Abstract
Insecticidal protein genes from the bacterium Bacillus thuringiensis (Bt) are expressed by transgenic Bt crops (Bt crops) for effective and environmentally safe pest control. The development of resistance to these insecticidal proteins is considered the most serious threat to the sustainability of Bt crops. Resistance in fall armyworm (Spodoptera frugiperda) populations from Puerto Rico to transgenic corn producing the Cry1Fa insecticidal protein resulted, for the first time in the United States, in practical resistance, and Bt corn was withdrawn from the local market. In this study, we used a field-collected Cry1Fa corn-resistant strain (456) of S. frugiperda to identify the mechanism responsible for field-evolved resistance. Binding assays detected reduced Cry1Fa, Cry1Ab, and Cry1Ac but not Cry1Ca toxin binding to midgut brush border membrane vesicles (BBMV) from the larvae of strain 456 compared to that from the larvae of a susceptible (Ben) strain. This binding phenotype is descriptive of the mode 1 type of resistance to Bt toxins. A comparison of the transcript levels for putative Cry1 toxin receptor genes identified a significant downregulation (>90%) of a membrane-bound alkaline phosphatase (ALP), which translated to reduced ALP protein levels and a 75% reduction in ALP activity in BBMV from 456 compared to that of Ben larvae. We cloned and heterologously expressed this ALP from susceptible S. frugiperda larvae and demonstrated that it specifically binds with Cry1Fa toxin. This study provides a thorough mechanistic description of field-evolved resistance to a transgenic Bt crop and supports an association between resistance and reduced Cry1Fa toxin binding and levels of a putative Cry1Fa toxin receptor, ALP, in the midguts of S. frugiperda larvae.
INTRODUCTION
Selected cry and/or vip insecticidal genes from the bacterium Bacillus thuringiensis are expressed in transgenic Bt crops, which provide relevant benefits to growers and the environment compared to synthetic pesticides (1). These benefits have supported a continuous increase in Bt crop adoption since their commercialization (2). These high levels of Bt crop adoption represent increased selection pressure for the evolution of insect resistance, which is currently considered the major threat to the sustainability of this technology. Almost 2 decades since the commercialization of Bt crops targeting lepidopteran pests, field control failures due to the evolution of insect resistance in lepidopteran pests have been documented for populations of Busseola fusca (maize stalk borer) in Cry1Ab corn in South Africa (3), Pectinophora gossypiella (pink bollworm) in Cry1Ac cotton in India (4), and Spodoptera frugiperda (fall armyworm) in Cry1Fa corn in Puerto Rico (5) and Brazil (6). More specifically, high levels of S. frugiperda resistance to Bt corn event TC1507 in Puerto Rico represented the first case in a U.S. territory of practical resistance (7) in a lepidopteran pest resulting in withdrawal of the Bt crop event from the local market (5). Recent reports support the evolution of resistance to the Bt corn event TC1507 in S. frugiperda from Brazil (8) and suggest the possibility that Bt corn-resistant S. frugiperda strains may have migrated to the southeastern United States (9).
Even though diverse aspects of resistance to TC1507 corn in S. frugiperda from Puerto Rico have thoroughly been studied (10–13), little information is available on the biochemical mechanisms responsible for field-evolved resistance in these insects. Theoretically, resistance to Bt corn may involve alterations in any of the steps in the modes of action of Bt toxins. Commonly accepted steps in the Cry intoxication model in lepidopteran larvae include solubilization and proteolysis in the gut digestive fluids of the target insect to yield an activated Cry toxin core, which then binds to receptors on the brush border membrane of the midgut epithelium and inserts into the cell membrane to form a pore responsible for osmotic cell lysis (14). Recent qualitative studies suggest an association between reduced binding and field-evolved resistance to TC1507 corn in S. frugiperda from Brazil (6). Reduced levels of diverse putative Cry1Fa toxin receptors were detected in strains of S. frugiperda from Puerto Rico with field-evolved resistance to TC1507 corn compared to the levels in susceptible insects (15, 16), yet the mechanistic relevance of these differences to resistance was not addressed.
The most commonly reported and well-studied Cry toxin receptors in lepidopteran insects include cadherin, aminopeptidase N (APN), alkaline phosphatase (ALP), and ATP binding cassette (ABC) proteins (17). The most reported resistance phenotype in laboratory-selected insect strains, the mode 1 type of resistance, is characterized by recessive inheritance, high levels (>500-fold) of resistance to at least one Cry1A toxin, lack of cross-resistance to Cry1Ca toxin, and reduced binding of at least one Cry1A toxin (18). This mode 1-type resistance has been linked genetically to mutations in cadherin genes in laboratory-selected strains of Heliothis virescens (19), P. gossypiella (20), and Helicoverpa armigera (21). In contrast, mode 1-type resistance to Bt sprays after field or greenhouse selection in Plutella xylostella and Trichoplusia ni, respectively, has been mapped to a multigene locus containing an ABC subfamily C2 transporter (ABCC2) gene (22). While no genetic linkage data are available, altered expression of ALP genes has been reported as being associated with resistance to Cry1 toxins in strains of H. virescens, H. armigera, Helicoverpa zea, and S. frugiperda (15, 16, 23). More recently, the finding that reduced expression of the mitogen-activated protein kinase 4 (MAPK4) gene trans-regulates reduced expression of the ALP and ABCC2 genes has been reported as being linked to resistance to Cry1Ac in diverse strains of P. xylostella (24).
The goal of this study was to identify the biochemical mechanism responsible for field-evolved resistance to corn event TC1507 in a strain of S. frugiperda (456) initiated from insects collected from corn fields in Puerto Rico (25). Our data provide a mechanistic description of field resistance to TC1507 corn and support the idea of ALP proteins being putative Cry1Fa toxin receptors, whose altered expression is associated with reduced Cry1Fa toxin binding in the midguts of resistant S. frugiperda larvae. This information is critical for an assessment of the effectiveness of resistance management practices, the development of methods to monitor resistance, and the design of improved Cry toxins and Bt crops.
MATERIALS AND METHODS
Insects.
Eggs of a Cry1Fa-susceptible strain of S. frugiperda (Ben) were purchased from Benzon Research (Carlisle, PA). The 456 strain of S. frugiperda was generated from a cross of two moths as part of a set of multiple single-pair crossings of insects obtained from corn fields in the Santa Isabel region of Puerto Rico. Before being submitted to selection, larvae of the 456 strain displayed high levels of field-evolved resistance to Cry1Fa (>7,500-fold) and Cry1Ac (>40-fold) toxins compared to those of susceptible S. frugiperda strains from diverse geographies (25). Larvae of the 456 and Ben strains were reared in the laboratory for about 24 generations and selected for four generations until they reached the third-instar stage on fresh corn leaf tissue from event TC1507 (for 456) or plants from TC1507's nontransgenic isoline, 2T777 (for Ben), before completion of the assays. The survivors from selection were transferred to an artificial diet (Bio-Serv) to complete their development. Previous work supports the ideas that the 456 strain is 100% homozygous for a single recessive resistance allele (12) and that these insects are susceptible to Bt pesticides (13). Adults from the Ben and 456 strains were maintained in plastic containers for mating and oviposition and fed with a 10% sucrose solution impregnated in cotton balls. Crosses between moths from the Ben and 456 strains were prepared by sexing pupae from the specific strain and mixing 20 females and 20 males in the same container for mating and oviposition. The offspring from a cross between 456 females and Ben males are designated the 456F strain, while larvae from the reciprocal cross are designated the 456M strain. All insect rearing and bioassays were performed in incubators at 24 ± 2°C, 50% relative humidity, and an 18-h light/6-h dark photoperiod.
Toxin production and purification.
B. thuringiensis strain HD-73 producing Cry1Ac and a recombinant Escherichia coli strain (ECE54) producing Cry1Ab were obtained from the Bacillus Genetic Stock Center (Columbus, OH, USA). A recombinant Bt strain containing the cry1Ca toxin gene from B. thuringiensis subsp. entomocidus strain 60.5 (26) was kindly provided by Jean Louis Schwartz (University of Montreal, Canada). Cry1Ac, Cry1Ab, and Cry1Ca protoxins were solubilized, activated, and purified as described elsewhere (27, 28). The full-length cry1Fa gene was expressed in Pseudomonas fluorescens and isolated from inclusion bodies that were washed with water and then solubilized in 20 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 11) plus 10 mM dithiothreitol (DTT). After the removal of insoluble material by centrifugation, the protoxin was processed to toxin by incubation in 0.5% (wt/vol) tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma, St. Louis, MO) for 1 h at room temperature. Activated toxin was then loaded onto a Pharmacia Mono Q 10/10 column equilibrated with 20 mM CAPS (pH 10.5) (buffer A) and eluted using a linear gradient of 0.5 M NaCl in buffer A. Fractions containing purified Cry1Fa toxin, as determined by SDS-PAGE examination, were pooled and purified further through a Superose 6 column (1.6-cm diameter, 60 cm long). Fractions comprising a single peak corresponding to the monomeric size of the Cry1Fa toxin were combined and concentrated. Protein concentration was determined using the Qubit protein assay kit in a Qubit fluorometer (Life Technologies), according to the manufacturer's instructions, and samples were maintained at −80°C until used (typically <2 months).
Bioassays.
For the leaf bioassays, we used transgenic Bt corn plants expressing Cry1Fa (event TC1507) or corn plants from their corresponding nontransgenic isoline (2T777). Excised leaf sections (approximately 2.5 cm in length and 1.5 cm in width) from the 6-7 leaf stage were placed in individual plastic cups containing 1% agar to prevent desiccation. A single S. frugiperda neonate larva was placed in each cup, with 32 larvae used per treatment and each treatment replicated in two independent bioassays. Leaf sections were replaced as needed during the bioassay, and mortality was recorded 7 days after larval inoculation. The percent mortality was calculated for each strain and tested for significance in SigmaPlot version 11 (Systat Software, San Jose, CA), using a one-way analysis of variance (ANOVA) with the post hoc Holm-Šidák test for multiple comparisons versus Ben as the control group (P < 0.01). Estimation of the dominance parameter (h) was performed, as previously described (29), except that the Schneider-Orelli formula (30) was used to correct for control mortality before estimating the fitness of each genotype.
