Abstract
Bifidobacteria represent one of the dominant microbial groups that occur in the gut of various animals, being particularly prevalent during the suckling period of humans and other mammals. Their ability to compete with other gut bacteria is largely attributed to their saccharolytic features. Comparative and functional genomic as well as transcriptomic analyses have revealed the genetic background that underpins the overall saccharolytic phenotype for each of the 47 bifidobacterial (sub)species representing the genus Bifidobacterium, while also generating insightful information regarding carbohydrate resource sharing and cross-feeding among bifidobacteria. The abundance of bifidobacterial saccharolytic features in human microbiomes supports the notion that metabolic accessibility to dietary and/or host-derived glycans is a potent evolutionary force that has shaped the bifidobacterial genome.
INTRODUCTION
The ecological relevance of bifidobacteria was immediately obvious when they were first isolated from stool samples of a breast-fed infant at the beginning of the last century (1). Members of the Bifidobacterium genus are Gram-positive bacteria that belong to the Actinobacteria phylum, and, together with the genera Aeriscardovia, Alloscardovia, Gardnerella, Parascardovia, and Scardovia, form the Bifidobacteriaceae family (2). The particular ecological role of the Bifidobacterium genus has been highlighted in recent years due to the identification of 47 different taxa, mostly isolated from the gut of social animals, which encompass mammals, birds, and insects (3), whose offspring are dependent on parental care. This appears to be a common ecological trademark of this bacterial genus and a feature that distinguishes it from other gut commensals like Bacteroides or Lactobacillus. This ecological habitat implies a special route of colonization, which is known as vertical transmission from mother to offspring and which in recent times has gained considerable scientific interest (4). In this context, it is widely accepted that the mammalian fetus develops in an essentially sterile environment within the amnion and that microbial colonization of the fetus commences as soon as the amnion breaks prior to delivery of the baby through the birth canal (4). Bifidobacteria are among the first bacterial colonizers of the human gut, and several (sub)species of this genus are genetically adapted to utilize the nourishment of infants through the metabolism of particular glycans present in human milk (5). Nevertheless, human milk not only represents an important reservoir of glycans that act as bifidogenic factors to specifically support growth of particular bifidobacteria but also acts as a repository of (bifido)bacteria for vertical transmission from mother to infant. This notion is corroborated by the isolation of bifidobacteria directly from human milk (6–8), although it is not clear how bifidobacteria reach this human bodily fluid (9). Similarly, bifidobacteria have been shown to be transferred from the gut lumen to tumors (and organs) by means of a route that is not fully understood (10). Only very recently, a study based on analysis of the gut microbiota of mothers and corresponding children by a combination of amplicon-based profiling and shotgun metagenomics demonstrated that mother and child share particular bifidobacterial strains, belonging to Bifidobacterium breve and Bifidobacterium longum subsp. longum, which is thus indicative of vertical transmission (11, 12). Mode of delivery (i.e., vaginally delivered versus delivered by cesarean section) and type of nutrition (i.e., breast-fed versus bottle-fed) are considered to be important factors that provide differential colonization opportunities, thereby impacting the composition of the neonatal gut microbiota, including the colonization level and species composition of bifidobacteria (13, 14).
As mentioned above, bifidobacteria have been reported to account on average for ∼80% of the total complement of the gut microbiota in breast-feeding infants (15). However, their relative abundance significantly decreases after weaning, although overall numbers of bifidobacteria are only moderately reduced (16, 17). Notably, individuals suffering from gastrointestinal diseases have been shown to display a marked reduction in bifidobacteria compared to healthy controls (16, 17). The latter finding suggests that bifidobacteria play a role in the establishment/maintenance of gut homeostasis through host-microbe interactions and/or their direct interplay with other members of the gut microbiota.
The aim of this review is to shed light on the saccharolytic behavior of bifidobacteria and how bifidobacterial carbohydrate metabolism influences the overall glycobiome of the gut microbiota and host and contributes to trophic relationships between members of gut microbiota.
Genomics of bifidobacteria.
The first fully decoded bifidobacterial genome was that of the human gut commensal Bifidobacterium longum subsp. longum NCC2705 (18). Since then, additional bifidobacterial strains have had their genomes sequenced, such as the adult human fecal isolate Bifidobacterium longum subsp. longum DJO10 (19) and the infant fecal isolates Bifidobacterium bifidum PRL2010 (20), Bifidobacterium breve UCC2003 (21), and Bifidobacterium longum subsp. infantis ATCC 15697 (5). Other bifidobacterial genomes that have been fully decoded include the human oral cavity isolate Bifidobacterium dentium Bd1 (22), as well as many strains belonging to the Bifidobacterium animalis subsp. lactis taxon, which due to their purported health-promoting activities have attracted a lot of commercial interest (23–25). Until 2014, genome sequences of only 10 of the 47 (sub)species assigned to the Bifidobacterium genus were available. Since then, all type strains of the currently recognized (sub)species belonging to this genus have been genomically decoded, thereby representing a genomic encyclopedia for the exploration of genetic variability within the genus Bifidobacterium (26). After the reclassification of Bifidobacterium stercoris as a junior synonym of Bifidobacterium adolescentis (27), the current number of (sub)species assigned to the genus Bifidobacterium is 47 (for which chromosomal sequences are available). Thus, all phylogenetic analyses that have recently been described in the study by Lugli et al. (28) have been reanalyzed in order to take the new Bifidobacterium genus layout into account. Characterization of the overall genetic content of members of the Bifidobacterium genus revealed genome sizes that ranged from 1.73 (Bifidobacterium indicum) to 3.25 Mb (Bifidobacterium biavatii), corresponding to 1,352 and 2,557 predicted protein-encoding open reading frames, respectively. Considering the close phylogenetic relationship between bifidobacteria (29), this substantial genomic size difference is reminiscent of an evolutionary pathway that has involved many gene loss and/or acquisition events. Functional classification of the overall genetic arsenal of the Bifidobacterium genus, representing the pan-genome of this taxon, revealed that 13.7% of the identified bifidobacterial genes encode enzymes involved in carbohydrate metabolism, which is higher than the percentage of such genes of many analyzed gut commensals (26). Among this large number of genes involved in the utilization of glycans, there is a notable presence of a genetic subset that is shared among all currently described 47 bifidobacterial (sub)species, thus being part of the bifidobacterial core genome, and of particular genes that are uniquely found in a specific taxon, and thus form part of the “truly unique genes” (TUGs) (26). Of particular note, among the bifidobacterial core genomic coding sequences, it is worth mentioning those genes that encode the enzymes that make up the bifid shunt (26). It is recognized that the evolutionary success of bifidobacteria may have been due to the fact that this particular metabolic pathway allows the generation of more ATP (per mole of glucose) than the other carbohydrate fermentative pathways, such as glycolysis or the pentose phosphate pathway (30). The evolutionary success of bifidobacteria is further compounded by the fact that they have specialized to metabolize either a very specific set or sometimes a broad range of dietary and/or host-derived glycans (20, 31, 32). Regarding the identified TUGs of the bifidobacterial pan-genome, it is estimated that 14.64% of these genes are encompassing proteins involved in carbohydrate metabolism, including glycosyl hydrolases (GHs) and proteins involved in carbohydrate uptake.
