Abstract
Morphological disintegration of neurons is coupled invariably to neural death. In particular, disruption of outer segments of photoreceptor neurons triggers photoreceptor death regardless of the pathological stressors. We show that Ranbp2−/−::Tg-Ranbp2CLDm mice with mutations in SUMO-binding motif (SBM) of cyclophilin-like domain (CLD) of Ranbp2 expressed in a null Ranbp2 background lack untoward effects in photoreceptors in the absence of light-stress. However, compared to wild type photoreceptors, light-stress elicits profound disintegration of outer segments of Ranbp2−/−::Tg-Ranbp2CLDm with paradoxical age-dependent resistance of photoreceptors to death and genotype-independent caspase activation. Ranbp2−/−::Tg-Ranbp2CLDm exhibit photoreceptor death-independent changes in ubiquitin-proteasome system (UPS), but death-dependent increase of ubc9 levels. Hence, insidious functional impairment of SBM of Ranbp2’s CLD promotes neuroprotection and uncoupling of photoreceptor degeneration and death against phototoxicity.
1. Introduction
Morphological disintegration of neurons invariably triggers neural death and is a hallmark feature of neurodegeneration [1–6]. Photoreceptors neurons are the primary photon-capturing neurons of the retina. In particular, the outer segment (OS) structures of photoreceptors undergo degeneration owing to intrinsic (e.g. genetic mutations) or extrinsic (e.g. environmental) pathological stimuli [5, 6]. Light-stress is a powerful and extrinsic deleterious stressor that promotes the dysplasia of the OS of photoreceptors and ultimately photoreceptor death [7–13]. However, the factors, molecular pathways, and mechanisms underpinning the degeneration of photoreceptors and their death are poorly understood and such understanding is needed to develop neuroprotective approaches against risk factors (e.g. light damage) that potentiate photoreceptor degeneration in disease conditions, such as age-related macular degeneration and retinitis pigmentosa [14–20].
The Ranbp2 is a vital, large, mosaic, pleiotropic and cytoplasmic peripheral nucleoporin, which among other functions, controls the nuclear-cytoplasmic trafficking and proteostasis of selective substrates in a cell-type dependent manner [21–27]. Despite the broad expression of Ranbp2, heterogeneous genetic changes in Ranbp2 underlie the insidious or pervasive expressions of diseases or traits affecting restricted cell types or tissues. For example, asymptomatic and selective semi-dominant mutations in RANBP2 cause necrotic encephalopathy upon exposure to selective infectious agents [28–31], whereas haploinsufficiency of Ranbp2 promotes carcinogenesis [24], deficits in glucose metabolism upon glucose challenge [27], MPTP-elicited parkinsonism [32], and neuroprotection of photoreceptor neurons to light damage in mice [10, 33]. However, the modular activities of Ranbp2 that contribute to stress-dependent and cell-type restricted expressions of these pathological traits remain to be defined.
Here, we show that mutations in a purported SUMO-binding motif (SBM) in the cyclophilin-like domain (CLD) of Ranbp2 [34, 35], a domain which associates with subunits of the 19S cap of the proteasome [36, 37], preordains outer segment of photoreceptors to profuse structural disruption upon light-stress, while causing the paradoxical suppression of death of photoreceptors by mechanisms independent of caspases’ activations and overall function of the ubiquitin-proteasome system (UPS).
2. Materials and methods
2.1. Transgenic mice
The generation of Ranbp2+/− (Ranbp2Gt(pGT0pfs)630Wcs/−) mice was previously described [27]. Ranbp2−/− are embryonic lethal [24, 27]. Tg-Ranbp2CLDm mice were generated by BAC recombineering with a BAC construct comprising the complete murine Ranbp2 gene and the I2471K and V2472A mutations in the sequence encoding its CLD as described elsewhere [26]. Ranbp2−/−::Tg-Ranbp2CLDm-HA mice were obtained by intercrossing Ranbp2+/− and Tg-Ranbp2CLDm-HA. Ranbp2−/−::Tg-Ranbp2CLDm-HA mice were pigmented, hemizygous for the transgene allele and they were in a heterozygous Rpe65L450/M450 background. Mice were reared in a pathogen-free transgenic barrier facility at <70 lux in the cages and 12 hr light-dark cycles (6:00 AM – 6:00 PM). Mice were given ad libitum access to water and chow diet 5LJ5 (Purina, Saint Louis, MO). The Institutional Animal Care and Use Committee at Duke University approved the animal protocols and all procedures adhered to the National Academy of Sciences and ARVO guidelines for the Use of Animals in Vision Research.
2.2. Light-exposure treatment
Light experiments were carried out as described elsewhere [26]. Briefly, mice were first dark-adapted for 18 hours. Then, mice pupils were dilated by applying 1% atropine sulfate and 10% phenylephrine hydrochloride (HUB Pharmaceuticals, LLC, Rancho Cucamonga, CA) onto their eyes. Mice were placed individually in white reflective cages with bedding, food, water, and they were exposed to the illuminance of 5,000 lux of continuous white light-emitting diode (LED) by a dimmable LED panel mounted at the top of the cage for 24 hours. Following the light-exposure treatment, mice were placed in the dark for 24 hrs, killed, and the eyeballs were immediately collected and processed for immunohistochemistry or biochemical analyses.
2.3. Semithin Sections and Transmission Electron Microscopy (TEM)
Semithin sections of posterior eyecups were processed for light and TEM of mice as described elsewhere [10, 38]. Briefly, mice were perfused with 2% paraformaldehyde/phosphate buffered saline (PBS); then, eyeballs were fixed with 2% glutaraldehyde-paraformaldehyde, 0.1% cacodylate buffer, pH 7.2, 0.5-μm sections along the vertical meridian were mounted on glass slides and stained with 1% methylene blue. Images were captured with an Axioplan-2 light microscope controlled by Axiovision release 4.6 and coupled to an AxioCam HRc digital camera (Carl Zeiss). For electron microscopy, specimens were postfixed in 2% osmium tetraoxide in 0.1% cacodylate buffer and embedded in Spurr resin. 60-nm-thick sections were cut with Leica Ultracut S (Leica Microsystems); stained with 2% uranyl acetate, 4% lead citrate and imaged with a JEM-1400 transmission electron microscope (JEOL) coupled with an ORIUS 1000CCD camera.
2.4. Antibodies
The following primary antibodies were used for immunohistochemistry: rabbit monoclonal anti-cleaved caspase-3 Asp175 (Cell Signaling), rabbit polyclonal anti-cleaved-caspase-7 Asp353 (Cell Signaling), rabbit anti-cleaved caspase-8 Asp391 (Cell Signaling), rabbit anti-cleaved caspase-9 Asp 330 (Cell Signaling), mouse anti-ubiquitin (Santa Cruz Biotechnology), rabbit anti-hsc70 (ENZO Life Science), rabbit anti-S1 subunit of the 19 S proteasome (Thermo Scientific); rabbit anti-HDAC4 (Santa Cruz Biotechnology); mouse anti-Ubc9 (BD Biosciences), Alexa-conjugated secondary antibodies (408, 488, 568 and Cy5) were from Invitrogen (Carlsbad, CA).
