Abstract
Mycoplasma bovis is considered a major contributor to respiratory diseases in young cattle. Resistant M. bovis isolates have increasingly been reported worldwide due to extensive use of antimicrobials to treat bovine pneumonia. The frequency of isolates resistant to fluoroquinolones varies considerably from one country to another. The MICs of isolates collected in France have only increased from “very low” to “low.” The present study was conducted to investigate whether alterations in the quinolone resistance-determining regions (QRDRs) could account for this slight modification in susceptibility. No correlation between QRDR alterations and increased MICs was evidenced in clinical isolates. In addition, all clinical isolates were subtyped, and the tendencies of the different sequence types to develop resistance through mutations in QRDRs under selective pressure in vitro were examined. In vitro, 3 hot spots for mutations in QRDRs (position 83 in GyrA and positions 80 and 84 in ParC) were associated with a high level of resistance when cumulated. We showed that the point mutations in the QRDRs observed in vitro were different (in location and selection rapidity) between the different subtypes. Our in vitro observations were corroborated by the recent detection of a clinical isolate highly resistant to fluoroquinolones (MIC ≥ 16 μg/ml) and belonging to the subtype which easily accumulates QRDR alterations in vitro. The current increased prevalence of this subtype in clinical isolates highlights the urgent need to control fluoroquinolone usage in veterinary medicine.
INTRODUCTION
Mycoplasma bovis is a wall-less bacterium responsible for severe infections in cattle, including pneumonia, mastitis, arthritis, and otitis (1). In young cattle, it is now recognized as a major contributor to economic losses associated with bovine respiratory diseases (BRD). The infection pressure of BRD usually peaks 2 to 3 weeks after calves are mingled in fattening units following transportation from their respective birth farms (2). Since no efficient vaccines are available and licensed for use outside the United States (3), efforts to control M. bovis infections often rely on antimicrobial treatments, administered either prophylactically or in the early stages of the disease. Antimicrobials used for the treatment or prevention of BRD usually include broad-spectrum cephalosporins (cefquinome and ceftiofur), extended-spectrum fluoroquinolones (enrofloxacin, danofloxacin, and marbofloxacin), florfenicol, and long-lasting macrolides (tulathromycin, gamithromycin, and tildipirosin) (4). This extensive use of antibiotics has predictably resulted in an increase in resistant isolates over time. In France, it was recently shown that contemporary M. bovis strains had become significantly less susceptible than archival strains to 9 of the 12 antimicrobials tested (5). With regard to fluoroquinolones, the decrease in susceptibility was limited (only 1 dilution of the MIC), and no highly resistant isolates were observed in a set of more than 90 strains. This was unexpected, since one of the two major medical indications of injectable extended-spectrum fluoroquinolones for cattle is BRD (6). However, in contrast to other antimicrobials, such as tulathromycin, tilmicosin, ceftiofur, and florfenicol, expanded-spectrum fluoroquinolones are not approved for metaphylaxis or prevention in herds (2, 7). This restricted use could have limited the acquisition of resistance in France. Moreover, we recently demonstrated that recent M. bovis isolates from France belong to the same unique subtype and suggested that this monoclonal spread on a country-wide scale could be linked to the acquisition and selection of multiresistance through therapeutic practices and prevention strategies (8). The reason for fluoroquinolones being spared in this selection process has yet to be clarified.
Elsewhere in the world, results for M. bovis susceptibility to fluoroquinolones are very varied. Three studies reported highly resistant isolates, with MICs of >8 μg/ml, in the United Kingdom, Japan, and Belgium (9–11), while several others indicated a low to no increase in resistance, except for a few isolates, in Japan, the United States, and several European countries (6, 12–15). Some of these studies involved isolates collected before 2005 (11, 13), but the majority referred to recent ones (6, 9, 10, 12, 14, 15), suggesting a recent phenomenon. In the United Kingdom, a shift of resistance from an MIC90 of 1 μg/ml to one of 32 μg/ml occurred during the last decade (9, 16). This significant increase over a relatively short period is indicative of highly efficient resistance mechanisms. Quinolones are synthetic bactericidal agents that are able to eliminate actively dividing bacteria by inhibiting the topoisomerases II and IV required for DNA replication. In mycoplasmas, resistance to fluoroquinolones generally results from several alterations in the so-called quinolone resistance-determining regions (QRDRs) of the genes encoding topoisomerases, namely, gyrA, gyrB, parC, and parE (17). No alterations related to resistance to fluoroquinolones have been found so far in gyrB or parE in any clinical isolates of mycoplasmas (17). In contrast, two hot spots for mutations, in ParC and GyrA, were described for M. bovis isolates and were associated with large increases of the MICs (2.5 to 16 μg/ml). These were the Asp84Asn substitution in ParC associated or not with the Ser83Phe mutation in GyrA (10, 12). Although target mutations are the main mechanisms conferring resistance to fluoroquinolones, an active efflux system has been described for the human pathogen M. hominis and suggested for another ruminant mycoplasma, M. mycoides subsp. capri, but has so far not been reported for M. bovis (18–20).
The present study was conducted to examine the presence of potential alterations in the QRDRs of M. bovis clinical isolates collected in France and whether these could account for the previously observed decrease in susceptibility to fluoroquinolones. In addition, clinical isolates were subtyped by single-locus sequence typing (8) and multilocus sequence typing (MLST) (21), and their ability to become resistant to fluoroquinolones through mutations of their QRDRs was explored in vitro. The results were analyzed in order to determine whether the process and rapidity of developing resistance to fluoroquinolones under selective pressure may differ by subtype.
MATERIALS AND METHODS
Mycoplasma isolates, growth, identification, and subtyping.
Fifty-two French M. bovis clinical isolates were included in this study (see Table S1 in the supplemental material). They originated from a collection maintained at ANSES Lyon and mostly derived from the French national surveillance network for mycoplasmoses of ruminants (VIGIMYC) (22). They were identified by membrane filtration dot-immunobinding tests (23) and by a species-specific PCR assay targeting the polC gene (24). Strain PG45T and two isolates from neighboring Switzerland (25) were added as controls. Isolates were grown in PPLO broth, modified as previously described (26), at 37°C in 5% CO2. Each isolate was subtyped by single-locus sequence typing (using a 486-bp region of the polC gene), which has been shown to provide relevant typing results (8). Clustering of the isolates was confirmed by applying the MLST scheme of Register et al. (21). In brief, the adh-1, gltX, gpsA, gyrB, pta-2, tdk, and tkt loci were amplified by PCR and sequenced. A polyfasta file comprising the 7 alleles was generated for each isolate and uploaded into a newly created MLST database (http://pubmlst.org/mbovis/) to be assigned a subtype. Individual subtypes were compared to those already present in the database.
