The pathogenesis of atrial fibrillation (AF), the most common sustained cardiac arrhythmia, involves multiple factors including dysfunction of a variety of ion channels, dysregulation of calcium handling proteins, developmental defects, etc.1 Emerging evidence has shown that genetic factors can increase the risk of AF. For example, genome-wide association studies (GWASs) have identified a number of single-nucleotide polymorphisms (SNPs) that are associated with AF.2–5 Among these AF-associated SNPs, two (rs2200733 and rs10033464) located on chromosome 4q25 are most significant and have been repeatedly found in AF patients of various ethnicities.2–4
It has been postulated that these 4q25 variants can increase AF susceptibility by modulating the activity of paired-like homeodomain transcription factor 2 (PITX2), since they are located in the vicinity (∼150 000 bp) of the cis-regulatory region of PITX2.2–4 Consistently, independent groups have demonstrated that the deficiency of PITX2c (the cardiac-specific isoform) predisposes to AF development in mice.6,7 However, it remains controversial whether and how human 4q25 variants regulate PITX2 transcription and whether the level of PITX2 is affected in AF patients.
Previous experimental work has revealed that PITX2 itself plays a crucial role in left atrium (LA)–right atrium (RA) patterning during cardiac development, and that the lack of PITX2c can alter LA–RA asymmetry leading to malformation of the pulmonary veins.6 The latter is a well-known site for ectopic activity promoting spontaneous AF induction.8 More recently, utilizing next generation sequencing and micro-array techniques, PITX2c was shown to potentially regulate a variety of ion transporters and gap junction proteins that are crucial for pace-making and cardiac conduction.7,9 Lozano-Velasco et al.10 and Pérez-Hernández et al.11 have separately investigated the impact of PITX2c on ion channels and calcium handling proteins.
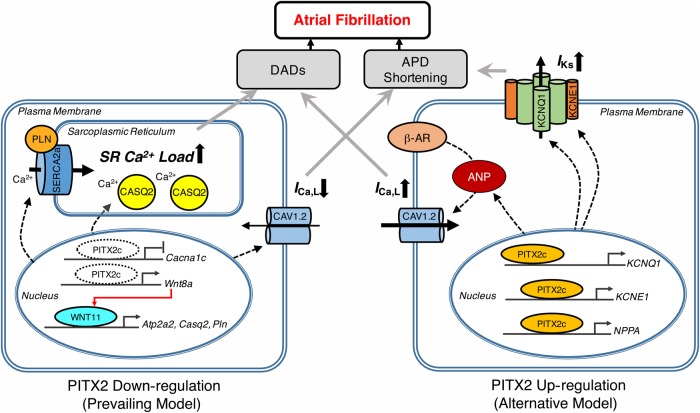
First, consistent with the majority of publications to date, Lozano-Velasco et al. demonstrated that down-regulation of PITX2 is arrhythmogenic (Figure 1, left panel). They utilized two distinct Pitx2 loss-of-function models, a conditional mouse line (Sox2-Cre-Pitx2) and a previously established atrial-specific knockout line (Nppa-Cre-Pitx2), to demonstrate potential transcriptional changes in key calcium handling proteins. This study revealed alterations in mRNA levels of calcium handling proteins such as SERCA2a, calsequestrin-2, and phospholamban; a reduction in ICa,L current density; and an increase in sarcoplasmic reticulum calcium content in Nppa-Cre-Pitx2 mice, but not in Sox2-Cre-Pitx2 mice. These differences explain the different phenotype in basal electrophysiology, as the former mouse line developed atrial arrhythmias spontaneously whereas the latter mouse strain exhibits normal sinus rhythm. Another novel finding from this study is that Wnt signalling may play a pivotal role in directing such differences, since (i) PITX2c is a negative regulator of Wnt11 via Wnt8a and (ii) overexpression of Wnt11 increases gene transcription of SERCA2a, calsequestrin-2, and phospholamban, similar to the effect of loss-of-function Pitx2c.