These data are a genetic reflection of the metabolic commitment of bifidobacteria to a saccharolytic lifestyle, which is a common genetic feature of many bacteria that make up the human gut microbiota (33).
Phylogenomics of the Bifidobacterium genus.
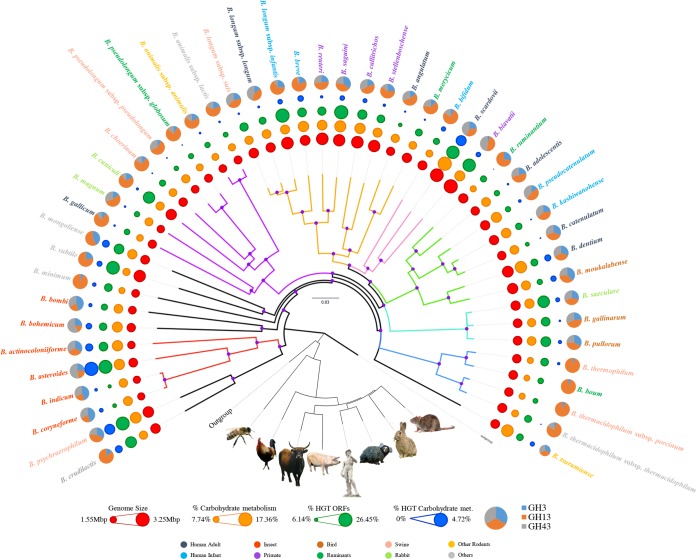
A widely recognized approach applied in modern microbial taxonomy is DNA sequencing followed by comparison of 16S rRNA gene sequences (34). However, a major limitation of this method is that there are cases in which two taxa belong to distant bacterial groups, yet show high identity levels of their 16S rRNA gene sequences (35, 36). Recently, bifidobacterial taxonomy has benefited from the use of a multilocus or multigene approach based on alternative molecular markers such as clpC, dnaB, dnaG, dnaJ1, purF, rpoC, and/or xfp (29). Such a multigene approach allows a high level of discriminatory resolution between closely related bifidobacterial taxa and provides a robust means to infer phylogenetic relationships among members of the Bifidobacterium genus (29). In bacterial taxonomy, thanks to the availability of a growing number of complete genome sequences, it has become possible to reconstruct phylogenetic relationships between taxa on the basis of a much larger set of sequence data per taxon, thus allowing a more reliable and representative inference of the tree of life. In this context, comparative genomic analyses involving the chromosomal sequences from each of the 47 bifidobacterial type strains resulted in the identification of 18,260 Bifidobacterium-specific clusters of orthologous genes (BifCOGs), which constitute the pan-genome of the genus Bifidobacterium (see below). Notably, analysis of the predicted BifCOGs allowed the identification of 459 COGs that were shown to be shared between all these genomes, thus representing the core of bifidobacterial genome coding sequences (core BifCOGs), which can be employed as alternative molecular markers to the 16S rRNA gene sequences (28). A concatenated protein sequence encompassing the protein products of 413 core genes (where these genes had a single representative for each bifidobacterial genome) was used to build a Bifidobacterium supertree, which was shown to be superior in discriminatory power and robustness to a corresponding 16S rRNA gene-based tree (Fig. 1) (28). Such a phylogenomic approach has also been successfully employed for the delineation of other genera, such as Lactobacillus and Streptococcus (37, 38). This Bifidobacterium supertree highlights the evolutionary positioning of all 47 bifidobacterial taxa and reveals the existence of seven phylogenetic groups within the genus, in contrast to the previously identified six groups (29), due to the existence of a particular B. bifidum phylogenetic cluster (28). When a similar analysis was performed by also including representatives of the other genera of the family of the Bifidobacteriaceae, bifidobacteria were shown to fit in the deepest branch of the resulting phylogenetic tree, clearly separating them from other genera within this family (26). Notably, the Bifidobacterium asteroides phylogenetic group is positioned closest to the root in this family-based supertree, therefore suggesting that members of this group most closely resemble the evolutionary ancestor of the Bifidobacterium genus, as had been noted previously (39).
FIG 1.