For Cry1Ca toxin bioassays, six purified toxin dilutions were prepared in phosphate-buffered saline (PBS) buffer (135 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1.7 mM KH2PO4 [pH 7.5]) and 30-μl aliquots were used to contaminate the surface of artificial diet previously poured on a bioassay tray well (EC International). A single S. frugiperda neonate larva was inoculated in each well, and 32 larvae were tested per toxin concentration, with the bioassay being performed twice. Mortality was scored on day 7 after larval inoculation. The 50% lethal concentration (LC50) value and slope of the dose-mortality curve were calculated using the PoloPlus software (31). Differences between LC50s were considered significant if there was no overlap between the corresponding fiducial limits.
BBMV preparation and specific enzyme activity assays.
Midguts were dissected from fourth-instar S. frugiperda larvae and used to prepare brush border membrane vesicles (BBMV) by the differential magnesium centrifugation method (32) but substituting mannose for sucrose in all solutions (33). The final BBMV pellets were suspended in ice-cold PBS buffer and their protein concentrations determined using the Coomassie Plus protein assay (Pierce) with bovine serum albumin (BSA) as a standard. To determine the purity of the BBMV preparations, the activities of aminopeptidase N (APN) and alkaline-phosphatase (ALP) were compared between initial midgut homogenates and final BBMV preparations, as described previously (34). Activities for APN and ALP were enriched 6- to 8- and 4- to 6-fold, respectively, in BBMV preparations compared to those in the initial midgut homogenates.
Specific activity in BBMV (1 μg) was quantified using p-nitrophenyl phosphate (pNPP) and leucine-p-nitroanilide as the substrates for ALP and APN, respectively, as described elsewhere (34). One enzymatic unit was defined as the amount of enzyme that would hydrolyze 1.0 μmol substrate to the chromogenic product per minute at the specific reaction pH and temperature. The experiments were performed in triplicate using at least three independent BBMV sample preparations. The significance of the differences in activity was tested using one-way analysis of variance (ANOVA) and a post hoc Holm-Šidák test for multiple pairwise comparisons (P < 0.01) in the SigmaPlot version 11.0 software. Since the APN activity data failed an equal-variance test, in this case, we used ANOVA on ranks (P < 0.01, Kruskal-Wallis test) to determine statistical significance.
Labeling of Cry toxins.
Purified Cry1Ab, Cry1Ac, and Cry1Ca toxins (1 μg) were radiolabeled with 0.5 mCi of iodine-125 (PerkinElmer) using the chloramine T method (35). The amount of labeled toxin was increased to 20 μg for Cry1Fa toxin to generate samples amenable to use in binding assays (36). The purity of labeled toxin preparations was monitored by 10% SDS-PAGE and autoradiography. Specific activity, calculated based on input toxin, for the labeled toxins was 3.49 mCi/pmol for Cry1Ab, 2.02 mCi/pmol for Cry1Ac, 0.37 mCi/pmol for Cry1Ca, and 0.04 mCi/pmol for Cry1Fa.
Biotinylation of purified activated Cry1Fa toxin (0.25 mg) was performed using the EZ-Link Sulfo-NHS-LC-Biotin kit (Life Technologies) and using a 1:30 molar ratio (Cry1Fa to biotin). Biotinylation was performed for 2 h at room temperature, and then excess biotin was removed by extensive dialysis against 20 mM carbonate buffer (pH 9.6) containing 200 mM NaCl.
BBMV-Cry1 toxin binding and competition assays.
Binding of radiolabeled Cry1 toxins to BBMV from Ben or 456 larvae was measured by incubating a constant amount of ligand (radiolabeled Cry1 toxin at 0.01 nM for Cry1Fa and 0.22 nM for Cry1Ab, Cry1Ac, and Cry1Ca) with increasing amounts of the receptor (BBMV) in binding buffer (PBS [pH 7.5] plus 0.1% BSA in a 0.1-ml final reaction volume) for 1 h at room temperature. Toxin bound to the BBMV was separated from unbound toxin by centrifugation (14,500 × g for 10 min), and the resulting pellets were washed with 0.5 ml of ice-cold binding buffer. This centrifugation-wash cycle was performed twice, and the radioactivity in the final BBMV pellet (total toxin binding) was measured in a Wizard2 gamma detector (PerkinElmer). To quantify the nonspecific toxin binding, a 300-fold excess of homologous unlabeled Cry toxin was included in the binding mixtures. Specific toxin binding was calculated as the difference between total minus nonspecific binding and is presented in the graphs as the percentage of the total input of labeled bound toxin. The data are the mean values calculated from the results of at least two independent experiments performed in duplicate, with their corresponding standard errors.
Competition experiments were performed by incubating 400 μg/ml S. frugiperda BBMV with 0.22 nM (Cry1Fa and Cry1Ab) or 0.57 nM (Cry1Ca) radioiodinated toxins for 1 h at room temperature in the presence of increasing concentrations of unlabeled competitor. After incubation, the samples were washed twice with 0.5 ml of ice-cold binding buffer, and the radioactivity in the final pellets was measured in a Wizard2 detector (PerkinElmer). The amount of toxin bound to BBMV in the absence of competitor was defined as 100% binding for each experiment and used as a reference to calculate the relative percentages of toxin bound in the presence of increasing competitor concentrations. Competition data from at least two replicated experiments performed in duplicate for each toxin were pooled and analyzed using the SigmaPlot version 11.0 software to obtain the apparent dissociation constant (Kd) and maximum concentration of binding sites (Bmax).
Molecular cloning.
Total RNA was extracted from pools of 30 midgut samples from late-third- and early-fourth-instar larvae from each strain using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For ALP fragments, mRNA was purified using the FastTrack 2.0 mRNA isolation kit (Invitrogen). A cDNA template was prepared from total RNA (5 μg) or mRNA (0.5 to 1 μg) with an oligo(dT) primer using the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer's protocol.
All primers used for cloning and quantitative PCR are shown in Table 1. To clone the 3′-cDNA end of the predicted Cry1 toxin receptors, we used the FirstChoice RLM rapid amplification of cDNA ends (RACE) kit (Ambion), with forward primers targeting a known or a highly conserved region and oligo(dT) as the reverse primer. To clone the 3′ cDNA end of the S. frugiperda cadherin, we designed primers to target the AX147205.1 sequence in GenBank previously identified as encoding an S. frugiperda cadherin protein that interacts with Cry1Fa toxin (37). In the case of the ABCC2 transporter gene, we designed a forward primer targeting the ABCC2 gene in H. virescens (GenBank accession no. GQ332571.1), which has been described to be involved in resistance to Cry1Ac (38). We named the amplified fragment SfABCC2 for S. frugiperda ATP binding cassette subfamily C member 2. Fragments of cDNAs encoding S. frugiperda APNs were cloned from a PCR using a forward primer targeting the GAMENWG fragment, which is highly conserved among available full-length APN protein sequences from Spodoptera litura (GenBank accession no. AAK69605.1) and Spodoptera exigua (GenBank accession no. AAT99437.1, AAP44964.1, AAP44965.1, AAP44966.1, AAP44967.1, AIK27005.1, and AIK27006), and oligo(dT) as the reverse primer. The amplicon obtained was cloned and sequenced, and four different APN sequences were detected and used to design sequence-specific reverse primers (Table 1), which were used for cloning of partial APN cDNAs. Fragments of cDNA sequences encoding the 3′ ends of putative ALP cDNAs from the S. frugiperda midgut were amplified using a degenerate primer (see Table S1 in the supplemental material, 3′-ALP) targeting conserved motifs identified in aligned lepidopteran ALP sequences.
TABLE 1.
List of primers used in this study
| Target | Forward primer (5′–3′) | Reverse primer (5′–3′) | Size (bp)a | Purpose |
|---|---|---|---|---|
| 3′-ALP | CTGTACGAGCAGAGCCAG | Oligo(dT) | 3′ RACE | |
| 3′-APN1 | TGGTCTTCGTGGTGG | Oligo(dT) | 3′ RACE | |
| 3′-APN2 | TGGTCTTCGTAATGG | Oligo(dT) | 3′ RACE | |
| 3′-APN3 | TGGTCTTCGCCAAGG | Oligo(dT) | 3′ RACE | |
| 3′-APN4 | TGGTCTTCGTCATGG | Oligo(dT) | 3′ RACE | |
| 3′-APN5 | TGGTCTTCGTGAGGG | Oligo(dT) | 3′ RACE | |
| 3′-ABCC2 | TATCAATAATTCCGCAAGAGCCAGTG | Oligo(dT) | 3′ RACE | |
| SfL18 | CGTATCAACCGACCTCCACT | AGGCACCTTGTAGAGCCTCA | 126 | qRT-PCR |
| Sfcadherin | GAGAGCTGAGGGTCACTTGG | AGCGGACTCGGTTGTAGAGA | 299 | qRT-PCR |
| SfmALP1 | CACTGCCGCTACTGTGCTG | CCTGTGCCTTATCATTCCAAA | 125 | qRT-PCR |
| SfmALP2 | GGCTTTCTGCCCAACTGT | TCTACGAGCCAATCAACG | 114 | qRT-PCR |
| SfsALP1 | ACGAGCGAGACGTGTATCACAA | CGCCCAGGAACATGACCAC | 177 | qRT-PCR |
| SfAPN1 | TCTCAGTTTCTTCACTTTGCTA | ACTTGGGCAAAGGTGTTC | 127 | qRT-PCR |
| SfAPN3 | TCTCAGGCAATGAAGCCAATA | CACCCATGCTTTGAAATCCTC | 167 | qRT-PCR |
| SfAPN4 | GAAGTGGTTCCCCTGCTA | CGAGACGACAACGACATG | 238 | qRT-PCR |
| SfAPN6 | ATCTTGGGACCGATTCTA | TTGTCATGGGACCTAACT | 255 | qRT-PCR |
| SfABCC2 | CCTCAGACGGATGCTTTG | GTCGCCTGTTTCCTTCAC | 210 | qRT-PCR |
| SfmALP1 | TTGTGGAAGGCGGTCGCATCGAC | Oligo(dT) | 3′ RACE | |
| SfmALP1 | Kit-specific adaptor primer | GCATGCCATCCCACGTGTCATTCG | 5′ RACE | |
| SfmALP1 | GGTACCATGGGGTCGTTGAGCTTGCTAT | AGATCTTCAGTGATGGTGGTGATGGTGTTTCACACTAAAGGAAGCCAAC | 1,635 | Full-length cloning |
| SfmALP2 | GTGGAGAACGATGGCTTCGACACGG | Oligo(dT) | 3′ RACE | |
| SfmALP2 | Kit-specific adaptor primer | GCGCTAGCTCTACCCAGTTGCGATGGT | 5′ RACE | |
| SfmALP2 | GGATCCACCATGAGGTCGCTACTGACTTA | CTCGAGTCAGTGGTGATGGTGATGATGTTGTTGGAATAGTGAAG | 1,626 | Full-length cloning |
| SfsALP1 | CACGCCTACCCCGGGGACCACTTCAA | Oligo(dT) | 3′ RACE | |
| SfsALP1 | Kit-specific adaptor primer | GGACATGACGTGCGCGTGGTCCG | 5′ RACE | |
| SfsALP1 | GGTACCATGTCTGCCCGCGCCCACGC | CTCGAGTCAGTGGTGATGGTGATGATGGTGTCGGCAGGCGTGTC | 1,572 | Full-length cloning |
The expected PCR product size is stated when available.