Figure 1.
Schematic representation of arrhythmic mechanisms associated with PITX2. In the prevailing model (left, Lozano-Velasco study), down-regulation of PITX2c (i) reduces the gene expression of α-subunit of L-type Ca2+ channel (ICa,L), a hallmark of electrical remodeling in AF (ii) increases the expression of Ca2+ handling proteins via Wnt signaling, which ultimately leads to Ca2+ overload in sarcoplasmic reticulum (SR). In an alternative model (right, Pérez-Hernández study), up-regulation of PITX2c, observed in chronic AF patients, can enhance the transcription of genes encoding the subunits of slow delayed rectifying K+ channel (IKs) and atrial natriuretic peptide (ANP). The latter is responsible for the augmented ICa,L in response to β-adrenergic stimulation. These cellular events will ultimately (i) promote the development of delayed afterdepolarizations (DADs), or (ii) abbreviate the action potential duration (APD), both of which are the well-established mechanisms contributing to pathogenesis of atrial fibrillation.
On the other hand, the study by Pérez-Hernández et al.11 has raised an alternative hypothesis that up-regulation of PITX2 is also associated with AF arrhythmogenesis (Figure 1, right panel). In this study, the investigators used freshly isolated human atrial cardiomyocytes from patients with long-standing persistent (chronic) AF (cAF) and HL-1 cells (murine atrial myocyte-derived cultured cells). In contrast to prior studies, they found an increased level of PITX2c mRNA in atrial cardiomyocytes of cAF patients. The expression levels of PITX2c correlated positively with IKs current density and negatively with ICa,L current density, respectively. The inward ICa,L and outward IKs are major currents that modulate phase 2 of the action potential of cardiomyocytes, and APD shortening is a hallmark of atrial electrical remodelling facilitating the induction and maintenance of AF. However, the putative correlation between PITX2c expression and APD was not determined in this study. Despite this limitation, this study provided the first evidence of PITX2c expression in atrial cardiomyocytes from cAF patients. Moreover, Pérez-Hernández et al. demonstrated that PITX2c can directly modulate the transcription of KCNQ1 and KCNE1 (genes encoding IKs channel) and that overexpression of PITX2c increases the IKs current in HL-1 cells. Although PITX2c does not exert direct transcriptional modulation on ICa,L subunits, Pérez-Hernández et al. showed that PITX2c can affect the function of ICa,L indirectly via an atrial natriuretic peptide-mediated transcriptional activity. The essential findings from these two studies are highlighted in Figure 1.
Data comparisons between humans and mice have been inconsistent at times. For example, the level of PITX2c in AF patients is either unaltered,12 increased in Pérez-Hernández et al.'s study,11 or decreased in a previous study by Dr. Franco's group.13 However, all data from animal models have shown consistently that deficiency of PITX2c increases the susceptibility to AF. A possible explanation is that human atrial cardiomyocytes were obtained from right atrial appendages of AF patients, which may not reflect properly the entire atrial landscape since PITX2c expression is higher in the LA. In addition, the history (duration and recurrence rate) of AF and clinical variability (age, comorbidities, drug therapy, etc.) among the patients may also complicate the results, although the higher PITX2 levels in cAF might be a compensatory mechanism counteracting the proarrhythmic effects of downregulated PITX2 at early stages. Finally, it remains to be established—for example in transgenic mouse models—whether increased PITX2c expression as observed by Pérez-Hernández et al.11 is causally linked to AF pathogenesis.
Despite the differences between these two studies, both groups have linked arrhythmia mechanisms attributed to PITX2 with other established molecular determinants of AF. With the findings from both cAF patients and conditional knockout mice, the role of PITX2 in AF arrhythmogenesis has been expanded to the adulthood stage.9–11,14 Finally, although we have witnessed a rapid discovery of genetic variants that could increase the risk of AF, it remains unclear whether and how these variants modulate nearby genes (directly vs. indirectly).12 Perhaps, with the assistance of cutting-edge technologies, such as Crispr gene-editing, iPS cells, and AAV-mediate gene transfer, future studies can focus on rapid validation of the causative variants associated with AF development. Even if these common variants revealed by GWASs are truly not causative genetic determinants, they may potentially act as genetic modifiers of other rare mutations or variants to alter the atrial electrophysiology.15 Last but not least, it deserves investigation to determine whether any of these common variants could serve as the genetic biomarker of AF.