Phylogenomic overview of the genus Bifidobacterium. A supertree based on the alignment of 413 core COGs (with a single representative identified for each bifidobacterial genome) was constructed in order to obtain a robust phylogenetic reconstruction. Phylogenetic clusters are highlighted with branches of the same color, and nodes with bootstrap values higher than 70% are marked with a purple dot. Circles surrounding the tree represent the approximate genome sizes (in red), relative percentage of genes predicted to be involved in carbohydrate metabolism and transport (in orange), relative percentage of genes predicted to have undergone horizontal gene transfer (in green), and relative percentage of genes predicted to have been subject to horizontal gene transfer or to be involved in carbohydrate metabolism and transport (in blue). The outermost layer represents the proportion of GH families (i.e., GH3, GH13, and GH43). E. coli, Escherichia coli; met., metabolism; ORFs, open reading frames. Bifidobacterial species names are colored based on their ecological origin. In addition, the tree in the lower part of the image represents the phylogeny of the host species from which bifidobacteria had been isolated. This tree was constructed with the Superfamily database and software (85).
Evolutionary development of bifidobacteria.
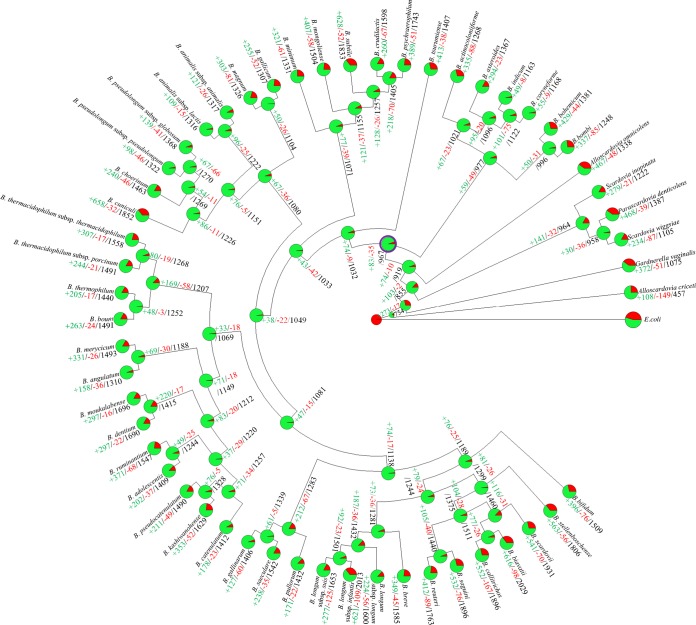
In silico analyses of the bifidobacterial genomes allowed the reconstruction of the genetic evolution of the (sub)species of this genus (26, 40). These analyses predict that the chromosome of the ancestor of the genus Bifidobacterium consisted of 967 COGs. This is just 196 COGs less than the number of COGs harbored by the B. indicum chromosome and 1,062 COGs less than the number in the B. biavatii genome, representing the smallest and largest genome, respectively (26). Thus, the evolutionary development of currently known bifidobacterial taxa appears to have involved a relatively small number of ancestral gene loss occurrences but a substantial number of gene acquisition events (Fig. 2). In contrast, the genomes of other examined gut commensals, such as those of lactic acid bacteria, are believed to have undergone extensive genomic simplification (41).
FIG 2.
Gene gain and loss events in a reconstruction of data representing the family Bifidobacteriaceae. A tree was constructed based on information regarding the presence or absence of COGs for the whole Bifidobacteriaceae pan-genome. Each node is represented by a pie diagram showing the acquired COGs (in red) and the COGs derived from the previous node (in green). Furthermore, additional information is displayed at each node as follows: number of acquired genes, number of lost genes, and total number of COGs. The predicted Bifidobacterium ancestor is highlighted with a thick purple circle surrounding the pie diagram.
In bacterial genomes, gene acquisition events that occur in the course of evolution are expected to facilitate adaptation to a novel ecological niche or to increase competitiveness in the existing ecological niche of the (micro)organism (29, 42). Investigation of gene families that are predicted to have been acquired by bifidobacterial genomes suggests that adaptation to an environment rich in complex glycans, like that of the animal gut, represented the main driving force responsible for speciation among members of the genus Bifidobacterium.
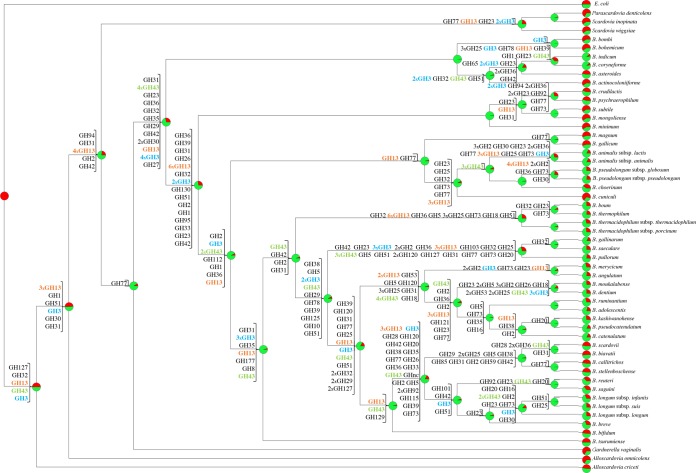
Genome-based analyses allowed a predicted reconstruction of gene acquisition as well as gene loss events that occurred in the course of evolution of bifidobacteria. In this context, eight COGs, including members of GH3 and GH43 families (GHs associated with the degradation of plant polysaccharides), were shown to have been acquired early in bifidobacterial speciation. In contrast, eight COGs encompassing members of the large GH13 family, representing α-amylases, were predicted to have been acquired during the evolution of the Bifidobacteriaceae family and prior to the acquisition of the eight aforementioned GH3 and GH43 members (Fig. 3). Moreover, several putatively acquired genes were identified with predicted carbohydrate uptake functions, including those that belong to the ATP-binding cassette (ABC), phosphoenolpyruvate-phosphotransferase system (PEP-PTS), and major facilitator superfamily (MFS) families of transporters (26).