MIC assays.
The MICs of enrofloxacin (ENR), danofloxacin (DAN), and marbofloxacin (MAR), all purchased from Sigma, were determined using the agar dilution method on modified PPLO agar as previously described (5, 27). In brief, 1-μl aliquots of each strain, diluted to 104 to 105 CFU/ml, were spotted onto agar plates containing serial 2-fold dilutions of each fluoroquinolone (0.125 to 64 μg/ml). MIC assays were performed at least twice for each isolate. The MIC was defined as the lowest fluoroquinolone concentration causing 100% inhibition of growth at 37°C in 5% CO2 for 72 h. The number of CFU per milliliter was determined by plating 2-μl aliquots of serial 10-fold broth dilutions onto agar plates. After incubation for 3 days, the colonies for several dilutions were counted using a stereomicroscope, and the mean final cell concentration was determined.
Selection of spontaneously ENR-resistant M. bovis variants.
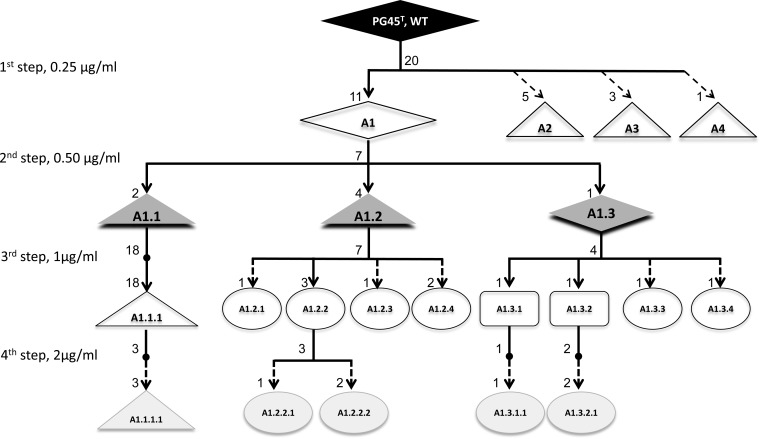
ENR-resistant clones were selected by plating 50-μl aliquots of parental cultures at 107 to 108 CFU/ml onto agar medium containing increasing inhibitory concentrations of enrofloxacin as described elsewhere (10, 28), with minor modifications. Each selection experiment involved four steps, using ENR concentrations of 1, 2, 4, and 8 times the MIC for the respective parent isolate (see Fig. 1 for details). At each selection step, resistant colonies were picked from the agar plates after incubation for 7 days and recultured in broth medium containing an equivalent amount of enrofloxacin before being inoculated onto another plate with twice that antimicrobial concentration. The recovery frequency was determined as the number of colonies appearing on the plate in the presence of enrofloxacin divided by the number of colonies contained in the inoculum. Several individual colonies were picked from the agar plate at each step in order to (i) analyze their QRDRs and (ii) determine their MICs.
FIG 1.
Experimental design of in vitro selection assays with enrofloxacin, using PG45T as an example. The selected clones are named by a letter followed by incremental numbers corresponding to successive experimental steps. The diamond symbols represent clones with no mutation, while clones with alterations in ParC, GyrA, and both loci are shown as triangles, squares, and circles, respectively. The numbers at nodes and arrow extremities indicate the number of clones isolated for each step and their distribution within the different genotypes. The enrofloxacin concentration for each step is indicated on the left. MIC assays were performed for all PG45T-derived clones except A2, A3, and A4. Dotted arrows indicate experimental steps with no further selection passages.
PCR amplification and sequence analysis of gyrA, gyrB, parE, and parC QRDRs.
Genomic DNAs were extracted from 20-ml logarithmic-phase broth cultures of M. bovis by using the phenol-chloroform method (29). QRDRs were amplified using previously described specific primers (12). For parE, the hybridization temperature in the original publication was modified from 56°C to 54°C. PCR products were sequenced using an external facility at Beckman Coulter Genomics. Sequence editing, consensus, and alignment construction were performed using Seaview software (http://doua.prabi.fr/software/seaview). For convenience, the amino acid numbering refers to the Escherichia coli numbering and is based on the E. coli K-12 sequences for GyrA (AAC75291.1), GyrB (AAT48201.1), ParC (AAC76055.1), and ParE (AAA69198.1).
RESULTS
An initial set of 31 clinical isolates of M. bovis, either old (1978 to 1983) or recent (2009 to 2012), was randomly chosen from our collection. These isolates were then subtyped by sequencing of the polC locus as previously described (8). As expected, all old strains and PG45T were homogeneously grouped into subtype 1 (ST1), while most (19/20 strains) of the recent strains were subtype 2 (ST2), which diverges from ST1 by one single nucleotide polymorphism (SNP) in the 486-bp polC locus. One of the recent isolates (strain 15527) belonged to ST3, an uncommon subtype characterized by 16 SNPs (see Table S1 in the supplemental material) (8).
The susceptibilities of the isolates to 3 fluoroquinolones, namely, enrofloxacin (ENR), danofloxacin (DAN), and marbofloxacin (MAR), were determined according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI), using the agar dilution method for MIC estimation (27). M. bovis PG45T was used as a control. Moderate increases in the MICs, i.e., 2-fold for ENR and 4-fold for DAN and MAR, were observed for all recent isolates in comparison to PG45T and the old isolates (Table 1). In contrast, 2 recent isolates from Switzerland were shown to be susceptible. These results are consistent with those of a previous study (5) and suggest an ongoing shift of isolates from the M. bovis population in France toward a low-level, non-clinically relevant quinolone resistance phenotype.
TABLE 1.