Amplicons for all genes evaluated by quantitative real-time reverse transcription-PCR (qRT-PCR) were subcloned into the pJET1.2 (Fermentas) or pCR2.1-TOPO (Invitrogen) cloning vectors and sequenced in the forward and reverse directions (sequencing facility at the University of Tennessee, Knoxville, TN, or Eurofins MWG/Operon, Louisville, KY). Once the identity of each fragment was confirmed, we amplified each from midgut cDNA from larvae of the Ben strain using AccuPrime SuperMix I (Invitrogen). Each PCR (10-μl final volume) contained 200 ng of cDNA and 0.2 μM forward and reverse primers. The conditions for the PCRs included an initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 68°C for 30 s. A final extension step at 72°C for 1 min was also included. Amplicons were purified from agarose gels using the PureLink gel extraction kit (Invitrogen), quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific), and used to prepare standard curves in qRT-PCR experiments, as described below.
For cloning the full-length ALP transcripts, we used the SMART-RACE cDNA kit (Clontech) according to the manufacturer's instructions and gene-specific primers (see Table S1 in the supplemental material for 3′-RACE and 5′-RACE primers). Amplicons were purified using the QIAquick kit (Qiagen) and cloned and sequenced as described above. Full-length open reading frames were assembled by aligning the sequences generated by degenerate PCR and the 5′- and 3′-RACE sequences. Primers (see Table S1 in the supplemental material for full-length primers) were then designed to amplify the open reading frames by RT-PCR. The resulting amplicons were ligated into pCR2.1-TOPO and transformed into E. coli for sequencing to confirm the open reading frames. Three full-length ALP isoforms were obtained, and the presence of glycosylphosphatidylinositol (GPI) anchor signals in the deduced protein sequences was evaluated using the big-Pi predictor (39), PredGPI (40), FragAnchor (41), and GPI-SOM (http://gpi.unibe.ch/) programs. All four programs predicted GPI anchors in two ALPs, which were named SfmALP1 and SfmALP2 for S. frugiperda membrane-bound ALP1 and -2, respectively. In contrast, only GPI-SOM predicted a potential GPI-anchoring site in the third ALP sequence, which we consequently named SfsALP to signify soluble S. frugiperda ALP.
Quantitative RT-PCR.
Midgut cDNA from fourth-instar larvae of the Ben and 456 strains was prepared as described above. Based on the sequences of the cloned 3′ cDNA fragments, we designed primer sets targeting each of the cloned cDNAs for use in qRT-PCR (listed in Table 1). Primer specificity was checked using the PrimerAnneal tool in JustBio. The reactions for qRT-PCR were performed using cDNA from three independent biological samples, each measured in triplicate in an ABI 7900HT fast real-time PCR system (Applied Biosystems) in standard mode using the SDS 2.3 and SDS RQ manager softwares to collect cycle threshold (CT) values. The final volume of each reaction mixture was 20 μl, containing 200 ng of cDNA, a 0.4 μM concentration of the primers, and 10 μl of Power SYBR green PCR master mix (Applied Biosystems). The cycling conditions included initial incubation at 55°C for 2 min and then denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 55°C for 30 s, 68°C for 30 s, and a final step at 95°C for 15 s. After the reaction was completed, amplification and dissociation curve analyses were performed using the SDS 2.3 and SDS RQ manager softwares. All dissociation curves showed a single amplification peak for each reaction, and the absolute value of the slope (CT value versus log quantity) for each primer set was <0.1. The amplification efficiencies were >90% for all primers, except for primer sets targeting SfAPN3 (89.6%) and SfsALP (87.8%), indicating a passing validation. Relative transcript amounts (CT values) were first normalized to the endogenous reference gene (S. frugiperda ribosomal protein L18, GenBank accession no. AF395587.1), and then the relative transcript levels were calculated using the 2−ΔΔCT method (42). Relative expression was standardized, with the level of gene transcripts detected in the Ben strain defined as 1. All comparisons for significance were performed using SigmaPlot version 11 (Systat Software, Inc.), using a one-way ANOVA with a post hoc Holm-Šidák test for multiple comparisons versus Ben as the control group (P < 0.01).
Heterologous expression and purification of SfmALP2.
The open reading frame (ORF) for SfmALP2 was excised from pCR2.1-TOPO and ligated into the pBacPAK9 plasmid (Clontech). To generate recombinant baculovirus, 8 × 105 S. frugiperda Sf9 cells were seeded into each well of 6-well plates and allowed to attach for 1 h at 27°C. Transfection mixtures (100-μl final volume in sterile water) containing 0.5 μg of transfer plasmid, 5 μl of linearized flashBACULTRA parental DNA (GenWay Biotech, San Diego, CA), and 4 μl of Cellfectin II (Invitrogen) were gently added to the wells, and the plates were incubated at 27°C for 5 h, after which an additional 0.75 ml of fresh medium was added. The cell cultures were incubated at 27°C in a humidified box for 5 days, and then recombinant virus was obtained from the medium after centrifugation. To amplify the virus, 30-ml flasks of Sf9 cells were seeded at 1 × 106 cells/ml and incubated overnight at 27°C and 135 rpm. The flasks were then infected with 0.5 ml of the transfection medium and incubated for 72 h at 27°C and 135 rpm. The culture was then centrifuged at 1,730 × g for 20 min and the medium filtered through a 0.2-μm-pore-size filter. A sample of each amplified virus (0.5 ml) was sent to Expression System LLC (Davis, CA) to determine the titers.
The production of SfmALP2 was performed in Sf9 cells seeded at 2 × 106 cells/ml. Cell cultures were incubated overnight at 27°C and 130 rpm before infection with baculovirus at a multiplicity of infection (MOI) of 1.2. The infected cells were incubated at 27°C and 130 rpm for 48 h and then centrifuged at 1,730 × g for 20 min. The medium was removed, and cells were suspended in 20 mM phosphate (pH 7.4), 0.1 M NaCl buffer (Sf9) containing a 1% (vol/vol) protease inhibitor cocktail (Sigma, St. Louis, MO), and 0.1% (vol/vol) phosphatidylinositol-specific phospholipase C (PI-PLC) (Invitrogen). The mixtures were incubated overnight on ice and then centrifuged at 50,000 × g for 1 h at 4°C. The solubilization of recombinant SfmALP2 was checked by comparing the ALP activities in the cells and supernatants, as described above. The supernatant containing solubilized SfmALP2 was concentrated using Amicon Ultra-15 centrifugal filter units (Millipore, Billerica, MA) and transferred into buffer (10 mM Tris [pH 8.0], 1 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF]) by dialysis. Recombinant protein was purified in a cold room through anion-exchange chromatography using a Source 15 Q Sepharose column preequilibrated in the respective dialysis buffer and connected to an Äkta explorer purification system (GE Healthcare, Piscataway, NJ). Protein was eluted using a gradient of buffer containing 1 M NaCl, and fractions containing ALP (based on ALP activity assays with pNPP as the substrate, as described above) were pooled, concentrated, and further purified by size exclusion chromatography in a Superdex 200 10/60 column equilibrated in 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 150 mM NaCl, 1 mM DTT, and 5% glycerol. Eluted protein was tested for purity using SDS-PAGE and Western blotting with antisera (as described below), and the total protein concentration was estimated using Bradford's method (43), with BSA as the standard.
Western and ligand blotting.
For the detection of ALP proteins in BBMV by Western blotting, total BBMV proteins (25 μg) or purified recombinant protein (0.5 μg) was solubilized in sample buffer (44) and heat denatured before separation on a 10% SDS-PAGE gel. Proteins were then transferred overnight at 4°C to a polyvinylidene difluoride (PVDF) filter in transfer buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, and 20% methanol) using constant voltage (20 V). After transfer, the filters were blocked in PBS containing 0.1% Tween 20 and 3% BSA for 1 h at room temperature and then probed for 1 h with primary antibody (1:10,000 dilution) against the membrane-bound form of ALP from Anopheles gambiae (a gift from Gang Hua and Mike Adang, University of Georgia, USA). The filters were washed six times for 10 min each with washing buffer (PBS, 0.1% Tween 20, and 0.1% BSA) and then probed with secondary antibody (goat anti-rabbit conjugated to horseradish peroxidase [HRP], 1:30,000 dilution) for 1 h at room temperature. After being washed, blots were developed using enhanced chemiluminescence (SuperSignal West Pico; Pierce), according to the manufacturer's instructions.
For ligand blots and the detection of recombinant SfmALP2, purified protein (0.5 μg) was separated using a 4 to 12% NuPAGE Bis-Tris gel (Invitrogen) and then transferred to a nitrocellulose filter using iBlot (Invitrogen). After blocking for 1 h in WesternBreeze solution (Invitrogen), the blot was probed for 2 h with 1 μg/ml activated Cry1Fa toxin in blocking solution. After being washed four times (5 min each) in the WesternBreeze wash solution, the blot was probed with a 1:10,000 dilution of rabbit anti-Cry1Fa or antisera to ALP (LifeSpan BioSciences, Inc.) in WesternBreeze solution at room temperature for 1 h. After being washed as described before, the blots were probed with a 1:5,000 dilution of goat anti-rabbit-HRP antibody for 1 h at room temperature and then washed and developed using enhanced-chemiluminescence (ECL) reagents (GE Healthcare).
Binding ELISAs.
