Abstract
In this study, we examined the role of neprilysin (NEP*), a key membrane-bound endopeptidase, in the inflammatory response induced by diesel exhaust emissions (DEE) in the airways through a number of approaches: in vitro, animal, and controlled human exposure. Our specific aims were (1) to examine the role of NEP in inflammatory injury induced by diesel exhaust particles (DEP) using Nep-intact (wild-type) and Nep-null mice; (2) to examine which components of DEP are associated with NEP downregulation in vitro; (3) to determine the molecular impact of DEP exposure and decreased NEP expression on airway epithelial cells’ gene expression in vitro, using a combination of RNA interference (RNAi) and microarray approaches; and (4) to evaluate the effects on NEP activity of human exposure to DEE. We report four main results: First, we found that exposure of normal mice to DEP consisting of standard reference material (SRM) 2975 via intratracheal installation can downregulate NEP expression in a concentration-dependent manner. The changes were accompanied by increases in the number of macrophages and epithelial cells, as well as proinflammatory cytokines, examined in bronchoalveolar lavage (BAL) fluid and cells. Nep-null mice displayed increased and/or additional inflammatory responses when compared with wild-type mice, especially in response to exposure to the higher dose of DEP that we used. These in vivo findings suggest that loss of NEP in mice could cause increased susceptibility to injury or exacerbate inflammatory responses after DEP exposure via release of specific cytokines from the lungs. Second, we found evidence, using in vitro studies, that downregulation of NEP by DEP in cultured human epithelial BEAS-2B cells was mostly attributable to DEP-adsorbed organic compounds, whereas the carbonaceous core and transition metal components of DEP had little or no effect on NEP messenger RNA (mRNA) expression. This NEP downregulation was not a specific response to DEP or its contents because the change also occurred after exposure to urban dust (SRM 1649a), which differs in physical and chemical composition from DEP. Third, we also collected the transcriptome profiles of the cells through a 2 × 3 factorial design. DEP exposure upregulated 151 genes and downregulated 59 genes. Cells with decreased NEP expression (accomplished by transfecting an NEP-specific small interfering RNA [siRNA]) substantially altered the expression of genes (upregulating 17 and downregulating 14) associated with DNA/protein binding, calcium channel activities, and the cascade of intracellular signaling by cytokines. Data generated from the combined RNAi and microarray approaches revealed that there is a complex molecular cascade mediated by NEP in different subcellular compartments, possibly influencing the inflammatory response. Fourth, in a controlled human exposure study, we observed significant increases in soluble NEP in sputum after acute exposure to DEE, with an average net increase of 31%. We speculate that the change in NEP activity in sputum, if confirmed in larger epidemiologic investigations at ambient exposure levels to DEE, may provide a useful endpoint and promote insight into the mechanism of DEE-induced airway alterations.
INTRODUCTION
UNCERTAINTY ABOUT UNDERLYING MECHANISMS OF HEALTH EFFECTS INDUCED BY AMBIENT PARTICULATE MATTER
Particulate matter (PM), especially with an aerodynamic diameter ::: 2.5 µm (PM2.5), is a major risk to human health. Its acute and chronic adverse effects at the ambient level, particularly the association between elevated levels of PM and morbidity or mortality, have been demonstrated epidemiologically (Dockery et al. 1992, 1994; Peters et al. 1997; Salvi et al. 1999; Samet et al. 2000a, b; Schwartz et al. 2002). Studies over a broad range of geographic areas indicate that with each increase of 10 µg/m3 in ambient particles, daily mortality increases approximately 1% to 5% (Pope et al. 2002; Schwartz et al. 2002). The adverse effects are especially apparent in the young or old and in those with preexisting cardiopulmonary disorders (Venn et al. 2001; Bateson and Schwartz 2004; Stenfors et al. 2004; McCreanor et al. 2007). Small short-term increases in PM2.5 levels have been associated with increases in symptoms of certain conditions such as asthma, bronchitis, and airway hyperresponsiveness. Although PM effects in the airways are commonly manifested by irritation, inflammation, and functional impairment (HEI Diesel Working Group 1995; Veronesi and Oortgiesen 2001; Pandya et al. 2002; Nel 2005), the precise mechanisms that control these responses to inhaled particulates are still poorly understood.
In vivo and in vitro experimental observations have suggested that the integrity of the targeted epithelium, especially its capacity to prevent or recover from oxidative injury, is central to the inflammatory reaction. Acute exposure to PM2.5, or its components, has been shown to induce release of mediators such as interleukin 6 (IL-6), the chemokine IL-8, epidermal growth factor, and granulocyte-macrophage colony-stimulating factor (Boland et al. 2000). Long-term exposure results in damage identified histopathologically, such as bronchioli wall thickening, concomitant with the presence of numerous particles in the lungs of exposed individuals (Churg et al. 2003). At the molecular level, the proinflammatory responses are in part mediated through the activation of many signaling transduction pathways. These may involve transcription factors (nuclear factor-kappa B [NFKB2], activator protein-1 [AP-1], and the signal transducer and activator of transcription 3 [Stat3]) (Takizawa et al. 1999, 2003; Zhang et al. 2004; Cao et al. 2007) and mitogen-activated protein kinases (MAPKs) (Fahy et al. 2000; Hashimoto et al. 2000) via reactive oxygen species–dependent mechanisms (Baulig et al. 2003). While substantial progress has been made in understanding inflammatory pathogenesis after PM-induced injury, other regulatory mechanisms remain to be studied.
Macrophages, another cell type found in the lung, are also part of the first line of host defense and play a critical role in the cellular response of the airways to inhaled PM. They are located largely on the epithelial surfaces and act as a barrier to inhaled matter by phagocytosing particles and intracellularly processing them (Bowden 1984; Kreyling 1992). It has been shown that there is a direct, positive relation between the extent of PM exposure and the number of macrophages present in the lungs (Brain 1992). However, the macrophages’ phagocytic and chemotactic functions are inhibited when the volume of the phagocytosed particles is greater than 60% of their total internal volume (Morrow 1988; MacNee and Donaldson 2003). In his 1988 study, Morrow observed that even if phagocytosed PM constituted only 6% of the total volume of macrophages, the ability of the airway macrophages to migrate to the mucociliary escalator was compromised (Morrow 1988). Phagocytosis of certain particles, such as silica and those from fire smoke, can damage or kill macrophages, releasing their contents, which then fuels an inflammatory response (Bowden 1984; Wong et al. 2004). Furthermore, activation of macrophages after PM phagocytosis releases copious amounts of oxygen radicals, neurokinins, proteases, proinflammatory cytokines, and growth-regulating proteins that may be involved in the pathogenesis of both acute and chronic lung inflammation (Oberdörster et al. 1992; Zhou et al. 2007). In addition, exposure to PM also compromises the macrophage response to infectious agents, possibly via an oxygen-radical–mediated process, by decreasing the cell’s ability to phagocytose bacteria (Becker and Soukup 1998; Yang et al. 1999; Mundandhara et al. 2006). These studies suggest that the functional capacity of macrophages is affected by exposure to PM in such a manner that host lung defenses and immune functions are decreased.
NEP AND ITS PHYSIOLOGIC IMPORTANCE
NEP (also known as neutral endopeptidase, EC 3.4.24.11, enkephalinase, and common acute lymphoblastic leukemia antigen) is a membrane-bound zinc-dependent endopeptidase. Structurally, NEP is a 90–110 kDa type II integral membrane glycoprotein consisting of a short N-terminal cytoplasmic domain, a transmembrane hydrophobic region, and a large extracellular domain that contains the catalytic site. The human NEP gene is located on chromosome 3 (q21–q27). NEP plays a central regulatory role in the maintenance of homeostasis and regulation of sensory reflexes including apnea, laryngeal narrowing, bronchoconstriction, sneezing, aspiration, and expiration reflexes (Martins et al. 1990; Nadel 1991). NEP is also involved in the pathogenesis of cardiopulmonary diseases, Alzheimer disease, and cancer (Bozic et al. 1996; El-Amouri et al. 2008; Iijima-Ando et al. 2008). The role of NEP in several organs has also been documented (Borson 1991; Joos 2000). The following is a brief summary of background information for the current study, culled from the large body of literature on NEP.
Cellular Distribution of NEP
Investigators have examined the cellular distribution of NEP by using specific antibodies, measuring its expression in different cell types. NEP is widely distributed in mammalian tissues. In the lung, NEP is abundantly expressed on airway or alveolar epithelial cells and is present in airway smooth muscle cells, submucosal gland cells, fibroblasts, postcapillary venules, and nerves (Painter et al. 1988; Baraniuk et al. 1995). Moreover, NEP is expressed on neutrophils and macrophages (Johnson et al. 1985a,b).
Physiologic Function of NEP
NEP is an enzyme with broad specificity. It effectively controls the bioavailability of peptide mediators. It maintains low levels of its substrates in the extracellular fluid under basal conditions. These substrates include tachykinins, endothelins, angiotensin II, bombesin, gastrin-releasing peptide, atrial natriuretic peptide, enkephalins, insulin B chain, and the chemotactic peptide N-formyl-met-leu-phe. Most of these substrates are neurotransmitters and proinflammatory mediators, which are released from sensory nerve terminals and activate immunoinflammatory cells such as neutrophils, eosinophils, lymphocytes, and macrophages (Nadel 1991). NEP substrates play important roles in numerous physiologic and pathophysiologic processes, including inflammatory processes (Lotz et al. 1988; Lilly et al. 1994; Di Maria et al. 1998), hyperresponsiveness (Dusser et al. 1989; Wu and Lee 1999), and carcinogenesis (Nanus 1998; Papandreou et al. 2000; Suzuki et al. 2001; Tomoda et al. 2003; Sumitomo et al. 2004). The NEP cytoplasmic tail plays a role in providing a scaffold for signaling proteins in the regulation of cell repair pathways and the organization of the membrane-associated cytoskeleton (Iwase et al. 2004; Sumitomo et al. 2004). The almost ubiquitous distribution of NEP, with its broad substrate specificity, suggests it has a role in the cleavage of different peptides involved in several functions. When NEP expression or activity is inhibited, its substrates are less rapidly inactivated and accumulate in tissue (Martins et al. 1990; Wong et al. 2004), thus contributing to an exaggerated response or increased susceptibility to environmental stressors.
Role of NEP in Lung Disease
NEP plays a key role in airway homeostasis and the development of acute lung injury (ALI) or acute respiratory distress syndrome (ARDS) (Wong et al. 2003), asthma (Lundberg et al. 1991; van Der Velden et al. 1999), chronic obstructive pulmonary disease (COPD) (Lotz et al. 1988), and lung cancer (D’Adamio et al. 1989; Shipp et al. 1991). In addition, several reviews (Borson 1991; Nadel 1991; Di Maria et al. 1998) of the role of NEP in lung airways suggest that upregulation of NEP gene expression may be one mechanism of the anti-inflammatory action of glucocorticoids.
NEP is a critical protective enzyme in limiting the activity of endogenously released substance P (SP), abnormal levels of which may be involved in the pathogenesis of ALI/ARDS. Loss of NEP activity clearly leads to a persistent increase in endogenous SP, which may in turn lead to exaggerated microvascular permeability, edema, and severe hypoxia. It is well known that SP, as a potent proinflammatory mediator, activates numerous signaling transduction pathways involving a complex network of chemokines, cytokines, reactive oxygen/nitrogen species, and other mediators. SP released in the lungs may signal immuno-inflammatory cells to generate these mediators through neurokinin (NK) receptors that may not be involved in the cellular responses to SP under normal physiologic conditions. The affected cell populations include neutrophils, eosinophils, lymphocytes, and macrophages, which express NK-1R on their cell surfaces. Once these cells are activated, an uncontrolled inflammatory cascade develops and progresses, with the involvement of multiple immuno-inflammatory cells and their mediators. This is similar to the early pathophysiologic processes of ALI/ARDS. A substantial disruption of NEP occurs, and high microvascular permeability and pulmonary edema may develop. This scenario is supported by previous experimental observations (Fine et al. 1989), which showed that removal of the airway epithelium cells or inhibition of NEP could induce airway hyperresponsiveness to SP.
Day and colleagues (2005) reported that NEP activity determines the severity of pancreatitis-associated lung injury. Nep-null and Nep-intact mice pretreated with the NEP antagonist phosphoramidon (10 mg/kg) had significant elevations of lung myeloperoxidase and worsened lung histology compared with Nep-intact mice given elastase to induce lung injury (Day et al. 2005). As observed in our animal models of ALI/ARDS–like injury, fire smoke inhalation induced a dose-dependent reduction in pulmonary NEP activity beginning as early as one hour after insult (Wong et al. 2004). The changes in NEP activity observed through immunohistochemistry were mainly attributable to damage of the epithelial lining, such as membrane disruption, necrosis, and sloughing of epithelial cells in the airways from the trachea to bronchioles of the smoke-exposed animals (Wong et al. 2004). Additionally, the causes of loss of NEP activity in this model could include inhibition of NEP activity by oxidants from combustion products such as nitrogen dioxide (NO2), ozone (O3), and particulates, but also from the thermal denaturing of plasma proteins and from inflammatory cell activation.
Bronchial asthma and COPD are conditions in which the tone of airway smooth muscle, airway secretions, bronchial circulation, and inflammatory and immune cells are affected. These responses are highly regulated by NEP, which is predominantly responsible for controlling levels of tachykinins in the lungs; uncontrolled or exaggerated responses due to decreased NEP show many similarities to the clinical symptoms of patients with bronchial asthma and COPD (Di Maria et al. 1998; Joos et al. 2000; Wick et al. 2011). Moreover, NEP may play a key role in the development of childhood asthma (Joos et al. 2000), not only because of the vulnerable nature of the children’s lungs, but also because the airway sensory innervations of a neonate develop rapidly during early postnatal life in parallel with the developing lung (Hislop et al. 1990). Additionally, NEP expression in airway epithelium is age-dependent and implicated in the regulation of peptides associated with normal lung growth in the fetus (Sunday et al. 1992).
Cohen and colleagues (1996) showed that NEP mRNA was not expressed or was at a low level in most lung cancer cell lines. Protein expression and activity for NEP were also reduced or undetectable in most small-cell lung carcinoma (SCLC), adenocarcinoma tumors, and lung adenocarcinoma cell lines. Similarly, NEP mRNA was undetectable in SCLC, adenocarcinoma, squamous cell carcinoma, and carcinoid tumors of the lung (Cohen et al. 1996). Genetically targeted disruption of the Nep locus in mice resulted in an enhanced lethality to endotoxin, indicating an important protective role for NEP in septic shock (Lu et al. 1996, 1997).
DEP TOXICITY ON NEP IN THE AIRWAYS
Expression of NEP varies widely in normal human lung tissue taken from different individuals (Cohen et al. 1999). This variation could be attributed to either environmental or individual genetic factors. Studies have shown that a variety of environmental factors, including exposure to viruses, allergens, cigarette smoke, and respiratory toxins, are able to reduce NEP activity, thus enhancing the effects of neuropeptides within the airways (Turner et al. 1993; Lilly et al. 1994; Sun et al. 2004; Wong et al. 2004). Therefore, we initially hypothesized that DEP exposure in rats could downregulate pulmonary NEP. Our studies have indicated that NEP activity in rat lungs was significantly reduced by exposure to DEP (Wong et al. 2003; Witten et al. 2005). Because of the high density of NEP expression in airway epithelium and NEP’s important regulatory role, it is not surprising that a reduction in NEP is accompanied by increases in bronchopulmonary plasma extravasation, vascular permeability, cytokine expression, and inflammatory mast cell infiltration, possibly evoked by abnormally high levels of peptides after DEP exposure. Reduced NEP activity may set up the airways to respond in an exaggerated fashion to irritation and other inflammatory mediators, thus producing a hyperresponsive state. Therefore, we speculate that a decrease in NEP activity after exposure to DEE may be a precursor in lungs for the ultimate loss of NEP expression. This may contribute to the DEE-related increase in health risk, including the risk of asthma, COPD, and lung cancer. This concept is strongly supported by numerous investigations reporting a decrease of NEP in first- and secondhand cigarette smokers and the loss of NEP due to lung cancer and in most cell lines derived from lung cancers (as well as many other human malignancies) (Shipp et al. 1988, 1991; Dusser et al. 1989; Cohen et al. 1996; Papandreou et al. 2000). Loss of NEP may be implicated in the broad, adverse effects observed in other organ diseases, such as cardiovascular diseases and Alzheimer disease. We believe that the inhalation of particles is an important environmental factor in the decrease or loss of NEP, which is directly or indirectly involved in the development of PM-related disorders.
Our pilot study (Wong et al. 2007 pilot study) sought to extend our earlier findings in rats (Wong et al. 2003; Witten et al. 2005). We exposed human airway epithelial cells to DEP (0–40 µg/mL, noncytotoxic) for 24 hours and observed the downregulation of both NEP expression and enzymatic activity in a concentration-dependent manner (Wong et al. 2007 pilot study). A substantial decrease (90%) in NEP mRNA expression occurred with exposure to 5 µg/mL DEP. A further stability test indicated that NEP downregulation by DEP occurs at the transcriptional level (Wong et al. 2007 pilot study). It is likely that the NEP gene promoter may have DEP responsive elements that are activated via signal transduction pathways after DEP challenge. Alternatively, the NEP gene may be directly affected by certain components of DEP. Using RNAi technique, we showed that the cell proliferation of NEP siRNA-infected cells was inhibited in a concentration-dependent manner with increased DEP concentrations (P < 0.01) (Wong et al. 2007 pilot study). The net proliferation inhibition rates at 5, 10, 20, and 40 µg/mL DEP were 1.4%, 7.6%, 10%, and 14.4%, respectively, when compared with their controls. This finding suggests that NEP per se is involved in cell proliferation in the presence of DEP. We interpreted our pilot study (Wong et al. 2007 pilot study) as indicating that DEP exposure resulted in the downregulation of NEP expression at the transcriptional level, which was associated with a change in cell proliferation, an important process in pathophysiology. However, the mechanisms of NEP downregulation by DEP and its relevance to these broad adverse health effects need to be investigated.
We believe that DEP is an important contributor to NEP downregulation in the airways. The decrease or loss of NEP expression and/or activity may cause a greater susceptibility to respiratory disorders due to dysregulated respiratory irritation, inflammatory response, and tissue repair or remodeling. To our knowledge, no previous cellular, animal, or human studies have been conducted on the effects of PM on airway epithelial NEP and its relevance to the development or exacerbation of airway disorders. Given that NEP is an important regulator of numerous pathophysiologic processes and is affected by inhaled particulates, the study of epithelial NEP is of considerable importance in the understanding of PM-induced adverse health effects.
SPECIFIC AIMS
In the current study, we had the following four specific aims:
To examine the role of NEP in DEP-induced inflammatory injury using Nep-intact and Nep-null mice.
To examine which components of DEP are associated with NEP downregulation in vitro.
To determine the molecular impact of DEP exposure and decreased NEP expression on airway epithelial cells’ gene expression in vitro, using a combination of RNAi and microarray approaches.
To evaluate the effects on NEP activity of human exposure to DEE.
HYPOTHESIS
Based on our previous study (Wong et al. 2007 pilot study), we hypothesize that certain components of DEP downregulate the functional expression of airway epithelial NEP via a transcriptional mechanism. Consequently, the affected cells’ metabolic, cellular, and regulatory functions in the maintenance of cellular homeostasis are compromised, leading to an exacerbated inflammatory response or increased susceptibility to injury.
There were five rationales for this hypothesis: First, NEP is abundantly expressed on airway epithelial cells and is directly targeted by DEP. Second, structurally NEP is a cell-surface metalloprotease with a large extracellular domain (700 amino acids), which contains six potential N-glycosylation sites and the pentapeptide consensus sequence (His-Glu-[Ile, Leu, Met]-X-His) of zinc-binding metalloproteases, in which the two histidines link up zinc and glutamic acid. NEP’s large extracellular domain containing the catalytic sequence may explain not only its critical structural ability to rapidly cleave substrates, but also its tendency to be highly susceptible to toxic insults. Third, NEP plays a central regulatory role in the maintenance of homeostasis and in the development of pathophysiologic processes involved in ALI or ARDS, asthma, COPD, and lung cancer. Fourth, a significant decrease in NEP activity in lung tissue has been demonstrated in rats exposed to ambient and occupational levels of diesel exhaust (DE) (Wong et al. 2003). Fifth and finally, exposure of human airway epithelial cells to DEP (0–40 µg/mL, noncytotoxic) for 24 hours downregulates NEP expression, as well as its enzymatic activity, in a concentration-dependent manner (Wong et al. 2007 pilot study).
METHODS AND STUDY DESIGN
AIM 1: TO EXAMINE THE ROLE OF NEP IN DEP-INDUCED INFLAMMATORY INJURY USING Nep-INTACT AND Nep-NULL MICE
Experimental Design
Specific-pathogen–free wild-type and Nep-null mice (from Dr. Craig Gerard of Harvard Medical School) on a C57BL/6 background (50/50 male/female, approximately 8 weeks old, weighing approximately 25 g) were used in this part of the study. The mice were bred and housed in the American Association of Animal Laboratory and Care (AAALAC)–approved animal facility at the University of Arizona Health Sciences Center. The mice were on a 12h:12h light–dark cycle and were given a standard mouse chow diet, tap water ad libitum, and filtered air. In order to simultaneously characterize the dose–response of DEP on respiratory NEP and examine the role of NEP in response to DEP exposure, we utilized a 2 × 3 factorial design (66 mice = 11 mice/group × 6 groups; wild-type vs. Nep-null mice exposed to control, low, and high doses of DEP). We examined the differences in DEP-induced pulmonary response between Nep-null and wild-type mice, as measured by BAL cell profile, cytokines, cell proliferation, and histopathologic evaluation. In addition, NEP protein expression in the lung tissues of wild-type mice was quantified by DuoSet (R&D Systems, Minneapolis, MN) enzyme-linked immunosorbent assay (ELISA) and localized by immunohistochemistry.
DEP Instillation
Considering that DEP composition is highly variable, depending on, among other factors, engine type and engine load (Madden et al. 2003), we utilized standard DEP (SRM 2975; National Institute of Standards and Technology, U.S. Department of Commerce) originally generated by a diesel-powered industrial forklift and whose physical aspects and chemical composition have been well characterized. SRM 2975 was instilled through an intratracheal cannula under anesthesia. This technique, although a non-physiologic method of administration, is useful for comparative studies in which collected samples cannot easily be used in inhalation exposures. The doses used in this study were 10 and 100 µg for the low and high exposure levels, respectively, of DEP instillation. These doses were chosen to simulate a continuous 7-day inhalation exposure of 50 and 500 µg/m3, using the following equation:
where 50 µg/m3 and 500 µg/m3 were the low and high levels of DEP exposure per day, respectively; 1/2 is the deposition coefficient of PM in the lung; and 0.06 m3/day is the daily inhalation volume for a mouse.
Briefly, we first anesthetized mice with an intramuscular injection mixture of ketamine hydrochloride (80 mg/kg; Parke-Davis, Morris Plains, NJ), xylene (10 mg/kg; Mobay, Shawnee, KS), and acepromazine maleate (3 mg/kg; Fermenta, Kansas City, MO). These animals were then intubated by a nonsurgical technique. Using a bulb-headed cannula inserted approximately 10 mm into the trachea, we instilled a suspension containing either 10 or 100 µg DEP in 100 µL phosphate-buffered saline (PBS) that was free of endotoxins and Ca2+/Mg2+, followed by 100 µL air. The suspension of DEP was sonicated on ice for 1 minute before instillation, using a model 250 Sonifier (Branson Ultrasonic Corporation, Danbury, CT) at a moderate level of 20 W. Sham control animals received 100 µL PBS only, followed by 100 µL air. In our experience, the instillation of 100 µL PBS does not cause any measurable stress effects, such as the expression of cytokines. Animals were treated humanely and with regard for alleviation of suffering. Compliance with AAALAC-approved animal protocol (#04-104) ensures that these animals were cared for and treated according to NIH guidelines.
Bronchoalveolar Lavage and Cell Count
At 7 days after DEP or PBS instillation, we euthanized the anesthetized animals (N = 7) by exsanguination of the abdominal aorta, and removed their lungs and cannulated them with a Teflon intravenous catheter. The lungs were lavaged three times with sterile isotonic saline (Baxter, Deerfield, IL) at a volume of 1 mL for each wash. BAL fluid was centrifuged at 4°C for 15 minutes at 500g. We stored the supernatant and lung tissue at −75°C for cytokine assays and NEP expression. We determined the number of cells using a hemocytometer and performed differential cell counts by counting 300 cells per slide on a Diff-Quik–stained (Dade Diagnostics, Aguada, PR) cytocentrifuged slide.
Protein Expression Quantification Using ELISA
We determined the interleukin (IL)-1[3, IL-6, and IL-10 concentrations in BAL fluid using the commercially available Enzyme Immunoassay Kits (R&D Systems). We quantified each sample in triplicate and then averaged it to obtain the final value. We used a BioTek ELx808IU microplate reader (BioTek Instruments, Winooski, VT) for spectrophotometer analyses at a reading wavelength of 450 nm and a reference wavelength of 570 nm for each analysis.
We followed the protocol for ELISA kits provided by the manufacturer to quantify NEP protein in lung tissue extracts of wild-type mice (R&D Systems). We coated a clear 96-well microplate with 100 µL of goat anti-mouse NEP antibody (as the capture antibody) per well at a concentration of 0.8 µg/mL in PBS and incubated it overnight at room temperature. We then washed the wells three times with 400 µL of wash buffer and blocked them with 300 µL of Reagent Diluent at room temperature for a minimum of 1 hour. The plates were washed three times with 400 µL of wash buffer. We added various dilutions (0–6000 pg/mL) of NEP and the sample (total volume, 100 µL) to the wells and incubated them for 2 hours at room temperature. After washing, biotin-conjugated donkey antigoat IgG secondary antibody (as the detection antibody) diluted at 400 ng/mL in Reagent Diluent was added to each well and incubated for 2 hours at room temperature. We then washed the wells again and incubated them at room temperature for 20 minutes with 100 µL streptavidin conjugated to horseradish-peroxidase diluted in Reagent Diluent. After the wells were washed, we added 100 µL freshly prepared substrate solution (0.55 mg of 2,2'-azino-bis- 3-ethylbenzothiazoline-6-sulfonic acid [ABTS]/mL and 0.001% H2O2 in 0.1-M citrate buffer, pH 4.3) to each well and incubated them at room temperature in the dark. After 20 minutes, we stopped the reaction by adding 50 µL 2N sulfuric acid to the wells. We read the absorbance at 450 and 570 nm with a BioTek ELx808IU microplate reader.
Immunohistochemistry
To directly analyze epithelial cell proliferation, we administered 100 mg/kg 5-bromo-2'-deoxyuridine (5-BrdU; Sigma, St. Louis, MO) intraperitoneally to mice 24 hours before euthanasia. Fixed tissue sections (4 µm) were deparaffinized, hydrated, and pretreated with blocking solution to decrease nonspecific antibody binding. We used a biotinylated monoclonal mouse anti-BrdU antibody (Zymed, South San Francisco, CA) according to the manufacturer’s protocol, followed by a streptavidin-peroxidase–conjugated secondary antibody and DAB substrate (Zymed). We counterstained sections with hematoxylin. We calculated a BrdU-labeling index for airways with epithelial cells incorporating BrdU by counting the number of BrdU-expressing nuclei relative to the total number of epithelial cells within 50 to 75 cell regions of the airways. We analyzed a total of 200 to 500 cells in each lung section.
We localized NEP protein in fixed lung tissue (N = 4/group) of wild-type mice using the protocol provided by Santa Cruz Biotechnology. Briefly, slides were deparaffinized three times using xylene for 5 minutes each and hydrated through graded ethanol solutions. We quenched the endogenous peroxidase activity on the slides by incubating them in 0.1% H2O2 for 5 minutes. We blocked non-specific binding with 10% horse or goat serum in PBS. After washing, we added goat antimouse NEP antibody (4 µg/mL in PBS) (R&D Systems) to each slide, and we incubated them for 2 hours at room temperature in a humidified chamber. The slides were rinsed in PBS and subsequently incubated with biotinconjugated donkey anti-goat IgG secondary antibody for 30 minutes. The slides were then incubated for 30 minutes in avidin-biotinylated horseradish peroxidase complex and substrate. After dehydration, we immediately mounted and observed the slides using light microscopy.
AIM 2: TO EXAMINE WHICH COMPONENTS OF DEP ARE ASSOCIATED WITH NEP DOWNREGULATION IN VITRO
Experimental Design
In order to clarify the contribution of major components of DEP in the downregulation of NEP, we compared the effects induced by (1) untreated DEP; (2) DEP treated with chelators to remove divalent cations, particularly transition metals (cDEP); and (3) DEP treated with dichloromethane to remove everything but the carbonaceous core, or “stripped DEP” (sDEP). We exposed BEAS-2B cells (described in the next section) to three noncytotoxic concentrations (0, 1, and 10 µg/cm2) of DEP (SRM 2975), cDEP, and sDEP for 24 hours. We ran parallel cell cultures with no DEP vehicle (see “Preparation of DEP,” below) as time-course controls. To determine whether the observed NEP downregulation was a specific response to DEP, we utilized standard urban dust (SRM 1649a) over the same concentration range as that of DEP (called here, oDEP). In all cases, the cells were monitored visually for viability and collected for measurements of NEP mRNA expression by using real-time polymerase chain reaction (RT-PCR) analysis or particle uptake by transmission electron microscopy. (Details on the methods used in the particle uptake portion of the experiment supporting Aim 2 can be found in Appendix A, available on the HEI Web site: www.healtheffects.org.)