FIG 3.
Reconstruction of gene gain and loss events regarding genes encoding members of the GH3, GH13, and GH43 families in the family Bifidobacteriaceae. A tree was constructed using information related to the presence or absence of COGs for the whole Bifidobacteriaceae pan-genome. For each node, a pie diagram shows the acquired COGs (in red) and the COGs derived from the previous node (in green). Furthermore, for each node the number of GH family members acquired is reported. (Gene decay events were omitted to allow readability of the figure.) Numbers are indicated when multiple COGs of the same GH family were acquired: otherwise only one COG was gained. GH3, GH13, and GH43 are colored in blue, orange, and green, respectively.
Even if gene gain events represent the key driving force for bifidobacterial genome evolution, gene decay as well as metabolic simplification may still represent another crucial trend for niche-specific adaptation. In silico analyses of bifidobacteria revealed many gene loss events, most of which affect genes encoding biosynthetic enzymes, presumably reflecting analogous environmental pressures. Concerning GHs, most GH13 family members, encompassing α-amylases, appear to have undergone a gene decay process in the genomes of the clade encompassing bifidobacteria isolated from social insects (e.g., B. asteroides, Bifidobacterium actinocoloniiforme, B. indicum, Bifidobacterim coryneforme, Bifidobacterium bombi, and Bifidobacterium bohemicum), possibly as a consequence of a dietary modification of their host or, alternatively, as a consequence of adaptation to a new host with a different diet.
Notably, bifidobacterial species isolated from social insects are predicted to possess a complete trehalose degradation IV pathway, which is absent in the majority of the other bifidobacterial taxa (43). Thus, this may reflect a genetic adaptation of bifidobacteria to the insect gut since trehalose is used as carbohydrate storage and blood sugar by many insects, including the honeybee (43).
A large proportion of the predicted gene acquisition events are believed to be due to horizontal gene transfer (HGT) from other bacteria. The donors of these putative alien genes appear to have preferentially originated from other members of the Actinobacteria class (28.5%), followed by Bacillus (11.7%), Gammaproteobacteria (8.7%), Clostridia (8.7%), or Alphaproteobacteria (5.9%) (26). Importantly and perhaps not unsurprisingly, members of these predicted bacterial donors are also commonly found in the gut environment (44).
The glycobiome of bifidobacteria.
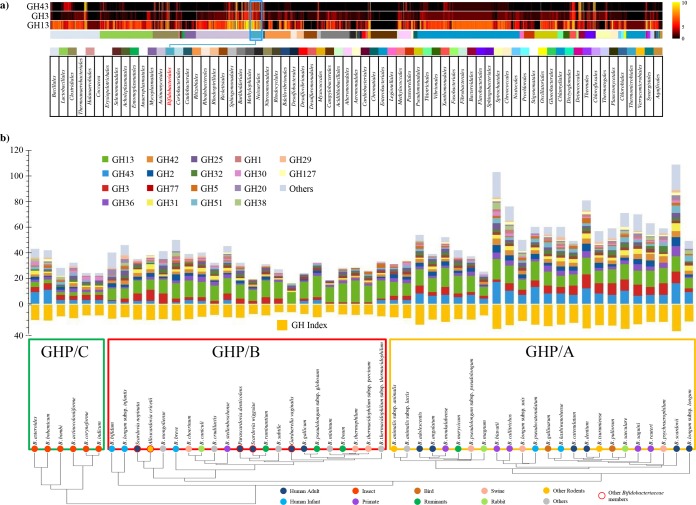
Classification according to the Carbohydrate Active Enzymes (CAZy) system (45) indicates that the pan-genome of the Bifidobacterium genus encompasses one of the largest predicted glycobiomes among known gut commensals, consisting of 3,385 genes that encode putative carbohydrate-active enzymes, including glycosyl hydrolases (GHs), glycosyl transferases (GTs), and carbohydrate esterases (CEs), which are distributed across 57 GH, 13 GT, and 7 CE families (Fig. 4). In contrast, no polysaccharide lyase (PL)-encoding genes were found in the pan-genome of the Bifidobacterium genus (43). GH13 represents the most dominant GH family identified in the glycobiome of the genus Bifidobacterium. Enzymes of this family are characterized by their common catalytic activities of hydrolysis of a wide range of complex carbohydrates, such as starch, glycogen, and related substrates (e.g., amylose, amylopectin, pullulan, maltodextrin, and cyclomaltodextrin), as well as palatinose, stachyose, raffinose, and melibiose (32), which represent dominant glycans found in the adult mammalian diet (46).
FIG 4.
The glycobiome of the Bifidobacterium genus and some additional members of the Bifidobacteriaceae family. Panel a shows a comparative analysis of the bifidobacterial GH3, GH13, and GH43 repertoire against that found in other gut bacteria. The heat maps show GH prediction data from 2,721 sequenced bacterial strains belonging to bacterial orders residing in the human gut, identified by different color codes as explained in the underlying table. Data regarding Bifidobacteriales are highlighted in blue. In panel b, GH family profiles identified by the CAZy database were used to construct a hierarchical clustering of all tested species of the Bifidobacterium genus and additional members of the Bifidobacteriaceae family. This clustering highlights the presence of three distinct clusters named GHP/A, GHP/B, and GHP/C displaying a different repertoire of GHs. GH arsenal prediction for each analyzed Bifidobacterium species is represented by a bar plot, and the GH index (the number of GHs predicted in each genome normalized by genome size expressed as megabase pairs) is illustrated as an orange bar plot.