2.5. Immunohistochemistry and confocal microscopy
The immunohistochemistry and confocal microscopy procedures employed were similar to those described elsewhere [26, 38]. Briefly, eyecups were fixed in 2% paraformaldehyde (PFA)/phosphate buffered saline (PBS), washed with 1xPBS and placed sequentially in 5% and 30% sucrose solutions until complete submersion of the eyecups was reached. The eyes were embedded in optimal cutting temperature (OCT) compound and immediately frozen on dry ice and stored at −80°C before sectioning. 12μm-thick cyrosections were collected for immunohistochemistry, blocked with blocking solution (1xPBS, 0.1% Triton X-100, and 10% normal donkey serum) at room temperature overnight followed by treatment with proteinase K (20μg/ml, Promega) for 8 minutes and standard immunostaining protocols. Specimens were viewed under 60x magnification using a Nikon C90i C1 Plus confocal laser-scanning microscope controlled by the EZ-C1.3.10 confocal software (v6.4). Two images were taken from the central and peripheral superior and inferior regions of the eyecup, for a total of six images per section. The central region was defined as the retinal area within a radius of 215μm of the optic nerve head, while the peripheral region was defined as any region outside the central region.
2.6. TUNEL assay
Sections were washed for 30 minutes with PBS, treated with proteinase K for 8 minutes and washed with PBS [38]. Apoptotic retinal neurons were labeled by the TdT-mediated dUTP nick end-labeling (TUNEL) assay (DeadEnd Fluorimetric TUNEL system; Promega) for 1 hour in a 37°C incubation chamber as described by the manufacturer and elsewhere [10, 38]. After washing with PBS for 30 minutes, the specimens were mounted with Fluoromount G (Southern Biotech) and a glass cover slip.
2.7. Morphometric and statistical analysis
Morphometric analyses of immunostained photoreceptors were carried out with Nikon Elements AR software (v4.0). TUNEL+-cell bodies in the outer nuclear layer (ONL) area were counted. Seven 12μm-thick cryosections were analyzed per eye globe. The total number of TUNEL+ and caspase+TUNEL+ cell bodies was counted throughout the entire retinal sections, converted to a percentage with the total number of TUNEL+ nuclei being 100%. Statistical Mann-Whitney U tests were performed with p≤ 0.05 considered significant.
2.8. Immunoblot and ubiquitin-proteasome assays
Retinas were frozen immediately on dry ice after sacrificing mice. The amount of poly-ubiquitylated substrates, diubiquitin, S1, ubc9 and HDAC4 were measured by western blot and densitometric analysis of RIPA-solubilized retinal homogenates resolved in premade 4–15% Criterion (BioRad) SDS-polyacrylamide gels and immunostained with respective antibodies as described elsewhere [26]. Blots were reprobed with hsc70 antibody. Band intensities were quantified with Metamorph v 7.0 (Molecular Devices), integrated density values (idv) of ubiquitylated smears or protein bands were normalized to the background and idv of hsc70. Data were analyzed by the two-tail t-test and a p value of ≤0.05 was considered significant. To measure the proteolytic activities of the 20S proteasome, retinas were re-suspended in 60 μl of homogenization buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, 5mM MgCl2, 2mM ATP,1mM DTT, 0.5mM EDTA and 0.0025% digitonin), incubated on ice for 5 min and then, they underwent standard protocols for 20S proteasome assays as described elsewhere [26]. Briefly, supernatants (3 μg) from retinal extracts were incubated with 100 μM 7-aminomethylcoumarin-conjugated substrates [Suc-LLVY-AMC (Boston Biochem), Ac-RLR-AMC (Boston Biochem) or Ac-nLPnLD-AMC (ENZO Life Science)] in the absence or presence of epoxomicin (20 μM, CalBiochem) for 45 minutes at 37°C. Proteolytic activities were determined by measuring free AMC at excitation/emission/cutoff =380/460/455 with a SpectraMax M5 spectrophotometer (Molecular Devices). Protein concentration for supernatant was measured by BCA method. Data were analyzed by the two-tail t-test and a p value of ≤0.05 was considered significant.
3. Results
3.1. The outer segments of photoreceptors of mice with mutations in SBM of CLD of Ranbp2 are preordained to profuse degeneration by light-stress
Ranbp2 has two internal repeats (IR1 and IR2) and a cyclophilin-like domain (CLD) with low homology to its C-terminal cyclophilin domain (CY) (Fig. 1) [34]. The first internal repeat (IR1) of Ranbp2 partially overlaps with CLD (Fig. 1) [34, 39]. The overlapping region of CLD and IR1 contains a purported SBM, V/I-X-V/I-V/I, and mutations in SBM reduce or abolish in vitro the interaction of SUMOylated RanGAP (SUMO-1-RanGAP) to Ranbp2 [35]. Prior studies showed that IR1 of Ranbp2 is sufficient to bind the E2-conjugating enzyme, ubc9 [39], and that the formation of the Ranbp2-SUMO-1-RanGAP-ubc9 complex induces the formation of a multisubunit E3 ligase in vitro and cell lines [40]. By contrast, CLD of Ranbp2 associates with the S1 (also called Rpn2 in yeast or Psmd1) and other subunits of the 19S cap of the 26S proteasome in cell lines and retinal extracts [36, 37]. Mutations in SBM of CLD selectively relieve the suppression of the degradation of a properly folded and ubiquitylated reporter substrate by ubiquitin-proteasome system (UPS) and without impairing the binding of subunits of the 19S cap of the 26S proteasome to CLD in cell lines [37].
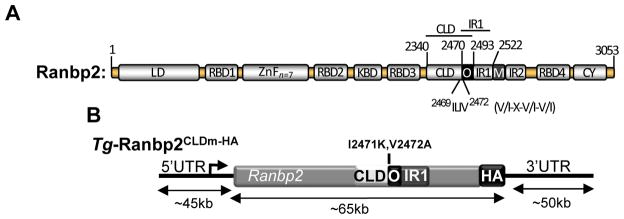
Fig. 1.
(A) Structural domains of Ranbp2. V/I-X-V/I-V/I is the consensus SUMO-binding motif (SBM) in the overlapping region (O) of CLD and IR1 of Ranbp2. Numbering refers to residues of the primary mouse sequence of Ranbp2 and positioning of selective domains and motifs of Ranbp2. Domains are not drawn to scale. (B) Transgenic BAC construct (~160 kb) of Ranbp2, Tg-Ranbp2CLDm-HA, with the mutations, I2471K andV2472A, in the SBM of CLD, and a C-terminal hemagglutinin (HA) tag insertion at the end of the terminal exon encoding the CY domain of Ranbp2. Note drawing not to scale. Legend: LD, leucine-rich domain; RBDn=1–4, Ran GTPase-binding domains, n=1–4; ZnFn =7, zinc finger-rich domains; KBD, kinesin-1-binding domain; CLD, cyclophilin-like domain; IR1 and IR2, internal repeats 1 and 2, respectively; M, middle domain between IR1 and IR2; O, overlapping region between CLD and IR1; CY, cyclophilin domain; UTR, untranslated region.
