Abstract
Viruses that infect the intestine include major human pathogens (retroviruses, noroviruses, rotaviruses, astroviruses, picornaviruses, adenoviruses, herpesviruses) constituting a major public health problem worldwide. These viral pathogens are members of a large, complex viral community inhabiting the intestine termed the enteric virome. Enteric viruses have intimate functional and genetic relationships with both the host and other microbial constituents that inhabit the intestine, like the bacterial microbiota, their associated phages, helminthes and fungi which together constitute the microbiome. Emerging data indicate that enteric viruses regulate, and are in turn regulated by, these other microbes through a series of processes termed transkingdom interactions. This represents a changing paradigm in intestinal immunity to viral infection. Here we review recent advances in the field and propose new ways in which to conceptualize this important area.
Overview
Enteric viruses include important human pathogens that spread in food, water and through contact. These viruses encounter an incredible environment on entry into the gastrointestinal tract, and have evolved their outer layers, either proteinaceous capsids or lipid envelopes, to both manage and leverage this complex milieu. Peristalsis moves enteric viruses through an assortment of pH gradients, digestive enzymes, and microbes before they penetrate the mucus layer or cross intestinal M cells to infect the host. Both direct and indirect interactions of enteric viruses with other microbes and the host immune system are increasingly recognized as a critical aspect of their infectivity, disease induction, and control (Figure 1). Perhaps this is not surprising since these viruses must complete their life cycle in the intestine which is one of the most complex microbial environments on earth (1–4).
Figure 1. The microbiome alters enteric virus infection by both direct and indirect mechanisms.
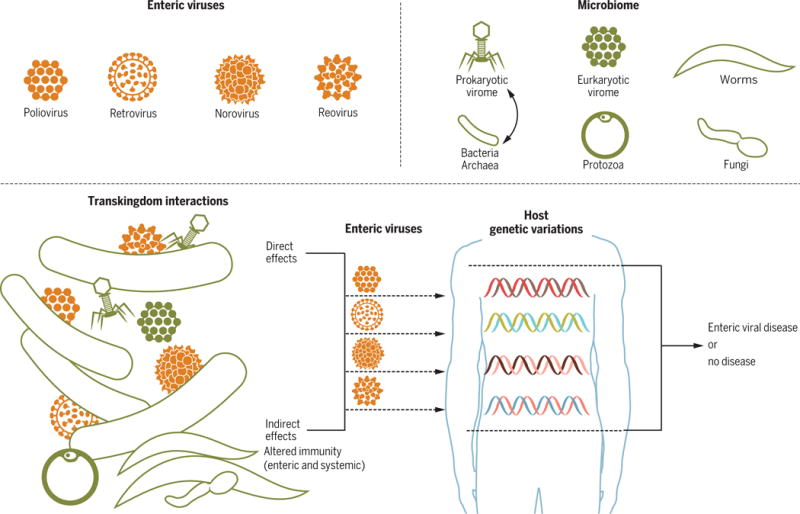
Shown is a schematic representation of major categories of organisms in the intestinal microbiome that, in aggregate, affect enteric infection with the four viral taxa shown. In some cases transkingdom interactions affect enteric viruses directly through physical interactions between viruses and organisms or components of organisms within the intestinal microbiome. In other cases the microbiome regulated viral infection indirectly by altering immunity. Systemic viral infection can also alter intestinal virus infection. Host genes also impact these transkingdom interactions, and the ultimate outcome of virus-triggered disease, as shown on the right portion of the figure.
Over the past decade many studies have examined the composition and, to a lesser extent, the function of microbes that inhabit the intestine and other body sites (5–7). In addition to bacteria, the intestine contains multiple other types of organisms that can have profound effects on mucosal and systemic immune responses including viruses (8–14), fungi (15, 16), and eukaryotes such as the protozoa Blastocystis (17) or helminths (18, 19). Together these organisms have been referred to as the microbiome or the microbiota. Given the emerging understanding of transkingdom interactions as determinants of host immunity and virus infection, it is possible that a significant proportion of the unexplained variation in mammalian responses to infection may be explained by inter-host variation in the microbiota (1, 3, 4, 9, 10, 20–27).
