Summary
Precise regulation of transcription is crucial for the cellular mechanisms underlying memory formation. However, the link between neuronal stimulation and the proteins that directly interact with histone modifications to activate transcription in neurons remains unclear. Brd4 is a member of the BET protein family, which binds acetylated histones and has a critical role in numerous cell types in regulating transcription, including in the response to external cues. Small molecule BET inhibitors are in clinical trials, yet almost nothing is known about Brd4 function in the brain. Here we show that Brd4 is a key player in neuronal function and mediates the transcriptional regulation underlying learning and memory. The loss of Brd4 function affects critical synaptic proteins, which results in memory deficits in mice but also decreases seizure susceptibility. Thus, Brd4 provides a critical, and previously uncharacterized, link between neuronal activation and the transcriptional responses that occur during memory formation.
The nervous system requires tight control of transcription in response to external signals. Rapid activation of immediate early genes (IEGs) in response to stimulation is critical for synaptic plasticity and is observed in vivo during learning and memory. Misregulation of gene expression in the brain results in neuronal deficits and neurodevelopmental disorders1,2, and inhibition of transcription immediately following neuronal stimulation blocks the mechanisms underlying memory formation3–6. This inducible transcription requires that transcription activators bind to promoters of target genes and recruit other proteins such as RNA Polymerase II (PolII)7,8. Recent work found that in several non-neuronal cell types, the protein Brd4 is critical in regulating the recruitment of protein complexes such as positive transcription elongation factor b (P-TEFb) to allow for PolII phosphorylation and the subsequent elongation of target genes in response to a signal9–12.
Brd4 is a member of the bromodomain and extra-terminal domain (BET) protein family and functions as a chromatin ‘reader’ that binds acetylated lysines in histones13,14. Knockout of Brd4 in mice is lethal15 and recent elegant work indicates that small molecule inhibitors of BET proteins represent a promising therapeutic strategy for several types of cancer16–18. Brd4 also regulates stimulus-dependent transcription in postmitotic cells by recruiting P-TEFb to target promoters in response to extracellular signals13,19. While P-TEFb recruitment is necessary for transcriptional elongation in neurons20, the link between neuronal stimulation and the proteins that directly interact with histone modifications to activate transcription remains unclear.
Brd4 is well-positioned to regulate transcription in neurons in response to neuronal activation. Acetyl marks are critical to brain function and are linked to memory formation and multiple neurological disorders21. Brd4 activity is regulated by casein kinase 2 (CK2)14, which is activated in response to neuronal stimulation22. In addition, a full understanding of if and how Brd4 functions in the brain is of particular importance now as multiple BET protein inhibitors are currently in clinical trials.
Here we show that Brd4 is critical to neuronal function and mediates the transcriptional regulation underlying learning and memory. We find that Brd4 regulates IEG transcription in neurons in response to activity and is regulated by CK2. Loss of Brd4 function affects critical synaptic proteins and the BET inhibitor Jq1 results in memory deficits and decreases seizure susceptibility in mice. These results provide the first demonstration of Brd4 function in the brain and provide a critical link between neuronal activity and transcriptional activation that underlies memory formation. In addition, our data call attention to the potential for small molecule inhibitors of BET proteins such as Jq1 to cause neuronal deficits. While BET protein inhibitors are a promising therapeutic strategy for several types of cancer17,18,23–25, modifications preventing blood-brain barrier penetrability may be necessary to prevent neurological side effects.
Results
Brd4 is expressed in neurons
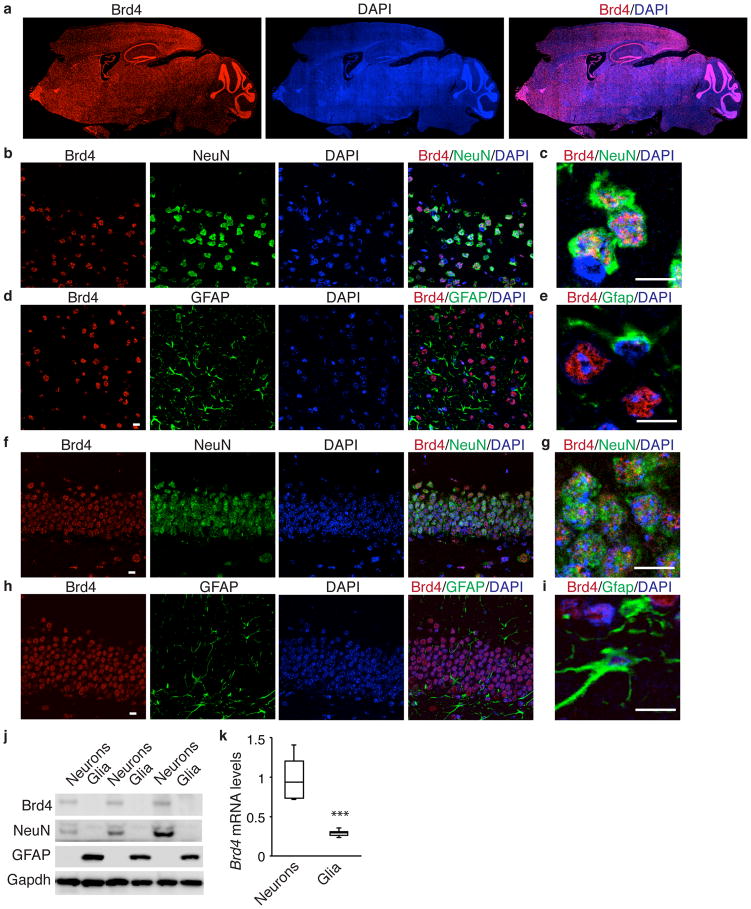
We examined Brd4 expression in adult mice using an antibody that detects the full-length form of Brd4 and found that it is expressed throughout the brain (Fig. 1a, Supplementary Fig. 1a). Brd4 positive cells typically express NeuN but not GFAP in both cortex and hippocampus (Fig. 1b–i) indicating that Brd4 is present in neurons while generally not seen in glial cells. In addition, we separately cultured cortical neurons and glia and found that neurons contain more Brd4 mRNA and protein than glial cells (Fig. 1j, k). Both CamKI-positive excitatory neurons and GABA-positive inhibitory neurons express Brd4 (Supplementary Fig. 1b, c). Finally, we treated cultured neurons with brain-derived neurotrophic factor (BDNF) to mimic physiological activation in the brain6, which resulted in small increases in Brd4 mRNA and protein (Supplementary Fig. 1d–f).