To interrogate the physiological role(s) of CLD of Ranbp2 and its SBM, we generated Ranbp2−/−::Tg-Ranbp2CLDm-HA mice with the mutations, I2471K and V2472A (CLDm), in SBM of CLD of the Ranbp2 gene in a bacterial artificial chromosome (BAC) and expressed in a null Ranbp2 background (Fig. 1) [26]. Ranbp2−/−::Tg-Ranbp2CLDm-HA mice present similar expression levels and subcellular localization of the transgenic protein, Tg-Ranbp2CLDm-HA, compared to endogenous Ranbp2 [26]. Ranbp2−/−::Tg-Ranbp2CLDm-HA mice reared in cyclic-light and low luminance harbor decreased steady-state levels of SUMO-1-RanGAP and of another substrate, HDAC4, which is prone to sumoylation and degradation by Ranbp2 [41, 42], but these mice do not harbor changes of S1 levels in the retina [26]. Although Ranbp2−/−::Tg-Ranbp2CLDm-HA mice present incomplete penetrance of embryonic lethality, surviving and adult Ranbp2−/−::Tg-Ranbp2CLDm-HA mice do not exhibit overt morphological, developmental or behavioral phenotypes [26].
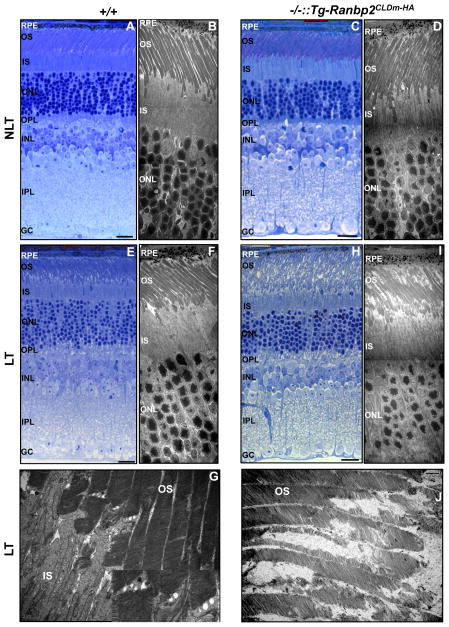
Ranbp2 haploinsufficiency confers neuroprotection to photoreceptors against light damage [10, 33] and light-stress is thought to induce proteotoxicity owing to increased oxidative stress and deregulation of the UPS and protein homeostasis (aka “proteostasis”) [10, 33, 43–46]. Hence, we investigated what domains of Ranbp2 conferred protection of photoreceptors to light-stress by ascertaining the role of CLD of Ranbp2 and its SBM in neuroprotection to phototoxicity. We compared by light and ultrastructural microscopy the overall morphologies of photoreceptors of Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild-type mice reared under cyclic/low light and light-stress conditions. As shown in figure 2, there were no overt differences in retinal morphology and photoreceptor structures by both light and ultrastructural microscopy between Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild-type mice reared under cyclic/low light conditions (Figs. 2A–D). Under light-stress however, the OS of photoreceptors of wild-type mice presented overall well-stacked disks but with the formation of duet vesicles which disrupted focally disk stacking, a feature that is typically observed in OS by phototoxicity (Figs. 2E–G) [10]. By contrast, Ranbp2−/−::Tg-Ranbp2CLDm-HA mice had broad disorganization of the OS of photoreceptors (Fig. 2H) with profuse structural erosion of their disks and some sparing of the rim structures (Figs. 2I, J).
Fig. 2.
Morphological changes of photoreceptor neurons between wild-type and Ranbp2−/−::Tg-Ranbp2CLDm-HA mice reared in the absence and presence of light-stress. There are no visible morphological changes under light (A, C) and ultrastructural microscopy (B, D) of photoreceptors of wild-type (A, B) and Ranbp2−/−::Tg-Ranbp2CLDm-HA mice (C, D) reared under non-light treatment (NLT) at 24 weeks of age (A–D). Under light-treatment (E–J), the OS of photoreceptors of 24-week-old wild-type mice show mild disruption primarily at the base of the OS as shown by light (E) and ultrastructure microscopy (F, G), whereas light (H) and ultrastructure microscopy (I, J) show profuse derangement and erosion of the OS of photoreceptors, respectively, of age-matched Ranbp2−/−::Tg-Ranbp2CLDm-HA mice. Inset picture is magnified dashed box in (G). Legend: OS, outer segments of photoreceptors; IS, inner segments of photoreceptors; ONL, outer nuclear (cell bodies) layer of photoreceptors; OPL, outer plexiform layer; INL, inner nuclear (cell bodies) layer of retinal neurons; IPL, inner plexiform layer; GC, ganglionic neurons; RPE, retinal pigment epithelium; NLT, non-light treatment; LT, light treatment (light-stress); +/+, wild type; −/−::Tg-Ranbp2CLDm-HA, Ranbp2−/−::Tg-Ranbp2CLDm-HA. Scale bars: 20 μm (A, C, E, H), 5 μm (B, D, F, I), 1 μm (G, J).
3.2. Photoreceptors of Ranbp2−/−::Tg-Ranbp2CLDm-HA undergo decreased apoptosis by light stress
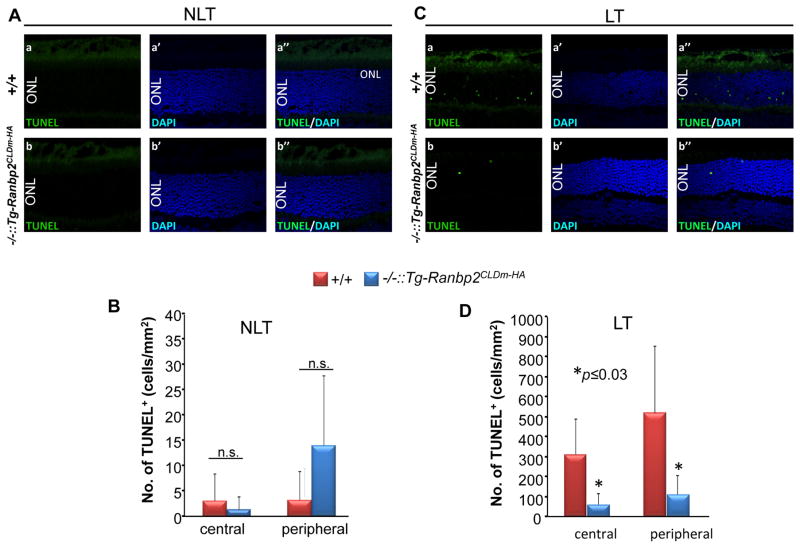
Light-stress elicits strongly the apoptosis of photoreceptors [10]. The effect of phototoxicity in photoreceptor cell death between genotypes was examined by TUNEL assays. We did not observed differences in the development of apoptotic cell bodies of photoreceptors between genotypes at 8 weeks of age in the absence and presence of light stress (data not shown). At 24 weeks of age, the amount of apoptotic photoreceptor cell bodies was very low in mice reared under low light conditions and no differences in cell death were detected between genotypes (Figs. 3A, B). However, light-stress elicited a strong increase of apoptotic cell bodies of photoreceptors in 24-week-old wild-type mice, whereas such increase in cell death was blunted in age-matched Ranbp2−/−::Tg-Ranbp2CLDm-HA mice (Figs. 3C, D).
