1/10, respectively (Fig. 5A and B). At the high ratio of 1/1, the nonglycosylated YrAPPA-L396V produced in E. coli retained 58.0% activity but the nonglycosylated wild-type YrAPPA expressed in E. coli showed only 27.9% activity (Fig. 5A). At a ratio of 1/20, the nonglycosylated wild-type YkAPPA expressed in E. coli lost all activity, whereas the nonglycosylated YkAPPA-L197V from the prokaryotic expression system retained 17.1% of the activity (Fig. 5B). The SDS-PAGE results indicated that the prokaryote-expressed YrAPPA-L396V and YkAPPA-L197V were degraded with pepsin treatment at a lower rate than the wild types produced in E. coli, indicating that nonglycosylated YrAPPA-L396V and YkAPPA-L197V were more stable against pepsin than the nonglycosylated wild types (Fig. 5C and D). After N-glycosylation of the phytases produced in P. pastoris, YrAPPA-L396V and YkAPPA-197V had activities similar to those of the wild types after pepsin treatment for 2 h at all the ratios (Fig. 5A and B). All these results indicated that L197V and L396V enhanced the pepsin resistance of the nonglycosylated phytase from E. coli but had no effect on the pepsin resistance of N-glycosylated phytases from P. pastoris; the N-glycosylation of P. pastoris YkAPPA and YrAPPA may therefore protect the peptic cleavage sites L197 and L396 from pepsin cleavage.
DISCUSSION
To optimize phosphorus bioavailability and reduce pollution of phosphorus, phytase used as animal feed additive must possess superior features, such as high specific activity, broad substrate specificity, good adaptability to low pH, high temperature resistance, and protease resistance. Proteases naturally occur in the gastrointestinal tracts of all organisms, so protease resistance becomes one of the most valued properties for feed enzymes. Two main strategies have been applied to improve the protease resistance of feed enzymes. One is to explore protease-resistant feed enzymes (41, 42), and the other is to screen engineered feed enzymes with improved protease resistance by rational design (43). Structure-based rational design has been less frequently used than naturally occurring protease-resistant enzymes (24, 44–46), especially with respect to pepsin resistance. The cleavage of enzymes by proteases involves a nucleophilic attack of the polarized carbonyl group of the substrate peptide bond, followed by stabilization of the oxygen in an oxyanion hole of proteases (47, 48). Therefore, glycans at the N-glycosylation site may sterically protect enzymes against proteases (24). In the present study, we employed a structure-based mutagenesis approach to introduce N-glycosylation sites into Yersinia phytases to determine the underlying mechanism of glycosylation-mediated enzyme resistance to pepsin.
The Pichia expression system is widely used for commercial-scale production of enzymes for industrial use (49). N-Glycosylation commonly occurs in this system (29) and is one of the most common posttranslational modification forms that have been used to modulate protein structure and function (50). In the present study, three Yersinia phytases were heterologously expressed in P. pastoris. Sequence analysis and comparison of molecular masses on SDS-PAGE and protease resistance indicated that both YrAPPA and YkAPPA are N-glycoproteins with trypsin and pepsin resistance, and YeAPPA that lacks an N-glycosylation site is moderately resistant to trypsin but sensitive to pepsin (Fig. 3A to C). Similar results have been reported with the P. pastoris-produced N-glycosylated Bacillus subtilis phytase that shows resistance to trypsin but not pepsin (41). In contrast, the N-glycosylated phytases derived from Aspergillus japonicus BCC18313, A. niger BCC18081, Eupenicillium parvum BCC17694, and Neosartorya spinosa BCC41923 are resistant to pepsin but not to trypsin (26–28). Our results indicated that the two functional N-glycosylation motifs, NXS/T, of Yersinia phytases contribute to the enzyme proteolytic resistance to pepsin but not trypsin (Fig. 3; see also Fig. S1 in the supplemental material). In order to verify the function of these N-glycosylation motifs, they were separately introduced into the corresponding sites on the exposed surface of wild-type YeAPPA. The resulting mutants, YeAPPA-A200S and YeAPPA-DMK397-399NLT, showed significantly enhanced tolerance to pepsin but not trypsin (Fig. 3C to F). The N-glycosylation motif NLT397-399 in YrAPPA was also introduced into YkAPPA to yield double N-glycosylation mutant YkAPPA-D397N/K399T, which had higher pepsin resistance than was observed for YkAPPA (Fig. 3B; see also Fig. S1C). The N-glycans of YeAPPA-A200S did not protect the nonglycosylated YeAPPA from enzyme proteolysis by pepsin (Fig. 3H). Considering that pepsin is the major protease in the stomach, a pepsin-resistant phytase would be a cost-effective method for reducing the use of dietary phytase in animal production.