Figure 1. Brd4 is expressed in neurons throughout the brain.
(a) Brd4 staining of a sagittal adult mouse brain section. (b, d) Brd4 and NeuN costaining of cortex (b) or hippocampus (d). (c, e) High magnification image of Brd4 and NeuN costaining of cortex (c) or hippocampus (e). (f, h) Brd4 and GFAP costaining of cortex (f) or hippocampus (h). (g, i) High magnification image of Brd4 and GFAP costaining of cortex (g) or hippocampus (i). (j) Western blot analysis of Brd4 protein from whole cell lysate of cultured cortical neurons or glia. (k) Brd4 mRNA from cultured cortical neurons or glia (n = 3 biological replicates, paired two-tailed t test, P = 0.0057, t = 4.195.) Full-length blots are presented in Supplementary Figure 10. Error bars represent standard error. ***, p<0.001. Scale bar is 10 μM.
Brd4 regulates IEG transcription in neurons
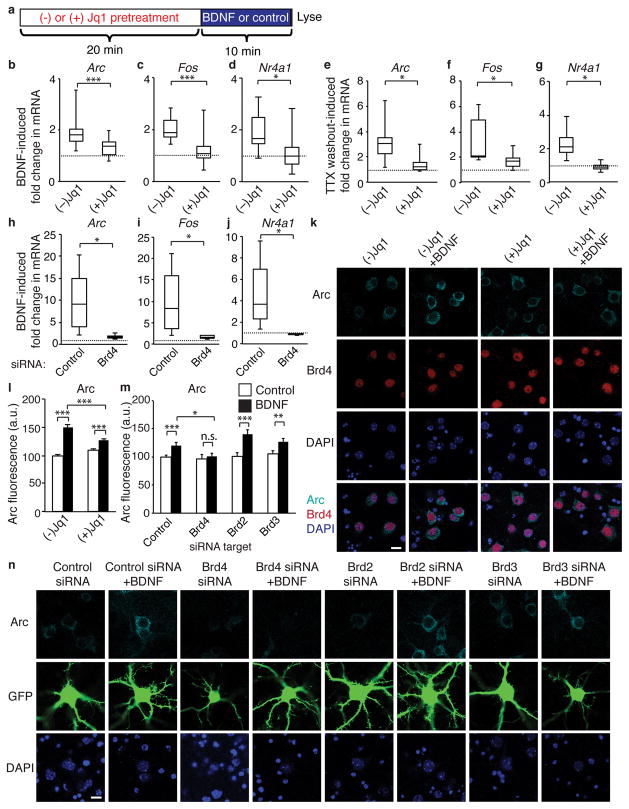
Similar to other post-mitotic cells that require Brd413,19, neurons activate a subset of genes (IEGs) in response to external signals. This rapid response is critical to the consolidation of synaptic modifications underlying synaptic plasticity and memory formation3–6. We examined whether Brd4 is involved in transcriptional activation in neurons using the small molecule inhibitor Jq1 which blocks BET proteins from binding to acetylated histones18. After pretreatment with Jq1 or the negative enantiomer (−)Jq1, cultured cortical neurons were stimulated with BDNF (Fig. 2a). As expected, BDNF caused a rapid increase in transcripts of IEGs Arc and Fos. However, pre-treatment with Jq1 blocked the BDNF-induced increase (Fig. 2b, c). Rapidly induced IEGS such as Arc and Fos have PolII poised on their promoters to allow for immediate activation, while other IEGs such as Nr4a1 must both recruit and phosphorylate PolII to activate transcription20. We found that Jq1 also prevented the activity-induced increase Nr4a1 (Fig. 2d) indicating that Jq1’s affects are not limited to IEGs with poised PolII.
Figure 2. Brd4 regulates IEG transcription in response to stimulation in neurons.
(a) Experimental paradigm for analysis of effects of Jq1 on activity-dependent transcription. (b, c, d) Jq1 pretreatment blocks the fold-increase in nascent RNA of Arc (b), Fos (c) and Nr4a1 (d) in cultured cortical neurons in response to 10 minutes of BDNF stimulation (n = 14 biological replicates, paired two-tailed t test for Arc P = 0.0014, t = 3.133, for Fos P = 1.86E-5, t = 4.064, for Nr4a1 P = 0.0178, t = 2.53). (e, f, g) Jq1 pretreatment blocks the fold-increase in nascent RNA of Arc (b), Fos (c) and Nr4a1 (d) in cultured cortical neurons 10 minutes after TTX withdrawal (paired two-tailed t test, for Arc n = 7 P = 0.0186, t = 2.475, for Fos P = 0.0264, t = 2.1998, for Nr4a1 P = 0.0406, t = 2.456). (h, i, j) Infection of neurons with a Brd4 siRNA lentivirus but not a srcrambled control virus blocks Arc (h), Fos (i) and Nr4a1 (j) induction in response to 10 minutes of BDNF stimulation (n = 6 biological replicates, paired two-tailed t test, for Arc P = 0.0337, t = 2.502, for Fos P = 0.0450, t = 2.327, for Nr4a1 P = 0.0311, t = 2.551). (k, l) Staining (k) and quantification (l) of Arc in neurons stimulated for 30 minutes with BDNF following pretreatment with Jq1 or the negative enantiomer (−)Jq1 (unpaired two-tailed t test, for control n = 232 neurons, for BDNF n = 185 neurons, for Jq1 n = 245, and for Jq1+BDNF n = 200 from 5 biological replicates per group, for control v. BDNF P = 8.52E-26, t = 11.447, Jq1 v Jq1+BDNF P = 0.00084, t = 3.363, for BDNF v. Jq1+BDNF P = 0.00013, t = 3.867). (m, n) Quantification (m) and staining (n) for Arc in neurons transfected with GFP and either a nontargeting siRNA pool or Brd4 siRNA (unpaired two-tailed t test, for control n = 105 neurons, for BDNF n = 80, for Brd2 siRNA n = 85, for Brd2+BDNF n = 71, for Brd3 n = 88, for Brd3+BDNF n = 63, for Brd4 n = 80, and for Brd4+BDNF n = 71 from 11 biological replicates, for control vs BDNF P = 0.0012, t = 3.296, for Brd4 siRNA vs Brd4 siRNA + BDNF P = 0.662, t = 0.438, for Brd2 siRNA vs Brd2 siRNA + BDNF P = 8.572E-5, t = 4.035, for Brd3 siRNA vs Brd3 siRNA + BDNF P = 0.167, t = 2.42, for BDNF vs Brd4 siRNA + BDNF P = 0.0157, t = 2.445. *, p<0.05. **, p<0.01 ***, p<0.001. n.s. nonsignificant. a.u. arbitrary units. Min, minutes. Error bars represent standard error. Scale bar is 10 μM.