Fig. 3.
Ranbp2−/−::Tg-Ranbp2CLDm-HA suppresses the development of apoptotic cell bodies by light-stress. The number of apoptotic cell bodies in 24-week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild type mice are scarce in the absence of light-stress and there is no difference between genotypes (A, B). In the presence of light stress (C, D), 24-week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA show strong suppression of TUNEL+-apoptotic cell bodies (arrowheads) of photoreceptors in the central and peripheral regions of the retina compared to age-matched wild type mice. (B) and (D) are quantitative analyses of representative images in (A) and (C), respectively. Retinal sections were counterstained with DAPI. Data shown represent the mean ± S.D., n=5–6. Legend: NLT, non-light treatment; LT, light treatment (light stress); ONL, outer nuclear (cell bodies) layer of photoreceptors; +/+, wild type; −/-::Tg-Ranbp2CLDm-HA, Ranbp2−/−::Tg-Ranbp2CLDm-HA; n.s., non-significant; Scale bar: 20 μm.
3.3. Genotype-independent caspases’ activations in photoreceptors by light-stress
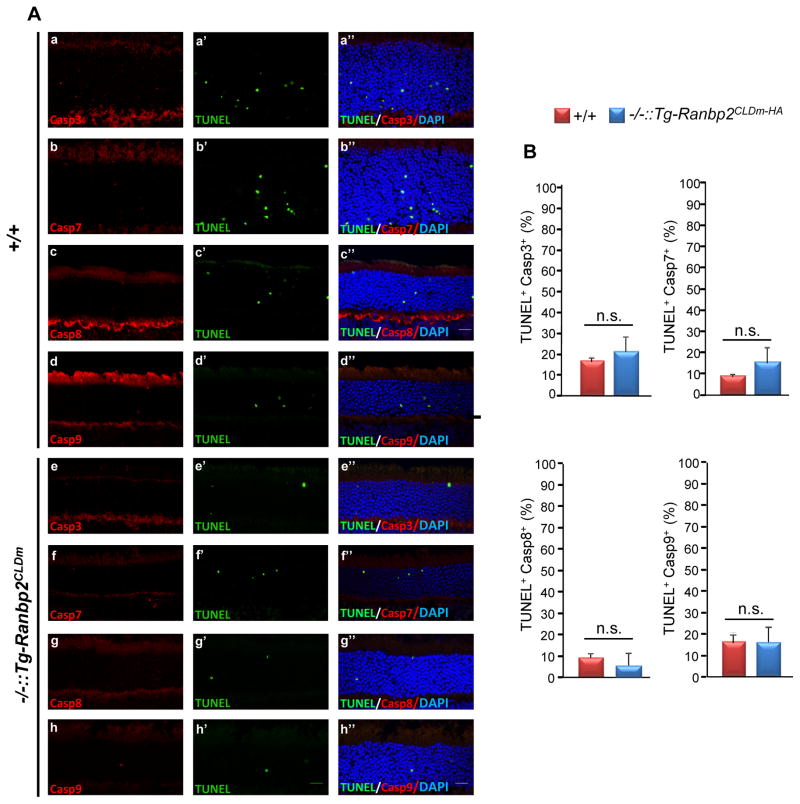
Intrinsic and extrinsic apoptotic stimuli are known to initiate the activation (cleavage) of caspases 9 and 8, respectively, and these promote the activations of downstream effector caspases (e.g. caspases 3, 6, 7) [47]. However, the conservation and modulation of these apoptotic pathways across pathophysiological stimuli and photoreceptor degenerations remain controversial [43, 48–53]. Hence, we extended the analysis of the modulatory role of CLDm of Ranbp2 in cell death by examining the activation of initiator and effector caspases in cell bodies of photoreceptors. Unlike the suppression of light-induced apoptosis in Ranbp2−/−::Tg-Ranbp2CLDm-HA, we found no differences of activated (cleaved) caspases 3, 7, 8 and 9 in apoptotic photoreceptors between Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild-type mice (Figs. 4A, B). Further, all caspase+-cell bodies were TUNEL+, but only a subset of TUNEL+ were caspase+ (Fig. 4A).
Fig. 4.
Apoptotic cell bodies of photoreceptors of 24-week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild type mice lack differences in caspases’ activations (cleaved caspases) induced by light-stress. There are no changes in TUNEL+Casp3+, TUNEL+Casp7+, TUNEL+Casp8+, TUNEL+Casp9+ in cell bodies of photoreceptors between age-matched genotypes by light stress (A, B). Arrows show TUNEL+ cell bodies without caspases’ activations, whereas arrowheads show TUNEL+Casp+ cell bodies. (B) are quantitative analyses of representative images in (A). Retinal sections were counterstained with DAPI. Data shown represent the mean ± S.D., n=3. Legend: +/+, wild type; −/−::Tg-Ranbp2CLDm-HA, Ranbp2−/−::Tg-Ranbp2CLDm-HA; n.s., non-significant; Scale bar: 20 μm.
3.4. Age-and genotype-dependent, but photoreceptor death-independent, modulation of the ubiquitin-proteasome system (UPS) by light stress
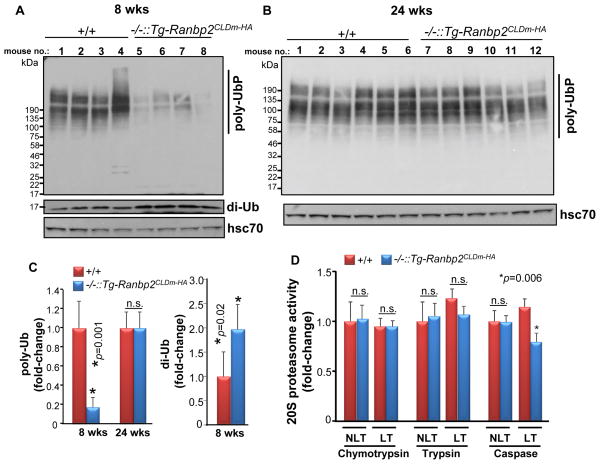
Light-stress is thought to induce proteotoxicity owing to increased oxidative stress and deregulation of the UPS [10, 33, 43–46]. In this regard, haploinsufficiency of Ranbp2 was shown to promote the age-dependent neuroprotection of photoreceptors to light damage [10] and elicit phototoxicity-dependent up-regulation of UPS activity and down-modulation of selective partners of Ranbp2, such as the S1 subunit of the 19S cap and ubc9 [33]. By contrast, Ranbp2−/−::Tg-Ranbp2CLDm-HA mice reared under cyclic-light and low luminance lack untoward retinal effects in the accumulation of ubiquitylated substrates, ubiquitin proteostasis and 20S proteasome activities except for a mild reduction in chymotrypsin-like activity [26]. To ascertain the role of CLD in UPS activity upon light-stress, we examined first the amount of ubiquitylated substrates between genotypes of 8 and 24-week old mice as an indicator of the activity of the UPS. As shown in figures 5A and C, 8-week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA compared to wild type mice had decreased and increased levels of ubiquitylated substrates and free diubiquitin, respectively, in retinal homogenates. However, there were no significant changes in the levels of ubiquitylated substrates between genotypes at 24 weeks of age, whereas free diubiquitin was undetectable in either genotype (Figs. 5B and C). Then, we determined the proteolytic activities of the 20S proteasome of Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild-type mice reared under light-stress. There were no changes in chymotrypsin and trypsin-like activities between genotypes; however, the caspase-like activity was mildly reduced in Ranbp2−/−::Tg-Ranbp2CLDm-HA compared with wild-type mice (Fig. 5D).