N-Glycosylation modification commonly occurs in eukaryotes, but with significant variance from enzyme to enzyme. YeAPPA that lacks an N-glycosylation site shows an apparent mass similar to the theoretical value when produced in P. pastoris (Fig. 2, lane 2), while YkAPPA and YrAPPA containing different N-glycosylation sites, NFS198-200 and NLT397-399, respectively, are homogeneously and heterogeneously N-glycosylated (Fig. 2, lanes 7 and 13, respectively). In contrast, YkAPPA and YrAPPA are nonglycosylated in E. coli (Fig. 5C and D). Thus, the N-glycosylation sites in YkAPPA and YrAPPA are presumed to be nonmodified in the native Yersinia strains. After introduction of the N-glycosylation sites, YkAPPA-D397N/K399T appeared as an incompletely N-glycosylated enzyme, and YeAPPA-DMK397-399NLT was heterogeneous, while YeAPPA-A200S was homogeneous after production in P. pastoris (Fig. 2, lanes 5, 9, and 3, respectively). This heterogeneity of N-glycosylation arises from the different glycosylation efficiencies of the N-glycosylation sites, which vary in the number of glycans on the proteins and the type of the glycans (51). Based on the SDS-PAGE pattern, we estimated that the glycosylation levels of NLT-, NFS-, and NLT/NFS-containing phytases are the lowest to highest (Fig. 2, lanes 5, 13, 3, 7, and 9, respectively; see also Table S2 in the supplemental material). Each NLT-containing monoglycosylated phytase, i.e., YeAPPA-DMK397-399NLT and YrAPPA, had significantly differential glycosylation levels of about 1.1 and 11.5% by the presumably altered size of the glycans of the N-glycoenzyme (Fig. 2, lanes 5 and 13, respectively; see also Table S2). This N-glycosylation variability of Yersinia phytases may account for their distinctive resistance to pepsin, thermostability, and stability at low pH.
As shown above, engineered Yersinia phytases with different N-glycosylation sites had altered resistances against pepsin and N-glycosylation levels, and the N-glycosylation motif NLT397-399 is more effective than NFS198-200 in enhancing pepsin resistance (Fig. 3C to G; see also Table S2). Although introduction of NLT397-399 resulted in similar glycosylation levels in the monoglycosylated YeAPPA-NLT397-399 and YrAPPA, its effect on pepsin resistance was quite variable (Fig. 3A, C, F, G, and I; see also Fig. S1A to D and Table S2). The 46.5-kDa forms of YeAPPA-NLT397-399 and YrAPPA had the lowest N-glycan contents of all engineered Yersinia phytases but exhibited most of the pepsin resistance of the 52-kDa N-glycoenzyme forms (Fig. 3F; see also Fig. S1A). After mixing of phytases, N-glycans of N-glycosylated YeAPPA-A200S failed to protect the nonglycosylated YeAPPA from enzyme proteolysis by pepsin (Fig. 3H). The N-glycans of the 52-kDa YrAPPA and YeAPPA-DMK397-399NLT forms were presumed to fail to be responsible for the resistance of the 46.5-kDa forms to pepsin. Thus, the 46.5-kDa forms of YrAPPA and YeAPPA-DMK397-399NLT were presumed to have a much smaller size for glycan, which plays a major role in the resistance to pepsin. A similar phenomenon has been reported, i.e., that the first two or three saccharides of glycans instead of the whole glycan make the major contribution to enzyme stabilization (52, 53). Moreover, the fully N-glycosylated YeAPPA-NLT397-399 and YrAPPA were more resistant against pepsin than the phytase mixtures of partially N-glycosylated YeAPPA and YeAPPA-NLT397-399 and of YrAPPA-NLT397-399DMK and YrAPPA, respectively (Fig. 3I; see also Fig. S1D). The proteolytic resistance of the double N-glycosylation mutant YkAPPA-D397N/K399T against pepsin can be improved by increasing the N-glycosylation levels (Fig. 3B; see also Fig. S1F and Table S2). In contrast, the nonglycosylated YeAPPA is sensitive to pepsin (Fig. 3C and E), while N-deglycosylated variants and enzymatically N-deglycosylated enzymes of YrAPPA and YkAPPA demonstrated significantly decreased resistance against pepsin (Fig. 3A and B; see also Fig. S1B, C, and E). Therefore, a minimal degree of glycosylation may be required for enhanced stability of phytases against pepsin degradation.