We similarly examined the effects of Jq1 on tetrodotoxin (TTX) withdrawal which rapidly increases neuronal activity. Neurons were treated with TTX for 2 days after which Jq1 or the negative enantiomer was added before TTX was removed from media. Jq1 again prevented rapid IEG induction (Fig. 2e–g). Interestingly, Jq1 cannot prevent IEG activation after long periods of BDNF stimulation, suggesting that Jq1 only affects the rapid increase in transcription, wherease at later times signaling may be robust enough to overcome BET inhibition (Supplementary Fig. 2a–c). Because Jq1 also inhibits other members of the BET protein family, we tested whether loss of Brd4 is sufficient to block IEG induction. Partial knockdown of Brd4 with a lentivirus also blocked BDNF-induced IEG expression but did not block upstream pathways such as MAPK signaling (Fig. 2h–j, Supplementary Fig. 2d, e). These data fit with a model similar to that observed in other cell types in which Brd4 recruits P-TEFb to promote PolII phosphorylation to allow for rapid transcriptional elongation.
To confirm that the loss of transcriptional activation results in a corresponding change in protein levels, we examined Arc protein expression at 30 minutes when newly transcribed mRNA has been translated into protein. As expected, Jq1-treated neurons exhibited less Arc protein induction than control neurons (Fig. 2k, l). Similarly, transfection of small interfering RNAs (siRNAs) targeted against Brd4 blocked the BDNF-induced increase in Arc protein whereas transfection with non-targeting siRNA or siRNAs targeted against the other BET family members did not (Fig. 2m, n, Supplementary Fig. 2f–i). BrdT is testes-specific so was not tested. To ensure that the loss of Arc induction was not due to off-target effects, we tested two distinct Brd4 siRNAs which also blocked BDNF-induced Arc expression (Supplementary Fig. 2j–l).
Finally, we sought to determine which of Brd4’s known functions is responsible for its effects in neurons. Full-length Brd4 can function by recruiting complexes such as pTEFb and Mediator to trigger elongation whereas both the long and short forms of Brd4 can promote PolII progression through acetylated nucleosomes after elongation begins26. We found that only the long form of Brd4 affects Arc expression (Supplementary Fig. 2m) indicating that Brd4 is likely functioning in neurons by recruiting co-activating complexes to target genes.
These data demonstrate that Brd4 regulates activity-induced IEG expression in neurons. However, inhibition of Brd4 may also disrupt transcriptional output in neurons even without a potent stimulation such as BDNF due to the cumulative loss of the response to endogenous signaling from other neurons over a long period of time. Indeed, we found that by 24 hours Jq1 decreased Arc, Fos, and Nr4a1 transcripts (Supplementary Fig. 2n). We also confirmed that long-term disruption of this BET function did not cause widespread disruption of chromatin acetylation (Supplementary Fig. 2o) as expected from its function as reader protein. Finally, we showed that Jq1 did not block upstream signaling pathways by demonstrating that MAP Kinase phosphorylation is intact (Supplementary Fig. 2p). Together, these data demonstrate that Brd4 is responsible for transcription of IEGs in neurons.