Specific binding of Cry1Fa toxin to recombinant purified SfmALP2 was tested using a competitive enzyme-linked immunosorbent assay (ELISA). Individual wells of 96-well ELISA plates (Immulon 2HB flat-bottom microtiter plates; Fisher Scientific) were coated overnight at 4°C with 1 μg/well of purified SfmALP2 in a final volume of 100 μl of PBS for each well. The plates were then washed three times with PBS and blocked with 200 μl of PBS containing 1% BSA for 90 min at room temperature. After this incubation, the plates were washed three times for 10 min per wash with PBS plus 0.1% Tween 20. Different concentrations (ranging from 0.26 to 8.33 nM) of biotinylated Cry1Fa toxin were added to each well in a final volume of 100 μl of PBS plus 0.1% BSA and incubated at room temperature with slow shaking for an hour. The wells were then washed three times (10 min each) using 150 μl of washing buffer (PBS, 0.1% Tween 20, and 0.1% BSA). For detection of the bound biotinylated toxin, the wells were incubated with a 1:1,000 dilution of streptavidin-HRP conjugate in a 100-μl final volume at room temperature for 1 h. After incubation, the plates were washed as described before and then incubated for 15 min at room temperature in 50 μl of the 1-Step Ultra 3,3′,5,5′-tetramethylbenzidine (TMB) ELISA substrate (Thermo Scientific). The reaction was terminated by adding 50 μl of 2 M H2SO4, and the absorbance at 450 nm was read with a Synergy HT microplate reader, with the associated Gen5 software used to analyze the data (BioTek, Winooski, VT). Nonspecific binding was determined in the presence of a 200-fold excess (1.6 μM) of unlabeled Cry1Fa toxin in the binding mixtures. Specific binding was calculated by subtracting nonspecific from total binding obtained in the absence of competitor. The data are the means and standard errors calculated from the results from two biological replicates measured in duplicate. The binding data were analyzed and plotted using the SigmaPlot version 11.0 software (Systat Software). Fitting of the specific binding data using nonlinear regression determined a one-site model to be the best fit to the data, which was used to obtain the apparent Cry1Fa dissociation constant (Kd).
Nucleotide sequence accession numbers.
The nucleotide sequences of cloned partial cDNAs for SfABCC2 (KT276302); the four different APNs named SfAPN1 (KT276298), SfAPN3 (KT276299), SfAPN4 (KT276300), and SfAPN6 (KT276301); and the three full-length ALPs named SfmALP1 (KT276303), SfmALP2 (KT276297), and SfsALP1 (KT276304) were deposited in GenBank with the indicated accession numbers.
RESULTS
Larvae of the 456 strain of S. frugiperda are not cross-resistant to Cry1Ca.
Larvae from the 456 strain were reported to be highly resistant to purified Cry1Fa, Cry1Ac, and Cry1Ab toxins (12, 13, 25). In agreement with these observations, <10% mortality was detected in the 456 strain in bioassays with leaf tissue from transgenic corn expressing the Cry1Fa toxin (Fig. 1A, event TC1507), while 100% mortality was observed for larvae from the Ben strain. No significant differences (P < 0.01) in susceptibility to TC1507 were detected when testing larvae from Ben × 456 crosses independently of the sex of the resistant parent in the cross, supporting the idea of autosomal resistance. As a control, survival on corn leaf tissue from the nontransgenic isoline was >90%, with no significant differences between the larvae from any of the strains and crosses tested (data not shown). Mortality in TC1507 for the 456F and 456M crosses (93 to 94%) was significantly different (P < 0.01) from the 100% mortality observed in the parental susceptible (Ben) strain (Fig. 1A). An estimation of dominance resulted in an h of 0.06, suggesting that susceptible heterozygous and homozygous larvae had almost identical levels of fitness and supporting the idea of incompletely recessive inheritance of resistance.
FIG 1.
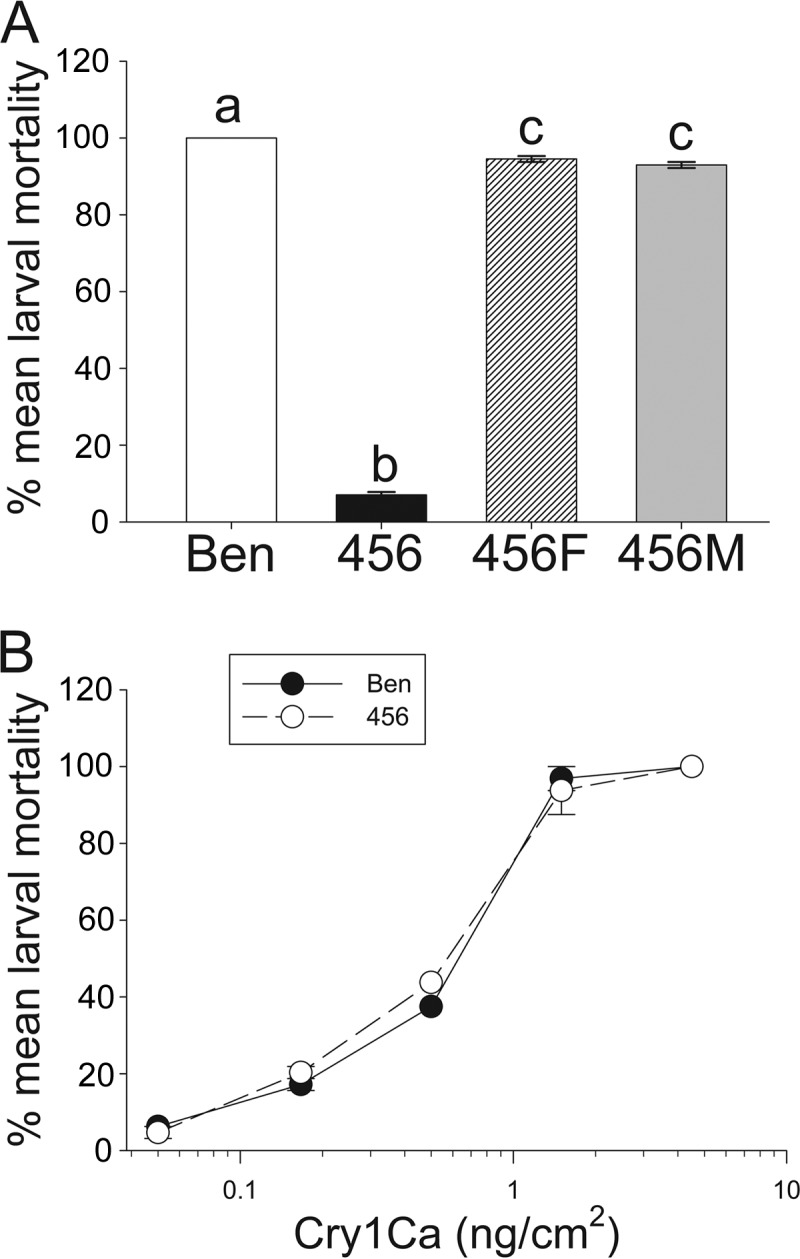
Larvae from the 456 strain of S. frugiperda are highly resistant to Cry1Fa toxin in Bt corn event TC1507 but not to Cry1Ca toxin compared to larvae from the Ben strain. Resistance in the 456 strain is transmitted as an autosomal recessive trait. Bioassays were conducted with fresh corn event TC1507 leaf tissue (A) or meridic diet contaminated with Cry1Ca toxin (B) and neonates from the Ben and 456 S. frugiperda strains. Neonates from 456♀ × Ben♂ (456F) and Ben♀ × 456♂ (456M) crosses were also used in the bioassay whose results are shown in panel A to test for genetic transmission of the resistance trait. The data presented are the mean mortality rates and corresponding standard errors calculated from the results from two bioassays with 64 larvae each per strain/cross (n = 2) for plant tissue bioassays or 32 larvae per Cry1Ca toxin concentration. Different letters represent significant differences between the mean mortalities (one-way ANOVA and post hoc Holm-Šidák test for multiple pairwise comparisons; P < 0.01).
In contrast to tests with Bt corn, bioassays with purified Cry1Ca toxin revealed no significant differences (overlapping 95% fiducial limits [FL]) in a comparison of susceptibilities of neonate larvae from the Ben (LC50, 25.0 ng/cm2; FL, 18.6 to 35.1) and 456 (LC50, 27.5 ng/cm2; FL, 20.2 to 33.8) strains (Fig. 1B).
Reduced Cry1Fa and Cry1A toxin binding is associated with resistance in S. frugiperda larvae.
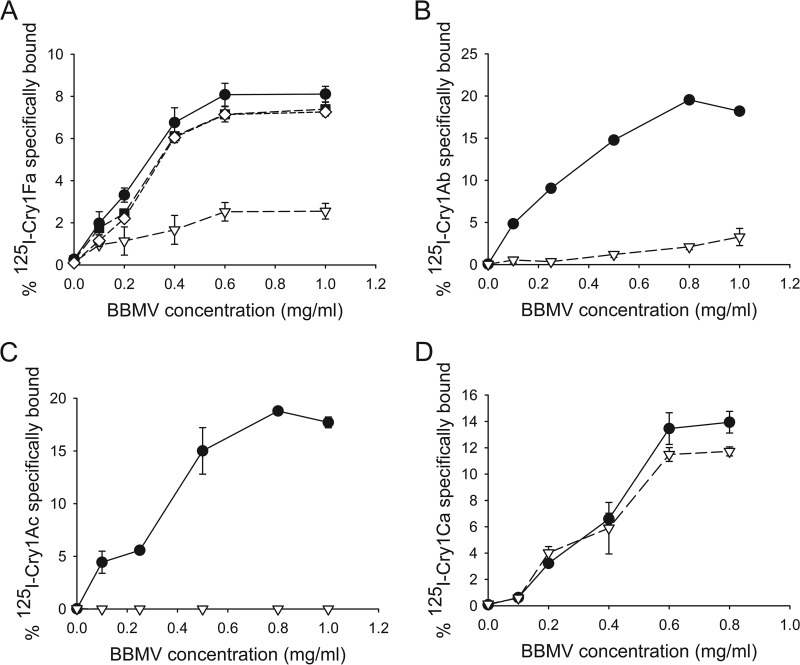
Analyses of radiolabeled Cry toxin binding to BBMV proteins revealed a >75% reduction in specific Cry1Fa toxin binding to BBMV from 456 larvae compared to that of Ben larvae (Fig. 2A). In contrast, Cry1Fa-specific levels of binding to BBMV from strains 456F and 456M were similar to that of the parental Ben strain, as expected from the autosomal and recessive inheritance patterns of resistance. Binding of Cry1Ab and Cry1Ac toxins to BBMV from 456 larvae was highly reduced (>80% in each case) compared to that of Ben larvae (Fig. 2B and C). However, in agreement with the bioassay data, similar levels of specific Cry1Ca toxin binding were detected in BBMV from the Ben and 456 strains (Fig. 2D).
