Abstract
Listeria monocytogenes is a saprophytic bacterium that thrives in diverse environments and causes listeriosis via ingestion of contaminated food. RsbX, a putative sigma B (σB) regulator, is thought to maintain the ready state in the absence of stress and reset the bacterium to the initial state in the poststress stage in Bacillus subtilis. We wondered whether RsbX is functional in L. monocytogenes under different stress scenarios. Genetic deletion and complementation of the rsbX gene were combined with survival tests and transcriptional and translational analyses of σB expression in response to stresses. We found that deletion of rsbX increased survival under secondary stress following recovery of growth after primary stress or following stationary-phase culturing. The ΔrsbX mutant had higher expression of σB than its parent strain in the recovery stage following primary sodium stress and in stationary-phase cultures. Apparently, increased σB expression had contributed to improved survival in the absence of RsbX. There were no significant differences in survival rates or σB expression levels in response to primary stresses between the rsbX mutant and its parent strain during the exponential phase. Therefore, we provide clear evidence that RsbX is a negative regulator of L. monocytogenes σB during the recovery period after a primary stress or in the stationary phase, thus affecting its survival under secondary stress.
INTRODUCTION
Listeria monocytogenes causes listeriosis in humans, mostly due to consumption of contaminated foods. As a saprophytic bacterium thriving in diverse environments, L. monocytogenes can survive and grow over a wide range of environmental conditions, including temperatures from −0.4 to 45°C, pH as low as 2.5, and high osmolarity (10% to 20% NaCl) (1–4). The general stress-responsive alternative sigma factor sigma B (σB), which was first identified in Bacillus subtilis (5), plays a pivotal role in its resistance to environmental stresses (6, 7).
σB is coexpressed with seven of its principal regulators (regulators of sigma B rsbR, rsbS, rsbT, rsbU, rsbV, rsbW, and rsbX) in B. subtilis (8) and L. monocytogenes (9–11). Partner switching upon phosphorylation and dephosphorylation is the main regulatory mechanism of this protein cluster in response to stresses. This has been studied mostly in B. subtilis (12–14) and seldom in L. monocytogenes. In unstressed B. subtilis, RsbW sequesters σB into an association that prevents it from interacting with RNA polymerase. RsbV is dephosphorylated by RsbU or RsbP in response to environmental or metabolic stress, respectively (15, 16). The dephosphorylated RsbV is capable of competing for RsbW, resulting in σB liberation. The upstream proteins RsbT, RsbS, and RsbR form a complex called the “stressosome” in B. subtilis (17–19). Environmental stresses stimulate the kinase activity of RsbT, which can then be released from the stressosome available to activate RsbU (20).
However, the interactions of these regulatory proteins in L. monocytogenes under stresses are not well understood, although there are a number of studies reporting the functional activity of some of the Rsb proteins as the turn-on mechanism upon stressing (6, 7). The RsbT and RsbV proteins in L. monocytogenes are considered to convey environmental and energy stress signals to σB (21), and RsbU is involved in the response to physical and antibiotic stresses (22). In B. subtilis, RsbX is considered a feedback phosphatase for resetting the stressosome poststress or for maintenance of the ready state in the absence of stress (14). Such a turn-off function could be important for conservation of energy and recovery of homeostasis of the bacteria upon withdrawal of the stressing factors or in transition from stationary phase to a new culture environment. This is because the bacteria have to mobilize their resources and energy for survival during stress or at different growth phases (23, 24). We wondered whether RsbX is functional in L. monocytogenes under stress conditions. We also hypothesized that L. monocytogenes RsbX might play a turn-off role in keeping the stressed state under check soon after removal of the stressing factor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
L. monocytogenes reference strain 10403S was used as the wild-type strain. The ΔsigB mutant was constructed and kept in our laboratory (25). Escherichia coli DH5α was employed as the host strain for plasmids pMD18-T (TaKaRa, Dalian, China), pET30a (+) (Merck), pERL3 (26), and pKSV7 (9, 27). L. monocytogenes was cultured in brain heart infusion (BHI) medium (Oxoid, Hampshire, England). E. coli DH5α and Rosetta (DE3) were grown at 37°C in LB broth (Oxoid). Stock solutions of ampicillin (50 mg/ml), erythromycin (30 mg/ml), kanamycin (50 mg/ml), and chloramphenicol (50 mg/ml) were added to the medium, where appropriate, at the required levels.
Construction of deletion mutants.
A homologous recombination strategy with splicing by overlap extension (SOE)-PCR was used for in-frame deletion to construct the rsbX deletion mutant according to the protocol described previously (27, 28). Genomic DNA of L. monocytogenes 10403S was extracted as described previously (29, 30). SOE-PCR primers were used to amplify the homologous arms upstream and downstream of rsbX (Table 1). The resulting product with deletion of rsbX was cloned into the temperature-sensitive shuttle vector pKSV7 and transformed into E. coli DH5α. After confirmation by sequencing, the recombinant vector containing the target gene deletion cassette was electroporated into L. monocytogenes 10403S (28). Transformants were selected on BHI agar plates containing chloramphenicol (10 μg/ml). A single transformant was serially passaged at a nonpermissive temperature (41°C) in BHI-chloramphenicol to promote chromosomal integration, which was confirmed by PCR. A single colony with chromosomal integration was successively passaged in BHI without chloramphenicol at a permissive temperature (30°C) and screened for loss of chloramphenicol resistance (31). Allelic exchange mutagenesis was confirmed by PCR and DNA sequencing. The mutant strain was designated the ΔrsbX mutant.
TABLE 1.