Genome-wide effects of Jq1
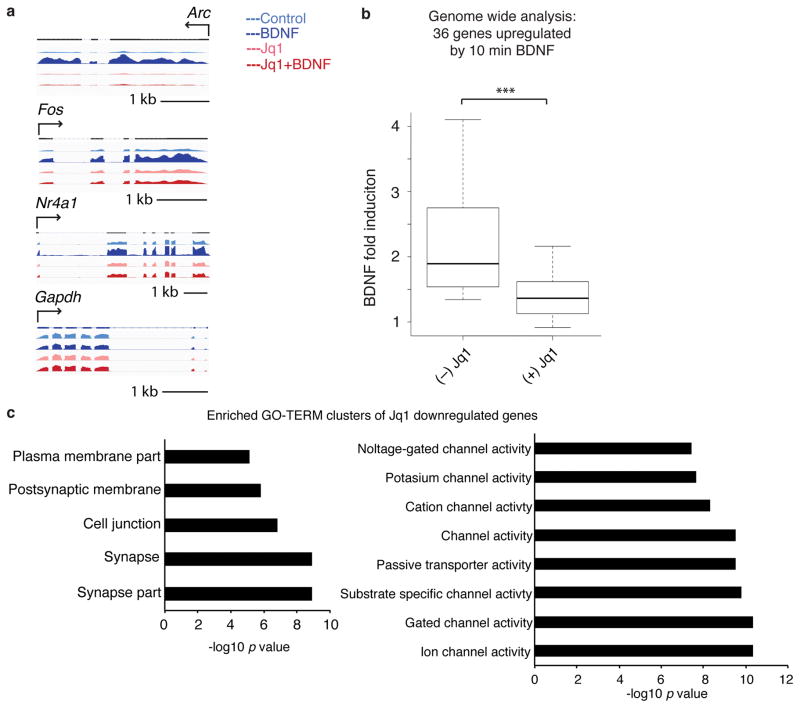
While Brd4 clearly regulates specific IEGs, inhibition of Brd4 likely also affects a wider range of genes. We used RNA-sequencing to examine BDNF induction of IEGs after Jq1 treatment and long-term effects of Jq1. An extended pretreatment of Jq1 followed by BDNF stimulation (Supplementary Fig. 3a) recapitulated the effects of Jq1 on induction of the specific IEGs previously examined but did not affect the housekeeping gene Gapdh (Fig. 3a). We expanded this comparison to all genes significantly induced by BDNF and found a consistent decrease in induction in the presence of Jq1 (Fig. 3b, Supplementary Fig. 3b), supporting a model in which Brd4 mediates the rapid response to neuronal activity. In addition, we also examined the effects of 24 hours of Jq1 treatment in the absence of exogenous stimulation. At this later time point, the effects of the loss of IEG transcription in response to basal levels of endogenous neuronal signaling will be apparent. We found a highly significant overlap between BDNF-induced genes and those regulated by Jq1 alone of p < 0.000005. We also separately examined genes up- and down-regulated by Jq1 as down-regulated genes are more likely to be direct targets of Brd4 due its function as a transcriptional activator. Gene clusters most significantly down-regulated by Jq1 included genes involved in ion channel regulation and synapse function (Fig. 3c). Jq1 treatment also increased genes involved in chromatin regulation and nuclear proteins (Supplementary Fig. 3c) which may be compensatory effects or indirect effects resulting from decreases in Brd4 target genes. To determine which histone modifications are responsible for recruiting Brd4 to target genes, we examined known Brd4 target acetyl marks and found that BDNF increased H3K14 and H4K16 acetylation at IEG promoters (Supplementary Fig. 3d) suggesting they may be involved in the stimulus dependent recruitment of Brd4. As expected, Jq1 did not affect these marks (Supplementary Fig. 3e).
Figure 3. Genome-wide analysis of effects of Jq1.
(a) Examples of RNA-sequencing of Arc, Fos, Nr4a1, and Gapdh in control neurons or in neurons after 24 hours of Jq1 treatment followed by a 10 minute stimulation. (b) Boxplot of RNA-sequencing data of BDNF-induced gene fold change after (−) or (+) Jq1 treatment in neurons for the 36 genes significantly upregulated by BDNF (paired two-tailed t test, n = 3 biological replicates, P = 2.98E-5, t = 3.357). (c) Top GO terms of genes clusters enriched in Jq1 down-regulated genes. Min, minutes. kb, kilobase. ***, p<0.001.
Brd4 is regulated by casein kinase II in neurons
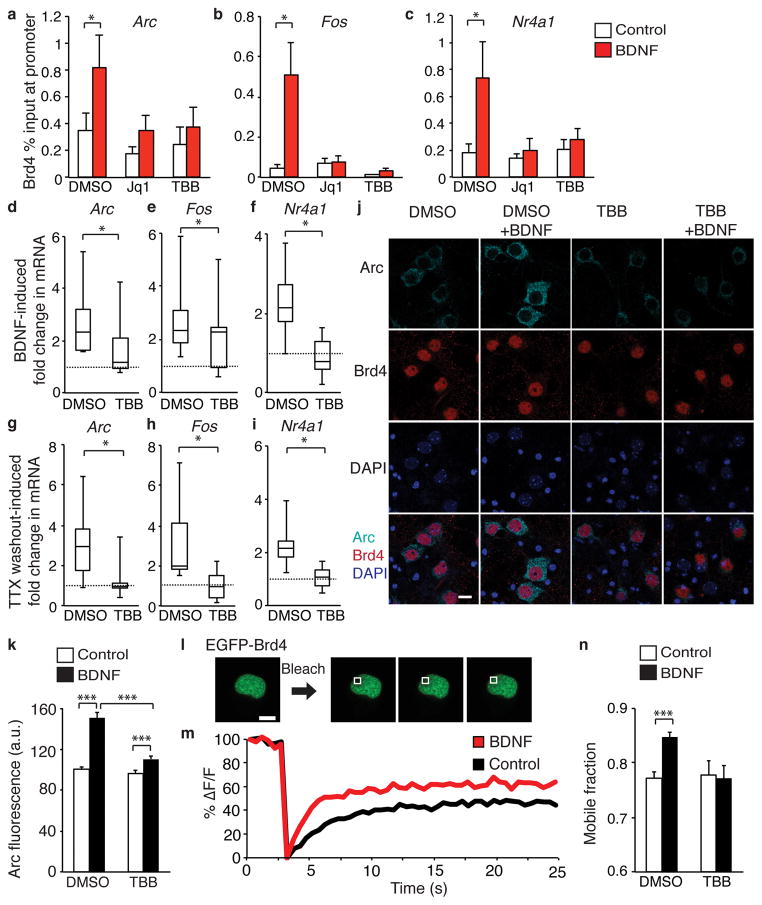
To better understand how Brd4 is targeted to chromatin in neurons, we next examined the mechanism underlying activation of Brd4 itself. We found that BDNF stimulation increased Brd4 association with promoter regions of IEGs suggesting that neuronal activity targets Brd4 to acetylated histones immediately after stimulation (Supplementary Fig. 4a–c). This effect was not observed for Brd2 or 3 (Supplementary Fig. 4d–i). To investigate how Brd4 is targeted to promoters, we explored the role of CK2. In HEK293 cells, CK2 phosphorylates Brd4, which triggers Brd4 binding to acetylated histones at target gene promoters to activate transcription14. In neurons, CK2 is important in regulating synaptic strength27,28 and is activated by BDNF stimulation22. We found that BDNF-induced targeting of Brd4 was blocked by pretreatment with the CK2 inhibitor TBB as well as Jq1 (Fig. 4a–c) suggesting that Brd4 is activated by CK2 in response to neuronal activity. Importantly, this short TBB pretreatment did not prevent MAPK phosphorylation demonstrating upstream signaling is intact (Supplementary Fig. 4j) and neither TBB nor Jq1 prevented BDNF-induced CREB Binding Protein (CBP) association with chromatin demonstrating other transcriptional cofactors are still recruited to target genes (Supplementary Fig. 4k–m). If Brd4 activation by CK2 is necessary for the activity-dependent transcription then CK2 inhibition should also block the activity-induced increase in IEGs. Fitting with this model, pretreatment with TBB blocked the increase in Arc, Fos, and Nr4a1 mRNA (Fig. 4d–i) as well as Arc protein levels (Fig. 4j–k). To control for off-target effects of TBB, we confirmed that transfection of Ck2 siRNA also blocks Arc induction (Supplementary Fig. 4n–p).