The bifidobacterial glycobiome also encompasses members of GH families that are pivotal in host glycan breakdown, such as those belonging to GH33 and GH34, which represent exo-sialidases, GH29 and GH30, which represent fucosidases, and GH20, which include hexosaminidase and lacto-N-biosidase activities (43).
Interestingly, comparison of the bifidobacterial GH repertoire with those encoded by representatives of the main bacterial families typically residing in the human gastrointestinal tract show that the Bifidobacteriaceae family possesses a very broad set of genes encoding GH families GH3, GH43, GH13, and GH51, similar to (relative to genome size) the numbers found in the genomes of Bacteroides spp. (Bacteroidales family), Clostridiales, Paenibacillus spp. (Bacillales family), and Streptomyces spp. (Actinomycetales family) (43).
In silico evaluation of the GH repertoire of the genus Bifidobacterium also included the clustering of bifidobacterial (sub)species based on their predicted GH and carbohydrate degradation pathway repertoire (Fig. 4), allowing the identification of three groups—designated GHP/A, GHP/B, and GHP/C (43). Notably, group GHP/A includes bifidobacterial (sub)species with a considerable array of predicted GH43 family members, representing enzymes involved in the breakdown of complex plant glycans such as xylan and arabinoxylans. This suggests adaptation of bifidobacterial (sub)species belonging to group GHP/A to hosts that enjoy a vegetarian or omnivorous diet. Bifidobacterial taxa isolated from social insects constitute the GHP/C group, as they encompass a discrete set of GH43 and GH3 family members and only a very limited number of GH13 members, which is in contrast to all other analyzed bifidobacteria (39). As described above, the GH13 family includes enzymes that are involved in the hydrolysis of poly- or oligosaccharides with α-glucosidic linkages such as starch, glycogen, and related substrates; thus, this finding can be explained by the paucity of such glycans in the (vegetarian) diet of honeybees and bumblebees. The remaining bifidobacterial (sub)species, not fitting in groups GHP/A or GHP/C, are included in group GHP/B, whose members are characterized by an underrepresentation of GH43 and GH3 enzymes (43).
As expected, part of the predicted glycobiome of bifidobacteria is extracellular, since it allows access to polysaccharides that are too large to be internalized. It is estimated that 10.9% of the total GH repertoire is located extracellularly, of which 32.9% are members of the GH13 family and annotated as pullulanases and α-amylases, and 24% are members of the GH43 family and predicted to act as β-xylosidases and α-l-arabinofuranosidases, while 12% are members of the GH51 family and classified as α-l-arabinofuranosidases (43).
Putative extracellular GHs were found in 43 (sub)species of the genus Bifidobacterium, with higher occurrence in the genomes of Bifidobacterium biavatii (predicted to encode 17 secreted GHs, including four GH43 and four GH13 members), Bifidobacterium scardovii (predicted to specify 11 secreted GHs, including three GH43 and three GH51 members, annotated as α-l-arabinofuranosidases), and B. bifidum (predicted to be endowed with 11 genes that encode secreted GHs, including two GH83 and two GH33 members, all four of which are putative sialidases) (43). The four secreted sialidases found in B. bifidum are clear evidence of its advanced genetic adaptation to the mammalian gut (47). In fact, sialidases are essential for the metabolism of human milk oligosaccharides (HMOs) and intestinal glycoconjugates such as mucin (see below) (20, 48, 49).
Host- or diet-derived glycans and bifidobacteria: an example of strict coevolution.
Gut commensals such as bifidobacteria display a saccharolytic behavior aimed at accessing carbohydrates as their sole carbon and energy sources (43). In silico analyses of gut commensals have shown that the genetic arsenal dedicated to the breakdown of complex, host-indigestible carbohydrates either (in)directly derived from the host (i.e., mucin and HMO) or from the diet has had a significant impact on shaping the gut microbiome composition (50).
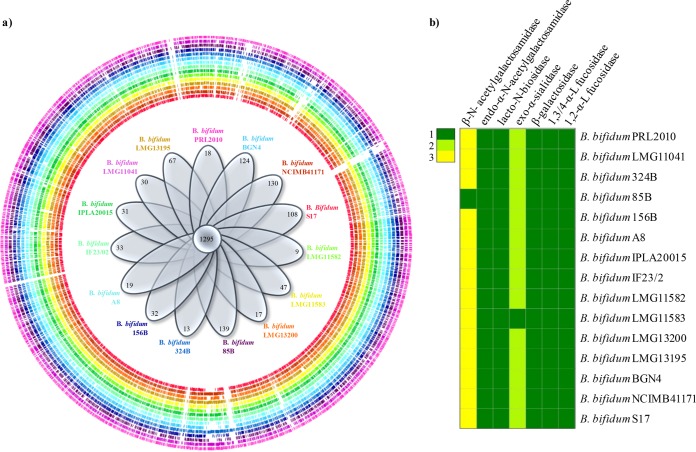
Mucins are host-produced glycans, secreted by intestinal goblet cells, that essentially make up the mucus layer covering the intestinal mucosa. The main monosaccharide components in mucin-derived glycoproteins are N-acetylglucosamine, N-acetylgalactosamine, and galactose, and these glycoproteins are decorated with fucose, sialic acid, and sulfate groups (51). Within the genus Bifidobacterium, only members of the B. bifidum species have been shown to efficiently degrade mucin (48, 52, 53). However, other bifidobacterial taxa, including B. longum subsp. infantis, B. biavatii, B. crudilactis, B. kashiwanohense, B. stellenboschense, and B. mongoliense, have recently been shown to metabolize host glycans, including mucin, though at a lower efficiency than the B. bifidum species (43). Further in silico analyses of the pan-genome of the B. bifidum species together with functional genome approaches revealed the existence of a gene set involved in mucin metabolism that was shown to be uniquely present in the genomes of members of this bifidobacterial species and thus constitutes the core genome sequences of the B. bifidum species (20, 49) (Fig. 5). Such findings represent an intriguing example of strict coevolution of a human gut commensal like B. bifidum to the human intestine, where the glycans produced by the host serve as carbon source for this bifidobacterial species (47). Another intriguing example of host-produced glycans that are fermented by bifidobacteria are the HMOs, which are present in human milk yet are not utilized by the (infant) host. The chromosome B. longum subsp. infantis, a typical fecal isolate from (breast-fed) infants, encompasses a gene cluster predicted to encode GHs and carbohydrate transporters necessary for the import and metabolism of HMOs (5). This 43-kb gene cluster encodes a variety of predicted or proven catabolic enzymes, such as fucosidases, sialidases, a β-hexosaminidase, and β-galactosidases, as well as extracellular solute binding proteins and permeases that are devoted to HMO metabolism (5, 54–56). Moreover, the genome of this microorganism contains a complete urease operon predicted to be involved in the utilization of urea, representing an important nitrogen source in milk (5).