PCR primers used in this study
| Name | Sequence (5′→3′)a | Purpose |
|---|---|---|
| rsbX-a | CGGAATTCGTAGAGTCCATCGCCCGAA | Construction of rsbX null mutant |
| rsbX-b | TTACTCCACTTCCTCATTCTGCAAC | |
| rsbX-c | AATGAGGAAGTGGAGTAACAAAAACACC | |
| rsbX-d | CGGGATCCATCATTCCGGCAACAAGTAAATCTTGG | |
| rsbX-up | ATGGCGATCAAGACGCCCA | Screening of positive clones of rsbX null mutant |
| rsbX-d | CGGGATCCATCATTCCGGCAACAAGTAAATCTTGG | |
| rsbX-w | CGGGATCCTATGGTTCAGCAGGAC | Complementation of rsbX deletion |
| rsbX-x | ATTCAACTGCCTTGTTCATCACTTCACCCCATCTAAT | |
| rsbX-y | ATGAACAAGGCAGTTGAATCAAATAATTTATTTGTATTT | |
| rsbX-z | CGAGCTCTTATTCCGGAAATTTCCC | |
| sigB-RT-F | GGTGTCACGGAAGAAGAAG | qRT-PCR |
| sigB-RT-R | TCCATCATCCGTACCACC | |
| gyrB-RT-F | AGACGCTATTGATGCCGATGA | |
| gyrB-RT-R | GTATTGCGCGTTGTCTTCGA | |
| sigB-pET-F | CGGGATCCATGCCAAAAGTATCTCA | Prokaryotic expression |
| sigB-pET-R | GCGTCGACTTACTCCACTTCCTCATTC | |
| gapdh-pET-F | CGCGGATCCATGACAGTTAAAGTTGGTAT | |
| gapdh-pET-R | CCCAAGCTTTTATTTAGCGATTTTTGCAA | |
| PsigB::gfp-a | CGGGATCCTGGCTTTGAGAGAGATTC | Construction of gfp reporter system |
| PsigB::gfp-b | TTTACTCATAAAATTTTCTCTCTTATT | |
| PsigB::gfp-c | AATTTTATGAGTAAAGGAGAAGAAC | |
| PsigB::gfp-d | CGAGCTCTTATTTGTATAGTTCATCCATGC |
Nucleotides introduced to create restriction sites are underlined.
Complementation of the rsbX deletion mutant.
rsbX complementation was conducted according to a previous protocol (26). The rsbX open reading frame (ORF) was fused with the sigB promoter in front of the rsbV gene by SOE-PCR (28) using primer pairs rsbX-w/x and rsbX-y/z (Table 1). The product containing BamHI and SacI sites was digested and ligated to pERL3. After confirmation by sequencing, the resulting plasmid was electroporated into the L. monocytogenes ΔrsbX strain. Transformants were selected on BHI agar plates containing erythromycin (10 μg/ml). The complemented strain was designated the ΔrsbX/rsbX strain.
Construction of PsigB::gfp gene fusion strains.
The green fluorescent protein gene (gfp) was amplified from the recombinant plasmid pFL251 kept in our laboratory and was fused with the stress-sensitive σB promoter in front of the rsbV gene by SOE-PCR. After digestion with BamHI and SacI, the PsigB::gfp fragment was cloned into pERL3. After confirmation by sequencing, the recombinant plasmid was electroporated into L. monocytogenes 10403S and the ΔrsbX derivative mutant strain as described earlier. Transformants were selected on BHI agar plates containing erythromycin (10 μg/ml).
Expression of SigB and GAPDH in E. coli for generation of polyclonal antibodies.
DNA sequences corresponding to the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene and sigB were PCR amplified from L. monocytogenes 10403S using primers listed in Table 1. Each fragment was then cloned into pET30a(+). The recombinant plasmids were confirmed by PCR and sequencing and then transformed into E. coli Rosetta(DE3). Overnight cultures of E. coli harboring the expression constructs were diluted 1:100 into 4 liters of LB broth supplemented with 50 μg/ml kanamycin and subcultured at 37°C until the cell growth reached an optical density at 600 nm (OD600) of 0.5 to 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) (TaKaRa) was added at a final concentration of 1 mM to induce expression at 37°C for 3 h. The cells were harvested, resuspended in phosphate-buffered saline (PBS) (pH 7.4), and then sonicated 50 times for 10 s each at 300 W. The sonicated mixtures were centrifuged to collect the supernatants. The proteins were purified with nickel-iminodiacetic acid (Ni-IDA) agarose (Weishi-Bohui Chromototech Co., Beijing, China) according to the manufacturer's directions. Protein purity was confirmed by 12% SDS-PAGE. The protein concentration was determined with a bicinchoninic acid (BCA) protein assay kit (MultiSciences, China). Antisera were generated by immunization, at 2-week intervals, of two female New Zealand White rabbits with each of the purified proteins emulsified with an equal volume of complete or incomplete Freund's adjuvant (100 μg protein per rabbit). One week after the fourth immunization, blood samples were collected for separation of serum samples as polyclonal antibodies. Preimmune serum samples were collected as negative controls. The animal experiment was approved by the Laboratory Animal Management Committee of Zhejiang University (approval no. 20140232).
Growth and survival assays.
L. monocytogenes wild-type, ΔrsbX mutant, and complemented strains were grown to exponential phase at 37°C in BHI with shaking. For growth assay, the cultures were collected and washed in PBS (10 mM, pH 7.4). Each of the cultures was then diluted 1:50 in BHI at pH 7.4 (BHI-pH 7.4), BHI-pH 4.8, or BHI–5% NaCl and incubated at 37°C. Growth was measured as OD600 at 1-h intervals up to 8 h. This test was performed in triplicate.
For survival assay, cultures were collected at exponential phase (∼4 h; OD600, ∼0.3) or stationary phase (∼16 h). After washing with PBS, 1 ml of each culture suspension was pelleted and resuspended in BHI-pH 3.5 or in BHI–15% NaCl and incubated at 37°C for 0.5 h. The viable bacterial cells were then plated onto BHI agar at appropriate dilutions. The relative survival of each strain is expressed as the ratio to the wild type. Percent survival is reported as the mean ± standard deviation (SD) from three independent experiments, each performed in duplicate.
Survival in the poststress stage.
L. monocytogenes wild-type, ΔrsbX mutant, and complemented strains were grown to exponential phase (∼4 h; OD600, ∼0.3) at 37°C in BHI-pH 7.4 with shaking. The cultures were pelleted and resuspended in stress medium (BHI-pH 4.8 or BHI–5% NaCl). After 0.5 h of stress incubation, the cultures were harvested, washed in PBS, resuspended in fresh BHI for 0.5 h of recovery growth, and then shifted to a harsher secondary stress medium (BHI-pH 3.5 or BHI–15% NaCl) for 0.5 h.
Under the overnight stress condition, the cultures were resuspended in a multistress medium (BHI containing 3% NaCl, 30% sucrose) (pH 5.5; water activity [aw], ∼0.96). After 12 h of incubation at 37°C, the bacterial strains were harvested, washed in PBS, and resuspended in BHI for 0.5 h or 1.5 h of recovery growth and then subjected to the same secondary stress medium as described above for 0.5 h. One milliliter of bacterial cells after recovery growth and secondary stress was plated onto BHI agar at appropriate dilutions to count the viable cells. The relative survival of each strain is expressed as the ratio of CFU of the secondary stress cultures to that at the end of recovery growth. Percent survival is reported as the mean ± SD from three independent experiments, each performed in duplicate. Statistically significant differences between the mutant or complemented strain and the wild type were determined.