Cell Culture
In our in vitro study, we used the BEAS-2B cell line—an immortalized human bronchial epithelial cell line transformed by an adenovirus 12-SV40 hybrid virus (American Type Culture Collection #CRL-9609, Manassas, VA). This cell line exhibits genotypic and phenotypic characteristics of human bronchial epithelial cells and so is broadly used to study in vitro molecular or cellular effects of agents that affect the airways. We cultured the cells as recommended by the suppliers. Briefly, BEAS-2B cells were used at passages 4–8 and maintained in complete keratinocyte growth medium (KGM) consisting of keratinocyte basal medium (KBM; Clonetics, San Diego, CA) and supplemented with bovine pituitary extract, human epidermal growth factor (5 ng/mL), hydrocortisone (0.5 mg/mL), ethanolamine (0.1 mM), phosphoethanolamine (0.1 mM), and insulin (5 mg/mL). We seeded cells on 6-well plates (2.5 × 104 cells/well) in 2 mL of complete KGM medium. Cells were grown to between 85% and 95% confluence in 6-well plates and then were treated with the different sets of particles described in the following section. We cultivated all cells under a humidified atmosphere of 5% carbon dioxide (CO2) and 95% air at 37°C. Cell toxicity was determined using the trypan blue-exclusion assay.
Preparation of DEP
We prepared a stock solution (2 mg/mL) of DEP by sonicating 10 mg SRM 2975 in 5 mL of 0.0025% culture solution containing Tween 80 (Sigma-Aldrich, St. Louis, MO) at 50 W for 2 minutes using a model 250 Sonifier (Branson Ultrasonic Corporation, Danbury, CT). We obtained sDEP by extracting 10 mg SRM 2975 in dichloromethane in a Soxhlet extractor (Sigma-Aldrich). The collected particles were extracted a second time to remove additional organic compounds and then suspended in 5 mL of 0.0025% culture solution containing Tween 80. For the cDEP preparation, we used a published method (Zhou and Kobzik 2007) that utilizes Chelex 100 (Bio-Rad, Hercules, CA), which preferentially removes divalent cations, particularly transition metals. We suspended 10 mg SRM 2975 in 2% Dulbecco’s modified Eagle’s medium (Invitrogen Corp., Carlsbad, CA) and centrifuged it at 8000 rpm for 5 minutes at 4°C. We recovered the supernatant (soluble fraction) and then added 50 mg/mL Chelex beads and mixed them on a rotating wheel for 4 hours at room temperature. After incubation, we centrifuged samples at 13,000 rpm for 5 minutes to convert the Chelex beads into pellets. The same amount of cDEP suspension as used with other DEP subtypes was mixed with culture solution. Finally, we added these resultant suspensions to cultured cells at a concentration of either 1 or 10 µg/cm2, based on the original mass of particles before extraction. The DEP suspension and control were added at equal volume to the cells.
RNA Preparation and RT-PCR
Cells were incubated with DEP (SRM 2975), cDEP, sDEP, or urban dust (SRM 1649a) for 24 hours. Using the Aurum Total RNA Mini Kit according to the manufacturer’s protocol (Bio-Rad), we isolated total RNA from BEAS-2B cells. We incubated all samples with ribonuclease-free deoxyribonuclease (DNase) (20 U/reaction) for 10 minutes at 37°C to eliminate DNA contamination. We quantified the RNA concentration by ultraviolet spectrophotometry at 260 nm and determined the purity by the 260-to-280-nm absorbance ratio (SpectraMax PLUS, Molecular Devices, Sunnyvale, CA). We verified the integrity of the RNA by electrophoresis on a 1.2% agarose gel containing form-aldehyde (2.2 mol/L) and ethidium bromide in 1 × 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (40 mmol/L MOPS [pH 7.0], 10 mmol/L sodium acetate, and 1 mmol/L ethylenediaminetetraacetic acid [pH 8.0]). One microgram of total RNA treated with DNase I was reverse-transcribed using the iScript complementary DNA (cDNA) synthesis kit (Bio-Rad), according to the manufacturer’s protocol. Subsequently, polymerase chain reactions (PCRs) were set up in 96-well plates, each containing 0.3 µM of oligonucleotide primer, 1 × SYBR Green Supermix (Bio-Rad), and 1 µL of cDNA synthesis reaction (in a total volume of 20 µL). All primer sequences designed specifically for Specific Aims 2 and 3 are listed in Table 1. Reactions were run and analyzed on a Bio-Rad iCycler iQ Real-Time PCR detection system. We determined the cycling parameters and analyzed the resulting data according to Applied Biosystems’ protocols. Briefly, data were analyzed using the comparative CT method as a means of relative quantitation, normalized to an endogenous reference (glyceraldehyde-3-phosphate dehydrogenase [GAPDH] and 18S ribosomal RNA [18S rRNA]) and relative to a calibrator (a normalized CT value obtained from vehicle-treated BEAS-2B cells) and expressed as 2−CT according to Applied Biosystems’ User Bulletin 2: Rev. B: Relative Quantitation of Gene Expression.
Table 1.
Primer Sequence Sets
| Target Gene | Primer / Probe | Sequence (5'→3') | Length (mer) |
|---|---|---|---|
| NEP | Forward | CAGCCGAACCTACAAGGAGTC | 21 |
| Reverse | TGCAATCAAATCCTCGACCAC | 21 | |
| IL6 | Forward | CAATCTGGATTCAATGAGGAGAC | 23 |
| Reverse | CTCTGGCTTGTTCCTCACTACTC | 23 | |
| IL8 | Forward | GAACTGAGAGTGATTGAGAGTGGA | 24 |
| Reverse | CTCTTCAAAAACTTCTCCACAACC | 24 | |
| EGFR | Forward | GGAGAACTGCCAGAAACTGACC | 22 |
| Reverse | GCCTGCAGCACACTGGTTG | 19 | |
| PTGS2 | Forward | GGAACACAACAGAGTATGCG | 20 |
| Reverse | AAGGGGATGCCAGTGATAGA | 20 | |
| BCL2L11 | Forward | ATCCCCGCTTTTCATCTTTA | 20 |
| Reverse | AGGACTTGGGGTTTGTGTTG | 20 | |
| 18S rRNA | Forward | ACGGACAGGATTGACAGATT | 20 |
| Reverse | GCCAGAGTCTCGTTCGTTAT | 20 | |
| GAPDH | Forward | ATCCCTCCAAAATCAAGTGG | 20 |
| Reverse | CAGAGATGATGACCCTTTTGG | 20 |
AIM 3: TO DETERMINE THE MOLECULAR IMPACT OF DEP EXPOSURE AND DECREASED NEP EXPRESSION ON AIRWAY EPITHELIAL CELLS’ GENE EXPRESSION IN VITRO, USING A COMBINATION OF RNAi AND MICROARRAY APPROACHES
Experimental Design
A test based on a 2 × 3 factorial design (NEP siRNA-transfected cells vs. mock controls, and controls vs. low and high DEP exposure) was conducted. This involved NEP siRNA–transfected BEAS-2B cells or mock control cells being exposed to 0 (control), 10, and 40 µg/cm2 DEP (SRM 2975) for 24 hours. We monitored the cells visually for cell viability and collected them for microarray analysis. With this experimental design, we aimed to dissect complex transcriptional responses mediated by (1) DEP exposure (concentration–effects) and (2) NEP downregulation.
NEP siRNA Assay
We seeded BEAS-2B cells in a 6-well tissue culture plate, at 3000 cells per square centimeter in 2 mL antibiotic-free normal KGM medium. We incubated the cells at 37°C in a CO2 incubator until the cells were 60% to 80% confluent. The NEP-specific siRNA or a control siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) was transfected using Transfection Reagent (Santa Cruz Biotechnology) according to the manufacturer’s instructions. Briefly, for each 6-well transfection, 3.6 µL of 10-µM siRNA duplex was diluted into 40 µL siRNA Transfection Medium (Solution A), and 2.4 µL of siRNA Transfection Reagent was diluted into 40 µL siRNA Transfection Medium (Solution B). Solution A was added directly to Solution B using a pipette and then mixed and incubated for 20 minutes at room temperature. After incubation, we added 0.32 mL siRNA Transfection Medium to each tube containing the siRNA–siRNA Transfection Reagent complex and mixed gently. The mixture was overlaid onto the washed cells and incubated for 5 to 7 hours at 37°C in a CO2 incubator. Then, we added 0.4 mL of normal growth medium containing 2 times the normal serum and antibiotics concentration (2 × normal growth medium) and incubated the cells for an additional 24 hours. Equal amounts of total protein (20 µg) from cultured plates were separated on 12% poly-acrylamide gels and transferred to a nitrocellulose membrane. We performed Western blotting (described below), using mouse monoclonal antibodies to human NEP (SN5c) and [3-actin, as a control (both from Santa Cruz Biotechnology), to confirm the knockdown efficiency of NEP siRNA.
Western Blotting
We harvested BEAS-2B cells after no treatment, treatment with control siRNA, and treatment with NEP siRNA and washed them with PBS. Then the cells were lysed with 200 µL of Mammalian Protein Extraction Reagent (M-PER) with Halt Protease Inhibitor Cocktail (Pierce, Rockford, IL) according to the manufacturer’s protocol. We performed protein quantification using the Coomassie Plus Kit (Pierce). Samples were resolved using electrophoresis on a 4% to 20% sodium dodecyl sulfate polyacrylamide gel and transferred to nitrocellulose. We quantified the signal intensity of the blots using densitometry. We scanned the film on a GS-700 Imaging Densitometer (Bio-Rad) and quantified it using Bio-Rad Quantity One software, version 4.1.1. NEP protein expression levels were normalized to [3-actin.
Microarray Analysis
BEAS-2B cells were treated with 0 (control), 10, or 40 µg/cm2 DEP (SRM 2975), for 24 hours. We then isolated total RNAs from these cultured cells using the Qiagen RNeasy minikit (Valencia, CA) according to the manufacturer’s instructions. The isolated total RNAs were used to produce a labeled target, hybridized to Affymetrix human U133 Plus 2.0 GeneChips, and read using the Agilent/Affymetrix 2500A scanner (Santa Clara, CA) according to the manufacturer’s protocol. We analyzed the raw data (CEL files) using the GC-RMA algorithm as implemented in GeneSpring software, version 7.0 (Silicon Genetics, Redwood City, CA) to produce a normalized transcript-level signal for further analysis. We loaded transcript-level signal data from GeneSpring into BRB-ArrayTools (linus.nci.nih.gov/BRB-ArrayTools.html) and filtered them based on the variance of each transcript across samples. This method of filtering provided an unbiased selection of transcripts, independent of the tissue classes. We compared the variance of the log ratios for each transcript to the median of all transcript variances and selected transcripts whose variance was significant (P < 0.05).
Five steps were used to identify transcripts of interest as potential NEP-related pathways. First, an analysis of variance (ANOVA) was performed to identify transcripts with significantly (P < 0.05) altered expression compared with background controls. Second, our power analysis showed that the number of samples analyzed in each class could reliably detect 5-fold differences in expression. Thus, transcripts with less than a 10-fold difference in expression between DEP exposure and controls were further filtered out of those that had already been identified by ANOVA. Third, we compared the transcript differences (a) after doses of 0 (control), 10, or 40 µg/cm2 DEP; and (b) between NEP siRNA–transfected and mock control cells. These analyses not only identified transcriptional responses mediated by NEP and DEP exposure, but also ruled out nonspecific responses to siRNA expression (using NEP siRNA–transfected cells vs. mock controls without DEP exposure). Fourth, we analyzed the data with the GeneSpring GX 9 software package, including unbiased cluster analyses (hierarchical clustering, k-means clustering, and self-organizing map). For each cluster, we analyzed the expression patterns and assigned the major functional categories using gene ontology (www.geneontology.org). Data on individual expression changes were sorted into potential biologic pathways using GenMAPP (www.genmapp.org), a recently developed tool for visualizing expression data in the context of biologic pathways. Last, we confirmed the expression of some key genes by using RT-PCR as stated earlier.
AIM 4: TO EVALUATE THE EFFECTS ON NEP ACTIVITY OF HUMAN EXPOSURE TO DEE
Experimental Design
This phase of the current study was approved by the University of Arizona (UA) Institutional Review Board. The study methods are described in greater detail in a previous publication (Burgess et al. 2007). Informed consent was obtained from all subjects volunteering to participate in the study. Mining students undergoing undergraduate and graduate training in mining engineering at UA were eligible for participation if they were 18 years or older. Students who were current or previous smokers, had existing lung diseases, and/or were taking inhaled steroids were excluded. None of the subjects were current asthmatics, and no subjects reported taking anti-inflammatory medications. Eleven subjects — 10 males and 1 female, ranging in age from 19 to 33 years (mean 23.7 ± 4.3 yr)—completed the study (Table 2). Seven (64%) of the subjects described themselves as White, 2 (18%) as Hispanic, and 1 each as Asian and other. At baseline and before the exposure experiments, none of the subjects reported having had a cold, flu, allergies, or any respiratory symptoms within the previous 6 days. These subjects had no record of having been exposed to very high levels of emissions exhaust, room dust, drilling mist, or other particulate sources within 1 month before baseline or exposure.
Table 2.
Characteristics of Subjects and Exposure Conditions
| Exposure Conditions | |||||
|---|---|---|---|---|---|
| Subject Identification |
Age (years) |
Sex | Racea | DEE (EC, mg/m3) |
Time (min) |
| 01 | 26 | M | 1 | 1.20 | 60 |
| 02 | 33 | M | 4 | 0.64 | 66 |
| 03 | 19 | M | 1 | 1.80 | 56 |
| 04 | 21 | M | 1 | 0.75 | 134 |
| 05 | 22 | M | 1 | 0.50 | 134 |
| 06 | 23 | M | 1 | 0.38 | 119 |
| 07 | 21 | F | 1 | 0.32 | 85 |
| 08 | 19 | M | 1 | 0.12 | 85 |
| 09 | 30 | M | 2 | 0.38 | 93 |
| 10 | 26 | M | 3 | 0.15 | 66 |
| 11 | 22 | M | 2 | 0.09 | 90 |
Race codes: 1, White; 2, Hispanic; 3, Asian; 4, Other.
The study was carried out at the San Xavier Underground Mining Laboratory, a research and training facility devoted to occupational health and safety in the mining and underground construction industries, operated under the auspices of the UA College of Engineering in collaboration with the UA College of Public Health. Mine access and ventilation are designed to simulate underground conditions found in an actual production facility. Respiratory protection was not worn during the exposures. Changes of cell numbers by type, protein, and NEP activity in the collected sputum of subjects were evaluated as the difference between baseline and after-exposure values.
Exposure Conditions
DEE exposure was characterized from a diesel-powered 1984 Jarvis Clark JS-220 load-haul-dump (LHD) vehicle (Mackwood Group, Elliot Lake, Ontario, Canada) with a 2-cubic-yard bucket and an 82-hp Deutz F6L-912W diesel engine fitted with a catalytic converter. Experiments using the LHD were conducted in a conventional 4 m × 4 m tunnel decline, and a 15-hp axial auxiliary fan provided ventilation during exposure. During baseline and the DEE exposure, two samples collected over a period of 66 to 68 minutes demonstrated background particulate matter (PM) concentrations of less than 10 µg/m3. DEP was collected on precleaned 37-mm open-face quartz fiber filters (SKC, Eighty Four, PA) with Escort ELF personal sampling pumps (Mine Safety Appliances, Pittsburgh, PA) and analyzed for elemental carbon according to National Institute for Occupational Safety and Health method 5040 by the Wisconsin State Laboratory of Hygiene (Madison, WI). Personal exposure to DEE, as measured by elemental carbon (N = 11), averaged 575 ± 512 µg/m3 (range, 91– 1800) (Table 2). Exposure times averaged 90 minutes (range, 56–134). NO2 and carbon monoxide (CO) concentrations were assessed with an MSA multi-gas detector (Mine Safety Appliances) during a single mucking shift (without concomitant rock removal operations) using the diesel-powered LHD. For a single experimental shift monitored for 60 minutes, peak concentrations for NO2 and CO were 1.5 ppm and 22 ppm, respectively. All subjects underwent two evaluations: a baseline evaluation on a nonexposure day and an evaluation after DEE exposure. These exposures were at least 1 week apart. Sputum induction, a health history, and an exposure questionnaire were completed on nonexposure days. On exposure days, groups of 1 to 3 subjects first completed an interim health history and then were exposed for a 1- to 2-hour period, depending on their individual class schedules. One hour after cessation of exposure, the subjects completed sputum induction. A 1-hour postexposure test time was chosen in order to take into account the timelines of both the acute response of the subjects’ airways and any possible change in soluble NEP activity.
Sputum Induction and Exposure
Induced sputum (based on methods in Djukanović et al. 2002) was collected using DeVilbiss Ultra-Neb 99HD ultra-sonic nebulizers (Somerset, PA) filled with 3% saline and set on maximum output. Sputum samples were diluted with 10% Sputolysin (Calbiochem, San Diego, CA) in PBS with penicillin–streptomycin and 0.5% bovine serum albumin (BSA). Supernatant was removed by centrifugation and frozen at −80°C for later analysis of NEP activity. The cellular pellet was reconstituted in 1 mL PBS in order to perform total cell counts with the use of a hemocytometer and trypan blue stain (Sigma Chemical Co.). A portion of the cell pellet was cytocentrifuged using a Shandon Cytospin (Thermo-Shandon, Pittsburgh, PA) onto a microscope slide and stained with Diff-Quik for cell number analysis. The protein concentration was determined using a Coomassie Plus Protein Assay (Pierce) with BSA as the standard.
Enzyme Activity Measurement
We measured cell-free NEP activity in sputum spectrophotometrically by a coupled assay, as described previously (Wong et al. 2004). Briefly, 5 µL of cell-free extract was incubated with 1 mM succinyl-Ala-Ala-Phe-p-nitroanilide (Suc-Ala-Ala-Phe-pNA) (Bachem Bioscience, King of Prussia, PA) as a substrate in 0.1 M of Tris-HCl (pH, 7.6) and 1 µL (0.14 units/µL) of porcine kidney aminopeptidase N (Sigma). We performed the reaction in duplicate in a 96-well microtiter plate. In this coupled activity assay, NEP cleaves Suc-Ala-Ala-Phe-pNA between Ala and Phe, yielding Phe-pNA. Aminopeptidase N (APN) then cleaves Phe-pNA, generating pNA as the final product. We determined the increase in specific absorbance (the accumulation of free pNA) at 405 nm using a plate reader (BioTek Instruments) after a 30-minute incubation at 37°C. We ran cell-free (substrate alone or substrate with APN) and substrate-free blanks in parallel. We determined the protein concentration by using a Coomassie Plus Protein Assay (Pierce) with BSA as the standard.
STATISTICAL METHODS AND DATA ANALYSES
We double-entered the laboratory and clinical data into a database and checked for miscoding of variables. Initially, standard descriptive statistics were run on the data to evaluate distributions, determine any needed transformations, and assess potential outliers or discrepancies in the data. First, we tested the data for homogeneity of variance using the Bartlett test and normalized them as appropriate, following a Gaussian distribution. Depending on the experimental designs and the size of the sample number (N), we used either paired or unpaired Student t tests or ANOVA for comparisons of mean concentrations between groups. Data were expressed as mean ± standard error of the mean (SEM), and we considered P < 0.05 to indicate significance. We performed statistical analyses using SPSS, version 17 (Chicago, IL). Duane L. Sherrill, professor of biostatistics and Associate Dean for Research at the University of Arizona College of Public Health, assisted us in all of the statistical analyses. The detailed statistical methods are specifically described for each aim as follows.
Aim 1
This aim examined the difference in DEP-induced inflammatory injury between Nep-null and wild-type mice, as measured by cell profiles and cytokines in BAL fluid, and epithelial proliferation. For this purpose, we normalized the data using log10 because they did not follow a Gaussian distribution. We used a factorial ANOVA for multiple comparisons of means in a 2 × 3 factorial design for each transformed measurement. Since the measures are independent variables, we evaluated mean changes for all groups using post hoc linear contrasts with adjustment for multiple comparisons, which were made using Bonferroni-corrected significance levels. Additionally, we used ANOVA for normalized measure comparisons for mRNA and protein expression of NEP in wild-type mice, comparing groups of controls and low and high DEP exposure. We calculated Pearson correlation coefficients to evaluate the strength of the linear relations for all of the dose–effect measurements in Nep-null mice versus wild-type mice.
Aim 2
For this aim, we assessed the concentration-dependent downregulatory effects of NEP induced by DEP and its contents (cDEP and sDEP); or (2) another type of particle. For this purpose, we normalized the data using log10 because they did not follow a Gaussian distribution. We used one-way factorial ANOVA for multiple comparisons of means, and we evaluated all mean changes using post hoc linear contrasts, with adjustments for multiple comparisons using Bonferroni-corrected significance levels. We performed additional statistical analyses to evaluate the strength of the linear relations for all of the concentration–effect measurements in the DEP, sDEP, and cDEP groups. We calculated the Pearson correlation coefficients.
Aim 3
First, we assessed the effects of NEP gene knockdown by RNAi using one-way ANOVA. Then, we compared the difference in transcripts in a 2 × 3 factorial design including (1) control versus 10 or 40 µg/cm2 DEP in NEP siRNA-transfected cells; (2) control versus 10 or 40 µg/cm2 DEP in mock controls; and (3) NEP siRNA-transfected cells versus mock controls in 0, 10, and 40 µg/cm2 DEP. We performed one-way factorial ANOVA using GeneSpring GX 9 software. We used the global error model (multisample interpretation), based on replicates of the samples, to estimate the variability in gene expressions within — and between — sample measurements. Correction for multiple testing was performed using the Benjamini-Hochberg-Yekutieli False Discovery Rate, set to 0.05% (i.e., a baseline of 5% of the genes identified as significant being false positives). The Tukey post hoc test was used to evaluate where statistically significant differences lay between the sample classes. We performed a power analysis, as described in Dobbin and Simon (2005), for single-label microarrays. We calculated within-dose variance using all data. We used the 90th percentile as a sample size estimate valid for 90% of the transcripts measured. Alpha was set at 0.05, and the power was set to 0.90.
Aim 4
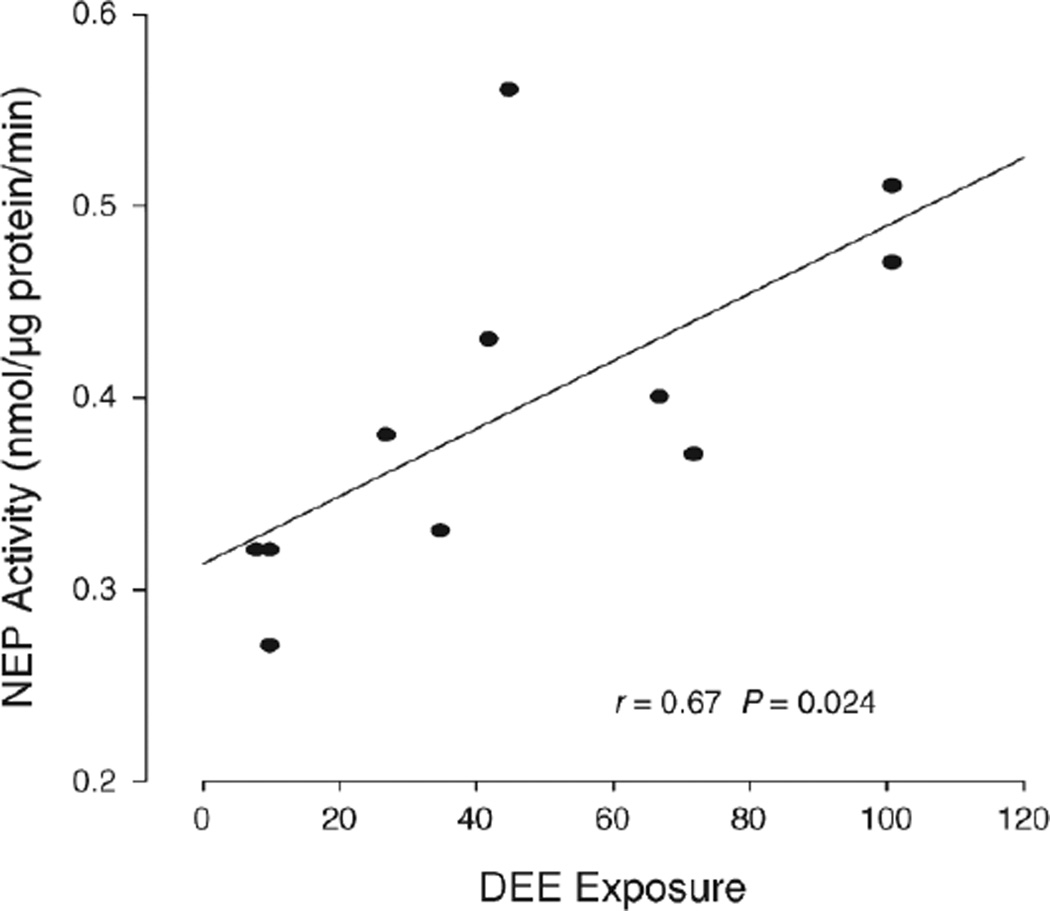
We assessed how DEE exposure affects NEP activity and cell numbers in sputum. For this purpose, we first normalized the data using log10 because they did not follow a Gaussian distribution. Then, we used the paired sample t tests to compare NEP activity between pre- and post-exposure sputum levels. We used one-way ANOVA for normalized measure comparisons of sputum levels of cells and total protein. Additionally, we calculated the Pearson correlation coefficients to identify linear relations between (1) NEP activity and the product of exposure concentration × exposure time; and (2) NEP activity and the number of total cells, macrophages, neutrophils, and epithelial cells in sputum.
RESULTS
AIM 1: TO EXAMINE THE ROLE OF NEP IN DEP-INDUCED INFLAMMATORY INJURY USING Nep-INTACT AND Nep-NULL MICE
Increased BAL Fluid Cell Numbers in Nep-Null Mice in Response to DEP
Total inflammatory cell numbers increased, depending on dose, in wild-type and Nep-null mice 7 days after DEP exposure (Table 3). After the low and high levels of DEP doses, the inflammatory cell numbers increased approximately 1.7- and 2.4-fold, respectively, in wild-type mice, and approximately 2.8- and 3.3-fold, respectively, in Nep-null mice. There were significant differences in total inflammatory cells between wild-type and Nep-null mice in response to both doses of DEP challenge. The changes in macrophage count followed the same trend as that of total inflammatory cells except in wild-type mice treated with the low DEP, where differences were not statistically significant. After the high dose of DEP, we observed significant increases in granulocytes and lymphocytes in Nep-null mice, but not in wild-type mice.
Table 3.
Cell Counts by Types (× 104 cells/mL) in BAL Fluid of Mice at 7 Days Following DEP Treatment
| 0 µg DEP | 10 µg DEP | 100 µg DEP | ||||
|---|---|---|---|---|---|---|
| Wild-type | Nep-null | Wild-type | Nep-null | Wild-type | Nep-null | |
| Inflammatory cells | ||||||
| Total cells | 41.17 ± 2.93 | 45.83 ± 3.66 | 70.29 ± 11.69a | 130.50 ± 17.09a,b,c | 100.00 ± 9.21a | 153.43 ± 10.61a,b,c |
| Macrophages | 37.86 ± 2.67 | 43.16 ± 3.57 | 65.43 ± 12.21 | 120.07 ± 18.47a,b,c | 88.90 ± 10.35a | 135.18 ± 5.6a,b,c |
| Granulocytes | 1.40 ± 0.33 | 1.21 ± 0.32 | 2.51 ± 0.44 | 4.80 ± 0.94 | 4.07 ± 0.67 | 6.24 ± 1.06b |
| Lymphocytes | 1.91 ± 0.22 | 1.47 ± 0.29 | 3.61 ± 0.87 | 5.63 ± 1.64 | 7.03 ± 1.79 | 10.09 ± 2.95b |
| Epithelial cells | 1.77 ± 0.65 | 1.95 ± 0.69 | 13.01 ± 2.34a | 19.50 ± 4.43a,b | 19.35 ± 3.98a | 37.71 ± 6.42a,b,c |
P < 0.05 compared to wild-type mice at 0 µg DEP exposure.
P < 0.05 compared to Nep-null mice at 0 µg DEP exposure.
P < 0.05 compared to wild-type mice given the same DEP dose.
Epithelial cell numbers in Nep-null mice treated with the high dose of DEP were significantly higher than those in wild-type mice treated with the high dose (Table 3).
Increased BAL Fluid Cytokine Response in Nep-Null Mice When Exposed to DEP
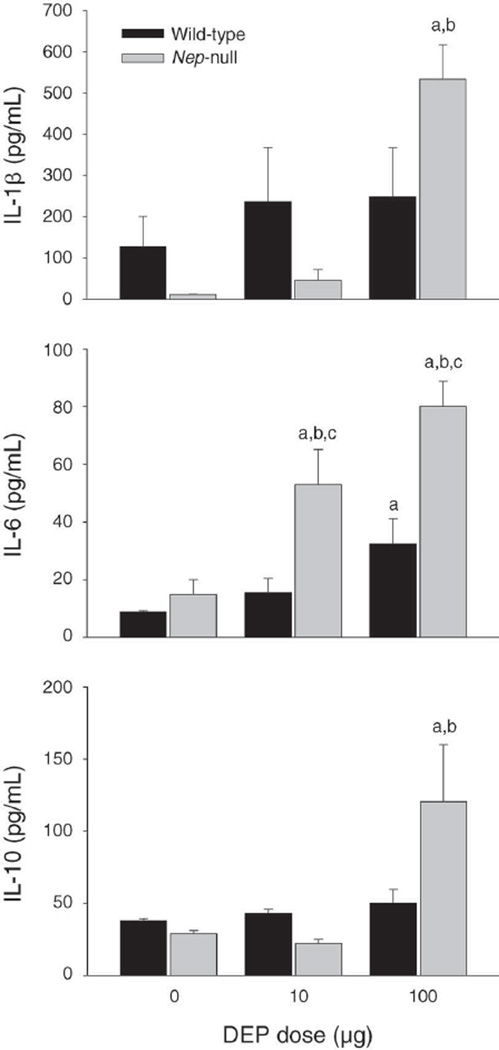
To determine whether the cytokine response induced by DEP was associated with NEP expression, we performed ELISA assays for IL-1[3, IL-6, and IL-10 with BAL fluid from wild-type and Nep-null mice 7 days after DEP exposure (Figure 1). In wild-type mice, the protein content of IL-6 was significantly elevated at the high dose, but not at the low dose, of DEP instillation. There were trends toward increased levels of IL-1[3 and IL-10 after both doses of DEP, but they were not statistically significant. In the Nep-null mice, the protein content for IL-6 was significantly elevated at both DEP dose levels and displayed a dose-dependent response to DEP. The levels of IL-1[3 and IL-10 proteins were significantly elevated at the high dose, but not at the low dose, in the Nep-null mice. Between wild-type and Nep-null mice, there was a statistically significant difference in IL-6 protein levels in response to both doses of DEP.
