Abstract
The success of mosquito-based malaria control is dependent upon susceptible bionomic traits in local malaria vectors. It is crucial to have accurate and reliable methods to determine mosquito species composition in areas subject to malaria. An unexpectedly diverse set of Anopheles species was collected in the western Kenyan highlands, including unidentified and potentially new species carrying the malaria parasite Plasmodium falciparum. This study identified 2,340 anopheline specimens using both ribosomal DNA internal transcribed spacer region 2 and mitochondrial DNA cytochrome oxidase subunit 1 loci. Seventeen distinct sequence groups were identified. Of these, only eight could be molecularly identified through comparison to published and voucher sequences. Of the unidentified species, four were found to carry P. falciparum by circumsporozoite enzyme-linked immunosorbent assay and polymerase chain reaction, the most abundant of which had infection rates comparable to a primary vector in the area, Anopheles funestus. High-quality adult specimens of these unidentified species could not be matched to museum voucher specimens or conclusively identified using multiple keys, suggesting that they may have not been previously described. These unidentified vectors were captured outdoors. Diverse and unknown species have been incriminated in malaria transmission in the western Kenya highlands using molecular identification of unusual morphological variants of field specimens. This study demonstrates the value of using molecular methods to compliment vector identifications and highlights the need for accurate characterization of mosquito species and their associated behaviors for effective malaria control.
Introduction
Malaria control methods focus largely on impacting the vector population to reduce transmission. Interventions such as use of indoor residual spray (IRS) and long-lasting insecticidal nets take advantage of species-specific vector behaviors that result in exposure to lethal insecticide.1–5 In many areas, multiple vector species contribute to malaria infection, which may prolong the transmission season through their different feeding behaviors and exploitation of the environment.3,6 There may also be more vector species in some areas than are presently known due to lack of regular in-depth descriptive surveys, the presence of cryptic or sibling species,7–9 or outdated keys to identify specimens, and inadequate descriptions used to morphologically identify species. Different species and even different members within species complexes can exhibit a variety of behaviors relevant to malaria epidemiology.10,11 For example, the Anopheles gambiae species complex is composed of at least eight distinct sibling species,8 some of which are generally nonhuman feeders, such as An. quadriannulatus,12,13 whereas others, such as An. gambiae s.s., An. coluzzii, and An. arabiensis, act as the primary malaria vectors in sub-Saharan Africa. An. gambiae s.s. is highly anthropophilic and endophagic, while in many areas An. arabiensis is considered to be more exophagic and exophilic.4,14–17 The outdoor feeding behavior of An. arabiensis and other vector species complicates control interventions, such as IRS and insecticide-treated bednets, which are used indoors, and some species can shift their behavioral patterns over time in response to interventions.18–22 Morphological identification cannot distinguish between these cryptic species and can be inaccurate even for morphologically distinct species, resulting in skewed interpretation of species compositions in behavioral or monitoring studies. Because of the limitations of morphological identification tools, there is a need for detailed molecular taxonomy and phylogenetic analyses to accurately identify specific vectors and to associate these with specific behaviors.23
The ribosomal DNA internal transcribed spacer region 2 (rDNA ITS2) and the mitochondrial DNA cytochrome oxidase subunit 1 (mtDNA CO1), two quickly evolving loci, have been used to differentiate members of species complexes, and to generate reference sequences and unique barcodes that identify and distinguish Anopheles species.9,10,24,25 Though an increasing number of Anopheles sequences at these two loci are available in databases, many common anophelines have not been part of molecular studies. There are approximately 200 Anopheles species represented by ITS2 and CO1 sequences in GenBank (National Center for Biotechnology Information [NCBI]), despite almost 500 recognized species in the genus.26 Genotyping of local anophelines, even presumed nonvector species, will enable accurate linking of bionomic traits with species, thereby allowing for the appropriate assessment of efficacy or limitations of interventions being implemented.27 Accurately identifying the bionomic vulnerabilities of specific populations or species that need to be targeted for malaria control also allows for more focused and potentially cost-effective intervention strategies.
In this study, a set of anopheline species collected in the western Kenyan highlands, Nyanza Province (Figure 1 ), was analyzed using rDNA ITS2 and mtDNA CO1 loci for accurate molecular species identification. Published studies from Kisii district, Nyanza Province, of the Kenyan highlands have focused on the presumed primary malaria vectors in the region, namely species belonging to the An. gambiae and An. funestus complexes.28–33 There are no reports of other anophelines in the area acting as major vectors and only limited descriptive vector studies from the highlands of Rachuonyo South, Nyanza Province. The western Kenyan highlands is an area of unstable, sometimes epidemic malaria34 where anopheline vector dynamics determine the spread and maintenance of malaria transmission.35–37 The aim of our study was to identify Anopheles species present in two neighboring districts in the western Kenyan highlands using both molecular and morphological tools. This study expands upon previously reported findings of unknown and potentially new vector species in Kisii district,38 detailing the rDNA ITS2 and mtDNA CO1 regions, and includes sequence analysis of two more recent sets of mosquito collections from an area in the neighboring district, Rachuonyo South.
