Abstract
Chikungunya is caused by the mosquito-borne arthrogenic alphavirus, chikungunya virus (CHIKV). Chikungunya was introduced into the Americas in late 2013 and Nicaragua in mid-2014. Here, we sequenced five imported and 30 autochthonous Nicaraguan CHIKV from cases identified in the first epidemic in the country between August 2014 and April 2015. One full-length and two partial genomic sequences were obtained by deep sequencing; Sanger methodology yielded 33 E1 sequences from five imported and 28 autochthonous cases. Phylogenetic analysis indicates that Nicaraguan CHIKV all belonged to the Asian genotype, Caribbean clade. Moreover, E1 gene sequences revealed accumulation of mutations in later months of the epidemic, including four silent mutations in 11 autochthonous cases and three non-synonymous mutations in three autochthonous cases. No mutations contributing to increased transmissibility by Aedes albopictus were identified in the E1 gene. This represents the most comprehensive set of CHIKV sequences available from the Americas to date.
Chikungunya is a reemerging infectious disease caused by a mosquito-borne arthrogenic alphavirus, chikungunya virus (CHIKV). CHIKV is an enveloped virus with a 12-kb positive-sense RNA genome that contains a 5′ untranslated region (UTR) followed by nonstructural protein genes (NS1–NS4), structural protein genes (C, E3, E2, 6K, and E1) under the control of a subgenomic promoter, and a 3′ UTR including a terminal poly-A tail. Often initially mistaken for dengue, chikungunya manifests clinically as fever and arthralgia involving intense pain and inflammation in joints typically lasting weeks or months but sometimes years.1 Other symptoms may include headache, muscle pain, or rash. Currently there is no licensed vaccine to prevent or medicine to treat CHIKV infection. Areas endemic for chikungunya include Africa and southeast Asia.2 Since 2004, CHIKV has expanded into the Indian and Pacific Oceans and Europe, (http://www.who.int/mediacentre/factsheets/fs327/en/), and since the end of 2013, it has caused explosive epidemics in the Caribbean and Latin America, with imported cases and some autochthonous transmission reported in the United States.3,4 However, limited numbers of CHIKV genomes are available from the Americas, despite the recent epidemic.4,5
CHIKV can be classified into three distinct genotypes: Asian, East/Central/South African (ECSA), and western African.2,4 Viral sequences from St. Martin Island, the point of introduction in the Americas, belonged to the CHIKV Asian genotype.4 Chikungunya is generally spread through the bite of Aedes aegypti mosquitoes, but transmission by A. albopictus has been reported.6–8 During the Reunion Island outbreak in 2005–2006, a single mutation, A226V, in the structural E1 gene of certain ECSA strains enhanced CHIKV transmissibility by Ae. albopictus, potentially aiding in the spread of CHIKV in unprecedented regions.9,10 In July 2014, CHIKV was introduced into Nicaragua. Here, we sequenced CHIKV from imported and autochthonous Nicaraguan cases to determine the origin of CHIKV strains circulating in Nicaragua, and we analyzed the viral envelope E1 gene for any variation that could provide insight into the introduction and dissemination of CHIKV across time in Nicaragua.
In Nicaragua, the first imported case was identified on July 9, 2014, and the first autochthonous case on September 23, 2014. Chikungunya cases were identified through a national surveillance program implemented by the Ministry of Health and two ongoing pediatric studies conducted in Managua, the capital city of Nicaragua: the Pediatric Dengue Cohort Study,11 a community-based cohort ongoing since 2004, and the Hospital-based Dengue Study, based at the National Pediatric Reference Hospital, ongoing since 1998.12 CHIKV testing was included in both studies in 2014. The studies were approved by the University of California, Berkeley, and Nicaraguan Ministry of Health institutional review boards. All samples were tested using CHIKV-specific real-time reverse-transcription polymerase chain reaction (RT-PCR) (Supplemental Methods). A total of 35 CHIKV-positive samples from five imported and 30 autochthonous cases, spanning the period from August 2014 to April 2015, were sequenced (Table 1). Four samples were sequenced directly from serum and for the rest, CHIKV was isolated in Vero cells and then sequenced (Table 1). Supplemental Table 1 lists detailed information about each strain sequenced, including the sample type (isolate or serum), case type (imported or autochthonous), source (Nicaraguan national surveillance, cohort study, or hospital study), date of collection, and mutations identified.