Relationships between MIC values and mutations in gyrA, gyrB, parC, and parE QRDRs of M. bovis clinical isolates
| Isolate name or characteristic | Origin, no. of isolates, yr of isolation | MIC (value or range) (μg/ml)a |
QRDR-encoded substitutionb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of isolates with phenotype/total no. of isolates | GyrA |
GyrB |
ParC |
||||||
| ENR | DAN | MAR | Ser83 | Asp362 | Ser80 | Asp84 | |||
| PG45T (control) | USA, 1, 1962 | 0.25 | 0.25 | 0.5 | 1/1 | ||||
| Old | France, 11, 1978–1983 | 0.125–0.25 | 0.125–0.25 | 0.25–1 | 11/11 | ||||
| Recent | France, 20, 2009–2012 | 0.25–0.5 | 0.5–1 | 0.5–2 | 17/20 | Asn | |||
| 0.5 | 1 | 2 | 1/20 | Asn | Iled | ||||
| 0.5–1 | 0.5–1 | 2c | 2/20 | Asn | Tyrd | ||||
| Contemporarye | France, 21, 2013–2014 | 0.25–0.5 | 0.5–1 | 0.5–2 | 20/21 | Asn | |||
| 16 | 16 | >64 | 1/21 | Phed | Asn | Iled | |||
| Control | Switzerland, 2, 2010–2011 | 0.125c | 0.25c | 0.5c | 2/2 | Asn | |||
ENR, enrofloxacin; DAN, danofloxacin; MAR, marbofloxacin.
The genotype of PG45T was used as a reference, and E. coli amino acid numbering was used. There were no mutations in ParE.
Same MIC values for both strains.
Data in bold refer to the recently detected clinical isolate with high-level resistance to fluoroquinolones.
In order to determine whether this shift was associated with mutations in the quinolone resistance-determining regions (QRDRs), we then sequenced the gyrA, gyrB, parC, and parE QRDRs. Apart from a silent mutation in ParC (position 84; GAC → GAT; Asp), all recent isolates harbored a single amino acid substitution in GyrB (Asp362Asn) that was absent from all old strains and PG45T. This mutation had already been described and linked to the recent M. bovis subtype ST2 (8). However, this mutation was also present in recent susceptible isolates from Switzerland and has never been described for highly resistant strains, so it is unlikely to be associated with resistance. No mutations in GyrA and ParE were evidenced, and only three isolates showed single mutations in ParC, either in codon 80 (Ser → Ile) or in codon 84 (Asp → Tyr), but without any marked difference in their MICs. Hence, the observed shift in fluoroquinolone susceptibility was not associated with mutations in QRDRs.
We also investigated whether clinical isolates were equally able to achieve higher, clinically relevant MICs through the acquisition of mutations in their QRDRs in vitro. The following three isolates, representing different genomic backgrounds (subtypes and QRDR genotypes), were chosen for the in vitro selection process: PG45T for subtype ST1, 15762 for ST2, and 15527 for ST3 (Table 2). In addition, two isolates belonging to ST2 and showing native alterations in ParC compared to 15762 were also selected (15488 and 15875). Laboratory-generated mutants were obtained by stepwise exposure to increasing concentrations of ENR, as illustrated in Fig. 1 for PG45T.
TABLE 2.
Characterization of the 1st-step enrofloxacin-resistant clones selected in vitro
| Isolate (ST) | Resistant coloniesa |
QRDR mutation at indicated positionb |
Genotype | MIC (μg/ml)c |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENR concn (μg/ml) | Recovery frequency (no. of clones) | Mutation distribution (no. of clones with mutation/total no. of clones) | GyrA |
GyrB |
ParC |
|||||||||
| 81 | 83 | 87 | 362 | 80 | 84 | 98 | ENR | DAN | MAR | |||||
| PG45 (ST1) | Gly | Ser | Glu | Asp | Ser | Asp | Thr | WT | 0.25 | 0.25 | 0.5 | |||
| 0.25 | 2E−05 (20) | 11/20 | GT1 | 0.25 | 1 | 0.5 | ||||||||
| 0.25 | 5/20 | Gly | NA | NA | NA | |||||||||
| 0.25 | 3/20 | Asn | NA | NA | NA | |||||||||
| 0.25 | 1/20 | Asn | Arg | NA | NA | NA | ||||||||
| 15762 (ST2) | Gly | Ser | Glu | Asn | Ser | Asp | Thr | WT | 0.25 | 0.5 | 1 | |||
| 0.25 | 3E−06 (20) | 20/20 | GT4 | 0.25 | 0.5 | 1 | ||||||||
| 15488 (ST2) | Gly | Ser | Glu | Asn | Ile | Asp | Thr | WT | 0.5 | 1 | 2 | |||
| 2 | 6E−07 (5) | 3/5 | Phe | 16 | 16 | >64 | ||||||||
| 2 | 1/5 | Tyr | NA | NA | NA | |||||||||
| 2 | 1/5 | Gly | NA | NA | NA | |||||||||
| 15875 (ST2) | Gly | Ser | Glu | Asn | Ser | Tyr | Thr | WT | 1 | 1 | 2 | |||
| 2 | 4E−07 (4) | 3/4 | Lys | 4 | 8 | 8 | ||||||||
| 2 | 1/4 | Phe | 4 | 8 | 8 | |||||||||
| 15527 (ST3) | Gly | Ser | Glu | Asn | Ser | Asp | Thr | WT | 0.5 | 0.5 | 2 | |||
| 0.5 | 7E−05 (12) | 1/12 | NA | NA | NA | |||||||||
| 0.5 | 6/12 | Val | NA | NA | NA | |||||||||
| 0.5 | 1/12 | Val | Gly | NA | NA | NA | ||||||||
| 0.5 | 2/12 | Phe | NA | NA | NA | |||||||||
| 0.5 | 1/12 | Phe | Asn | GT2 | 4 | 4 | 8 | |||||||
| 0.5 | 1/12 | Phe | Asn | Arg | GT3 | 4 | 2 | 8 | ||||||
| 8619 (ST3) | Gly | Ser | Glu | Asn | Ser | Asp | Thr | WT | 0.25 | 0.5 | 0.5 | |||
| 0.25 | 2E−06 (10) | 8/10 | NA | NA | NA | |||||||||
| 0.25 | 2/10 | Phe | GT5 | 1 | 2 | 1 | ||||||||
Resistant colonies were selected on plates containing enrofloxacin at the indicated concentrations. The recovery frequency was determined as the number of colonies appearing on the plate in the presence of enrofloxacin divided by the number of colonies contained in the inoculum.