Figure 1. Differences in the amounts of cytokines IL-1(3, IL-6, and IL-10 in BAL fluid of C57BL/6 wild-type mice and Nep-null mice following DEP instillation.
Data expressed as mean ± SEM (N = 7). Letter “a” indicates significantly higher compared with the wild-type control group; “b” indicates significantly higher compared with Nep-null control group; and “c” indicates significant difference between wild-type and Nep-null mice with the same dose of DEP (P < 0.05). The DEP exposure was meant to approximate an accumulated dose of 7 days.
Downregulation of NEP in Wild-Type Mice by DEP
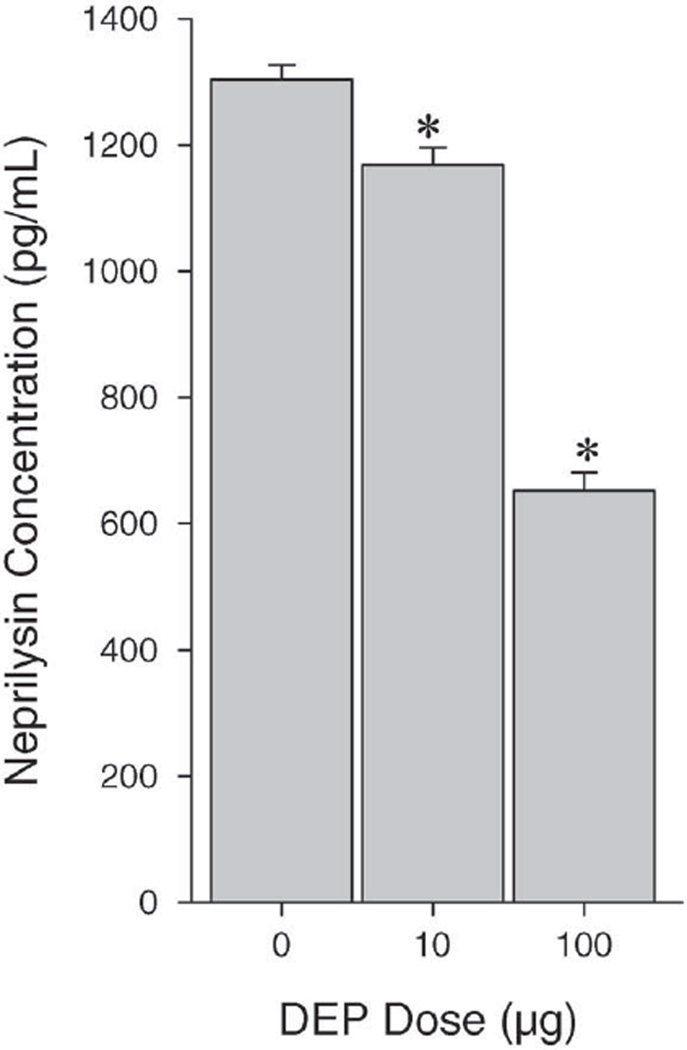
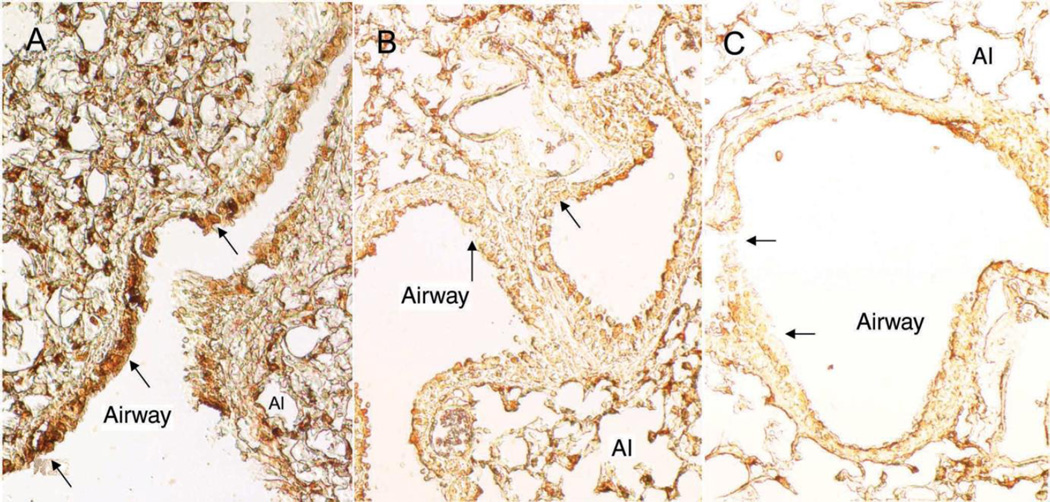
Our previous study (Wong et al. 2007 pilot study) showed that exposure of human airway epithelial cells to DEP (0– 40 µg/mL) for 24 hours downregulated NEP expression as well as its enzymatic activity in a concentration-dependent manner. We noted in that study that a substantial decrease (90%) in NEP mRNA expression occurred at 5 µg/mL DEP. To confirm these in vitro findings, we quantified the concentration of NEP protein in lung tissue in the current study. Exposure of wild-type mice to DEP significantly decreased NEP protein levels (Figure 2) in a dose-dependent manner. Both levels of DEP exposure resulted in significant reductions in NEP protein when compared with that of controls. The protein levels were reduced by 10.4% and 50.0% of the control at the low and high DEP doses, respectively, suggesting that the changes may be attributable to DEP exposure. Using immunohistochemistry, we observed NEP expression on epithelial cells, alveolar type II cells, and macrophages (Figure 3A). Low and high levels of DEP (Figures 3B and 3C, respectively) appeared to reduce NEP protein expression in the epithelial cells of small airways.
Figure 2. Reduced expression of NEP protein in lung tissue following DEP instillation.
Wild-type mice received 0.85% saline vehicle, or 10 or 100 µg DEP in saline. Lung tissue was harvested 7 days after instillation. Data expressed as mean ± SEM (N = 7). Asterisk (*) indicates statistically significantly higher compared with control group (P < 0.05).
Figure 3. Immunohistochemical micrographs of lung tissue showing localization and reduction of NEP protein expression after DEP instillation of 0 µg (control); (B) 10 µg; and (C) 100 µg.
NEP stain (brown color) was mostly detected in airway epithelial cells, alveolar type II cells (Al), and macrophages. NEP stain density appeared to be reduced in small airway epithelial cells (arrows) after exposure to DEP. Magnification: ×40.
Effects of DEP on Epithelial Proliferation in Wild-Type and Nep-Null Mice
We monitored the incorporation of labeled BrdU into airway epithelial cell DNA as a marker of epithelial proliferation. In wild-type mice, we observed no significant changes in airway epithelial proliferation after the high dose of DEP when compared with its control (2.67 ± 0.58 positive cells/102 cells vs. 2.00 ± 1.41 positive cells/102 cells; P = 0.976). Also, we did not observe any significant changes in airway epithelial proliferation in Nep-null mice treated with the high dose of DEP when compared with control (2.04 ± 1.00 positive cells/102 cells vs. 2.00 ± 0.71 positive cells/102 cells, P = 0.627). DEP exposure did not result in significant changes in epithelial proliferation between wild-type and Nep-null mice (P = 0.667). Either there was no detectable wound-repair process in the acute phase of epithelial inflammatory response to DEP exposure 7 days after exposure or the sample size was not large enough to detect the presumably mild changes resulting from the current exposure conditions.
AIM 2: TO EXAMINE WHICH COMPONENTS OF DEP ARE ASSOCIATED WITH NEP DOWNREGULATION IN VITRO
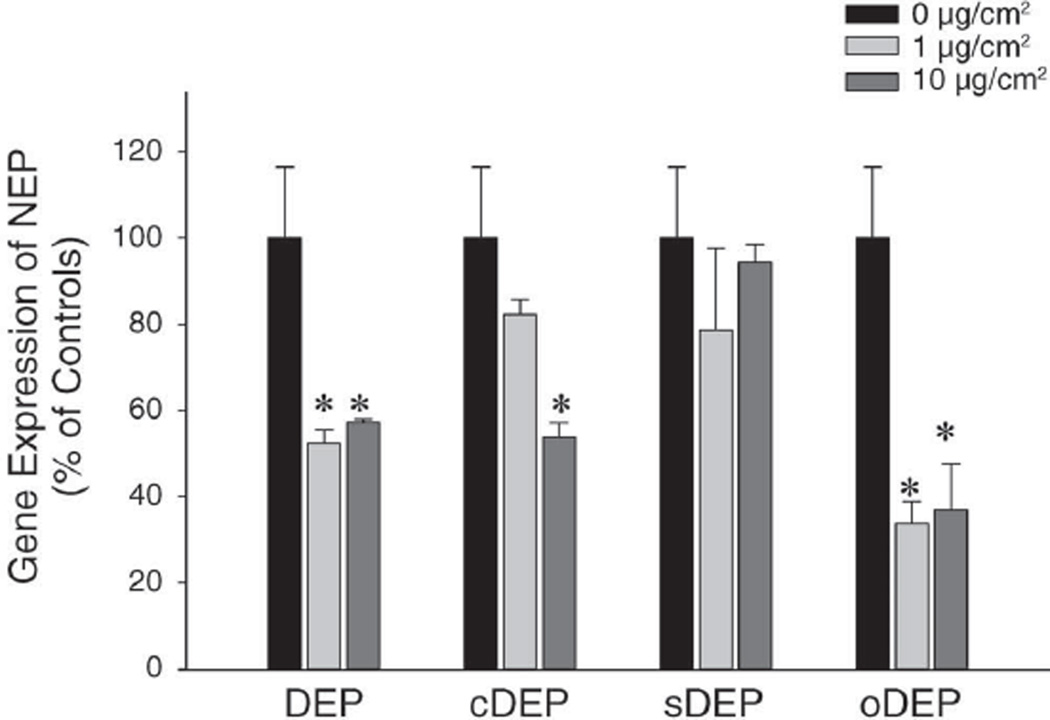
Our previous study indicated that NEP downregulation by DEP occurs at the transcriptional level (Wong et al. 2007 pilot study). To clarify the contribution of the major components of DEP in the downregulation of NEP, we compared the effects induced by (1) DEP; (2) cDEP; and (3) sDEP. We also examined another type of particle— oDEP, or standard urban dust (SRM 1649a) — to test whether NEP downregulation is a nonspecific response to PM. We conducted parallel cell cultures with the same concentration–response designs (using concentrations of 0, 1, and 10 µg/cm2) in the same period of time (24 hr). The changes in NEP were not associated with cell proliferation because the cells stopped growing in the culture wells before exposure to the different levels of DEP. The results showed that BEAS-2B cells cultured with DEP statistically significantly downregulated NEP mRNA expression at 1 µg/cm2 (50%) and 10 µg/cm2 (45%), as measured by RT-PCR (Figure 4). Cells treated with 1 and 10 µg/cm2 cDEP expressed approximately 80% and 55% NEP mRNA, respectively, of control cells. When compared with controls, the decrease in NEP mRNA expression was significant at the higher, but not the lower, level of cDEP, suggesting that cDEP has less effect than DEP due to removal of the divalent cations, particularly transition metals, by chelators. Moreover, cells treated with sDEP were not significantly affected at either concentration, indicating that DEP, after organic stripping, lost its ability to downregulate NEP mRNA expression. Collectively, these findings suggest that downregulation of NEP by DEP was mostly attributable to DEP-adsorbed organic compounds, whereas the carbonaceous core of NEP has little or no effect on NEP mRNA expression under the current experimental conditions.
Figure 4. Comparisons of downregulated NEP mRNA expression by DEP and its components in cultured BEAS-2B cells.
Cells were treated with (1) DEP (SRM 2975); (2) DEP without transition metals, using chelators (cDEP); (3) carbonaceous core, represented by stripped DEP (sDEP); and (4) another type of particle, oDEP, or standard urban dust (SRM 1649a), at 0, 1, and 10 µg/cm2 for 24 hours. The mRNA levels were determined by RT-PCR and normalized to endogenous references (GAPDH and 18S rRNA). Values represent the means ± SEM (% of controls) of 3 independent experiments (*P < 0.05).
NEP mRNA expression in cells treated with oDEP followed the same pattern as that of DEP (Figure 4). This suggests that the observed NEP downregulation is a nonspecific response to PM, because the two kinds of particles (DEP and standard urban dust) have different chemical characteristics (Table 4). SRM 1649a contains additional trace metals, pesticides, and a number of components from multiple sources of air pollutants in addition to DEP.
Table 4.
The Key Characteristics of DEP and Standard Urban Dust
| SRM 2975 | SRM 1649a | |
|---|---|---|
| Sources | DEP (from industrial forklift) | Urban dust |
| Particle diameter (µm) | 31.9 | 20.8 |
| Particle distribution (µm) | ||
| 90% | 70 | 48.6 |
| 10% | 5.3 | 2.2 |
| Elemental carbon | +++a | + |
| Trace metals | + | +++ |
| Pesticides | [not available] | Heptachlor, levoglucosan, dibenzo-p-dioxin, dibenzofuran congeners |
| Selected PAHsb | ||
| Phenanthrene | 17.0 ± 2.8 | 4.14 ± 0.37 |
| Anthracene | 0.038 ± 0.008c | 0.432 ± 0.082 |
| Fluoranthene | 26.6 ± 5.1 | 6.45 ± 0.18 |
| Pyrene | 0.90 ± 0.24 | 5.29 ± 0.25 |
| Benz[a]anthracene | 0.317 ± 0.066 | 2.208 ± 0.073 |
| Chrysene | 4.56 ± 0.16 | 3.049 ± 0.060 |
| Triphenylene | 5.22 ± 0.20 | 1.357 ± 0.054 |
| Benzo[b]fluoranthene | 11.5 ± 3.6c | 6.45 ± 0.64 |
| Benzo[k]fluoranthene | 0.678 ± 0.076 | 1.913 ± 0.031 |
| Benzo[a]fluoranthene | 0.06 ± 0.02c | 0.409 ± 0.035 |
| Benzo[e]pyrene | 1.11 ± 0.10 | 3.09 ± 0.19 |
| Benzo[a]pyrene | 0.0522 ± 0.0053 | 2.509 ± 0.087 |
| Perylene | 0.054 ± 0.009c | 0.646 ± 0.075 |
| Anthanthrene | 0.038 ± 0.008c | 0.450 ± 0.067 |
| Benzo[g,h,i]perylene | 0.498 ± 0.044 | 4.01 ± 0.91 |
| Indeno[1,2,3-cd]pyrene | 1.4 ± 0.2c | 3.18 ± 0.72 |
| Dibenz[a,j]anthracene | 0.37 ± 0.07c | 0.310 ± 0.034 |
| Dibenz[a,c/a,h]anthracene | 0.52 ± 0.08c | 0.488 ± 0.025 |
| Pentaphene | 0.038 ± 0.007c | 0.151 ± 0.035 |
| Benzo[b]chrysene | 0.08 ± 0.03c | 0.315 ± 0.013 |
| Picene | 1.0 ± 0.2c | 0.426 ± 0.022 |
| Dibenzo[b,k]fluoranthene | 2.7 | 0.724 ± 0.076 |
| Dibenzo[a,e]pyrene | 0.57 | 0.565 ± 0.060 |
Plus signs (+) indicate levels relative to each other.
Values are mass fractions (expressed in mg/kg) of a mean of means from two or more analytical methods ± expanded uncertainty.
Referenced concentrations for selected PAHs.
AIM 3: TO DETERMINE THE MOLECULAR IMPACT OF DEP EXPOSURE AND DECREASED NEP EXPRESSION ON AIRWAY EPITHELIAL CELLS’ GENE EXPRESSION IN VITRO, USING A COMBINATION OF RNAi AND MICROARRAY APPROACHES
In Vitro Model of NEP Knockdown
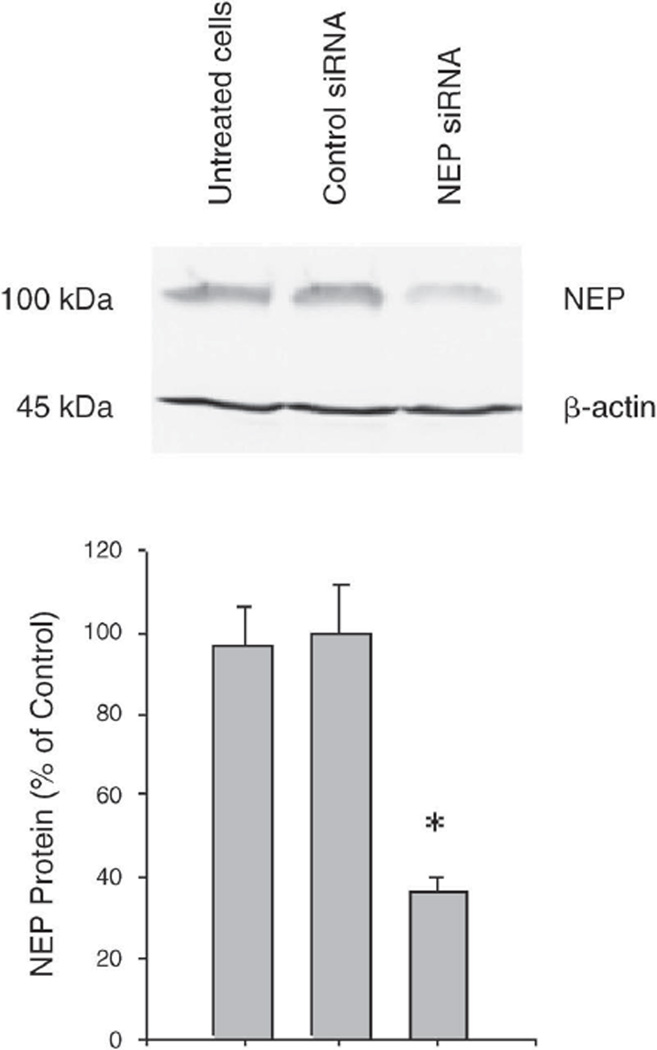
To determine the impact of NEP downregulation due to DEP exposure, we knocked down NEP gene expression in BEAS-2B cells using the RNAi technique. The controls for our experiments were untreated (uninfected) cells and cells infected with a vector carrying siRNA against lacZ (mock), which has little or no homology to any human gene. The effects of NEP siRNA transfection, compared with controls, were measured 30 hours later by Western blot analysis of NEP protein expression with anti-NEP antibody. We used [3-actin levels as internal controls. The results of densitometric quantification of NEP/[3-actin ratios are shown in Figure 5. Efficient reduction (63.5% when compared with mock control) of protein levels was confirmed. There was no difference between untreated and mock transfected controls.
Figure 5. NEP transcript knockdown in BEAS-2B cells with NEP-specific siRNA.
Cells (at 3000 cells/cm2) were transfected with control siRNA (middle column) or NEP siRNA (right column). Cells were harvested 30 hours post-transfection, and the effect of siRNA transfection was determined by Western blotting. NEP protein expression levels were normalized to [3-actin (*P < 0.05).
Gene Expression Profile
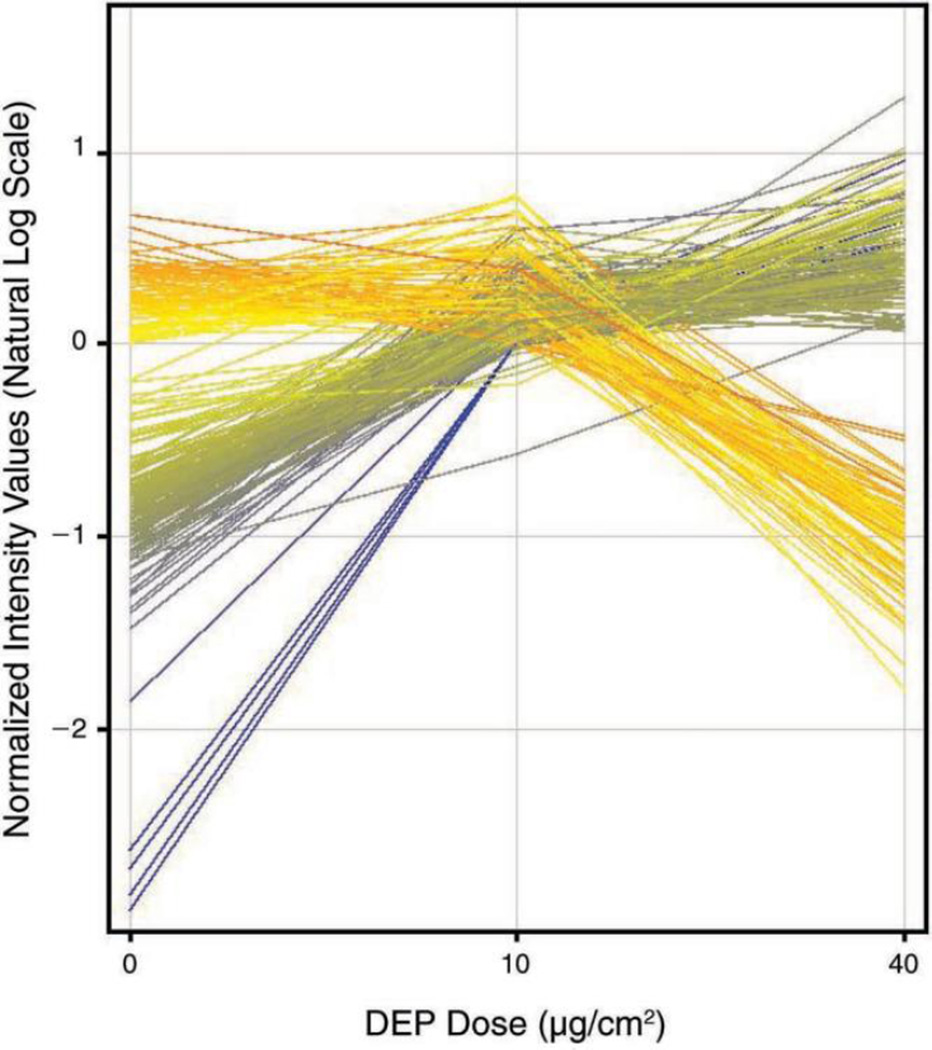
Twenty-four hours after transfection with siRNA, we treated the NEP-knockdown cells and mock controls with zero, low, and high concentrations of DEP for an additional 24 hours for the microarray study. Figure 6 illustrates that hybridization signal intensities varied statistically significantly as a function of NEP knockdown, DEP exposure, and a combination of both. Neither NEP siRNA transfection nor DEP exposure induced statistically significant changes in cell viability. Labeled complementary RNA (cRNA) from mock or NEP siRNA–transfected cells after incubation with 0, 10, and 40 µg/cm2 DEP was evaluated on two different Affymetrix U133 Plus human genome GeneChips. Overall, about 2158 probe sets, out of a total of 54,675, were detected above background and shown to be statistically different (P < 0.01) in the NEP siRNA experimental group when compared with the mock controls.
Figure 6. Hybridization signal intensities, which indicate the changes in gene expression in both mock-control and NEP siRNA-transfected cells after DEP exposure.
Intensities varied statistically significantly as a function of NEP knockdown and DEP exposure. Labeled cRNA from mock or NEP siRNA-transfected cells was tested using two different Affymetrix U133 Plus human genome GeneChips, following 0, 10, 40 µg/cm2 DEP incubation. About 2158 probe sets, out of a total of 54,675, were detected above background and shown to be statistically different (P < 0.01) in the NEP siRNA experimental group when compared with mock controls.
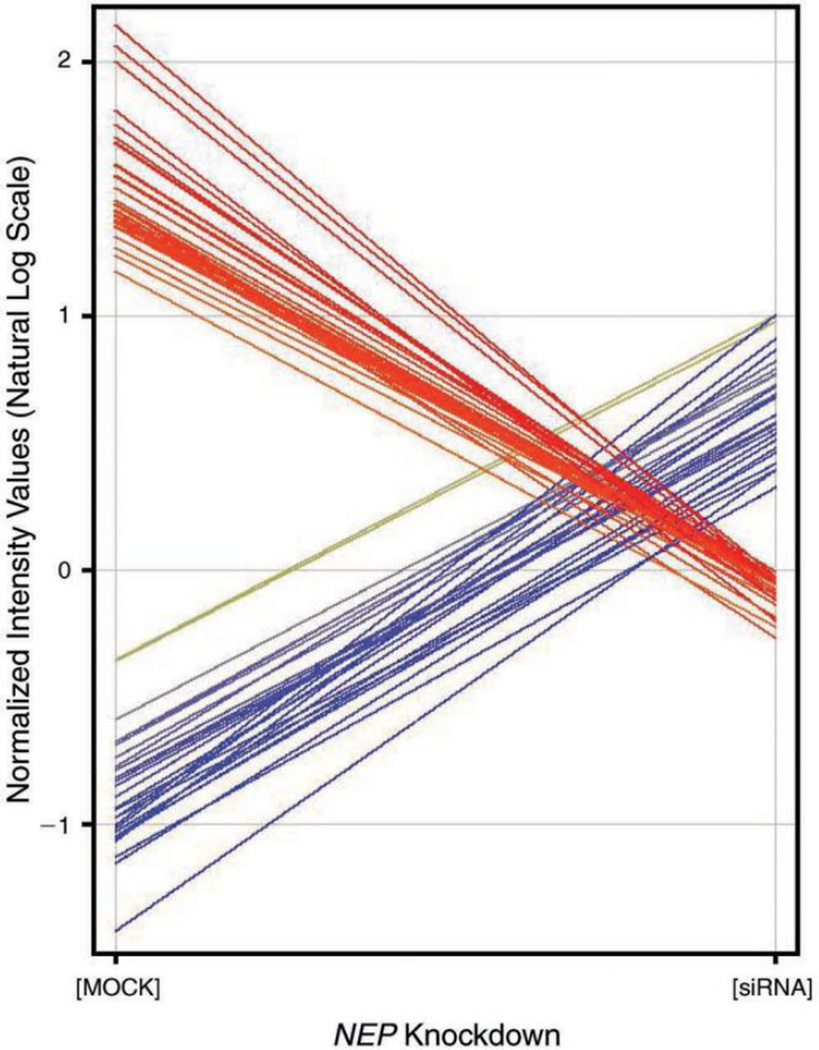
Genes Associated with NEP Knockdown
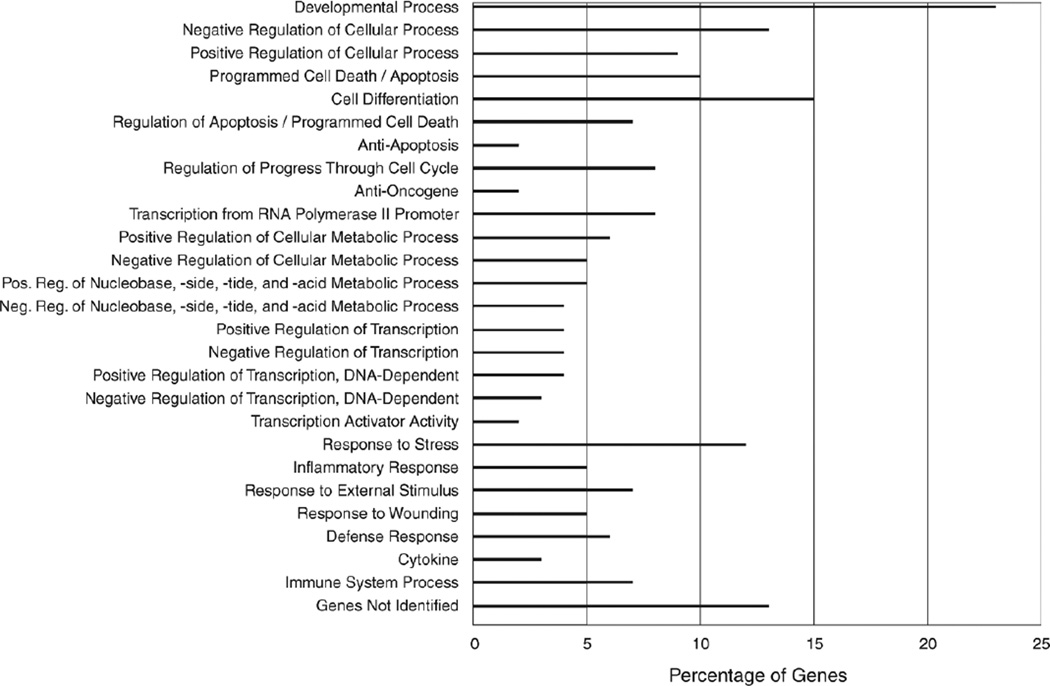
In order to identify genes that were specifically upregulated or downregulated and associated with decreased NEP expression in BEAS-2B cells, we employed hierarchical clustering (a functional annotation tool that is part of the GeneSpring GX 9 software, which measures the relations among the annotation terms based on the degree of co-association of genes into the group) in order to evaluate the 121 genes identified in the statistical analysis. Table 5 lists 31 genes with a high fold change when compared with mock controls; the normalized (to MOCKp0) fluorescent-spot intensity values are shown in Figure 7. Initial analysis suggests that the depletion of NEP expression (by siRNA) in epithelial cells induced upregulation (17 genes) or downregulation (14 genes) of genes involved in DNA/protein binding, calcium channel activities, and the intracellular signaling cascade of cytokines. The identified genes included IL6, IL8, and epidermal growth factor receptor (EGFR). The depletion of NEP expression also disturbed the transcription of genes involved in cellular monooxygenase, guanosine triphosphatase (GTPase), and protein kinase activities. These data suggest that NEP is involved in a complex molecular cascade in different subcellular compartments, possibly influencing normal cellular functions and inflammatory conditions.