Table 1.
Nicaraguan CHIKV samples used in this study
| Total | Case type* | Types of sample | Source | August 2014 | October 2014 | November 2014 | December 2014 | January 2015 | February 2015 | March 2015 | April 2015 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nicaraguan CHIKV samples (35) | Imported (5) | Virus isolates (5) | National surveillance (5) | 5 | – | – | – | – | – | – | – |
| Autochthonous (30) | Virus isolates (26) | Pediatric cohort study (13) | – | 3 | 3 | 2 | 1 | 2 | 1 | 1 | |
| Pediatric hospital study (13) | – | – | 2 | 5 | 2 | 4 | – | – | |||
| Serum samples (4) | National surveillance (2) | – | 2 | – | – | – | – | – | – | ||
| Pediatric cohort study (2) | 2 | – | – | – | – | – | – |
CHIKV = chikungunya virus.
Autochthonous case: if the individual had not traveled; imported case: if the patient returned ill from a country with a current CHIKV epidemic.
To obtain complete genomic sequence information and determine the origin of CHIKV strains circulating in Nicaragua, whole-genome amplification of viral RNA from a subset of samples (three serum samples from autochthonous cases in October 2014), combined with Nextera technology, was used to generate libraries for deep sequencing (Supplemental Methods). In brief, complementary DNA for each sample was synthesized using random hexamers and Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and then amplified using multiple displacement amplification with phi29 DNA Polymerase (New England Biolabs, Ipswich, MA). Amplified DNA was purified using the Qiagen PCR Purification Kit (Qiagen, Valencia, CA) and prepared for high-throughput sequencing using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA). The libraries were pooled in equimolar ratios and sequenced on the HiSeq2000 sequencer (Illumina) to generate 100-bp reads. Reads for each sample were mapped to full-length CHIKV sequences from the National Center for Biotechnology Information (NCBI), using the “Bowtie2” software.13 “Samtools”14 and in-house Python (Python Software Foundation, Beaverton, OR; http://www.python.org) scripts were used to generate pileups and consensus nucleotide sequences.
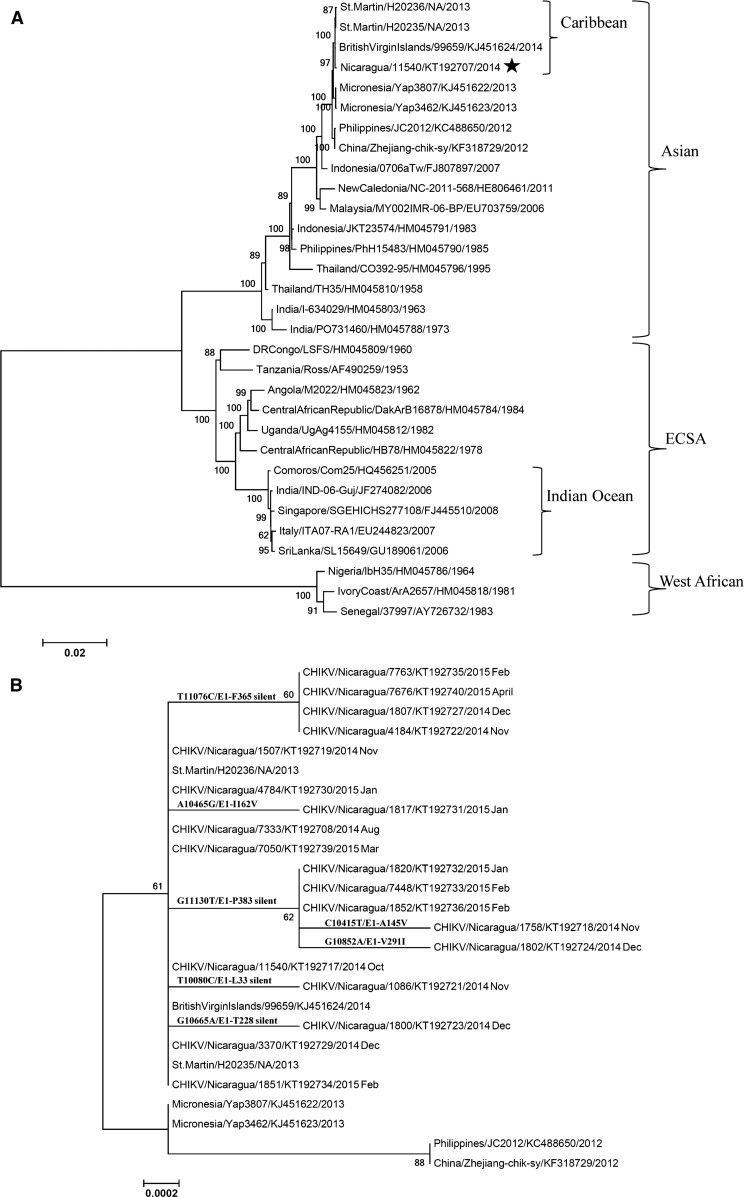
Complete full-length sequence from one individual (Nicaragua/11540/KT192707/2014) (Figure 1A ) was obtained, along with partial genome sequences from two other samples (Nicaragua/4916/NA/2014 and Nicaragua/11519/NA/2014; data not shown) because of lower-than-expected coverage across the CHIKV genome. Two of the sequences cover the NSP3 region (Nicaragua/11540/KT192707/2014 and Nicaragua/4916/NA/2014), and both share the same four amino acid deletion in NSP3 previously reported in Indonesia/0706aTw/FJ807897/2007 (Figure 1A) and related Asian genotype strains.16