FIG 5.
The pan-genome of Bifidobacterium bifidum. Panel a shows a genome atlas representation of all publicly available genomes of the species B. bifidum in which each circle represents a different strain identified by a different color. Inside the genome atlas, a Venn diagram illustrates the number of identified core and unique genes. Panel b displays a heat map that summarizes the presence and number of particular genes predicted to be involved in mucin degradation in the analyzed B. bifidum genomes.
In contrast, in silico analyses of the genomes of two other members of the B. longum phylogenetic group (i.e., B. longum subsp. longum and B. longum subsp. suis) revealed a higher genomic capacity to utilize plant-derived glycans, including arabinoxylan (57, 58).
Another important sign of genetic adaptation of bifidobacteria to the human gut is the specific utilization of various complex glycans, such as resistant starches, which are derived from the diet and which escape host-mediated digestion. Starch consists of amylose and amylopectin moieties, with the former being a linear α-(1,4) glucose chain with a plant-specific degree of polymerization of 200 to 6,000, while the latter represents short linear α-(1,4)-glucose-linked chains with α-(1,6)-linked glucose side chains (59). Natural derivatives of starch are maltodextrin, maltotriose, and maltose (59). The breakdown of these complex carbohydrates is operated by gut commensals through the combined action of amylases (EC 3.2.1.1, EC 3.2.1.2, and EC 3.2.1.3) and amylopullulanases (APU [EC 3.2.1.41]). Starch is metabolized by various members of the gut microbiota, such as Ruminococcus bromii (60), Bacteroides thetaiotaomicron (61), and Roseburia inulinivorans (62), whose genomes encode various amylases. Even if bifidobacteria are nondominant members of the adult gut microbiota, their biological roles in the metabolism of dietary and host-derived glycans have only recently been appreciated (63). Analyses of the genome sequences of the various type strains representing each of the 47 (sub)species of the genus Bifidobacterium revealed the widespread occurrence of the above-mentioned starch/starch derivative-degrading enzymes (26), especially in the genomes of the adult-type Bifidobacterium adolescentis (31, 64) and B. breve (64, 65). The prediction of the glycobiome of the B. adolescentis species revealed, compared to most other bifidobacterial gut commensals, a much larger set of GH13 enzymes, which include amylase, pullulanase, and cyclomaltodextrinase activities, thus suggesting superior growth performance of this species on particular plant-derived carbohydrates (60). Such findings were substantiated by the analyses of fermentation profiles of members of the B. adolescentis taxon, which highlighted a preference for the utilization of different esose-containing sugars (e.g., galactose, mannose, and glucose), as well as plant-derived glycans that are typically present in the human diet, such as starch (31).
Among bifidobacterial species used as probiotics, B. animalis subsp. lactis deserves a special mention (25). Members of this bifidobacterial species can transit and impact resident microbial communities even if their overall effects are still not well defined (66–68). Notably, members of the B. animalis subsp. lactis taxon can only hydrolyze/metabolize very limited number of carbohydrates, perhaps underlining a rather high level of genetic adaptation to an ecological niche or perhaps due to massive genome decay as a result of its industrial exploitation, which has involved long-term cultivation of B. animalis subsp. lactis on synthetic media (25, 43).
A bifidobacterial species possessing carbohydrate breakdown capabilities toward both dietary glycans as well as host-derived glycan is represented by Bifidobacterium breve (65, 69–72). The reconstruction of the pan-genome of this taxon revealed a wide genetic variability for genes previously characterized as being involved in the utilization of the carbohydrates ribose, sucrose, and raffinose, as well as the plant-derived polysaccharides starch, galactan, and cellodextrin (73).
Notably, the core genome of B. breve encompasses genes that are predicted to encode enzymes involved in the uptake and utilization of host-derived mono/oligosaccharides, in particular those derived from mucin and HMOs (73). Examples include gene clusters predicted to be involved in the metabolism of sialic acid, lacto-N-biose, fucose, and N-linked glycans. While B. breve is not known to be able to grow on mucin or HMOs, host-derived mono/oligosaccharides may become available through hydrolytic activities of other (bifido)bacteria present in the gut (e.g., B. bifidum and B. longum subsp. infantis), allowing B. breve strains to utilize such liberated carbohydrates through cross-feeding activities (74–76).
Cross-feeding activities by bifidobacteria and the effect on the gut microbiota.