Experimental procedure for σB expression analysis.
For sigB transcription analysis during the recovery phase, L. monocytogenes wild-type, mutant, and complemented strains were grown to exponential phase (∼4 h; OD600, ∼0.3) at 37°C in BHI with shaking. Cultures were then collected and resuspended in BHI–5% NaCl for 0.5 h. Three aliquots of the stressed cultures were taken; one was used as a primary stress control for direct total RNA isolation, and the other two were pelleted and resuspended in BHI-pH 7.4 for 0.5 h or 1.5 h of recovery growth. At the end of each recovery growth, the cultures were pelleted for total RNA isolation. The same protocol was used for reporter assay or quantification of ATP in the recovery cultures. For analysis of SigB protein expression, the time period for stress and recovery was extended to 45 min. For comparison of σB expression between exponential-phase and stationary-phase cultures, bacterial strains were subcultured to exponential phase (∼4 h; OD600, ∼0.3) or stationary phase (∼16 h) before being collected for total RNA isolation, protein extraction, and ATP quantification. Expression of σB at the transcriptional and translational levels in the above-described treatments was analyzed by quantitative reverse transcription-PCR (qRT-PCR), Western blotting, and GFP reporter assay as described below.
qRT-PCR.
Total RNAs from the above-described treatments were prepared using the SV total RNA isolation system (Promega, Inc., WI), and cDNAs were synthesized with reverse transcriptase (Toyobo, Japan). Transcription of the sigB gene (with primer pair sigB-RT-F/sigB-RT-R [Table 1]) was quantified in 20-μl reaction mixtures containing SYBR qPCR mix (Toyobo) on an iCycler iQ5 real-time PCR detection system (Bio-Rad, USA). The housekeeping gene gyrB was selected as an internal control for normalization as previously described (26). All reactions were conducted in triplicate, and the target gene mRNA expression in each mutant was calculated using the 2−ΔΔCT method (32) and shown as fold changes relative to the wild type (mean ± SD). Statistically significant differences between the mutant or complemented strain and the wild type were determined.
Western blotting.
All bacterial cells from the above-described treatments were harvested and resuspended in 1 ml protein extraction buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1% SDS, and 2% Triton X-100) for crude protein extraction. The bacterial suspensions were homogenized 3 times for 20 s each in a Precellys 24 homogenizer (Bertin, France) in the presence of 0.5-mm ceramic beads at 6,500 rpm. The supernatant samples were collected after centrifugation as the whole-protein samples for further analysis. The protein concentration was measured with a BCA protein assay kit. Equal amount of protein samples were subjected to 12% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked for 2 h in Tris-buffer saline with 0.05% Tween 20 (TBST) containing 5% skim milk and were incubated for 1 h with each of the antibodies. The blots were washed and incubated for another hour with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (MultiSciences, China). Protein bands were revealed using the ECL Plus detection system under conditions recommended by the manufacturer (Thermo, USA). Images were captured in a Gel 3100 chemiluminescent imaging system (Sagecreation, China), and the densities of the protein bands were normalized to the GAPDH signal and quantified using Quantity One software (Bio-Rad, USA).
Quantification of GFP fluorescence.
The L. monocytogenes wild-type strain and its rsbX deletion mutant harboring the recombinant plasmid carrying PsigB::gfp were either subjected to primary stress followed by recovery growth for 0.5 h and 1.5 h or grown to the required growth phases as described above. The bacterial cultures were then pelleted, resuspended in PBS, and adjusted to equal OD600s. To quantify the fluorescence due to σB promoter-driven GFP expression, 200 μl of adjusted suspensions was transferred to white flat-bottom 96 wells (Corning, NY) for measurement of fluorescence with excitation at 488 nm and emission at 507 nm on a SpectraMax M2 spectrophotometer (Molecular Devices, USA). Relative fluorescence was expressed as the ratio of that of the ΔrsbX mutant to that of the wild-type strain at a specific recovery time or a particular growth phase. Each experiment was repeated three times, each in triplicate wells for each strain.
Quantification of extracellular and intracellular ATP.
Quantification of ATP in the bacterial strains from the above-described treatments followed the protocol of Hironaka et al. (33). The OD600 of the treated cultures was measured. Briefly, either the treated suspension or the 0.22-μm-filtered supernatant (100 μl each) was mixed with an equal volume of BacTiter-Glo ATP reagent (Promega, Inc., WI) for measurement of total ATP or extracellular ATP. The amount of intracellular ATP was calculated by subtracting the amount of extracellular ATP from that of the total ATP (from uncentrifuged bacterial cultures). The bioluminescence response was detected (500 ms) with a luminometer (MLX luminometer; Dynex Technologies, USA). The ATP concentration was determined from the calibration curve using reference ATP dilutions (Sigma-Aldrich, St. Louis, MO, USA). The cell lysis time for these bacteria was empirically determined to be 8 min. Corresponding media were used as the negative controls. Bioluminescence measurements for each sample were obtained in four replicate wells. Data are expressed as mean (nM/0.1 OD) ± SD for replicate wells or repeated experiments. ATP levels in the ΔrsbX strain at two different growth phases were calculated relative to those in the wild-type at corresponding phases, which were normalized to 100%.
Statistical analysis.
All data comparisons were analyzed using the two-tailed Student t test. Differences with P values of <0.05 were considered statistically significant, and those with P values of <0.01 were considered markedly statistically significant.
RESULTS
Listeria monocytogenes rsbX does not affect growth under mild acidic or sodium stress.
To investigate whether deletion of rsbX would affect responses of L. monocytogenes to mild or acute stresses, the ΔrsbX mutant strain and its parent strain were exposed to BHI broth at pH 4.8 or 3.5 or to BHI broth (pH 7.4) containing 5% NaCl or 15% NaCl to examine their growth potential or survival. With mild stresses (pH 4.8 or 5% NaCl), the ΔrsbX mutant strain had growth similar to that of its parent strain as shown by average OD600 at hour 8 (Table 2). Deletion of rsbX did not affect listerial survival upon brief exposure (0.5 h) in 15% NaCl-supplemented BHI broth, since the ΔrsbX mutant and its parent strain had similar levels of surviving cells. However, the ΔrsbX mutant did show significant reduction of survival compared to that of its parent strain upon 0.5 h of exposure to BHI at pH 3.5 (P < 0.05) (Table 2).
TABLE 2.