Figure 1.
Phylogenetic analysis of Nicaraguan chikungunya viruses (CHIKV). (A) Nicaraguan CHIKV belongs to the Caribbean clade of the Asian genotype. Phylogenetic analysis included 37 full-length genomic nucleotide sequences (one Nicaraguan strain and 36 reference strains), with the format of country/strain/GenBank no./year. The three major genotypes and two clades are labeled. Bootstrap values are shown on major branches. (B) Mutations in the E1 gene of Nicaraguan CHIKV. This analysis included E1 gene sequences of 19 representative monthly Nicaraguan CHIKV strains, with the format of country/strain/GenBank no./year month, and E1 sequences of seven reference strains in the format of country/strain/GenBank no./year. For simplicity, each month only one sample identical to and all the samples different from the BritishVirginIslands/99659/KJ451624/2014 strain were included in the subtree, resulting in elimination of 14 duplicate identical Nicaraguan CHIKV sequences from the subtree. The relevant mutations are indicated at each branch point. Both trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values are shown on major branches. All positions containing gaps and missing data were eliminated. Phylogenetic analyses were conducted in MEGA6.15
Phylogenetic analysis was performed to identify the origin of the Nicaraguan CHIKV. Full-length genomic nucleotide sequences of Nicaragua/11540/KT192707/2014 and 36 reference strains were available from GenBank and the European Virus Archive. Nicaragua/11540 is a sample from October 2014 (first month of autochthonous cases used in the study); thus, it is representative of CHIKV strains circulating early in the epidemic in Nicaragua. FASTA format alignment was performed by MAFFT FFT-NS-2 (v7.221). The evolutionary history was inferred using the maximum likelihood method based on the Tamura–Nei model.17 The tree with the highest log likelihood is shown in Figure 1A. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood approach.
Our phylogenetic analysis identified that Nicaraguan strains belong to the Caribbean clade5 of the Asian genotype, most similar to BritishVirginIslands/99659/KJ451624/2014, St.Martin/H20235/NA/2013, and St.Martin/H20236/NA/2013 (Figure 1A).
To investigate the molecular evolution of CHIKV through its emergence and spread in Nicaragua, primers for Sanger sequencing were designed according to BritishVirginIslands/99659/KJ451624/2014 to amplify and sequence the E1 gene (Supplemental Table 2). The PCR product of ∼2.4 kb was sequenced using Sanger methodology (Supplemental Methods).