Recently, several studies have revealed that bifidobacteria play an ecological role in shaping the gut murine microbiome toward a saccharolytic microbiota by means of cross-feeding activities (43, 74–79). Cross-feeding activities target polysaccharides that reach the gut intact, where they may undergo extracellular hydrolysis by enteric bacteria like bifidobacteria, thus generating simple glycans (i.e., monosaccharides and oligosaccharides) that may become available to other microbial gut inhabitants (80). In this context, various studies involving simple bifidobacterial communities have shown how saccharolytic bacteria may cooperate in order to obtain access to complex diet carbohydrates (e.g., starch, xylan, or arabinoxylan) (26, 43, 79) or host-derived carbohydrate (e.g., mucin and HMOs) (75, 76, 79). In such scenario, the cross-feeding activities exerted by bifidobacteria ultimately influence the gut microbiota composition as well as its functionality by enhancing the production of (certain) short-chain fatty acids (SCFA) directly or indirectly though the production of acetate, which is then converted to butyrate by a species of eubacteria (77–79).
Overall, the ability of these mutualistic/commensal activities of bifidobacteria to specifically target carbohydrates in such a sophisticated manner suggests the existence of some simplistic form of social intelligence that is aimed at regulating the dynamics of the gut microbiota relationships (43, 79).
Functional contribution of bifidobacteria to the human gut.
As mentioned above, bifidobacteria are commonly isolated from the mammalian gut, with a higher prevalence during the suckling stages of life (15). However, their functional contribution to the human gut is still largely ignored. Surprisingly, in various microbiome-based studies their presence is not even detected (81) or is severely underestimated. This is likely to be linked to methodological inadequacies related to primer design and/or to sample processing (82). Nevertheless, a recent investigation of currently available metagenomic data sets did indeed identify, as expected, a variable presence of bifidobacteria, with the highest prevalence in infant-associated data sets (43). Notably, an extensive repertoire of GH-encoding genes, specifying genes encoding GH3, GH13, GH43, GH51, and GH77 members that are involved in hydrolysis of complex plant carbohydrates commonly present in the adult diet, are among the most frequently identified bifidobacterial genes in metagenomic data sets obtained from (adult) human fecal samples. This highlights a crucial aspect of the functionality contributed by bifidobacteria in the adult gut: despite the relative paucity of these bacteria in the adult human gut, their functional contribution to the human gut microbiome is important in terms of expanding the overall glycobiome of the large intestine and may thus affect the overall gut physiology. In contrast, in the microbiomes from infant fecal samples, the high prevalence of bifidobacteria is also reflected by the abundance of bifidobacterial GH-encoding genes, such as those specifying members of the GH2, GH20, GH42, GH112, and GH129 families (43). Notably, all of these GHs are involved in the breakdown of milk-related carbohydrates such as lactose and HMOs, which represent the majority of carbohydrates present in the diet of a breast-fed infant.
CONCLUSIONS
Genome sequencing of bifidobacteria started in 2002 with the decoding of the first bifidobacterial chromosome of B. longum subsp. longum NCC2705 (18). Since then, there have been a rapidly growing number of bifidobacterial strains whose genome sequences have been decoded, providing a detailed overview of the genetic diversity among members of the Bifidobacterium genus, as well as generating information concerning the mechanisms by which they colonize and persist in the gastrointestinal tract. Furthermore, their relative abundance in the human gut microbiome has been substantially underestimated due to various technical issues (11, 15, 82, 83). It is expected that metagenomic studies directed to explore the composition of the microbial consortia will discover novel representatives of the Bifidobacterium genus. Despite such findings, many aspects of bifidobacterial biology have yet to be explored (84). This is due to the lack of molecular tools to facilitate efficient exploration of their genetic arsenal, although significant progress has been made in recent years. The continued development of tools for bifidobacteria will allow the discovery of genes as well as gene products involved in gut colonization and other host-microbe interactions, including those from which we as human beings may benefit.
ACKNOWLEDGMENTS
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. D.V.S. is a member of the APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (grant no. SFI/12/RC/2273).
REFERENCES
- 1.Tissier H. 1900. Recherches sur la flore intestinale des nourrissons: (état normal et pathologique). University of Paris, Paris, France. [Google Scholar]
- 2.Goodfellow M, Kampfer P, Busse H-J, Trujillo M, Suzuki K-I, Ludwig W, Whitman W (ed). 1989. Bergey's manual of systematic bacteriology, vol 5 Springer, New York, NY. [Google Scholar]
- 3.Ventura M, Turroni F, Lugli GA, van Sinderen D. 2014. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric 94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 4.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol 11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turroni F, Foroni E, Serafini F, Viappiani A, Montanini B, Bottacini F, Ferrarini A, Bacchini PL, Rota C, Delledonne M, Ottonello S, van Sinderen D, Ventura M. 2011. Ability of Bifidobacterium breve to grow on different types of milk: exploring the metabolism of milk through genome analysis. Appl Environ Microbiol 77:7408–7417. doi: 10.1128/AEM.05336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, Rodriguez JM. 2009. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol 75:965–969. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arboleya S, Ruas-Madiedo P, Margolles A, Solis G, Salminen S, de Los Reyes-Gavilan CG, Gueimonde M. 2011. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int J Food Microbiol 149:28–36. doi: 10.1016/j.ijfoodmicro.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Wassenaar TM, Panigrahi P. 2014. Is a foetus developing in a sterile environment? Lett Appl Microbiol 59:572–579. doi: 10.1111/lam.12334. [DOI] [PubMed] [Google Scholar]