Growth and survival of Listeria monocytogenes wild-type strain 10403S and its ΔrsbX mutant under stress conditions and sigB transcription by the strains stressed with 5% NaCla
| Strain | OD600 at 8 h |
Survival (%) |
sigB mRNA (%) with 5% NaCl | |||
|---|---|---|---|---|---|---|
| pH 7.4 | pH 4.8 | 5% NaCl | pH 3.5 | 15% NaCl | ||
| WT | 0.56 ± 0.03 | 0.09 ± 0.007 | 0.39 ± 0.01 | 100 ± 3.82 | 100 ± 1.30 | 100 ± 15.7 |
| ΔrsbX mutant | 0.53 ± 0.01 | 0.09 ± 0.009 | 0.39 ± 0.03 | 92.9 ± 2.49* | 100 ± 8.53 | 105 ± 19.6 |
| P value | 0.188 | 0.978 | 0.986 | 0.026 | 0.551 | 0.649 |
Data are expressed as means ± SD from triplicate experiments. Growth was measured as OD600 up to 8 h. Maximum OD values at hour 8 are shown. Stress durations for transcription analysis and survival tests were 0.5 h. Survival of and sigB transcription by the ΔrsbX mutant were calculated relative to those for the wild-type strain (WT), which were normalized to 100%. *, P < 0.05.
Deletion of rsbX increases survival of prestressed Listeria monocytogenes in secondary stress media.
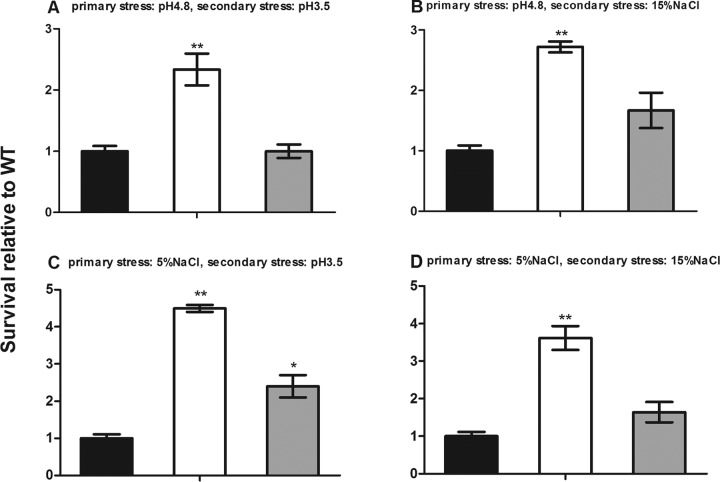
We further examined whether the rsbX gene functions in response to secondary stresses. Bacterial strains were first exposed to BHI at acidic pH 4.8 for 0.5 h, allowed to recover for 0.5 h in BHI at pH 7.4, and then subjected to 0.5 h of secondary stresses. Figure 1A and B show that the ΔrsbX mutant was more tolerant to secondary stress than its parent strain, with a 2.3-fold increase in survival rate in pH 3.5 medium (P < 0.01) and a 2.7-fold increase in 15% NaCl (P < 0.01) compared to that of the parent strain. With BHI containing 5% NaCl as the primary stress medium, the survival rate of the ΔrsbX mutant strain was 4.5-fold higher than that of its parent strain when restressed in BHI at pH 3.5 (Fig. 1C) (P < 0.01) and 3.6-fold higher when restressed in 15% NaCl-supplemented BHI (Fig. 1D) (P < 0.01). The ΔrsbX/rsbX complemented strain, however, showed survival rates close to those of the wild-type strain.
FIG 1.
Survival of Listeria monocytogenes wild-type (WT) strain 10403S (black bars), its ΔrsbX mutant (white bars), and the complemented strain (gray) in response to secondary stress after 0.5 h of recovery growth. The durations of primary stress, recovery, and secondary stress were 0.5 h. Data are the means ± SDs from triplicate experiments. Statistically significant differences between the mutant or complemented strain and the wild type were determined (n = 3; *, P < 0.05; **, P < 0.01).
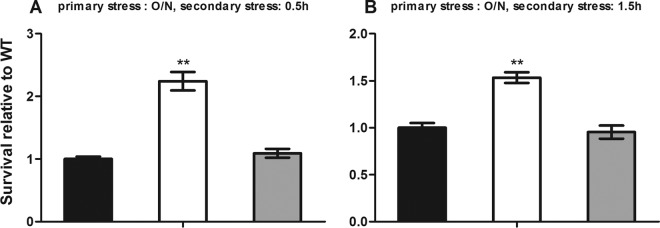
We then sought to determine if prolonged primary stress (∼12 h) could affect the listerial response to secondary stress after a 0.5-h recovery period. The primary multistress medium was prepared with a water activity of 0.96 and a pH of 5.5. After a 0.5 h or 1.5 h of recovery growth, the survival rate of the ΔrsbX mutant strain was 1.5 to 2.2-fold higher than that of its parent strain at both time points of the secondary stress (Fig. 2A and B) (P < 0.01). Complementation of the rsbX gene reduced survival to the wild-type level. The above results indicate that the RsbX plays a role in the response to secondary stress after a short recovery growth.
FIG 2.
Survival of Listeria monocytogenes wild-type strain 10403S (black bars), its ΔrsbX mutant (white bars), and the complemented strain (gray bars) in response to a multistress medium (BHI–3% NaCl, 30% sucrose) (pH 5.5; aw, ∼0.96) after 0.5 h of recovery growth. The primary stress was an overnight (O/N) (∼12-h) stress in a multistress medium, the recovery growth was for 0.5 h, and the secondary stress was also multistress medium for 0.5 h (A) or 1.5 h (B). Data are the means ± SDs from triplicate experiments. Statistically significant differences between the mutant or complemented strain and the wild type were determined (n = 3; **, P < 0.01).
Deletion of RsbX increases σB expression in response to secondary stresses.