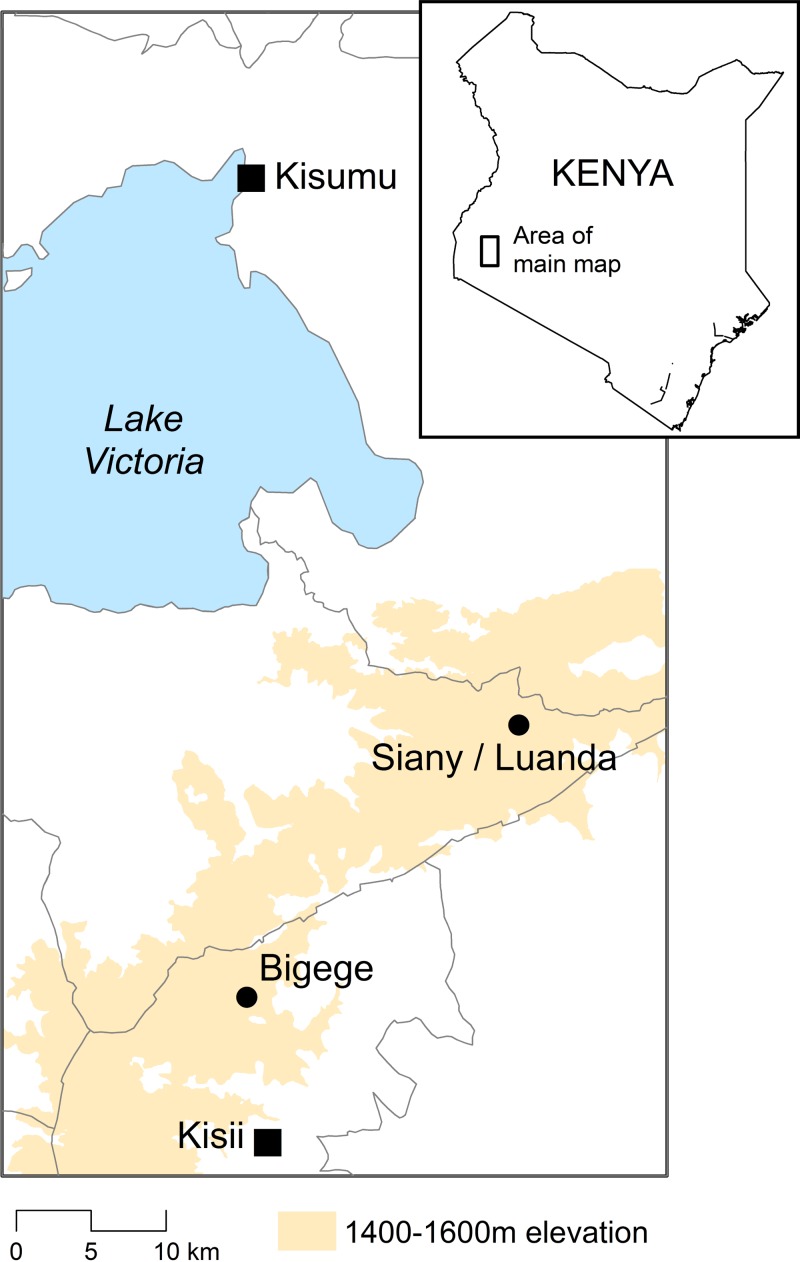
Figure 1.
Map of the Kenya highlands. Malaria Transmission Consortium sites where entomological collections took place. Adult mosquitoes were collected in Bigege village in Kisii Central District and Luanda and Siany villages in Rachonyo South District (Luanda and Siany villages overlap), in Nyanza Province using Centers for Disease Control and Prevention miniature light traps placed indoors and outdoors, and a small number of immature stages were collected by larval dipping from May 2010 to December 2011.
Materials and Methods
Study sites.
Mosquitoes were collected in Bigege village, Kisii Central District, and Luanda and Siany villages, Rachuonyo South District, Nyanza Province, western Kenya (Figure 1). The global positioning system coordinates for these villages are the following: Bigege 0°35.576′ S, 34°44.975′ E; Luanda 0°25.909′ S, 34°55.687′ E; and Siany 0°25.619′ S, 34°54.707′ E. This is a rural area in the highlands (1,400–1,600 m above sea level) dominated by the Kisii and Luo ethnic groups, who are mainly subsistence farmers. The landscape is hilly with many small streams and rivers and households distributed on the sides of the valley. There are two seasonal peaks of malaria transmission reflecting a bimodal rainfall pattern, with the heaviest rainfall typically occurring between March and June with a smaller peak in October and November. Previous studies in Kisii have reported the primary vectors to be An. gambiae and An. funestus.39
Mosquito collections.
Mosquitoes were collected from consenting households between May 2010 and December 2011 from a number of different entomological studies as part of the Malaria Transmission Consortium (MTC) (institutional review board approval Kenya Medical Research Institute (KEMRI) no. SSC 1399, 2007). This project sought to improve tools for evaluating malaria transmission and control in diverse transmission environments.37 The MTC studies in the Kenya highlands have included entomological surveys that evaluate biting behavior, compare trapping techniques, and assess the impact of interventions in the highlands of Ranchuonyo and Kisii districts, western Kenya.40 Adult mosquitoes were collected using Centers for Disease Control and Prevention (CDC) miniature light traps set indoors and outdoors with either standard or ultraviolet bulbs. Indoor traps were either set next to a human under a bednet or baited with CO2. In Bigege, limited larval collections were conducted in the vicinity of surveyed houses using the standard dipping method and a small number (N = 95) of these immature stages also underwent molecular analyses for species identification.41 Water bodies were mapped and sampling undertaken at the same time as adult collections in the peak transmission season. In addition to these collections, larval collections were made from fish ponds in multiple locations in Rachuonyo. The larvae were brought to a field insectary and raised to adults. Larval and pupal skins and adults were identified using standard morphological keys. Mosquitoes were identified in the field using standard morphological keys,7,42 scored visually as blood-fed or unfed, and dried on silica gel until molecular processing.
Infectivity.
Female mosquitoes were analyzed for Plasmodium infection using circumsporozoite enzyme-linked immunosorbent assay (CS ELISA) for Plasmodium falciparum43 at KEMRI/CDC laboratories in Kisumu, western Kenya. Positive samples were sent to the University of Notre Dame, Indiana, for subsequent polymerase chain reaction (PCR) confirmation of infection using a nested PCR assay for P. falciparum and P. vivax.44
Blood meal analysis.
Abdomens of mosquitoes found to be blood-fed were analyzed using a blood meal diagnostic PCR assay based on vertebrate mitochondrial cytochrome b DNA sequences.45 Blood meals that did not amplify in the diagnostic PCR were sequenced and blasted against the NCBI nr database to identify the source46 of the blood meal.
Amplification by PCR and sequencing of the ITS2 and CO1 regions.
Genomic DNA was isolated from female anophelines using a CTAB technique or simple 70% alcohol precipitation. The rDNA ITS2 was amplified from genomic DNA using the ITS2A (5′-TGTGAACTGCAGGACACAT-3′) and ITS2B (5′-TATGCTTAAATTCAGGGGGT-3′) primers.47 The 25-μL PCR mixture contained 2.5 μL of 10× buffer, 0.2 mM of each deoxynucleotide triphosphate (dNTP), 1.2 mM MgCl2, 0.5 units of Taq DNA polymerase, 0.75 μL of 10 pmol/μL each of forward and reverse primers, and 1 μL of DNA template prepared as above. The thermocycling conditions were as follows: 94°C for 5 minutes, 30 cycles of denaturation at 94°C for 1 minute, annealing at 52°C for 1 minute, and extension at 72°C for 2 minutes, with a final extension at 72°C for 5 minutes.