- 10.Cronin M, Morrissey D, Rajendran S, El Mashad SM, van Sinderen D, O'Sullivan GC, Tangney M. 2010. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol Ther 18:1397–1407. doi: 10.1038/mt.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 12.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 14.Fanaro S, Vigi V, Chierici R, Boehm G. 2003. Fecal flora measurements of breastfed infants using an integrated transport and culturing system. Acta Paediatr 92:634–635. doi: 10.1111/j.1651-2227.2003.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 15.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Ventura M, Margolles A, Gueimonde M. 2015. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr 166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 17.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1):S4578–S4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O'Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O'Toole PW, van Sinderen D. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet 5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrangou R, Briczinski EP, Traeger LL, Loquasto JR, Richards M, Horvath P, Coute-Monvoisin AC, Leyer G, Rendulic S, Steele JL, Broadbent JR, Oberg T, Dudley EG, Schuster S, Romero DA, Roberts RF. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J Bacteriol 191:4144–4151. doi: 10.1128/JB.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bottacini F, Dal Bello F, Turroni F, Milani C, Duranti S, Foroni E, Viappiani A, Strati F, Mora D, van Sinderen D, Ventura M. 2011. Complete genome sequence of Bifidobacterium animalis subsp. lactis BLC1. J Bacteriol 193:6387–6388. doi: 10.1128/JB.06079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killer J, Sedlacek I, Rada V, Havlik J, Kopecny J. 2013. Reclassification of Bifidobacterium stercoris Kim et al. 2010 as a later heterotypic synonym of Bifidobacterium adolescentis. Int J Syst Evol Microbiol 63:4350–4353. doi: 10.1099/ijs.0.054957-0. [DOI] [PubMed] [Google Scholar]
- 28.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura M, Canchaya C, Del Casale A, Dellaglio F, Neviani E, Fitzgerald GF, van Sinderen D. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int J Syst Evol Microbiol 56:2783–2792. doi: 10.1099/ijs.0.64233-0. [DOI] [PubMed] [Google Scholar]
- 30.Scardovi V, Trovatelli LD. 1965. The fructose-6-phosphate shunt as peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann Microbiol Enzimol 15:19–29. [Google Scholar]
- 31.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura M. 2014. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol 80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (ed). 2009. Bergey's manual of systematic bacteriology, 2nd ed. Springer-Verlag, New York, NY. [Google Scholar]
- 35.Fox GE, Wisotzkey JD, Jurtshuk P Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 37.Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeno-Tarraga AM, Parkhill J, Flynn S, O'Sullivan GC, Collins JK, Higgins D, Shanahan F, Fitzgerald GF, van Sinderen D, O'Toole PW. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci U S A 103:6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anisimova M, Bielawski J, Dunn K, Yang Z. 2007. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol Biol 7:154. doi: 10.1186/1471-2148-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lugli GA, Duranti S, Milani C, Turroni F, Viappiani A, Mangifesta M, van Sinderen D, Ventura M. 2014. The genome sequence of Bifidobacterium moukalabense DSM 27321 highlights the close phylogenetic relatedness with the Bifidobacterium dentium taxon. Genome Announc 2:e00048-14. doi: 10.1128/genomeA.00048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milani C, Andrea Lugli G, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active Enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 47.Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol 5:437. doi: 10.3389/fmicb.2014.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 49.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 50.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. 2008. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol 74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruiz L, Gueimonde M, Coute Y, Salminen S, Sanchez JC, de los Reyes-Gavilan CG, Margolles A. 2011. Evaluation of the ability of Bifidobacterium longum to metabolize human intestinal mucus. FEMS Microbiol Lett 314:125–130. doi: 10.1111/j.1574-6968.2010.02159.x. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 55.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sela DA. 2011. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol 149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 57.O'Callaghan A, Bottacini F, O'Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaplin AV, Efimov BA, Smeianov VV, Kafarskaia LI, Pikina AP, Shkoporov AN. 2015. Intraspecies genomic diversity and long-term persistence of Bifidobacterium longum. PLoS One 10:e0135658. doi: 10.1371/journal.pone.0135658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. 2002. Properties and applications of starch-converting enzymes of the alpha-amylase family. J Biotechnol 94:137–155. doi: 10.1016/S0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 60.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D'Elia JN, Salyers AA. 1996. Contribution of a neopullulanase, a pullulanase, and an alpha-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol 178:7173–7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer CD, Young P, Rucklidge G, Ramsay AG, Flint HJ. 2011. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci U S A 108(Suppl 1):S4672–S4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. 2011. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duranti S, Turroni F, Milani C, Foroni E, Bottacini F, Dal Bello F, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2013. Exploration of the genomic diversity and core genome of the Bifidobacterium adolescentis phylogenetic group by means of a polyphasic approach. Appl Environ Microbiol 79:336–346. doi: 10.1128/AEM.02467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonnenburg JL, Chen CT, Gordon JI. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol 4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Derrien M, van Hylckama Vlieg JE. 2015. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 68.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokusaeva K, Neves AR, Zomer A, O'Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol 3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pokusaeva K, O'Connell-Motherway M, Zomer A, Fitzgerald GF, van Sinderen D. 2009. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol 75:1135–1143. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Connell Motherway M, Fitzgerald GF, van Sinderen D. 2011. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol 4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pokusaeva K, O'Connell-Motherway M, Zomer A, Macsharry J, Fitzgerald GF, van Sinderen D. 2011. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol 77:1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bottacini F, O'Connell Motherway M, Kuczynski J, O'Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falony G, Calmeyn T, Leroy F, De Vuyst L. 2009. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl Environ Microbiol 75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Egan M, O'Connell Motherway M, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de Los Reyes-Gavilan CG. 2015. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362:fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 78.Riviere A, Gagnon M, Weckx S, Roy D, De Vuyst L. 2015. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol 81:7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 81.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ventura M, Turroni F, Canchaya C, Vaughan EE, O'Toole PW, van Sinderen D. 2009. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci 14:3214–3221. [DOI] [PubMed] [Google Scholar]
- 84.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, Motherway MO, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. 2009. SUPERFAMILY—sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res 37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]