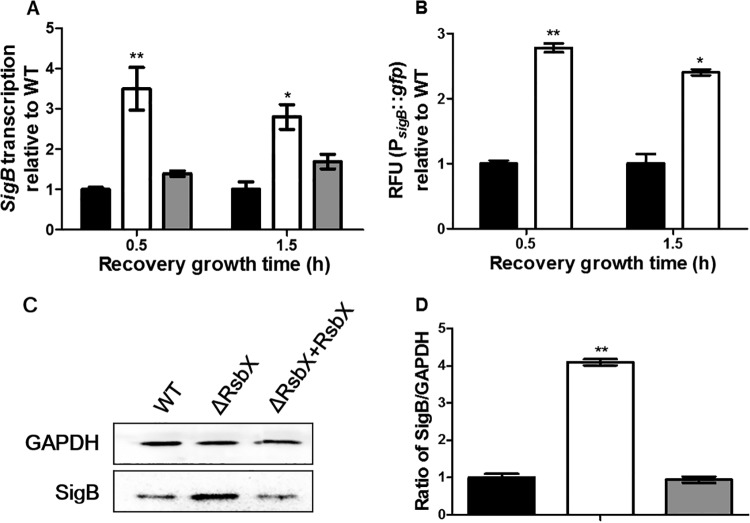
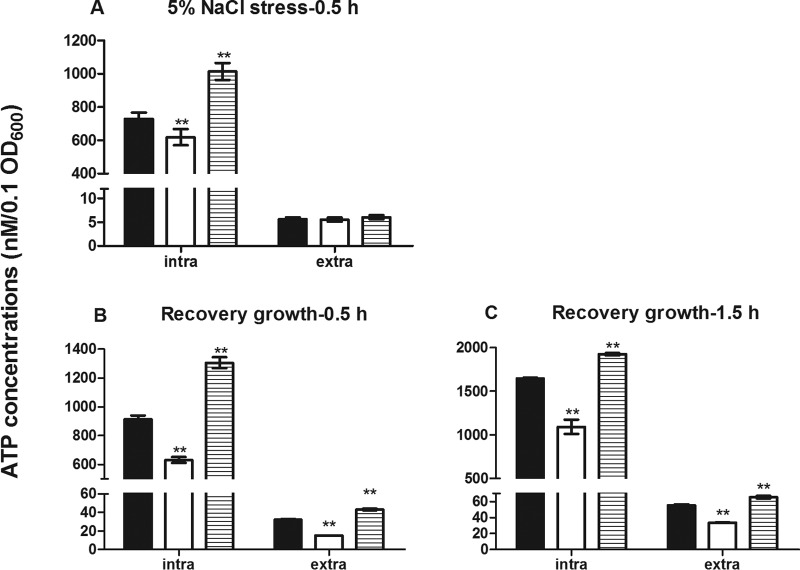
In B. subtilis, RsbX is known to regulate σB (34, 35). We hypothesized that RsbX might modulate survival of L. monocytogenes under secondary stress by regulating σB expression following primary stress. As shown in Fig. 3A, the sigB transcriptional level of the ΔrsbX mutant was 3.5-fold higher than that of its parent strain after 0.5 h of recovery growth (P < 0.01) and 2.8-fold higher after 1.5 h of recovery growth following primary stress (BHI–5% NaCl) (P < 0.05), which could be reversed by genetic complementation. The σB promoter-driven GFP reporter system was used as an additional support to qRT-PCR. The ΔrsbX mutant showed significantly higher levels of GFP fluorescence than its parent strain at both time points of recovery growth (Fig. 3B). At the protein level, σB expression in the ΔrsbX mutant was about 4-fold higher than that of its parent or complemented strain (Fig. 3C and D) (P < 0.01). All these findings reveal that increased survival of the ΔrsbX mutant under secondary stress resulted from elevated σB expression during the recovery stage following primary stress. Using a luciferase-based ATP assay, the ΔrsbX mutant showed significantly lower intracellular and extracellular ATP levels than its parent strain during the recovery period (Fig. 4). The ΔsigB mutant, however, had significantly higher intracellular and extracellular ATP levels than its parent strain and the ΔrsbX mutant subjected to sodium stress or during the recovery period (Fig. 4). Deletion of rsbX also led to a reduced intracellular ATP level right after the primary stress. Therefore, we suggest that RsbX might act as a negative σB regulator during the recovery period following primary stress and that such repression seems to be energy saving.
FIG 3.
Expression of σB of Listeria monocytogenes wild-type strain 10403S (black bars), its ΔrsbX mutant (white bars), and the complemented strain (gray bars) after recovery growth following primary stress with BHI containing 5% NaCl. (A) sigB transcription after 0.5 h and 1.5 h of recovery growth. (B) Relative fluorescence units (RFU) of PsigB::gfp reporter fusion after 0.5 h and 1.5 h of recovery growth. (C and D) Western blotting and quantification of expression of SigB and GAPDH (as an internal control) after 45 min of primary stress followed by 45 min of recovery growth. Data are reported as means ± SDs. Statistically significant differences between the mutant or complemented strain and the wild type were determined (n = 3; *, P < 0.05; **, P < 0.01).
FIG 4.
Comparison of intracellular and extracellular ATP levels at the end of sodium stress or after recovery growth of Listeria monocytogenes wild-type strain 10403S (black bars) and its ΔrsbX (white bars) or ΔsigB (hatched bars) mutant. Intracellular (intra) and extracellular (extra) ATP concentrations were quantified as nM/0.1 OD600 unit at the end of 0.5 h of 5% NaCl stress (A) and after a subsequent 0.5 h of recovery growth (B) or 1.5 h of recovery growth (C). Data are means ± SDs for four replicate wells of one typical experiment. Statistically significant differences between the wild-type strain 10403S and its ΔrsbX mutant or ΔsigB mutant were determined (n = 4; **, P < 0.01).
RsbX is involved in σB activity in stationary phase.