Figure 4. CK2 regulates Brd4 in neurons.
(a, b, c) Brd4 ChIP-qPCR analysis in cultured neurons pretreated with vehicle or CK2 inhibitor TBB and stimulated with BDNF for 10 minutes to measure Brd4 at promoters of Arc (a), Fos (b), and Nr4a1 (c) (paired two-tailed t test, for Arc dmso treatments n = 10 biological replicates, P = 0.00955, t = 1.693, for TBB treatments n = 6, and for Jq1, n = 3, for Fos n = 8 biological replicates, P = 0.0303, t = 2.791, for TBB n = 6 and for Jq1 treatments, n = 3, for Nr1A1, dmso treatments n = 9 biological replicates, P = 0.0411 t = 1.987, for TBB treatments n = 5, and for Jq1, n = 3). (d, e, f) TBB pretreatment blocks increased Arc (d), Fos (e), and Nr4a1 (f) mRNA after 10 minute BDNF stimulation. (paired two-tailed t test, for Arc n = 14 biological replicates, P = 0.0215, t = 2.505, for Fos n = 14 biological replicates, P = 0.0175, t = 2.496, and for Nr4a1 n = 13 biological replicates, P = 0.0122, t = 2.926). (g, h, i) TBB pretreatment blocks increased Arc (g), Fos (h), and Nr4a1 (i) mRNA 10 minute after TTX withdrawal (paired two-tailed t test, for Arc n = 8 biological replicates, P = 0.0451, t = 2.485, for Fos n = 8 biological replicates, P = 0.0179, t = 2.728, and for Nr4a1 n = 6 biological replicates, P = 0.0437, t = 2.983). (j, k) Arc and Brd4 staining (j) and quantification (k) after TBB or vehicle pretreatment and 30-minute BDNF stimulation. (unpaired two-tailed t test, for control n = 108 neurons, for BDNF n = 131, for TBB n = 117, for TBB+BDNF n = 118 from 2 biological replicates, for control vs bdnf P = 7.42E-14, t = 7.962, for TBB vs TBB+BDNF P = 0,001, t = 3.331, for BDNF vs TBB+BDNF P = 7.598E-10, t = 6.408). (l, m) Example of images (l) and recovery curves (m) for FRAP of EGFP-Brd4 expressed in neurons. (n) Mobile fraction quantification of EGFP-Brd4. (unpaired two-tailed t test, for control n = 120 neurons, for BDNF n = 113, for TBB n = 39, for TBB+BDNF n = 50 from 6 biological replicates, P = 9.98E-7, t = 5.02). *, p<0.05. ***, p<0.001. a.u. arbitrary units. s, seconds. Error bars represent standard error. Scale bar is 10 μM.
To further support our proposed mechanism of Brd4 phosphorylation induced chromatin targeting, we examined the movement of Brd4 after neuronal stimulation using FRAP. Using live neurons, we photobleached a region of the nucleus of expressing EGFP-Brd4, and observed the recovery of the signal in the bleached region over time to measure the mobile fraction of Brd4. As expected, Jq1 increased the mobile fraction (Supplementary Fig. 4q). The mobile fraction of Brd4 also increases after BDNF treatment, presumably as it is activated and relocates to acetylated chromatin. However, the enhanced mobility of Brd4 was blocked by TBB (Fig. 4l–n) indicating CK2 is responsible for this effect.