The mitochondrial DNA cytochrome oxidase subunit 1 (CO1) gene was amplified using LCO and HCO primers.48 The primers used were LCO 1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO 2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′). The 25-μL PCR mixture contained 2.5 μL of 10× buffer, 0.2 mM of each dNTP, 1.2 mM MgCl2, 0.5 units of Taq DNA polymerase, 0.75 μL of 10 pmol/μL each of forward and reverse primers, and 1 μL of DNA template prepared as above. The thermocycling conditions were as follows: 94°C for 5 minutes, five cycles of denaturation at 94°C for 40 seconds, annealing at 45°C for 1 minute, and extension at 72°C for 1.5 minutes; then 30 cycles of denaturation at 94°C for 40 seconds, annealing at 51°C for 1 minute, and extension at 72°C for 1.5 minutes, with a final extension at 72°C for 5 minutes.
The amplified fragments were visualized by electrophoresis on a 1% agarose gel. The PCR product was purified using an enzyme cleanup; 2 units of exonuclease 1 (USB Corporation, Cleveland, OH), 1 units of Shrimp Alkaline Phosphatase (USB Corporation), and 1.8 μL of double distilled H2O were added to 8 μL of PCR product. This mixture was incubated at 37°C for 15 minutes, followed by 15 minutes at 80°C to inactivate the enzymes. The PCR products were sequenced directly (with one of the PCR primers) using Sanger sequencing on ABI 3730xl DNA Analyzer platform (PE Applied Biosystems, Warrington, England). A subset of specimens were independently confirmed using species-diagnostic PCRs for An. funestus and An. gambiae.9,24
Sequence analysis.
Raw ITS2 sequences were initially aligned using the Seqman pro assembler (Lasergene v 10.1.1, DNASTAR Inc., Madison, WI) with a minimum match of 90%. Assembled contigs were examined for repeats and further divided into subcontigs based on consistent single nucleotide polymorphisms (SNPs). A limit of 98% identity was used to assemble ITS2 sequences into final “species groups.” Single sequence contigs, low quality, or contaminated sequences were not included in the analysis. The consensus sequences of these ITS2 contigs were compared (Basic Local Alignment Search Tool nucleotide [BLASTn]) to the NCBI nr database for confirmation of species identities. High sequence identity (99% or greater) to voucher specimen sequences in the database or a combination of sequence and morphological identification was used for final species confirmation. High sequence similarity to non-voucher specimens was noted but not used for final species identity.
The CO1 sequences were similarly assembled and compared with the NCBI nr database for confirmation of species identities. Sequence groups were merged (minimum identity of 94%) when CO1 BLAST results indicated that they belonged to the same species for a final minimum match of > 95%. Single sequence contigs were not included in this analysis.
Before phylogenetic analysis, ITS2 sequences were initially annotated in web interface for ITS2 delimitation accessible at the ITS2-DB (http://its2.bioapps.biozentrum.uni-wuerzburg.de). This database utilizes comprehensive Hidden Markov Model approach to define the boundaries (start and end positions) of the ITS2 region by comparing to a conserved structural motif at 5.8S/28S ribosomal RNA regions. The ITS2 sequences were then aligned in MAFT49 using X-INS-i50 strategy. This alignment method detects conserved secondary structures in noncoding RNA sequences and is based on the Four-way Consistency objective function to build a multiple alignment by combining Stem Candidate Aligner for RNAs (SCARNA) algorithm for the initial pairwise alignments.50
Separate analyses for ITS2 sequences were done using Bayesian approach in MrBayes v3.1.251 using a general time reversible substitution model. Each analysis was performed with two independent runs with four chains, and each run was carried out for 10,000,000 generations with a sample frequency of 1,000. The first 25% of trees were discarded as burn-in and the posterior probabilities were estimated from the remaining trees to infer branch support.
Results
The ITS2 sequences representing 2,340 anopheline mosquitoes were aligned into 17 distinct groups with > 98% sequence identity within each group. These groups were arbitrarily designated Anopheles species A through Q. The consensus sequences of each group were compared (BLASTn) to the NCBI nr database to identify similarity to any sequence present in the database. Anopheles species B had a high (99.8%) sequence identity with ITS2 sequences of An. arabiensis, and those of Anopheles species D were identical to ITS2 sequences of An. funestus. The ITS2 consensus sequences from Anopheles species C, E, H, I, J, and L matched recently added non-voucher ITS2 sequences in the database. These included An. coustani (species C), An. maculipalpis (species E), An. pretoriensis (species H), An. theileri (species I), An. rufipes (species J), and An. leesoni (species L). The species' letter (B, C, D, E, H, I, J, and L) designations are henceforth referred to by their identified species' names. The consensus sequences from the other eight groups (A, F, G, K, M, N, O, P, and Q) did not share > 90% identity with any nr database sequence(s) (Table 1).
Table 1.
Summary of ITS2 and CO1 sequence groups, molecular and morphological identifications, and circumsporozoite ELISA and PCR positivity of anopheline specimens caught in the Kenyan highlands
| Sequence species group | Field-based morphological identification | Morphological identification | ITS2 no. sequenced | ITS2 sequence homology | CO1 no. sequenced | CO1 sequence homology | Tentative species ID | Final species ID | ELISA P.f. spz +ve/no. tested | PCR +ve/no. tested |
|---|---|---|---|---|---|---|---|---|---|---|
| Species A | (multiple species) | An. demeilloni group | 529 | – | 78 | – | Subgenus Cellia, Myzomyia series | Unknown | 11/318 | 5 P.f. 1 P./343 |
| Species B | An. gambiae s.l. | An. gambiae s.l. | 272 | An. arabiensis | 126 | An. arabiensis | An. arabiensis | An. arabiensis | 0/102 | 0/105 |
| Species C | An. coustani | An. coustani | 216 | An. coustani | 25 | An. coustani | An. coustani | An. coustani | 0/169 | 0/174 |
| (multiple species) | ||||||||||
| Species D | (multiple species) | An. funestus s.l. | 724 | An. funestus | 21 | An. funestus | An. funestus | An. funestus | 10/549 | 16 P.f. 7 P./687 |