The enteric virome
Analysis of the intestinal virome has revealed a dynamic environment featuring both bacteriophages and multiple eukaryotic viruses. Viruses that infect systemic tissues or other mucosal surfaces such as the lung also influence the intestine in important ways including via control of tissue homeostasis, healing, and the outcome of infection with bacterial pathogens (23, 28). Viruses can even benefit the host by promoting immune development and influencing tissue architecture (10, 20, 22, 23). It is now possible to understand, albeit incompletely, the taxonomic structure of the viruses present in the intestine. The task of identifying the individual viruses present is in its infancy because many sequences obtained in random or ’shot gun’ libraries of DNA and RNA from intestinal contents or mucosae cannot be effectively annotated due to weaknesses in databases and the lack of easy-to-use software. A particular problem has been a focus on sequencing only entities with DNA genomes when so many enteric viruses have RNA genomes.
It is nevertheless clear that the viral contents of the intestine are remarkably complex (2, 4, 13, 29–31). It has been estimated that the intestine contains about 100-trillion prokaryotic cells (32) many of which carry temperate bacteriophages in their genomes. Bacteriophages that infect these prokaryotic organisms may be ~10-fold more abundant than their host cells (1, 4, 33). In addition, there is a eukaryotic virome in the asymptomatic host, with humans averaging at least 10 permanent systemic eukaryotic viral infections (3, 4, 20). A variety of enteric viruses infect humans, including members of the Retroviridae, Adenoviridae, Astroviridae, Caliciviridae, Reoviridae, and Picornaviridae, Picobirnaviridae, Annelloviridae and Circoviridae families. Some of these infections are very common (e.g., norovirus and reovirus), while others are rare (e.g., poliovirus, a picornavirus). Infections range from asymptomatic to severe acute disease to severe chronic disease. Asymptomatic people can shed noroviruses for prolonged periods (34), and some strains of murine norovirus (MNoV) establish life-long intestinal infection of mice (34–36). Several studies have identified enteric viruses in feces from asymptomatic people or other mammals (3, 37–42); the intestine of healthy children can contain various eukaryotic viruses including picobirnaviruses, adenoviruses, anelloviruses, astroviruses, caliciviruses, bocaviruses, enteroviruses, circoviruses, rotaviruses, and sapoviruses (13, 43–47). Together these data indicate that the enteric virome has been vastly underestimated. Thus, the number of prokaryotic and eukaryotic viruses that might contribute to, or be regulated by, transkingdom interactions relevant to the intestine is daunting.
Replication and transmission of mammalian enteric viruses
For successful propagation and transmission, enteric viruses must navigate several unique niches within the gastrointestinal tract. Less than 100 viral particles are frequently sufficient for infection with human enteric viruses such as norovirus, rotavirus, and poliovirus (48–50). Enteric viruses encounter low pH, proteolytic enzymes, microbiota, and intestinal mucus prior to replication in the intestine. Therefore, initiation of viral replication from the intestinal lumen is likely to be far more difficult than subsequent cycles of viral replication initiated by spread from previously infected intestinal cells or between cells in tissue culture. Different enteric viruses replicate in different cell types, ranging from enterocytes to lymphocytes to myeloid cells. Following viral replication, progeny viruses are shed into the lumen leading to transmission to new hosts.
Promotion of enteric viral infection by bacteria
To examine how intestinal microbes influence viral infection and transmission many investigators use mouse models of infection including antibiotic treated animals or germ-free mice; each approach has pros and cons (51, 52). Antibiotic treatment is relatively simple and inexpensive, but depletion of bacteria is incomplete and selective depending on the antibiotic(s) used. Germ-free mice, while providing a “clean slate” devoid of all microbes, require specialized facilities and have immature intestinal architecture and immune responses compared with conventional mice; the organ targeted by enteric viruses is therefore abnormal. Mice are relatively resistant to a variety of human enteric pathogens, including many viruses and bacteria (53), rarely develop diarrhea and cannot vomit. As a result, the general approach has been to examine human pathogens in immune-deficient and/or young mice and to examine mouse pathogens that are closely related to human pathogens in their natural host.