Point mutations detected in QRDRs of gyrA, parC, and gyrB are presented as the corresponding amino acid substitutions in individual clones. For each isolate, the WT phenotype is shown first. The GT1, GT2, GT3, and GT4 genotypes were used to perform further selection steps. When the phenotype of an isolate differs from that of PG45T, the mutation is indicated in bold. Newly acquired mutations are underlined.
MIC of an ENR-resistant mutant representing the genotype. NA, not applicable (mutant not available in broth culture).
The first selection step consisted of plating the wild-type (WT) isolate onto agar medium containing ENR at a concentration corresponding to its MIC. Table 2 shows the frequencies at which ENR-resistant clones were recovered and the distribution of randomly selected colonies between the different types of QRDR mutants. In general, the recovery frequency of ENR-resistant clones was low (6 × 10−7 to 2 × 10−5) regardless of the parental subtype. Eight different amino acid substitutions were identified as being encoded in the QRDRs of mutants: five in GyrA, three in ParC, and none in GyrB. No ParE sequences were analyzed, since previous studies had shown that they were seldom altered and that the rare mutations obtained were not associated with fluoroquinolone resistance (10, 12, 17). Two mutations (Ser83Phe in GyrA and Asp84Asn/Gly in ParC) recurred in several mutants derived from different parental isolates. The lowest rate of mutation acquisition was observed for isolate 15762 (ST2), with no mutation at all during the 1st selection step, and also for PG45T (ST1), for which 45% of the selected colonies were mutated (clones A2, A3, and A4 in Fig. 1). Another ST2 isolate, 9072, was also included in the first step of ENR selection but failed to grow under antibiotic pressure, despite repeated attempts. Since the 2nd step of in vitro selection was carried out before sequences were available, many of the generated ENR-resistant colonies were not put into broth culture and hence were not available for susceptibility tests. However, several of the selected ENR-resistant colonies showed 2- to 32-fold increases in the fluoroquinolone MICs (Table 2). Resistant colonies that had not acquired any mutations, such as strains PG45T (clone A1 in Fig. 1) and 15762, showed at most a 2-fold increase in the MIC. In contrast, mutants with alterations in both GyrA and ParC, the latter either inherited from parental strains (isolates 15488 and 15875) or acquired during the selection process (isolate 15527 and clones of genotype 2 [GT2] and GT3), attained MICs as high as 64 μg/ml.
Four ENR-resistant clones with different genotypes (subtypes and QRDRs) were selected during the 1st step for further selection. These were clones of GT1 from PG45T (ST1), GT4 from 15762 (ST2), and GT2 and GT3 from 15527 (ST3) (Table 2).
Additional point mutations were identified in GyrA and ParC during the successive selection steps (Table 3; Fig. 1). The substitutions varied, but their positions in the QRDRs were always the same. The most frequently altered codons were those for Ser83 in GyrA and Asp84 in ParC, and their concomitant mutation was systematically associated with high MICs (Table 3). Another mutation in ParC, namely, Ser80Ile, was associated with a high MIC for PG45 mutants (ST1). The Gly81Asp/Asn mutation in GyrA was observed after the 3rd and 4th selection steps, and only in PG45 mutants. Interestingly, the frequencies of recovery of ENR-resistant clones varied only slightly (10−6 to 10−4) for PG45T (ST1) and 15762 (ST2), regardless of the selection step, but varied a lot (10−7 to 1) for clones derived from 15527 (ST3). More specifically, when the selection pressure was lower than the MIC attained in the 1st selection step, no new mutation was acquired and, as expected, the recovery frequency reached about 1. Furthermore, once the two key mutations (GyrA Ser83Phe and ParC Asp84Asn) had been acquired, the enrofloxacin concentrations could be increased rapidly without influencing the mutant's ability to grow. The switch from Asn to Lys at position 84 of ParC, which corresponds to a change of the amino acid R-group charge from neutral to positive, contributed to further increasing the MIC. In contrast, for isolates belonging to the ST2 subtype, it was either impossible (9072) (data not shown) or difficult (15762) to maintain viable mycoplasma cells under increasing selective pressure, and no mutation in ParC was acquired over the whole selection process.
TABLE 3.
Characteristics of ENR-resistant clones obtained in vitro under increased selective pressure, point mutations in QRDRs, and MIC values
| Clonea | ENR concn (μg/ml)b | Mutation distributionc |
Mutation at indicated position |
MIC (μg/ml)d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA |
ParC |
||||||||||||
| 2nd step | 3rd step | 4th step | Gly81 | Ser83 | Glu87 | Ser80 | Asp84 | Thr98 | ENR | DAN | MAR | ||
| PG45 (GT1 [ST1], GyrB Asp362, 0.25) | 0.5 | 2/7 | Ile | 4 | 8 | 8 | |||||||
| 1 | 18/18 | Ile | 4 | 8 | 16 | ||||||||
| 2 | 3/3 | Ile | 8 | 8 | 16 | ||||||||
| 0.5 | 4/7 | Tyr | 1 | 2 | 2 | ||||||||
| 1 | 1/7 | Tyr | Tyr | 4 | 8 | 8 | |||||||
| 1 | 2/7 | Lys | Tyr | 4 | 8 | 4 | |||||||
| 1 | 1/7 | Phe | Tyr | 4 | 8 | 8 | |||||||
| 1 | 3/7* | Asp | Tyr | 4 | 8 | 4 | |||||||
| 2 | 1/3 | Asn | Tyr | 4 | 8 | 4 | |||||||
| 2 | 2/3 | Asp | Tyr | 4 | 4 | 4 | |||||||
| 0.5 | 1/7 | 1 | 2 | 1 | |||||||||
| 1 | 1/4* | Tyr | 2 | 2 | 4 | ||||||||
| 1 | 1/4* | Phe | 1 | 2 | 2 | ||||||||
| 1 | 1/4 | Tyr | Tyr | 4 | 8 | 8 | |||||||
| 1 | 1/4 | Phe | Tyr | 4 | 8 | 8 | |||||||
| 2 | 1/1 | Tyr | Ile | 16 | 16 | 16 | |||||||
| 2 | 2/2 | Phe | Tyr | 16 | 16 | 16 | |||||||
| 15762 (GT4 [ST2], GyrB Asn362, 0.25) | 0.5 | 2/7 | Lys | ND | ND | ND | |||||||
| 0.5 | 1/7 | 0.5 | 1 | 2 | |||||||||
| 0.5 | 4/7* | 0.5 | 1 | 2 | |||||||||
| 1 | 5/8 | Tyr | ND | ND | ND | ||||||||
| 1 | 1/8 | Phe | ND | ND | ND | ||||||||
| 1 | 1/8 | Lys | ND | ND | ND | ||||||||
| 1 | 1/8 | ND | ND | ND | |||||||||
| 15527 (GT2 [ST3], GyrB Asn362, 4) | 1 | 12/12 | Phe | Asn | 8 | 8 | 16 | ||||||