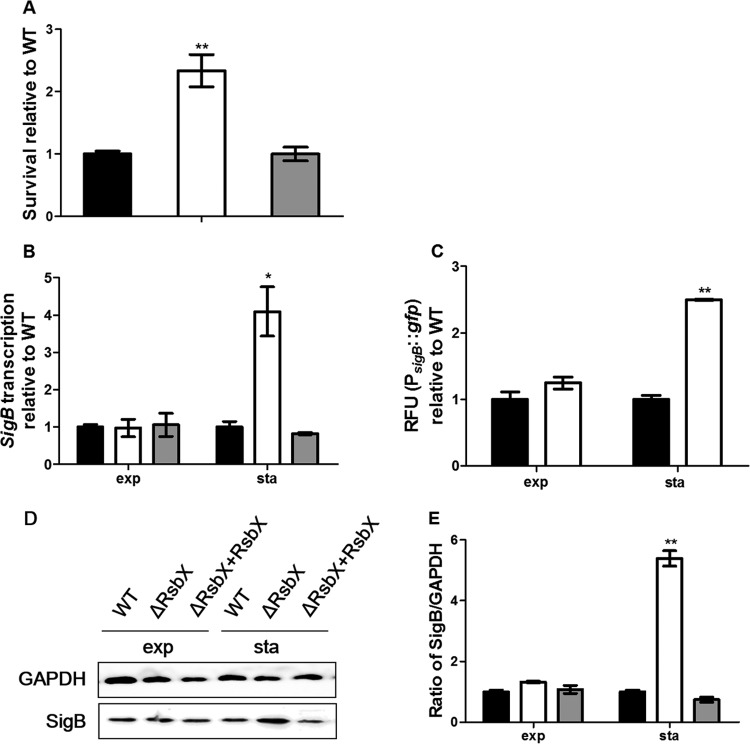
Expression of σB in B. subtilis is known to differ with its growth phases (35). We wondered whether RsbX in L. monocytogenes could be involved in regulating bacterial activity particularly in the stationary phase, since there was no difference in survival between the ΔrsbX mutant and its parent strain in the exponential phase. We found that the survival rate of the ΔrsbX mutant was markedly increased compared to that of its parent strain when their stationary-phase cultures were subjected to sodium (15% NaCl) stress (Fig. 5A) (P < 0.01). Deletion of the rsbX gene increased transcription of sigB by nearly 4-fold compared to that of its parent strain (Fig. 5B) (P < 0.05) in the stationary phase, while there was no difference in sigB transcription in the exponential phase. The rsbX deletion mutant also had higher σB promoter-driven GFP fluorescence than its parent strain in the stationary-phase culture but not in the exponential-phase culture (Fig. 5C) (P < 0.01). Western blotting showed that σB expression in the ΔrsbX mutant was significantly higher than that in its parent strain (Fig. 5D and E) (P < 0.01).
FIG 5.
Survival and expression of σB of Listeria monocytogenes wild-type strain 10403S (black bars), its ΔrsbX mutant (white bars) and the complemented strain (gray bars) in the exponential (exp) and stationary (sta) phases. (A) Survival of the stationary-phase cultures in BHI–15% NaCl. (B) sigB transcription. (C) Relative fluorescence units (RFU) of PsigB::gfp reporter fusion. (D and E) Western blotting and quantification of expression of SigB and GAPDH (as an internal control). Data are reported as means ± SDs. Statistically significant differences between the mutant or complemented strain and the wild type were determined (n = 3; *, P < 0.05; **, P < 0.01).
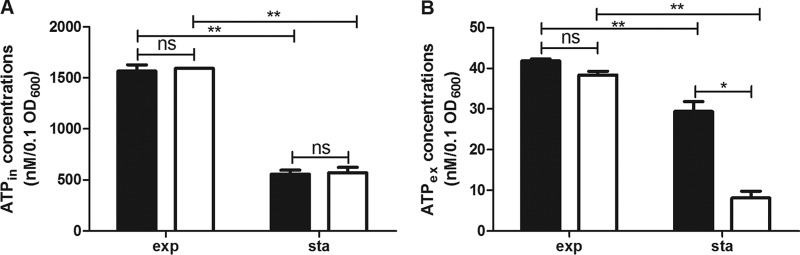
The above results suggest that RsbX might function to exert a negative regulatory effect in the stationary phase. This prompted us to postulate that downregulation of σB by RsbX might be an energy-saving strategy for the bacteria to cope with potential incoming stimuli in the stationary phase. We found that there were significant differences in intracellular ATP levels between the exponential and stationary phases of L. monocytogenes culture, with those in the exponential phase being 2-fold higher (Fig. 6) (P < 0.01). ATP was also secreted to the extracellular medium but at a lower percentage (about 3% of the total ATP in exponential-phase culture and nearly 5% in stationary-phase culture). Deletion of rsbX did not affect the level of intracellular ATP in either phase, but the extracellular ATP level was reduced by one-third compared with that for parent strain in the stationary-phase culture (Fig. 6) (P < 0.05).
FIG 6.
Intracellular and extracellular ATP levels in exponential- and stationary-phase cultures of Listeria monocytogenes wild-type strain 10403S (black bars) and its ΔrsbX mutant (white bars). Intracellular ATP (ATPin) (A) and extracellular ATP (ATPex) (B) concentrations in the exponential (exp) and stationary (sta) phases were quantified as nM/0.1 OD600 unit. The experiment was repeated four times. Data are means ± SDs for four replicate wells of one typical experiment. Statistically significant differences between the wild-type strain and the ΔrsbX mutant were determined (n = 4; **, P < 0.01; not significant [ns], P > 0.05).
DISCUSSION
In B. subtilis, RsbX was found to dephosphorylate RsbS and/or RsbR, thus preventing RsbT from activating RsbU (36, 37). Therefore, RsbX is considered a feedback phosphatase to reset the general stress response (14). So far there has been no study on its roles in responses of L. monocytogenes to stresses. Here we reveal that RsbX contributes to repressed growth by negative regulation of σB activity when L. monocytogenes is exposed to secondary stress after a short recovery period or when the culture is in the stationary phase. This suggests that RsbX functions to downregulate the general stress response after the primary stress or in the stationary phase, a complex condition of high osmosis, reduced pH, and inhibition by its own metabolites.
Listeria monocytogenes RsbX does not function in response to primary stress.
Studies using L. monocytogenes ΔsigB mutants have demonstrated that σB contributes to bacterial survival under acid and sodium stresses (6, 10). The gene rsbX is the last one of the sigB operon that is transcriptionally regulated by sigB (11). We supposed that RsbX could be functional as it is in B. subtilis, although there is only 30.2% amino acid sequence identity between the two RsbX proteins. By genetic deletion, we found that RsbX did not affect growth in BHI medium at pH 4.8, in BHI with 5% NaCl, or in BHI with high sodium (15% NaCl) stress, although it did have some negative effect on listerial survival at sublethal acidic pH. Initially we found that sodium stress induced stronger σB activation than low pH and that the window of lethal and sublethal pH was narrow, so we chose NaCl as a model stressor to study the effect of RsbX on σB expression upon primary stress. There was virtually no difference in sigB transcription between the ΔrsbX mutant and its parent strain in response to 5% NaCl stress. These findings suggest that RsbX does not have direct involvement in primary environmental stresses and sigB transcription, at least under sodium stress. This is similar to the case for B. subtilis, where RsbX does not have direct involvement in σB ethanol stress induction and some stress-induced σB activation can occur in the absence of RsbX (34, 38).
RsbX is a negative regulator of σB in Listeria monocytogenes during the recovery period following primary stress or in stationary-phase culture.