A combination of experiments using germ-free and antibiotic-treated mice infected with human and murine viruses have provided compelling evidence that the bacterial microbiota, through transkingdom interactions, influences viral infection (Figure 2). Intestinal bacteria promote replication and transmission of enteric viruses from four different families in orally infected mice. Mouse mammary tumor virus (MMTV), a member of the Retroviridae, is spread from mother to offspring through milk. Experiments in germ-free mice demonstrate that intestinal bacteria are required for efficient MMTV transmission (54). Poliovirus, a member of the Picornaviridae, is spread by the fecal-oral route and can disseminate to the central nervous system. Intestinal bacteria enhance poliovirus replication, systemic pathogenesis, and fecal-oral transmission in mice (55, 56). Intestinal microbes enhance replication and pathogenesis of reovirus and rotavirus, both members of the Reoviridae, in mice (55, 57). MNoV, a member of the norovirus genus within the Caliciviridae, is commonly present in mouse facilities and is spread by the fecal-oral route. Several groups have shown that intestinal bacteria promote MNoV replication and persistence (10, 58, 59). Importantly, the beneficial effects of the bacterial microbiota on enteric viruses are not observed when viruses are delivered by intraperitoneal injection that bypasses the natural oral infection route (55, 59, 60). This may explain why effects of the microbiota on enteric viruses were unrecognized until recently; many previous studies delivered virus by injection rather than the natural oral route.
Figure 2. Transkingdom interactions and mechanisms by which bacteria enhance enteric virus replication and transmission.
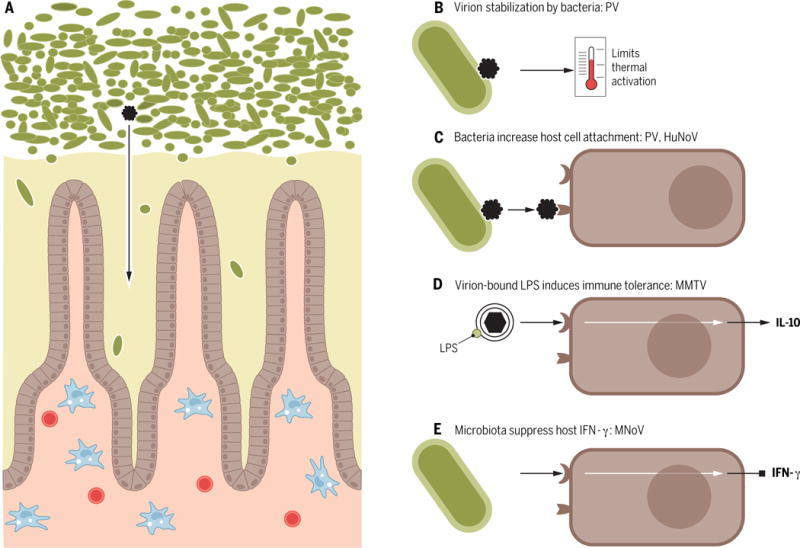
(A) The intestinal environment includes bacteria (green) and other components of the microbiome, the mucus layer (yellow), host enterocytes (brown), and underlying immune cells (blue and red). Enteric viruses (black) are exposed to bacteria prior to initiating replication in host cells (enterocytes or immune cells, depending on the virus type). Bacteria promote enteric virus replication and transmission by several mechanisms. (B) Poliovirus (PV) binds bacteria, which stabilizes virions and limits premature RNA release to promote transmission. (C) PV and human norovirus (HuNoV) bind bacteria, which increases viral attachment to host cells. (D) Mouse mammary tumor virus (MMTV) binds LPS, which induces host TLR signaling and IL10-mediated immune tolerance. (E) Murine norovirus (MNoV) replication is enhanced by bacteria, likely through regulation of IFNλ responses.