Table 5.
Up- and Downregulated Genes in NEP-Knockdown Human Airway Epithelial Cells
| Probe Set ID | Unigene IDa |
Gene Symbol |
Gene Title | Fold (log) ([siRNA]/[MOCK]) |
|---|---|---|---|---|
| 231578_at | Hs.62661 | GBP1 | Guanylate binding protein 1 | 1.27 |
| 1554997_a_at | Hs.196384 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1.28 |
| 204897_at | Hs.199248 | PTGER4 | Prostaglandin e receptor 4 (subtype ep4) | 1.81 |
| 202393_s_at | Hs.435001 | KLF10 | Kruppel-like factor 10 | 1.48 |
| 219995_s_at | Hs.653124 | ZNF750 | Zinc finger protein 750 | 1.44 |
| 229450_at | Hs.47338 | IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | 1.52 |
| 211506_s_at | Hs.624 | IL8 | Interleukin 8 | 1.89 |
| 205207_at | Hs.654458 | IL6 | Interleukin 6 | 1.73 |
| 201565_s_at | Hs.180919 | ID2 | Inhibitor of dna binding 2 | 1.43 |
| 202672_s_at | Hs.460 | ATF3 | Activating transcription factor 3 | 1.64 |
| 202241_at | Hs.444947 | TRIB1 | Tribbles homolog 1 (drosophila) | 1.75 |
| 238688_at | Hs.133892 | TPM1 | Tropomyosin 1 (alpha) | 1.66 |
| 1555372_at | Hs.469658 | BCL2L11 | Bcl2-like 11 (apoptosis facilitator) | 1.67 |
| 222662_at | Hs.458513 | PPP1R3B | Protein phosphatase 1 | 1.74 |
| 222162_s_at | Hs.643357 | ADAMTS1 | Adam metallopeptidase with thrombospondin type 1 motif | 2.12 |
| 234608_at | Hs.436367 | LAMA3 | Laminin 3 | 2.27 |
| 206115_at | Hs.534313 | EGR3 | Early growth response 3 | 3.81 |
| 238177_at | Hs.585128 | SLC6A19 | Solute carrier family 6 (neutral amino acid transporter) | −1.09 |
| 1558814_s_at | Hs.482873 | TMED5 | Transmembrane emp24 protein transport domain containing 5 | −1.04 |
| 1565484_x_at | Hs.488293 | EGFR | Epidermal growth factor receptor | −1.06 |
| 1560878_at | Hs.696346 | SYT15 | Synaptotagmin xv | −1.03 |
| 236033_at | Hs.56281 | ASB12 | Ankyrin repeat and socs box-containing 12 | −1.02 |
| 1565537_at | Hs.526396 | NKX1-1 | Nk1 homeobox 1 | −1.04 |
| 228691_at | Hs.505202 | BICD1 | Bicaudal d homolog 1 (drosophila) | −1.04 |
| 1553657_at | Hs.10697 | VWA3A | von Willebrand factor A domain containing 3A | −1.03 |
| 243534_at | Hs.652240 | CC2D2B | Coiled-coil and C2 domain containing 2B | −1.05 |
| 1565483_at | Hs.488293 | EGFR | Epidermal growth factor receptor | −1.03 |
| 1554400_at | Hs.584808 | TCTE3 | T-complex-associated-testis-expressed 3 | −1.03 |
| 220421_at | Hs.189109 | BTNL8 | Butyrophilin-like 8 | −1.06 |
| 1568513_x_at | Hs.654496 | IL23A | Interleukin 23 | −1.03 |
| 214414_x_at | Hs.654744 | HBA2 | Hemoglobin | −1.01 |
Per Avadis software platform (Strand Life Sciences, San Francisco, CA).
Figure 7. The changes in fluorescent-spot intensity values between NEP knockdown and controls.
NEP knockdown resulted in 17 upregulated and 14 downregulated genes in in vitro cultures of human airway epithelial BEAS-2B cells.
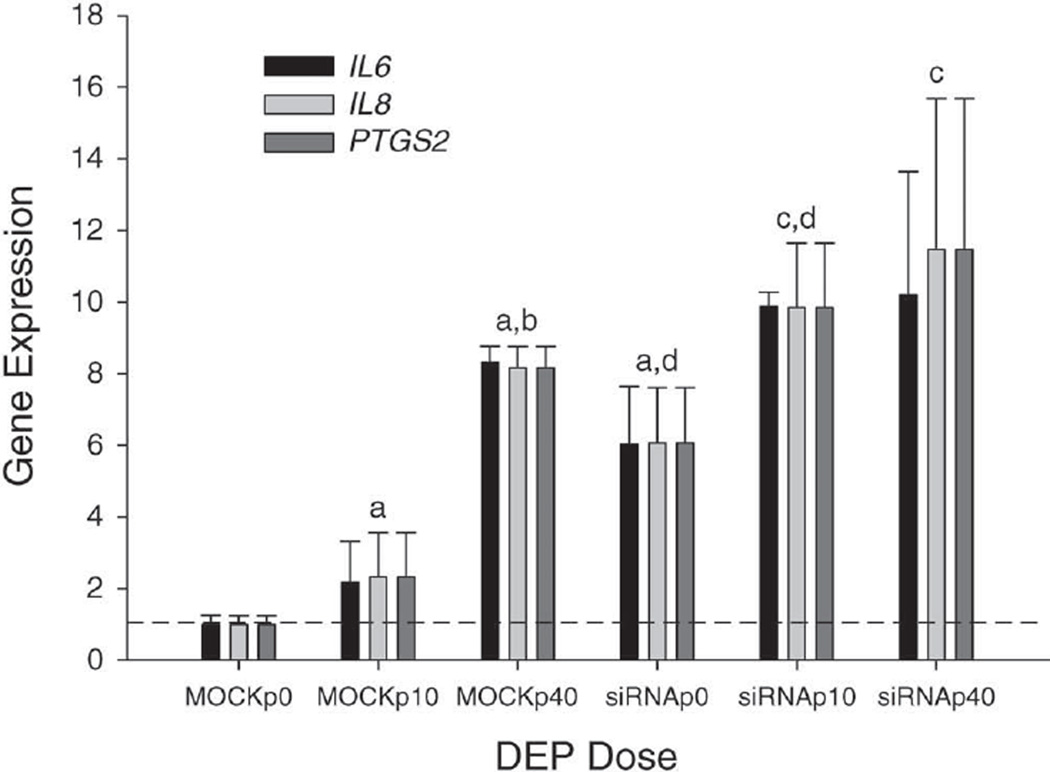
Using hierarchical clustering, we consistently observed five genes in the top three ranking pathways delineated by the GeneSpring GX 9 software — “The Role of Cytokines in Mediating Communication between Immune Cells,” “IL-17 Signaling,” and “Colorectal Cancer Metastasis Signaling” — and in the top twelve pathways, including “IL-8 Signaling,” “IL-6 Signaling,” and “HGF Signaling.” These five genes (IL6, IL8, EGFR, prostaglandin-endoperoxide synthase 2 [PTGS2], and BCL2L11) were chosen due to their known association with inflammation. Increased expression of IL-6, IL-8, and PTGS2 was confirmed by RT-PCR to be associated with NEP knockdown (Figure 8).
Figure 8. Confirmation of selected upregulated genes (normalized to MOCKp0) either by DEP exposure (0, 10, or 40 µg/cm2) or by NEP knockdown using RT-PCR.
Total RNAs isolated from BEAS-2B cells were the same as those of the microarray analyses. Comparisons are as follows: a and b indicate significantly higher compared with the MOCKp0 and MOCKp10 groups, respectively; c indicates significantly higher compared with the siRNAp0 group; and d indicates a significant difference between siRNA and MOCK groups with the same level of DEP (P < 0.05).
RT-PCR assays for IL-6, IL-8, and PTGS2 were performed with cDNAs generated from the microarray study (Figure 8). A comparison analysis showed that mRNA expression of IL6, IL8, and PTGS2 in the siRNAp0 group was approximately 6.1-fold greater than that of the MOCKp0 group, suggesting the knockdown of NEP significantly enhanced the expression of these genes. However, the degree of increased expression of the measured genes became less with DEP exposures of 10 µg/cm2 (4.8-fold, siRNAp10 vs. MOCKp10) and 40 µg/cm2 (1.3-fold, siRNAp40 vs. MOCKp40), suggesting that DEP exposure covers the effect of NEP knockdown on gene expression. In both siRNA and MOCK groups, moreover, the expression of IL6, IL8, and PTGS2 mRNA was significantly elevated in a dose-dependent manner in response to DEP exposure. Thus these data further confirmed the findings observed in the microarray and in vivo studies.
Genes Associated with DEP Exposure
Tables 6 and 7 represent the up- and downregulated genes, respectively, after exposure of BEAS-2B cells to DEP. The normalized fluorescent-spot intensity values are shown in Figure 9. There were 151 upregulated and 59 downregulated genes with in vitro exposure to DEP. Of the up- or downregulated genes, 89.5% and 46.6%, respectively, were changed in a DEP concentration-dependent manner (data not shown). The results showed that DEP incubation increased the mRNA levels of metabolic enzymes cytochrome P450 CYP1A1 and CYP1B1, cytokines IL-6 and IL-8, PTGS2, and chemokine (C-X-C motif) ligand 1 and 2 (CCL1 and CCL2), death inducer-obliterator 1 (DIDO1), heat shock 70kDa protein 1B (HSPA1B), aquaporin 3 (AQP3), early growth response 2 and 3 (EGR2 and EGR3), among others. In contrast, DEP decreased the expression of many enzymes in BEAS-2B cells including phosphoinositide-3-kinase (PIK3C2A), phosphoribosylglycinamide formyl-transferase (GART), and threonyl-tRNA synthetase (TARS), indicating that DEP may potentially influence a multitude of biologic processes.
Table 6.
Gene Upregulation After DEP Exposure in Cultured Human Airway Epithelial Cells
| Probe Set ID | Unigene IDa |
Gene Symbol |
Gene Title | Fold (log)b | |
|---|---|---|---|---|---|
| 10 µg/cm2 | 40 µg/cm2 | ||||
| 202437_s_at | Hs.154654 | CYP1B1 | Cytochrome P450 | 1.00 | 1.22 |
| 202435_s_at | Hs.154654 | CYP1B1 | Cytochrome P450 | 1.00 | 1.19 |
| 205749_at | Hs.72912 | CYP1A1 | Cytochrome P450 | 1.00 | 1.29 |
| 202436_s_at | Hs.154654 | CYP1B1 | Cytochrome P450 | 1.00 | 1.21 |
| 202393_s_at | Hs.435001 | KLF10 | Kruppel-like factor 10 | 1.05 | 1.51 |
| 208937_s_at | Hs.504609 | ID1 | Inhibitor of DNA binding 1 | 1.08 | 1.47 |
| 208892_s_at | Hs.298654 | DUSP6 | Dual specificity phosphatase 6 | 1.15 | 1.29 |
| 202241_at | Hs.444947 | TRIB1 | Tribbles homolog 1 (Drosophila) | 1.19 | 1.56 |
| 201631_s_at | Hs.591785 | IER3 | Immediate early response 3 | 1.13 | 1.30 |
| 222168_at | Hs.675520 | 10q21 mRNA sequence | 1.46 | 1.59 | |
| 208891_at | Hs.298654 | DUSP6 | Dual specificity phosphatase 6 | 1.18 | 1.41 |
| 225133_at | Hs.298658 | KLF3 | Kruppel-like factor 3 (basic) | 1.25 | 1.38 |
| 234608_at | Hs.436367 | LAMA3 | Laminin | 1.36 | 1.85 |
| 202859_x_at | Hs.624 | IL8 | Interleukin 8 | 1.12 | 1.42 |
| 233413_at | Hs.210390 | CDNA FLJ13457 fis | 1.06 | 1.38 | |
| 231578_at | Hs.62661 | GBP1 | Guanylate binding protein 1 | 0.48 | 1.13 |
| 213848_at | Hs.595184 | MRNA; cDNA DKFZp586F2224 | 1.09 | 1.27 | |
| 227335_at | Hs.517172 | DIDO1 | Death inducer-obliterator 1 | 1.00 | 1.73 |
| 238909_at | Hs.143873 | S100A10 | S100 calcium binding protein A10 | 1.00 | 1.29 |
| 235803_at | Hs.658293 | Transcribed locus | 1.22 | 1.40 | |
| 208893_s_at | Hs.298654 | DUSP6 | Dual specificity phosphatase 6 | 1.24 | 1.32 |
| 232947_at | Hs.661499 | CDNA FLJ11476 fis | 1.05 | 1.49 | |
| 202768_at | Hs.590958 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 0.98 | 2.20 |
| 202581_at | Hs.274402 | HSPA1B | Heat shock 70kDa protein 1B | 1.00 | 1.58 |
| 228280_at | Hs.512833 | ZC3HAV1L | Zinc finger CCCH-type | 1.21 | 1.43 |
| 203394_s_at | Hs.250666 | HES1 | Hairy and enhancer of split 1 | 0.99 | 1.50 |
| 213680_at | KRT6B | Keratin 6B | 1.12 | 1.57 | |
| 204014_at | Hs.417962 | DUSP4 | Dual specificity phosphatase 4 | 1.44 | 1.35 |
| 232164_s_at | Hs.200412 | EPPK1 | Epiplakin 1 | 1.38 | 1.46 |
| 212665_at | Hs.12813 | TIPARP | TCDD-inducible poly(ADP-ribose) polymerase | 1.21 | 1.19 |
| 214696_at | Hs.597755 | C17orf91 | Chromosome 17 open reading frame 91 | 1.07 | 1.71 |
| 1558220_at | Hs.599259 | MUC20 | Mucin 20 | 1.12 | 1.32 |
| 213765_at | Hs.512842 | MFAP5 | Microfibrillar associated protein 5 | 1.23 | 1.22 |
| 205207_at | Hs.654458 | IL6 | Interleukin 6 (interferon) | 1.19 | 1.40 |
| 227280_s_at | Hs.471234 | CCNYL1 | Cyclin Y-like 1 | 1.22 | 1.33 |
| 212099_at | Hs.502876 | RHOB | Ras homolog gene family | 1.00 | 1.72 |
| 235629_at | Hs.658355 | Transcribed locus | 1.36 | 1.15 | |
| 200769_s_at | Hs.516157 | MAT2A | Methionine adenosyltransferase II | 1.00 | 1.35 |
| 227697_at | Hs.527973 | SOCS3 | Suppressor of cytokine signaling 3 | 1.29 | 1.10 |
| 227949_at | Hs.473218 | PHACTR3 | Phosphatase and actin regulator 3 | 1.03 | 1.38 |
| 203395_s_at | Hs.250666 | HES1 | Hairy and enhancer of split 1 | 1.02 | 1.51 |
| 242471_at | Hs.191475 | Clone HLS_IMAGE_238756 mRNA sequence | 1.21 | 1.13 | |
| 232174_at | Hs.655763 | CDNA: FLJ21635 fis | 1.43 | 1.12 | |
| 218559_s_at | Hs.702085 | MAFB | v-maf Musculoaponeurotic fibrosarcoma oncogene | 1.06 | 1.82 |
| 205476_at | Hs.75498 | CCL20 | Chemokine (C-C motif) ligand 20 | 1.49 | 1.12 |
| 228955_at | Hs.280387 | Transcribed locus | 1.28 | 1.40 | |
| 216997_x_at | Hs.444213 | TLE4 | Transducin-like enhancer of split 4 | 1.18 | 1.07 |
| 211506_s_at | Hs.624 | IL8 | Interleukin 8 | 1.31 | 1.51 |
| 204614_at | Hs.594481 | SERPINB2 | Serpin peptidase inhibitor | 1.58 | 1.28 |
| 226734_at | Hs.292026 | EIF4E2 | Eukaryotic translation initiation factor 4E-2 | 1.00 | 1.46 |
| 223441_at | Hs.597422 | SLC17A5 | Solute carrier family 17 (anion/sugar transporter) | 1.15 | 1.09 |
| 209774_x_at | Hs.590921 | CXCL2 | Chemokine (C-X-C motif) ligand 2 | 1.40 | 1.43 |
| 1557094_at | Hs.463110 | LOC653110 | Hypothetical LOC653110 | 1.00 | 1.73 |
| 232181_at | Hs.483816 | LOC153346 | Hypothetical protein LOC153346 | 1.32 | 1.32 |
| 209684_at | Hs.472270 | RIN2 | Ras and Rab interactor 2 | 1.08 | 1.45 |
| 212993_at | Hs.531457 | MRNA; cDNA DKFZp667B1718 | 1.00 | 1.77 | |
| 221840_at | Hs.127022 | PTPRE | Protein tyrosine phosphatase | 1.16 | 1.35 |
| 205064_at | Hs.1076 | SPRR1B | Small proline-rich protein 1B (cornifin) | 1.02 | 1.16 |
| 234594_at | Hs.612888 | C14orf85 | Chromosome 14 open reading frame 85 | 1.24 | 1.28 |
| 212660_at | Hs.483419 | PHF15 | PHD finger protein 15 | 1.00 | 1.65 |
| 208712_at | Hs.523852 | CCND1 | Cyclin D1 | 1.00 | 1.51 |
| 212200_at | Hs.654628 | KIAA0692 | KIAA0692 | 1.26 | 1.23 |
| 226982_at | Hs.592742 | ELL2 | Elongation factor | 1.39 | 1.17 |
| 232165_at | Hs.200412 | EPPK1 | Epiplakin 1 | 1.40 | 1.55 |
| 201466_s_at | Hs.525704 | JUN | Jun oncogene | 1.22 | 1.63 |
| 226860_at | Hs.688627 | TMEM19 | Transmembrane protein 19 | 1.16 | 1.46 |
| 244354_at | Hs.128434 | ELISC-1 | 1.12 | 1.30 | |
| 241950_at | Hs.680156 | Transcribed locus | 1.07 | 1.24 | |
| 230653_at | Hs.671710 | LOC728555 | Hypothetical protein LOC728555 | 1.13 | 1.48 |
| 244447_at | Hs.666767 | Transcribed locus | 1.13 | 1.24 | |
| 205463_s_at | Hs.705381 | PDGFA | Platelet-derived growth factor alpha polypeptide | 1.00 | 1.54 |
| 39248_at | Hs.234642 | AQP3 | Aquaporin 3 (Gill blood group) | 1.16 | 1.57 |
| 243431_at | Hs.674461 | Transcribed locus | 1.13 | 1.54 | |
| 219016_at | FASTKD5 | FAST kinase domains 5 | 1.01 | 1.69 | |
| 232541_at | Hs.664233 | CDNA FLJ20099 fis | 1.18 | 1.24 | |
| 202936_s_at | Hs.700579 | SOX9 | SRY (sex determining region Y)-box 9 | 0.96 | 1.46 |
| 238477_at | Hs.634167 | CDNA clone IMAGE:4830091 | 1.00 | 1.52 | |
| 209189_at | Hs.25647 | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 1.00 | 1.58 |
| 227140_at | Hs.28792 | CDNA FLJ11041 fis | 1.73 | 1.50 | |
| 202016_at | Hs.270978 | MEST | Mesoderm specific transcript homolog (mouse) | 1.19 | 1.58 |
| 229126_at | Hs.688627 | TMEM19 | Transmembrane protein 19 | 1.10 | 1.30 |
| 221768_at | Hs.355934 | SFPQ | Splicing factor proline/glutamine-rich | 1.13 | 1.27 |
| 226974_at | Hs.594057 | cDNA clone CS0DF038YD07 of Fetal brain | 1.36 | 1.39 | |
| 213051_at | Hs.133512 | ZC3HAV1 | Zinc finger CCCH-type | 1.06 | 1.42 |
| 212239_at | Hs.132225 | PIK3R1 | Phosphoinositide-3-kinase | 1.15 | 1.24 |
| 227034_at | Hs.355455 | ANKRD57 | Ankyrin repeat domain 57 | 1.15 | 1.47 |
| 212168_at | RBM12 | RNA binding motif protein 12 | 1.00 | 1.49 | |
| 200768_s_at | Hs.516157 | MAT2A | Methionine adenosyltransferase II | 1.00 | 1.37 |
| 232235_at | Hs.124673 | DSEL | Dermatan sulfate epimerase-like | 1.18 | 1.59 |
| 223159_s_at | Hs.197071 | NEK6 | NIMA (never in mitosis gene a)-related kinase 6 | 1.01 | 1.35 |
| 229549_at | Hs.592258 | Transcribed locus | 1.06 | 1.34 | |
| 225262_at | Hs.220971 | FOSL2 | FOS-like antigen 2 | 1.00 | 1.55 |
| 208691_at | Hs.529618 | TFRC | Transferrin receptor (p90) | 1.01 | 1.57 |
| 204326_x_at | Hs.374950 | MT1X | Metallothionein 1X | 1.00 | 1.32 |
| 1554250_s_at | Hs.661254 | TRIM73 | Tripartite motif-containing 73 | 1.22 | 1.62 |
| 221215_s_at | Hs.517310 | RIPK4 | Receptor-interacting serine-threonine kinase 4 | 0.84 | 1.79 |
| 227964_at | Hs.578433 | FRMD8 | FERM domain containing 8 | 1.20 | 1.38 |
| 226360_at | Hs.655242 | ZNRF3 | Zinc and ring finger 3 | 1.28 | 1.49 |
| 204470_at | Hs.789 | CXCL1 | Chemokine (C-X-C motif) ligand 1 | 1.45 | 1.40 |
| 223474_at | Hs.179260 | C14orf4 | Chromosome 14 open reading frame 4 | 1.00 | 1.49 |
| 211538_s_at | Hs.432648 | HSPA2 | Heat shock 70kDa protein 2 | 1.00 | 1.95 |
| 223218_s_at | Hs.319171 | NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer | 1.43 | 1.31 |
| 219476_at | Hs.32417 | C1orf116 | Chromosome 1 open reading frame 116 | 1.21 | 1.88 |
| 225601_at | Hs.693708 | HMGB3 | High-mobility group box 3 | 1.19 | 1.37 |
| 213139_at | Hs.360174 | SNAI2 | Snail homolog 2 (Drosophila) | 1.17 | 1.38 |
| 229830_at | Hs.535898 | Transcribed locus | 1.00 | 1.44 | |
| 238431_at | Hs.593044 | Transcribed locus | 1.29 | 1.39 | |
| 225634_at | Hs.133512 | ZC3HAV1 | Zinc finger CCCH-type | 1.00 | 1.81 |
| 202935_s_at | Hs.700579 | SOX9 | SRY (sex determining region Y)-box 9 | 0.94 | 1.38 |
| 201465_s_at | Hs.525704 | JUN | Jun oncogene | 1.14 | 1.64 |
| 242434_at | Hs.529514 | CDNA FLJ31093 fis | 1.08 | 1.51 | |
| 239448_at | Hs.658524 | Transcribed locus | 1.33 | 1.81 | |
| 209446_s_at | C7orf44 | Chromosome 7 open reading frame 44 | 1.04 | 1.40 | |
| 222243_s_at | Hs.474978 | TOB2 | Transducer of ERBB2 | 1.21 | 1.39 |
| 200799_at | Hs.520028 | HSPA1A | Heat shock 70kDa protein 1A | 1.00 | 2.02 |
| 229492_at | Hs.515130 | VANGL1 | Vang-like 1 (van gogh) | 1.04 | 1.57 |
| 211985_s_at | Hs.282410 | CALM1 | Calmodulin 1 (phosphorylase kinase) | 1.00 | 1.86 |
| 228977_at | Hs.130652 | LOC729680 | Hypothetical protein LOC729680 | 1.11 | 1.44 |
| 208161_s_at | Hs.463421 | ABCC3 | ATP-binding cassette | 1.00 | 1.44 |
| 209360_s_at | Hs.149261 | RUNX1 | Runt-related transcription factor 1 | 1.01 | 1.55 |
| 218051_s_at | Hs.84753 | NT5DC2 | 5'-Nucleotidase domain containing 2 | 1.00 | 2.04 |
| 223217_s_at | Hs.319171 | NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer | 1.38 | 1.49 |
| 233506_at | Hs.102941 | Full length insert cDNA clone ZB81B12 | 1.17 | 1.53 | |
| 226632_at | Hs.95120 | CYGB | Cytoglobin | 1.00 | 1.67 |
| 222450_at | Hs.517155 | TMEPAI | Transmembrane | 1.00 | 2.58 |
| 209260_at | Hs.523718 | SFN | Stratifin | 1.00 | 1.69 |
| 235086_at | Hs.164226 | THBS1 | Thrombospondin 1 | 1.47 | 2.40 |
| 205027_s_at | Hs.432453 | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 1.68 | 1.85 |
| 201150_s_at | Hs.701968 | TIMP3 | TIMP metallopeptidase inhibitor 3 | 1.00 | 1.76 |
| 200880_at | Hs.445203 | DNAJA1 | DnaJ (Hsp40) homolog | 1.00 | 1.66 |
| 234730_s_at | Hs.517310 | RIPK4 | Receptor-interacting serine-threonine kinase 4 | 1.13 | 2.04 |
| 219371_s_at | Hs.107740 | KLF2 | Kruppel-like factor 2 (lung) | 0.76 | 1.77 |
| 214702_at | Hs.203717 | FN1 | Fibronectin 1 | 1.74 | 1.64 |
| 242539_at | Hs.657550 | LOC730069 | Nuclear receptor binding factor 2 pseudogene | 1.06 | 2.26 |
| 226574_at | Hs.213198 | PSPC1 | Paraspeckle component 1 | 1.00 | 1.88 |
| 202434_s_at | Hs.154654 | CYP1B1 | Cytochrome P450 | 1.00 | 1.87 |
| 208711_s_at | Hs.523852 | CCND1 | Cyclin D1 | 1.00 | 2.07 |
| 229978_at | Hs.130661 | LOC440338 | Hypothetical gene supported by AJ002784 | 1.00 | 2.28 |
| 208153_s_at | Hs.591255 | FAT2 | FAT tumor suppressor homolog 2 (Drosophila) | 1.00 | 2.29 |
| 215462_at | Hs.632415 | PLK3 | Polo-like kinase 3 (Drosophila) | 1.00 | 2.08 |
| 1557049_at | Hs.700947 | LOC149478 | Hypothetical protein LOC149478 | 1.40 | 2.32 |
| 207332_s_at | Hs.529618 | TFRC | Transferrin receptor (p90) | 1.00 | 2.12 |
| 211984_at | Hs.282410 | CALM1 | Calmodulin 1 (phosphorylase kinase) | 1.00 | 2.33 |
| 200800_s_at | Hs.520028 | HSPA1 | Heat shock 70kDa protein 1A | 1.00 | 2.80 |
| 222853_at | Hs.41296 | FLRT3 | Fibronectin leucine rich transmembrane protein 3 | 1.00 | 3.14 |
| 205249_at | Hs.1395 | EGR2 | Early growth response 2 (Krox-20 homolog) | 1.64 | 2.78 |
| 222895_s_at | Hs.699440 | BCL11B | B-cell CLL/lymphoma 11B (zinc finger protein) | 1.00 | 3.69 |
| 1555847_a_at | Hs.436426 | LOC284454 | Hypothetical protein LOC284454 | 0.33 | 3.34 |
| 206393_at | Hs.523403 | TNNI2 | Troponin I type 2 (skeletal) | 1.00 | 3.64 |
| 206115_at | Hs.534313 | EGR3 | Early growth response 3 | 2.52 | 3.70 |
| 228523_at | Hs.591918 | NANOS1 | Nanos homolog 1 (Drosophila) | 1.00 | 5.08 |
Per Avadis software platform (Strand Life Sciences, San Francisco, CA).
Compared with control (0 µg/cm2 DEP).
Table 7.