| Species E | An. maculipalpis (multiple species) | An. maculipalpis | 138 | An. maculipalpis | 16 | – | Subgenus Cellia, Neocellia series, likely An. maculipalpis | An. maculipalpis | 0/97 | 0/108 |
| Species F | An. gambiae s.l. | – | 51 | – | 10 | – | Subgenus Cellia, Myzomyia series, near An. theileri | Unknown | 2/37 | 2 P.f./38 |
| Species G | (multiple species) | – | 38 | – | 5 | – | Subgenus Cellia, Myzomyia series, near An. theileri | Unknown | 1/23 | 1 P.f./23 |
| Species H | An. pretoriensis (multiple species) | – | 38 | An. pretoriensis | 9 | – | Subgenus Cellia, Neocellia series, possibly An. pretoriensis | An. pretoriensis | 0/28 | 0/28 |
| Species I | An. gambiae s.l. (multiple species) | – | 198 | An. theileri | 28 | – | Subgenus Cellia, Myzomyia series, possibly An. theileri | Unknown | 4/164 | 1 P./168 |
| Species J | (multiple species) | – | 30 | An. rufipes | 4 | – | Subgenus Cellia, Neocellia series, possibly An. rufipes | An. rufipes | 0/20 | 0/24 |
| Species K | (multiple species) | – | 29 | – | 3 | – | Subgenus Cellia, Cellia series | Unknown | 0/25 | 0/25 |
| Species L | An. funestus s.l. | An. funestus s.l. | 62 | An. leesoni | 2 | – | Subgenus Cellia, Myzomyia series, likely An. leesoni | An. leesoni | 0/59 | 1 P./60 |
| Species M | (multiple species) | An. squamosus | 4 | – | 2 | An. squamosus | An. squamosus | An. squamosus | 0/2 | 0/2 |
| Species N | – | – | 3 | – | 2 | – | Subgenus Cellia, Myzomyia series, near An. theileri | Unknown | 0/1 | 0/1 |
| Species O | An. coustani | – | 5 | – | 2 | An. coustani | Subgenus Anopheles, possibly an An. coustani sibling | Unknown | 0/3 | 0/3 |
| Species P | (multiple species) | – | 3 | – | 3 | An. christyi | Subgenus Cellia, Cellia series, possibly An. christyi | Unknown | 0/3 | 0/3 |
| Species Q | – | – | 2 | – | 2 | An. coustani | Subgenus Anopheles, possibly an An. coustani sibling | Unknown | 0/2 | 0/2 |
CO1 = cytochrome oxidase subunit 1; ELISA = enzyme-linked immunosorbent assay; ITS2 = internal transcribed spacer region 2; mtDNA = mitochondrial DNA; PCR = polymerase chain reaction; rDNA = ribosomal DNA.
Sequence groups were determined based on sequence identity of > 98% based on ribosomal DNA ITS2 sequences. The composition of the groups was confirmed with mitochondrial CO1 sequences. In total, 2,340 and 336 specimens were sequenced for rDNA ITS2 and mtDNA CO1, respectively. Detailed morphological identifications were carried out on a subsample of specimens by at least three entomologists in reference laboratories. P.f. refers to Plasmodium falciparum PCR confirmation of P.f. circumsporozoite ELISA positives. P. refers to PCR positives for Plasmodium that could not be confirmed to species using a second species-specific Plasmodium PCR. Positive specimens are bolded. The total number of each sequence group tested by ELISA and PCR are listed in the last two columns. Voucher sequences are underlined.
The CO1 sequences representing 336 mosquito specimens were aligned and the resulting distinct sequence groups were named as they correlated to the ITS2 groupings (again, A through Q). These included An. arabiensis (species B), An. coustani (species C), An. funestus (species D), An. squamosus (species M), and An. christyi (species P). Anopheles species O and Q sequences matched An. coustani sequences in the Barcode of Life Database (BOLD) database, though the ITS2 did not match An. coustani. The CO1 consensus sequences from Anopheles species groups A, E, F, G, H, I, J, K, and N did not share > 90% identity with any database sequence(s) (Table 1), indicating that these CO1 regions have not been previously sequenced.
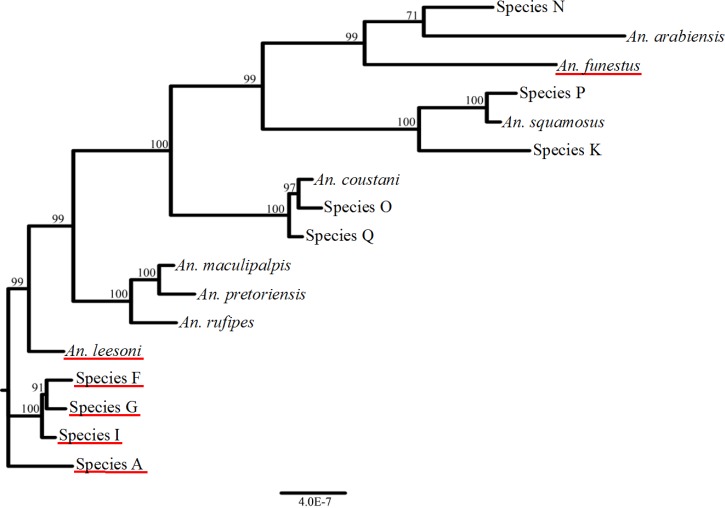
Consensus ITS2 sequences were structurally examined and aligned to construct a phenogram (Figure 2 ). This phenogram is meant only to demonstrate sequence divergence based on ITS2 sequences rather than represent the exact phylogenetic relationships of these groups. Insertions seen in group-specific sequences might increase the divergence between similar sequences. The distinct clustering and sequence divergence between molecular sequence groups indicate that many very distinct species of anophelines are present in these collections from a small geographic area. Notably, Plasmodium-positive Anopheles species A, I, F, and G cluster together, as do An. maculipalpis, An. pretoriensis, and An. rufipes, and Anopheles species K, P, and An. squamosus. Both the ITS2 and CO1 sequence of species O, Q, and An. coustani are very different than the other species analyzed and reference sequences. These are An. coustani and a potential An. coustani sibling species, which belong to subgenus Anopheles, whereas the rest of the species are members of subgenus Cellia. The grouping of An. maculipalpis, An. pretoriensis, and An. rufipes is consistent with other ITS2 phylogenies.52 Anopheles species B (An. arabiensis) and Anopheles species D (An. funestus) cluster closely at the top of the tree.
Figure 2.
Majority rule consensus tree from Bayesian analysis of internal transcribed spacer region 2 (ITS2) sequences of anopheline specimens collected in the Kenyan highlands. Bayesian posterior probabilities are shown above branches. The ITS2 sequence groups are named by their formal species name or by unknown Species A–Q. Red underscoring indicates groups that were found positive for Plasmodium infection. Note that Plasmodium positive unknown species A, F, G, and I group together.
As ITS2 sequence groups were eventually used to characterize the number of species, a stringent threshold of 98% identity was used to determine the maximum number of possible clusters and final number of species. Because of a typically higher rate of sequence divergence, a lower threshold of 94% sequence identity, after a higher initial threshold, was used for the assembly of CO1 groups, resulting in lower sequence similarity in the final species groupings.53,54 The CO1 group sequences that matched the same anopheline species reference sequence were collapsed into one group. This was reflected in the lower CO1 sequence similarity within a final species group when compared with that within the same species ITS2 sequence. To validate the proper assembly of these sequences, contigs were manually inspected for SNPs and repeat regions that would inflate divergence and decrease identity scores. These sequences are available in GenBank with accession numbers KJ522813–KJ544843.