FIG 2.
Resistance to TC1507 corn in strain 456 is associated with reduced Cry1A and Cry1Fa but not Cry1Ca toxin binding to midgut brush border membrane vesicles (BBMV). Binding of radiolabeled, purified, activated Cry1Fa (A), Cry1Ab (B), Cry1Ac (C), or Cry1Ca (D) toxins to BBMV from larvae of the Ben (black circles), 456 (open inverted triangles), 456F (black squares), and 456M (open diamonds) strains was tested. Shown are the percentages of specifically bound input toxin, calculated by subtracting nonspecific binding in the presence of excess unlabeled homologous competitor from total toxin binding. Each data point represents the mean and corresponding standard error calculated from the results from at least two independent experiments performed in duplicate (n = 2).
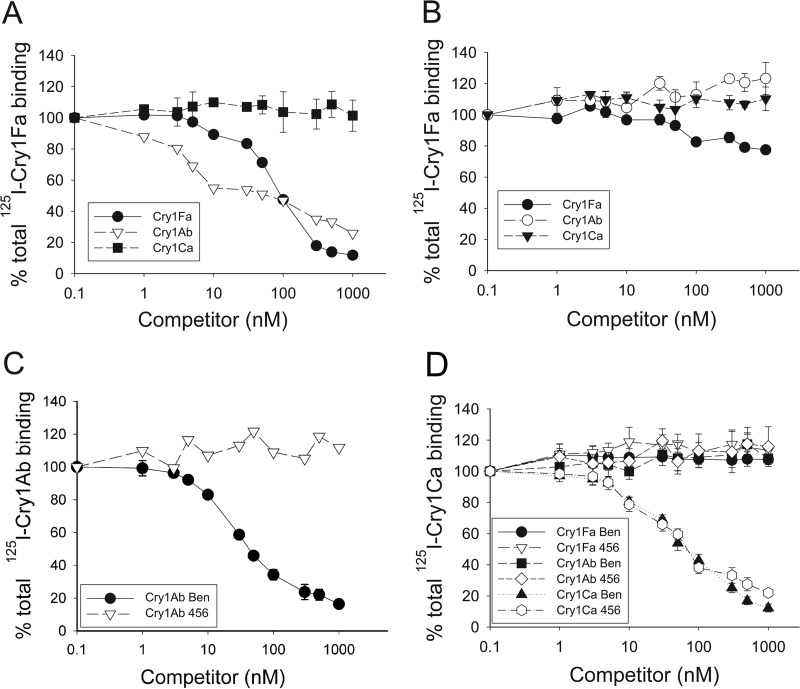
In the competition assays (Fig. 3A to D), 90% of 125I-Cry1Fa binding was competed by unlabeled Cry1Fa, while up to 75% of 125I-Cry1Fa binding was competed by Cry1Ab in BBMV from the Ben strain (Fig. 3A). In contrast, in BBMV from 456 larvae, only marginal competition (20%) of 125I-Cry1Fa binding with unlabeled Cry1Fa (and no competition with Cry1Ab) was detected (Fig. 3B). These results were suggestive of BBMV from 456 larvae lacking a relevant Cry1Fa binding site also recognized by Cry1Ab. The binding parameters, dissociation constant (Kd), and maximum concentration of binding sites (Bmax) from an analysis of the homologous competition assay results (Table 2) demonstrated high-affinity Cry1Ab (58.6 nM) and Cry1Fa (42.6 nM) binding to BBMV from the Ben strain. In contrast, only low-affinity Cry1Fa binding (>500 nM) was detected in BBMV from larvae of the 456 strain. Moreover, the Bmax for Cry1Fa was reduced >7-fold in 456 compared to that in Ben BBMV (Table 2), further supporting the unavailability of a population of high-affinity Cry1Fa binding sites in 456 larvae. As expected from the bioassays with purified toxin, we did not find differences in Cry1Ca's affinity of binding to BBMV from the Ben or 456 strain and only a 2-fold reduction in the concentration of binding sites was detected (Fig. 3C and Table 2). The binding of Cry1Ca was not competed by Cry1Fa or Cry1Ab toxin, supporting the idea that Cry1Ca binding sites are not recognized by these proteins.
FIG 3.
Competition binding assays demonstrate reduced numbers of Cry1Fa and Cry1Ab binding sites in 456 compared to those in Ben larvae. Graphs show the competition of 125I-Cry1Fa binding (A and B) to BBMV from larvae of the Ben (A) and 456 (B) strains by unlabeled Cry1Fa, Cry1Ab, and Cry1Ca competitors, as indicated in the figure. (C) Homologous competition of 125I-Cry1Ab binding to BBMV from the Ben and 456 strains, as indicated, by unlabeled Cry1Ab. (D) Competition of 125I-Cry1Ca binding to BBMV from the Ben and 456 strains by unlabeled Cry1Fa, Cry1Ab, and Cry1Ca competitors, as indicated in the figure. Binding for each data point in all graphs is the total binding percentage calculated relative to the total binding observed in the absence of a competitor. Each data point represents the mean percentage of bound labeled toxin calculated from the results of at least two independent experiments performed in duplicate.
TABLE 2.
Binding parameters calculated from the Cry1 toxin binding competition assays whose results are shown in Fig. 3a
| Strain | Toxin | Kd ± SE (nM) | Bmax ± SE (nmol/mg BBMV protein units) | R2 |
|---|---|---|---|---|
| Ben | Cry1Ab | 58.6 ± 4.2 | 5.2 ± 0.2 | 0.9978 |
| 456 | Cry1Ab | NA | NA | NA |
| Ben | Cry1Fa | 42.6 ± 6.9 | 570.1 ± 25.1 | 0.9428 |
| 456 | Cry1Fa | 534.6 ± 336.1 | 77.7 ± 20.7 | 0.9198 |
| Ben | Cry1C | 13.1 ± 2.4 | 8.4 ± 0.7 | 0.9538 |
| 456 | Cry1C | 13.4 ± 2.1 | 4.4 ± 0.3 | 0.9391 |
No competition was observed when labeled Cry1Ab toxin and BBMV from the 456 strain were used. The statistic for the goodness of fit for each curve (R2) is also presented. NA, not available.
Reduced Cry1Fa toxin binding is associated with reduced alkaline phosphatase expression.
The binding data supported the absence of a critical Cry1Fa binding site in BBMV from 456 larvae, in agreement with previous reports suggesting reduced levels of diverse midgut proteins in larvae from 456 compared to those from susceptible insects (15, 16). To identify specific Cry1Fa toxin receptors potentially involved in resistance, we cloned partial fragments of putative Cry1 receptor genes (17) from Ben larvae. Sequence information was then used to design primers and perform quantitative PCR assays comparing transcript levels of putative Cry1 receptor genes in Ben and 456 larvae.
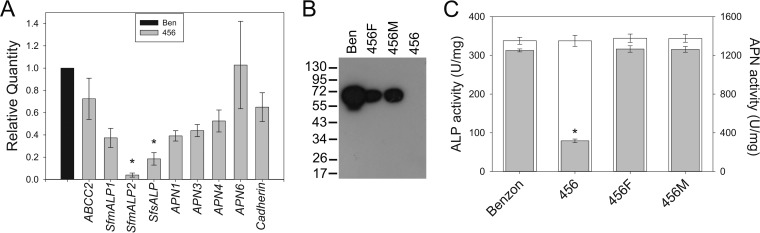
Transcript levels for S. frugiperda cadherin and ABCC2 transporter genes homologous to genes linked with resistance to Cry1Ac in H. virescens (19, 38) were unaltered (P > 0.01) in a comparison of Ben and 456 larvae (Fig. 4A). We cloned the fragments of four APNs, which we named based on their homologies to described APN genes. Thus, SfAPN1 and SfAPN3 displayed the highest sequence identity (86%; E value, 5e−90) to APN1 (GenBank accession no. AAP44964.1) and APN3 (GenBank accession no. AAP44966.1) from S. exigua, respectively. The cloned SfAPN4 fragment displayed the highest identity (86%; E value, 0.0) to APN4 from S. litura (GenBank accession no. AAK69605.1), and SfAPN6 was most similar to APN6 from H. armigera (GenBank accession no. ACA35025.1). The transcription of any of these four different APN genes did not differ significantly (P > 0.01) between the Ben and 456 larvae (Fig. 4A). In contrast, the transcript levels for a membrane-bound alkaline phosphatase gene (SfmALP2) and a predicted soluble alkaline phosphatase (SfsALP) were significantly (P < 0.01) reduced in larvae from the 456 strain compared to those from the Ben strain (Fig. 4A). The highest downregulation (>90%) was detected for SfmALP2, which is an ortholog (87% identity; E value, 0.0) of ALP2 from S. litura (GenBank accession no. JN687589.1). The SfsALP protein is most identical (68%; E value, 0.0) to the soluble ALP from Bombyx mori (GenBank accession no. BAB62746.2). Unlike the transcript levels of SfmALP2 and SfsALP, the transcript levels for an alternative membrane-bound ALP (SfmALP1) did not significantly differ (P > 0.01) among larvae from the Ben and 456 strains (Fig. 4A). The SfmALP1 protein is highly identical (89%; E value, 0.0) to ALP1 from S. litura (GenBank accession no. AFJ04289.1).
FIG 4.