Gene Downregulation After DEP Exposure in Cultured Human Airway Epithelial Cells
| Probe Set ID | Unigene IDa |
Gene Symbol |
Gene Title | Fold (log)b | |
|---|---|---|---|---|---|
| 10 µg/cm2 | 40 µg/cm2 | ||||
| 219270_at | Hs.155569 | CHAC1 | ChaC | −0.11 | −23.60 |
| 206085_s_at | Hs.19904 | CTH | Cystathionase (cystathionine gamma-lyase) | 3.12 | −18.20 |
| 1555788_a_at | Hs.516826 | TRIB3 | Tribbles homolog 3 (Drosophila) | 1.78 | −16.33 |
| 217168_s_at | Hs.146393 | HERPUD1 | Homocysteine-inducible | 2.05 | −14.18 |
| 238760_at | Hs.213264 | YARS | Tyrosyl-tRNA synthetase | 0.65 | −13.95 |
| 223062_s_at | Hs.494261 | PSAT1 | Phosphoserine aminotransferase 1 | 1.41 | −11.09 |
| 220892_s_at | Hs.494261 | PSAT1 | Phosphoserine aminotransferase 1 | 0.22 | −13.13 |
| 209383_at | Hs.505777 | DDIT3 | DNA-damage-inducible transcript 3 | 0.49 | −11.42 |
| 241905_at | Hs.175343 | PIK3C2A | Phosphoinositide-3-kinase | 1.16 | −8.36 |
| 213672_at | Hs.632707 | MARS | Methionyl-tRNA synthetase | 2.43 | −7.78 |
| 1554008_at | Hs.120658 | OSMR | Oncostatin M receptor | 0.22 | −7.09 |
| 205047_s_at | Hs.489207 | ASNS | Asparagine synthetase | 0.14 | −10.87 |
| 223195_s_at | Hs.469543 | SESN2 | Sestrin 2 | 0.78 | −6.36 |
| 1554933_at | Hs.658434 | PSIP1 | PC4 and SFRS1 interacting protein 1 | 0.00 | −5.86 |
| 243299_at | Hs.666703 | Transcribed locus | 1.71 | −5.78 | |
| 202388_at | Hs.78944 | RGS2 | Regulator of G-protein signaling 2 | −1.00 | −7.41 |
| 239451_at | Hs.658060 | CDNA FLJ26407 fis | 0.06 | −6.45 | |
| 222763_s_at | Hs.620490 | WDR33 | WD repeat domain 33 | −0.40 | −5.31 |
| 220195_at | Hs.458312 | MBD5 | Methyl-CpG binding domain protein 5 | 0.18 | −5.04 |
| 238311_at | Hs.668598 | Transcribed locus | 1.19 | −5.11 | |
| 204203_at | Hs.429666 | CEBPG | CCAAT/enhancer binding protein (C/EBP) | 0.03 | −5.48 |
| 232017_at | Hs.50382 | TJP2 | Tight junction protein 2 (zona occludens 2) | −0.54 | −5.95 |
| 206884_s_at | Hs.534699 | SCEL | Sciellin | 0.77 | −4.23 |
| 244822_at | Hs.473648 | GART | Phosphoribosylglycinamide formyltransferase | 0.24 | −5.74 |
| 243543_at | Hs.655159 | Transcribed locus | −0.49 | −4.88 | |
| 1556602_at | Hs.663643 | CDNA FLJ36293 fis | 0.91 | −3.93 | |
| 233406_at | Hs.667905 | CDNA FLJ12038 fis | 1.43 | −6.13 | |
| 218943_s_at | Hs.190622 | DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −0.03 | −4.83 |
| 203438_at | Hs.233160 | STC2 | Stanniocalcin 2 | 1.94 | −4.76 |
| 230097_at | Hs.473648 | GART | Phosphoribosylglycinamide formyltransferase | −0.30 | −6.62 |
| 244075_at | Hs.380900 | Transcribed locus | −0.15 | −4.79 | |
| 244803_at | Hs.658791 | Transcribed locus | 0.74 | −4.64 | |
| 229999_at | Hs.432355 | Full length insert cDNA clone ZE12A08 | −0.84 | −3.77 | |
| 238875_at | Hs.656795 | Clone HLS_IMAGE_731119 mRNA sequence | 0.48 | −6.35 | |
| 243332_at | Hs.670186 | Transcribed locus | 0.82 | −4.46 | |
| 236244_at | Hs.166463 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | −1.00 | −3.59 |
| 215470_at | General transcription factor IIH | −0.98 | −3.67 | ||
| 240206_at | Hs.481860 | TARS | Threonyl-tRNA synthetase | −0.78 | −3.39 |
| 236149_at | Hs.677583 | Transcribed locus | 0.77 | −2.96 | |
| 242784_at | Hs.665271 | Transcribed locus | −0.37 | −3.31 | |
| 241433_at | Hs.696152 | RCOR3 | REST corepressor 3 | −1.00 | −3.32 |
| 221577_x_at | Hs.616962 | GDF15 | Growth differentiation factor 15 | −0.30 | −4.77 |
| 206411_s_at | Hs.159472 | ABL2 | v-abl Abelson murine leukemia viral oncogene homolog 2 | −0.75 | −3.41 |
| 218986_s_at | Hs.591710 | FLJ20035 | Hypothetical protein FLJ20035 | −0.66 | −3.33 |
| 233436_at | Hs.657656 | MTBP | Mdm2 | −0.16 | −2.88 |
| 242712_x_at | Hs.652430 | RANBP2 | RAN binding protein 2 | −0.30 | −3.59 |
| 243631_at | Hs.659894 | LOC642333 | Similar to M-phase phosphoprotein | −0.06 | −3.84 |
| 237839_at | Hs.672300 | Transcribed locus | 0.02 | −3.39 | |
| 238348_x_at | Hs.667738 | Transcribed locus | −0.67 | −4.14 | |
| 230375_at | Hs.520287 | SFRS18 | Splicing factor | 0.21 | −4.04 |
| 238357_at | Hs.282901 | RBM39 | RNA binding motif protein 39 | −1.00 | −2.82 |
| 230516_at | Hs.87385 | C7orf30 | Chromosome 7 open reading frame 30 | 0.01 | −3.04 |
| 238666_at | Hs.659573 | Transcribed locus | −0.37 | −3.75 | |
| 239358_at | Hs.655048 | Transcribed locus | −0.65 | −3.06 | |
| 236696_at | Hs.596572 | SR140 | U2-associated SR140 protein | 0.38 | −2.36 |
| 237379_at | Hs.325838 | KIAA1542 | CTD-binding SR-like protein rA9 | −1.00 | −1.93 |
| 217616_at | Hs.613488 | Transcribed locus | −1.00 | −1.78 | |
| 1559691_at | Hs.684424 | CDNA clone IMAGE:3869664 | −0.41 | −2.12 | |
| 203434_s_at | Hs.307734 | MME (NEP) | Membrane metallo-endopeptidase (neprilysin) | −0.53 | −0.76 |
Per Avadis software platform (Strand Life Sciences, San Francisco, CA).
Figure 9. The changes in fluorescent-spot intensity values for genes after DEP exposure.
Exposure to either 10 or 40 µg/cm2 DEP for 24 hours resulted in 151 upregulated and 59 downregulated genes in in vitro cultures of human airway epithelial BEAS-2B cells.
Utilizing the DAVID bioinformatics database (david.abcc.ncifcrf.gov) — which annotates, visualizes, and integrates lists of genes — we analyzed 243 preselected genes based on dose–response changes in expression as a result of DEP exposure. We obtained DAVID identification for 229 genes. Our annotative analysis indicated that 74% of the genes map to biologic processes listed as Gene Ontology (GO) terms in the DAVID database, 73% map to a cellular component, and 78% map to a molecular function. Of the cellular components, 118 genes map to intracellular; 84, to intracellular membrane-bound; 92, to intracellular organelle; 71, to the cytoplasm; and 65, to the nucleus. Among the genes distributed to GO terms that were associated with molecular function, 149 were to binding terms: 107, to protein binding; 47, to nucleic acid binding; 38, to nucleotide binding; and 13, to protein kinase activity. Interpro (part of the DAVID system) identified 83% of the genes and indicated that 11 genes were associated with a protein kinase core. The Self-Monitoring, Analysis, and Reporting Technology system identified 52% of the genes and indicated that 8 genes were associated with basic-leucine zipper transcription factors, and 9 genes were associated with the serine or threonine-specific kinase subfamily. The Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa Laboratories, Kyoto, Japan) software mapped 9 genes to focal adhesion and 9 genes to MAPK signaling pathways. BioCarta (also part of the DAVID system) mapped 3 genes to IL-6 signaling pathways and 3 genes to thrombospondin-1 (TSP-1)–induced apoptosis in microvascular endothelial cell pathways. A clustering analysis at medium stringency produced 27 significant clusters, the most frequently occurring categories of which are listed in Figure 10.
Figure 10. Functional annotation clustering of changed genes in BEAS-2B cells following DEP exposure.
Cells were treated with 0, 10, 40 µg/cm2 DEP for 24 hours. Categories of mode of action are represented on the y-axis according to the GO category of cell function (DAVID database). The bars in the histogram indicate the percentage of genes in each category that were up- or downregulated in a dose-dependent manner.
AIM 4: TO EVALUATE THE EFFECTS ON NEP ACTIVITY OF HUMAN EXPOSURE TO DEE
Cell Profile
The exposure of subjects to DEE induced an increase of inflammatory cells (Table 8), as shown by a statistically significant increase in the total number of cells in sputum. The concentration of macrophages— but not neutrophils and lymphocytes — increased after DEE exposure. The data suggest that DEE exposure resulted in a macrophage-related proinflammatory or inflammatory response in the airways.
Table 8.
Sputum Cell Profiles in Human Subjects Exposed to DEE
| Cell Numbera | ||
|---|---|---|
| Cell Type | Baseline | After DEE Exposure |
| Inflammatory cells (× 106/mL) | ||
| Total | 7.98 ± 1.68 | 20.29 ± 7.65b |
| Macrophages | 6.53 ± 1.32 | 16.10 ± 5.57b |
| Neutrophils | 1.29 ± 0.49 | 4.05 ± 2.32 |
| Lymphocytes | 0.15 ± 0.05 | 0.21 ± 0.08 |
| Epithelial cells (× 103/mL) | 2.80 ± 0.82 | 12.43 ± 7.98b |
Average for 11 subjects ± SEM.
P < 0.05 when compared with baseline (N = 11).
The increase (4.44×) in sputum epithelial cell counts after exposure of subjects to DEE was statistically significant (Table 8), indicating that the exposure resulted in acute airway epithelial cell shedding.
Total Protein and NEP Activity
We found that DEE exposure (0.88 ± 0.08) did not induce a statistically significant change in total sputum protein when compared with baseline control (0.82 µg/µL ± 0.09) (Table 9). We conducted an analysis of soluble NEP activity in sputum for all 11 healthy nonsmoking volunteers. The results showed a range of baseline values from 0.115 to 0.445 nmol/µg protein/min, averaging 0.303 ± 0.036 (mean ± SEM). We observed individual increases in NEP activity in 9 out of 11 subjects after DEE exposure (Table 9). When compared with baseline control, the average net increase after DEE exposure was 31%, with a maximum change of 0.26 nmol/µg protein/min. The change indicates that an increase of NEP activity, observed in sputum, occurred after the acute exposure of subjects to approximately 575 µg/m3 of elemental carbon (DEE). A Pearson correlation analysis indicated that NEP activity in individual sputum after DEE exposure was slightly associated with the product of exposure concentration × exposure time (Figure 11). DEE exposure was not corrected for pre-exposure levels of NEP activity (data not shown).
Table 9.
Induced Sputum Protein Level and NEP Activity in Human Subjects Before and After Exposure to DEE
| Total protein (µg/µL) |
NEP (nmol/µ protein/min) |
|||
|---|---|---|---|---|
| Baseline | After DEE Exposure |
Baseline | After DEE Exposure |
|
| 1.05 | 1.07 | 0.364 | 0.369 | |
| 0.66 | 0.99 | 0.445 | 0.431 | |
| 1.00 | 0.97 | 0.436 | 0.507 | |
| 1.27 | 1.21 | 0.258 | 0.465 | |
| 0.41 | 0.84 | 0.395 | 0.399 | |
| 0.56 | 0.43 | 0.294 | 0.558 | |
| 0.82 | 1.13 | 0.413 | 0.272 | |
| 1.16 | 0.79 | 0.159 | 0.382 | |
| 0.74 | 0.92 | 0.289 | 0.332 | |
| 0.94 | 1.00 | 0.164 | 0.324 | |
| 0.41 | 0.32 | 0.115 | 0.320 | |
| Mean | 0.82 | 0.88 | 0.303 | 0.396 |
| SEM | 0.09 | 0.08 | 0.036 | 0.026 |
| P valuea | 0.42 | 0.035 | ||
P values based on a two-tailed, paired sample t test (N = 11).
Figure 11. Statistically significant correlations of soluble NEP activity in subject sputum (N = 11) with DEE exposure (concentration [mg/m3] × time [min]).
Associations of Sputum NEP Activity with Cell Types
To further evaluate the association between changes in sputum cell type and NEP activity, we conducted Pearson correlation analyses. Sputum NEP activity was not significantly associated with the changes in total cell number or with the changes in macrophages, neutrophils, or epithelial cells (data not shown).
DISCUSSION AND CONCLUSIONS
Using a number of approaches (in vitro, animal, and controlled human exposure), we examined the role of NEP in DEE-induced airway inflammatory response, taking the following steps:
To simultaneously characterize the dose–response of DEP on respiratory NEP and the role of NEP in the inflammatory response to DEP exposure, we conducted an animal study with a 2 × 3 factorial design. We observed that pulmonary NEP protein expression in wild-type mice at 7 days after exposure to DEP via intratracheal installation was significantly downregulated in a concentration-dependent manner, decreasing by 10.4% and 50.0% at the low and high levels of DEP exposure compared with control. The changes were accompanied by increases in macrophages, epithelial cells, and the proinflammatory cytokine IL-6 (but not IL-1[3 or IL-10) in BAL fluid. Nep-null mice displayed enhanced increases in these markers of the inflammatory response when compared with wild-type mice and showed increases in IL-1[3 and IL-10 in BAL fluid, especially in response to the high dose of DEP. These in vivo findings suggest that loss of NEP in mice could cause an increased susceptibility to injury or exacerbate the inflammatory response to DEP exposure by allowing the release of higher levels of specific cytokines from the lungs.
-
To clarify the contribution of major components of DEP (SRM 2975) in the downregulation of NEP in BEAS-2B cells, we compared the effects induced by DEP, cDEP, and sDEP. We also examined oDEP (or standard urban dust [SRM 1649a]), another type of particle, to test whether NEP downregulation was a specific response to DEP. Parallel BEAS-2B cell cultures were conducted with 0, 1, and 10 µg/cm2 DEP for 24 hours. As measured by RT-PCR, cells treated with 1 and 10 µg/cm2 DEP had approximately 50% and 45% NEP mRNA expression, respectively, of control. Cells treated with cDEP at 10 µg/cm2, but not at 1 µg/cm2, also downregulated NEP mRNA expression by about 45% compared with control, suggesting that cDEP has less effect than DEP due to removal of chelated divalent cations, particularly transition metals. Moreover, NEP mRNA expression was not affected in cells exposed to sDEP, indicating that DEP after organic stripping lost its downregulation effect. Collectively, these findings suggest that downregulation of NEP by DEP was mostly attributable to DEP-adsorbed organic compounds, whereas the carbonaceous core and transition metal components of DEP had little or no effect on NEP mRNA expression at the current experimental condition. This NEP downregulation in vitro was not a specific response to DEP or its contents because the change also occurred after urban dust exposure, which has a different composition.
To further understand the molecular impact of NEP loss in response to DEP and the effect of DEP on epithelial NEP in vitro, we collected transcriptome profiles of the concentration–effects of SRM 2975 in cultured BEAS-2B cells through a 2 × 3 factorial design. Microarray data showed that there were 151 upregulated and 59 downregulated genes in response to DEP exposure. Of the up- or downregulated genes, 89.5% and 46.6%, respectively, changed in a concentration-dependent manner. These genes included metabolic enzymes CYP1A1 and CYP1B1, IL6, IL8, CCL1 and CCL2, DIDO1, HSPA1B, AQP3, and EGR2 and EGR3. Utilizing the DAVID bioinformatics database, we analyzed 243 preselected genes based on concentration-dependent changes in expression as a result of DEP exposure. The KEGG pathways mapped 9 genes to focal adhesion and 9 genes to MAPK signaling pathways. BioCarta mapped 3 genes to IL-6 signaling pathways and 3 genes to TSP-1–induced apoptosis in the microvascular endothelial cell pathway. Cells depleted of NEP expression (by siRNA) showed numerous up-regulated (17) or downregulated (14) genes, involving DNA/protein binding, calcium channel activities, and intracellular signaling cascade of cytokines. Genes identified included IL6, IL8, and EGFR. Knockdown of NEP expression also disturbed the transcriptions of many genes relevant to cellular monooxygenase, GTPase, and protein kinase activities. Increased expression of IL6, IL8, and PTGS2 was confirmed by RT-PCR to be associated with both DEP exposure and NEP knockdown. The data generated from a combined RNAi–microarray approach revealed that there is a complex molecular cascade mediated by NEP in different subcellular compartments, possibly influencing inflammatory response.
In the controlled human exposure study, DEE induced significant increases in the number of macrophage, neutrophil, and airway epithelial cells. Moreover, we observed statistically significant increases in soluble NEP activity in sputum after DEE exposure (0.40 ± 0.03 nmol/µg protein/min; P = 0.035) when compared with the baseline control (0.30 ± 0.04 nmol/µg protein/min), with a 31% average net increase. Based on this, we speculate that the change in NEP activity in sputum may possibly be a new endpoint to measure the effects of DEE exposure. Change in NEP activity may also provide insight into the mechanism of airway effects after DEE exposure, if it can be confirmed in human investigation with a larger sample size and at ambient exposure levels.
We discuss these summarized results in detail along with our previous findings in the pilot study (Wong et al. 2007 pilot study) in the following sections.
LOSS OF NEP ENZYMATIC ACTIVITY IN RAT AND HUMAN LUNG FOLLOWING DEE EXPOSURE
Our previous in vivo study showed that NEP activity in rat lung was significantly reduced when rats were exposed to DEP (35 µg/m3) (Wong et al. 2003; Witten et al. 2005). The change in NEP activity was accompanied by increases in bronchopulmonary plasma extravasation, vascular permeability, and cytokine expression, as well as inflammatory/mast cell infiltration, possibly evoked by abnormally high levels of peptides after DEP exposure. Reduced NEP activity could induce the airways to respond in an exaggerated fashion to peptide substrates, thus producing a hyperresponsive or abnormal response state. The fact that diesel components enhance the vasoconstrictive effects of endothelin-1 and reduce the dilatory response to sodium nitroprusside (Campen et al. 2005) indirectly suggests the diesel-mediated loss of NEP activity in the response to endothelin-1 in vivo. Thus, this reduction in NEP activity may affect the bioavailability of many peptide mediators (such as neurokinins, endothelins, and bombesin) released from sensory nerve terminals or immunoinflammatory cells (Joos et al. 2000). To evaluate loss or decrease in human airway NEP activity in Aim 4, we took advantage of a collected sputum sample from an earlier controlled exposure study (Burgess et al. 2007). The results showed that there was a rapid increase in soluble NEP activity in sputum shortly after the exposure of human volunteers to DEE. Increased NEP activity in sputum may reflect a local inflammatory response, with subsequent shedding of membrane-bound enzymes in response to lung injury (Soleilhac et al. 1996). We also found a similar change in sputum NEP activity after exposure of these subjects to mining dust particulates, suggesting that the change was not a specific response to DEE exposure (data not shown). These results were consistent with the findings from exposure to viruses, fire smoke, and cigarette smoke (Lilly et al. 1994; Wong et al. 2004). We believe that the current investigation provides additional environmental evidence for the finding that NEP expression varies widely in human lung tissue from different individuals (Cohen et al. 1999). Although this effect remains to be confirmed in a large population, our data suggest that DEE-induced effects may be linked mechanistically to changes in NEP activity and/or expression.
The cellular origin of soluble NEP activity in the sputum of humans exposed to DEE remains to be characterized. The results of this study suggest that airway epithelial cells could be one of the major sources of soluble NEP activity found in sputum due to exposure-induced epithelial membrane damage. In addition, inflammatory cells, especially airway macrophages, might be another major source of soluble NEP activity in sputum as a result of DEE exposure. Our speculation is based on our findings that both epithelial and macrophage cells, which abundantly express NEP, were affected by DEP exposure in this study. Since, structurally, NEP is a cell-surface metalloprotease with an extracellular domain of 700 amino acids, its large extracellular domain containing the catalytic sequence may explain its critical ability to rapidly cleave substrates. However, this structure is fully exposed to an extracellular environment that is possibly susceptible to toxic insults. Therefore, the likely mechanism to explain the increase of NEP activity in sputum is the shedding of affected cells into the airways after DEE exposure, with membrane-bound proteins being released with portions of plasma membrane or as proteolipid aggregates.
We note that the dose (575 µg/m3 of elemental carbon) used in the human exposure part of our study was too high to reflect a possible change in NEP activity after exposure to an ambient level of DEP. Future studies with large sample sizes are needed to test whether exposure to real-world levels of DEP and other particulates causes a change in NEP activity with functional consequences for airway biology. However, we hope that soluble NEP activity in sputum could serve as a potential early endpoint in identifying DEP-mediated health risks if our results are confirmed by animal investigations and by studies using a larger human population at an ambient exposure level.
DOWNREGULATION OF NEP EXPRESSION BY DEP IN HUMAN EPITHELIAL CELL AND MOUSE MODELS
Our previous in vitro experiments have shown that DEP could downregulate NEP at the levels of gene and protein expression (Wong et al. 2007 pilot study). In human airway epithelial cells (BEAS-2B), we observed that exposure to DEP for 24 hours significantly repressed NEP expression, as well as its enzymatic activity, in a concentration-dependent manner. Furthermore, experiments with DEP in primary human airway epithelial cells confirmed the results in BEAS-2B cells; an approximate 90% decrease in NEP mRNA expression at 5 µg/mL DEP concentration was observed in our 2007 pilot study when compared with controls. Therefore, downregulation of NEP expression after DEP exposure was not a cell-line–specific effect. These findings were confirmed in the current in vitro study. Moreover, in the current study, we also demonstrated the down-regulation of NEP expression in mouse lung after acute exposure to DEP. The mechanisms underlying downregulation of NEP expression by DEP remain to be investigated.
DOWNREGULATION OF NEP BY DEP-ADSORBED COMPOUNDS IN VITRO
DEP is composed of a carbonaceous core with adsorbed transition metals and various organic substances including polycyclic aromatic hydrocarbons (PAHs), nitro-aromatic hydrocarbons, quinones, and acids (McClellan 1987). To examine the contribution of the major classes of DEP components in downregulation of NEP expression, we compared the effects induced by whole DEP; sDEP, with everything removed but the carbonaceous core; and cDEP, without transition metals. Our data showed that the organic compounds attached to particles might play a major role in any downregulation effects of NEP. It has been reported that the treatment of diesel exhaust that diminished the adsorbed organic compounds could reduce the DEP-induced inflammatory response and in vivo toxicity (Boland et al. 1999). Therefore, the current data provide additional evidence that DEP may pose a particularly high risk of adverse effects, possibly because of the high fraction of potentially toxic substances found on the relatively large surface of the particles (Rudell et al. 1999; Cohen et al. 2002; Kulkarni et al. 2006).
Downregulation of NEP mRNA expression may not be a specific effect of DEP. In the same culture conditions, epithelial cells exposed to urban dust (SRM 1649a) had reduced NEP expression that was similar to the reduction after exposure to SRM 2975. Certified analyses have indicated that the two types of particles have certain common or similar organic compounds (NIST Certificate of Analysis 2000, 2001). A comparison between the two particles showed that the concentrations of many nitro-PAHs in SRM 2975 were an order of magnitude higher than concentrations measured in SRM 1649a, due to dilution of DEP by other non-diesel particles in the urban air sample (Bamford et al. 2003). 1-Nitropyrene was pointed out to be the dominant nitro-PAH present in the diesel particulate samples, while 2-nitrofluoranthene was the highest nitro-PAH concentration measured in the urban air sample (Bamford et al. 2003). However, additional in vitro experiments are needed to further demonstrate that organic compounds of SRM 1649a play a major role in the downregulation of NEP expression.
IMPACT OF NEP DOWNREGULATION IN DEP-INDUCED INFLAMMATORY INJURY IN VIVO AND IN VITRO
The current study showed that the DEP-induced inflammatory response in wild-type mice was dramatically enhanced in Nep-null mice, as measured by BAL fluid cell profiles, proinflammatory cytokines, and alterations identified histopathologically. In addition, in vitro experiments showed that human airway epithelial cells depleted of NEP expression displayed higher expression of proinflammatory cytokines IL-6 and IL-8 than those of control cells. These findings suggest that these inflammatory responses to DEP exposure were, at least in part, associated with either NEP expression or NEP activity or both. These findings also suggest new insights into the mechanisms underlying the DEP-induced inflammatory response. Since IL-6 and IL-8 are not substrates for NEP, the exacerbated inflammation and injury responses were likely mediated by diminished degradation of NEP substrates and/or nonenzymatic functional loss, such as injury repair, of NEP protein, or both.
In addition, our previous in vitro study (Wong et al. 2007 pilot study) suggested that NEP expression, rather than its catalytic activity, may possibly affect the regulatory machinery involved in cell proliferation. Other studies have also shown that NEP is involved in cell growth (Ganju et al. 1994). In our pilot study, NEP knockdown by RNAi in airway epithelial cells resulted in a concentration-dependent reduction of epithelial proliferation in response to DEP exposure. The change in epithelial proliferation may possibly be attributable to the nonenzymatic interaction of NEP protein through its cytoplasmic tail with several intracellular proteins. For example, it is known that the NEP cytoplasmic domain contains putative sites for phosphorylation by casein kinase II (Ganju et al. 1996). The casein kinase II-mediated phosphorylation of specific cellular proteins has been linked with proliferation (Ganju et al. 1996). Also, NEP can associate with ezrin/radixin/moesin (ERM) proteins and inhibit the association of ERM proteins with CD44, and may thus play a role in restricting CD44-mediated cell motility (Iwase et al. 2004). In addition, NEP directly associates with and stabilizes PTEN tumor suppressor, leading to dephosphorylation of phosphatidylinositol (3,4,5)-trisphosphate to its less active form. This results in regulation of downstream cell growth and cell survival pathways such as those regulated by Akt/PKB kinase (Sumitomo et al. 2004). It is likely that these mechanisms participate in epithelial repair in vitro through NEP-mediated signal transduction pathways.
However, in the current study, we were unable to detect the effects of DEP on epithelial proliferation in wild-type or Nep-null mice, possibly because there were limited numbers of BrdU-positive cells observed in slides of dissected mouse lung tissue. This suggests that the extent of the injury was too mild to be observed using the immunohistochemical quantification of incorporated BrdU as a measure of epithelial proliferation. It is possible that the degree of cell injury caused by DEP did not result in cell death, which would ordinarily result in synthesis of new cells. It is known that there are two cell populations that proliferate in response to airway injury (Rawlins et al. 2007): basal cells in the trachea and primary bronchi, and Clara cells in the more distal bronchi, bronchioles, and bronchoalveolar duct junction. Under normal conditions, epithelial cell turnover in the lung is relatively low. Nevertheless, if these cells are extensively damaged, the lung can replace lost cells quickly. In the current study, however, histopathologic examination showed that instillation of DEP at a level of 10 or 100 µg resulted in mild lung injury, which did not destroy the majority of the Clara cells in either the proximal or distal conducting airways. It may be necessary to design additional studies with mice exposed to the higher dose of DEP in order to determine whether NEP has a role in epithelial proliferation and repair.
NEP-MEDIATED MOLECULAR MECHANISM IN DEP-INDUCED INFLAMMATORY INJURY
With three techniques (RNAi, microarrays, and RT-PCR), we dissected the complicated molecular networks mediated by NEP. A comparison of expression files between normal and NEP-knockdown cells reveals highly NEP-connected pathways. The analysis showed that genes associated with the induction of apoptosis (BCL2L11), signal transduction (EGFR, SLC6A19), and inflammatory processes (IL6, IL8, PTGS2, PTGER4) were significantly over-represented. To our knowledge, this is the first report of array-based “genetic unmasking” in combination with NEP knockdown, and it reveals the potential of this strategy in identifying particular signaling transduction pathways affected by NEP. For example, previous in vivo, in vitro, and human studies showed that proinflammatory cytokines IL-6 and IL-8 frequently increase after exposure to DEP, leading to immuno-inflammatory injury (Diaz-Sanchez et al. 1994; Bayram et al. 1998; Nightingale et al. 2000; Li et al. 2002; Gong et al. 2003). The changes in cytokine expression are mediated in part through the activation of many signaling transduction pathways, involving transcription factors such as NFKB2, AP-1, and STAT3 (Takizawa et al. 2003; Zhang et al. 2004; Cao et al. 2007), MAPKs (Fahy et al. 2000; Hashimoto et al. 2000) via reactive oxygen species–dependent mechanisms (Baulig et al. 2003). Our study suggested that the abnormal response pattern of a cascade of cellular and molecular events brought on by a depletion of NEP induced by DEP contributed not only to inflammatory processes, but also to apoptosis and abnormal signal transduction.
In addition, our study showed that depletion of NEP expression (by siRNA) in epithelial cells was associated with many pathways, including DNA/protein binding, calcium channel activities, and intracellular signaling cascades. The proteins identified included cellular monooxygenase, GTPase, PTGS2, and protein kinase. It is noted that PTGS2, also known as cyclooxygenase, is the key enzyme in prostaglandin biosynthesis and acts both as a dioxygenase and as a peroxidase. PTGS2 is regulated by specific stimulatory events and is responsible for the prostanoid biosynthesis involved in inflammation and mitogenesis. Collectively, our data from the microarray experiments suggest that there is a complex molecular cascade mediated by NEP in different subcellular compartments in response to DEP challenge, possibly influencing the metabolic, cellular, and regulatory functions of epithelial cells.
We conclude that (1) exposure to DEP may downregulate NEP expression in a concentration-dependent manner, and we attribute this effect mostly to DEP-associated organic compounds; (2) Nep-null mouse and cellular models with NEP knockdown helped us discover that NEP plays an important role in the inflammatory response to DEP exposure in vivo and in vitro; (3) there exists a complex molecular cascade mediated by NEP in different subcellular compartments, further influencing normal cellular response to DEP; and (4) exposure of human subjects to DEE resulted in changes in airway NEP activity. We speculate that downregulation of airway NEP may be one of the mechanisms that links DEE exposure to airway inflammatory injury or airway disease susceptibility.
