Abstract
The vast majority of microbial life remains uncatalogued due to the inability to cultivate these organisms in the laboratory. This “microbial dark matter” represents a substantial portion of the tree of life and of the populations that contribute to chemical cycling in many ecosystems. In this work, we leveraged an existing single-cell genomic data set representing the candidate bacterial phylum “Calescamantes” (EM19) to calibrate machine learning algorithms and define metagenomic bins directly from pyrosequencing reads derived from Great Boiling Spring in the U.S. Great Basin. Compared to other assembly-based methods, taxonomic binning with a read-based machine learning approach yielded final assemblies with the highest predicted genome completeness of any method tested. Read-first binning subsequently was used to extract Calescamantes bins from all metagenomes with abundant Calescamantes populations, including metagenomes from Octopus Spring and Bison Pool in Yellowstone National Park and Gongxiaoshe Spring in Yunnan Province, China. Metabolic reconstruction suggests that Calescamantes are heterotrophic, facultative anaerobes, which can utilize oxidized nitrogen sources as terminal electron acceptors for respiration in the absence of oxygen and use proteins as their primary carbon source. Despite their phylogenetic divergence, the geographically separate Calescamantes populations were highly similar in their predicted metabolic capabilities and core gene content, respiring O2, or oxidized nitrogen species for energy conservation in distant but chemically similar hot springs.
INTRODUCTION
The vast majority of the diversity of microbial life on Earth remains undiscovered; the core metabolisms of yet-uncultivated species, interguild interactions within natural and managed ecosystems, and the contributions of microbial populations to the geochemistry of the environment remain poorly understood (1, 2). There are currently over 60 bacterial and archaeal phylum-level groups that have been observed through the use of 16S rRNA gene sequencing and phylogenetics, with over half containing no cultivated representatives (3). This so-called microbial dark matter comprises a substantial proportion of the tree of life and of microbial communities that likely play significant roles in biogeochemical cycles in a variety of environments (4–9).
Metagenomic analyses of low-diversity microbial communities have yielded robust, near-complete genomic assemblies representative of the abundant populations, expanding the knowledge of the metabolic potential of predominant organisms in these ecosystems (10–14). Nucleotide word frequencies calculated from metagenomic contiguous assembled sequences (contigs) have been used to separate population-specific clusters, or “bins,” from the community DNA pool (15–17), which has greatly advanced our understanding of the genomic diversity in natural environments and how populations differ between chemically distinct environments and along environmental gradients (18, 19). Even using modern sequencing techniques, however, it can be challenging to confidently separate and assemble genomes of community members using metagenomic data alone due to problems such as low population abundance, high community diversity, or similarities in nucleotide word frequencies between phylogenetically distant taxa.
High-throughput, semiautomated isolation and sequencing of single cells representing candidate phyla has opened the door for systematic analysis of environmental DNA that can be unambiguously assigned to these uncultured organisms (4, 20). Great Boiling Spring (GBS), located in the U.S. Great Basin, harbors abundant populations of several candidate phyla, and the low biological diversity in high-temperature sediments facilitates access to their genomes (4, 21–24). Previously, 10 separate single cells representing a deeply branching lineage in the bacterial domain, candidate phylum EM19, were isolated from GBS sediments, sequenced, and assembled into draft single amplified genomes (SAGs) ranging from 0.3 to 1.9 Mbp in length (4). The coassembly of eight of the 10 SAGs yielded a draft genome of 2.24 Mbp that was estimated to be 94% complete based on the proportion of 139 single-copy conserved bacterial markers (SCMs) observed in the assembly (4). Phylogenomic analyses confirmed the independence of EM19 from other bacterial phyla, justifying a proposal to name the lineage more formally as candidate phylum “Calescamantes” and the genus and species as “Candidatus Calescibacterium nevadense” (4). Although core metabolic genes were identified, this work was part of a much larger single-cell genomic survey, and did not describe the metabolic capabilities of “Ca. Calescibacterium nevadense” or encompass Calescamantes genomes from other locations.
The goals of the current study were to obtain greater phylogenetic, geographic, and genomic coverage of members of the Calescamantes. Although metagenomic read binning utilizing read abundances (25) and k-mer frequencies (25, 26) have been utilized previously in whole-community analyses, none have used SAGs as environmentally relevant anchors to improve clustering and genome quality. Here, we assess the effectiveness of a SAG-assisted, read-based binning approach for identifying Calescamantes sequences in metagenomes prior to assembly and to use the resulting genomic data sets to predict the metabolic potential and conserved features of the Calescamantes in detail. Nucleotide word frequencies obtained from SAGs representative of several candidate phyla abundant in GBS and other environmentally relevant genomes were used as anchors to confidently separate Calescamantes sequence reads from environmental metagenomes prior to assembly using a multilayer perceptron (MLP) machine learning approach. Because this approach of read-binning prior to metagenomic assembly has the potential advantage of avoiding chimeric artifacts that can occur during assembly, we compared it with other commonly used assembly-based binning methods on the GBS metagenome (15, 27, 28). As read-binning outperformed assembly-based technologies on the GBS metagenome, these read-binning methods were also used to confidently predict taxonomic bins from all available metagenomes that contained relatives of “Ca. Calescibacterium nevadense,” including metagenomes from geothermal springs in Yellowstone National Park (YNP) (Bison Pool and Octopus Spring) and Yunnan Province, China (Gongxiaoshe Spring), expanding both the geographic and phylogenetic coverage of Calescamantes genomic data sets. Comparative analysis of these distinct Calescamantes populations identified a predicted core metabolism that couples the oxidation of proteins to the reduction of oxygen or oxidized nitrogen compounds, suggesting niche conservatism among these phylogenetically distinct and geographically distant populations.
MATERIALS AND METHODS
SAG data sets, metagenome sampling, and metagenomic DNA extraction.
Ten individual Calescamantes SAGs and a combined SAG assembly obtained from GBS, described previously (4) were retrieved from the IMG system. Previously published (14, 19) metagenomes from Octopus Spring and Bison Spring in YNP were accessed from the IMG system as summarized in Table S1 in the supplemental material. Associated sampling sites, dates, and temperatures are shown in Table S1 in the supplemental material.
Sediment samples for metagenomic analyses were collected from the north edge of the source pool of GBS (site A [80°C] in reference 23) on 2 December 2008 and from the bottom of Gongxiaoshe Spring (Tengchong, China) on 10 January 2011 as described by Hou et al. (14). DNA was extracted from both sediments using the FastDNA spin kit for soil (MP Biomedicals, Solon, OH) according to the manufacturer's protocol.
DNA sequencing and assembly of whole metagenomes.
Library preparation and sequencing of the GBS metagenome using the 454 FLX platform with Titanium chemistry (Roche, Branford, CT) was performed at the Joint Genome Institute (JGI). GBS metagenomic reads were assembled at the JGI with Newbler 2.4 using a minimum nucleotide identity of 98% and a minimum overlap of 80 bp (29).
For the Gongxiaoshe metagenome, the extracted DNA was sheared to 500 bp using a Covaris ultrasonicator (Covaris Inc., Woburn, MA) according to the manufacturer's recommendations. The sheared DNA was end repaired, adaptor ligated with multiplexing, and purified using the Ovation SP ultralow DR multiplex system (NuGEN Technologies Inc., CA), and the resulting libraries were sequenced (2 by 250) using the Illumina MiSeq platform (Illumina, San Diego, CA). Gongxiaoshe metagenome reads with no ambiguous bases, quality score of >20, and length of >74 bases were assembled using the CLC Genomic Workbench, version 6.0, de Bruijn graph assembler (CLC bio, Boston, MA), taking into account paired reads. After binning, only contigs of at least 500 bases long were included in the final curated assembly.