All phytases were also evaluated for their resistance to trypsin. After trypsin treatment at ratios from 1/1,000 to 1/1, N-glycosylated YkAPPA and YrAPPA had unchanged activity, and nonglycosylated YeAPPA showed decreased activity (22.0%) (Fig. 3A to C). Introduction or deletion of N-glycosylation sites to Yersinia phytases did not change their resistance to trypsin (Fig. 3A to C). Moreover, the nonglycosylated YrAPPA and YkAPPA produced in E. coli showed activities similar to that of the N-glycosylated forms upon expression in P. pastoris (data not shown). Thus, the N-glycosylation motifs NFS198-200 and NLT397-399 and the N-glycans of the N-glycosylated phytases produced in P. pastoris had no effect on the trypsin resistance of phytases.
Phytate hydrolysis catalyzed by microbial phytases mostly takes place in the stomach. The pH of gastric acid in the stomach is usually from 1.5 to 3.5 (54). Maximal biocatalytic capability of phytase in the stomach therefore requires that the enzyme have high activity and good stability under acidic conditions. The N-glycosylated mutants of YeAPPA had lower pH optima and better stabilities at low pH than those of the native nonglycosylated counterpart, as shown in Table 1 and Fig. 4. Thus, the glycoproteins likely confer pepsin resistance partially through regulation of acidophilic stability. The acidophilic feature of the N-glycosylated phytases facilitates their survival in the acidic environment of the digestive tract and increases their usefulness in the feed industry.
Excellent thermostability can allow an enzyme to survive the high pelleting temperatures used in feed production. N-Glycosylation of Yersinia phytases significantly increased the optimal temperature (by 5°C) and thermal stability (Table 1). The combination of two N-glycosylation motifs had an additive effect on the improvement of thermal properties (Table 1). The tolerance for high temperature is probably ascribed to the enhanced structural rigidity of the proteins with glycan and decreased dynamic fluctuations throughout the molecule (55). For example, in comparison to the wild-type nonglycosylated YeAPPA, which had the highest activity at pH 5.0 and 45°C, was weakly active at pH 2.0 and at 42°C, and was highly sensitive to pepsin, the YeAPPA mutants containing the N-glycosylation motif NFS198-200 or NLT397-399 demonstrated pronounced shifts in pH optima (0.5 and 1.5 pH units downward, respectively), improved stability at acidic pH (83.2 and 98.8% activities at pH 2.0 for 1 h, respectively) and thermostability (19.6 and 36.5% residual activities at 42°C for 2 h, respectively), and increased resistance to pepsin (∼44-fold and ≥50.6-fold, respectively) (Fig. 3C and Table 1; see also Table S1). Hence, introducing the N-glycosylation motifs benefits YeAPPA in terms of its resistance to pepsin, thermostability, and stability at acidic pH. The NLT397-399 mutant was more effective than NFS198-200 in terms of improving the enzyme properties.