Comparison of ITS2 and CO1 sequence groups.
The final description of 17 ITS2 sequence groups from the entire set of mosquitoes was validated by their 1:1 association with 15 separate CO1 sequence groups, except for two groups, Anopheles species O and Q, which matched the An. coustani CO1 sequence, but diverged at the ITS2 locus. This may be due to the presence of sibling species or introgression, which would need to be determined through further collections.
The molecular species identification was compared with morphological identification and many specimens could not be morphologically identified to terminal species in multiple keys. There was a low degree of concordance between molecular identification and field-based morphological identification of most groups (Table 1). For example, only half (51%) of the specimens morphologically identified as An. funestus s.l. belonged to that group when compared with well-characterized ITS2 reference sequences.9,55 Those not belonging to the An. funestus group fell into 12 other sequence groups. Results were similar for An. gambiae s.l. In most cases, the morphological identifications were mixed and not predictive of sequence groupings.
Preliminary morphological identification suggested that some of the specimens were not of the more commonly reported species in the area, and putatively identified two of the species groups—Anopheles species E was putatively identified as An. maculipalpis and Anopheles species I as possibly An. harperi. Anopheles species A, which was the second most abundant species in the collections, shared morphological features with members of the An. demeilloni group, for which there are no published ITS2 or CO1 sequences. The inconclusive nature of the morphological observations was primarily due to limitations in using available morphological keys in identifying some species, though several field specimens were also damaged. High-quality specimens representing the unknown groups could not be identified when compared with museum voucher specimens or identified to species using currently available keys or species descriptions. Only adult females were compared with morphological keys and museum voucher specimens.
Some molecular species identifications that were supported by morphological identifications and could be paired with high sequence identity to non-voucher sequences in the database were factored into the final species identification presented in Table 1. For example, Anopheles species M and E had morphological features supporting their ID to one species and had CO1 and ITS2 sequences matching with a high similarity to published sequences, it was concluded these were An. maculipalpis and An. squamosus, respectively. Since many of the sequences in NCBI and BOLD are incorrectly morphologically identified, species with matches in the databases to only non-voucher specimens, and not confirmed by any other method, could not be resolved to species in our final identification (Table 1). This is particularly true for sequence matches only within the BOLD database, such as Anopheles species O, P, and Q when the sequence alignment and source of the match could not be confirmed. The lack of species resolution of some sequence groups is based on the absence of any similar ITS2 or CO1 datasets.38
Several species were confirmed using both molecular and morphological tools, such as Anopheles species L, which morphologically matched with the An. funestus group, had high ITS2 sequence homology to a non-voucher sequence, and several specimens were tested using a species-diagnostic PCR for members of that group,9,24,56 indicating that the specimens were An. leesoni. Similarly, An. maculipalpis, An. squamosus, and An. coustani were confirmed through multiple indicators for their identification.
Sporozoite analysis.
Of 1,601 mosquitoes tested for the presence of P. falciparum sporozoites using CS ELISA, 28 were found positive. Eleven belonged to the Anopheles species A sequence group, 10 to An. funestus, two to Anopheles species F, one to Anopheles species G, and four to Anopheles species I (Table 1). All 1,601 specimens tested by ELISA, including those found positive, were tested by PCR. In addition, a further 193 specimens of those species shown to be positive for sporozoite by ELISA, that is, specimens with sequences matching Anopheles species A, An. funestus, Anopheles species F, Anopheles species G, and Anopheles species I with positive CS ELISA from the samearea were confirmed to be positive for Plasmodium infection by PCR.44 A total of five specimens of Anopheles species A, 16 of An. funestus, two of Anopheles species F, and one of Anopheles species G were positive for P. falciparum DNA by PCR (Table 1). These PCR positives were confirmed by sequencing. In addition, one Anopheles species A, seven An. funestus, one Anopheles species I, and one An. leesoni were positive for Plasmodium DNA but could not be confirmed to be Plasmodium species in a secondary species-specific PCR.44 Anopheles species A, F, G, and I were species that could not be definitively identified using morphology or molecular tools to known species. No mosquitoes of the third most abundant sequence group, An. arabiensis, a presumed primary vector in the area, were found to be positive for Plasmodium infections in these collections.
Blood meal analysis.
Of 42 blood-engorged mosquitoes, 28 analyzed from the light trap collections were identified to specific blood meal hosts by PCR. Specimens that fell into the Anopheles species A group were found to contain human blood mixed with cow blood in two blood meals, as well as eight blood meals from cow, one from a dog, and one from a donkey. One specimen of An. pretoriensis had a mixed human and cow blood meal. Of four blood-fed Anopheles species I, one had a human blood meal and three had cow blood meals. These blood meal identifications confirm only that these species had fed on human and other blood hosts. An. maculipalpis, An. species F, and Anopheles species G had two, three, and three cow blood meals, respectively. No blood meals were found in females of the known vector species An. arabiensis and An. funestus available for analysis.
Discussion
Downstream associations of bionomic characteristics, vectorial capacity, and entomological inoculation rates based solely on morphologically identified mosquitoes may differ substantially from those obtained from the more precise types of molecular identifications presented here. Molecular analysis of divergence at both rDNA ITS2 and mtDNA CO1 loci of over 2,300 mosquitoes collected over an 18-month period in three villages in the Kenyan highlands of Nyanza Province indicated the presence of P. falciparum sporozoite–infected mosquitoes that are not members of the commonly known vector species in Kenya. More than half of the specimens, 63%, were assigned to known vector taxa and the remaining 37% fell into groups that could not be conclusively identified using morphology or multiple molecular markers. The latter contained most of the sporozoite-positive specimens and females that had fed on humans. Of those specimens for which there are no published ITS2 and CO1 genetic sequences, CS ELISA positive specimens of Anopheles species A, F, and G were confirmed positive for parasite DNA by PCR and sequencing. The specimens of Anopheles species I that were ELISA positive for P. falciparum sporozoites could not be confirmed with PCR.57
The sequences were assembled into 17 (ITS2) and 15 (CO1) groups. These sequence groups had a 1:1 correspondence, that is, all specimens in an ITS2 sequence group had a matching set of CO1 sequences, except for one group (Anopheles species O) that had a different ITS2 sequences but a matching CO1 sequence. A large number of sequences were not present in the NCBI database. This result alone does not necessarily represent a novel (i.e., unnamed) species, but rather that the ITS2 or CO1 sequences may belong to formally described species which have not been sequenced previously.