Binning and assembly of Calescamantes reads from GBS based on machine learning.
For read-binning prior to assembly, metagenomic reads were binned by nucleotide trimer frequencies using the MLP machine learning package in WEKA, version 3.6, with default parameters (30). This package uses back-propagation to facilitate the training of nonlinear networks. The MLP initially was trained using nucleotide frequencies of clipped genomic segments (2,000 bp; also see Fig. S1 in the supplemental material) from the GBS Calescamantes SAG coassembly. SAGs and isolate genomes representing other abundant populations in GBS included Calescamantes (JGI_2527291514), “Fervidibacteria” (JGI_0000001-G10), a novel crenarchaeota, “Geoarchaeota” (JGI_AAA471-L13), “Aigarchaeota” (JGI_2264867219), and the Thermoflexus hugenholtzii strain JAD2 draft genome (JGI_2140918011) (31). This approach provides multiple points of reference during training of the MLP algorithm. Metagenomic reads were assigned to the GBS Calescamantes population if their MLP confidence score was ≥0.9 (a score of 1 indicates 100% confidence), assessed as the point at which false positives were minimized while maximizing true positives (data not shown). This cutoff was designed to be relatively liberal in order to be inclusive of divergent or novel sequences with the intent to curate postassembly. To augment the MLP-based Calescamantes bin and provide access to tRNA/rRNA (32) and other genomic regions with anomalous nucleotide frequency, predicted coding regions from the GBS Calescamantes SAG coassembly were queried against an unassembled GBS metagenomic nucleotide database using BLASTN (33), and matches with an E value of ≤1E−15 were selected for further incorporation into the assembly. All reads belonging to the Calescamantes bin were assembled with SPAdes using the “careful” setting and step k-mer values of 77, 99, 111, and 127. Sequence reads that did not assemble were removed (38,340 of 129,215), and assembled contigs were further curated as described below.
Binning and assembly of Calescamantes reads from Gongxiaoshe Spring, Bison Pool, and Octopus Spring.
The above-described procedure was repeated for sequence reads from all other sites using MLP genomic anchors determined by the closest matching genomes in NCBI (those which had BLAST hits with the highest identity to metagenomic reads) and were representative of predominant populations in those environments. In addition to the Calescamantes SAG coassembly, the MLP was trained using Thermoflexus hugenholtzii (JGI_2140918011), Acetothermus autotrophicum (AP011800.1 to AP011803.1), Thermus aquaticus (NZ_ABVK00000000.2), and the Fervidibacteria SAG (JGI_0000001-G10) for Gongxiaoshe Spring; Thermus aquaticus (NZ_ABVK00000000.2), Thermoflexus hugenholtzii (JGI_2140918011), Pyrobaculum islandicum (CP000504.1), Thermocrinis ruber (CP007028.1), and the Aigarchaeota SAG (JGI_2264867219) for Octopus Spring (JGI_2264867219); and Thermus aquaticus (NZ_ABVK00000000.2), Hydrogenobacter thermophilus (AP011112), Pyrobaculum islandicum (CP000504.1), and Thermoflexus hugenholtzii (JGI_2140918011) for Bison Pool. Calescamantes bins from Gongxiaoshe and Octopus Spring were assembled using SPAdes, as described above. Bison Pool metagenome reads were assembled using Dragon (SequentiX, Germany) using default settings, as SPAdes had difficulty assembling small numbers of Sanger sequences. Accession numbers for all SAGs and metagenomes used are also reported in Table S1 in the supplemental material.
Postassembly contig binning.
Postassembly binning was used as a comparison tool to determine whether MLP-based binning was consistent with established techniques in terms of content, genome completeness, and SCM redundancy. Binning of assembled metagenomic contigs was performed using either MaxBin 2.1.1 (34, 35), Metawatt, version 1.7 (15), or emergent self-organizing mapping (ESOM) (12, 28). The JGI assembly of the GBS metagenome was used as the input sequence for all tested assembly-based binning methods. For Metawatt, the assembled contigs representing Calescamantes were identified separately with a medium sensitivity setting using all GBS SAG assemblies (Calescamantes, Fervidibacteria, Geoarchaeota, and Aigarchaeota) (4), Thermoflexus hugenholtzii draft genome (JGI_2140918011), and BLASTN references, as previously described by Nobu et al. (36). For MaxBin, the assembled contigs greater than 1,000 bp were binned as previously described (34, 35). Resulting bins, including Calescamantes, were identified with BLASTN using the SAG assemblies (as described for Metawatt). For ESOM, assembled GBS metagenome and SAG contigs were trimmed to a window size of 2,500 to 5,000 bp, and training was completed using 124 rows and 556 columns. Sequences were separated along topological boundaries generated by ESOM, and nonredundant reads identified by BLASTN were included as described above. Metagenomic contigs and unassembled reads from GBS were also binned by nucleotide trimer frequencies using the MLP machine learning package, including nonredundant reads identified by BLASTN, as described above. GBS assembled contigs were curated as previously described for preassembly binning.
16S rRNA gene analysis.
Partial 16S rRNA gene sequences identified by RAST in the GBS Calescamantes metagenomic assembly and SAG coassembly were used to query the GenBank NCBI-nr database by BLASTN to identify the nearest publically available related sequences. All Calescamantes 16S rRNA sequences and a reference data set of distantly related taxa were aligned using the SILVA package (SINA version 1.2.11) (37, 38). Maximum-likelihood phylogenies were generated using MEGA 6.0 using the Tamura-Nei model and complete deletion with 100× bootstrapping (39).
Analysis of SAGs and metagenome assemblies.
Assembled contigs meeting the following conservative criteria were uploaded to RAST (40) for gene calling and KAAS (41) for gene annotation and metabolic mapping: (i) sequences between 500 and 2,000 bp in length with any BLASTN matches with an E value lower than 1E−30 (representing small contigs containing confident gene matches), and (ii) sequences ≥2,000 bp in length with any BLASTN matches to the Calescamantes SAG coassembly with an E value of lower than 1E−50 (to keep only high-quality contigs).
The relative abundance of Calescamantes populations was calculated by mapping the sequence reads to the final assemblies using the Burrows Wheel Aligner (42) at default settings and counting the number of reads that were mapped from each site. Genome alignments were conducted on assembled contigs with the software package MAUVE using default settings (43) (see Fig. S2 in the supplemental material). For all assemblies, open reading frames were identified using MetaGeneMark (44) and Prodigal (45), and SCMs were identified as described by Rinke et al. (4) to estimate genome completeness and binning fidelity. KEGG ortholog identifiers (KO numbers) obtained from RAST-identified gene regions of assembled metagenome bins were used to construct metabolic pathway maps in iPath2 (46). PFAM domains of proteins involved in bacterial outer membrane assembly (47) were identified in RAST-annotated metagenome bins using hmmsearch (HMMER v3.1b1; http://hmmer.janelia.org/). The average nucleotide identity (ANI) for SAGs and metagenomic assemblies was calculated using the Kostas Konstantinidis laboratory's ANI calculator (http://enve-omics.ce.gatech.edu/ani/) under default settings (48).
RESULTS
Read-based binning of Calescamantes in GBS.