The aspartic protease pepsin is an endopeptidase with broad substrate specificity. Pepsin prefers to cleave C-terminal F and L residues, unless they are adjacent to H and K residues (56, 57). To further study the exact function of N-glycosylation in pepsin resistance, we mutated the cleavage sites L197 and L396, which are adjacent to N at the NFS198-200 and NLT397-399 sites on the exposed surface of YkAPPA and YrAPPA, respectively. Compared with nonglycosylated phytases produced in E. coli (29), the mutation of L197 in YkAPPA and L396 in YrAPPA to V enhanced the pepsin resistance but did not change the pepsin resistance of N-glycosylated YkAPPA and YrAPPA produced in yeast (Fig. 5A to D). These mutations did not affect resistance to trypsin at all tested ratios in E. coli and P. pastoris (data not shown). We infer that the N-glycan linkage at N198 and N397 may sterically hinder pepsin from the pepsin cleavage sites of Yersinia phytases but has no effect on trypsin cleavage sites. In future studies, we will resolve the crystal structures of the glycoenzyme to verify the role of glycans in proteolytic resistance of phytases to pepsin.
In conclusion, this study reveals that N-glycosylation occurred at two particular sites, NFS198-200 and NLT397-399, and markedly increased the proteolytic resistance of Yersinia phytases to pepsin but not to trypsin. By using site-directed mutagenesis, the N-glycosylation sites were introduced into other Yersinia counterparts and also improved the resistance to pepsin but not to trypsin, as well as elevating the stability at acidic pH and high temperature. Mutagenesis of the pepsin cleavage sites in close proximity to the N-glycosylation motifs promoted the resistance of nonglycosylated phytases to pepsin digestion. The pepsin resistance of glycoenzymes would therefore appear to occur through enhanced acid tolerance and by steric hindrance between the glycans and the pepsin active site.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Fund for Distinguished Young Scholars of China (31225026), the National Natural Science Foundation of China (31370044), the National High Technology Research and Development Program of China (2012AA022208), and the China Modern Agriculture Research System (CARS-42).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02881-15.
REFERENCES
- 1.Ravindran V, Bryden WL, Kornegay ET. 1995. Phytates: occurrence, bioavailability and implications in poultry nutrition. Avian Poult Biol Rev 6:125–143. [Google Scholar]
- 2.Cowieson AJ, Acamovic T, Bedford MR. 2004. The effects of phytase and phytic acid on the loss of endogenous amino acids and minerals from broiler chickens. Br Poult Sci 45:101–108. doi: 10.1080/00071660410001668923. [DOI] [PubMed] [Google Scholar]
- 3.Cowieson AJ, Ravindran V, Selle PH. 2008. Influence of dietary phytic acid and source of microbial phytase on ileal endogenous amino acid flows in broiler chickens. Poult Sci 87:2287–2299. doi: 10.3382/ps.2008-00096. [DOI] [PubMed] [Google Scholar]
- 4.Bitar K, Reinhold JG. 1972. Phytase and alkaline phosphatase activities in intestinal mucosae of rat, chicken, calf and man. Biochim Biophys Acta 68:442–452. [DOI] [PubMed] [Google Scholar]
- 5.Correll DL. 1999. Phosphorus: a rate limiting nutrient in surface waters. Poult Sci 78:674–682. doi: 10.1093/ps/78.5.674. [DOI] [PubMed] [Google Scholar]
- 6.Wyss M, Brugger R, Kronenberger A, Rémy R, Fimbel R, Oesterhelt G, Lehmann M, van Loon AP. 1999. Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl Environ Microbiol 65:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N, Liu G, Li F, Sands J, Zhang S, Zheng A, Ru Y. 2007. Efficacy of phytases on egg production and nutrient digestibility in layers fed reduced phosphorus diets. Poult Sci 86:2337–2342. doi: 10.3382/ps.2007-00079. [DOI] [PubMed] [Google Scholar]
- 8.Selle PH, Ravindran V. 2007. Microbial phytase in poultry nutrition. Anim Feed Sci Technol 135:1–41. doi: 10.1016/j.anifeedsci.2006.06.010. [DOI] [Google Scholar]