The Sanger methodology yielded sequences from five imported CHIKV cases from August 2014 and 28 autochthonous cases from October 2014 to April 2015, including one already sequenced by deep sequencing (Nicaragua/11540/KT192707/2014) (Supplemental Table 1). For sample Nicaragua/11540, E1 sequences obtained from full-genome deep sequencing and Sanger methodology were 100% identical. E1 sequences from all five imported cases (August 2014) and five autochthonous cases (October 2014) from the early months of the epidemic shared 100% identity with the BritishVirginIslands/99659/KJ451624/2014 reference genome (Supplemental Table 1). However, later in the epidemic season (November 2014 to April 2015), 14 of the 23 autochthonous cases showed silent mutations at four sites (T10080C/E1-L33, G10665A/E1-T228, T11076C/E1-F365, or G11130T/E1-P383) or non-synonymous mutations at three sites (C10415T/E1-A145V, A10465G/E1-I162V, or G10852A/E1-V291I), with nucleotide numbering according to BritishVirginIslands/99659/KJ451624/2014 (Figure 1B, Supplemental Table 1). Accumulation of substitutions over the course of an epidemic is not surprising. Of the mutations in E1 gene that appeared in later months of the epidemic, five mutations (T10080C/E1-L33 silent, C10415T/E1-A145V, A10465G/E1-I162V, G10665A/E1-T228 silent, and G10852A/E1-V291I) appeared only once in certain samples, whereas the other two mutations (G11130T/E1-P383 and T11076C/E1-F365) were identified in several cases. G11130T/E1-P383 appeared five times, spanning sequences from cases from November 2014 to February 2015, two of which also contained a second mutation (either C10415T/E1-A145V or G10852A/E1-V291I). T11076C/E1-F365 appeared in four samples spanning cases from November 2014 to April 2015, as can be seen in the small branch in Figure 1B. Interestingly, this mutation appears in numerous distinct E1 sequences, including several from Brazil in 2014.18 It has not yet been determined whether these genetic variations play a role in viral infectivity or transmission, and further studies are needed to investigate their implications. Furthermore, consistent with other Asian genotype viruses, all 35 Nicaraguan samples sequenced thus far contain E1-A226 and E1-T9819 and therefore do not display the mutations that have been associated with adaptation to Ae. albopictus (data not shown). It should be noted that Figure 1B is intended to demonstrate the Caribbean origin of the Nicaraguan samples and the extent of mutations accumulated, but not the phylogenetic relationship among these Nicaraguan strains, given the limited number of mutations identified.
In summary, Nicaraguan CHIKV samples (five imported and 30 autochthonous cases) from national surveillance and two ongoing pediatric studies from the first epidemic in the country were sequenced. One full-length sequence obtained by deep sequencing demonstrated the origin of the Nicaraguan strains to be the Caribbean clade5 of the Asian genotype, containing the four amino acid deletion in NSP3.16 Sanger methodology yielded 33 E1 sequences from five imported cases from August 2014 and 28 autochthonous cases from October 2014 to April 2015. The imported cases were observed in August 2014 (the first month of epidemic included in this study), with E1 sequences 100% homologous to the BritishVirginIslands/99659/KJ451624/2014 strain. Beginning in October 2014, all cases used in this study were autochthonous, with either 100% homology to the BritishVirginIslands/99659/KJ451624/2014 strain or with accumulation of mutations (starting November 2014). The accumulated mutations included four silent mutations in 11 autochthonous cases and three non-synonymous mutations (E1-A145V, E1-I162V, or E1-V291I) in three autochthonous cases. All 35 sequences contained E1-A226 and E1-T98 mutations, and thus do not display the mutations suggestive of enhanced CHIKV transmissibility in Ae. albopictus.19 It should be noted that E1-A226V alone only affects increased infectivity and transmissibility in Ae. albopictus when it occurs in the ECSA lineage but not in the Asian lineage strains unless accompanied by additional mutations.20 Altogether, this study represents the most comprehensive set of CHIKV sequences available from the Americas to date.
Supplementary Material
ACKNOWLEDGMENTS
We thank Juan Carlos Mercado and Andrea Núñez for assistance with sample selection, processing, and shipping; Shannon Bennett for advice on phylogenetic analysis; Edwina Beryl Tran for technical assistance; our colleagues involved in the national surveillance program for dengue and chikungunya at the Nicaraguan Ministry of Health; and the entire study team at the National Virology Laboratory, Hospital Infantil Manuel de Jesús Rivera, and Centro de Salud Sócrates Flores Vivas. Finally, we are extremely grateful to the study participants and their families.