The analysis pipeline for binning metagenome reads based on nucleotide trimer frequency using the MLP approach is shown in Fig. 1. Calescamantes SAG nucleotide trimer frequencies from GBS were similar to those of an archaeal population (Geoarchaeota) present in GBS at ∼5% of the population (average Pearson correlation of ∼0.92), complicating the recovery of Calescamantes sequences from the metagenome (4, 21, 49). This coincidental similarity in nucleotide frequency between these distant taxa confounded binning by principal component analysis alone. However, the availability of multiple SAGs from both Calescamantes and Geoarchaeota allowed us to train the MLP algorithm for confident bin assignment of the metagenome reads. The MLP classified 148,637 of 1,203,155 reads in the metagenome, and BLASTN separately identified an additional 14,098 nonredundant reads, for a total of 162,735 GBS metagenomic reads belonging to the Calescamantes population. After quality-control filtering, the SPAdes assembly of the bin containing reads identified by MLP and BLASTN consisted of 319 contigs assembled from 50,774 sequence reads. Reads that were included in this assembly totaled 4.2% of the metagenome, whereas Calescamantes had a relative abundance of 1.21% in a previous 16S rRNA gene analysis of the same DNA sample from which the GBS metagenome was derived (23). It is likely that the abundance based on metagenome reads more closely reflects the abundance of Calescamantes, because differences in ribosomal copy number and amplification bias can skew organisms' relative abundances (50).
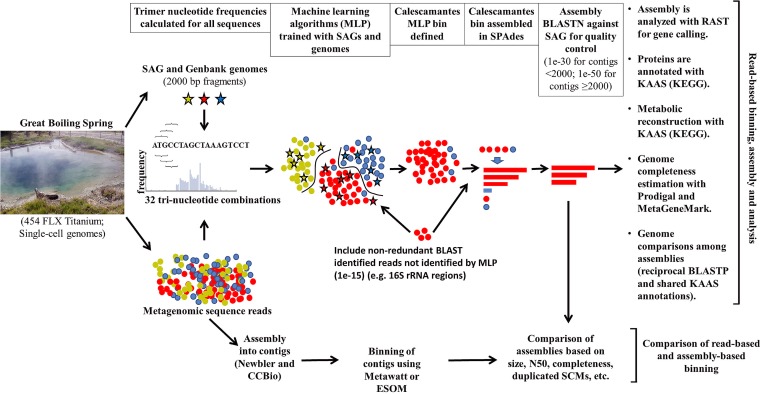
FIG 1.
Flow chart of the progression of data analysis, showing (left to right) sample collection; sequencing of single-cell genomes and reference GenBank genomes (stars) and metagenomes (circles); nucleotide trimer frequency calculation; MLP model training; and read-binning, assembly, and postassembly analyses (annotation, metabolic reconstruction, and estimation of genome completeness). The MLP pipeline (top and right) subsequently was conducted on all geographic sites analyzed in this study.
The Calescamantes MLP assembly from the GBS metagenomic reads binned using SAGs as training models, but without the inclusion of any SAG contigs in the assembly, shared a similar nucleotide trimer composition (average Pearson correlation of >0.98) with the GBS SAG coassembly and had near-identical G+C content (34.1% and 34.2%, respectively). An updated SAG coassembly provided by JGI and the MLP assembly were annotated on the RAST online server (40), identifying 2,205 coding regions in the SAG coassembly, 46% of which were classified as hypothetical proteins, and 2,721 coding regions in the GBS MLP assembly, of which 47% were classified as hypothetical proteins (Table 1 and Fig. 2A). The higher number of gene regions in the MLP assembly compared to that of the SAG coassembly at GBS most likely is due to the presence of approximately 200 more contigs in the MLP assembly, which typically contained gene fragments on their discontinuous ends, supported by the fact that 64% of these additional truncated genes are duplicated elsewhere in the MLP assembly. The GBS MLP assembly contained 367 predicted genes that were not present in the SAG coassembly, although it is important to note that the MLP (and any metagenome) assembly is an assemblage of population-level diversity in the environmental sample. In contrast, there were 292 predicted coding regions in the SAG coassembly that were not detected in the GBS MLP assembly. The GBS MLP assembly contained several predicted coding regions that closed some apparent metabolic gaps in the annotated SAG coassembly (namely, cases where some, but not all, genes in a pathway were present), including genes encoding proteins involved in fatty acid biosynthesis (fabZ), NADH-quinone oxidoreductase subunit F (nuoF), and DNA gyrase subunit A (gyrA). Likewise, the SAG coassembly identified metabolic gaps in the GBS MLP assembly: protein biosynthesis gene 3-dehydroquinate dehydratase I (aroE), flagellar basal body rod gene (flgB), and glycyl-tRNA synthetase (glyQ). Both the MLP and SAG assemblies contained ribosomal proteins that were not detected in the other assembly (see Table S5 in the supplemental material).
TABLE 1.
Statistics for the Calescamantes SAG coassembly and GBS, Gongxiaoshe Spring, Octopus Spring, and Bison Pool metagenome MLP assemblies
| Parameter | Value for: |
||||
|---|---|---|---|---|---|
| GBS SAG coassembly | GBS | Gongxiaoshea | Octopus Springa | Bison Pool | |
| Total no. of reads | NAc | 1,203,155 | 9,408,748 | 322,871,342 | 164,182 |
| MLP binned reads (no.) | NA | 162,735 | 641,824 | 8,525,286 | 4,772 |
| Assembly size (Mbp) | 2.25 | 2.21 | 2.27 | 2.15 | 1.22 |
| No. of contigs | 138 | 319 | 294 | 799 | 727 |
| Largest contig (bp) | 103,150 | 49,870 | 46,853 | 26,269 | 6,728 |
| N50 | 25,634 | 11,262 | 16,080 | 4,260 | 1,749 |
| G+C content | 34.1 | 34.2 | 34.7 | 34.2 | 38.6 |
| No. of coding sequences (RAST) | 2,205 | 2,721 | 2,131 | 2,040 | 1,869 |
| Assigned KO no. (KAAS) | 851 | 827 | 809 | 695 | 367 |
| No. (%) of hypothetical genes (RAST) | 945 (46) | 1,270 (47) | 1,056 (53) | 789 (40) | 1,163 (62) |
| tRNAs (RAST) | 49 | 46 | 48 | 53 | 23 |
| tRNAs (KAAS) | 49 | 44 | 45 | 53 | 23 |
| 23S | 1 | 1 | 1 | 2 | 0 |
| 16S | 1 | 1 | 1 | 2 | 1 |
| 5S | 1 | 1 | 1 | 0 | 0 |
| SCM duplicatesb | 5 | 0 | 3 | 15 | 8/6 |
| Estimated completenessb (%) | 94 | 89/90 | 99 | 87 | 30/31 |
Gongxiaoshe and Octopus Spring metagenomes contained paired-end sequences.
Conserved gene copy number and genome completeness were estimated with MetaGeneMark and Prodigal using single-conserved markers (SCMs) described by Rinke et al. (4). If the values differed between programs, both values are given.
NA, not applicable.
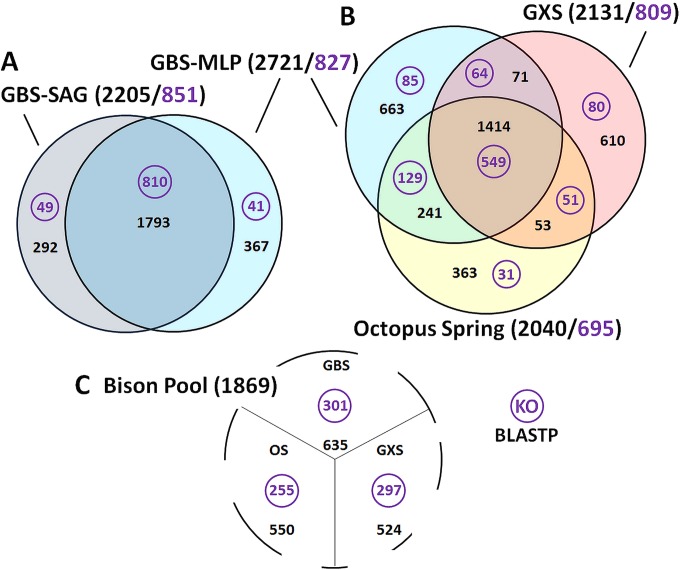
FIG 2.