A large number of studies have examined how bacteria survive under stress conditions, namely, the turn-on mechanisms upon stress. Expression of σB could be readily induced in L. monocytogenes subjected to various stresses as part of its survival strategy (6, 21, 25). Few have paid attention to the turn-off switch upon removal of primary stress and the subsequent responses to secondary stress. In B. subtilis, RsbX is thought to complete a negative feedback loop that returns the stressed cells to their initial state without direct participation of the stress induction process (34, 36). This type of turn-off function might be important for energy conservation and maintenance of homeostasis of the bacteria. We thought that if L. monocytogenes RsbX plays a turn-off role in keeping the stressed state under check soon after withdrawal of the stressing factor, deletion of rsbX would exhibit a rebound phenomenon with a phenotype more resistant to secondary stress. We designed a stress-recovery-restress approach to examine the effects of primary stress on the recovery survival upon secondary stress.
After a 0.5-h recovery period, the survival rate of the ΔrsbX mutant increased significantly compared to that of its parent strain, no matter what primary or secondary stress factor was used. Extended overnight multistress showed similar results. We speculated that increased survival of the ΔrsbX mutant in response to secondary stress might be related to elevated σB expression in the recovery period following primary stress. Analyses by quantitative PCR, PsigB::gfp reporter assay, and Western blotting revealed that σB was significantly increased at the transcriptional and translational levels in the ΔrsbX mutant compared with its parent strain. This is in contrast with finding that there was no difference in sigB transcription between the two strains in response to primary stresses. These results indicate that the RsbX protein in L. monocytogenes is not directly involved in stress-induced σB activation but rather is involved in negative regulation of σB of stressed cells shifted to a new favorable environment (i.e., in the recovery medium used in this study). This is similar to the case for B. subtilis RsbX, which is believed to participate in poststress recovery of σB activity to prestress levels (35).
We also found that RsbX, though not affecting σB activity in exponential-phase cultures, played a role in restricting the σB activity in stationary phase, as shown by more resistance and higher σB expression in the ΔrsbX mutant than its parent strain when the stationary-phase cultures were shifted to sodium stress. Therefore, we believe that RsbX negatively regulates σB of the stationary cultures of L. monocytogenes. In B. subtilis, Smirnova et al. also found that the wild-type strain had higher σB activity at the onset of stationary phase, while the RsbX-negative suppressor mutants (due to mutations in RsbT and RsbU that suppressed RsbX expression) did not show elevated σB activity until entering the stationary phase, where it was higher than that in their congenic RsbX-positive strains (35). They concluded that RsbX is required to restrict RsbU activity in the absence of obvious stress stimulation.
Listeria monocytogenes RsbX might serve to save energy by downregulating σB.
Under stress conditions, the bacteria should attempt to mobilize their resources for survival, which requires energy consumption (39, 40). Resetting or turning off the σB activity (to maintain low σB activity) after removal of stressors or shifting from stationary-phase cultures to fresh medium might be a strategy of the bacteria to save energy. If the σB turn-off mechanism is meant to save energy, we would see increased ATP in the parent strain as opposed to the ΔrsbX mutant. Thus, we tested intracellular and extracellular ATP levels of bacterial cultures of the ΔrsbX mutant and its parent strain subjected to a brief sodium stress followed by recovery in normal BHI. We did see reduced intracellular and extracellular ATP levels in the ΔrsbX mutant cultures at the end of stress or during the recovery period, suggesting that RsbX might be acting to save energy by downregulating σB. This is supported by the fact that the ΔsigB mutant had markedly higher levels of intracellular ATP than the wild-type strain.
However, we did not observe any differences in intracellular ATP levels between the ΔrsbX mutant and its parent strain in either exponential or stationary phase, although there were significant differences in the intracellular and extracellular ATP levels of both strains between exponential- and stationary-cultures. The intracellular ATP at the stationary phase was only one-third of that in the exponential-phase cultures. Alper et al. reported a close correlation between the concentration of ATP required for efficient RsbW-mediated phosphorylation of RsbV, inhibition of RsbW-RsbV complex formation, and inhibition of σB-directed transcription (41). Lower ATP levels in the stationary-phase cultures could be an inducing factor for σB activity (42).
We show that L. monocytogenes is able to secrete ATP outside the cells, but at a lower percentage. Recent reports showed that some bacterial species, including Gram-positive Staphylococcus aureus and Enterococcus faecalis, could release ATP to the culture supernatants (33, 43). E. coli and Salmonella were able to hydrolyze the extracellular ATP on the bacterial surface (43). The only significant difference we observed was reduced extracellular ATP in the ΔrsbX mutant compared with that in its parent strain in the stationary phase. It is known that the class III heat shock protein ATPase ClpC shows σB-dependent induction in response to energy stresses (44). The stationary-phase cultures are energy deficient with low ATP, which could induce σB activity (42). Therefore, it is tempting to speculate that reduced extracellular ATP in the ΔrsbX mutant could be due to hydrolysis of ATP crossing the cell wall by ATP-dependent ClpC as a result of σB activation.
In conclusion, we clearly show that RsbX is a negative regulator of L. monocytogenes σB during the recovery period after the primary stress or in the stationary phase, thus affecting its survival under secondary stress. This is probably a strategy of the bacterium for energy conservation. Further research on the mechanisms by which RsbX operates to downregulate σB and on the relationship between energy metabolism and RsbX-σB interaction is required.