The comparison of specimens within the sequence groups to voucher sequences in public databases, to each other, and detailed morphological criteria has enabled the identification of reference sequences for several Kenyan species: ITS2 for An. squamosus and An. christyi, and CO1 for An. rufipes, An. pretoriensis, and An. maculipalpis. Several specimens that shared some features, but not all, with those of species of the An. gambiae complex were later shown to be An. arabiensis because of high sequence similarity (100%) to published sequences. A match of a single non-voucher sequence to either ITS2 or CO1 was not considered in the final species identification because many unpublished or non-voucher sequences that are deposited in sequence databases are misidentified. This issue underlies the need for sequences of voucher specimens of both vector and nonvector species to be represented in the database.
Anopheles species A, F, G, and I are described here as species that have not previously been identified as malaria vectors in the study area, and which may or may not be species of Anopheles new to science. These four Plasmodium positive species also cluster together in the ITS2 phenogram, indicating that they could be closely related or sibling species (Figure 2). Specimens with ITS2 sequences matching those of Anopheles species A have been found in multiple sites in Zambia (N. Lobo, unpublished data), indicating that this species may be distributed more widely in Africa and could be playing a role in malaria transmission elsewhere. This study represents only an important first step in identifying any potentially new species. Detailed descriptions of adult female mosquitoes need to be further validated with careful inspection of a greater number of high-quality adult female and male specimens and other life stages. The specific behaviors of Anopheles species A, one of the most abundant species in our study sites with a P. falciparum infection rate comparable to that previously published for An. funestus in western Kenya,58,59 are currently being characterized.
This study emphasizes the importance of combining molecular tools with morphological identifications, particularly in areas with diverse species. Available adult female anopheline morphological keys and species descriptions may be suitable for identifying common and well-known species but do not include more recently described species, some life stages or sexes are missing from keys, descriptions can be incomplete and open to misinterpretation, and captured mosquitoes may be damaged and missing key morphological features. Members of the An. gambiae and An. funestus complexes are the most studied mosquitoes in Africa. Studies that focus on primarily identifying these mosquito species, often based on the assumptions that they are the most common anophelines or the only vectors, might result in misidentification or discarding of unexpected or unknown vectors. Even within these well-studied species complexes, new species are being discovered using molecular techniques, such as a new species in the An. funestus complex that was found indoors and could not be identified using the common species diagnostic PCR.55 Anopheles species A, for example, thought by the authors to belong to the An. demeilloni group, might be mistaken for An. funestus. An initial or periodic molecular characterization of species composition, using sequencing or available PCR diagnostics, could be used to compliment morphological approaches for more cost-effective monitoring of vector species. This would also enable the identification of field specimens that are often too damaged to identify morphologically and enable quick processing of large numbers of specimens.
The presence of new or unknown vectors with uncharacterized behaviors will impact the efficiency of interventions and hints at the complex nature of the malaria transmission paradigm. The two most common interventions, indoor residual spraying and use of insecticide-treated bednets, target indoor behaviors exhibited by members of the An. gambiae and An. funestus complexes that may not be characteristic of other species involved in malaria transmission. Detailed descriptions of vector species, their behaviors, vectorial capacity, insecticide resistance, and other characteristics relevant to transmission are vital to our understanding of local transmission dynamics and for the deployment of effective interventions.60
ACKNOWLEDGMENTS
We are grateful to the community of Bigege village, Kisii Central, and Luanda and Siany villages, Rachuonyo South, Nyanza Province for their cooperation and participation in the study. We thank Ralph Harbach and Lorna Culverwell at the Department of Life Sciences, Natural History Museum, London, for providing taxonomic advice and assistance for field collections, identifying mosquitoes, and contributing to the manuscript. We thank all of the skilled field entomologists for their hard work and dedication and also thank the Division of Vector Borne Diseases (DVBD). We pay a special tribute to one of the key entomologists at DVBD, Mr. Peter Karanja, who sadly passed away. We are also grateful for the support of our partners in the Kenya Medical Research Institute/U.S. Centers for Disease Control and Prevention, Kisumu, Kenya.
Footnotes
Financial support: This project was funded by the Bill & Melinda Gates Foundation under the Malaria Transmission Consortium grant no. 45114. This article has been approved by the Director of the Kenya Medical Research Institute.
Authors' addresses: Brandyce St. Laurent, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN, and Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, E-mail: brandyce.stlaurent@nih.gov. Mary Cooke, Chris Drakeley, and Jonathan Cox, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom, E-mails: mary_cooke@hotmail.com, chris.drakeley@lshtm.ac.uk, and jonathan.cox@lshtm.ac.uk. Sindhu M. Krishnankutty, Western Triangle Research Center, Montana State University, Conrad, MT, E-mail: oriolealis@gmail.com. Puji Asih, Department of Malaria, Eijkman Institute for Molecular Biology, Jakarta, Indonesia, E-mail: puji@eijkman.go.id. John D. Mueller, Julie Thumloup, Frank H. Collins, and Neil F. Lobo, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN, E-mails: jmuelle8@alumni.nd.edu, juliethumloup@hotmail.com, frank@nd.edu, and nlobo@nd.edu. Samuel Kahindi, Elizabeth Ayoma, and Robin M. Oriango, Centre for Global Health Research, Kenya Medical Research Institute/Centers for Disease Control and Prevention, Kisumu, Kenya, E-mails: kahindisamuel@gmail.com, bettynyawera@yahoo.com, and robinmigiro@rocketmail.com. Jennifer C. Stevenson, Johns Hopkins School of Public Health, Malaria Research Institute, E-mail: jennyc.stevenson@macharesearch.org.