Comparison of Calescamantes assemblies. The total numbers of predicted proteins (black) and KEGG orthologs (KO) (purple) are shown in parentheses. Best reciprocal BLASTP hits using an E value cutoff of ≤1E−15 were calculated between all analyzed assemblies. (A) Comparison of predicted proteins in the GBS SAG coassembly (dark gray) compared to the GBS MLP metagenome assembly (light blue). (B) Venn diagram of GBS MLP metagenome assembly (light blue), the Octopus Spring MLP metagenome assembly (yellow), and the Gongxiaoshe MLP metagenome assembly (red). Unique genes had no BLAST hit to any other assembly. (C) Number of shared protein-coding regions between the incomplete Calescamantes assembly from Bison Pool and other assembled genomes (E value of ≤1E−15).
Assessment of MLP-based binning methods compared to traditional binning tools.
The Calescamantes GBS MLP assembly using MLP read-binning methods developed for this work, combined with unique BLASTN-identified reads, resulted in 319 assembled contigs, with a largest contig of 49,870 bp and an estimated genome completeness of 89 to 90% based on SCM content. The GBS MLP assembly without the inclusion of BLASTN-identified reads resulted in a total of 528 assembled contigs, with a largest contig of 30,007 bp and an estimated genome completeness of 83% (Table 2). Additionally, assembly-based binning of GBS contigs, using MLP and additional contigs identified by BLASTN, resulted in 435 contigs, with a largest contig of 44,489 bp and an estimated genome completeness of 90 to 91% based on SCM content, although the assembly contained 5 duplicated SCM genes not present as duplicates in the read-based bin assembly.
TABLE 2.
SAG coassembly statistics compared to those of different binning methods on Calescamantes populations from GBS
| Binning method | N50 | Contig size (bp) |
Genome size (Mbp) | No. of contigs | %cova |
No. of SCMs at >1 copy | |||
|---|---|---|---|---|---|---|---|---|---|
| Maximum | Minimum | Avg contig | M | P | |||||
| SAG | 40,523 | 204,003 | 320 | 16,262 | 2.24 | 138 | 95 | 93 | 5 |
| MLP plus BLASTb | 11,262 | 49,870 | 513 | 6,937 | 2.21 | 319 | 90 | 89 | 0 |
| MLP contig bin | 11,223 | 44,489 | 129 | 5,375 | 2.33 | 435 | 90 | 90 | 5 |
| MLPc | 5,510 | 30,007 | 135 | 3,843 | 2.03 | 528 | 83 | 83 | 0 |
| ESOM plus BLASTb | 4,968 | 22,905 | 100 | 1,867 | 2.19 | 1,177 | 84 | 83 | 0 |
| MaxBin plus BLASTb | 4,057 | 22,905 | 103 | 2,509 | 2.13 | 805 | 84 | 83 | 3 |
| Metawatt | 4,023 | 22,905 | 900 | 3,076 | 2.04 | 138 | 81 | 81 | 0 |
| BLASTN | 4,109 | 14,181 | 501 | 2,832 | 1.89 | 668 | 79 | 77 | 0 |
Percent total genome coverage (%cov) was estimated with MetaGeneMark (M) and Prodigal (P) using single conserved markers (SCMs) as described by Rinke et al. (4).
BLAST indicates the inclusion of nonredundant SAG-identified BLAST reads in the Calescamantes bin prior to assembly.
MLP-only assembly without including nonredundant SAG-identified BLAST reads in the assembly.
Binning of assembled GBS metagenome contigs was also performed using several well-known methods in order to assess the fidelity of MLP binning. Metawatt binning, based on contig tetramer word frequency (10), yielded a Calescamantes bin consisting of 138 contigs (the largest contig was 22,905 bp) and an estimated genome completeness of 81%. Emergent self-organizing maps (ESOM) (9), also based on tetramer word frequency, identified 313 metagenome contigs that clustered with the Calescamantes SAG. The largest contig identified was 22,905 bp (the same contig as that from the Metawatt bin), and the genome was estimated to be 79% complete. The confident inclusion of shorter-length contigs was a reason for increased contig numbers in the MLP binning, as assembly-based binning methods typically limit binning to contigs over 2 kbp. Finally, the BLASTN assembly, assembled using reads identified only by their BLASTN homology to the Calescamantes SAG (E values of ≤1E−15), consisted of 668 contigs, with a largest contig of 14,181 bp and an estimated genome completeness of 77%.
Binning of Calescamantes populations in metagenomes from YNP and China.
The MLP read-binning approach was used to recruit Calescamantes reads from geographically distinct geothermal springs where Calescamantes populations were identified. The Calescamantes genome assembly based on MLP-binned and nonredundant BLASTN reads from Gongxiaoshe consisted of 294 contigs totaling 2.26 Mbp, with a longest contig of 46,853 bp and an estimated genome completeness of 99%. RAST identified a total of 2,131 predicted coding regions, 53% of which were classified as hypothetical and 593 of which were unique to this assembly (Fig. 2B). Despite the high number of genes shared with other Calescamantes assemblies, the Gongxiaoshe MLP assembly was nonsyntenous compared with the GBS assemblies (see Section I and Fig. S2 in the supplemental material). The more complete assembly was likely the result of the higher relative abundance of Calescamantes populations at Gongxiaoshe (5.8%), deeper sequencing, and the absence of Geoarchaeota in this spring.
MLP-binned and nonredundant BLASTN Calescamantes reads were also assembled from Bison Pool and Octopus Spring, Yellowstone National Park. A total of 4,772 Sanger reads from Bison Pool were assembled into 727 contigs, with a longest contig of 6,728 bp. The final genome assembly was estimated to be ∼31% complete. The small assembly size likely was due to the limited sequence depth (162,984 reads averaging between 889 and 1,111 bp in length) of the Sanger metagenome and the lower relative abundance of Calescamantes in Bison Pool (1.8%), yielding only an estimated 1- to 2-fold sequencing depth for this bin. RAST identified a total of 1,869 predicted coding regions, 62% of which were classified as hypothetical and 53 of which were unique to this assembly (Fig. 2C).
The Octopus Spring MLP binning and assembly yielded a draft genome larger than 4 Mb in length, containing multiple duplicate SCMs, indicating that this assembly was composed of two distinct but closely related Calescamantes populations (Table 1). A comparison of 16S rRNA gene sequences recovered by BLASTN from the Octopus metagenome to the 16S rRNA gene in the corresponding assembled genome exhibited a bimodal pattern of sequence identity (see Fig. S3 in the supplemental material), lending further evidence to the concept of the presence of two very closely related Calescamantes populations in Octopus Spring. To obtain a higher-fidelity assembly, contigs from the Calescamantes populations in Octopus Spring were separated by top BLASTN hits to the predicted gene-coding regions identified in the GBS SAG. The resulting Octopus Spring assembly consisted of 799 contigs, with a largest contig of 26,269 bp, and was estimated to be 87% complete. RAST identified a total of 2,040 predicted coding regions, 40% of which were classified as hypothetical proteins, and 363 genes were unique to this assembly, 94% of which were classified as hypothetical. However, the Octopus Spring assembly still contained duplicates of 15 SCMs, as well as two distinct 16S and 23S rRNA regions, indicating that this genome is most likely an amalgam of the two genotypes (Table 1). Efforts at separating the two genotypes (by k-means and single-nucleotide polymorphism pattern separation) were confounded by the relatively low abundance of Calescamantes (∼4% of the community) and the lack of available site-specific SAG references for training the MLP algorithm.