Resistance to corn event TC1507 and reduced Cry1Fa toxin binding are associated with reduced levels of alkaline phosphatase transcript and protein levels. (A) Transcript levels for putative Cry1 toxin receptors were quantified and compared between larvae from the Ben and 456 strains using quantitative real-time PCR (qRT-PCR). Shown are the relative transcript levels for S. frugiperda ABCC2 transporter, cadherin, three ALPs, and four APN genes calculated using the ribosomal L18 gene as an internal reference, according to the 2−ΔΔCT method and relative to the transcript levels in Ben larvae (black bar). The data in each bar are the mean with the corresponding standard error calculated from three independent biological samples, each measured in triplicate (n = 3). Asterisks above a column indicate significantly reduced transcripts levels in larvae from the 456 strain compared to those in the Ben strain (ANOVA and post hoc Holm-Šidák test for multiple comparisons to Ben as a control; P < 0.01). (B) Alkaline phosphatase (ALP) protein levels detected in Western blots of BBMV from the Ben, 456F, 456M, and 456 strains. The detection of ALP was accomplished using antisera against A. gambiae mALP, as described in Materials and Methods. Numbers on the left are molecular masses (in kilodaltons). (C) Specific alkaline phosphatase (ALP, gray bars) and aminopeptidase-N (APN, white bars) activity levels in BBMV from the Ben, 456, 456F, and 456M strains, as indicated in the graph. One enzymatic unit was defined as the amount of enzyme that would hydrolyze 1.0 μmol substrate to the chromogenic product per minute at the specific reaction pH and temperature. The data shown are the mean specific activities obtained from the results from at least three independent BBMV batches for each strain measured at least in triplicate. Asterisks above a column indicate significant differences in specific activity for an enzyme among strains (ANOVA and post hoc Holm-Šidák test for multiple pairwise comparisons, P < 0.01). Since APN activity data failed an equal-variance test, in this case, we used ANOVA on ranks (Kruskal-Wallis test, P < 0.01) to determine statistical significance.
As previously reported (15), the transcriptional downregulation of selected ALP genes in larvae from strain 456 translated to reduced ALP protein amounts in BBMV compared to those from Ben larvae, as detected in Western blots (Fig. 4B) and specific activity assays (Fig. 4C). Moreover, levels of ALP protein in heterozygous strains detected in the Western blots were intermediate between those of susceptible and resistant parents, supporting the codominance of both alleles at the biochemical level. However, while specific ALP activity was highly reduced (75%) in BBMV from larvae of the 456 strain, activities did not differ (P > 0.01) between BBMV from the larvae of Ben and 456M or 456F larvae under the conditions used to measure enzymatic activity. Unlike with ALP, no differences in APN-specific activities were detected among the tested BBMV samples (Fig. 4C).
Alignment of the SfmALP1 and SfmALP2 protein sequences indicated 73% identity, with the most diversity detected at the N termini of the proteins and with the last 53 amino acids at the C termini, which include the predicted GPI-anchoring sites (see Fig. S1 in the supplemental material), being identical. Interestingly, the predicted N-glycosylation sites in SfmALP1 and SfmALP2 were unique and located in different parts of the protein sequences, while the only predicted N-glycosylation site in SfsALP1 was in relative proximity to the site in SfmALP2 (see Fig. S1). Alignment of the SfmALP2 sequences from Ben and 456 (see Fig. S2 in the supplemental material) detected 9 amino acid substitutions that did not affect predicted N-glycosylation or GPI-anchoring sites. Only 3 of these substitutions were conservative; nonconservative changes included asparagine (Ben) to threonine (456) at position 321, phenylalanine (Ben) to lysine (456) at position 374, proline (Ben) to serine (456) at position 384, proline (Ben) to leucine (456) at position 389, glycine (Ben) to aspartic acid (456) at position 453, and glycine (Ben) to arginine (456) at position 455.
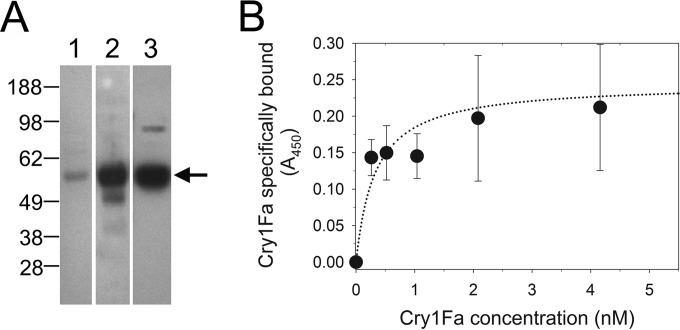
Cry1Fa toxin binds to SfmALP2.
To test for specific interactions between Cry1Fa toxin and the most downregulated ALP gene in the resistant S. frugiperda larvae, we cloned the ALP gene's full-length cDNA and expressed and purified it from insect cell cultures. The resulting affinity-purified protein of about 62 kDa in size (Fig. 5A, lane 1) was detected with antisera in Western blots (Fig. 5A, lane 2), and bound Cry1Fa toxin was detected in ligand blots (Fig. 5A, lane 3). The specificity of Cry1Fa binding to purified SfmALP2 was tested using saturation binding assays using SfmALP2 bound to wells in ELISA plates (Fig. 5B). The resulting specific-binding data (total minus nonspecific binding) fitted to a one-site saturation binding curve as the most significant fit. The calculated dissociation constant (Kd) was 0.245 ± 0.02 nM.
FIG 5.
Heterologously expressed and purified SfmALP2 specifically binds Cry1Fa toxin. The full-length cDNA encoding SfmALP2 was expressed in insect cell cultures and purified. (A) Purified protein (0.5 μg) was separated by electrophoresis and stained with Coomassie blue for total protein (lane 1) or transferred to nitrocellulose filters that were probed with antisera to ALP (lane 2) or with Cry1Fa toxin (lane 3). The arrow indicates the approximate position of the recombinant ALP proteins. Numbers at the left are molecular masses (in kilodaltons). (B) Saturation of specific binding detected in an ELISA with purified SfmALP2 bound to the plate and probing with biotinylated Cry1Fa toxin. Shown are the mean specific binding values calculated by subtracting nonspecific binding (in the presence of a 300-fold excess of unlabeled Cry1Fa) from total binding (in the absence of the competitor) from two independent experiments, each performed in duplicate. The curve shows the results from the fitting of the specific binding data to a one-site saturation model as the best fit obtained using the SigmaPlot version 11 global curve fit wizard.
DISCUSSION
The evolution of resistance to Bt crops in field pest populations is considered the foremost threat to the future utility of this technology, and while there are diverse reported cases of field-evolved resistance resulting in extensive field control failures (3–5), there are only limited data available on the resistance mechanisms involved. In this study, we report data describing a mechanism of field-evolved resistance in S. frugiperda to Bt corn producing the Cry1Fa toxin (event TC1507). Since corn event TC1507 produces a truncated form of the Cry1Fa protein that appears to be processed by plant enzymes to the activated Cry1Fa toxin core (45), we did not test for alterations in toxin activation in resistant insects and concentrated on potential alterations in toxin binding to midgut cells.
Previous reports on field-evolved resistance mechanisms against Bt cotton in P. gossypiella have shown differential splicing of cadherin transcripts (46) and estimates of reduced Cry1Ac toxin binding in field-collected resistant P. gossypiella insects (47). Although not demonstrated directly, since pink bollworm cadherin binds Cry1Ac (48) and alterations in cadherin are linked to resistance against that toxin (20, 49), it is highly plausible that the alterations in cadherin transcripts detected in field-collected P. gossypiella strains may be responsible for reduced Cry1Ac binding and resistance to Bt cotton. However, alterations in cadherin transcripts that are unrelated to resistance have also been reported in Cry1Ab-resistant Ostrinia nubilalis strains (50). In addition, levels of nonspecific and specific binding and the participation of alternative Cry1Ac-binding proteins were not considered in reports with small numbers of field-collected P. gossypiella insects. Additional screening of high numbers of field-collected individuals and binding assays with susceptible and resistant forms of cadherin would be necessary to establish a direct mechanistic linkage with resistance to Bt cotton.
Strains of S. frugiperda from Brazil with field-evolved resistance have been reported to display reduced Cry1Fa toxin binding using semiquantitative methods (6), although the receptors involved were not documented. Moreover, while Cry1Aa and Cry1Ab binding was also reduced in resistant compared to susceptible S. frugiperda, Cry1Ac binding was not affected. These results are difficult to explain when considering that Cry1Ab, Cry1Fa, and Cry1Ac share all their binding sites in the BBMV of S. frugiperda (51). Sensitivity limitations of semiquantitative binding assays or differences among S. frugiperda strains (52) may explain this discrepancy. In contrast, we present quantitative data and binding estimates supporting an association between reduced Cry1Fa and Cry1A toxin binding. Moreover, visual inspection of the competition curve of 125I-Cry1Fa binding by Cry1Ab is suggestive of the existence of at least two populations of Cry1Fa binding sites recognized with different affinities (high and low) by Cry1Ab. The lack of a similar shape in the homologous Cry1Fa competition curve is explained by Cry1Fa displaying similar affinities of binding to both populations of binding sites. Since Cry1Ab did not compete all 125I-Cry1Fa binding sites, our data support the existence of at least three populations of binding sites for Cry1Fa in S. frugiperda. The highly reduced 125I-Cry1Fa and 125I-Cry1A binding in BBMV from the 456 strain supported alterations in the populations of the binding sites. Our transcript quantification and ALP activity data support the idea that the reduced expression of selected ALP genes is associated with field-evolved resistance to corn event TC1507 and cross-resistance to Cry1Ab and Cry1Ac in the 456 strain of S. frugiperda.
The cross-resistance and reduced Cry1A toxin binding phenotype, together with maintained Cry1Ca binding and susceptibility detected in our resistant S. frugiperda strain, identify practical resistance to Bt corn event TC1507 (7) as being of the mode 1 type (18). This type of resistance was also described in field populations of P. xylostella (53) and greenhouse populations of Trichoplusia ni (54) resistant to commercial Bt pesticides. In those cases, resistance was initially mapped to a multigene locus containing an ABCC2 gene (22). However, more-recent reports demonstrate that in P. xylostella, the reduced expression of a membrane-bound ALP gene (pxmALP) serving as a functional Cry1Ac receptor is linked to resistance against that toxin (24). Downregulation of the pxmALP gene was concomitant with alterations in ABCC gene expression trans-regulated by a mitogen-activated protein kinase (MAPK) gene. While not yet tested, based on the genetic synteny among the species (22), it is plausible that the same MAPK gene is responsible for resistance in alternative lepidopteran pests. The current work is aimed at identifying the specific molecular mechanism responsible for the downregulation of selected ALP genes in 456 larvae to demonstrate linkage with resistance. While the presence of this mechanism is speculative at this point, it is possible that gene proximity, which occurs with the genes for membrane-bound ALP (mALP) and soluble ALP (sALP) in B. mori larvae (55), may explain the selective downregulation of the SfmALP2 and SfsALP genes compared to the expression of the gene for SfmALP1, which may be the product of an alternative gene at a distant locus. This selective downregulation of ALP genes may explain the lack of significant fitness costs, as reported for larvae from the 456 strain (12). The relevance of this selective ALP downregulation for resistance is highlighted by high-affinity specific Cry1Fa binding to heterologously expressed SfmALP2. Since SfsALP is predicted to be a soluble ALP isoform, Cry1Fa binding to this protein would probably not be conducive to toxicity, and thus downregulation of SfsALP is possibly irrelevant to resistance.