Fig. 5.
Age-dependent accumulation of ubiquitylated substrates and diubiquitin, and 20S proteasome activities, between Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild type mice exposed to light-stress. In comparison to wild type mice, Ranbp2−/−::Tg-Ranbp2CLDm-HA mice present decreased and increased levels of ubiquitylated substrates and diubiquitin, respectively, at 8 weeks of age (A, C), but their levels are similar between genotypes at 24 weeks of age (B, C). (D) Retinal extracts of 24 week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild type mice have similar chymotrypsin and trypsin-like activities of the 20S proteasome, but Ranbp2−/−::Tg-Ranbp2CLDm-HA have modestly lower caspase-like activity than wild type mice. Data shown represent the mean ± S.D., n=4–6. Legend: +/+, wild type; −/−::Tg-Ranbp2CLDm-HA, Ranbp2−/−::Tg-Ranbp2CLDm-HA; di-Ub, diubiquitin; poly-UbP, poly-ubiquitylated proteins; hsc70, heat shock cytosolic protein 70; n.s., non-significant.
3.5. Age-and genotype-dependent modulation of the proteostasis of the S1 subunit of the 19S cap and ubc9 by light-stress
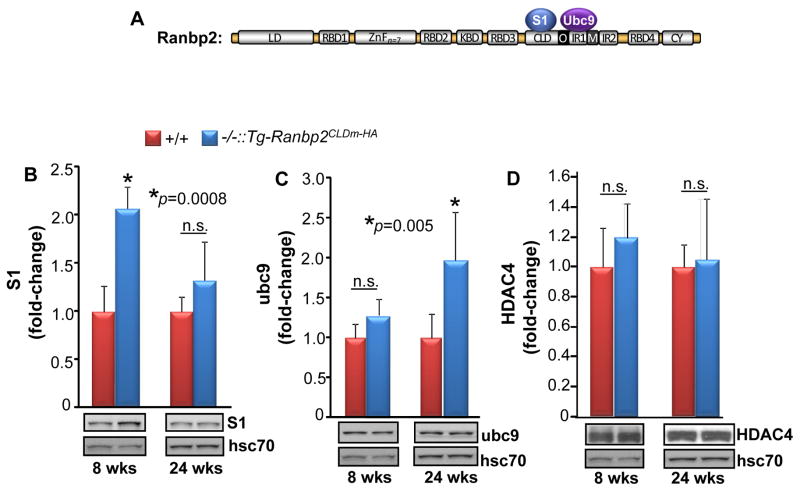
The CLD sequence overlaps partially with the IR1 of Ranbp2 [34]. CLD and IR1 associate with the S1 subunit of the 19S cap of the 26S proteasome and the E2 SUMO-conjugating enzyme, ubc9, respectively (Fig. 6A), and SBM mutations have a limited effect in CLD binding to subunits of the 19S cap in cell culture [36, 37, 39]. Physiologically and compared with wild type mice, light-stress down-modulates the levels of S1 and ubc9 in Ranbp2 haploinsufficient mice [33]. Hence, we examined the effect of CLDm of Ranbp2 and light-stress on the steady-state levels of S1 and ubc9 (Figs. 6B, C). As shown in figure 6B, Ranbp2−/−::Tg-Ranbp2CLDm-HA had increased levels of S1 subunit than wild type mice at 8 weeks of age, but by 24 weeks of age, there were no differences between genotypes. By contrast and in comparison to wild type mice, 24-week-old Ranbp2−/−::Tg-Ranbp2CLDm-HA mice had increased levels of ubc9, but ubc9 levels were similar between genotypes at 8 weeks of age (Fig. 6C).
Fig. 6.
Age-dependent changes in S1 subunit and ubc9 proteostasis between Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild type mice exposed to light-stress. (A) Diagram of the S1 and ubc9 partners of CLD and IR1 of Ranbp2, respectively. In comparison to wild type mice, Ranbp2−/−::Tg-Ranbp2CLDm-HA mice have increased levels of S1 subunit of the 19S cap of the 26S proteasome (B) and ubc9 (C) selectively at 8 and 24 weeks of age, respectively, whereas the levels of HDAC4 remain unchanged between genotypes at 8 and 24 weeks of age (D). Data shown represent the mean ± S.D., n=4. Legend: +/+, wild type; −/−::Tg-Ranbp2CLDm-HA, Ranbp2−/−::Tg-Ranbp2CLDm-HA; hsc70, heat shock cytosolic protein 70; n.s., non-significant.
We also examined HDAC4 proteostasis, because its levels are decreased in retinas of Ranbp2−/−::Tg-Ranbp2CLDm-HA in the absence of light stress [26]. In cell lines, HDAC4 is also a substrate for degradation by Ranbp2 [42] and sumoylation by a Ranbp2 construct comprising the IR1+2 and the C-terminal region of CLD (~103 residues) [41]. Further, over-expression and pharmacological inhibition of HDAC4 in rd1 mice paradoxically lead to the protection of photoreceptors [54, 55], whereas HDAC4 nuclear accumulation promotes neurodegeneration in ataxia telangiectasia (ATM) [56]. However, as shown in Fig. 6D, comparisons of HDAC4 levels between Ranbp2−/−::Tg-Ranbp2CLDm-HA and wild-type mice of 8 and 24 weeks of age showed that light-stress, genotype and age lacked untoward effects in HDAC4 proteostasis.
4. Discussion
This study uncovers that mice with insidious functional expression of I2471K and V2472A mutations (CLDm) in SBM of CLD of Ranbp2 lead to two contrasting and counterintuitive manifestations of photoreceptors upon exposure to phototoxicity: the photoreceptors undergo massive disruption of their outer segments, while the cell bodies of photoreceptors become strongly resistant to apoptosis in a manner which is independent of activations of prodeath caspases and overall UPS functions.
The paradoxical disruption of OS and suppression of apoptosis by the light stress-dependent insidious function of CLDm of Ranbp2 is counter to expectation. Hence, the molecular underpinnings linked to these manifestations will shed light to novel targets and biological mechanisms that are critical to sustain the integrity of OS of photoreceptors and their survival against deleterious stressors. Two alternative models for the light-dependent insidious expression of CLDm function of Ranbp2 in photoreceptors are worth further investigation. The first model is based on the findings that Ranbp2 localizes to two subcellularly and functionally distinct regions of the photoreceptors: the connecting cilium and the cytoplasmic fibrils emerging from nuclear pores [26, 57, 58]. These distinct subcellular structures are thought to share substrates and transport mechanisms that are regulated by Ranbp2 [59–61]. In this regard, CLDm of Ranbp2 may impair the intercompartmental trafficking of limiting substrate(s) at the cilium that are highly prone to oxidative damage and essential to the maintenance of OS, while suppressing the nucleocytoplasmic transport and signaling of selective light-induced factors which promote photoreceptor death. The second model entails the existence of a crosstalk mechanism mediated by the nucleocytoplasmic shuttling of light-induced and pleiotropic factor(s), whose pathological functions hinge on the Ranbp2-dependent partitioning of such factors(s) between the nuclear and cytosolic or ciliary compartments. In this regard, it is possible that light-stress activates nuclear signaling and photoreceptor death by factor(s), whose delocalization(s) in the cytosol by CLDm of Ranbp2 promotes the activation (e.g. posttranslational) of proteolytic factors which cause the acute erosion of the OS of photoreceptors. Regardless, the elucidation of the insidious functions of CLD of Ranbp2 under light-stress will likely rest on the identification of CLD partner(s) whose light-dependent function(s) and association to CLD are impaired by SBM mutations in CLD of Ranbp2.