16S rRNA phylogenetic analysis.
The Calescamantes 16S rRNA gene sequences represented a distinct phylum-level lineage that diverged early from other Bacteria (Fig. 3) along with the Aquificae, and it shared <80% nucleotide identity with other phyla (4). This branching order is consistent with a previous analysis of Calescamantes phylogeny using 38 concatenated protein-coding marker genes, with Aquificae branching closest to the Calescamantes (4).
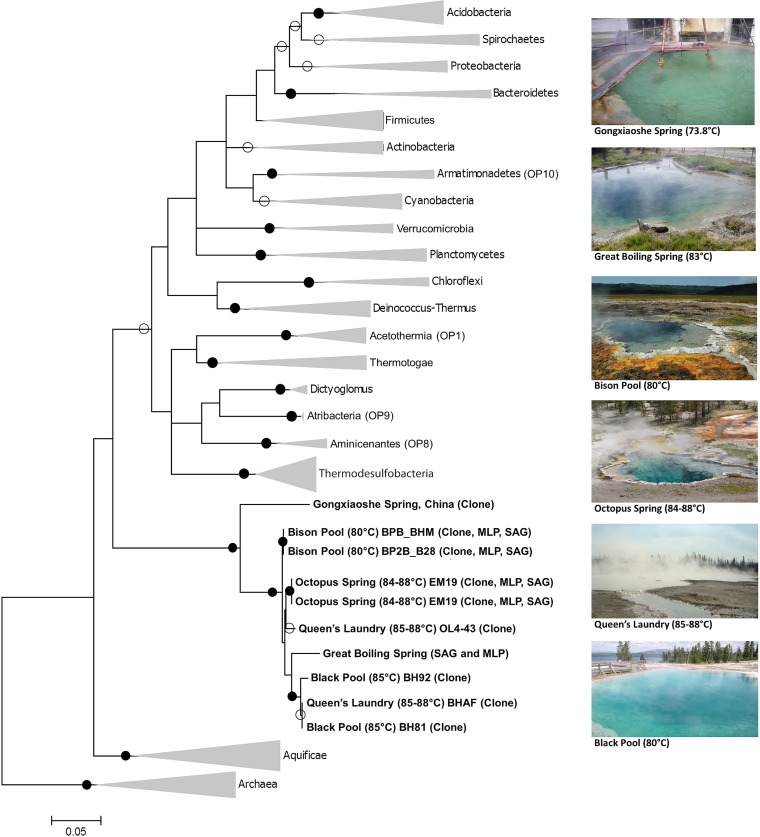
FIG 3.
Maximum-likelihood phylogeny based on partial 16S rRNA genes (953 bp) representing all sequenced members within the Calescamantes phylum (≥85% identity), as well as members of other well-known bacterial phyla. Hot spring locations from which Calescamantes sequences were obtained are shown on the right. Black dots indicate bootstrap values of ≥90, and white dots indicate values of ≥50. The SAG coassembly and MLP assembly 16S rRNA gene sequences are shown together on the same branch, as they were 100% identical. 16S rRNA gene sequences from Cole et al. (23), which branch as a close sister to the SAG/MLP 16S rRNA gene (99.2 to 100% identical), were not included in the phylogeny due to their short length (200 to 400 bp). The scale bar indicates 0.05 substitutions per site. Sequences originating from MLP assemblies (MLP), SAGs, and/or amplified 16S rRNA gene clone libraries (Clone) are indicated in parentheses.
All nearly full-length 16S rRNA gene sequences for Calescamantes were analyzed to determine the evolutionary relationship between Calescamantes from different geographic locations (Fig. 3). BLASTN of the Calescamantes SAG coassembly 16S rRNA gene sequence against GenBank and IMG yielded only five other unique sequences (eight total) at >85% nucleotide identity, in addition to the metagenome sequences discussed in this work, all of which were recovered from circumneutral to alkaline terrestrial geothermal springs (14, 51–53). The recovery of 16S and 23S rRNA gene sequences from the GBS metagenome was accomplished using BLASTN, and assembled full-length rRNA sequences were 100% identical to the SAG coassembly (see Section II in the supplemental material). Additionally, 16S rRNA gene sequences from both the GBS MLP assembly and the SAG coassembly were 99.2 to 100% identical to 16S rRNA gene tags (200 to 400 bp in length) identified by Cole et al. (23), although these reads were omitted from the tree due to their short length. The 16S rRNA gene sequence from the Gongxiaoshe metagenomic assembly was the most divergent sequence in the Calescamantes clade, branching basally to the North American sequences and having 90% nucleotide identity to sequences recovered from GBS. Calescamantes 16S rRNA gene sequences recovered from hot springs in Yellowstone generally clustered by location, with the Great Basin sequences forming a monophyletic clade embedded within the Yellowstone cluster.
Average nucleotide identity also supported the Calescamantes phylogenetic structure, as GBS SAG and MLP assemblies shared 99.5% ANI, while all other genomic assemblies shared ANIs below the suggested species-level cutoff (95%) (see Table S2 in the supplemental material) (54, 55). Calescamantes populations at Octopus Spring and Bison Pool were the most closely related (92.5% ANI) and shared 85.5% and 88.6% ANI with the GBS population, respectively, suggesting that they are in the same genus (55). The population in Gongxiaoshe shared 76.4% ANI with the GBS SAG coassembly, with too few hits to accurately calculate ANI with all other assemblies (48). The low ANI and 16S rRNA gene identity to North American genotypes below median interfamily values (92.2%) suggests the population at Gongxiaoshe represents a separate family (56).
Calescamantes core metabolism and genomic properties.
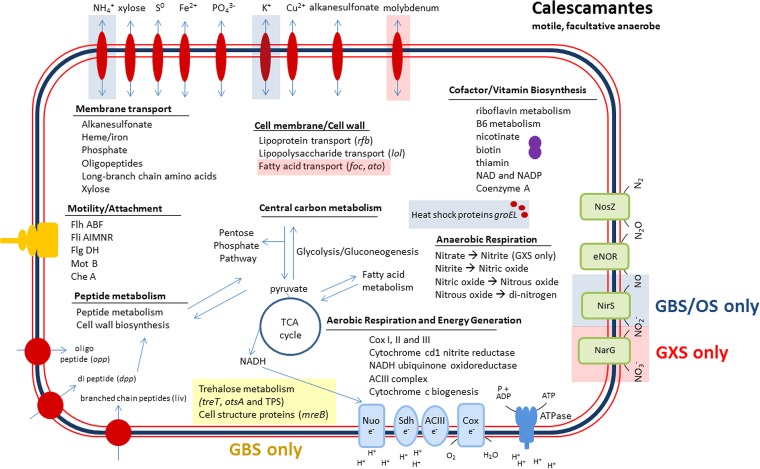
The Calescamantes assemblies are predicted to code for a complete glycolytic pathway, tricarboxylic acid (TCA) cycle (see Table S3 in the supplemental material), and pentose phosphate pathway (Fig. 4; also see Fig. S4 and S5). Proteins predicted to be involved in aerobic respiration included components of the NADH dehydrogenase complex I (nuo), succinate/fumarate dehydrogenase complex II (sdh), and the cytochrome c oxidase complex IV (cox). Whereas bc complex III proteins were not annotated, an alternative complex III (ACIII) that performs the same function, despite being evolutionarily distinct, was identified (57). Genes encoding an A-type oxygen reductase and all but an epsilon subunit of a putative F-type ATP synthase were also annotated. The Gongxiaoshe assembly contained nitrate reductase (narG) and nitrous oxide reductase (nosZ) genes, but nitrite reductase (nirS or nirK) was not identified. North American assemblies contained putative oxygen limitation-sensing genes (fixL), as well as nitrite reductase (nirS), nitric oxide reductase (eNOR family), and nitrous oxide reductase (nosZ) genes. The nirS gene was predicted to encode an enzyme using a heme-heme binding site (58). nosZ and the eNOR gene both contained copper-copper (CUA/CUB) binding sites (59, 60). The putative eNOR gene was not closely related to anything in NCBI GenBank, with a closest BLAST hit to Hydrogenobacter thermophilus TK-6 (E value of 1E−22). North American assemblies did not contain narG. The gene encoding nitric oxide reductase (norB), catalyzing the reduction of nitric oxide to nitrous oxide, was not detected in any of the SAG or metagenomic assemblies, although the eNOR gene, an energy-conserving alternative nitric oxide reductase, was identified in both the GBS and Gongxiaoshe assemblies (61).