- 9.Fu D, Huang H, Luo H, Wang Y, Yang P, Meng K, Bai Y, Wu N, Yao B. 2008. A highly pH-stable phytase from Yersinia kristensenii: cloning, expression, and characterization. Enzyme Microb Tech 42:499–505. doi: 10.1016/j.enzmictec.2008.01.014. [DOI] [Google Scholar]
- 10.Huang H, Shi P, Wan Y, Luo H, Shao N, Wang G, Yang P, Yao B. 2009. Diversity of beta-propeller phytase genes in the intestinal contents of grass carp provides insight into the release of major phosphorus from phytate in nature. Appl Environ Microbiol 75:1508–1516. doi: 10.1128/AEM.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dionisio G, Madsen CK, Holm PB, Welinder KG, Jørgensen M, Stoger E, Arcalis E, Brinch-Pedersen H. 2011. Cloning and characterization of purple acid phosphatase phytases from wheat, barley, maize and rice. Plant Physiol 156:1087–1100. doi: 10.1104/pp.110.164756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haefner S, Knietsch A, Scholten E, Braun J, Lohscheidt M, Zelder O. 2005. Biotechnological production and applications of phytases. Appl Microbiol Biotechnol 68:588–597. doi: 10.1007/s00253-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 13.Andreeva NS. 1994. How and why is pepsin stable and active at pH 2? Mol Biol (Mosk) 28:1400–1406. [PubMed] [Google Scholar]
- 14.Iliadis G, Zundel G, Brzezinski B. 1994. Aspartic proteinases-Fourier transform IR studies of the aspartic carboxylic groups in the active site of pepsin. FEBS Lett 352:315–317. doi: 10.1016/0014-5793(94)00979-1. [DOI] [PubMed] [Google Scholar]
- 15.Andreeva NS, Rumsh LD. 2001. Analysis of crystal structures of aspartic proteinases: on the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes. Protein Sci 10:2439–2450. doi: 10.1110/ps.ps.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn J, Cao M, Yu Y, Engen JR. 2013. Accessing the reproducibility and specificity of pepsin and other aspartic proteases. Biochim Biophys Acta 1834:1222–1229. doi: 10.1016/j.bbapap.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi W, Su R, He Z, Zhang Y, Jin F. 2007. Pepsin-induced changes in the size and molecular weight distribution of bovine casein during enzymatic hydrolysis. J Dairy Sci 90:5004–5011. doi: 10.3168/jds.2007-0230. [DOI] [PubMed] [Google Scholar]
- 18.Saliha BH, Louis LC, Farida MM, Saliha SA, Nasma M, Elkhir SO, Abderrahmane M. 2011. Comparative study of milk clotting activity of crude gastric enzymes extracted from camels' abomasum at different ages and commercial enzymes (rennet and pepsin) on bovine and camel milk. Emir J Food Agric 23:301–310. [Google Scholar]
- 19.Schmelzer CEH, Getie M, Neubert RHH. 2005. Mass spectrometric characterization of human skin elastin peptides produced by proteolytic digestion with pepsin and thermitase. J Chromatogr A 1083:120–126. doi: 10.1016/j.chroma.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Monogioudi E, Faccio G, Lille M, Poutanen K, Buchert J, Mattinen ML. 2011. Effect of enzymatic cross-linking of β-casein on proteolysis by pepsin. Food Hydrocolloid 25:71–81. doi: 10.1016/j.foodhyd.2010.05.007. [DOI] [Google Scholar]
- 21.Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Bichim Biophys Acta 1473:4–8. doi: 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 22.Jafari-Aghdam J, Khajeh K, Ranjbar B, Nemat-Gorgani M. 2005. Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Bichim Biophys Acta 1750:61–68. doi: 10.1016/j.bbapap.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Rudd PM, Joao HC, Coghill E, Fiten P, Saunders MR, Opdenakker G, Dwek RA. 1994. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry 33:17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 24.Zou S, Huang S, Kaleem I, Li C. 2013. N-Glycosylation enhances functional and structural stability of recombinant β-glucuronidase expressed in Pichia pastoris. Biotechnol J 164:75–81. doi: 10.1016/j.jbiotec.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Sano M, Korekane H, Ohtsubo K, Yamaguchi Y, Kato M, Shibukawa Y, Tajiri M, Adachi H, Wada Y, Asahi M, Taniguchi N. 2012. N-Glycans of SREC-I (scavenger receptor expressed by endothelial cells): essential role for ligand binding, trafficking and stability. Glycobiology 22:714–724. doi: 10.1093/glycob/cws010. [DOI] [PubMed] [Google Scholar]