The SBM of Ranbp2 overlaps with the C-and the N-terminal sequences of CLD and IR1, respectively [34, 35, 39]. CLD and IR1 associate with subunits of the 19S cap of the proteasome and ubc9, respectively [36, 37, 39]. Further, haploinsufficiency of Ranbp2 promotes the down-regulation of the S1 subunit of the 19S cap, ubc9, and ubiquitylated substrates in retinas exposed to light-stress [33]. However, this and other studies showed that there were no differences in the levels of ubiquitylated substrates and free diubiquitin between wild-type and Ranbp2−/−::Tg-Ranbp2CLDm-HA mice at 24 weeks of age, when photoreceptors become neuroprotected to light-stress in Ranbp2−/−::Tg-Ranbp2CLDm-HA and haploinsufficient Ranbp2 mice [10, 33]. Hence, these data support that the light-dependent control of proteostasis of ubiquitylated substrates by Ranbp2 is likely not a major contributor to neuroprotection against phototoxicity and that insidious function(s) of SBM of CLD of Ranbp2 exerts a neuroprotective role via another partner(s), whose function is stress-elicited and unrelated to the overall UPS activity. This notion is in apparent contrast to recent studies suggesting that there is induction of broad cytoprotective mechanisms by the modest down-regulation of the proteostasis of 19S cap subunits against chemical toxic insults targeting the 20S proteasome [62] and that SUMOylation of S1 subunit (also called Psmd1) of the 19S cap, a modification which has not been observed in our physiological studies, may control 26S proteasome activity and composition [63]. These apparent discrepancies may be caused by differences in regulatory signaling activities converging to the 26S proteasome owing to differences in the nature of stress stimuli, biological systems, and/or developmental stages employed between these studies.
Our study suggests that the age-dependent up-regulation of ubc9 proteostasis by insidious SBM function(s) of CLD of Ranbp2 may contribute to the age-dependent neuroprotection of photoreceptors against selective stressors, such as phototoxicity. Notably, ubc9 is reported to have dual activities. Ubc9 acts as an E2 enzyme by modifying Ranbp2 and some of its partners, such as RanGAP and Ran GTPase [64–66], with SUMO-1, but ubc9 also acts as a co-transcriptional regulator independently of its E2 activity [67–70]. Contrary to ubc9−/− mice [71], loss of SBM of CLD of Ranbp2 and SUMO-1 do not produce overt morphological and physiological phenotypes in adult mice [26, 72, 73], although loss of SBM of Ranbp2 cannot be fully compensated during development [26]. Further, ubc9 controls cell death [74] and its activity is exquisitely sensitive to oxidative stress [75]. Hence, ubc9 proteostasis and functions are likely under tight regulation by Ranbp2, aging and oxidative stress. Selective deregulation of ubc9 by SBM loss of Ranbp2 and phototoxicity may underlie paradoxical and insidious manifestations, such as the OS disruption and death resistance of Ranbp2−/−::Tg-Ranbp2CLDm-HA mice. In this regard, the Ranbp2−/−::Tg-Ranbp2CLDm-HA mice emerge as unique physiological platforms to enable the discovery of novel factors and mechanisms that control the maintenance of critical subcellular structures and the age-dependent survival of selective neural-cell types, such as photoreceptors, against light damage.
Highlights.
SBM mutations in CLD of Ranbp2 cause photoreceptor dysmorphology by light-stress
SBM mutations in CLD of Ranbp2 suppress photoreceptor death by light-stress
Caspase activation by light-stress is independent of Ranbp2 CLD function(s)
Light and Ranbp2 CLD-dependent photoreceptor death lack untoward effects in the UPS
Insidious loss of SBM function of CLD of Ranbp2 confers neuroprotection
Acknowledgments
We thank Nomingerel Tserentsoodol for help with processing some light microscopy images. This work was supported by the National Institutes of Health Grants EY019492 and GM083165 to P.A.F.
Abbreviations
- CLD
cyclophilin-like domain
- CLDm
cyclophilin-like domain mutant
- SBM
SUMO-binding motif
- Ranbp2
Ran-binding protein 2
- Tg
transgenic
- UPS
ubiquitin-proteasome system
- ubc9
ubiquitin carrier protein 9
- SUMO
small ubiquitin-like modifier
- RanGAP
Ran GTPase-activating protein
- HDAC4
histone deacetylase 4
- HA
hemagglutinin
- BAC
bacterial artificial chromosome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu HY, Hudry E, Hashimoto T, Kuchibhotla K, Rozkalne A, Fan Z, Spires-Jones T, Xie H, Arbel-Ornath M, Grosskreutz CL, et al. Amyloid beta induces the morphological neurodegenerative triad of spine loss, dendritic simplification, and neuritic dystrophies through calcineurin activation. J Neurosci. 2010;30:2636–2649. doi: 10.1523/JNEUROSCI.4456-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Looi JC, Walterfang M. Striatal morphology as a biomarker in neurodegenerative disease. Mol Psychiatry. 2013;18:417–424. doi: 10.1038/mp.2012.54. [DOI] [PubMed] [Google Scholar]
- 3.Luebke JI, Weaver CM, Rocher AB, Rodriguez A, Crimins JL, Dickstein DL, Wearne SL, Hof PR. Dendritic vulnerability in neurodegenerative disease: insights from analyses of cortical pyramidal neurons in transgenic mouse models. Brain Struct Funct. 2010;214:181–199. doi: 10.1007/s00429-010-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 5.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 6.Sancho-Pelluz J, Arango-Gonzalez B, Kustermann S, Romero FJ, van Veen T, Zrenner E, Ekstrom P, Paquet-Durand F. Photoreceptor cell death mechanisms in inherited retinal degeneration. Mol Neurobiol. 2008;38:253–269. doi: 10.1007/s12035-008-8045-9. [DOI] [PubMed] [Google Scholar]
- 7.Noell WK, Walker VS, Kang BS, Berman S. Retinal damage by light in rats. Invest Ophthalmol. 1966;5:450–473. [PubMed] [Google Scholar]
- 8.Grimm C, Reme CE. Light damage as a model of retinal degeneration. Methods Mol Biol. 2013;935:87–97. doi: 10.1007/978-1-62703-080-9_6. [DOI] [PubMed] [Google Scholar]
- 9.Wenzel A, Grimm C, Samardzija M, Reme CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog Retin Eye Res. 2005;24:275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Cho KI, Yi H, Yeh A, Tserentsoodol N, Cuadrado L, Searle K, Hao Y, Ferreira PA. Haploinsufficiency of RanBP2 is neuroprotective against light-elicited and age-dependent degeneration of photoreceptor neurons. Cell Death Differ. 2009;16:287–297. doi: 10.1038/cdd.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grignolo A, Orzalesi N, Castellazzo R, Vittone P. Retinal damage by visible light in albino rats. An electron microscope study. Ophthalmologica. 1969;157:43–59. doi: 10.1159/000305619. [DOI] [PubMed] [Google Scholar]