Data from mode 1 resistance cases in S. frugiperda, T. ni, and P. xylostella emphasize the importance of alterations in Cry toxin binding for practical field-evolved resistance in lepidopteran larvae, regardless of the Bt technology (pesticides or transgenic crops) used as a selective agent. This conclusion has important implications for resistance prevention and monitoring, since it suggests that only genetic alterations affecting Cry toxin binding may provide resistance levels sufficient to cause field control failures in cases of highly susceptible pests for which the high-dose requirement is fulfilled for the crop. So far, resistance due to reduced Cry toxin binding has been linked to recessive alleles in autosomal genes. While nonrecessive (56) and dominant (57) resistance alleles have also been detected in field populations of H. armigera from China, their relevance to Cry toxin binding has not been evaluated. Our data and previous reports (5, 10, 25) provide support for Cry1Fa-resistant S. frugiperda from Puerto Rico, fulfilling the assumption of monogenic resistance transmitted as a recessive autosomal trait, considered in insect resistance management (IRM) practices for Bt crops (58). The lack of significant differences in susceptibility or Cry1Fa toxin binding between homozygous susceptible and heterozygous larvae observed in this study is analogous to Cry1Ac resistance in strains of H. virescens linked to mutations in cadherin genes (19). This observation was explained by cadherin representing a “lethal target” receptor facilitating susceptibility, unless its expression was reduced to a critical level. Accordingly, Cry1Fa toxin binding to slightly reduced levels of SfmALP2 as a lethal receptor in S. frugiperda would result in sufficient midgut epithelial damage in heterozygotes to cause mortality similar to that of homozygous susceptible insects. Interestingly, Cry1Ab and Cry1Ac binding was more drastically affected than Cry1Fa binding in BBMV from 456 larvae, suggesting that SfmALP2 may represent the main functional Cry1A receptor in S. frugiperda. In support of this hypothesis, midgut ALPs have been reported as receptors for Cry1Ab (59) and Cry1Ac (34, 60) toxins in alternative lepidopteran species. More importantly, the downregulation of ALP gene expression (15, 24, 34) and increased release of ALP from the midgut epithelium into the gut fluids (23) have been proposed as mechanisms of resistance against Cry1Ac in laboratory-selected strains of lepidopteran insects. It is remarkable that neither Cry1Ab nor Cry1Ac is a highly effective toxin against S. frugiperda relative to Cry1Fa (61). Diverse susceptibilities to these toxins in S. frugiperda larvae may be explained by differences in the interactions between Cry1A or Cry1Fa toxins with SfmALP2.
The characterization of field-evolved S. frugiperda resistance to transgenic corn producing the Cry1Fa toxin represents a unique opportunity to evaluate IRM strategies aimed at delaying and overcoming resistance to Bt crops. Based on the results from this study, the use of stacked Bt proteins (pyramids) with high-dose/refuge tactics would be effective as management plans to prevent and overcome Cry1Fa resistance. Crops expressing pyramided toxins that do not share binding sites with Cry1Fa or Cry1A toxins should be effective in controlling resistant S. frugiperda, as demonstrated in our bioassays with Cry1Ca or Vip3A and Cry2Ab bioassays reported elsewhere (10, 62). Since Cry1Ab and Cry1Ac share all their binding sites with Cry1Fa (51), it is highly plausible that Cry1Fa and modified Cry1AMod toxins share binding sites and thus may not be suitable for forming pyramids with Cry1Fa. Unexpectedly, recent reports suggest that Cry1AMod toxins remain active against S. frugiperda from Brazil (6), which may suggest that Cry1AMod has binding sites not recognized by Cry1Fa or Cry1A toxins. Further work to test the sharing of binding sites between Cry1AMod and Cry1Fa in susceptible S. frugiperda and their activities against alternative resistant strains of S. frugiperda would be necessary to assess the effectiveness of Cry1Fa-Cry1AMod combinations to delay resistance in Bt corn.
The data in this study provide a first mechanistic model for field-evolved resistance in S. frugiperda, with practical consequences for pest control (practical resistance) (7) that resulted in the withdrawal of the crop from the Puerto Rican market (5). Further work, including identification of the gene responsible for ALP downregulation and screens of high numbers of field-collected S. frugiperda, would be necessary to extrapolate our results to field populations. However, our data provide information that allows an assessment of current resistance management practices and advances the identification of putative receptor genes to target in crop improvement and resistance monitoring programs. Once the gene responsible for the downregulation of ALP is identified, it will allow for the development of sensitive resistance monitoring methods and subsequent detection of resistant insects and preliminary estimates of the frequency of resistance alleles before introduction of a Bt crop technology in new geographies. For instance, testing for reduced Cry1Fa toxin binding and ALP gene expression in field-resistant S. frugiperda strains recently detected in the southeastern United States(63) will help determine if resistance in Puerto Rico and the mainland United States is a result of insect migration or represents independent resistance events.
Supplementary Material
Funding Statement
DuPont provided funding to J.L.J.-F. through a Young Professor Award. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02871-15.
REFERENCES
- 1.Betz FS, Hammond BG, Fuchs RL. 2000. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul Toxicol Pharmacol 32:156–173. doi: 10.1006/rtph.2000.1426. [DOI] [PubMed] [Google Scholar]
- 2.James C. 2013. Brief 46: global status of commercialized biotech/GM crops: 2013. International Service for the Acquisition of Agri-Biotech Applications (ISAAA), Ithaca, NY: http://www.isaaa.org/resources/publications/briefs/46/. [Google Scholar]
- 3.van Rensburg JBJ. 2007. First report of field resistance by stem borer, Busseola fusca (Fuller) to Bt-transgenic maize. S Afr J Plant Soil 24:147–151. doi: 10.1080/02571862.2007.10634798. [DOI] [Google Scholar]
- 4.Dhurua S, Gujar GT. 2011. Field-evolved resistance to Bt toxin Cry1Ac in the pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manag Sci 67:898–903. doi: 10.1002/ps.2127. [DOI] [PubMed] [Google Scholar]
- 5.Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW, Huckaba RM. 2010. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J Econ Entomol 103:1031–1038. doi: 10.1603/EC10040. [DOI] [PubMed] [Google Scholar]
- 6.Monnerat R, Martins E, Macedo C, Queiroz P, Praça L, Soares CM, Moreira H, Grisi I, Silva J, Soberon M, Bravo A. 2015. Evidence of field-evolved resistance of Spodoptera frugiperda to Bt corn expressing Cry1F in Brazil that is still sensitive to modified Bt toxins. PLoS One 10:e0119544. doi: 10.1371/journal.pone.0119544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabashnik BE, Mota-Sanchez D, Whalon ME, Hollingworth RM, Carrière Y. 2014. Defining terms for proactive management of resistance to Bt crops and pesticides. J Econ Entomol 107:496–507. doi: 10.1603/EC13458. [DOI] [PubMed] [Google Scholar]
- 8.Farias JR, Andow DA, Horikoshi RJ, Sorgatto RJ, Fresia P, dos Santos AC, Omoto C. 2014. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot 64:150–158. doi: 10.1016/j.cropro.2014.06.019. [DOI] [Google Scholar]
- 9.Huang F, Qureshi JA, Meagher RL Jr, Reisig DD, Head GP, Andow DA, Ni X, Kerns D, Buntin GD, Niu Y, Yang F, Dangal V. 2014. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. PLoS One 9:e112958. doi: 10.1371/journal.pone.0112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vélez AM, Spencer TA, Alves AP, Moellenbeck D, Meagher RL, Chirakkal H, Siegfried BD. 2013. Inheritance of Cry1F resistance, cross-resistance and frequency of resistant alleles in Spodoptera frugiperda (Lepidoptera: Noctuidae). Bull Entomol Res 103:700–713. doi: 10.1017/S0007485313000448. [DOI] [PubMed] [Google Scholar]
- 11.Vélez AM, Spencer TA, Alves AP, Crespo ALB, Siegfried BD. 2013. Fitness costs of Cry1F resistance in fall armyworm, Spodoptera frugiperda. J Appl Entomol 138:315–325. [Google Scholar]
- 12.Jakka SR, Knight VR, Jurat-Fuentes JL. 2014. Fitness costs associated with field-evolved resistance to Bt maize in Spodoptera frugiperda (Lepidoptera: Noctuidae). J Econ Entomol 107:342–351. doi: 10.1603/EC13326. [DOI] [PubMed] [Google Scholar]
- 13.Jakka SR, Knight VR, Jurat-Fuentes JL. 2014. Spodoptera frugiperda (J. E. Smith) with field-evolved resistance to Bt maize are susceptible to Bt pesticides. J Invertebr Pathol 122:52–54. [DOI] [PubMed] [Google Scholar]
- 14.Vachon V, Laprade R, Schwartz JL. 2012. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: a critical review. J Invertebr Pathol 111:1–12. doi: 10.1016/j.jip.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Jurat-Fuentes JL, Karumbaiah L, Jakka SRK, Ning CM, Liu CX, Wu KM, Jackson J, Gould F, Blanco C, Portilla M, Perera O, Adang M. 2011. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to cry toxins from Bacillus thuringiensis. PLoS One 6:e17606. doi: 10.1371/journal.pone.0017606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu YC, Blanco CA, Portilla M, Adamczyk J, Luttrell R, Huang F. 2015. Evidence of multiple/resistance to Bt and organophosphate insecticides in Puerto Rico population of the fall armyworm, Spodoptera frugiperda. Pestic Biochem Physiol 122:15–21. doi: 10.1016/j.pestbp.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Adang M, Crickmore N, Jurat-Fuentes JL. 2014. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action, p 39–87. In Dhadialla TS, Gill S (ed), Advances in insect physiology, vol 47. Insect midgut and insecticidal proteins. Academic Press, San Diego, CA. [Google Scholar]
- 18.Tabashnik BE, Liu Y, Malvar T, Heckel DG, Masson L, Ferré J. 1998. Insect resistance to Bacillus thuringiensis: uniform or diverse. Philos Trans R Soc Lond B Biol Sci 353:1751–1756. doi: 10.1098/rstb.1998.0327. [DOI] [Google Scholar]