Several enteric viruses that benefit from the bacterial microbiota bind bacterial surface polysaccharides resulting in enhanced viral infectivity and pathogenesis. MMTV, a virus with a lipid bilayer envelope, binds lipopolysaccharide (LPS), a glycan on the surface of Gram-negative bacteria (54). As a consequence, viral infection initiates innate immune responses that culminate in host tolerance, viral replication and transmission (Figure 2). Poliovirus, a non-enveloped virus, binds LPS as well as peptidoglycan, a major component of the bacterial cell wall, enhancing viral attachment to host cells and promoting transmission by stabilizing viral particles and limiting thermal inactivation (55, 56) (Figure 2). Human norovirus, a non-enveloped virus, binds carbohydrate histo-blood group antigens (HBGAs) present on host cells and certain bacterial cells (58, 61), facilitating human norovirus infection of B cells, likely though enhanced viral attachment to host cells (58) (Figure 2). Collectively, these studies indicate that diverse viruses have evolved to bind to and benefit from bacterial surface glycans.
Regulation of intestinal immunity to viruses by the microbiota
The intestinal immune system has been extensively reviewed (62, 63). Here we focus on recent data relevant to the role of immunity in transkingdom interactions regulating virus infection.
The bacterial microbiota influences immunity to a variety of viruses. Recently it has been shown that pre-existing antibodies to enteric bacteria can skew vaccine responses to cross-reacting HIV-1 antigens, arguably rendering a vaccine less protective (64). Enteric bacterial components regulate vaccine responses to influenza in mice through activation of the innate immune receptor, Toll like receptor (TLR)5 (65). Antibiotic treatment of mice has profound effects on antiviral immunity at another mucosal surface, the lung, since antibiotic treatment prevents normal innate and adaptive immune responses to influenza resulting in death of the host (66–68). These results underscore the importance of the bacterial microbiota for antiviral immune responses.
Members of the enteric microbiota in addition to bacteria, such as helminthic worms, also have profound effects on intestinal antiviral immunity. Helminth infection of mice inhibits intestinal antiviral immune CD8 T cell responses to MNoV (69), an effect that is independent of other members of the microbiota as it is observed in germ free mice. Intestinal helminth infection also affects the control of systemic herpesvirus infection in mice (70). Here, helminths induce secretion of cytokines such as interleukin (IL)4 or IL13, which triggers transcription from specific viral promoters and subsequent reactivation from latency. This effect is evolutionarily conserved between murine and human γ-herpesviruses. Interestingly in both of these studies, the effect of helminth infection was associated with worm-induced changes in macrophage differentiation. The role of macrophages in the control of intestinal biology is also suggested by the fact that systemic virus infection results in the production of type 1 interferons (IFN), which through their effects on macrophages, significantly increase proliferation of intestinal epithelial cells resulting in improved wound healing (28).
Bacterial molecules also have striking effects on immune signaling pathways relevant to enteric virus infection. Virus-associated LPS induces TLR4 signaling culminating in IL10-mediated immune tolerance and increased transmission of MMTV (54). Antibiotic treatment of mice prevents persistent enteric MNoV infection, an effect that is reversed by fecal transplantation (59). This action of antibiotics required the presence of the IFNλ receptor, indicating that this signaling pathway is involved in transkingdom regulation of persistent enteric virus infection (Figure 2). Together these data indicate that, in addition to direct binding of bacterial components to viruses, the indirect effects of the enteric microbiota on immunity can have profound effects on viral infection.
One of the most interesting findings to come from studies of transkingdom interactions and viral infection is the discovery of sterilizing innate immunity in the intestine (71, 72). Treatment of mice with IFNλ effectively cured persistent MNoV infection in mice lacking T cells and B cells and therefore unable to mount an antigen-specific adaptive immune response. It is intriguing that enteric bacteria also control persistent enteric norovirus infection in a manner requiring the IFNλ receptor, although the mechanisms connecting these two observations remain to be elucidated (59). In the case of rotavirus infection, treatment of mice with bacterial flagellin cured virus infection via a mechanism involving TLR signaling and induction of the cytokines IL22 and IL18. Again, viral clearance did not require adaptive immunity. It has recently been reported that IL22 and IFNλ can synergize to clear enteric rotavirus infection, suggesting that there may be a common pathway for sterilizing innate immunity in the intestine (73). Strategies to enhance these innate immune mechanisms, perhaps by manipulating the enteric bacterial microbiota, may be useful for treating enteric virus infection independent of vaccine responses. These studies indicate the importance of analyzing transkingdom interactions controlling enteric viral infection as a window into fundamental mechanisms of immunity.