- 12.Kuwabara T, Gorn RA. Retinal damage by visible light. An electron microscopic study. Arch Ophthalmol. 1968;79:69–78. doi: 10.1001/archopht.1968.03850040071019. [DOI] [PubMed] [Google Scholar]
- 13.O’Steen WK, Shear CR, Anderson KV. Retinal damage after prolonged exposure to visible light. A light and electron microscopic study. Am J Anat. 1972;134:5–21. doi: 10.1002/aja.1001340103. [DOI] [PubMed] [Google Scholar]
- 14.Dunaief JL, Dentchev T, Ying GS, Milam AH. The role of apoptosis in age-related macular degeneration. Arch Ophthalmol. 2002;120:1435–1442. doi: 10.1001/archopht.120.11.1435. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki M, Tsujikawa M, Itabe H, Du ZJ, Xie P, Matsumura N, Fu X, Zhang R, Sonoda KH, Egashira K, et al. Chronic photo-oxidative stress and subsequent MCP-1 activation as causative factors for age-related macular degeneration. J Cell Sci. 2012;125:2407–2415. doi: 10.1242/jcs.097683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White DA, Fritz JJ, Hauswirth WW, Kaushal S, Lewin AS. Increased sensitivity to light-induced damage in a mouse model of autosomal dominant retinal disease. Invest Ophthalmol Vis Sci. 2007;48:1942–1951. doi: 10.1167/iovs.06-1131. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Lam TT, Tso MO, Naash MI. Expression of a mutant opsin gene increases the susceptibility of the retina to light damage. Vis Neurosci. 1997;14:55–62. doi: 10.1017/s0952523800008750. [DOI] [PubMed] [Google Scholar]
- 18.Organisciak DT, Darrow RM, Barsalou L, Kutty RK, Wiggert B. Susceptibility to retinal light damage in transgenic rats with rhodopsin mutations. Invest Ophthalmol Vis Sci. 2003;44:486–492. doi: 10.1167/iovs.02-0708. [DOI] [PubMed] [Google Scholar]
- 19.Budzynski E, Gross AK, McAlear SD, Peachey NS, Shukla M, He F, Edwards M, Won J, Hicks WL, Wensel TG, et al. Mutations of the opsin gene (Y102H and I307N) lead to light-induced degeneration of photoreceptors and constitutive activation of phototransduction in mice. J Biol Chem. 2010;285:14521–14533. doi: 10.1074/jbc.M110.112409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naash ML, Peachey NS, Li ZY, Gryczan CC, Goto Y, Blanks J, Milam AH, Ripps H. Light-induced acceleration of photoreceptor degeneration in transgenic mice expressing mutant rhodopsin. Invest Ophthalmol Vis Sci. 1996;37:775–782. [PubMed] [Google Scholar]
- 21.Walde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, Wiemann S, Kehlenbach RH. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo-and transport receptor-specific manner. Traffic. 2012;13:218–233. doi: 10.1111/j.1600-0854.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 22.Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, Joseph J, Kehlenbach RH, van Deursen JM. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol. 2011;194:597–612. doi: 10.1083/jcb.201102018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patil H, Saha A, Senda E, Cho KI, Haque M, Yu M, Qiu S, Yoon D, Hao Y, Peachey NS, et al. Selective Impairment of a Subset of Ran-GTP-binding Domains of Ran-binding protein 2 (Ranbp2) Suffices to Recapitulate the Degeneration of the Retinal Pigment Epithelium (RPE) Triggered by Ranbp2 Ablation. J Biol Chem. 2014;298:29767–29789. doi: 10.1074/jbc.M114.586834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho KI, Patil H, Senda E, Wang J, Yi H, Qiu S, Yoon D, Yu M, Orry A, Peachey NS, et al. Differential Loss of Prolyl Isomerase or Chaperone Activity of Ran-binding Protein 2 (Ranbp2) Unveils Distinct Physiological Roles of Its Cyclophilin Domain in Proteostasis. J Biol Chem. 2014;289:4600–4625. doi: 10.1074/jbc.M113.538215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslanukov A, Bhowmick R, Guruju M, Oswald J, Raz D, Bush RA, Sieving PA, Lu X, Bock CB, Ferreira PA. RanBP2 Modulates Cox11 and Hexokinase I Activities and Haploinsufficiency of RanBP2 Causes Deficits in Glucose Metabolism. PLoS Genet. 2006;2:e177. doi: 10.1371/journal.pgen.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neilson DE, Adams MD, Orr CM, Schelling DK, Eiben RM, Kerr DS, Anderson J, Bassuk AG, Bye AM, Childs AM, et al. Infection-Triggered Familial or Recurrent Cases of Acute Necrotizing Encephalopathy Caused by Mutations in a Component of the Nuclear Pore, RANBP2. Am J Hum Genet. 2009;84:44–51. doi: 10.1016/j.ajhg.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf K, Schmitt-Mechelke T, Kollias S, Curt A. Acute necrotizing encephalopathy (ANE1): rare autosomal-dominant disorder presenting as acute transverse myelitis. J Neurol. 2013;260:1545–1553. doi: 10.1007/s00415-012-6825-7. [DOI] [PubMed] [Google Scholar]
- 30.Singh RR, Sedani S, Lim M, Wassmer E, Absoud M. RANBP2 mutation and acute necrotizing encephalopathy: 2 cases and a literature review of the expanding clinico-radiological phenotype. Eur J Paediatr Neurol. 2015;19:106–113. doi: 10.1016/j.ejpn.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Denier C, Balu L, Husson B, Nasser G, Burglen L, Rodriguez D, Labauge P, Chevret L. Familial acute necrotizing encephalopathy due to mutation in the RANBP2 gene. J Neurol Sci. 2014;345:236–238. doi: 10.1016/j.jns.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 32.Cho KI, Searle K, Webb M, Yi H, Ferreira PA. Ranbp2 haploinsufficiency mediates distinct cellular and biochemical phenotypes in brain and retinal dopaminergic and glia cells elicited by the Parkinsonian neurotoxin, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Cell Mol Life Sci. 2012;69:3511–3527. doi: 10.1007/s00018-012-1071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho KI, Yi H, Tserentsoodol N, Searle K, Ferreira PA. Neuroprotection resulting from insufficiency of RANBP2 is associated with the modulation of protein and lipid homeostasis of functionally diverse but linked pathways in response to oxidative stress. Dis Model Mech. 2010;3:595–604. doi: 10.1242/dmm.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira PA, Hom JT, Pak WL. Retina-specifically expressed novel subtypes of bovine cyclophilin. J Biol Chem. 1995;270:23179–23188. doi: 10.1074/jbc.270.39.23179. [DOI] [PubMed] [Google Scholar]
- 35.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira PA, Yunfei C, Schick D, Roepman R. The cyclophilin-like domain mediates the association of Ran-binding protein 2 with subunits of the 19 S regulatory complex of the proteasome. J Biol Chem. 1998;273:24676–24682. doi: 10.1074/jbc.273.38.24676. [DOI] [PubMed] [Google Scholar]