References
- 1.Lindblade KA, Eisele TP, Gimnig JE, Alaii JA, Odhiambo F, ter Kuile FO, Hawley WA, Wannemuehler KA, Phillips-Howard PA, Rosen DH, Nahlen BL, Terlouw DJ, Adazu K, Vulule JM, Slutsker L. Sustainability of reductions in malaria transmission and infant mortality in western Kenya with use of insecticide-treated bednets: 4 to 6 years of follow-up. JAMA. 2004;291:2571–2580. doi: 10.1001/jama.291.21.2571. [DOI] [PubMed] [Google Scholar]
- 2.Gimnig JE, Kolczak MS, Hightower AW, Vulule JM, Schoute E, Kamau L, Phillips-Howard PA, ter Kuile FO, Nahlen BL, Hawley WA. Effect of permethrin-treated bed nets on the spatial distribution of malaria vectors in western Kenya. Am J Trop Med Hyg. 2003;68:115–120. [PubMed] [Google Scholar]
- 3.Gimnig JE, Vulule JM, Lo TQ, Kamau L, Kolczak MS, Phillips-Howard PA, Mathenge EM, ter Kuile FO, Nahlen BL, Hightower AW, Hawley WA. Impact of permethrin-treated bed nets on entomologic indices in an area of intense year-round malaria transmission. Am J Trop Med Hyg. 2003;68:16–22. [PubMed] [Google Scholar]
- 4.Miller JE, Lindsay SW, Armstrong JR. Experimental hut trials of bednets impregnated with synthetic pyrethroid or organophosphate insecticide for mosquito control in the Gambia. Med Vet Entomol. 1991;5:465–476. doi: 10.1111/j.1365-2915.1991.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 5.Bayoh M, Walker E, Kosgei J, Ombok M, Olang G, Githeko A, Killeen G, Otieno P, Desai M, Lobo N, Vulule J, Hamel M, Kariuki S, Gimnig J. Persistently high estimates of late night, indoor exposure to malaria vectors despite high coverage of insecticide treated nets. Parasit Vectors. 2014;7:380. doi: 10.1186/1756-3305-7-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, Fontenille D. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 2006;43:1215–1221. doi: 10.1603/0022-2585(2006)43[1215:cotmvs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Gillies MT, de Meillon B. The Anopheline of Africa South of the Sahara. Johannesburg, South Africa: South African Institute of Medical Research; 1968. [Google Scholar]
- 8.Coetzee M, Hunt RH, Wilkerson R, Torre AD, Coulibaly MB, Besansky NJ. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- 9.Koekemoer L, Kamau L, Hunt R, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;6:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 10.Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5:1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- 11.Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16:74–77. doi: 10.1016/s0169-4758(99)01563-x. [DOI] [PubMed] [Google Scholar]
- 12.Pates HV, Takken W, Curtis CF, Jamet H. Zoophilic Anopheles quadriannulatus species B found in a human habitation in Ethiopia. Ann Trop Med Parasitol. 2006;100:177–179. doi: 10.1179/136485906X86374. [DOI] [PubMed] [Google Scholar]
- 13.Fettene M, Hunt R, Coetzee M, Tessema F. Behaviour of Anopheles arabiensis and An. quadriannulatus sp. B mosquitoes and malaria transmission in southwestern Ethiopia. Afr Entomol. 2004;12:83–87. [Google Scholar]
- 14.Petrarca V, Beier JC, Onyango F, Koros J, Asiago C, Koech DK, Roberts CR. Species composition of the Anopheles gambiae complex (Diptera: Culicidae) at two sites in western Kenya. J Med Entomol. 1991;28:307–313. doi: 10.1093/jmedent/28.3.307. [DOI] [PubMed] [Google Scholar]
- 15.Githeko AK, Adungo NI, Karanja DM, Hawley WA, Vulule JM, Seroney IK, Ofulla AV, Atieli FK, Ondijo SO, Genga IO, Odada PK, Situbi PA, Oloo JA. Some observations on the biting behavior of Anopheles gambiae s.s., Anopheles arabiensis, and Anopheles funestus and their implications for malaria control. Exp Parasitol. 1996;82:306–315. doi: 10.1006/expr.1996.0038. [DOI] [PubMed] [Google Scholar]
- 16.Sinka M, Bangs M, Manguin S, Coetzee M, Mbogo C, Hemingway J. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Killeen G, Seyoum A, Sikaala C, Zomboko A, Gimnig J, Govella N. Eliminating malaria vectors. Parasit Vectors. 2013;6:172. doi: 10.1186/1756-3305-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lwetoijera D, Harris C, Kiware S, Dongus S, Devine G, McCall P, Majambere S. Increasing role of Anopheles funestus and Anopheles arabiensis in malaria transmission in the Kilombero Valley, Tanzania. Malar J. 2014;13:331. doi: 10.1186/1475-2875-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent RJ, Thuma PE, Mharakurwa S, Norris DE. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am J Trop Med Hyg. 2007;76:267–274. [PMC free article] [PubMed] [Google Scholar]
- 20.Rishikesh N, Di Deco MA, Petrarca V, Coluzzi M. Seasonal variations in indoor resting Anopheles gambiae and Anopheles arabiensis in Kaduna, Nigeria. Acta Trop. 1985;42:165–170. [PubMed] [Google Scholar]
- 21.Mnzava AE, Rwegoshora RT, Wilkes TJ, Tanner M, Curtis CF. Anopheles arabiensis and An. gambiae chromosomal inversion polymorphism, feeding and resting behaviour in relation to insecticide house-spraying in Tanzania. Med Vet Entomol. 1995;9:316–324. doi: 10.1111/j.1365-2915.1995.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 22.Durnez L, Coosemans M. Residual Transmission of Malaria: An Old Issue for New Approaches. Rijeka, Croatia: InTech; 2013. [Google Scholar]
- 23.Pates H, Curtis C. Mosquito behavior and vector control. Annu Rev Entomol. 2005;50:53–70. doi: 10.1146/annurev.ento.50.071803.130439. [DOI] [PubMed] [Google Scholar]
- 24.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 25.Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) J Med Entomol. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Harbach RE. The phylogeny and classification of Anopheles. In: Manguin S, editor. Anopheles Mosquitoes—New Insights into Malaria Vectors. Rijeka, Croatia: InTech; 2013. pp. 3–55. [Google Scholar]
- 27.Kelly-Hope L, Ranson H, Hemingway J. Lessons from the past: managing insecticide resistance in malaria control and eradication programmes. Lancet Infect Dis. 2008;8:387–389. doi: 10.1016/S1473-3099(08)70045-8. [DOI] [PubMed] [Google Scholar]