HEI STATEMENT Synopsis of Research Report 159.
Role of Neprilysin in Airway Inflammation Induced by Diesel Exhaust Emissions
BACKGROUND
Neprilysin — also known as neutral endopeptidase (NEP) — is an enzyme that degrades multiple peptides that affect airway blood vessels. The expression of NEP on the surface of cells varies in a number of airway injury conditions and in several types of cancers. HEI periodically issues a Request for Preliminary Applications for novel research on the health effects of air pollutants derived from motor vehicle emissions. In response to Request for Preliminary Applications 05-3, “Health Effects of Air Pollution,” issued in 2005, Dr. Simon Wong at the University of Arizona and colleagues submitted an application to study whether exposure to diesel exhaust affected the airway expression or function of NEP. Some prior studies had shown that exposure of humans and laboratory animals to diesel exhaust particles (DEP) or whole diesel exhaust emissions (DEE) affected the airways. Dr. Wong and colleagues hypothesized that components of diesel exhaust downregulate the function or expression of NEP in the airways and that this may lead to disorders in airway function. They also hypothesized that in conditions in which NEP expression was decreased, responses to diesel exhaust would be increased.
APPROACH
Wong and colleagues evaluated airway inflammatory responses and NEP expression and activity in the fluid and cells obtained from bronchoalveolar lavage and the lung tissue of mice genetically deficient in NEP (Nep-knockout mice) or wild-type mice (control). The mice were instilled with 10 or 100 µg resuspended DEP, and their BAL fluid was analyzed 7 days later; the investigators used National Institute of Standards and Technology SRM 2975 particles, which were originally generated by a diesel-powered industrial forklift. The investigators also measured airway inflammatory responses and NEP activity in the induced sputum of 11 healthy human volunteers (ages 19–33 yr) 1 hour after exposure to DEE in a staged mining environment. Individual exposure concentrations ranged from 0.09 to 1.80 mg/m3 elemental carbon (a component of DEE), and duration ranged from 56 to 134 minutes. A baseline measurement of NEP activity was taken at least 1 week before volunteers were exposed to DEE.
In addition, the investigators used a transformed human airway epithelial cell line, BEAS-2B, to evaluate whether exposure to DEP decreased the expression of NEP messenger RNA (mRNA). They used 1 or 10 µg/cm2 untreated DEP (SRM 2975); DEP treated with chelating agents to remove divalent cations, particularly transition metals; DEP treated with dichloromethane to remove all but the carbonaceous core; or a control particle (standard urban dust SRM 1649a). Using microarray and real-time polymerase chain reaction approaches on extracts of these cells, the investigators also evaluated the expression of genes affected by exposure to DEP. Similar gene expression studies were conducted in DEP-exposed BEAS-2B cells with depleted levels of NEP by incubating the cells with a small interfering RNA specific for NEP.
RESULTS
The investigators found airway inflammatory effects in response to DEP instillation— specifically, increases, found in bronchoalveolar lavage fluid, of the numbers of macrophage and epithelial cells and levels of cytokines that these cells synthesize— that were greater in Nep-knockout mice than in wild-type mice that express NEP. They also found a 50% decrease in NEP protein expression in lung tissue and decreased NEP protein expression in airway epithelial cells and macrophages after DEP instillation into wild-type mice.
In human volunteers, the investigators observed that baseline levels of NEP activity in induced sputum varied considerably among the volunteers. Exposure to a high concentration of DEE via inhalation resulted in airway inflammatory effects, measured as increased numbers of macrophages and epithelial cells in the induced sputum, and, in some participants at least, increased NEP activity.
In addition, the investigators observed changes (either up or down) in the expression of several genes in the airway epithelial cell line in response to DEP in normal control cells as well as in cells in which NEP expression was decreased.
NEP mRNA expression in BEAS-2B cells was decreased by approximately 45% after exposure to untreated DEP or to DEP with transition metals removed and by approximately 60% after exposure to the control standard urban dust particle. DEP that was stripped down to its carbonaceous core did not affect NEP mRNA levels significantly.
CONCLUSIONS
In its independent review of the study, the HEI Health Review Committee considered that Wong and colleagues had made a comprehensive attempt to explore the role of NEP in response to exposure to diesel exhaust components, using an appropriate animal model (an Nep-knockout mouse), human studies, and in vitro models using a human cell line. For most of the experiments performed, the study design was strong and the statistical analyses appropriate. The human exposure study, however, had the limitation that the investigators used samples and data that had already been collected in an earlier study, and so only a limited number of assays could be performed.
The investigators found that DEP instillation into the airways of mice resulted in stronger airway inflammatory effects in mice genetically deficient in Nep than in mice expressing Nep and was accompanied by a significant decrease in NEP expression in lung tissue. The Committee concluded that these findings plausibly suggested that expression of NEP in some way damps down inflammatory responses in the airways — supporting one of the hypotheses for the study — but noted that the mechanism by which this might occur was not addressed in this study. The Committee thought that other important study findings were that exposure of human volunteers to a high concentration of DEE via inhalation resulted in airway inflammatory effects and, in some participants at least, increased NEP activity. However, this was at odds with the investigators’ other hypothesis for the study, that exposure to DEP would decrease NEP levels in the airways.
The Committee noted that it was challenging to compare the responses in the mouse and human airways because the exposures were of different levels, durations, and routes, and the effects were measured at different times after the exposures. Nonetheless, the Committee thought that the study’s human and mouse diesel-exhaust–associated findings could be interpreted in a consistent fashion — that is, that the observed changes in NEP levels result from a response to injury in the airways, measured as the shedding of airway epithelial cells and an increase in macrophage numbers in induced sputum (human) or bronchoalveolar lavage fluid (mouse). Since both epithelial cells and macrophages express NEP, this could explain the increase in NEP activity found in the induced sputum of some of the human volunteers.
The in vitro studies in the transformed human airway epithelial cell line provided some useful information about DEP constituents that affect NEP expression — namely, removing metals did not change NEP expression, but removing organic components did. The studies also provided information about which genes’ expression in a transformed airway epithelial cell line may be affected by exposure to DEP. These data may help in determining the pathways involved in airway response to DEP and any possible NEP role in that response.
The investigators speculated in the report that changes in NEP in sputum might be a useful early marker of DEE-induced injury in humans. However, the Committee thought that changes in NEP activity or levels in the airways are unlikely to be useful biomarkers of exposure to diesel components, because the observed effects in airway cells were not specific to diesel exposures, and because baseline levels of airway NEP activity differed markedly in different people. Thus, although changes in NEP function and activity have been noted in airway conditions, particularly after injury, the role of NEP is still not resolved.
Acknowledgments
We thank R. Clark Lantz (University of Arizona) for covering the cost of the pathology core service as part of NIH/NIEHS center grant ES06694. We are very grateful to HEI for funding and to the HEI Health Review Committee for their critical assessment of the final report. We especially thank the external reviewers and HEI science staff for their expert comments, as well as Hilary Selby Polk, HEI senior science editor, for her excellent editing.
This Statement, prepared by the Health Effects Institute, summarizes a research project funded by HEI and conducted by Dr. Simon S. Wong at the University of Arizona Health Science Center, Tucson, Arizona, and colleagues. Research Report 159 contains both the detailed Investigators’ Report and a Critique of the study prepared by the Institute’s Health Review Committee.
This Investigators’ Report is one part of Health Effects Institute Research Report 159, which also includes a Critique by the Health Review Committee and an HEI Statement about the research project.
Although this document was produced with partial funding by the United States Environmental Protection Agency under Assistance Award CR–83234701 to the Health Effects Institute, it has not been subjected to the Agency’s peer and administrative review and therefore may not necessarily reflect the views of the Agency, and no official endorsement by it should be inferred. The contents of this document also have not been reviewed by private party institutions, including those that support the Health Effects Institute; therefore, it may not reflect the views or policies of these parties, and no endorsement by them should be inferred.
ABBREVIATIONS AND OTHER TERMS
- 18S rRNA
18S ribosomal RNA
- AAALAC
American Association for Animal Laboratory and Care
- ABTS
2,2'-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid
- ALI
acute lung injury
- ANOVA
analysis of variance
- AP-1
activator protein-1
- AQP3
aquaporin 3
- ARDS
acute respiratory distress syndrome
- BAL
bronchoalveolar lavage
- BrdU
5-bromo-2'-deoxyuridine
- BSA
bovine serum albumin
- cDEP
DEP with divalent cations, particularly transition metals removed using a chelator
- cDNA
complementary DNA
- CO
carbon monoxide
- CO2
carbon dioxide
- COPD
chronic obstructive pulmonary disease
- cRNA
complementary RNA
- CYP
cytochrome P450
- DAB
3,3'-diaminobenzidine tetrahydrochloride
- DAVID
Database for Annotation, Visualization and Integrated Discovery (U.S. NIAID)
- DEE
diesel exhaust emissions
- DEP
diesel exhaust particles
- DIDO
death inducer-obliterator
- DNase
deoxyribonuclease
- EC
elemental carbon
- EGFR
epidermal growth factor receptor
- EGR2&3
early growth response 2&3
- ELISA
enzyme-linked immunosorbent assay
- ERM
ezrin/radixin/moesin
- FEV1
forced expiratory volume in one second
- FVC
forced vital capacity
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GTPase
guanosine triphosphatase
- GO
Gene Ontology (DAVID database)
- H2O2
hydrogen peroxide
- hp
horsepower
- IL
interleukin
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KGM
keratinocyte growth medium
- LHD
load-haul-dump
- MAPK
mitogen-activated protein kinase
- MOPS
3-(N-morpholino) propanesulfonic acid
- mRNA
messenger RNA
- NEP
neprilysin, or neutral endopeptidase
- NFKB2
nuclear factor-kappa B
- NIST
National Institute of Standards and Technology
- NK
neurokinin
- NO2
nitrogen dioxide
- NOx
nitrogen oxides
- O3
ozone
- oDEP
standard urban dust (another type of diesel exhaust particle)
- PAH
polycyclic aromatic hydrocarbon
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PM
particulate matter
- PM2.5
PM ::: 2.5 µm in aerodynamic diameter
- PTGS2
prostaglandin-endoperoxide synthase 2
- RNAi
RNA interference
- RT-PCR
real-time polymerase chain reaction
- SAEC
small airway epithelial cell
- SCLC
small-cell lung carcinoma
- sDEP
“stripped DEP” carbonaceous core of DEP
- SEM
standard error of the mean
- siRNA
small interfering RNA
- SP
substance P
- SRM
standard reference material
- TSP-1
thrombospondin-1
- UA
University of Arizona
Biographies
Simon S. Wong is currently working in the Division of Respiratory Medicine at the University of California–San Diego. Before his current appointment, he was a research associate professor in the Department of Pediatrics at the University of Arizona Health Science Center, Tucson, Arizona. His recent research interests include the pulmonary neural pathogenesis of acute and chronic lung injury, inhalational toxicology and pharmacology, and establishment of respiratory disease models. He also has more than 10 years of involvement in epidemiologic and experimental investigations of airborne particulate pollution in China. He is the principal investigator (PI) for multiple current grant and research projects characterizing the pathogeneses of ARDS, as well as a co-PI or co-investigator for several research projects evaluating hydrocarbon, cigarette smoke, and ambient particle exposure. He received his M.D. and M.P.H. in environmental and occupational health from China Medical University, Shenyang, China, in 1982 and 1988, respectively.
Nina N. Sun is currently a senior research specialist at the University of Arizona BIO5 Institute in Tucson, Arizona. She was an assistant research scientist in the Department of Pediatrics at the University of Arizona College of Medicine from 2000 to 2006. She also worked as a research associate for the Department of Physiology at the University of Texas Medical Branch in Galveston from 1998 to 1999, where she concentrated on molecular biology studies. She has more than 23 years of hands-on experience in interdisciplinary research projects. She received her B.S. in biochemistry at Peking University, Beijing, China, in 1986 and a master’s degree in Molecular Nutrition at the University of Arizona, Tucson, Arizona, in 1998.
Cynthia D. Fastje is a research technician at the University of Arizona Children’s Research Center, Tucson, Arizona. She received a B.S. from the University of Arizona in psychology in 1986. She joined the Lung Injury Laboratory in the Pediatrics Department of the University of Arizona College of Medicine as a volunteer in 2002, worked as a laboratory assistant from 2003 to 2005, and as a research technician from 2005 until the present. Her research interest is in identifying the etiology of leukemia, and currently she is specifically focused on characterizing the effect that prenatal exposure to tungsten has on the post-natal immune response to a viral agent.
Mark L. Witten is a research professor and director of the Joan B. and Donald Diamond Lung Injury Laboratory in the Department of Pediatrics at the University of Arizona College of Medicine, Tucson, Arizona. He received his Ph.D. in physiology and exercise physiology at Indiana University in 1983. He has been involved for more than 20 years in studies of environmental pollutants, including jet fuel emissions, cigarette smoke, fire smoke, and airborne particles. He has more than 200 publications including conference abstracts and book chapters in toxicology, environmental health, and lung physiology.
R. Clark Lantz is a professor and associate head of the Department of Cell Biology and Anatomy at the University of Arizona. He is also currently deputy director of the Southwest Environmental Health Science Center, a National Institute of Environmental Health Sciences Center of Excellence at the University of Arizona. He received his B.S. in physics from Juniata College in 1970 and his doctorate in physiology and biophysics from West Virginia University in 1975. After postdoctoral research fellowships at Rockefeller University and Emory University, he took a position as assistant professor of anatomy at West Virginia University in 1981. In 1987, he moved to the University of Arizona, where he holds his current academic position. Over the past 30 years, he has concentrated his research in the area of pulmonary toxicology.
Bao Lu is an assistant professor of pediatrics at the Harvard Medical School Children’s Hospital, Boston, Massachusetts. He received a B.S. at Guangdong Medical College in 1977 and an M.D. at Sun Yat-Sen University of Medical Sciences in China in 1987. After his postdoctoral training in the Pulmonary Division of the Harvard Medical School Children’s Hospital (1990–1991), he was a member of Craig Gerard’s research group. His current research activities involve NIH-supported projects in the biology and biochemistry of the endopeptidase neprilysin; the molecular pathology of chronic lung disease; asthma, airway inflammation, and beta chemokine receptors; and the roles of PI3K gamma in host defense.
Duane L. Sherrill is a professor of biostatistics and associate dean for research at the Mel and Enid Zuckerman College of Public Health, University of Arizona. He received a B.S. in mathematics at the Metropolitan State College in 1976 and an M.S. and Ph.D. in biometrics at the University of Colorado Health Science Center in 1982 and 1987, respectively. His research interests include the development of measurement and analytical techniques for estimating the spatial relationships between lung function and diseases.
Craig J. Gerard is a professor of pediatrics, chief and director of research at the Division of Respiratory Diseases, Children’s Hospital, Harvard Medical School, Boston, Massachusetts. His research focuses on the molecular mechanisms, including tachykinins and chemokines, of leukocyte trafficking in host defense and inflammatory responses. He has used both mouse genetic models (Nep-null mice) and in vitro transfection and expression analyses to understand the complex balance between defense and untoward inflammation associated with these mediators and receptors. He received a B.S. at the University of Connecticut in 1976, an M.S. and Ph.D. in chemistry at the University of California, San Diego, California, in 1981, and an M.D. at the Bowman Gray School of Medicine, Wake Forest University, Winston-Salem, North Carolina, in 1985. He received postdoctoral training at the Harvard Medical School, Boston, Massachusetts, in 1988. He has published more than 154 journal articles.
Jefferey L. Burgess is a professor and director of the Community, Environment and Policy Division at the University of Arizona Mel and Enid Zuckerman College of Public Health in Tucson, Arizona. His research includes respiratory toxicology in firefighters and smoke inhalation victims; reduction of mining-related injuries and exposures; environmental arsenic exposure; and hazardous materials exposures, including in methamphetamine laboratories. He is the principal investigator at the Arizona Center for Public Health Preparedness, which is funded by the Centers for Disease Control and Prevention, as well as for multiple research projects evaluating fire smoke, mining injury surveillance, and arsenic exposure. He received his M.D. at the University of Washington in 1988, an M.S. in toxicology/industrial hygiene at the University of Arizona in 1993, and an M.P.H. at the University of Washington in 1996.
APPENDIX AVAILABLE ON THE WEB
The following material may be obtained from HEI’s Web site, www.healtheffects.org:
Appendix A
Uptake of DEP into Human BEAS-2B Cells Observed Using Transmission Electron Microscopy.
CRITIQUE Health Review Committee
Research Report 159, Role of Neprilysin in Airway Inflammation Induced by Diesel Exhaust Emissions, S.S. Wong et al.
INTRODUCTION
Diesel engines emit less carbon dioxide (CO2*) and carbon monoxide (CO) than gasoline engines, but until recently, they have released higher amounts of particulate matter (PM) and nitrogen oxides (NOx). Concerns about the health effects of diesel exhaust emissions (DEE) and the particulate component of these emissions (diesel exhaust particles, or DEP) have resulted in the funding of numerous epidemiologic and toxicologic studies (reviewed in HEI 2007).
HEI periodically issues a request for preliminary applications (RFPA) for novel research on the health effects of air pollutants derived from motor vehicle emissions. In response to RFPA 05–3, “Health Effects of Air Pollution,” issued in 2005, Dr. Simon Wong of the University of Arizona and colleagues submitted an application for a study titled “The Molecular Effects of Diesel Exhaust Particulates on Respiratory Neutral Endopeptidase.” Dr. Wong proposed to study whether the exposure of human airways (in vivo and in vitro) to diesel exhaust affects the expression or activity of neprilysin (also known as neutral endopeptidase, or NEP), an enzyme that degrades multiple vasoactive peptides in the airways (discussed in more detail in the next section). Dr. Wong hypothesized that components of diesel exhaust downregulate the function or expression of NEP in the airways and that this may lead to disorders in airway function; he proposed to evaluate the effects of DEP on airway epithelial cells in vitro and of DEE in human volunteers.
Previously, HEI had funded a one-year pilot study by Dr. Wong and colleagues, in which they had obtained preliminary data on the effects of DEP on NEP expression and activity in airway epithelial cells in vitro (Wong et al. 2007 pilot study). The HEI Research Committee considered the proposed study a logical extension of the pilot study and invited Dr. Wong to submit a full application. After reviewing the application, the Committee suggested that Dr. Wong include in vivo studies in an animal model to complement the proposed human studies. Dr. Wong submitted a revised application that included experiments in mice, and the Research Committee recommended the study for funding.
SCIENTIFIC BACKGROUND
NEPRILYSIN AND ITS FUNCTION
NEP is a widely distributed endopeptidase expressed on the outer surface of several cell types in a variety of tissues and organs (Ronco et al. 1988). For example, in the lung it is expressed on the surface of cells including epithelium, endothelium, nerve cells, macrophages, and neutrophils. In the airways, the extracellular portion of NEP cleaves a wide range of physiologically important vasoactive peptides that include substance P (SP), bradykinin, and endothelin-1, -2, and -3 (Koehne et al. 1998). NEP substrates such as bombesin and endothelin-1 also induce cell migration (Sumitomo et al. 2000). Inside the cell, NEP is involved in many signaling pathways through interactions with the tumor suppressor gene PTEN, the activation of several kinases (focal adhesion kinase, phosphatidylinositol-3, and Rho), and degradation of phosphokinase C delta.
Because of NEP’s ability to catabolize vasoactive peptides that affect airway function, there have been a number of studies investigating changes in NEP expression in several airway conditions in humans as well as animal models. Older studies suggested that NEP levels are elevated in the serum of patients with adult respiratory distress syndrome and cardiogenic pulmonary edema, and in a subset of patients with chronic obstructive pulmonary disease (COPD) (Johnson et al. 1985). However, NEP levels were not different in the serum or bronchoalveolar lavage (BAL) fluid of asthma patients compared with those of healthy individuals (van der Velden et al. 1999). At the time the current study was funded, the role of NEP in human inflammatory lung conditions such as COPD and asthma was uncertain (Barnes 2001).
In animal studies, pharmacologic inhibition of NEP activity or genetic deletion of Nep resulted in enhanced injury in an elastase-induced mouse lung injury model (Day et al. 2005). Decreases in NEP expression or activity have also been noted in a variety of laboratory animals exposed to factors such as cigarette smoke, allergens, and respiratory irritants (Di Maria et al. 1998). In a rat hypoxia model, animals deprived of oxygen had significantly reduced NEP expression in the lungs and increased vascular leak (Carpenter and Stenmark 2001).
Before the current study, Wong and colleagues had conducted several studies of the effects of exposure to different types of air pollutants on NEP in animals. In one study, they exposed rats via nose-only inhalation to DEE (specifically, to either 669 or 35.3 µg/m3 of PM) for 4 hr/d, 5 d/wk, for 3 weeks. They found that exposed rats had an approximately 30% decrease in NEP activity in lung tissue, as well as airway inflammatory responses and changes in SP levels and SP signaling (Wong et al. 2003; Witten et al. 2005). In another study (Wong et al. 2004), rats were exposed through the nose only to fire smoke (1.24 ppm CO2, 2.0 ppm NOx, and 99.5 mg/L PM) for 20 minutes as a model of acute respiratory distress. Wong and colleagues reported increases in endogenous SP levels and decreases in NEP activity and expression in lungs of rats from 1 to 24 hours after the exposure.
It is also of note that NEP expression is altered in many neoplasias — downregulated in some (e.g., prostate and small-cell lung tumors [Cohen et al. 1996; Sumitomo et al. 2004]), but upregulated in others (e.g., the progression of colorectal tumors and diffuse large B-cell lymphomas [Bai et al. 2003, Fujimoto et al. 2005]).
In summary, these results indicate that changes in NEP activity or expression are found in a variety of lung and airway conditions, especially those involving injury, but that these changes do not appear to be specific or causal.
EFFECTS OF CONTROLLED EXPOSURES TO DEE AND ITS COMPONENTS
Emissions from diesel engines are important components of urban air pollution and contribute significantly to traffic-derived ambient PM concentrations (HEI Panel on the Health Effects of Traffic-Related Air Pollution 2010). Increasingly stringent emissions standards, as well as advances in engine technology, have resulted in decreasing concentrations of PM and gaseous pollutants in the emissions from diesel engines. However, PM and NOx generated from older engines still in use (both on-road and off-road) remain a public health concern. Additionally, engine type and age affect the levels of PM and other pollutants emitted, with older engines contributing larger amounts of PM and NOx than newer engines.
Diesel emissions are a complex mixture of particulate and gaseous components. Diesel exhaust particles are composed of a carbon core to which nitrate, sulfate, metals, and organic compounds adhere. Thus, DEP is often characterized by the level of elemental carbon (EC) and the presence of multiple polycyclic aromatic hydrocarbons. The principal gaseous components are CO2, nitrogen and sulfur compounds (including nitrogen dioxide [NO2]), and low-molecular-weight hydrocarbons.
Some studies in humans have suggested that administration of DEP enhances features characteristic of an allergic pattern of immune response. For example, administering 300 µg of DEP into the noses of healthy and asthmatic individuals enhanced the total number of immunoglobulin E–secreting B cells in the upper airways (Diaz-Sanchez et al. 1994), and administering DEP in combination with ragweed allergen enhanced the production of messenger RNA (mRNA) specific for interleukin (IL)-4 and IL-13 — cytokines associated with the development of an allergic response (Diaz-Sanchez et al. 1997). Studies in rodents (e.g., Fujimaki et al. 1997; Takano et al. 1997) have reported that the administration of DEP with allergens such as ovalbumin enhanced key features of the allergic response.
However, the effects on airway responses and pulmonary function resulting from short-term controlled exposure to DEE of either healthy or asthmatic subjects have been less clear-cut. Salvi and colleagues (1999, 2000) reported some changes in immunologic and inflammatory endpoints in healthy and asthmatic volunteers exposed to DEE (300 µg/m3 PM); lung function in healthy volunteers was not affected, however (Salvi et al. 1999; Nightingale et al. 2000). Nordenhäll and colleagues (2001) found no changes in airway inflammatory markers, except in cytokine IL-6 (a molecule that is “proinflammatory” — i.e., activates the inflammatory response), in 14 asthmatic human subjects exposed to DEE (300 µg/m3 PM) in an exposure chamber for 1 hour. They found changes in some measures of airway and pulmonary function (specifically, increased airway hyperresponsiveness and airway resistance) but no change in forced vital capacity (FVC) or forced expiratory volume in one second (FEV1).
Zhang and colleagues evaluated asthmatic volunteers who walked for 1 to 2 hours at two sites in Central London: a street on which a high proportion of diesel vehicles travelled (Oxford Street) and a control site less than a mile away with far less traffic (Hyde Park). Compared with walking in Hyde Park, walking on Oxford Street resulted in mild changes in some markers of airway inflammation and respiratory parameters (a 3–4% decrease in FEV1 and FVC), but no changes in self-reported symptoms (McCreanor et al. 2007; Zhang et al. 2009).
In summary, although the administration of DEP — either to the upper airways of humans or via instillation in rodents — enhances features of the allergic-type immune response, controlled exposures to DEE in healthy and asthmatic humans appear to have some effects on markers of inflammation in the airways, but little effect on pulmonary function. Because Wong and colleagues obtained preliminary data to suggest that exposure to DEP decreased NEP mRNA levels in BEAS-2B cells (a transformed human airway epithelial cell line) (Wong et al. 2007 pilot study), they proposed to investigate further the connection between exposure to diesel exhaust and changes in NEP expression and activity in airway cells.
TECHNICAL EVALUATION
SPECIFIC AIMS
The goals of Dr. Wong’s study were to evaluate (a) whether exposure to diesel exhaust components in vivo in mice and humans and in vitro in a human cell line affected the activity or expression of NEP, and (b) whether a decrease in NEP expression affected responses induced by diesel exhaust. Wong and colleagues hypothesized that exposure to diesel exhaust would decrease the activity or expression of NEP in airway cells, and that decreased expression of NEP would enhance responses induced by diesel exhaust.
Their specific aims and general approaches were as follows:
To examine the role of NEP in DEP-induced inflammatory injury using Nep-intact and Nep-null mice. The investigators evaluated whether airway inflammatory responses after the instillation of DEP were greater in mice genetically deficient in Nep (Nep-null mice) than responses in wild-type, control mice.
To examine which components of DEP are associated with NEP downregulation in vitro. Wong and colleagues evaluated whether untreated DEP, DEP treated to remove certain components, or a control particle affected the expression of NEP in the transformed human airway epithelial cell line BEAS-2B.
To determine the molecular impact of DEP exposure and decreased NEP expression on airway epithelial cells’ gene expression in vitro, using a combination of RNA interference (RNAi) and microarray approaches. The investigators assessed the effects of exposure to DEP on gene expression in two sets of BEAS-2B cells: control cells, which express NEP, and cells in which NEP expression had been decreased — “knocked down” — by treating BEAS-2B cells with an siRNA specific for NEP.
To evaluate the effects on NEP activity of human exposure to DEE. Wong and colleagues measured NEP activity and markers of inflammation in the airways of volunteers who had been exposed to DEE in a controlled mining scenario.
STUDY DESIGN AND METHODS
Key features of the study design are summarized in Critique Table 1 and in the following sections.
Diesel Exhaust, Control Particles, Emissions, and Exposures
DEP
In Aims 1, 2, and 3 the investigators evaluated the effects of resuspended DEP in the form of SRM 2975 particles from the National Institute of Standards and Technology (NIST), a set of particles originally generated by a diesel-powered industrial forklift. Physical and chemical characteristics of SRM 2975, including particle size distribution and concentration of selected polycyclic aromatic hydrocarbons, are shown in the Investigators’ Report (IR) Table 4.
In Aim 2, the investigators treated DEP with the chelator Chelex-100, which preferentially removes divalent cations, particularly transition metals (“cDEP”), or dichloromethane to strip all components except for the carbonaceous core (“sDEP”).
Control Particles
In Aim 2, the investigators also evaluated the effects of a second particle, SRM 1649a, obtained from NIST, as a control for the DEP (SRM 2975). The physical and chemical characteristics of SRM 1649a, described as a “standard urban dust,” are also shown in IR Table 4.
Emissions
In Aim 4, DEE were generated from a 1984 Jarvis Clark JS-220 load-haul-dump vehicle commonly used in mining. The vehicle was equipped with a 2-cubicyard bucket and an 82-hp Deutz F6L-912W diesel engine fitted with a catalytic converter.
Exposures to DEP or DEE
In Aim 1, resuspended particles were instilled intratracheally in mice. In Aims 2 and 3 resuspended particles were incubated with the cell line BEAS-2B. In Aim 2, uptake of SRM 2975 particles into BEAS-2B cells was assessed by transmission electron microscopy. In reviewing the study, HEI’s Review Committee thought these uptake studies were ancillary to the main part of the study on biologic effects; thus these experiments are described in IR Appendix A (available on the Web at www.healtheffects.org).
Critique Table 1.
Study Designa
| Specific Aim |
Test System | Exposures: Concentration and Duration |
Endpoints (Timing of Post-Exposure Measurement) |
|---|---|---|---|
| 1 | NEP knockout and control (wild-type) mice | Intratracheally instilled with 0, 10, or 100 µg resuspended DEP (SRM 2975) | NEP and cytokine levels in BAL fluid, NEP expression in lung (in wild-type only), and airway epithelial cell proliferation (day 7) |
| 2 | BEAS-2B (transformed epithelial human cell line) | Treated for 24 hr with 0, 1, or 10 µg/cm2 particles: DEP (SRM 2975), cDEP, sDEP, control (SRM 1649a) | NEP mRNA levels (24 hr) |
| 3 | BEAS-2B human cells: untreated or treated with NEP-specific siRNA to knock down NEP expression | Treated for 24 hr with 0, 10, or 40 µg/cm2 DEP (SRM 2975) | RNA expression, measured by microarray and RT-PCR, of multiple genes (and molecular pathways) affected by either DEP treatment or NEP knockdown (24 hr) |
| 4 | 11 Healthy human volunteers (ages 19–33 yr) | Single inhalation exposure to DEE. Individual exposure concentrations ranged from 0.09 to 1.80 mg/m3 EC and 56 to 134 min duration | NEP activity, and differential cell count and protein content in induced sputum (baseline pre-exposure and 1 hr post-exposure [at least 1 wk after baseline]) |
cDEP indicates SRM 2975 treated with the chelator Chelex-100; sDEP indicates SRM 2975 treated with dichloromethane; RT-PCR indicates real-time polymerase chain reaction.