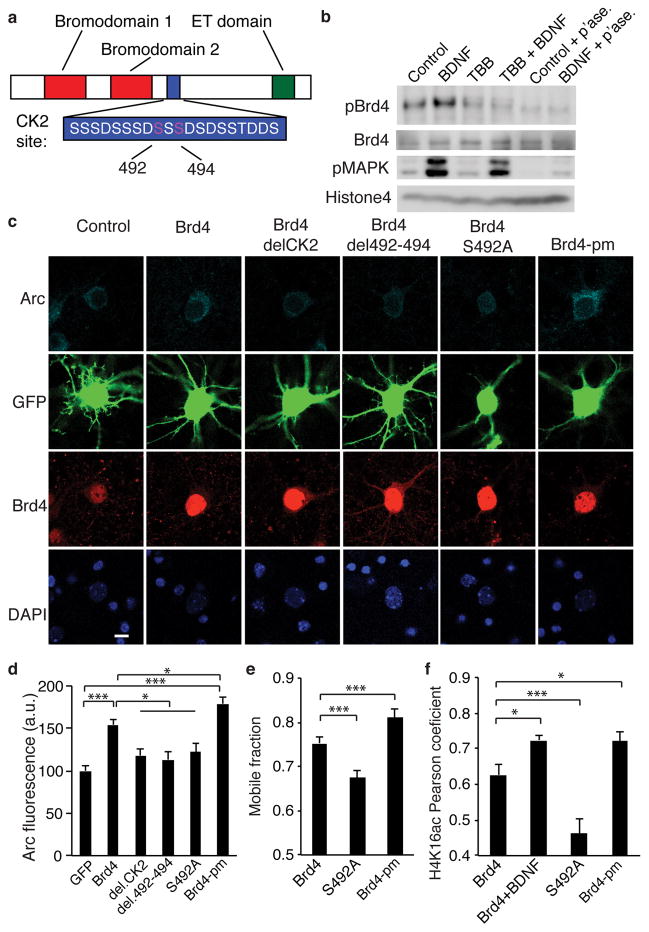
To better understand the mechanism of Brd4 activation in neurons, we examined the specific serine residues in the CK2 site in Brd4, S492 and S494, that are critical to Brd4 activation14 (Fig. 5a). We developed a novel, site-selective antibody against a peptide containing the Brd4 CK2 site with phosphorylated S492. Using a dot blot assay, we found that the affinity-purified antibody specifically bound a peptide phosphorylated at S492 as well as a peptide phosphorylated at both S492 and S494, but failed to recognize the unphosphorylated peptide (Supplementary Fig. 5a). Next we used neuronal lysates to determine the specificity of the antibody in cells and observed a band matching the size of full length Brd4 that was not present in lysates treated with phosphatase, although other small molecular weight bands were also observed at high exposures (Supplementary Fig. 5b). We observed a robust BDNF-induced increase in the phospho-Brd4 signal that was lost with TBB pretreatment (Fig. 5b).
Figure 5. Phosphorylation of Brd4 is critical for its function.
(a) Model of Brd4 and the critical amino acids in the CK2 phosphorylation site. (b) Western blot for phosphorylated Brd4 shows an increase with BDNF but not after TBB pretreatment or phosphatase treatment of lysates. Representative of 3 biological replicates. (c, d) Staining (c) and quantification (d) for Arc and Brd4 in neurons transfected with GFP and Brd4 with deletions or mutations in the CK2 site (unpaired two-tailed t test, for GFP n = 68, for Brd4n = 61, for CK2 deletion n = 46, for deletion 492-494 n = 44, for S492A n = 54, for Brd4-pm n = 51 from 5 biological replicates, for GFP vs Brd4 P = 1.223E-8, t = 6.09, for GFP vs Brd4-pm P = 2.00E-13, t = 2.353, for Brd4 vs CK2 deletion P = 0.0011, t = 3.36, for Brd4 vs deletion 492-494 P = 0.00037, t = 3.689, for Brd4 vs S492A P = 0.0075, t = 2.724, for Brd4 vs Brd4-pm P = 0.0204, t = 2.353). (e) Quantification of the mobile fraction of FRAP performed on Brd4 with mutations in the CK2 domain (unpaired two-tailed t test, for Brd4 n = 57 neurons, for S492A n = 52 and for Brd4-pm n = 38 from 3 biological replicates, for Brd4 vs S492A P = 0.0001, t = 4.042, for Brd4 vs Brd4-pm P = 0.0085, t = 2.488). (f) Pearson correlation coefficient for H4K16acetyl colocalization with Brd4 with CK2 site mutations (two-sided two-tailed t test for Brd4 n = 35 neurons, for Brd4 + BDNF n = 21, for S492A n = 29, and for Brd4-pm n = 18 from 3 biological replicates, for Brd4 vs Brd4 + BDNF P = 0.0164, t = 2.477, for Brd4 vs S492A P = 0.00167, t = 3.286, for Brd4 vs SSS492A P = 0.035, t = 2.166. *, p<0.05. ***, p<0.001. a.u. arbitrary units. Full-length blots are presented in Supplementary Figure 11. p’ase, phosphatase. Error bars represent standard error. Scale bar is 10 μM.
Next, to ensure that the effects of CK2 inhibition and knockdown are due to its effects on Brd4 and not an indirect effect of other CK2 targets, we tested the critical target residues in the CK2 phosphorylation site in Brd4. We created a full (delCK2) and partial deletion (del4920494) and a point mutation (S492A) in the CK2 site in Brd4. Transfection of wildtype Brd4 increased Arc expression even in the absence of exogenous stimulation. However, this effect was greatly reduced when the CK2 site was mutated or deleted. Conversely, a phosphomimic (Brd4-pm) at the key serines in Brd4 (S492E and S494E) resulted in an even greater increase in Arc expression (Fig. 5c, d). This demonstrates that phosphorylation of the CK2 site within Brd4 is necessary for its ability to activate transcription of Arc. We also repeated our FRAP assay and found the S492A mutant decreases Brd4 mobility, presumably by preventing activation of Brd4 from endogenous signaling while the phospho-mimic Brd4 behaved similarly to BDNF-stimulated Brd4 showing an increased mobile fraction (Fig. 5e). Finally, we sought to confirm that this increase in the mobile fraction corresponds to a translocation to active chromatin. We focused on acetylated H4K16, which recruits Brd4 and increases in response to BDNF (Supplementary Fig. S3d). We found moderate colocalization of H4K16acetyl and wildtype Brd4 under basal conditions that was enhanced with BDNF stimulation as expected. We then tested the phospho- mutant and mimic forms of Brd4 and determined that the S492A mutation decreased H4K16acetyl colocalization while Brd4-pm mutations increased colocalization as measured by Pearson’s coefficient (Fig. 5f and Supplementary Fig. 5d–e). Together, these data support a model in which CK2 phosphorylates Brd4 in response to neuronal activity resulting in Brd4 binding to target promoter regions and increased transcription of target IEGs.