REFERENCES
- 1.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev 55:476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan-Thanh L, Mahouin F, Alige S. 2000. Acid responses of Listeria monocytogenes. Int J Food Microbiol 55:121–126. doi: 10.1016/S0168-1605(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 3.Cole MB, Jones MV, Holyoak C. 1990. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J Appl Bacteriol 69:63–72. doi: 10.1111/j.1365-2672.1990.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 4.Sleator RD, Gahan CGM, O'Driscoll B, Hill C. 2000. Analysis of the role of betL in contributing to the growth and survival of Listeria monocytogenes LO28. Int J Food Microbiol 60:261–268. doi: 10.1016/S0168-1605(00)00316-0. [DOI] [PubMed] [Google Scholar]
- 5.Boylan SA, Redfield AR, Brody MS, Price CW. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J Bacteriol 175:7931–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira A, O'Byrne CP, Boor KJ. 2001. Role of sigma(B) in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol 67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira A, Sue D, O'Byrne CP, Boor KJ. 2003. Role of Listeria monocytogenes sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Environ Microbiol 69:2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise AA, Price CW. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J Bacteriol 177:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker LA, Cetin MS, Hutkins RW, Benson AK. 1998. Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol 180:4547–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira A, Gray M, Wiedmann M, Boor KJ. 2004. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other Gram-positive bacteria. Curr Microbiol 48:39–46. doi: 10.1007/s00284-003-4020-x. [DOI] [PubMed] [Google Scholar]
- 12.Samina I, Brenner J, Moalem U, Berenstein M, Cohen A, Peleg BA. 1997. Enhanced antibody response in cattle against Leptospira hardjo by intradermal vaccination. Vaccine 15:1434–1436. doi: 10.1016/S0264-410X(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 13.Kim TJ, Gaidenko TA, Price CW. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 186:6124–6132. doi: 10.1128/JB.186.18.6124-6132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eymann C, Schulz S, Gronau K, Becher D, Hecker M, Price CW. 2011. In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis sigmaB. Mol Microbiol 80:798–810. doi: 10.1111/j.1365-2958.2011.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang CM, Brody MS, Akbar S, Yang X, Price CW. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor sigma(b) in response to environmental stress. J Bacteriol 178:3846–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijay K, Brody MS, Fredlund E, Price CW. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol 35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 17.Delumeau O, Chen CC, Murray JW, Yudkin MD, Lewis RJ. 2006. High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 188:7885–7892. doi: 10.1128/JB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TJ, Gaidenko TA, Price CW. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J Mol Biol 341:135–150. doi: 10.1016/j.jmb.2004.05.043. [DOI] [PubMed] [Google Scholar]
- 19.Marles-Wright J, Grant T, Delumeau O, van Duinen G, Firbank SJ, Lewis PJ, Murray JW, Newman JA, Quin MB, Race PR, Rohou A, Tichelaar W, van Heel M, Lewis RJ. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92–96. doi: 10.1126/science.1159572. [DOI] [PubMed] [Google Scholar]
- 20.Hardwick SW, Pane-Farre J, Delumeau O, Marles-Wright J, Murray JW, Hecker M, Lewis RJ. 2007. Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis. J Biol Chem 282:11562–11572. doi: 10.1074/jbc.M609733200. [DOI] [PubMed] [Google Scholar]
- 21.Chaturongakul S, Boor KJ. 2004. RsbT and RsbV contribute to sigmaB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol 70:5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin JH, Brody MS, Price CW. 2010. Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology 156:2660–2669. doi: 10.1099/mic.0.041202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serrazanetti DI, Ndagijimana M, Sado-Kamdem SL, Corsetti A, Vogel RF, Ehrmann M, Guerzoni ME. 2011. Acid stress-mediated metabolic shift in Lactobacillus sanfranciscensis LSCE1. Appl Environ Microbiol 77:2656–2666. doi: 10.1128/AEM.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haddix PL, Jones S, Patel P, Burnham S, Knights K, Powell JN, LaForm A. 2008. Kinetic analysis of growth rate, ATP, and pigmentation suggests an energy-spilling function for the pigment prodigiosin of Serratia marcescens. J Bacteriol 190:7453–7463. doi: 10.1128/JB.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng C, Yang Y, Dong Z, Wang X, Fang C, Yang M, Sun J, Xiao L, Fang W, Song H. 2015. Listeria monocytogenes varies among strains to maintain intracellular pH homeostasis under stresses by different acids as analyzed by a high-throughput microplate-based fluorometry. Front Microbiol 6:15. doi: 10.3389/fmicb.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Cheng C, Xia Y, Zhao H, Fang C, Shan Y, Wu B, Fang W. 2011. Lmo0036, an ornithine and putrescine carbamoyltransferase in Listeria monocytogenes, participates in arginine deiminase and agmatine deiminase pathways and mediates acid tolerance. Microbiology 157:3150–3161. doi: 10.1099/mic.0.049619-0. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Jiang L, Chen Q, Zhao H, Luo X, Chen X, Fang W. 2009. lmo0038 is involved in acid and heat stress responses and specific for Listeria monocytogenes lineages I and II, and Listeria ivanovii. Foodborne Pathog Dis 6:365–376. doi: 10.1089/fpd.2008.0207. [DOI] [PubMed] [Google Scholar]
- 28.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Jiang L, Chen X, Luo X, Chen Y, Yu Y, Tian G, Liu D, Fang W. 2009. Listeria monocytogenes serovar 4a is a possible evolutionary intermediate between L. monocytogenes serovars 1/2a and 4b and L. innocua. J Microbiol Biotechnol 19:238–249. [PubMed] [Google Scholar]
- 30.Jiang L, Chen J, Xu J, Zhang X, Wang S, Zhao H, Vongxay K, Fang W. 2008. Virulence characterization and genotypic analyses of Listeria monocytogenes isolates from food and processing environments in eastern China. Int J Food Microbiol 121:53–59. doi: 10.1016/j.ijfoodmicro.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Camilli A, Tilney LG, Portnoy DA. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Hironaka I, Iwase T, Sugimoto S, Okuda K, Tajima A, Yanaga K, Mizunoe Y. 2013. Glucose triggers ATP secretion from bacteria in a growth-phase-dependent manner. Appl Environ Microbiol 79:2328–2335. doi: 10.1128/AEM.03871-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voelker U, Luo T, Smirnova N, Haldenwang W. 1997. Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J Bacteriol 179:1980–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smirnova N, Scott J, Voelker U, Haldenwang WG. 1998. Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulator RsbX. J Bacteriol 180:3671–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X, Kang CM, Brody MS, Price CW. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev 10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 37.Chen CC, Yudkin MD, Delumeau O. 2004. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J Bacteriol 186:6830–6836. doi: 10.1128/JB.186.20.6830-6836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott JM, Mitchell T, Haldenwang WG. 2000. Stress triggers a process that limits activation of the Bacillus subtilis stress transcription factor sigma(B). J Bacteriol 182:1452–1456. doi: 10.1128/JB.182.5.1452-1456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duche O, Tremoulet F, Glaser P, Labadie J. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl Environ Microbiol 68:1491–1498. doi: 10.1128/AEM.68.4.1491-1498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metris A, George SM, Mulholland F, Carter AT, Baranyi J. 2014. Metabolic shift of Escherichia coli under salt stress in the presence of glycine betaine. Appl Environ Microbiol 80:4745–4756. doi: 10.1128/AEM.00599-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alper S, Dufour A, Garsin DA, Duncan L, Losick R. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol 260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 42.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang WG. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol 177:3771–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mempin R, Tran H, Chen C, Gong H, Kim Ho K, Lu S. 2013. Release of extracellular ATP by bacteria during growth. BMC Microbiol 13:301. doi: 10.1186/1471-2180-13-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaturongakul S, Boor KJ. 2006. SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol 72:5197–5203. doi: 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]