In Aim 4, 11 human volunteers were exposed to DEE at the University of Arizona’s San Xavier Underground Mining Laboratory, an underground facility in which the effects of mining can be studied (www.mge.arizona.edu/templates/standard/index.php?ID=31). The original exposure study (Burgess et al. 2007) was led by a collaborator on the current study, Dr. Jefferey Burgess. The investigators expressed the exposure concentration in terms of EC, which had been collected on a quartz filter during the exposure. Individual exposures to DEE ranged in concentration from 0.09 to 1.80 mg/m3 of EC and in duration from 56 to 134 minutes.
Biologic Endpoints
Inflammatory Markers
In the experiment supporting Aim 1, at 7 days after instillation of DEP or control, the investigators measured levels of the cytokines IL-1[3, IL-6, and IL-10 in BAL fluid, and BAL total and differential cell counts. The DEP effects on the proliferation of airway epithelial cells were assessed by evaluating the number of cells that had incorporated 5-bromo-2'-deoxyuridine (5-BrdU) injected intraperitoneally 24 hours before euthanasia.
In Aim 4, Wong and colleagues measured changes in differential cell count and protein content in the induced sputum of 11 healthy individuals (see IR Table 2 for details) exposed to DEE. Each subject underwent two sputum inductions: the first on a nonexposure day to establish a baseline, and the second— at least 1 week later — 1 hour after the DEE exposure.
NEP Expression: Levels and Activity of Protein in Tissue, and Levels of mRNA
In Aim 1, the investigators assessed levels of NEP protein in the lung tissue extracts of wild-type mice by enzyme-linked immunosorbent assay (ELISA). Expression of NEP protein in the lungs of these mice was localized by immunohistochemistry using light microscopy. In Aim 2, NEP mRNA levels in BEAS-2B cells were measured after a 24-hour incubation period with the different types of particles. In Aim 4, cell-free NEP activity in induced sputum of human volunteers was measured spectrophotometrically.
To assess gene expression changes by microarray analysis in Aim 3, the investigators extracted total RNA from the following:
BEAS-2B cells exposed for 24 hours to DEP (0, 10, or 40 µg/cm2 SRM 2975). This experiment was to determine which genes were affected by exposure to DEP.
BEAS-2B cells transfected with an siRNA specific for NEP and subsequently exposed to DEP (0, 10, or 40 µg/cm2 SRM 2975). This experiment was to determine which genes were affected by exposure to DEP in a cell with decreased expression of NEP. The investigators established that NEP siRNA transfection decreased expression of the NEP protein by about 63% compared with untreated cells or cells transfected with a control siRNA (IR Figure 5).
The investigators organized gene expression changes into major functional categories and potential biologic pathways, using different types of software described in the IR. To determine whether changes in expression in the gene microarray assay corresponded to changes in RNA levels, the investigators used an alternative technique — real-time polymerase chain reaction (RT-PCR) — to evaluate some genes whose expression was found to differ from control cells by microarray.
RESULTS
Aim 1: Effects of DEP in Mice
Airway Inflammatory Responses
Instillation of either 10 or 100 µg of DEP (SRM 2975) resulted in increases in several markers of the inflammatory response in the BAL fluid and cells of both Nep-null and wild-type mice measured 7 days after the treatment (IR Figure 1 and Table 3). At the same doses of DEP, increases in total, macrophage, and epithelial cell counts, as well as IL-6 levels, were greater in the Nep-null than in the wild-type mice. The higher, but not the lower, dose of DEP increased IL-1[3 and IL-10 levels in Nep-null mice but not in wild-type mice.
NEP Protein Level
Instillation of DEP reduced NEP protein level in lung tissue by approximately 10% at 10 µg and 50% at 100 µg (IR Figure 2). The investigators showed that DEP instillation reduced NEP protein expression in lung sections, concluding that the reduction in NEP expression was localized to small airway epithelial cells (IR Figure 3). In the absence of DEP, NEP was expressed on macrophages and alveolar type II cells, as well as on epithelial cells.
Epithelial Cell Proliferation
Exposure to high-dose DEP did not result in increased epithelial cell proliferation in either wild-type or Nep-null mice.
Aim 2: DEP Effects on NEP mRNA Expression in BEAS-2B Cells In Vitro
DEP (SRM 2975 particles) at either 1 or 10 µg/cm2 decreased expression of NEP mRNA in BEAS-2B cells by approximately 45% compared with control cells after a 24-hour culture period (IR Figure 4). Similarly, cDEP (DEP with transition metals removed) decreased expression of NEP mRNA by 45% at 10 µg/cm2 (but not at 1 µg/cm2). On the other hand, sDEP (DEP stripped down to the carbonaceous core) did not affect NEP mRNA levels significantly at doses of either 1 or 10 µg/cm2. As a comparison, standard urban dust (SRM 1649a) decreased NEP mRNA expression by approximately 60% at both levels (1 and 10 µg/cm2).
Aim 3: Effects of DEP Exposure on Gene Expression in Control BEAS-2B Cells and BEAS-2B Cells with Decreased NEP Expression In Vitro
Genes Associated with DEP Exposure
Exposure to DEP upregulated the expression of 151 genes and downregulated the expression of 59 genes (IR Tables 6 and 7) in multiple intracellular pathways (IR Figure 10). Upregulated genes included the metabolic enzymes cytochrome P450 CYP1A1 and CYP1B1, cytokines IL6 and IL8, chemokine ligand 1 and 2 (CCL1 and CCL2), death inducer-obliterator 1 (DIDO1), heat shock 70kDa protein 1B (HSPA1B), aquaporin 3 (AQP3), and early growth response 2 and 3 (EGR2 and EGR3). Downregulated genes included phosphoinositide-3-kinase (PIK3C2A), phosphoribosylglycinamide formyltransferase (GART), and threonyl-tRNA synthetase (TARS).
Genes Associated with NEP Knockdown
The investigators showed that transfecting a NEP-specific siRNA into BEAS-2B cells decreased expression of NEP protein approximately 63% compared with control cells. In BEAS-2B cells with reduced NEP expression, the investigators identified 31 genes with the greatest changes in expression (17 upregulated and 14 downregulated) compared with control, mock-infected cells (IR Table 5 and Figure 7). The genes included IL6, IL8, and epidermal growth factor receptor, and pathways that involved DNA/protein binding, calcium channel activity, cytokine signaling, cellular monooxygenase, guanosine triphosphatase (GTPase), and protein kinases.
Of the 5 genes whose expression was upregulated in the microarray and selected for further investigation of expression by RT-PCR, only 3 (specifically, IL6, IL8, and PTGS2) showed increased expression by the RT-PCR technique (IR Figure 8).
Aim 4: DEE Effects on Airway Endpoints in Human Volunteers
In induced sputum evaluated 1 hour after exposure to DEE, the total number of cells increased approximately 2.5-fold (mostly macrophages, but neutrophil numbers also increased concomitantly), and epithelial cell numbers increased approximately 4.5-fold (IR Table 8). Total protein level was not affected.
The group mean of the NEP activity in induced sputum after DEE exposure increased 31% over baseline (IR Table 9). Exposure to DEE — calculated as the product of the EC concentration during the exposure and the duration of exposure — was highly correlated with NEP activity in sputum after the exposure period (IR Figure 11).
HEI REVIEW COMMITTEE EVALUATION OF THE STUDY
In its independent evaluation of the study, the HEI Review Committee considered that Wong and colleagues had made a comprehensive attempt to explore the role of NEP in response to exposure to components of DEE, using an appropriate animal model (an Nep-null mouse), human studies, and in vitro models employing a human cell line. The Committee thought that the study design and statistical analyses for the diverse sets of experiments were appropriate. The human exposure study, however, had the limitation that the investigators used samples and data that had already been collected in an earlier study, and so only a limited number of assays could be performed.
Key results were that airway inflammatory effects in response to DEP instillation (i.e., increases in BAL fluid of the numbers of macrophage and epithelial cells and the levels of cytokines that these cells synthesize) were greater in mice genetically deficient in Nep than in wild-type mice that express Nep. In addition, the fact that DEP instillation (at 100 µg) into wild-type mice resulted in a decrease in NEP protein expression both in lung tissue extracts and in airway epithelial cells and macrophages suggests at first glance that exposure to DEP reduces NEP expression in the lung. Other important findings were that exposure of human volunteers to a high concentration of DEE via inhalation resulted in airway inflammatory effects measured in induced sputum — in particular increased numbers of macrophages and epithelial cells and, in some participants at least, increased NEP activity. However, this finding is at odds with the investigators’ hypothesis for the study, that exposure to DEE would decrease NEP levels in the airways.
The Committee noted that responses in the mouse lung were measured 7 days postexposure to DEP, whereas responses in the exposed human volunteers were measured at 1 hour postexposure. Thus, making comparisons between the human responses to DEE and mouse responses to DEP is challenging, since the investigators did not establish an optimal time for measuring effects. Nonetheless, the Committee thought that the study’s human and mouse findings could be interpreted in a consistent fashion— that is, that the observed diesel-exhaust–induced decreases in NEP levels in lung tissue are the result of an injury response in the airways. As a consequence, airway epithelial cells are shed and macrophage numbers are increased, and hence both changes are measurable in either induced sputum (human) or BAL fluid (mouse). Since both epithelial cells and macrophages express NEP, this would explain the increase in NEP activity in the induced sputum of some of the human volunteers. The investigators did not measure NEP levels in the BAL fluid of DEP-exposed, NEP-expressing (i.e., wild-type) mice, so they do not know whether the decrease in NEP expression in the mouse lung is accompanied by an increase in NEP expression or activity in the BAL fluid.
The finding that mice genetically lacking Nep showed greater inflammatory response than wild-type mice after exposure to DEP suggests that in this model NEP plays some role in damping down inflammatory responses to agents introduced into the airways. This supports one of the hypotheses of the study, namely, that decreased expression of NEP would enhance responses induced by diesel exhaust. Because the Nep-null mice exposed to DEP synthesized more IL-6, as seen in BAL fluid, than did wild-type mice, the investigators not unreasonably interpret these findings as suggesting that the loss of NEP resulted in enhanced production of this proinflammatory cytokine. This enhancement of IL-6 synthesis was also supported by the results of the gene expression studies supporting Aims 2 and 3. How NEP expression and IL-6 synthesis may be linked was not part of the current study. It is also likely that, in the absence of NEP, levels of neuropeptides such as SP may also increase, which may also play a role in enhancing inflammatory effects, as Wong and colleagues have previously suggested (Wong et al. 2003; Witten et al. 2005).
Wong and colleagues’ findings of heightened effects in Nep-null mice after challenges to the airways are consistent with other findings in these mice. For example, Dempsey and colleagues (2009) found that, after the induction of chronic hypoxia, Nep-null mice showed increases in pulmonary hypertension, proliferation of smooth muscle cells from pulmonary arteries, and muscularization of vessels compared with wild-type, control mice. Dempsey and colleagues (2009) also found that in hypoxic wild-type mice, NEP expression was decreased early in distal pulmonary arteries, the site of prominent vascular remodeling. In follow-up studies, these investigators found that COPD patients had decreased NEP expression in the alveolar walls and distal vessels of their lungs and that pulmonary arterial smooth muscle cells exposed to cigarette smoke extract or hypoxia showed decreased expression of NEP protein and mRNA levels (Wick et al. 2011).
In this report Wong and colleagues speculate that changes in NEP in sputum may be a useful early marker of diesel-exhaust–associated responses in humans. The Review Committee noted that the DEP and DEE exposures used in the current study were unlikely to reflect the properties of emissions generated by the current fleet of diesel-powered, on- or off-road vehicles, but recognized that the diesel exhaust exposures used were intended to offer a proof of principle. Nonetheless, the Committee held that changes in NEP activity or levels are unlikely to be useful human biomarkers of exposure to diesel exhaust for two reasons. First, in the limited sample of 11 volunteers, only 5 showed changes greater than 15% in NEP activity in induced sputum in response to DEE, and only after exposure to a high level of DEE (PM in the milligram/m3 range). Second, the observed effects on NEP activity are not specific to diesel exhaust exposures: The results of Aim 2 in the current study show that standard urban dust particles, which contain lower levels of EC and thus presumably lower levels of DEE components, decrease NEP mRNA levels in BEAS-2B cells as effectively as DEP. In addition, Wong and colleagues have published elsewhere that the same volunteers who were exposed to DEE in the current study were also exposed separately to high levels of particles in “overshot mucking”— a blasting process in mining (Wong et al. 2010). Overshot mucking exposures contain little or no diesel exhaust components, but the correlation between exposure (concentration × duration) to overshot mucking particles and NEP activity was very similar to the correlation between exposure to DEE and NEP activity (Wong et al. 2010).
Regarding the correlation between DEE exposure and NEP activity in sputum that the investigators reported in the current study (IR Figure 11) and their recent paper (Wong et al. 2010, Figure 1), the Committee noted that it was with NEP activity measured after the exposure period (i.e., not corrected for an individual’s baseline NEP activity). The Committee thought it would have been more convincing to show a correlation between DEE exposure and the difference in NEP activity before and after the exposure period (i.e., show the change in NEP activity resulting from the DEE exposure). The investigators have indicated that they had evaluated that correlation but had not found it to be significant (S. Wong, personal communication, October 2010).
The investigators’ in vitro studies indicated that organic components of particles, rather than metals, were likely to be important in the downregulation of NEP expression in BEAS-2B cells. Because both standard urban dust particles and DEP contain organic components such as polycyclic aromatic hydrocarbons, this may explain why both sets of particles were equally effective at decreasing NEP mRNA levels in these cells.
Based on preliminary in vitro data from their pilot study, Wong and colleagues hypothesized that exposure to DEP would affect the proliferation of airway epithelial cells in mice. No effects were found, however, in either wild-type or Nep-null mice. Thus, the investigators could not find evidence to support a role for DEP in affecting the proliferation of airway epithelial cells that either did or did not express NEP.
The investigators provided some useful information about changes in gene expression after exposure to DEP in a transformed airway epithelial cell line. The Committee thought that the investigators’ use of NEP knockdown by a specific siRNA was an innovative way to approach the role that NEP may play in gene expression changes in response to DEP. The studies also provided information about which genes’ expression in a transformed airway epithelial cell line may be affected by exposure to DEP; the results may be helpful in trying to determine what pathways are involved in the lungs’ response to DEP and how NEP may play a role.
These findings will need to be confirmed or refuted in future studies. However, the Committee noted that some questions remain about the interpretation of the gene expression studies (supporting Aims 2 and 3). First, BEAS-2B cells exposed to DEP did not show significant changes in NEP mRNA levels by microarray (see last line of IR Table 7), but did in a different assay (RT-PCR; see IR Figure 4). Second, of the 5 genes the investigators selected to submit to RT-PCR testing in order to confirm the microarray findings, confirmatory results were shown for only 3 (IR Figure 8). If extended to a broader set of genes, this would suggest that 2 out of 5 genes (i.e., 40%) identified by the microarray method may be false positives.
SUMMARY AND CONCLUSIONS
Wong and colleagues explored the role of NEP in response to DEE exposure, using an appropriate animal model (an Nep-null mouse), a human study, and in vitro models. The investigators hypothesized that (a) DEE exposure would decrease NEP expression in airway cells and that (b) decreased expression of NEP would enhance responses induced by diesel exhaust. Key findings were that (1) DEP instillation into the airways of mice resulted in stronger airway inflammatory effects (i.e., increases in macrophage and epithelial cell numbers and in levels of cytokines that these cells synthesize, as observed in BAL fluid) in mice genetically deficient in Nep than in mice expressing Nep; and (2) DEP instillation into wild-type mice resulted in a 50% decrease in NEP expression in lung tissue and particularly in airway epithelial cells. The HEI Review Committee agreed with the investigators that these findings suggest that expression of NEP in some way damps down inflammatory responses in the airways, supporting one of the hypotheses for the study.
Other important findings were that exposure of human volunteers to a high concentration of DEE via inhalation resulted in airway inflammatory effects measured in the induced sputum, particularly increased numbers of macrophages, neutrophils, and epithelial cells and, in some participants at least, increased NEP activity. However, the Committee thought that this last result was at odds with the investigators’ other hypothesis for the study— that exposure to DEE would decrease NEP levels in the airways.
The Review Committee noted that it was challenging to compare the responses in mouse airways with those in human airways because the exposures were of different levels, durations, and routes of exposure, and the effects were measured at different times after the exposures. Nonetheless, the Committee thought that the study’s human and mouse diesel-exhaust–associated findings could be interpreted in a consistent fashion — that is, that the observed changes in NEP levels result from a response to injury in the airways, measured as the shedding of airway epithelial cells and as an increase in macrophage numbers in the induced sputum (human) or BAL fluid (mouse). Since both epithelial cells and macrophages express NEP, this could explain the increase in NEP activity found in the induced sputum of some of the human volunteers.
The in vitro studies in the transformed human airway epithelial cell line also provided some useful information about DEP constituents that affect NEP expression — namely, that removing metals did not change NEP expression but removing organic components did. The studies also provided information about which genes’ expression in a transformed airway epithelial cell line may be affected by exposure to DEP. These data may help in determining the pathways involved in the airway response to DEP and any possible NEP role in that response.
The investigators speculated in the report that changes in NEP in sputum might be a useful early marker of DEE-induced injury in humans. However, the Committee thought that changes in NEP activity or levels in the airways are unlikely to be useful biomarkers of exposure to diesel components, because the observed effects in airway cells were not specific to diesel exposures and because baseline levels of airway NEP activity differed markedly in different people. Thus, although changes in NEP function and activity have been noted in airway conditions, particularly after injury, the role of NEP is still not resolved.
ACKNOWLEDGMENTS
The Health Review Committee thanks the ad hoc reviewers for their help in evaluating the scientific merit of the Investigators’ Report. The Committee is also grateful to Maria Costantini for her oversight of the study, to Geoffrey Sunshine and Morgan Younkin for their assistance in preparing its Critique, to Hilary Selby Polk for science editing of this Report and its Critique, and to Suzanne Gabriel, Fred Howe, Bernard Jacobson, and Flannery Carey McDermott for their roles in preparing this Research Report for publication.
REFERENCES
- Bai M, Agnantis NJ, Skyrlas A, Tsanou E, Kamina S, Galani V, Kanavaros P. Increased expression of the bcl6 and CD10 proteins is associated with increased apoptosis and proliferation in diffuse large B-cell lymphomas. Mod Pathol. 2003;16:471–480. doi: 10.1097/01.MP.0000067684.78221.6E. [DOI] [PubMed] [Google Scholar]
- Barnes PJ. Neurogenic inflammation in the airways. Respir Physiol. 2001;125:145–154. doi: 10.1016/s0034-5687(00)00210-3. [DOI] [PubMed] [Google Scholar]
- Burgess JL, Fleming JE, Mulenga EM, Josyula A, Hysong TA, Joggerst PJ, Kurzius-Spencer M, Miller HB. Acute changes in sputum IL-10 following underground exposure to diesel exhaust. Clin Toxicol. 2007;45:255–260. doi: 10.1080/15563650601072142. [DOI] [PubMed] [Google Scholar]
- Carpenter TC, Stenmark KR. Hypoxia decreases lung neprilysin expression and increases pulmonary vascular leak. Am J Physiol Lung Cell Mol Physiol. 2001;281:L941–L948. doi: 10.1152/ajplung.2001.281.4.L941. [DOI] [PubMed] [Google Scholar]
- Cohen AJ, Bunn PA, Franklin W, Magill-Solc C, Hartmann C, Helfrich B, Gilman L, Folkvord J, Helm K, Miller YE. Neutral endopeptidase: Variable expression in human lung, inactivation in lung cancer, and modulation of peptide-induced calcium flux. Cancer Res. 1996;56:831–839. [PubMed] [Google Scholar]
- Day AL, Wick E, Jordan TH, Jaffray CE, Bunnett NW, Grady EF, Kirkwood KS. Neutral endopeptidase determines the severity of pancreatitis-associated lung injury. J Surg Res. 2005;128:21–27. doi: 10.1016/j.jss.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Dempsey EC, Wick MJ, Karoor V, Barr EJ, Tallman DW, Wehling CA, Walchak SJ, Laudi S, Le M, Oka M, Majka S, Cool CD, Fagan KA, Klemm DJ, Hersh LB, Gerard NP, Gerard C, Miller YE. Neprilysin null mice develop exaggerated pulmonary vascular remodeling in response to chronic hypoxia. Am J Pathol. 2009;174:782–796. doi: 10.2353/ajpath.2009.080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maria GU, Bellofiore S, Geppetti P. Regulation of airway neurogenic inflammation by neutral endopeptidase. Eur Respir J. 1998;12:1454–1462. doi: 10.1183/09031936.98.12061454. [DOI] [PubMed] [Google Scholar]
- Diaz-Sanchez D, Dotson AR, Takenaka H, Saxon A. Diesel exhaust particles induce local IgE production in vivo and alter the pattern of IgE messenger RNA isoforms. J Clin Invest. 1994;94:1417–1425. doi: 10.1172/JCI117478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Sanchez D, Tsien A, Fleming J, Saxon A. Combined diesel exhaust particulate and ragweed allergen challenge markedly enhances human in vivo nasal ragweed-specific IgE and skews cytokine production to a T helper cell 2-type pattern. J Immunol. 1997;158:2406–2413. [PubMed] [Google Scholar]
- Fujimaki H, Saneyoshi K, Shiraishi F, Imai T, Endo T. Inhalation of diesel exhaust enhances antigen-specific IgE antibody production in mice. Toxicology. 1997;116:227–233. doi: 10.1016/s0300-483x(96)03539-1. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Nakanishi Y, Sekine S, Yoshimura K, Akasu T, Moriya Y, Shimoda T. CD10 expression in colorectal carcinoma correlates with liver metastasis. Dis Colon Rectum. 2005;48:1883–1889. doi: 10.1007/s10350-005-0141-6. [DOI] [PubMed] [Google Scholar]
- HEI Air Toxics Review Panel. Mobile-Source Air Toxics: A Critical Review of the Literature on Exposure and Health Effects. Boston, MA: Health Effects Institute; 2007. Special Report 16. [Google Scholar]
- HEI Panel on the Health Effects of Traffic-Related Air Pollution. Traffic-Related Air Pollution: A Critical Review of the Literature on Emissions, Exposure, and Health Effects. Boston, MA: Health Effects Institute; 2010. Special Report 17. [Google Scholar]
- Johnson AR, Coalson JJ, Ashton J, Larumbide M, Erdös EG. Neutral endopeptidase in serum samples from patients with adult respiratory distress syndrome: Comparison with angiotensin-converting enzyme. Am Rev Respir Dis. 1985;132:1262–1267. doi: 10.1164/arrd.1985.132.6.1262. [DOI] [PubMed] [Google Scholar]
- Koehne P, Schäper C, Graf K, Kunkel G. Neutral endopeptidase 24.11: Its physiologic and possibly pathophysiologic role in inflammation with special effect on respiratory inflammation. Allergy. 1998;53:1023–1042. doi: 10.1111/j.1398-9995.1998.tb03812.x. [DOI] [PubMed] [Google Scholar]
- McCreanor J, Cullinan P, Nieuwenhuijsen MJ, Stewart-Evans J, Malliarou E, Jarup L, Harrington R, Svartengren M, Han IK, Ohman-Strickland P, Chung KF, Zhang J. Respiratory effects of exposure to diesel traffic in persons with asthma. N Engl J Med. 2007;357:2348–2358. doi: 10.1056/NEJMoa071535. [DOI] [PubMed] [Google Scholar]
- Nightingale JA, Maggs R, Cullinan P, Donnelly LE, Rogers DF, Kinnersley R, Chung KF, Barnes PJ, Ashmore M, Newman-Taylor A. Airway inflammation after controlled exposure to diesel exhaust particulates. Am J Respir Crit Care Med. 2000;162:161–166. doi: 10.1164/ajrccm.162.1.9908092. [DOI] [PubMed] [Google Scholar]
- Nordenhäll C, Pourazar J, Ledin MC, Levin JO, Sandström T, Adelroth E. Diesel exhaust enhances airway responsiveness in asthmatic subjects. Eur Respir J. 2001;17:909–915. doi: 10.1183/09031936.01.17509090. [DOI] [PubMed] [Google Scholar]
- Ronco P, Pollard H, Galceran M, Delauche M, Schwartz JC, Verroust P. Distribution of enkephalinase (membrane metalloendopeptidase, E.C. 3.4.24.11) in rat organs: Detection using a monoclonal antibody. Lab Invest. 1988;58:210–217. [PubMed] [Google Scholar]
- Salvi S, Blomberg A, Rudell B, Kelly F, Sandström T, Holgate ST, Frew A. Acute inflammatory responses in the airways and peripheral blood after short-term exposure to diesel exhaust in healthy human volunteers. Am J Respir Crit Care Med. 1999;159:702–709. doi: 10.1164/ajrccm.159.3.9709083. [DOI] [PubMed] [Google Scholar]
- Salvi SS, Nordenhall C, Blomberg A, Rudell B, Pourazar J, Kelly FJ, Wilson S, Sandström T, Holgate ST, Frew AJ. Acute exposure to diesel exhaust increases IL-8 and GRO-alpha production in healthy human airways. Am J Respir Crit Care Med. 2000;161:550–557. doi: 10.1164/ajrccm.161.2.9905052. [DOI] [PubMed] [Google Scholar]
- Sumitomo M, Iwase A, Zheng R, Navarro D, Kaminetzky D, Shen R, Georgescu MM, Nanus DM. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- Sumitomo M, Shen R, Walburg M, Dai J, Geng Y, Navarro D, Boileau G, Papandreou CN, Giancotti FG, Knudsen B, Nanus DM. Neutral endopeptidase inhibits prostate cancer cell migration by blocking focal adhesion kinase signaling. J Clin Invest. 2000;106:1399–1407. doi: 10.1172/JCI10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano H, Yoshikawa T, Ichinose T, Miyabara Y, Imaoka K, Sagai M. Diesel exhaust particles enhance antigen-induced airway inflammation and local cytokine expression in mice. Am J Respir Crit Care Med. 1997;156:36–42. doi: 10.1164/ajrccm.156.1.9610054. [DOI] [PubMed] [Google Scholar]
- van Der Velden VH, Naber BA, Van Hal PT, Overbeek SE, Hoogsteden HC, Versnel MA. Peptidase activities in serum and bronchoalveolar lavage fluid from allergic asthmatics: Comparison with healthy non-smokers and smokers and effects of inhaled glucocorticoids. Clin Exp Allergy. 1999;29:813–823. doi: 10.1046/j.1365-2222.1999.00550.x. [DOI] [PubMed] [Google Scholar]
- Wick MJ, Buesing EJ, Wehling CA, Loomis ZL, Cool CD, Zamora MR, Miller YE, Colgan SP, Hersh LB, Voelkel NF, Dempsey EC. Decreased neprilysin and pulmonary vascular remodeling in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:330–340. doi: 10.1164/rccm.201002-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten M, Wong S, Sun N, Keith I, Kweon C-B, Foster D, Schaer J, Sherrill D. Neurogenic Responses in Rat Lungs After Nose-Only Exposure to Diesel Exhaust. Boston, MA: Health Effects Institute; 2005. Research Report 128. [PubMed] [Google Scholar]
- Wong SS, Sun NN, Keith I, Kweon CB, Foster DE, Schauer JJ, Witten ML. Tachykinin substance P signaling involved in diesel exhaust–induced bronchopulmonary neurogenic inflammation in rats. Arch Toxicol. 2003;77:638–650. doi: 10.1007/s00204-003-0485-4. [DOI] [PubMed] [Google Scholar]
- Wong SS, Sun NN, Lantz RC, Witten ML. Substance P and neutral endopeptidase in development of acute respiratory distress syndrome following fire smoke inhalation. Am J Physiol Lung Cell Mol Physiol. 2004;287:L859–L866. doi: 10.1152/ajplung.00388.2003. [DOI] [PubMed] [Google Scholar]
- Wong SS, Sun NN, Miller HB, Witten ML, Burgess JL. Acute changes in sputum collected from exposed human subjects in mining conditions. Inhal Toxicol. 2010;22:479–485. doi: 10.3109/08958370903464185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SS, Sun NN, Witten ML, Lu B, Sherrill DL, Gerard C. The molecular effects of diesel exhaust particles on respiratory neutral endopeptidase. Boston, MA 02110, USA: 101 Federal Street, Suite 500; 2007. Unpublished pilot study. Available on request from Health Effects Institute. [Google Scholar]
- Zhang J, McCreanor JE, Cullinan P, Chung KF, Ohman-Strickland P, Han I-K, Järup L, Nieuwenhuijsen MJ. Health Effects of Real-World Exposure to Diesel Exhaust in Persons with Asthma. Boston, MA: Health Effects Institute; 2009. Research Report 138. [PubMed] [Google Scholar]
Footnotes
OTHER PUBLICATIONS RESULTING FROM THIS RESEARCH
Wong SS, Sun NN, Hugh BM, Witten ML, Burgess JL. 2010. Acute changes in sputum collected from exposed human subjects in mining conditions. Inhal Toxicol 22: 479–485.