| 2 | 36/36 | Phe | Asn | ND | ND | ND | |||||||
| 8–16 | 5/6 | Phe | Asn | 16 | 8 | 16 | |||||||
| 8–16 | 1/6 | Phe | Lys | 32 | 8 | 16 | |||||||
| 15527 (GT3 [ST3], GyrB Asn362, 4) | 1 | 12/12 | Phe | Asn | Arg | 8 | 8 | 16 | |||||
| 2 | 36/36 | Phe | Asn | Arg | ND | ND | ND | ||||||
| 8–16 | 1/2 | Phe | Asn | Arg | 16 | 8 | 16 | ||||||
| 8–16 | 1/2 | Phe | Lys | Arg | 16 | 16 | 32 | ||||||
| 8619 (GT5 [ST3], GyrB Asn362, 1)e | 0.5 | 22/22 | Phe | 1 | 2 | 2 | |||||||
| 1 | 3/4 | Phe | ND | ND | ND | ||||||||
| 1 | 1/4* | Phe | Ile | 32 | 16 | >64 | |||||||
| 2 | 2/2 | Phe | Ile | 32 | 16 | >64 | |||||||
| 0.5 | 22/22 | Phe | 1 | 2 | 2 | ||||||||
| 1 | 12/28 | Phe | ND | ND | ND | ||||||||
| 1 | 2/28 | Phe | Gly | 8 | 8 | 16 | |||||||
| 1 | 4/28* | Phe | Asn | 8 | 8 | 16 | |||||||
| 1 | 10/28* | Phe | Tyr | 8 | 8 | 16 | |||||||
| 2 | 2/2 | Phe | Asn | 8 | 8 | 16 | |||||||
| 2 | 2/2 | Phe | Tyr | 8 | 8 | 16 | |||||||
Clones are presented by the name of the parental strain, the subtype, the GyrB phenotype, and the enrofloxacin MIC (μg/ml) at the 1st selection step.
Concentration of ENR used to select the different resistant clones during individual passages.
Asterisks indicate clones used to perform the following selection step.
MICs were evaluated on at least one clone of the selected genotype. When no clone was available in broth culture, MIC assays were not performed. ND, not done.
Step 3 was performed using two different clones (same genotype) randomly selected at step 2.
Because the ability to develop resistance to fluoroquinolones through mutations in the QRDRs under selective pressure in vitro was different for the different subtypes, we investigated the evolution of clinical isolates by screening a more recent set of isolates. The susceptibilities to fluoroquinolones and the subtypes of 21 clinical isolates collected in 2013 and 2014 were determined (Table 1; see Table S1 in the supplemental material). The poorly abundant subtype ST3, which represented only 5% of the recent (2009 to 2012) M. bovis population, was found to have become more prevalent (20%) in the contemporary (2013 and 2014) M. bovis population. The MIC distributions were identical to those for the 2009-2012 isolates, except for one isolate, namely, 8428, that was shown to be highly resistant to all 3 fluoroquinolones (MICs of ≥16 μg/ml). These high MIC values were associated with the Ser83Phe mutation in GyrA and the Ser80Ile mutation in ParC, i.e., two of the mutations that had been observed in mutants selected in vitro. Furthermore, this isolate was demonstrated to be of the ST3 molecular subtype (see Table S1).
The clustering of isolates into different subtypes based on single-gene polymorphism (polC) was further confirmed using the MLST scheme proposed by Register et al. (21). The polC-defined ST1, ST2, and ST3 subtypes were grouped under ST17, ST18, and ST5, respectively, in the MLST tree of Register et al., with ST17 and ST18 belonging to the same clade, thereby confirming the distribution of isolates between different genotypes. Moreover, another isolate from the ST3 subtype, namely, 8619, was randomly chosen to confirm our in vitro selection results obtained with isolate 15527, originally chosen as representative of ST3. Twenty percent of the isolate 8619 colonies selected in the 1st step were shown to have gained a mutation in GyrA (Ser83Phe) (Table 2) associated with a moderate increase in MICs. In the 3rd selection step, as soon as some of the ParC codons had been altered (Ser80Ile or Asp84Gly/Asn/Tyr), the MICs reached very high values. This result confirms the ease with which ST3 isolates acquire and select key point mutations in QRDRs that lead to a high level of resistance to fluoroquinolones.
DISCUSSION
Most often, a low level of resistance to fluoroquinolones, associated with a narrow MIC distribution in the range of 0.25 to 4 μg/ml, is considered to represent a single homogeneous population of isolates (6). However, in the French M. bovis population, the MIC distribution of enrofloxacin, in the range of 0.125 to 1 μg/ml, was previously shown to be bimodal, with a very moderate shift of MICs between old and recent isolates (5). This shift was further associated with a modification of the molecular subtype from ST1 (old isolates) to the predominant subtype ST2 (recent isolates) (8). In the present work, we confirmed the existence of two groups of clinical isolates with different MIC levels. Old strains, isolated between 1978 and 1983, were highly susceptible (MIC range of 0.125 to 1 μg/ml), while recent strains, isolated between 2009 and 2014, were moderately susceptible or intermediate to fluoroquinolones (MIC range of 0.25 to 2 μg/ml). However, no mutations in the QRDR region were evidenced that could explain this loss of susceptibility. The only recurrent mutation that was present in all recent strains and absent from old ones was Asp362Asn, encoded in the gyrB QRDR. However, alterations in the gyrB gene have rarely been associated with a loss of susceptibility to fluoroquinolones, except in M. gallisepticum, where the Asp362Asn substitution was detected once in mutants selected in vitro (30). Furthermore, two isolates from Switzerland, included in this study as controls, harbored the same mutation but still remained very sensitive to fluoroquinolones (0.125 to 0.25 μg/ml). These findings suggest that the Asp362Asn mutation in gyrB is more likely a molecular marker that discriminates old from recent isolates, as already suggested (8), but does not play a role in isolate susceptibility. Two other mutations, in parC, were observed in 3 clinical isolates but were associated with MICs that did not differ from the range for the overall isolate population. These parC mutations had previously been described for mutants of nonruminant mycoplasmas selected in vitro (28, 30, 31) and had been identified in clinical isolates of M. bovis (10, 12). However, only in the case of a concomitant alteration of the GyrA codon for position 83 had they been associated with high resistance (4 to 16 μg/ml).