Brd4 affects neuronal receptor proteins
In neurons, IEGs regulate the response to activity by changing the receptor content of synapses both by directly modifying synaptic proteins and altering gene expression of these proteins29,30. Thus, the prolonged loss of IEG activation resulting from Jq1 treatment may affect neuronal function by changing critical synaptic proteins. In addition, RNA-sequencing data suggest that Jq1 treatment affects transcription of synaptic proteins and receptors in neurons (Fig. 2d). We therefore examined the GluA1 subunit of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR), the major excitatory receptor in neurons. Jq1 treatment decreased transcript levels of Gria1 (Fig. 6a), the gene encoding GluA1, and decreased total GluA1 protein (Fig. 6b). Jq1 did not affect Gria2, which unlike Gria1, lacks activity-responsive promoter regions31,32. These changes, as well as those observed on IEGs, occurred slowly over the course of several hours of Jq1 treatment (Supplementary Fig. 6a). To confirm that decreasing the total pool of available GluA1 results in decreased surface expression, we used a surface-staining assay that specifically stains receptors expressed on the exterior of dendrites. We found that both Jq1 and Brd4 siRNA, but not Brd2 or Brd3 siRNA, decreased GluA1 surface expression in both cortical and hippocampal neurons without affecting spine number (Fig. 6c–g, Supplementary Fig. 6b–e). Increasing Brd4 expression resulted in a small, but significant, increase in GluA1 surface expression (Fig. 6g–i). We again compared the long and short forms of Brd4 and found that full length Brd4 GluA1 increases surface expression while the short isoform does not (Supplementary Fig. 6f), indicating that Brd4 functions by recruiting co-activating complexes to the promoter of target genes. Gria1 may be a direct target of Brd4 because Brd4 ChIP assays show a high basal level of Brd4 at Gria1 regulatory elements in the promoter region and the small BDNF-induced increase in Brd4 binding is not seen with Jq1 (Supplementary Fig. 6g). In addition, we observed a non-significant increase in histone acetyl marks at the Gria1 promoter following BDNF treatment (Supplementary Fig. 6h). These data provide support for RNA-sequencing data showing that Jq1 affects synaptic proteins and demonstrate that Brd4 affects the expression of a critical subunit of a major excitatory receptor in neurons.
Figure 6. Brd4 regulates synaptic receptor subunit GluA1.
(a) 24-hour Jq1 treatment decreases Gria1 but not Gria2 mRNA in the absence of exogenous stimulation in cultured cortical neurons. (one sample t test, for Gria1 n = 12 biological replicates, P = 6.852E-6, t = 7.959, for Gria2 n = 6 biological replicates). (b) 24 or 48-hour Jq1 treatment decreased gluA1 protein in neurons. Representative of 3 biological replicates. (c, d) GluA1 surface staining (c) and quantification (d) in neurons transfected with GFP and treated with Jq1 or the negative enantiomer for 24 hours (unpaired two-tailed t test, for control n = 33 neurons and for Jq1 n = 26 neurons from 4 biological replicates, P = 4.242E-8, t = 6.322). (e, f) Surface GluA1 staining (e) and quantification (f) in neurons transfected with GFP and either a nontargeting siRNA pool or Brd4, Brd2, or Brd3 siRNA (unpaired two-tailed t test with Bonferroni correction, n = 33 neurons for control siRNA, for Brd2 siRNA n = 31, for Brd3 siRNA n = 28, and for Brd4 siRNA n = 28 from 5 biological replicates, for control vs Brd4 P = 0.00958, t = 2.667). (g) Brd4 staining in neurons transfected with GFP and a construct expressing Brd4 under a constitutively active promoter. (h, i) GluA1 surface staining (h) and quantification (i) 2 days after transfection with GFP and Brd4 (unpaired two-tailed t test, for GFP n = 40 neurons and for GFP-Brd4 n = 35 neurons from 5 biological replicates, P = 0.00167, t = 3.264). ###, p<0.001 with univariate analysis. **, p<0.01, ***, p<0.001. ROI, region of interest. a.u. arbitrary units. h, hours. Full-length blots are presented in Supplementary Figure 11. Error bars represent standard error. Scale bar is 10 μM.
Jq1 treatment affects memory formation
Based on the importance of Brd4 in controlling critical neuronal proteins, we examined whether Jq1 affects brain function in WT adult mice. We injected adult male mice with Jq1 (50mg/kg) daily for one week or three weeks before performing behavioral tests. Jq1 has excellent blood brain permeability33 and similar to previous reports, we found that Jq1 is well tolerated in mice at this dose and schedule17,18,22,23 (Supplementary Fig. 7a). In an open field test Jq1 did not affect distance travelled or zone preference indicating Jq1 does not cause problems with mobility or anxiety (Fig. 7a, Supplementary Fig. 7b–d).
Figure 7. Jq1 affects mouse behavior.
(a) Time spent in the inner or outer zone of an open field in mice treated with vehicle or with Jq1 for 1 or 3 weeks (for DMSO n = 10 mice, for 1 week Jq1 n = 9 mice, and for 3 weeks Jq1 n = 10 mice, two-way ANOVA (zone) P = 2E16, F = 598.57, two-way ANOVA (treatment) P = 0.986, F = 0.015, df = 54). (b) Novel object recognition paradigm. (c) Discrimination index of time spent with a novel vs familiar object one day after initial exposure to the objects (unpaired two-tailed t test, for DMSO n = 10 mice, for 1 week Jq1 n = 9 mice, and for 3 weeks Jq1 n = 10 mice, for DMSO vs 1 week Jq1 P = 0.0107, t = 2.88, for DMSO vs 3 weeks Jq1 P = 0.0094, t = 2.927, one sample t test for DMSO P = 0.00127, t = 4.85)..(d) Discrimination index of time spent with a novel vs familiar object one day after initial exposure after a single dose of Jq1 (unpaired two-tailed t test, n = 10 mice per group, for DMSO vs post-learning Jq1 P = 0.0376, t = 2.25, univariate analysis for DMSO P = 0.00262, t = 4.11). (e) Fear conditioning paradigm. (f) Percent of time spent freezing in a new context after fear conditioning (for DMSO n = 10 mice, for 1 week Jq1 n = 9 mice, and for 3 weeks Jq1 n = 10 mice, two-way ANOVA (treatment) P = 0.0135, F = 4.67 and unpaired two-tailed t test for DMSO vs 3 weeks Jq1 P = 0.0036, t = 3.34, df = 54). Discrimination index on a scale of −1 to 1: (% time with novel object − % time with familiar object)/(% time with novel object + % time with familiar object). ###, p<0.001 with univariate analysis. *, p<0.05. ***, p<0.001. n.s., nonsignificant. s, seconds. Error bars represent standard error.