- 37.Yi H, Friedman JL, Ferreira PA. The cyclophilin-like domain of Ran-binding protein-2 modulates selectively the activity of the ubiquitin-proteasome system and protein biogenesis. J Biol Chem. 2007;282:34770–34778. doi: 10.1074/jbc.M706903200. [DOI] [PubMed] [Google Scholar]
- 38.Cho KI, Haque M, Wang J, Yu M, Hao Y, Qiu S, Pillai IC, Peachey NS, Ferreira PA. Distinct and atypical intrinsic and extrinsic cell death pathways between photoreceptor cell types upon specific ablation of Ranbp2 in cone photoreceptors. PLoS Genet. 2013;9:e1003555. doi: 10.1371/journal.pgen.1003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT-nor RING-type. Nat Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- 40.Werner A, Flotho A, Melchior F. The RanBP2/RanGAP1(*)SUMO1/Ubc9 Complex Is a Multisubunit SUMO E3 Ligase. Mol Cell. 2012;46:287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scognamiglio A, Nebbioso A, Manzo F, Valente S, Mai A, Altucci L. HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochim Biophys Acta. 2008;1783:2030–2038. doi: 10.1016/j.bbamcr.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Organisciak DT, Vaughan DK. Retinal light damage: mechanisms and protection. Prog Retin Eye Res. 2010;29:113–134. doi: 10.1016/j.preteyeres.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louie JL, Kapphahn RJ, Ferrington DA. Proteasome function and protein oxidation in the aged retina. Exp Eye Res. 2002;75:271–284. [PubMed] [Google Scholar]
- 45.Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- 46.Naash MI, Al-Ubaidi MR, Anderson RE. Light exposure induces ubiquitin conjugation and degradation activities in the rat retina. Invest Ophthalmol Vis Sci. 1997;38:2344–2354. [PubMed] [Google Scholar]
- 47.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troy CM, Akpan N, Jean YY. Regulation of caspases in the nervous system implications for functions in health and disease. Prog Mol Biol Transl Sci. 2011;99:265–305. doi: 10.1016/B978-0-12-385504-6.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, Gorman A, Zhou X, Sandra C, Chen E. Involvement of caspase-3 in photoreceptor cell apoptosis induced by in vivo blue light exposure. Invest Ophthalmol Vis Sci. 2002;43:3349–3354. [PubMed] [Google Scholar]
- 50.Zeiss CJ, Neal J, Johnson EA. Caspase-3 in postnatal retinal development and degeneration. Invest Ophthalmol Vis Sci. 2004;45:964–970. doi: 10.1167/iovs.03-0439. [DOI] [PubMed] [Google Scholar]
- 51.Doonan F, Donovan M, Cotter TG. Caspase-independent photoreceptor apoptosis in mouse models of retinal degeneration. J Neurosci. 2003;23:5723–5731. doi: 10.1523/JNEUROSCI.23-13-05723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donovan M, Carmody RJ, Cotter TG. Light-induced photoreceptor apoptosis in vivo requires neuronal nitric-oxide synthase and guanylate cyclase activity and is caspase-3-independent. J Biol Chem. 2001;276:23000–23008. doi: 10.1074/jbc.M005359200. [DOI] [PubMed] [Google Scholar]
- 53.Grimm C, Wenzel A, Groszer M, Mayser H, Seeliger M, Samardzija M, Bauer C, Gassmann M, Reme CE. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat Med. 2002;8:718–724. doi: 10.1038/nm723. [DOI] [PubMed] [Google Scholar]
- 54.Sancho-Pelluz J, Alavi MV, Sahaboglu A, Kustermann S, Farinelli P, Azadi S, van Veen T, Romero FJ, Paquet-Durand F, Ekstrom P. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell death & disease. 2010;1:e24. doi: 10.1038/cddis.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, Herrup K. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2012;18:783–790. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delphin C, Guan T, Melchior F, Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mavlyutov TA, Cai Y, Ferreira PA. Identification of RanBP2-and Kinesin-Mediated Transport Pathways with Restricted Neuronal and Subcellular Localization. Traffic. 2002;3:630–640. doi: 10.1034/j.1600-0854.2002.30905.x. [DOI] [PubMed] [Google Scholar]
- 59.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan S, Whiteman EL, Hurd TW, McIntyre JC, Dishinger JF, Liu CJ, Martens JR, Verhey KJ, Sajjan U, Margolis B. Induction of Ran GTP drives ciliogenesis. Mol Biol Cell. 2011;22:4539–4548. doi: 10.1091/mbc.E11-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsvetkov P, Mendillo ML, Zhao J, Carette JE, Merrill PH, Cikes D, Varadarajan M, van Diemen FR, Penninger JM, Goldberg AL, et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. Elife. 2015;4 doi: 10.7554/eLife.08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryu H, Gygi SP, Azuma Y, Arnaoutov A, Dasso M. SUMOylation of Psmd1 controls Adrm1 interaction with the proteasome. Cell reports. 2014;7:1842–1848. doi: 10.1016/j.celrep.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saitoh H, Sparrow DB, Shiomi T, Pu RT, Nishimoto T, Mohun TJ, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 65.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 66.Sakin V, Richter SM, Hsiao HH, Urlaub H, Melchior F. Sumoylation of the GTPase Ran by the RanBP2 SUMO E3 Ligase Complex. J Biol Chem. 2015;290:23589–23602. doi: 10.1074/jbc.M115.660118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurtzman AL, Schechter N. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc Natl Acad Sci U S A. 2001;98:5602–5607. doi: 10.1073/pnas.101129698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi S, Shibata H, Kurihara I, Yokota K, Suda N, Saito I, Saruta T. Ubc9 interacts with chicken ovalbumin upstream promoter-transcription factor I and represses receptor-dependent transcription. J Mol Endocrinol. 2004;32:69–86. doi: 10.1677/jme.0.0320069. [DOI] [PubMed] [Google Scholar]
- 69.Kurihara I, Shibata H, Kobayashi S, Suda N, Ikeda Y, Yokota K, Murai A, Saito I, Rainey WE, Saruta T. Ubc9 and Protein Inhibitor of Activated STAT 1 Activate Chicken Ovalbumin Upstream Promoter-Transcription Factor I-mediated Human CYP11B2 Gene Transcription. J Biol Chem. 2005;280:6721–6730. doi: 10.1074/jbc.M411820200. [DOI] [PubMed] [Google Scholar]
- 70.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 72.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 74.Nowak M, Hammerschmidt M. Ubc9 regulates mitosis and cell survival during zebrafish development. Mol Biol Cell. 2006;17:5324–5336. doi: 10.1091/mbc.E06-05-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]