FIG 4.
Schematic diagram of the core metabolic potential of Calescamantes populations analyzed from GBS, GXS, and Octopus Spring (OS). GBS-specific genes are shaded green, GBS- and OS-specific genes are shaded blue, and GXS-specific genes are shaded red.
The SAG coassembly and the GBS, Gongxiaoshe, and Octopus Spring metagenomic assemblies all code for complete or near-complete pathways for purine and pyrimidine nucleotide metabolism, fatty acid biosynthesis and degradation, peptidoglycan and cell membrane biosynthesis, and crucial genes involved in the cell cycle (e.g., ftsZ). Genes predicted to encode ABC transporters involved in lipopolysaccharide and lipoprotein transport across the cytoplasmic membrane used for outer membrane and lipopolysaccharide (LPS) assembly were present (e.g., rfb, lbtB and lol), as were other outer membrane proteins typically associated with Gram-negative (diderm) organisms (see Table S4 in the supplemental material) (47). Genes involved in DNA replication, nucleotide excision and mismatch repair, and homologous recombination were also present (e.g., DNA polIII, uvrA, mutA, and recA). With the exception of assemblies from Bison Pool, putative genes encoding all tRNA synthetases and at least one tRNA for each amino acid were identified in all assemblies.
Calescamantes contained near-complete pathways for amino acid biosynthesis and peptide ABC transporters (e.g., pst and liv) and the putative capability to import long branched-chain amino acids and oligopeptides (e.g., opp). These transport proteins could be the primary mechanism for carbon uptake in these organisms. Besides being a probable electron donor for respiration, peptides and amino acids could also be an important resource for anabolic reactions, as no key enzymes for autotrophic metabolism were identified (e.g., RubisCO, ATP citrate lyase, pyruvate:ferredoxin oxidoreductase, acetyl-coenzyme A synthase, or carbon monoxide dehydrogenase). Calescamantes also contained a putative d-xylose transporter (xyl) but no other known xylose utilization enzymes, suggesting that any xylose utilization would have to be through novel means. Additional membrane transport proteins putatively identified in Calescamantes assemblies include those specific to heme (ccm), potassium (kdp), copper (cus), phosphate (pst), and alkanesulfonate (ssu) uptake, and all assemblies had complete assimilatory sulfate reduction pathways (cys), indicating the ability to transport and anabolize trace elements and cofactors necessary for enzymatic activity.
The Calescamantes populations also appear capable of chemotactic motility, as they code for near-complete bacterial chemotaxis and flagellar assembly systems. Putative genes were identified for methyl-accepting chemotaxis proteins (mcp) and two-component sensor kinases (che), flagellum motor switch genes (fli), ATP motor proteins (mot), and chemotaxis motor proteins (mot). In addition, multiple flagellar structural assembly genes were identified (fli, flh, and flg), indicating that these organisms have the capability to both sense and respond to a changing chemical environment, although fliC propeller filament genes were absent from all annotated genome assemblies. Annotated genes are listed in Tables S5A to E in the supplemental material.
Gene content differences between geographically separated populations.
Many of the coding regions in the Gongxiaoshe and GBS MLP assemblies were hypothetical (53 and 47%, respectively), and most genes unique to the Calescamantes populations in each location (610 and 663, respectively) were also hypothetical (92 and 94%, respectively). However, the Gongxiaoshe metagenome assembly did contain unique genes with annotated functions that may be ecologically important. Although the nosZ and eNOR genes were present in the Gongxiaoshe assembly, nirS was not identified, suggesting that these populations lack the ability to reduce nitrite to nitric oxide. However, narG (large α subunit) was annotated, suggesting that the Gongxiaoshe populations have the capability to reduce nitrate to nitrite, supported by the presence of site-specific genes that code for the transport of molybdenum (modA and modB), which is used as a cofactor in nitrate reductase (NarG) (62, 63), although narH (small β subunit) was not identified. Additionally, formate and short-chain fatty acid transporters were identified (foc and ato) as well as an acetate kinase (ack), indicating that the Calescamantes populations in Gongxiaoshe have more capabilities for carbon assimilation and metabolism.
The North American assemblies (GBS, Bison Pool, and Octopus Spring) all contained nirS as well as nosZ, which clustered phylogenetically with sequences from members of the Aquificae (see Fig. S6 in the supplemental material). The GBS assembly also contained the eNOR gene, which reduces nitric oxide to nitrous oxide, completing the denitrification pathway from nitrite to dinitrogen (61). In addition to nirS, GBS and Octopus Spring assemblies also contained uniquely annotated genes for glutamate symport (glt), carbon starvation (cst and dps), heat shock chaperone proteins (groEL), protein metabolism (panD), alcohol dehydrogenase (adh), potassium uptake (kup), and ammonia transport (amt), which could give insight into important environmentally specific metabolisms; the spring at Gongxiaoshe, for instance, is cooler in temperature and ammonia was not detected (14). Additional trehalose metabolism genes (treT and otsA) were annotated exclusively at GBS, which may enable the use of trehalose as an alternative carbon source for these organisms; however, no trehalose-specific transport proteins were annotated. GBS assemblies also exclusively contained unique CRISPR regions (cas 1 and 3), indicating more viral activity at this spring. The Octopus Spring assembly exclusively contained the cell structure gene mreB, suggesting these Calescamantes are rod-shaped.
DISCUSSION
Calescamantes 16S rRNA gene and protein-coding sequences are representative of a yet-uncultivated bacterial phylum that is very poorly represented in sequence databases (4). To date, Calescamantes 16S rRNA gene sequences have been identified and recovered from only a small number of chemically similar, circumneutral to alkaline hot springs in North America and Tengchong, China (14, 18, 29, 51, 53).
As DNA sequencing technology advances at an extremely rapid pace, it has become possible to assemble nearly complete genomes from uncultured candidate phylum populations in environmental metagenomes. However, even with these advances, small population sizes and assembly artifacts can make analysis difficult for rare community members. Single-cell genomics allows for confident anchoring and binning of metagenomic reads by facilitating the identification and analysis of genomic data from low-abundance populations in diverse environmental metagenomes.