In the absence of associated alterations of the QRDRs, active efflux is the only known alternative mechanism in mycoplasmas that could lead to acquired resistance to fluoroquinolones and might explain the moderate shift in susceptibility in our set of isolates. It has been described for M. hominis strains selected on ethidium bromide in vitro (19, 20). M. hominis is a human urogenital mycoplasma that belongs to the same phylogenetic group as M. bovis, the so-called hominis group. Also, M. hominis active efflux has been linked to the overexpression of 2 genes, namely, md1 and md2, encoding multidrug resistance ATP-binding cassette (ABC) transporters that are constitutively expressed in the reference strain (20). In another ruminant mycoplasma, M. mycoides subsp. capri, it was recently shown that orthovanadate, an inhibitor of ABC efflux pumps, was able to induce a 2-fold decrease of the MICs of 3 fluoroquinolones in both clinical and in vitro mutants, suggesting once again the contribution of an efflux mechanism to the overall resistance patterns of isolates (18).
The efflux hypothesis for M. bovis is consistent with (i) the presence in the genome of PG45T of genes encoding several ABC transporters with a predicted role in drug resistance and a multidrug efflux transporter of the MatE family; (ii) the moderate increase of the MICs observed between the recent and old M. bovis populations, as efflux systems usually confer low levels of resistance (32, 33); and (iii) the slight differences in the MICs of different fluoroquinolones, as efflux system efficiency usually depends on the hydrophobicity profiles and molecular masses of the extruded fluoroquinolones (34, 35). However, in our set of isolates, we failed to experimentally demonstrate the efflux hypothesis, as we were unable to accumulate enrofloxacin inside mycoplasma cells in a reproducible manner.
Under increased antimicrobial pressure, the selected M. bovis clones were able to gain resistance and to achieve higher MICs than those of their parental clinical isolates. In most cases, concomitant alterations were observed in the QRDRs, with the exception of some clones derived from PG45T (1st step) (Table 2) and 15762 (2nd step) (Table 3) that showed slightly higher MICs despite their WT QRDR genotype. The latter observation is also in favor of a native efflux system that can be upregulated to better allow tolerance of increased antimicrobial concentrations. The point mutations observed in the present study were identical to hot spots for resistance to fluoroquinolones previously described for various mycoplasma species (10, 12, 19, 30, 36, 37). These substitutions included 5 in GyrA (2 in codon 83 [Ser83Phe/Tyr] and 3 in codon 87 [Glu87Val/Gly/Lys]) and 4 in ParC (1 in codon 80 [Ser80Ile], 1 in codon 98 [Thr98Arg], and 2 in codon 84 [Asp84Asn/Tyr]). The most frequently observed substitutions were Ser83Phe in GyrA and Asp84Asn/Tyr in ParC, leading to 8- to 16-fold increases in the MICs, consistent with results from previous studies of M. pneumoniae in vitro (28) and M. bovis in vivo (12). Another combination of mutations was observed less frequently (only 3 of 72 selected clones), namely, Ser83Phe in GyrA and Ser80Ile in ParC, and was associated with 16- to 128-fold increases in the MICs. In contrast to our results, this combination of mutations was reported to occur very frequently in vitro for M. hominis (28) and for Japanese and Chinese clinical isolates of M. bovis (10, 38). Hence, our investigations of in vitro resistance selection clearly confirmed the existence of hot spots for mutations conferring high resistance levels and the cumulative effects of mutations in GyrA and ParC on the MICs. We also showed that the sequence in which mutations were accumulated by an isolate could vary as a function of the strain's molecular subtype. The 1st mutation appeared (i) at ParC position 84 for PG45T (ST1), (ii) at GyrA position 83 for strain 15762 (ST2), (iii) at GyrA when ParC was already mutated in the WT genotype (15488; ST2), and (iv) equally in both genes for strain 15527 or 8619 (both ST3).
Furthermore, the frequency of resistant clone selection was also shown to vary between isolates. The recovery of PG45T (ST1) and 15762 (ST2) was low during all the selection steps, whereas that of ST3 clones (15527 and 8619) could become high once they had acquired the double mutation in GyrA83 and ParC80/84. ST2 mutants had difficulty in acquiring alterations in QRDRs and failed to mutate in parC, and hence to accumulate the parC and gyrA mutations associated with high-level resistance. In contrast, the ST3 clones easily accumulated mutations leading to high levels of resistance. These results suggest that ST2 isolates are somehow blocked in a configuration which does not facilitate a gain in resistance and could have helped to limit the emergence of resistance in clinical isolates in France, most of which belong to ST2 (8). In contrast, ST3 isolates seemed able to rapidly and efficiently gain a high level of resistance. It is tempting to speculate that ST3 isolates are able to counteract the fitness loss associated with topoisomerase mutations. Whether they have increased mutation rates due to deficient DNA repair systems, which could facilitate the acquisition of compensatory mutations, has yet to be investigated. This hypothesis does not exclude the contribution of efflux systems to the early steps toward resistance.
The recent increased prevalence of ST3 isolates, from 5 to 20%, in the French M. bovis population supports this hypothesis. Indeed, the selective pressure imposed by the use of antimicrobials might result in the selection of this subtype that used to be rare (8). Furthermore, the highly resistant clinical isolate detected in 2014 (isolate 8428), with an MIC of 16 μg/ml for ENR and DAN and of >64 μg/ml for MAR, harbored the same mutations as the in vitro mutants (Ser83Phe in GyrA and Ser80Ile in ParC) and was of the ST3 subtype. This nicely emphasizes the need to survey the selection and spread of ST3 strains in France. Actually, this subtype was already described in Switzerland and Austria (8) and in the United States, as early as 2000 and 2002 (13, 21). Whether it was imported into France through international cattle trade has yet to be ascertained. Direct importation is unlikely, since according to the history of U.S. cattle trade (http://www.ers.usda.gov/data-products/livestock-meat-international-trade-data.aspx), only approximately 1,000 heads of cattle have been exported from the United States to Europe since 2002, and never to France.