Complex genetic interactions dictate outcomes
Given that transkingdom interactions govern enteric virus infection, it is not surprising that viral disease is dependent on interaction of microbes and viruses with host genes (Figure 1). In one example, the combination of MNoV and a murine mutation in a human Crohn’s disease susceptibility gene, ATG16L1, results in pathological abnormalities in intestinal Paneth cells when neither the host gene mutation nor the virus infection alone can induce these abnormalities (9, 74). A similar process has been shown in studies of MNoV in combination with mutation of the immunoregulatory cytokine IL10 (24). Importantly virus-triggered colonic pathology in each of these studies is dependent on intestinal bacteria. In the case of MNoV + ATG16L1 interactions, treatment of mice with antibiotics ameliorated pathology, while in studies of MNoV + IL10 interactions pathology was ameliorated in germ free mice. Thus, virus-induced pathology in the intestine can be driven by a transkingdom interaction with bacteria.
Viruses may also control intestinal bacteria by transferring genes between bacteria and through predator-prey relationships in which bacteriophages alter the taxonomic structure of the bacterial microbiota (1, 8, 14, 47). These types of transkingdom interactions may have disease relevance. For example in human Crohn’s disease and ulcerative colitis, enteric bacteriophages become more taxonomically complex even as the bacterial microbiota becomes less diverse and taxonomically rich (8). This is consistent with disease-associated changes in predatory-prey relationships between viruses and bacteria. Taken together, these studies underscore the importance of transkingdom interactions, and the association of these events with host genetic variation, in dictating responses to infectious agents and inflammatory disease.
Future challenges
While we have expanded our understanding of the virome and transkingdom control of enteric virus infection, several key challenges remain. First, a more thorough examination of the enteric virome, incliding both DNA and RNA viruses, and how it changes over time is essential. We do not fully understand all of the players in the complex dance of transkingdom interactions. Further, comparatively few studies have evaluated the intestinal mycobiome, the archaea or the meiofauna. Characterizing the virome and other members of the enteric microbiota will allow investigators to more thoroughly explore the phenomenology of transkingdom control of viral infection, setting the stage for more mechanistic studies and therapeutic translation of novel concepts that arise.
Another challenge is making sense of multi-factorial interactions that frequently have so many components related in non-linear ways that they seem to defy analysis. It is likely that there will be more examples of mechanisms revealed only upon combined analysis of host genetics, components of the microbiota, and viral infection (9, 24, 54–56). Furthermore, environmental factors such as diet and circadian rhythms are likely to influence enteric viral infections (75–77). We are only taking the first steps on a long road into a new area of research in virology, but it is clear that we need to embrace this complexity if we are to fully understand viral pathogenesis and immunity.
Finally, we need to address whether new knowledge about enteric viruses gained by studies of transkingdom interactions can be used therapeutically. While recent work implies that antibiotic treatment can have antiviral effects, this is clearly not a viable treatment option for enteric virus infections due to logistical issues and side effects, not the least of which is disruption of the many positive effects the bacterial microbiota has for the host. However, targeted therapies based on mechanistic insights into transkingdom interactions are more realistic. For example, treatment with IFNλ or bacterial flagellin could have significant effects on enteric virus infection (71, 72). Furthermore, it is possible that some enteric viruses may benefit the host by providing immunoregulatory cues (4, 10, 20). Alterations of the infectivity of viruses by binding of bacterial or other microbiota components might alter virulence and immunogenicity. Identification of beneficial human enteric or systemic viruses may inform fecal transplant therapies.
Conclusion
The pro-viral effect of other microbes for enteric virus infection was revealed only recently, which is surprising considering that the technology required for most of the experiments has existed for many years. A simple conceptual advance, examining enteric viruses in the intestine following natural oral infection, was key for driving the field forward. Therefore, while reductionist experiments are important and aid our understanding of viral replication and pathogenesis, removing the complexity of the intestinal environment has apparently masked important biology.