Compared to commonly used postassembly binning methods, the MLP read-binning approach used in this study yielded a slightly more complete genomic assembly from metagenome sequences in each case tested without increasing SCM redundancy or decreasing the N50 value, although SAG-assisted postassembly binning with MLP yielded the most comparable results to read-first binning with MLP. The coordinated analysis of metagenomic assemblies and SAG coassemblies has allowed us to expand upon the limitations of each technique, fill apparent gaps in metabolic pathways, and confidently predict and contrast the metabolic potential of the Calescamantes populations in several geothermal environments, even when the genomes used for MLP training were genetically divergent from the populations in those environments, as was the case at Gongxiaoshe. While 16S rRNA gene sequencing, single-cell genome sequencing, and community metagenomics are informative on their own, the intersection of these sequencing approaches is greater than the sum of their parts. As sequencing continues to expand at exponential rates, it has become increasingly important to adopt a multifaceted approach in order to improve read-binning, assembly, and annotation for the ultimate goal of accurately reconstructing metabolic potential for yet-uncultivated microbial populations from the natural environment. The integrative MLP read-based binning approach represents an additional tool for organism-based, cultivation-independent genomic analysis and can potentially enhance the understanding of uncultivated microbes beyond what can be obtained by any individual approach.
Calescamantes populations in GBS, Yellowstone, and Gongxiaoshe are predicted to be heterotrophic, motile bacteria that have the capability to respire aerobically and incorporate carbon into biomass through protein transport and metabolism (Fig. 4; also see Fig. S4 and S5 in the supplemental material). Additionally, these Calescamantes populations are putative facultative anaerobes, capable of reducing oxidized nitrogen sources in the absence or limitation of oxygen. The sediment environment in GBS is likely to be either hypoxic or anoxic, and the bulk water in GBS has a relatively low oxygen tension (25 to 50 μM), with nitrate and nitrite concentrations of 1.8 to 16 and 0.79 to 10.2 μM, respectively (22). Previous analysis at this spring has shown that chemolithotrophic ammonia oxidation and denitrification are active processes in GBS sediments (64). Although chemolithotrophic nitrite oxidation has not been measured directly in GBS, enrichment cultures for nitrite-oxidizing bacteria (NOB) in GBS and other geothermal springs failed to show evidence of nitrite oxidation above ∼65°C (65), and evidence of NOB above these temperatures is absent from 16S rRNA gene censuses (23, 24, 64) and metagenomes (data not shown). These data are consistent with a model in which ammonia from the source pool is oxidized to nitrite by ammonia-oxidizing archaea in the genus “Nitrosocaldus,” followed by anaerobic respiration of nitrite to nitrous oxide or dinitrogen (22, 64). In addition to the predicted capacity for nitrite reduction to dinitrogen by Calescamantes, Thermus thermophilus isolates from GBS are also capable of reduction of nitrate to nitrous oxide but lack the ability to reduce nitrous oxide to dinitrogen (22). As such, Thermus thermophilus and Calescamantes could be important consumers of the nitrite produced by Nitrosocaldus, where the combined activities of these organisms may result in complete denitrification to dinitrogen. The colocalization of the ACIII complex gene with the nirS gene provides further evidence of the potential ability of Calescamantes to reduce oxidized nitrogen sources. As it is unlikely that the ACIII complex conserves as much energy as the bc complex, the eNOR gene likely conserves more energy in the reduction of nitric oxide to nitrous oxide, which could make up for this loss (57, 61).
Octopus Spring (66) and Gongxiaoshe (14) also harbored abundant populations of Thermus and Nitrosocaldus, making up 10 to 20% and 5 to 10% of the microbial community, respectively, in the two springs (13, 14) and indicating similar cometabolic roles in these communities. The oxygen tension in all studied springs was between 10 and 50 μM at or near the source. Although ammonia and nitrate were below detectable limits at Gongxiaoshe, nitrite was present (∼26 μM). Ammonium and nitrate were present in Octopus Spring (2.3 and 6 μM, respectively) (13), although nitrite was not directly measured. Low ammonium and nitrite levels at Bison Pool (not detected and 0.4 μM, respectively) may contribute to the low abundance of Calescamantes in this spring.
The phylogenetic position of Calescamantes in the 16S rRNA and concatenated protein trees reaffirms its identity as a candidate phylum in the Bacteria, and it is most closely related to the Aquificae and Thermodesulfobacteria (67, 68). Based on the recent recommendation to allow the expansion of Candidatus status to nearly complete genomes conforming to genomic species delineations based on ANI values and other criteria (5), we propose to expand the candidate taxonomy within the phylum Calescamantes (4) to include the lineage inhabiting Gongxiaoshe Spring.
Taxonomy.
We propose the taxonomic epithet “Candidatus Calescimonas tengchongensis.” The taxonomic description is “Calescimonas” (Cal.es.ci.mo'nas. L. v. calesco, to become warm, grow hot; N.L. n. monas a unit; N.L. n. Calescimonas; a rod-shaped bacterium from an extremely hot environment), “tengchongensis” (teng.chong.en'sis. N.L. fem. adj. tengchongensis of or pertaining to Tengchong County, Yunnan province, China).
Supplementary Material
ACKNOWLEDGMENTS
We thank Carrine Blank and D'Arcy Meyer-Dombard for the use of images of Black Pool hot spring and Queen's Laundry hot spring, respectively. We thank James Hemp for stimulating discussions on the eNOR and ACIII complex.
This work was supported by NASA exobiology grant EXO-NNX11AR78G, U.S. National Science Foundation grant OISE 0968421, and U.S. Department of Energy grant DE-EE-0000716. B.P.H. acknowledges generous support from Greg Fullmer through the UNLV Foundation, and W.S. acknowledges Northern Illinois University for funding. B.P.H and S.K.M. acknowledge support from an Amazon Web Services Education Research Grant award.
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03140-15.