Footnotes
Financial support: Funding for this study came from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (R01AI099631 to Angel Balmaseda), with support for deep sequencing assay development from NIAID/PSWRCE grant U54AI065359 (Poornima Parameswaran and Andrew Fire).
Authors' addresses: Chunling Wang, Meghana Eswarappa, Poornima Parameswaran, and Eva Harris, Division of Infectious Diseases and Vaccinology, School of Public Health, University of California Berkeley, Berkeley, CA, E-mails: chunlingwang@berkeley.edu, meghana.eswarappa@berkeley.edu, nima.param@gmail.com, and eharris@berkeley.edu. Saira Saborio and Angel Balmaseda, Laboratorio Nacional de Virología, Centro Nacional de Diagnóstico y Referencia, Ministry of Health, Managua, Nicaragua, E-mails: ysaborio@minsa.gob.ni and abalmaseda@minsa.gob.ni. Lionel Gresh, Sustainable Sciences Institute, Managua, Nicaragua, E-mail: lionel.gresh@gmail.com. Diane Wu and Andrew Fire, Department of Genetics, Stanford University, Stanford, CA, and Department of Pathology, Stanford University, Stanford, CA, E-mails: diwu@alumni.stanford.edu and afire@stanford.edu.
References
- 1.Halstead SB. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis. 2015;21:557–561. doi: 10.3201/eid2104.141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88:2363–2377. doi: 10.1099/vir.0.82858-0. [DOI] [PubMed] [Google Scholar]
- 3.Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerg Infect Dis. 2014;20:1400–1402. doi: 10.3201/eid2008.140268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 5.Diaz Y, Carrera JP, Cerezo L, Arauz D, Guerra I, Cisneros J, Armien B, Botello AM, Arauz AB, Gonzalez V, Lopez Y, Moreno L, Lopez-Verges S, Moreno BA. Chikungunya virus infection: first detection of imported and autochthonous cases in Panama. Am J Trop Med Hyg. 2015;92:482–485. doi: 10.4269/ajtmh.14-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatun S, Chakraborty A, Rahman M, Nasreen Banu N, Rahman MM, Hasan SM, Luby SP, Gurley ES. An outbreak of chikungunya in rural Bangladesh, 2011. PLoS Negl Trop Dis. 2015;9:e0003907. doi: 10.1371/journal.pntd.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delisle E, Rousseau C, Broche B, Leparc-Goffart I, L'Ambert G, Cochet A, Prat C, Foulongne V, Ferre JB, Catelinois O, Flusin O, Tchernonog E, Moussion IE, Wiegandt A, Septfons A, Mendy A, Moyano MB, Laporte L, Maurel J, Jourdain F, Reynes J, Paty MC, Golliot F. Chikungunya outbreak in Montpellier, France, September to October 2014. Euro Surveill. 2015;20 doi: 10.2807/1560-7917.es2015.20.17.21108. pii 21108. [DOI] [PubMed] [Google Scholar]
- 8.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. CHIKV Study Group Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 9.de Lamballerie X, Leroy E, Charrel RN, Ttsetsarkin K, Higgs S, Gould EA. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virol J. 2008;5:33. doi: 10.1186/1743-422X-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nunez A, Coloma J, Balmaseda A, Harris E. The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol. 2009;170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narvaez F, Gutierrez G, Perez MA, Elizondo D, Nunez A, Balmaseda A, Harris E. Evaluation of the traditional and revised WHO classifications of dengue disease severity. PLoS Negl Trop Dis. 2011;5:e1397. doi: 10.1371/journal.pntd.0001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang JH, Yang CF, Su CL, Chang SF, Cheng CH, Yu SK, Lin CC, Shu PY. Imported chikungunya virus strains, Taiwan, 2006–2009. Emerg Infect Dis. 2009;15:1854–1856. doi: 10.3201/eid1511.090398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 18.Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, Azevedo Rdo S, da Silva DE, da Silva EV, da Silva SP, Carvalho VL, Coelho GE, Cruz AC, Rodrigues SG, Vianez JL, Jr, Nunes BT, Cardoso JF, Tesh RB, Hay SI, Pybus OG, Vasconcelos PF. Emergence and potential for spread of chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, Forrester N, Perng GC, Sreekumar E, Leal G, Huang J, Mukhopadhyay S, Weaver SC. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. doi: 10.1038/ncomms5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.