Dr. Simon S. Wong’s 2-year study, “The Molecular Effects of Diesel Exhaust Particulates on Respiratory Neutral Endopeptidase,” began in September 2007. Total expenditures were $190,289. The draft Investigators’ Report from Wong and colleagues was received for review in January 2010. A revised report, received in May 2010, was accepted for publication in June 2010. During the review process, the HEI Health Review Committee and the investigators had the opportunity to exchange comments and to clarify issues in both the Investigators’ Report and the Review Committee’s Critique.
This document has not been reviewed by public or private party institutions, including those that support the Health Effects Institute; therefore, it may not reflect the views of these parties, and no endorsements by them should be inferred.
A list of abbreviations and other terms appears at the end of the Investigators’ Report.
REFERENCES
- Bamford HA, Bezabeh DZ, Schantz MM, Wise SA, Baker JE. Determination and comparison of nitrated-polycyclic aromatic hydrocarbons measured in air and diesel particulate reference materials. Chemosphere. 2003;50:575–587. doi: 10.1016/s0045-6535(02)00667-7. [DOI] [PubMed] [Google Scholar]
- Baraniuk JN, Ohkubo K, Kwon OJ, Mak J, Ali M, Davies R, Twort C, Kaliner M, Letarte M, Barnes PJ. Localization of neutral endopeptidase (NEP) mRNA in human bronchi. Eur Respir J. 1995;8:1458–1464. [PubMed] [Google Scholar]
- Bateson TF, Schwartz J. Who is sensitive to the effects of particulate air pollution on mortality? A case-crossover analysis of effect modifiers. Epidemiology. 2004;15:143–149. doi: 10.1097/01.ede.0000112210.68754.fa. [DOI] [PubMed] [Google Scholar]
- Baulig A, Garlatti M, Bonvallot V, Marchand A, Barouki R, Marano F, Baeza-Squiban A. Involvement of reactive oxygen species in the metabolic pathways triggered by diesel exhaust particles in human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L671–L679. doi: 10.1152/ajplung.00419.2002. [DOI] [PubMed] [Google Scholar]
- Bayram H, Devalia JL, Sapsford RJ, Ohtoshi T, Miyabara Y, Sagai M, Davies RJ. The effect of diesel exhaust particles on cell function and release of inflammatory mediators from human bronchial epithelial cells in vitro. Am J Respir Cell Mol Biol. 1998;18:441–448. doi: 10.1165/ajrcmb.18.3.2882. [DOI] [PubMed] [Google Scholar]
- Becker S, Soukup JM. Decreased CD11b expression, phagocytosis, and oxidative burst in urban particulate pollution-exposed human monocytes and alveolar macrophages. J Toxicol Environ Health A. 1998;55:455–477. doi: 10.1080/009841098158278. [DOI] [PubMed] [Google Scholar]
- Boland S, Baeza-Squiban A, Fournier T, Houcine O, Gendron MC, Chevrier M, Jouvenot G, Coste A, Aubier M, Marano F. Diesel exhaust particles are taken up by human airway epithelial cells in vitro and alter cytokine production. Am J Physiol. 1999;275:L604–L613. doi: 10.1152/ajplung.1999.276.4.L604. [DOI] [PubMed] [Google Scholar]
- Boland S, Bonvallot V, Fournier T, Baeza-Squiban A, Aubier M, Marano F. Mechanisms of GM-CSF increase by diesel exhaust particles in human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2000;278:L25–L32. doi: 10.1152/ajplung.2000.278.1.L25. [DOI] [PubMed] [Google Scholar]
- Borson DB. Roles of neutral endopeptidase in airways. Am J Physiol. 1991;260:L212–L225. doi: 10.1152/ajplung.1991.260.4.L212. [DOI] [PubMed] [Google Scholar]
- Bowden DH. The alveolar macrophage. Environ Health Perspect. 1984;55:327–341. doi: 10.1289/ehp.8455327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic CR, Lu B, Höpken UE, Gerard C, Gerard NP. Neurogenic amplification of immune complex inflammation. Science. 1996;273:1722–1725. doi: 10.1126/science.273.5282.1722. [DOI] [PubMed] [Google Scholar]
- Brain JD. Mechanisms, measurement, and significance of lung macrophage function. Environ Health Perspect. 1992;97:5–10. doi: 10.1289/ehp.92975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess JL, Fleming JE, Mulenga EM, Josyula A, Hysong TA, Joggerst PJ, Kurzius-Spencer M, Miller HB. Acute changes in sputum IL-10 after underground exposure to diesel exhaust. Clinical Toxicology. 2007;45:255–260. doi: 10.1080/15563650601072142. [DOI] [PubMed] [Google Scholar]
- Campen MJ, Babu NS, Helms GA, Pett S, Wernly J, Mehran R, McDonald JD. Nonparticulate components of diesel exhaust promote constriction in coronary arteries from ApoE−/− mice. Toxicol Sci. 2005;88:95–102. doi: 10.1093/toxsci/kfi283. [DOI] [PubMed] [Google Scholar]
- Cao D, Tal TL, Graves LM, Gilmour I, Linak W, Reed W, Bromberg PA, Samet JM. Diesel exhaust particulate-induced activation of Stat3 requires activities of EGFR and Src in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2007;292:L422–L429. doi: 10.1152/ajplung.00204.2006. [DOI] [PubMed] [Google Scholar]
- Churg A, Brauer M, del Carmen Avila-Casado M, Fortoul TI, Wright JL. Chronic exposure to high levels of particulate air pollution and small airway remodeling. Environ Health Perspect. 2003;111:714–718. doi: 10.1289/ehp.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AJ, Bunn PA, Franklin W, Magill-Solc C, Hartmann C, Helfrich B, Gilman L, Folkvord J, Helm K, Miller YE. Neutral endopeptidase: Variable expression in human lung, inactivation in lung cancer, and modulation of peptide-induced calcium flux. Cancer Res. 1996;56:831–839. [PubMed] [Google Scholar]
- Cohen AJ, Franklin WA, Magill C, Sorenson J, Miller YE. Low neutral endopeptidase levels in bronchoalveolar lavage fluid of lung cancer patients. Am J Respir Crit Care Med. 1999;159:907–910. doi: 10.1164/ajrccm.159.3.9806062. [DOI] [PubMed] [Google Scholar]
- Cohen HJ, Borak J, Hall T, Sirianni G, Chemerynski S. Exposure of miners to diesel exhaust particulates in underground nonmetal mines. AIHA J. 2002;63:651–658. doi: 10.1080/15428110208984753. [DOI] [PubMed] [Google Scholar]
- D’Adamio L, Shipp MA, Masteller EL, Reinherz EL. Organization of the gene encoding common acute lymphoblastic leukemia antigen (neutral endopeptidase 24.11): Multiple miniexons and separate 5' untranslated regions. Proc Natl Acad Sci U S A. 1989;86:7103–7107. doi: 10.1073/pnas.86.18.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AL, Wick E, Jordan TH, Jaffray CE, Bunnett NW, Grady EF, Kirkwood KS. Neutral endopeptidase determines the severity of pancreatitis-associated lung injury. J Surg Res. 2005;128:21–27. doi: 10.1016/j.jss.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Di Maria GU, Bellofiore S, Geppetti P. Regulation of airway neurogenic inflammation by neutral endopeptidase. Eur Respir J. 1998;12:1454–1462. doi: 10.1183/09031936.98.12061454. [DOI] [PubMed] [Google Scholar]
- Diaz-Sanchez D, Dotson AR, Takenaka H, Saxon A. Diesel exhaust particles induce local IgE production in vivo and alter the pattern of IgE messenger RNA isoforms. J Clin Invest. 1994;94:1417–1425. doi: 10.1172/JCI117478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanović R, Sterk PJ, Fahy JV, Hargreave FE. Standardised methodology of sputum induction and processing. Eur Respir J. 2002;37:1s–2s. doi: 10.1183/09031936.02.00000102. [DOI] [PubMed] [Google Scholar]
- Dobbin K, Simon R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics. 2005;6:27–38. doi: 10.1093/biostatistics/kxh015. [DOI] [PubMed] [Google Scholar]
- Dockery DW, Pope CA., III Acute respiratory effects of particulate air pollution. Annu Rev Public Health. 1994;15:107–132. doi: 10.1146/annurev.pu.15.050194.000543. [DOI] [PubMed] [Google Scholar]
- Dockery DW, Schwartz J, Spengler JD. Air pollution and daily mortality: Associations with particulates and acid aerosols. Environ Res. 1992;59:362–373. doi: 10.1016/s0013-9351(05)80042-8. [DOI] [PubMed] [Google Scholar]
- Dusser DJ, Djokic TD, Borson DB, Nadel JA. Cigarette smoke induces bronchoconstrictor hyperresponsiveness to substance P and inactivates airway neutral endopeptidase in the guinea pig: Possible role of free radicals. J Clin Invest. 1989;84:900–906. doi: 10.1172/JCI114251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amouri SS, Zhu H, Yu J, Marr R, Verma IM, Kindy MS. Neprilysin: An enzyme candidate to slow the progression of Alzheimer’s disease. Am J Pathol. 2008;172:1342–1354. doi: 10.2353/ajpath.2008.070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy O, Hammond H, Senechal S, Pestel J, Tonnek AB, Wallaert B. Synergistic effect of diesel organic extracts and allergen Der p1 on the release of chemokines by peripheral blood mononuclear cells from allergic subjects: Involvement of the MAP kinase pathway. Am J Respir Cell Mol Biol. 2000;23:247–254. doi: 10.1165/ajrcmb.23.2.4116. [DOI] [PubMed] [Google Scholar]
- Fine JM, Gordon T, Sheppard D. Epithelium removal alters responsiveness of guinea pig trachea to substance P. J Appl Physiol. 1989;66:232–237. doi: 10.1152/jappl.1989.66.1.232. [DOI] [PubMed] [Google Scholar]
- Ganju RK, Shpektor RG, Brenner DG, Shipp MA. CD10/neutral endopeptidase 24.11 is phosphorylated by casein kinase II and coassociates with other phosphoproteins including the lyn src-related kinase. Blood. 1996;88:4159–4165. [PubMed] [Google Scholar]
- Ganju RK, Sunday M, Tsarwhas DG, Card A, Shipp MA. CD10/NEP in non-small cell lung carcinomas: Relationship to cellular proliferation. J Clin Invest. 1994;94:1784–1791. doi: 10.1172/JCI117526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Jr, Sioutas C, Linn WS. Controlled Exposures of Healthy and Asthmatic Volunteers to Concentrated Ambient Particles in Metropolitan Los Angeles. Boston, MA: Health Effects Institute; 2003. Research Report 118. [PubMed] [Google Scholar]
- Hashimoto S, Gon Y, Takeshita I, Matsumoto K, Jibiki I, Takizawa H, Kudoh S, Horie T. Diesel exhaust particles activate p38 MAP kinase to produce interleukin 8 and RANTES by human bronchial epithelial cells and N-acetylcysteine attenuates p38 MAP kinase activation. Am J Respir Crit Care Med. 2000;161:280–285. doi: 10.1164/ajrccm.161.1.9904110. [DOI] [PubMed] [Google Scholar]
- HEI Diesel Working Group. Diesel Exhaust: Critical Analysis of Emissions, Exposure, and Health Effects. Cambridge, MA: Health Effects Institute; 1995. Special Report. [Google Scholar]
- Hislop AA, Wharton J, Allen KM, Polak JM, Haworth SG. Immunohistochemical localization of peptide-containing nerves in human airways: Age-related changes. Am J Respir Cell Mol Biol. 1990;3:191–198. [PubMed] [Google Scholar]
- Iijima-Ando K, Hearn SA, Granger L, Shenton C, Gatt A, Chiang HC, Hakker I, Zhong Y, Iijima K. Overexpression of neprilysin reduces alzheimer amyloid-beta42 (Abeta42)-induced neuron loss and intraneuronal Abeta42 deposits but causes a reduction in cAMP-responsive element-binding protein-mediated transcription, age-dependent axon pathology, and premature death in Drosophila. J Biol Chem. 2008;283:19066–19076. doi: 10.1074/jbc.M710509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Shen R, Navarro D, Nanus DM. Direct binding of neutral endopeptidase 24.11 to ezrin/radixin/moesin (ERM) proteins competes with the interaction of CD44 with ERM proteins. J Biol Chem. 2004;279:11898–11905. doi: 10.1074/jbc.M212737200. [DOI] [PubMed] [Google Scholar]
- Johnson AR, Asthon J, Schulz WW, Erdös EG. Neutral metalloendopeptidase in human lung tissue and cultured cells. Am Rev Resp Dis. 1985a;132:564–568. doi: 10.1164/arrd.1985.132.3.564. [DOI] [PubMed] [Google Scholar]
- Johnson AR, Coalson JJ, Ashton J, Larumbide M, Erdös EG. Neutral endopeptidase in serum samples from patients with adult respiratory distress syndrome: Comparison with angiotensin-converting enzyme. Am Rev Respir Dis. 1985b;132:1262–1267. doi: 10.1164/arrd.1985.132.6.1262. [DOI] [PubMed] [Google Scholar]
- Joos GF, Germonpre PR, Pauwels RA. Role of tachykinins in asthma. Allergy. 2000;55:321–337. doi: 10.1034/j.1398-9995.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- Kreyling WG. Intracellular particle dissolution in alveolar macrophages. Environ Health Perspect. 1992;97:121–126. doi: 10.1289/ehp.9297121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni N, Pierse N, Rushton L, Grigg J. Carbon in airway macrophages and lung function in children. N Engl J Med. 2006;355:21–30. doi: 10.1056/NEJMoa052972. [DOI] [PubMed] [Google Scholar]
- Li N, Wang M, Oberley TD, Sempf JM, Nel AE. Comparison of the pro-oxidative and proinflammatory effects of organic diesel exhaust particle chemicals in bronchial epithelial cells and macrophages. J Immunol. 2002;169:4531–4541. doi: 10.4049/jimmunol.169.8.4531. [DOI] [PubMed] [Google Scholar]
- Lilly CM, Kobzik L, Hall AE, Drazen JM. Effects of chronic airway inflammation on the activity and enzymatic inactivation of neuropeptides in guinea pig lungs. J Clin Invest. 1994;93:2667–2674. doi: 10.1172/JCI117280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M, Vaughan JH, Carson DA. Effect of neuropeptides on production of inflammatory cytokines by human monocytes. Science. 1988;241:1218–1221. doi: 10.1126/science.2457950. [DOI] [PubMed] [Google Scholar]
- Lu B, Figini M, Emanueli C, Geppetti P, Grady EF, Gerard NP, Ansell J, Payan DG, Gerard C, Bunnett N. The control of microvascular permeability and blood pressure by neutral endopeptidase. Nat Med. 1997;3:904–907. doi: 10.1038/nm0897-904. [DOI] [PubMed] [Google Scholar]
- Lu B, Gerard NP, Kolakowski LF, Jr, Finco O, Carroll MC, Gerard C. Neutral endopeptidase modulates septic shock. Ann N Y Acad Sci. 1996;780:156–163. doi: 10.1111/j.1749-6632.1996.tb15119.x. [DOI] [PubMed] [Google Scholar]
- Lundberg JM, Alving K, Karlsson JA, Matran R, Nilsson G. Sensory neuropeptide involvement in animal models of airway irritation and of allergen-evoked asthma. Am Rev Respir Dis. 1991;143:1429–1430. doi: 10.1164/ajrccm/143.6.1429. [DOI] [PubMed] [Google Scholar]
- MacNee W, Donaldson K. Mechanism of lung injury caused by PM10 and ultrafine particles with special reference to COPD. Eur Respir J Suppl. 2003;40:47s–51s. doi: 10.1183/09031936.03.00403203. [DOI] [PubMed] [Google Scholar]
- Madden MC, Dailey LA, Stonehuerner JG, Harris BD. Responses of cultured human airway epithelial cells treated with diesel exhaust extracts will vary with the engine load. J Toxicol Environ Health A. 2003;66:2281–2297. doi: 10.1080/15287390390244292. [DOI] [PubMed] [Google Scholar]
- Martins MA, Shore SA, Gerard NP, Gerard C, Drazen JM. Peptidase modulation of the pulmonary effects of tachykinins in tracheal superfused guinea pig lungs. J Clin Invest. 1990;85:170–176. doi: 10.1172/JCI114408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan RO. Health effects of exposure to diesel exhaust particles. Ann Rev Pharmacol Toxicol. 1987;27:279–300. doi: 10.1146/annurev.pa.27.040187.001431. [DOI] [PubMed] [Google Scholar]
- McCreanor J, Cullinan P, Nieuwenhuijsen MJ, Stewart-Evans J, Malliarou E, Jarup L, Harrington R, Svartengren M, Han IK, Ohman-Strickland P, Chung KF, Zhang J. Respiratory effects of exposure to diesel traffic in persons with asthma. N Engl J Med. 2007;357:2348–2358. doi: 10.1056/NEJMoa071535. [DOI] [PubMed] [Google Scholar]
- Morrow PE. Possible mechanisms to explain dust overloading of the lungs. Fundam Appl Toxicol. 1988;10:369–384. doi: 10.1016/0272-0590(88)90284-9. [DOI] [PubMed] [Google Scholar]
- Mundandhara SD, Becker S, Madden MC. Effects of diesel exhaust particles on human alveolar macrophage ability to secrete inflammatory mediators in response to lipopolysaccharide. Toxicol In Vitro. 2006;20:614–624. doi: 10.1016/j.tiv.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Nadel JA. Neutral endopeptidase modulates neurogenic inflammation. Eur Respir J. 1991;4:745–754. [PubMed] [Google Scholar]
- Nanus DM. Neutral endopeptidase 24.11 loss in metastatic human prostate cancer contributes to androgen-independent progression. Nat Med. 1998;4:50–57. doi: 10.1038/nm0198-050. [DOI] [PubMed] [Google Scholar]
- National Institute of Standards and Technology Certificate of Analysis. Standard Reference Material (SRM) 2975, Diesel Particulate Matter (Industrial Forklift) Gaithersburg, MD: NIST; 2000. [Google Scholar]
- National Institute of Standards and Technology Certificate of Analysis. Standard Reference Material (SRM) 1649a, Urban Dust. Gaithersburg, MD: NIST; 2001. [Google Scholar]
- Nel A. Air pollution-related illness: Effects of particles. Science. 2005;308:804–806. doi: 10.1126/science.1108752. [DOI] [PubMed] [Google Scholar]
- Nightingale JA, Maggs R, Cullinan P, Donnelly LE, Rogers DF, Kinnersley R, Chung KF, Barnes PJ, Ashmore M, Newman-Taylor A. Airway inflammation after controlled exposure to diesel exhaust particulates. Am J Respir Crit Care Med. 2000;162:161–166. doi: 10.1164/ajrccm.162.1.9908092. [DOI] [PubMed] [Google Scholar]
- Oberdörster G, Ferin J, Gelein R, Soderholm SC, Finkelstein J. Role of the alveolar macrophage in lung injury: Studies with ultrafine particles. Environ Health Perspect. 1992;97:193–199. doi: 10.1289/ehp.97-1519541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RG, Dukes R, Sullivan J, Carter R, Erdös EG, Johnson AR. Function of neutral endopeptidase on the cell membrane of human neutrophils. J Biol Chem. 1988;263:9456–9461. [PubMed] [Google Scholar]
- Pandya RJ, Solomon G, Kinner A, Balmes JR. Diesel exhaust and asthma: Hypotheses and molecular mechanisms of action. Environ Health Perspect. 2002;110:103–112. doi: 10.1289/ehp.02110s1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou CN, Giancotti FG, Knudsen B, Nanus DM. Neutral endopeptidase inhibits prostate cancer cell migration by blocking focal adhesion kinase signaling. J Clin Invest. 2000;106:1399–1407. doi: 10.1172/JCI10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou CN, Usmani B, Geng YP, Bogenrieder T, Freeman RH, Wilk S, Finstad CL, Reuter VE, Powell CT, Scheinberg D, Magill C, Scher HI, Albino AP, Peters A, Dockery DW, Heinrich J, Wichmann HE. Short-term effects of particulate air pollution and respiratory morbidity in asthmatic children. Eur Respir J. 1997;10:872–879. [PubMed] [Google Scholar]
- Peters A, Wichmann HE, Tuch T, Heinrich J, Heyder J. Respiratory effects are associated with the number of ultrafine particles. Am J Respir Crit Care Med. 1997;155:1376–1383. doi: 10.1164/ajrccm.155.4.9105082. [DOI] [PubMed] [Google Scholar]
- Pope CA, III, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: Contribution of the ciliated lineage. Proc Natl Acad Sci U S A. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudell B, Blomberg A, Helleday R, Ledin MC, Lundbäck B, Stjernberg N, Hörstedt P, Sandström T. Bronchoalveolar inflammation after exposure to diesel exhaust: Comparison between unfiltered and particle trap filtered exhaust. Occup Environ Med. 1999;56:527–534. doi: 10.1136/oem.56.8.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S, Blomberg A, Rudell B, Kelly F, Sandstrom T, Holgate ST, Frew A. Acute inflammatory responses in the airways and peripheral blood after short-term exposure to diesel exhaust in healthy human volunteers. Am J Respir Crit Care Med. 1999;159:702–709. doi: 10.1164/ajrccm.159.3.9709083. [DOI] [PubMed] [Google Scholar]
- Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. Fine particulate air pollution and mortality in 20 U.S. cities, 1987–1994. N Engl J Med. 2000a;14(343):1742–1749. doi: 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- Samet JM, Zeger SL, Dominici F, Curriero F, Coursac I, Dockery DW, Schwartz J, Zanobetti A. The National Morbidity, Mortality, and Air Pollution Study. Cambridge, MA: Health Effects Institute; 2000b. Part II. Morbidity and mortality from air pollution in the United States. Research Report 94. [PubMed] [Google Scholar]
- Schwartz J, Laden F, Zanobetti A. The concentration–response relation between PM(2.5) and daily deaths. Environ Health Perspect. 2002;110:1025–1029. doi: 10.1289/ehp.021101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp MA, Richardson NE, Sayre PH. Molecular cloning of the common acute lymphoblastic leukaemia antigen (CALLA) identifies a type II integral membrane protein. Proc Natl Acad Sci U S A. 1988;85:4819–4823. doi: 10.1073/pnas.85.13.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp MA, Tarr GE, Chen CY, Switzer SN, Hersh LB, Stein H, Sunday ME, Reinherz EL. CD10/neutral endopeptidase 24.11 hydrolyzes bombesin-like peptides and regulates the growth of small cell carcinomas of the lung. Proc Natl Acad Sci U S A. 1991;88:10662–10666. doi: 10.1073/pnas.88.23.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleilhac JM, Lafuma C, Porcher JM, Auburtin G, Roques BP. Characterization of a soluble form of neutral endopeptidase-24.11 (EC 3.4.24.11) in human serum: Enhancement of its activity in serum of underground miners exposed to coal dust particles. Eur J Clin Invest. 1996;26:1011–1017. doi: 10.1046/j.1365-2362.1996.2420580.x. [DOI] [PubMed] [Google Scholar]
- Stenfors N, Nordenhall C, Salvi SS, Mudway I, Soderberg M, Blomberg A, Helleday R, Levin JO, Holgate ST, Kelly FJ, Frew AJ, Sandstrom T. Different airway inflammatory responses in asthmatic and healthy humans exposed to diesel. Eur Respir J. 2004;23:82–86. doi: 10.1183/09031936.03.00004603. [DOI] [PubMed] [Google Scholar]
- Sumitomo M, Iwase A, Zheng R, Navarro D, Kaminetzky D, Shen R, Georgescu MM, Nanus DM. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- Sun NN, Wong SS, Keith I, Witten ML. Tachykinin substance P depletion by capsaicin exacerbates inflammatory response to sidestream cigarette smoke in rats. Toxicology. 2004;201:39–50. doi: 10.1016/j.tox.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Sunday ME, Hua J, Torday JS, Reyes B, Shipp MA. CD10/neutral endopeptidase 24.11 in developing human fetal lung: Patterns of expression and modulation of peptide-mediated proliferation. J Clin Invest. 1992;90:2517–2525. doi: 10.1172/JCI116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kikkawa F, Ino K, Nagasaka T, Tamakoshi K, Mizutani S. Imbalance between neutral endopeptidase 24.11 and endothelin-1 expression in human endometrial carcinoma. Oncology. 2001;60:258–267. doi: 10.1159/000055327. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Abe S, Okazaki H, Kohyama T, Sugawara I, Saito Y, Ohtoshi T, Kawasaki S, Desaki M, Nakahara K, Yamamoto K, Matsushima K, Tanaka M, Sagai M, Kudoh S. Diesel exhaust particles upregulate eotaxin gene expression in human bronchial epithelial cells via nuclear factor-kappa B-dependent pathway. Am J Physiol Lung Cell Mol Physiol. 2003;284:L1055–L1062. doi: 10.1152/ajplung.00358.2002. [DOI] [PubMed] [Google Scholar]
- Takizawa H, Ohtoshi T, Kawasaki S, Kohyama T, Desaki M, Kasama T, Kobayashi K, Nakahara K, Yamamoto K, Matsushima K, Kudoh S. Diesel exhaust particles induce NF-kappa B activation in human bronchial epithelial cells in vitro: Importance in cytokine transcription. J Immunol. 1999;162:4705–4711. [PubMed] [Google Scholar]
- Tomoda C, Kushima R, Takeuti E, Mukaisho K, Hattori T, Kitano H. CD10 expression is useful in the diagnosis of follicular carcinoma and follicular variant of papillary thyroid carcinoma. Thyroid. 2003;13:291–295. doi: 10.1089/105072503321582105. [DOI] [PubMed] [Google Scholar]
- Turner CR, Stow RB, Hubbs SJ, Gomes BC, Williams JC. Acrolein increases airway sensitivity to substance P and decreases NEP activity in guinea pigs. J Appl Physiol. 1993;74:1830–1839. doi: 10.1152/jappl.1993.74.4.1830. [DOI] [PubMed] [Google Scholar]
- van Der Velden VH, Naber BA, Van Hal PT, Overbeek SE, Hoogsteden HC, Versnel MA. Peptidase activities in serum and bronchoalveolar lavage fluid from allergic asthmatics — comparison with healthy non-smokers and smokers and effects of inhaled glucocorticoids. Clin Exp Allergy. 1999;29:813–823. doi: 10.1046/j.1365-2222.1999.00550.x. [DOI] [PubMed] [Google Scholar]
- Venn AJ, Lewis SA, Cooper M, Hubbard R, Britton J. Living near a main road and the risk of wheezing illness in children. Am J Respir Crit Care Med. 2001;164:2177–2180. doi: 10.1164/ajrccm.164.12.2106126. [DOI] [PubMed] [Google Scholar]
- Veronesi B, Oortgiesen M. Neurogenic inflammation and particulate matter (PM) air pollutants. Neurotoxicology. 2001;22:795–810. doi: 10.1016/s0161-813x(01)00062-6. [DOI] [PubMed] [Google Scholar]
- Wick MJ, Buesing EJ, Wehling CA, Loomis ZL, Cool CD, Zamora MR, Miller YE, Colgan SP, Hersh LB, Voelkel NF, Dempsey EC. Decreased neprilysin and pulmonary vascular remodeling in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183:330–340. doi: 10.1164/rccm.201002-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten ML, Wong SS, Sun NN, Keith I, Kweon C-B, Foster DE, Schauer JJ. Neurogenic Responses in Rats Lungs after Nose-Only Exposure to Diesel Exhaust. Cambridge, MA: Health Effects Institute; 2005. Research Report 128. [PubMed] [Google Scholar]
- Wong SS, Sun NN, Keith I, Kweon CB, Foster DE, Schauer JJ, Witten ML. Tachykinin substance P signaling involved in diesel exhaust-induced bronchopulmonary neurogenic inflammation in rats. Arch Toxicol. 2003;77:638–650. doi: 10.1007/s00204-003-0485-4. [DOI] [PubMed] [Google Scholar]
- Wong SS, Sun NN, Lantz RC, Witten ML. Substance P and neutral endopeptidase in development of ARDS following fire smoke inhalation. Am J Physiol Lung Cell Mol Physiol. 2004;277:L859–L866. doi: 10.1152/ajplung.00388.2003. [DOI] [PubMed] [Google Scholar]
- Wong SS, Sun NN, Witten ML, Lu B, Sherrill DL, Gerard C. The molecular effects of diesel exhaust particles on respiratory neutral endopeptidase. Boston, MA 02110, USA: 101 Federal Street, Suite 500; 2007. Unpublished pilot study (not peer-reviewed). Available on request from Health Effects Institute. [Google Scholar]
- Wu ZX, Lee LY. Airway hyperresponsiveness induced by chronic exposure to cigarette smoke in guinea pigs: Role of tachykinins. J Appl Physiol. 1999;87:1621–1628. doi: 10.1152/jappl.1999.87.5.1621. [DOI] [PubMed] [Google Scholar]
- Yang HM, Barger MW, Castranova V, Ma JK, Yang JJ, Ma JY. Effects of diesel exhaust particles (DEP), carbon black, and silica on macrophage responses to lipopolysaccharide: Evidence of DEP suppression of macrophage activity. J Toxicol Environ Health A. 1999;58:261–278. doi: 10.1080/009841099157232. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Kleeberger SR, Reddy SP. DEP-induced fra-1 expression correlates with a distinct activation of AP-1-dependent gene transcription in the lung. Am J Physiol Lung Cell Mol Physiol. 2004;286:L427–L436. doi: 10.1152/ajplung.00221.2003. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kobzik L. Effect of concentrated ambient particles on macrophage phagocytosis and killing of Streptococcus pneumoniae. Am J Respir Cell Mol Biol. 2007;36:460–465. doi: 10.1165/rcmb.2006-0293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]