Conclusions.
This study attempted to decipher the molecular mechanisms responsible for the moderate decrease in susceptibility of recent French clinical isolates of M. bovis to fluoroquinolones. No direct link was established between QRDR polymorphisms and the shift in MIC values between old and recent isolates. Although an efflux system appears to be the most probable mechanism behind the observed phenotypes, our investigations in vitro were inconclusive. However, we showed that different clinical isolates, with different initial MICs and different genetic subtypes, were not equal in the ability to gain resistance in vitro. Notably, isolates belonging to the subtype 3 cluster were more likely to rapidly accumulate mutations in their QRDRs under selective pressure in vitro and hence to become resistant. These results are congruent with in vivo observations, as the first resistant clinical isolate reported in France was shown to belong to subtype 3. The relative potential contributions of (i) active efflux pumps and (ii) increased mutation rates leading to compensatory mutations to the process of developing resistance to fluoroquinolones in ST3 have yet to be explored. Lastly, the increasing prevalence of subtype 3 observed in the French population of M. bovis isolates strongly indicates an urgent need to control the veterinary usage of fluoroquinolones in France in order to preserve the favorable epidemiological situation there, where the level of resistance to fluoroquinolones is only moderate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Agnès Tricot for her excellent technical assistance, Patrice Cuchet for performing membrane filtration-dot identification of isolates, François Poumarat for providing clinical isolates with known MICs, and all the members of the VIGIMYC network for supplying several M. bovis isolates with documented clinical histories. We are also grateful to Paola Pilo, who kindly provided isolates from Switzerland.
Dima Khalil's Ph.D. thesis is jointly funded by the French Agency for Food, Environmental and Occupational Health & Safety (ANSES) and the Institute of Higher Education in Food Science, Animal Health, Agricultural and Environmental Sciences (VetAgro Sup).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03280-15.
REFERENCES
- 1.Maunsell FP, Woolums AR, Francoz D, Rosenbusch RF, Step DL, Wilson DJ, Janzen ED. 2011. Mycoplasma bovis infections in cattle. J Vet Intern Med 25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 2.Rerat M, Albini S, Jaquier V, Hussy D. 2012. Bovine respiratory disease: efficacy of different prophylactic treatments in veal calves and antimicrobial resistance of isolated Pasteurellaceae. Prev Vet Med 103:265–273. doi: 10.1016/j.prevetmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Soehnlen MK, Aydin A, Lengerich EJ, Houser BA, Fenton GD, Lysczek HR, Burns CM, Byler LI, Hattel AL, Wolfgang DR, Jayarao BM. 2011. Blinded, controlled field trial of two commercially available Mycoplasma bovis bacterin vaccines in veal calves. Vaccine 29:5347–5354. doi: 10.1016/j.vaccine.2011.05.092. [DOI] [PubMed] [Google Scholar]
- 4.O'Connor AM, Coetzee JF, da Silva N, Wang C. 2013. A mixed treatment comparison meta-analysis of antibiotic treatments for bovine respiratory disease. Prev Vet Med 110:77–87. doi: 10.1016/j.prevetmed.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Gautier-Bouchardon AV, Ferre S, Le Grand D, Paoli A, Gay E, Poumarat F. 2014. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS One 9:e87672. doi: 10.1371/journal.pone.0087672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer S, Galland D, Guerin-Faublee V, Giboin H, Woehrle-Fontaine F. 2012. Survey of marbofloxacin susceptibility of bacteria isolated from cattle with respiratory disease and mastitis in Europe. Vet Rec 170:53. doi: 10.1136/vr.100246. [DOI] [PubMed] [Google Scholar]
- 7.Nickell JS, White BJ. 2010. Metaphylactic antimicrobial therapy for bovine respiratory disease in stocker and feedlot cattle. Vet Clin North Am Food Anim Pract 26:285–301. doi: 10.1016/j.cvfa.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Becker CA, Thibault FM, Arcangioli MA, Tardy F. 2015. Loss of diversity within Mycoplasma bovis isolates collected in France from bovines with respiratory diseases over the last 35 years. Infect Genet Evol 33:118–126. doi: 10.1016/j.meegid.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Ayling RD, Rosales RS, Barden G, Gosney FL. 2014. Changes in antimicrobial susceptibility of Mycoplasma bovis isolates from Great Britain. Vet Rec 175:486. doi: 10.1136/vr.102303. [DOI] [PubMed] [Google Scholar]
- 10.Sato T, Okubo T, Usui M, Higuchi H, Tamura Y. 2013. Amino acid substitutions in GyrA and ParC are associated with fluoroquinolone resistance in Mycoplasma bovis isolates from Japanese dairy calves. J Vet Med Sci 75:1063–1065. doi: 10.1292/jvms.12-0508. [DOI] [PubMed] [Google Scholar]
- 11.Thomas A, Nicolas C, Dizier I, Mainil J, Linden A. 2003. Antibiotic susceptibilities of recent isolates of Mycoplasma bovis in Belgium. Vet Rec 153:428–431. doi: 10.1136/vr.153.14.428. [DOI] [PubMed] [Google Scholar]
- 12.Lysnyansky I, Mikula I, Gerchman I, Levisohn S. 2009. Rapid detection of a point mutation in the parC gene associated with decreased susceptibility to fluoroquinolones in Mycoplasma bovis. Antimicrob Agents Chemother 53:4911–4914. doi: 10.1128/AAC.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbusch RF, Kinyon JM, Apley M, Funk ND, Smith S, Hoffman LJ. 2005. In vitro antimicrobial inhibition profiles of Mycoplasma bovis isolates recovered from various regions of the United States from 2002 to 2003. J Vet Diagn Invest 17:436–441. doi: 10.1177/104063870501700505. [DOI] [PubMed] [Google Scholar]
- 14.Sulyok KM, Kreizinger Z, Fekete L, Hrivnak V, Magyar T, Janosi S, Schweitzer N, Turcsanyi I, Makrai L, Erdelyi K, Gyuranecz M. 2014. Antibiotic susceptibility profiles of Mycoplasma bovis strains isolated from cattle in Hungary, Central Europe. BMC Vet Res 10:256. doi: 10.1186/s12917-014-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uemura R, Sueyoshi M, Nagatomo H. 2010. Antimicrobial susceptibilities of four species of Mycoplasma isolated in 2008 and 2009 from cattle in Japan. J Vet Med Sci 72:1661–1663. doi: 10.1292/jvms.10-0165. [DOI] [PubMed] [Google Scholar]