- 26.Promdonkoy P, Tang K, Sornlake W, Harnpicharnchai P, Kobayashi RS, Ruanglek V, Upathanpreecha T, Vesaratchavest M, Eurwilaichitr L, Tanapongpipat S. 2009. Expression and characterization of Aspergillus thermostable phytase in Pichia patoris. FEMS Microbiol Lett 290:18–24. doi: 10.1111/j.1574-6968.2008.01399.x. [DOI] [PubMed] [Google Scholar]
- 27.Fugthong A, Boonyapakron K, Sornlek W, Tanapongpipat S, Eurwilaichitr L, Pootanakit K. 2010. Biochemical characterization in vitro digestibility assay of Eupenicillium parvum (BCC17694) phytase expressed in Pichia pastoris. Protein Expr Purif 70:60–67. doi: 10.1016/j.pep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Pandee P, Summpunn P, Wiyakrutta S, Isarangkul D, Meevootisom V. 2011. A thermostable phytase from Neosartorya spinosa BCC 41923 and its expression in Pichia pastoris. J Microbiol 49:257–264. doi: 10.1007/s12275-011-0369-x. [DOI] [PubMed] [Google Scholar]
- 29.Wyss M, Pasamontes L, Friedlein A, Rémy R, Tessier M, Kronenberger A, Middendorf A, Lehmann M, Schnoebelen L, Röthlisberger U, Kusznir E, Wahl G, Müller F, Lahm HW, Vogel K, van Loon AP. 1999. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl Environ Microbiol 65:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo M, Hang H, Zhu T, Zhuang Y, Chu J, Zhang S. 2008. Effect of glycosylation on biochemical characterization of recombinant phytase expressed in Pichia pastoris. Enzyme Microb Technol 42:340–345. doi: 10.1016/j.enzmictec.2007.10.013. [DOI] [Google Scholar]
- 31.Wu T, Chen C, Cheng Y, Ko T, Lin C, Lai H, Huang T, Liu J, Guo R. 2014. Improving specific activity and thermostability of Escherichia coli phytase by structure-based rational design. J Biotechnol 175:1–6. doi: 10.1016/j.jbiotec.2014.01.034. [DOI] [PubMed] [Google Scholar]
- 32.Zou S, Xie L, Liu Y, Kaleem I, Zhang G, Li C. 2012. N-Linked glycosylation influences on the catalytic and biochemical properties of Penicillium purpurogenum β-d-glucuronidase. Biotechnol J 157:399–404. doi: 10.1016/j.jbiotec.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Qi F, Zhang W, Zhang F, Chen G, Liu W. 2014. Deciphering the effect of the different N-glycosylation sites on the secretion, activity and stability of cellobiohydrolase I from Trichoderma reesei. Appl Environ Microbiol 80:3962–3971. doi: 10.1128/AEM.00261-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller-Steffner H, Kuhn I, Argentini M, Schuber F. 2010. Identification of the N-glycosylation sites on recombinant bovine CD38 expressed in Pichia pastoris: their impact on enzyme stability and catalytic activity. Protein Expr Purif 70:151–157. doi: 10.1016/j.pep.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar A, Wintrode PL. 2011. Effects of glycosylation on the stability and flexibility of a metastable protein: the human serpin α1-antitrypsin. Int J Mass Spectrom 302:69–75. doi: 10.1016/j.ijms.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H, Luo H, Wang Y, Fu D, Shao N, Wang G, Yang P, Yao B. 2008. A novel phytase from Yersinia rohdei with high phytate hydrolysis activity under low pH and strong pepsin conditions. Microbiol Biotechnol 80:417–426. doi: 10.1007/s00253-008-1556-5. [DOI] [PubMed] [Google Scholar]
- 37.Fu D, Li Z, Huang H, Yuan T, Shi P, Luo H, Meng K, Yang P, Yao B. 2011. Catalytic efficiency of HAP phytases is determined by a key residue in close proximity to the active site. Appl Microbiol Biotechnol 90:1295–1302. doi: 10.1007/s00253-011-3171-0. [DOI] [PubMed] [Google Scholar]