Funding
This work was supported by National Institutes of Health (R01-HL089598, R01-HL091947, R01-HL117641, and R41-HL129570 to X.H.T.W.), American Heart Association (13EIA14560061 to X.H.T.W., and 14SDG20080008 to N.L.), and DZHK (German Center for Cardiovascular Research, to D.D.).
References
- 1.Andrade J, Khairy P, Dobrev D, Nattel S. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res 2014;114:1453–1468. [DOI] [PubMed] [Google Scholar]
- 2.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 3.Kaab S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, Schnabel R, Makino S, Sinner MF, Kannankeril PJ, Beckmann BM, Choudry S, Donahue BS, Heeringa J, Perz S, Lunetta KL, Larson MG, Levy D, MacRae CA, Ruskin JN, Wacker A, Schomig A, Wichmann HE, Steinbeck G, Meitinger T, Uitterlinden AG, Witteman JC, Roden DM, Benjamin EJ, Ellinor PT. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J 2009;30:813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME, Dorr M, Ozaki K, Smith AV, Muller-Nurasyid M, Walter S, Agarwal SK, Bis JC, Brody JA, Chen LY, Everett BM, Ford I, Franco OH, Harris TB, Hofman A, Kaab S, Mahida S, Kathiresan S, Kubo M, Launer LJ, Macfarlane PW, Magnani JW, McKnight B, McManus DD, Peters A, Psaty BM, Rose LM, Rotter JI, Silbernagel G, Smith JD, Sotoodehnia N, Stott DJ, Taylor KD, Tomaschitz A, Tsunoda T, Uitterlinden AG, Van Wagoner DR, Volker U, Volzke H, Murabito JM, Sinner MF, Gudnason V, Felix SB, Marz W, Chung M, Albert CM, Stricker BH, Tanaka T, Heckbert SR, Jukema JW, Alonso A, Benjamin EJ, Ellinor PT. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol 2014;63:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA 2010;107:9753–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet 2011;4:123–133. [DOI] [PubMed] [Google Scholar]
- 8.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res 2007;101:902–909. [DOI] [PubMed] [Google Scholar]
- 9.Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, Kaminski HJ, Martin JF. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet 2014;7:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozano-Velasco E, Hernandez-Torres F, Daimi H, Serra SA, Herraiz A, Hove-Madsen L, Aranega A, Franco D. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc Res 2016;109:55–66. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Hernandez M, Matamoros M, Barana A, Amoros I, Gomez R, Nunez M, Sacristan S, Pinto A, Fernandez-Aviles F, Tamargo J, Delpon E, Caballero R. Pitx2c increases in atrial myocytes from chronic atrial fibrillation patients enhancing IKs and decreasing ICa,L. Cardiovasc Res 2016;109:431–441. [DOI] [PubMed] [Google Scholar]
- 12.Gore-Panter SR, Hsu J, Hanna P, Gillinov AM, Pettersson G, Newton DW, Moravec CS, Van Wagoner DR, Chung MK, Barnard J, Smith JD. Atrial Fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS One 2014;9:e86245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpon E, Tamargo J, Cinca J, Hove-Madsen L, Aranega AE, Franco D. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet 2011;4:269–279. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, Martin JF. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci USA 2014;111:9181–9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie MD, Rowan S, Kucera G, Stubblefield T, Blair M, Carter S, Roden DM, Darbar D. Chromosome 4q25 variants are genetic modifiers of rare ion channel mutations associated with familial atrial fibrillation. J Am Coll Cardiol 2012;60:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]