- 16.Ayling RD, Baker SE, Peek ML, Simon AJ, Nicholas RA. 2000. Comparison of in vitro activity of danofloxacin, florfenicol, oxytetracycline, spectinomycin and tilmicosin against recent field isolates of Mycoplasma bovis. Vet Rec 146:745–747. doi: 10.1136/vr.146.26.745. [DOI] [PubMed] [Google Scholar]
- 17.Waites KB, Lysnyansky I, Bébéar CM. 2014. Emerging antimicrobial resistance in mycoplasmas of humans and animals, p 289–322. In Browning GF, Citti C (ed), Mollicutes—molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 18.Antunes NT, Assuncao P, Poveda JB, Tavio MM. 2015. Mechanisms involved in quinolone resistance in Mycoplasma mycoides subsp. capri. Vet J 204:327–332. doi: 10.1016/j.tvjl.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Raherison S, Gonzalez P, Renaudin H, Charron A, Bebear C, Bebear CM. 2002. Evidence of active efflux in resistance to ciprofloxacin and to ethidium bromide by Mycoplasma hominis. Antimicrob Agents Chemother 46:672–679. doi: 10.1128/AAC.46.3.672-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raherison S, Gonzalez P, Renaudin H, Charron A, Bebear C, Bebear CM. 2005. Increased expression of two multidrug transporter-like genes is associated with ethidium bromide and ciprofloxacin resistance in Mycoplasma hominis. Antimicrob Agents Chemother 49:421–424. doi: 10.1128/AAC.49.1.421-424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Register KB, Thole L, Rosenbush RF, Minion FC. 2015. Multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison. Vet Microbiol 175:92–98. doi: 10.1016/j.vetmic.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Chazel M, Tardy F, Le Grand D, Calavas D, Poumarat F. 2010. Mycoplasmoses of ruminants in France: recent data from the national surveillance network. BMC Vet Res 6:32. doi: 10.1186/1746-6148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poumarat F, Perrin B, Longchambon D. 1991. Identification of ruminant mycoplasmas by dot immunobinding on membrane filtration (MF dot). Vet Microbiol 29:329–338. doi: 10.1016/0378-1135(91)90140-B. [DOI] [PubMed] [Google Scholar]
- 24.Marenda MS, Sagne E, Poumarat F, Citti C. 2005. Suppression subtractive hybridization as a basis to assess Mycoplasma agalactiae and Mycoplasma bovis genomic diversity and species-specific sequences. Microbiology 151:475–489. doi: 10.1099/mic.0.27590-0. [DOI] [PubMed] [Google Scholar]
- 25.Aebi M, Bodmer M, Frey J, Pilo P. 2012. Herd-specific strains of Mycoplasma bovis in outbreaks of mycoplasmal mastitis and pneumonia. Vet Microbiol 157:363–368. doi: 10.1016/j.vetmic.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Poumarat F, Longchambon D, Martel JL. 1992. Application of dot immunobinding on membrane filtration (MF dot) to the study of relationships within “M. mycoides cluster” and within “glucose and arginine-negative cluster” of ruminant mycoplasmas. Vet Microbiol 32:375–390. [DOI] [PubMed] [Google Scholar]
- 27.Hannan PC. 2000. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International Research Programme on Comparative Mycoplasmology. Vet Res 31:373–395. [DOI] [PubMed] [Google Scholar]
- 28.Gruson D, Pereyre S, Renaudin H, Charron A, Bebear C, Bebear CM. 2005. In vitro development of resistance to six and four fluoroquinolones in Mycoplasma pneumoniae and Mycoplasma hominis, respectively. Antimicrob Agents Chemother 49:1190–1193. doi: 10.1128/AAC.49.3.1190-1193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen WP, Kuo TT. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res 21:2260. doi: 10.1093/nar/21.9.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhardt AK, Bebear CM, Kobisch M, Kempf I, Gautier-Bouchardon AV. 2002. Characterization of mutations in DNA gyrase and topoisomerase IV involved in quinolone resistance of Mycoplasma gallisepticum mutants obtained in vitro. Antimicrob Agents Chemother 46:590–593. doi: 10.1128/AAC.46.2.590-593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bebear CM, Renaudin J, Charron A, Renaudin H, de Barbeyrac B, Schaeverbeke T, Bebear C. 1999. Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob Agents Chemother 43:954–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob Agents Chemother 44:2595–2599. doi: 10.1128/AAC.44.10.2595-2599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrandiz MJ, Oteo J, Aracil B, Gomez-Garces JL, De La Campa AG. 1999. Drug efflux and parC mutations are involved in fluoroquinolone resistance in viridans group streptococci. Antimicrob Agents Chemother 43:2520–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaatz GW, Seo SM, Ruble CA. 1993. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother 37:1086–1094. doi: 10.1128/AAC.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piddock LJ, Jin YF, Griggs DJ. 2001. Effect of hydrophobicity and molecular mass on the accumulation of fluoroquinolones by Staphylococcus aureus. J Antimicrob Chemother 47:261–270. doi: 10.1093/jac/47.3.261. [DOI] [PubMed] [Google Scholar]
- 36.Bebear CM, Grau O, Charron A, Renaudin H, Gruson D, Bebear C. 2000. Cloning and nucleotide sequence of the DNA gyrase (gyrA) gene from Mycoplasma hominis and characterization of quinolone-resistant mutants selected in vitro with trovafloxacin. Antimicrob Agents Chemother 44:2719–2727. doi: 10.1128/AAC.44.10.2719-2727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bebear CM, Renaudin H, Charron A, Bove JM, Bebear C, Renaudin J. 1998. Alterations in topoisomerase IV and DNA gyrase in quinolone-resistant mutants of Mycoplasma hominis obtained in vitro. Antimicrob Agents Chemother 42:2304–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mustafa R, Qi J, Ba X, Chen Y, Hu C, Liu X, Tu L, Peng Q, Chen H, Guo A. 2013. In vitro quinolones susceptibility analysis of Chinese Mycoplasma bovis isolates and their phylogenetic scenarios based upon QRDRs of DNA topoisomerases revealing a unique transition in ParC. Pak Vet J 33:364–369. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.