REFERENCES
- 1.Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Torsvik V, Ovreas L, Thingstad TF. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 3.Hugenholtz P, Kyrpides NC. 2009. A changing of the guard. Environ Microbiol 11:551–553. doi: 10.1111/j.1462-2920.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 4.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 5.Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T. 2014. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter.” Extremophiles 18:865–875. doi: 10.1007/s00792-014-0664-7. [DOI] [PubMed] [Google Scholar]
- 6.Gies EA, Konwar KM, Beatty JT, Hallam SJ. 2014. Illuminating microbial dark matter in meromictic Sakinaw Lake. Appl Environ Microbiol 80:6807–6818. doi: 10.1128/AEM.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WE, Song Y, Xu J. 2015. Single cell biotechnology to shed a light on biological “dark matter” in nature. Microb Biotechnol 8:15–16. doi: 10.1111/1751-7915.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Garcia M, Santos F, Moreno-Paz M, Parro V, Anton J. 2014. Unveiling viral-host interactions within the “microbial dark matter.” Nat Commun 5:4542. doi: 10.1038/ncomms5542. [DOI] [PubMed] [Google Scholar]
- 9.Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu WT. 2015. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J doi: 10.1038/ismej.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 11.Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, Gihring TM, Lapidus A, Lin LH, Lowry SR, Moser DP, Richardson PM, Southam G, Wanger G, Pratt LM, Andersen GL, Hazen TC, Brockman FJ, Arkin AP, Onstott TC. 2008. Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- 12.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, Banfield JF. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol 10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inskeep WP, Jay ZJ, Tringe SG, Herrgard MJ, Rusch DB, YNP Metagenome Project Steering Committee and Working Group Members . 2013. The YNP Metagenome Project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front Microbiol 4:67. doi: 10.3389/fmicb.2013.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou W, Wang S, Dong H, Jiang H, Briggs BR, Peacock JP, Huang Q, Huang L, Wu G, Zhi X, Li W, Dodsworth JA, Hedlund BP, Zhang C, Hartnett HE, Dijkstra P, Hungate BA. 2013. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province, China, using 16S rRNA gene pyrosequencing. PLoS One 8:e53350. doi: 10.1371/journal.pone.0053350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. 2012. The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol 3:410. doi: 10.3389/fmicb.2012.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrighton KC. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 17.Klatt CG, Inskeep WP, Herrgard MJ, Jay ZJ, Rusch DB, Tringe SG, Parenteau MN, Ward DM, Boomer SM, Bryant DA, Miller SR. 2013. Community structure and function of high-temperature chlorophototrophic microbial mats inhabiting diverse geothermal environments. Front Microbiol 4:106. doi: 10.3389/fmicb.2013.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swingley WD, Meyer-Dombard DR, Shock EL, Alsop EB, Falenski HD, Havig JR, Raymond J. 2012. Coordinating environmental genomics and geochemistry reveals metabolic transitions in a hot spring ecosystem. PLoS One 7:e38108. doi: 10.1371/journal.pone.0038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, Grossman AR, Bhaya D, Cohan FM, Kuhl M, Bryant DA, Ward DM. 2011. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J 5:1262–1278. doi: 10.1038/ismej.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepanauskas R. 2012. Single cell genomics: an individual look at microbes. Curr Opin Microbiol 15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Kozubal MA, Romine M, Jennings R, Jay ZJ, Tringe SG, Rusch DB, Beam JP, McCue LA, Inskeep WP. 2013. Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J 7:622–634. doi: 10.1038/ismej.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedlund BP, McDonald AI, Lam J, Dodsworth JA, Brown JR, Hungate BA. 2011. Potential role of Thermus thermophilus and T. oshimai in high rates of nitrous oxide (N2O) production in ~80°C hot springs in the US Great Basin. Geobiology 9:471–480. doi: 10.1111/j.1472-4669.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 23.Cole JK, Peacock JP, Dodsworth JA, Williams AJ, Thompson DB, Dong H, Wu G, Hedlund BP. 2013. Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. ISME J 7:718–729. doi: 10.1038/ismej.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP. 2009. Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13:447–459. doi: 10.1007/s00792-009-0230-x. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Leung HC, Yiu SM, Chin FY. 2012. MetaCluster 5.0: a two-round binning approach for metagenomic data for low-abundance species in a noisy sample. Bioinformatics 28:i356–i362. doi: 10.1093/bioinformatics/bts397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Hu H, Li X. 2015. MBBC: an efficient approach for metagenomic binning based on clustering. BMC Bioinformatics 16:36. doi: 10.1186/s12859-015-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHardy AC, Martin HG, Tsirigos A, Hugenholtz P, Rigoutsos I. 2007. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods 4:63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 28.Ultsch A, Moerchen F. 2005. ESOM-Maps: tools for clustering, visualization, and classification with emergent SOM. Technical report 46. Department of Mathematics and Computer Sciences, University of Marburg, Marburg, Germany. [Google Scholar]
- 29.Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PS, Scholz MB, Lo CC, Raymond J, Quake SR, Hedlund BP. 2013. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun 4:1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witten IH, Frank E, Hall MA. 2011. Data mining: practical machine learning tools and techniques, 3rd ed Morgan Kaufmann, Burlington, MA. [Google Scholar]
- 31.Dodsworth JA, Gevorkian J, Despujos F, Cole JK, Murugapiran SK, Ming H, Li WJ, Zhang G, Dohnalkova A, Hedlund BP. 2014. Thermoflexus hugenholtzii gen. nov., sp. nov., a thermophilic, microaerophilic, filamentous bacterium representing a novel class in the Chloroflexi, Thermoflexia classis nov., and description of Thermoflexaceae fam. nov. and Thermoflexales ord. nov. Int J Syst Evol Microbiol 64:2119–2127. doi: 10.1099/ijs.0.055855-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang HC, Hickey DA. 2002. Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes. Nucleic Acids Res 30:2501–2507. doi: 10.1093/nar/30.11.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Wu YW, Simmons BA, Singer SW. 25 October 2015. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 35.Wu YW, Tang YH, Tringe SG, Simmons BA, Singer SW. 2014. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. doi: 10.1186/2049-2618-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu W-T. 2015. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9:1710–1722. doi: 10.1038/ismej.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 46.Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. 2011. iPath2.0: interactive pathway explorer. Nucleic Acids Res 39:W412–W415. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutcliffe IC. 2010. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol 18:464–470. doi: 10.1016/j.tim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 49.Guy L, Spang A, Saw JH, Ettema TJ. 2014. “Geoarchaeote NAG1” is a deeply rooting lineage of the archaeal order Thermoproteales rather than a new phylum. ISME J 8:1353–1357. doi: 10.1038/ismej.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kembel SW, Wu M, Eisen JA, Green JL. 2012. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput Biol 8:e1002743. doi: 10.1371/journal.pcbi.1002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blank CE, Cady SL, Pace NR. 2002. Microbial composition of near-boiling silica-depositing thermal springs throughout Yellowstone National Park. Appl Environ Microbiol 68:5123–5135. doi: 10.1128/AEM.68.10.5123-5135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reysenbach AL, Wickham GS, Pace NR. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol 60:2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer-Dombard DR, Swingley W, Raymond J, Havig J, Shock EL, Summons RE. 2011. Hydrothermal ecotones and streamer biofilm communities in the Lower Geyser Basin, Yellowstone National Park. Environ Microbiol 13:2216–2231. doi: 10.1111/j.1462-2920.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- 54.Chun J, Rainey FA. 2014. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol 64:316–324. doi: 10.1099/ijs.0.054171-0. [DOI] [PubMed] [Google Scholar]
- 55.Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 56.Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer KH, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 57.Refojo PN, Teixeira M, Pereira MM. 2012. The alternative complex III: properties and possible mechanisms for electron transfer and energy conservation. Biochim Biophys Acta 1817:1852–1859. doi: 10.1016/j.bbabio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Simon J, Klotz MG. 2013. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta 1827:114–135. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemp J, Gennis RB. 2008. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ 45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 61.Yanyushin MF, del Rosario MC, Brune DC, Blankenship RE. 2005. New class of bacterial membrane oxidoreductases. Biochemistry 44:10037–10045. doi: 10.1021/bi047267l. [DOI] [PubMed] [Google Scholar]
- 62.Hille R. 1996. The mononuclear molybdenum enzymes. Chem Rev 96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 63.Mendel RR. 2007. Biology of the molybdenum cofactor. J Exp Bot 58:2289–2296. doi: 10.1093/jxb/erm024. [DOI] [PubMed] [Google Scholar]
- 64.Dodsworth JA, Hungate B, de la Torre JR, Jiang H, Hedlund BP. 2011. Measuring nitrification, denitrification, and related biomarkers in terrestrial geothermal ecosystems. Methods Enzymol 486:171–203. doi: 10.1016/B978-0-12-381294-0.00008-0. [DOI] [PubMed] [Google Scholar]
- 65.Edwards TA, Calica NA, Huang DA, Manoharan N, Hou W, Huang L, Panosyan H, Dong H, Hedlund BP. 2013. Cultivation and characterization of thermophilic Nitrospira species from geothermal springs in the US Great Basin, China, and Armenia. FEMS Microbiol Ecol 85:283–292. doi: 10.1111/1574-6941.12117. [DOI] [PubMed] [Google Scholar]
- 66.Bateson MM, Thibault KJ, Ward DM. 1990. Comparative-analysis of 16S ribosomal-RNA sequences of Thermus species. Syst Appl Microbiol 13:8–13. doi: 10.1016/S0723-2020(11)80173-6. [DOI] [Google Scholar]
- 67.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.