- 28.Okara RM, Sinka ME, Minakawa N, Mbogo CM, Hay SI, Snow RW. Distribution of the main malaria vectors in Kenya. Malar J. 2010;9:69. doi: 10.1186/1475-2875-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndenga B, Githeko A, Omukunda E, Munyekenye G, Atieli H, Wamai P, Mbogo C, Minakawa N, Zhou G, Yan G. Population dynamics of malaria vectors in western Kenya highlands. J Med Entomol. 2006;43:200–206. doi: 10.1603/0022-2585(2006)043[0200:pdomvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 30.Carlson JC, Byrd BD, Omlin FX. Field assessments in western Kenya link malaria vectors to environmentally disturbed habitats during the dry season. BMC Public Health. 2004;4:33. doi: 10.1186/1471-2458-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ototo EN, Githeko AK, Wanjala CL, Scott TW. Surveillance of vector populations and malaria transmission during the 2009/10 El Nino event in the western Kenya highlands: opportunities for early detection of malaria hyper-transmission. Parasit Vectors. 2011;4:144. doi: 10.1186/1756-3305-4-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou G, Afrane YA, Vardo-Zalik AM, Atieli H, Zhong D, Wamae P, Himeidan YE, Minakawa N, Githeko AK, Yan G. Changing patterns of malaria epidemiology between 2002 and 2010 in western Kenya: the fall and rise of malaria. PLoS One. 2011;6:e20318. doi: 10.1371/journal.pone.0020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minakawa N, Omukunda E, Zhou G, Githeko A, Yan G. Malaria vector productivity in relation to the highland environment in Kenya. Am J Trop Med Hyg. 2006;75:448–453. [PubMed] [Google Scholar]
- 34.Wanjala CL, Waitumbi J, Zhou G, Githeko AK. Identification of malaria transmission and epidemic hotspots in the western Kenya highlands: its application to malaria epidemic prediction. Parasit Vectors. 2011;4:81. doi: 10.1186/1756-3305-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atieli HE, Zhou G, Lee MC, Kweka EJ, Afrane Y, Mwanzo I, Githeko AK, Yan G. Topography as a modifier of breeding habitats and concurrent vulnerability to malaria risk in the western Kenya highlands. Parasit Vectors. 2011;4:241. doi: 10.1186/1756-3305-4-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minakawa N, Munga S, Atieli F, Mushinzimana E, Zhou G, Githeko AK, Yan G. Spatial distribution of anopheline larval habitats in western Kenyan highlands: effects of land cover types and topography. Am J Trop Med Hyg. 2005;73:157–165. [PubMed] [Google Scholar]
- 37.Stuckey EM, Stevenson JC, Cooke MK, Owaga C, Marube E, Oando G, Hardy D, Drakeley C, Smith TA, Cox J, Chitnis N. Simulation of malaria epidemiology and control in the highlands of western Kenya. Malar J. 2012;11:357. doi: 10.1186/1475-2875-11-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson J, St Laurent B, Lobo NF, Cooke MK, Kahindi SC, Oriango RM, Harbach RE, Cox J, Drakeley C. Novel vectors of malaria parasites in the western highlands of Kenya. Emerg Infect Dis. 2012;18:1547–1549. doi: 10.3201/eid1809.120283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ototo EN, Mbugi JP, Wanjala CL, Zhou G, Githeko AK, Yan G. Surveillance of malaria vector population density and biting behaviour in western Kenya. Malar J. 2015;14:244. doi: 10.1186/s12936-015-0763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooke MK, Kahindi SC, Oriango RM, Owaga C, Ayoma E, Mabuka D, Nyangau D, Abel L, Atieno E, Awuor S, Drakeley C, Cox J, Stevenson J. “A bite before bed”: exposure to malaria vectors outside the times of net use in the highlands of western Kenya. Malar J. 2015;14:259. doi: 10.1186/s12936-015-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver JB. Mosquito Ecology: Field Sampling Methods. Dordrecht, The Netherlands: Springer Science and Business Media; 2007. [Google Scholar]
- 42.Gillies T, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara (Afrotropical Region) Johannesburg, South Africa: South African Institute for Medical Research; 1987. Vol. 55. [Google Scholar]
- 43.Burkot TR, Zavala F, Gwadz RW, Collins FH, Nussenzweig RS, Roberts DR. Identification of malaria-infected mosquitoes by a two-site enzyme-linked immunosorbent assay. Am J Trop Med Hyg. 1984;33:227–231. doi: 10.4269/ajtmh.1984.33.227. [DOI] [PubMed] [Google Scholar]
- 44.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 45.Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- 46.Hostetter G, Collins E, Varlan P, Edewaard E, Harbach PR, Hudson EA, Feenstra KJ, Turner LM, Berghuis BD, Resau JH, Jewell SD. Veterinary and human biobanking practices: enhancing molecular sample integrity. Vet Pathol. 2014;51:270–280. doi: 10.1177/0300985813510532. [DOI] [PubMed] [Google Scholar]
- 47.Beebe NW, Saul A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction–restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995;53:478–481. doi: 10.4269/ajtmh.1995.53.478. [DOI] [PubMed] [Google Scholar]
- 48.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 49.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 52.Norris LC, Norris DE. Phylogeny of anopheline (Diptera: Culicidae) species in southern Africa, based on nuclear and mitochondrial genes. J Vector Ecol. 2015;40:16–27. doi: 10.1111/jvec.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubinoff D, Cameron S, Will K. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J Hered. 2006;97:581–594. doi: 10.1093/jhered/esl036. [DOI] [PubMed] [Google Scholar]
- 54.Lin CP, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 2004;30:686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 55.Spillings BL, Brooke BD, Koekemoer LL, Chiphwanya J, Coetzee M, Hunt RH. A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am J Trop Med Hyg. 2009;81:510–515. [PubMed] [Google Scholar]
- 56.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 57.Durnez L, Van Bortel W, Denis L, Roelants P, Veracx A, Trung HD, Sochantha T, Coosemans M. False positive circumsporozoite protein ELISA: a challenge for the estimation of the entomological inoculation rate of malaria and for vector incrimination. Malar J. 2011;10:195. doi: 10.1186/1475-2875-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fillinger U, Ndenga B, Githeko A, Lindsay SW. Integrated malaria vector control with microbial larvicides and insecticide-treated nets in western Kenya: a controlled trial. Bull World Health Organ. 2009;87:655–665. doi: 10.2471/BLT.08.055632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawada H, Dida GO, Sonye G, Njenga SM, Mwandawiro C, Minakawa N. Reconsideration of Anopheles rivulorum as a vector of Plasmodium falciparum in western Kenya: some evidence from biting time, blood preference, sporozoite positive rate, and pyrethroid resistance. Parasit Vectors. 2012;5:230. doi: 10.1186/1756-3305-5-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brady OJ, Godfray HCJ, Tatem AJ, Gething PW, Cohen JM, McKenzie FE, Alex Perkins T, Reiner RC, Tusting LS, Scott TW, Lindsay SW, Hay SI, Smith DL. Adult vector control, mosquito ecology and malaria transmission. Int Health. 2015;7:121–129. doi: 10.1093/inthealth/ihv010. [DOI] [PMC free article] [PubMed] [Google Scholar]