- 38.Tran TT, Mamo G, Búxo L, Le NN, Gaber Y, Mattiasson B, Hatti-Kaul R. 2011. Site-directed mutagenesis of an alkaline phytase: influencing specificity, activity and stability in acidic milieu. Enzyme Microb Technol 49:177–182. doi: 10.1016/j.enzmictec.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Holman WI. 1943. A new technique for the determination of phosphorus by the molybdenum blue method. Biochem J 37:256–259. doi: 10.1042/bj0370256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerrero-Olazarán M, Rodríguez-Blanco L, Carreon-Treviño JG, Gallegos-López JA, Viader-Salvadó JM. 2010. Expression of a Bacillus phytase C gene in Pichia pastoris and properties of the recombinant enzyme. Appl Environ Microbiol 76:5601–5608. doi: 10.1128/AEM.00762-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan P, Meng K, Wang Y, Luo H, Shi P, Huang H, Bai Y, Yang P, Yao B. 2012. A protease-resistant exo-polygalacturonase from Klebsiella sp. Y1 with good activity and stability over a wide pH range in the digestive tract. Bioresour Technol 123:171–176. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Hu F, Wang X, Cao H, Liu D, Yao D. 2013. A rational design for trypsin-resistant improvement of Armillariella tabescens β-mannanase MAN47 based on molecular structure evaluation. J Biotechnol 163:401–407. doi: 10.1016/j.jbiotec.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Yang P, Shi P, Wang Y, Luo H, Meng K, Zhang Z, Wu N, Yao B, Fan Y. 2007. Purification and characterization of a novel protease-resistant α-galactosidase from Rhizopus sp. F78 ACCC 30795. Enzyme Microb Technol 41:835–841. doi: 10.1016/j.enzmictec.2007.07.005. [DOI] [Google Scholar]
- 45.Luo H, Wang Y, Li J, Wang H, Yang J, Yang Y, Huang H, Fan Y, Yao B. 2009. Cloning, expression and characterization of a novel acidic xylanase, XYL11B, from the acidophilic fungus Bispora sp. MEY-1. Enzyme Microb Technol 45:126–133. [Google Scholar]
- 46.Zhou J, Zhang R, Gao Y, Li J, Tang X, Mu Y, Wang F, Li C, Dong Y, Huang Z. 2012. Novel low-temperature-active, salt-tolerant and proteases-resistant endo-1,4-β-mannanase from a new Sphingomonas strain. J Biosci Bioeng 113:568–574. doi: 10.1016/j.jbiosc.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 47.van der Hoorn RA. 2008. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- 48.Dionisio G, Madsen CK, Holm PB, Welinder KG, Jørgensen M, Stoger E, Arcalis E, Shi P, Dunn BM. 2001. Determination of protease mechanism, p 77–104. In Beynon R, Bond JS (ed), Proteolytic enzymes: a practical approach, 2nd ed Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 49.Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. 2005. Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 50.Kornfeld R, Kornfeld S. 1985. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 51.Helenius A, Aebi M. 2001. Intracellular functions of N-linked glycans. Science 291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 52.Shental-Bechor D, Levy Y. 2009. Folding of glycoproteins: toward understanding the biophysics of the glycosylation code. Curr Opin Struct Biol 19:524–533. doi: 10.1016/j.sbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Lisowska E, Jaskiewicz E. 2012. Protein glycosylation, an overview, p 1–7. In Finazzi-Agrò A, Clarke A, Zheng Y, Tickle C, Jansson R, Kehrer-Sawatzki H, Cooper DN, Delves P, Battista J, Melino G, Perkel DJ, Hetherington AM, Bynum WF, Valpuesta JM, Harper D (ed), eLS. John Wiley & Sons Ltd., Chichester, United Kingdom. doi: 10.1002/9780470015902.a0006211.pub3. [DOI] [Google Scholar]
- 54.Marieb EN, Hoehn K. 2010. Human anatomy & physiology. Benjamin Cummings, San Francisco, CA. [Google Scholar]
- 55.Shental-Bechor D, Levy Y. 2008. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci U S A 105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brier S, Maria G, Carginale V, Capasso A, Wu Y, Taylor RM, Borotto NB, Capasso C, Engen JR. 2007. Purification and characterization of pepsins A1 and A2 from the Antarctic rock cod Trematomus bernacchii. FEBS J 274:6152–6166. doi: 10.1111/j.1742-4658.2007.06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. 2008. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun Mass Spectrom 22:1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.