- 19.Gahan LJ, Gould F, Heckel DG. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857–860. doi: 10.1126/science.1060949. [DOI] [PubMed] [Google Scholar]
- 20.Morin S, Biggs RW, Sisterson MS, Shriver L, Ellers-Kirk C, Higginson D, Holley D, Gahan LJ, Heckel DG, Carrière Y, Dennehy TJ, Brown JK, Tabashnik BE. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc Natl Acad Sci U S A 100:5004–5009. doi: 10.1073/pnas.0831036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Yu L, Wu Y. 2005. Disruption of a cadherin gene associated with resistance to Cry1Ac δ-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl Environ Microbiol 71:948–954. doi: 10.1128/AEM.71.2.948-954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baxter SW, Badenes-Perez FR, Morrison A, Vogel H, Crickmore N, Kain W, Wang P, Heckel DG, Jiggins CD. 2011. Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics 189:675–679. doi: 10.1534/genetics.111.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caccia S, Moar WJ, Chandrashekhar J, Oppert C, Anilkumar KJ, Jurat-Fuentes JL, Ferre J. 2012. Association of Cry1Ac toxin resistance in Helicoverpa zea (Boddie) with increased alkaline phosphatase levels in the midgut lumen. Appl Environ Microbiol 78:5690–5698. doi: 10.1128/AEM.00523-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z, Kang S, Chen D, Wu Q, Wang S, Xie W, Zhu X, Baxter SW, Zhou X, Jurat-Fuentes JL, Zhang Y. 2015. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet 11:e1005124. doi: 10.1371/journal.pgen.1005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco CA, Portilla M, Jurat-Fuentes JL, Sanchez JF, Viteri D, Vega-Aquino P, Teran-Vargas AP, Azuara-Dominguez A, Lopez JDJ, Arias R, Zhu Y-C, Lugo-Barrera D, Jackson R. 2010. Susceptibility of isofamilies of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Cry1Ac and Cry1Fa proteins of Bacillus thuringiensis. Southwest Entomol 35:409–415. doi: 10.3958/059.035.0325. [DOI] [Google Scholar]
- 26.Visser B, van der Salm T, van den Brink W, Folkers G. 1988. Genes from Bacillus thuringiensis entomocidus 60.5 coding for insect-specific crystal proteins. Mol Gen Genet 212:219–224. doi: 10.1007/BF00334688. [DOI] [Google Scholar]
- 27.Perera OP, Willis JD, Adang MJ, Jurat-Fuentes JL. 2009. Cloning and characterization of the Cry1Ac-binding alkaline phosphatase (HvALP) from Heliothis virescens. Insect Biochem Mol Biol 39:294–302. doi: 10.1016/j.ibmb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Lee MK, Milne RE, Ge AZ, Dean DH. 1992. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis delta-endotoxin. J Biol Chem 267:3115–3121. [PubMed] [Google Scholar]
- 29.Liu YB, Tabashnik BE. 1997. Inheritance of resistance to the Bacillus thuringiensis toxin Cry1C in the diamondback moth. Appl Environ Microbiol 63:2218–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Püntener W. 1981. Manual for field trials in plant protection, 2nd ed Agricultural Division, Ciba-Geigy Ltd, Basel, Switzerland. [Google Scholar]
- 31.Robertson JL, Russell RM, Preisler HK, Savin NE. 2007. Binary quantal response: data analyses, p 36–54. In Bioassays with arthropods. CRC Press, Boca Raton, FL. [Google Scholar]
- 32.Wolfersberger M, Luethy P, Maurer A, Parenti P, Sacchi VF, Giordana B, Hanozet GM. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp Biochem Physiol 86A:301–308. [Google Scholar]
- 33.Jurat-Fuentes JL, Gould FL, Adang MJ. 2002. Altered glycosylation of 63- and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins. Appl Environ Microbiol 68:5711–5717. doi: 10.1128/AEM.68.11.5711-5717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurat-Fuentes JL, Adang MJ. 2004. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem 271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 35.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. 1989. Specificity of Bacillus thuringiensis delta-endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur J Biochem 186:239–247. [DOI] [PubMed] [Google Scholar]
- 36.Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. 2012. Specific binding of radiolabeled Cry1Fa insecticidal protein from Bacillus thuringiensis to midgut sites in lepidopteran species. Appl Environ Microbiol 78:4048–4050. doi: 10.1128/AEM.07591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahman K, Abdullah MA, Ambati S, Taylor MD, Adang MJ. 2012. Differential protection of Cry1Fa toxin against Spodoptera frugiperda larval gut proteases by cadherin orthologs correlates with increased synergism. Appl Environ Microbiol 78:354–362. doi: 10.1128/AEM.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gahan LJ, Pauchet Y, Vogel H, Heckel DG. 2010. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet 6:e1001248. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenhaber B, Bork P, Eisenhaber F. 1999. Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292:741–758. doi: 10.1006/jmbi.1999.3069. [DOI] [PubMed] [Google Scholar]
- 40.Pierleoni A, Martelli P, Casadio R. 2008. PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poisson G, Chauve C, Chen X, Bergeron A. 2007. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics 5:121–130. doi: 10.1016/S1672-0229(07)60022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 45.Dow AgroSciences LLC. 2000. Petition for the determination of non-regulated status B.t. Cry1F insect resistant, glufosinate tolerant maize line. Dow AgroSciences, Indianapolis, IN: http://www.aphis.usda.gov/brs/aphisdocs/00_13601p.pdf. [Google Scholar]
- 46.Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ, Li X, Carriere Y, Tabashnik BE. 2014. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS One 9:e97900. doi: 10.1371/journal.pone.0097900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ojha A, Sree KS, Sachdev B, Rashmi MA, Ravi KC, Suresh PJ, Mohan KS, Bhatnagar RK. 2014. Analysis of resistance to Cry1Ac in field-collected pink bollworm, Pectinophora gossypiella (Lepidoptera: Gelechiidae), populations. GM Crops Food 5:280–286. doi: 10.4161/21645698.2014.947800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabrick JA, Tabashnik BE. 2007. Binding of Bacillus thuringiensis toxin Cry1Ac to multiple sites of cadherin in pink bollworm. Insect Biochem Mol Biol 37:97–106. doi: 10.1016/j.ibmb.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Tabashnik BE, Biggs RW, Higginson DM, Henderson S, Unnithan DC, Unnithan GC, Ellers-Kirk C, Sisterson MS, Dennehy TJ, Carriere Y, Morin S. 2005. Association between resistance to Bt cotton and cadherin genotype in pink bollworm. J Econ Entomol 98:635–644. doi: 10.1603/0022-0493-98.3.635. [DOI] [PubMed] [Google Scholar]
- 50.Bel Y, Siqueira HA, Siegfried BD, Ferré J, Escriche B. 2009. Variability in the cadherin gene in an Ostrinia nubilalis strain selected for Cry1Ab resistance. Insect Biochem Mol Biol 39:218–223. doi: 10.1016/j.ibmb.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. 2013. Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS One 8:e68164. doi: 10.1371/journal.pone.0068164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monnerat R, Martins E, Queiroz P, Ordúz S, Jaramillo G, Benintende G, Cozzi J, Real MD, Martinez-Ramirez A, Rausell C, Cerón J, Ibarra JE, Del Rincon-Castro MC, Espinoza AM, Meza-Basso L, Cabrera L, Sánchez J, Soberon M, Bravo A. 2006. Genetic variability of Spodoptera frugiperda Smith (Lepidoptera: Noctuidae) populations from Latin America is associated with variations in susceptibility to Bacillus thuringiensis Cry toxins. Appl Environ Microbiol 72:7029–7035. doi: 10.1128/AEM.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferré J, Real MD, Van Rie J, Jansens S, Peferoen M. 1991. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci U S A 88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang P, Zhao JZ, Rodrigo-Simón A, Kain W, Janmaat AF, Shelton AM, Ferré J, Myers J. 2007. Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni. Appl Environ Microbiol 73:1199–1207. doi: 10.1128/AEM.01834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh M, Inoue T, Kanamori Y, Nishida S, Yamaguchi M. 2003. Tandem duplication of alkaline phosphatase genes and polymorphism in the intergenic sequence in Bombyx mori. Mol Genet Genomics 270:114–120. doi: 10.1007/s00438-003-0880-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Tian W, Zhao J, Jin L, Yang J, Liu C, Yang Y, Wu S, Wu K, Cui J, Tabashnik BE, Wu Y. 2012. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc Natl Acad Sci U S A 109:10275–10280. doi: 10.1073/pnas.1200156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin L, Wei Y, Zhang L, Yang Y, Tabashnik BE, Wu Y. 2013. Dominant resistance to Bt cotton and minor cross-resistance to Bt toxin Cry2Ab in cotton bollworm from China. Evol Appl 6:1222–1235. doi: 10.1111/eva.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gould F. 1998. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu Rev Entomol 43:701–726. doi: 10.1146/annurev.ento.43.1.701. [DOI] [PubMed] [Google Scholar]
- 59.Arenas I, Bravo A, Soberón M, Gómez I. 2010. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem 285:12497–12503. doi: 10.1074/jbc.M109.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ning C, Wu K, Liu C, Gao Y, Jurat-Fuentes JL, Gao X. 2010. Characterization of a Cry1Ac toxin-binding alkaline phosphatase in the midgut from Helicoverpa armigera (Hübner) larvae. J Insect Physiol 56:666–672. doi: 10.1016/j.jinsphys.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 61.van Frankenhuyzen K. 2009. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol 101:1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Storer NP, Kubiszak ME, King ED Jr, Thompson GD, Santos AC. 2012. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J Invertebr Pathol 110:294–300. doi: 10.1016/j.jip.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Huang F, Qureshi JA, Meagher RL Jr, Reisig DD, Head GP, Andow DA, Ni X, Kerns D, Buntin GD, Niu Y, Yang F, Dangal V. 2014. Cry1F resistance in fall armyworm Spodoptera frugiperda: single gene versus pyramided Bt maize. PLoS One 9:e112958. doi: 10.1371/journal.pone.0112958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.