Enteric viruses do not exist in a vacuum. They have evolved in the unique environment of the intestine for optimal replication and transmission. A complex interplay between viruses, bacteria, the host, and the environment determines the efficiency of viral replication, disease, and transmission. Recent work has shed light on factors that influence enteric virus infection. However, there are still major gaps in our understanding of enteric virus infection. Future studies on the mammalian virome and transkingdom factors that influence enteric viral infection may inspire new therapeutic approaches.
Supplementary Material
Acknowledgments
We thank members of our laboratories for useful discussions and comments on the manuscript. J.K.P. is supported by NIH grants R01 AI074668 and R21 AI114927 and a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases Award. H.W.V. is supported by U19 AI109725, R24 OD019793, R01 AI111918, R01 DK 101354, Kenneth Rainin Foundation grant #14H16, and Juvenile Diabetes Research Foundation grant 2-SRA-2015-305-Q-R.
Contributor Information
Julie K. Pfeiffer, Email: Julie.Pfeiffer@UTSouthwestern.edu.
Herbert W. Virgin, Email: virgin@wustl.edu.
References
- 1.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nature immunology. 2013 Jul;14:654. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norman JM, Handley SA, Virgin HW. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 2014 May;146:1459. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nature reviews Microbiology. 2011 Apr;9:254. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014 Mar 27;157:142. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naik S, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015 Apr 2;520:104. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. The ISME journal. 2012 May;6:915. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014 Oct 2;514:59. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015 Jan 29;160:447. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010 Jun 25;141:1135. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kernbauer E, Ding Y, Cadwell K. An enteric viral infection can functionally replace the beneficial cues provided by commensal bacteria. Nature. 2014;516:94. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2012 Apr 06;109:3962. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010 Jul 15;466:334. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes A, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proceedings of the National Academy of Sciences of the United States of America. 2015 Sep 8; doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proceedings of the National Academy of Sciences of the United States of America. 2013 Dec 10;110:20236. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science (New York, NY) 2012 Jul 08;336:1314. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nature reviews Immunology. 2014 Jun;14:405. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher SM, Stark D, Harkness J, Ellis J. Enteric Protozoa in the Developed World: a Public Health Perspective. Clinical microbiology reviews. 2012 Jul 03;25:420. doi: 10.1128/CMR.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McSorley HJ, Hewitson JP, Maizels RM. Immunomodulation by helminth parasites: defining mechanisms and mediators. International journal for parasitology. 2013 Apr;43:301. doi: 10.1016/j.ijpara.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Stelekati E, Wherry EJ. Chronic bystander infections and immunity to unrelated antigens. Cell Host & Microbe. 2012 Oct 18;12:458. doi: 10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009 Jul 10;138:30. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Moon C, et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature. 2015 May 7;521:90. doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007 May 17;447:326. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 23.MacDuff DA, et al. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife. 2015 Jan 15;4 doi: 10.7554/eLife.04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basic M, et al. Norovirus Triggered Microbiota-driven Mucosal Inflammation in Interleukin 10-deficient Mice. Inflammatory bowel diseases. 2014;20:431. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- 25.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011 May 26;473:523. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293. doi: 10.1038/nm.1935. 3/2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furman D, et al. Cytomegalovirus infection enhances the immune response to influenza. Science translational medicine. 2015 Apr 1;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun L, et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell host & microbe. 2015 Jan 14;17:85. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon C, Stappenbeck TS. Viral interactions with the host and microbiota in the intestine. Curr Opin Immunol. 2012 Aug;24:405. doi: 10.1016/j.coi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804. doi: 10.1038/nature06244. 10/18/2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647. doi: 10.1126/science.1155725. 6/20/2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proceedings of the …. 1998 doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minot S, et al. Rapid evolution of the human gut virome. Proceedings of the National Academy of Sciences of the United States of America. 2013 Jul 23;110:12450. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in norovirus biology. Cell host & microbe. 2014 Jun 11;15:668. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thackray LB, et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. Journal of virology. 2007 Oct;81:10460. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nice TJ, Strong DW, McCune BT, Pohl CS, Virgin HW. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. Journal of virology. 2013 Jan;87:327. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, et al. The fecal viral flora of California sea lions. Journal of virology. 2011 Oct;85:9909. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkbeiner SR, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS pathogens. 2008 Feb;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handley SA, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012 Oct 12;151:253. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan T, et al. The fecal virome of pigs on a high-density farm. Journal of virology. 2011 Nov;85:11697. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan TG, et al. The fecal viral flora of wild rodents. PLoS pathogens. 2011 Sep;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith I, Wang LF. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 2013 Feb;3:84. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. 2012 Nov;50:3427. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phan TG, et al. A third gyrovirus species in human faeces. The Journal of general virology. 2012 Jul;93:1356. doi: 10.1099/vir.0.041731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Victoria JG, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. Journal of virology. 2009 Jun;83:4642. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux S, Hallam SJ, Woyke T, Sullivan MB. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. eLife. 2015;4 doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim ES, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nature medicine. 2015 Sep 14; doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward RL, et al. Human rotavirus studies in volunteers: determination of infectious dose and serological response to infection. The Journal of infectious diseases. 1986 Nov;154:871. doi: 10.1093/infdis/154.5.871. [DOI] [PubMed] [Google Scholar]
- 49.Katz M, Plotkin SA. Minimal infective dose of attenuated poliovirus for man. Am J Public Health Nations Health. 1967 Oct;57:1837. doi: 10.2105/ajph.57.10.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teunis PF, et al. Norwalk virus: how infectious is it? J Med Virol. 2008 Aug;80:1468. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 51.Robinson CM, Pfeiffer JK. Viruses and the Microbiota. Annual review of virology. 2014;1:55. doi: 10.1146/annurev-virology-031413-085550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012 Jun 8;336:1268. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabin AB. Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science. 1956 Jun 29;123:1151. doi: 10.1126/science.123.3209.1151. [DOI] [PubMed] [Google Scholar]
- 54.Kane M, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011 Oct 14;334:245. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuss SK, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011 Oct 14;334:249. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell host & microbe. 2014 Jan 15;15:36. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. The Journal of infectious diseases. 2014 Jul 15;210:171. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jones MK, et al. Enteric bacteria promote human and murine norovirus infection of B cells. Science. 2014;346:755. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldridge MT, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015 Jan 16;347:266. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaffer J, Beamer PR, Trexler PC, Breidenbach G, Walcher DN. Response of germ-free animals to experimental virus monocontamination. I. Observation on Coxsackie B virus. Proc Soc Exp Biol Med. 1963 Mar;112:561. doi: 10.3181/00379727-112-28105. [DOI] [PubMed] [Google Scholar]
- 61.Miura T, et al. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J Virol. 2013;87:9441. doi: 10.1128/JVI.01060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annual review of immunology. 2012 May 23;30:759. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell host & microbe. 2012 Oct 18;12:496. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams WB, et al. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015 Jul 30; doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014 Sep 18;41:478. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dolowy WC, Muldoon RL. Studies of Germfree Animals. I. Response of Mice to Infection with Influenza a Virus. Proc Soc Exp Biol Med. 1964 Jun;116:365. doi: 10.3181/00379727-116-29249. [DOI] [PubMed] [Google Scholar]
- 67.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011 Mar 29;108:5354. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012 Jul 27;37:158. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osborne LC, et al. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science. 2014;345:578. doi: 10.1126/science.1256942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reese TA, et al. Helminth infection reactivates latent y-herpesvirus via cytokine competition at a viral promoter. Science. 2014;345:573. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014 Nov 14;346:861. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nice TJ, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015 Jan 16;347:269. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hernandez PP, et al. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nature immunology. 2015 Jul;16:698. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008 Nov 13;456:259. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hickman D, et al. The effect of malnutrition on norovirus infection. mBio. 2014;5:e01032. doi: 10.1128/mBio.01032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu X, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013 Nov 8;342:727. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014 Oct 23;159:514. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