We next used a novel object-recognition task (Fig. 7b) in which mice were briefly exposed to 2 identical objects and later presented with one familiar and one novel object. If mice remember the previous objects they will subsequently spend more time with a novel object34. All groups behaved similarly during habituation and the initial exposure although mice receiving Jq1 for 3 weeks explored less during testing (Supplementary Fig. 7e–g). Strikingly, while control mice preferred the novel object as expected, Jq1-treated mice showed no preference (Fig. 7c). However, when mice were tested immediately after the initial exposure, control and Jq1 treated mice performed equally well (Supplementary Fig. 7h–j) suggesting that Jq1 does not disrupt learning or short-term memory but instead affects long-term memory. To control for possible health issues resulting from long-term treatments, we injected mice with a single dose of Jq1 or DMSO either 6 hours before or within 30 minutes after their initial exposure to objects and tested the following day. We found a complete loss of preference in mice that received a single dose after training compared to control mice (Fig. 7d). This suggests that Jq1 given during the process of memory consolidation can block long-term memory formation. The smaller effect observed after a dose given before training may be due to a smaller amount of Jq1 remaining in the brain during the consolidation process several hours later.
We also tested a fear-conditioning paradigm of 3 tones paired with shocks to determine the extent of the memory deficits (Fig. 7e). All mice learned both cued conditioning and the context-dependent conditioning (Supplementary Fig. 7k, l) demonstrating that Jq1 did not affect this simple behavior dependent on the amygdala. However, mice given Jq1 for 3 weeks froze more in a new context suggesting they were less able to distinguish between the training context and a new context. This indicates that the more difficult hippocampal-dependent test of context discrimination may also require BET protein function (Fig. 7f). Together, these data provide in vivo support of our cell-based data demonstrating that Jq1 disrupts the transcriptional responses that are critical to neuronal function.
Jq1 treatment decreases seizure susceptibility
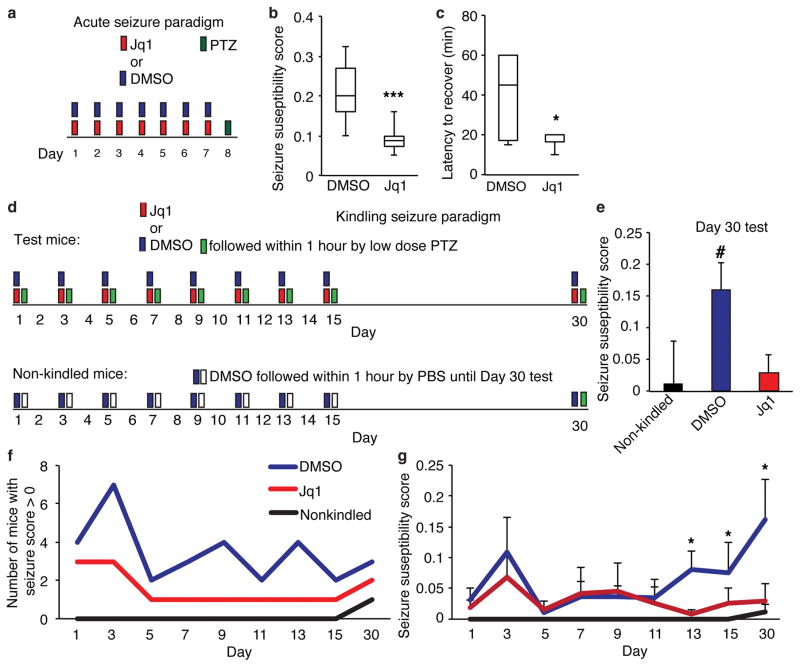
To confirm that Jq1 has similar effects on neurons in vivo as it does in vitro we examined tissue from mice after behavioral testing. Despite the heterogeneity of cortical tissue we found either trends or significant decreases in IEGs, Gria1 and GluA1 protein (Supplementary Fig. 8a, b). Long-term decreases in Gria1 and other synaptic proteins (Fig 3c) may dampen of synaptic strength which also has implications for other aspects of mouse behavior. We hypothesized that if Jq1 effectively decreases neuronal firing though regulation of synaptic proteins, then Jq1 treatment might decrease seizure susceptibility because seizures result from excess synaptic excitability. We injected adult male mice with Jq1 or DMSO for one week and then induced seizures with pentylenetetrazol (PTZ) (50mg/kg) (Fig. 8a), which inhibits GABA-A receptors resulting in increased excitatory activity. Jq1 treated mice showed decreased seizure susceptibility, as measured by a modified Racine scale which measures both severity of seizure and latency to onset of each seizure stage35,36. In addition, while approximately 30% of control mice died after seizure induction, all Jq1 treated mice survived and recovered faster as measured by a return to normal movement (Fig. 8c, d, Supplementary Fig. 8c, d). Similar but more variable effects were observed in female mice (Supplementary Fig. 8e, f).
Figure 8. Jq1 decreases seizure susceptibility.
(a) Model of seizure induction paradigm. (b) Seizure susceptibility score of mice treated for one week with either DMSO or Jq1 and given pentylenetetrazol (PTZ) to induce seizures (unpaired two-tailed t test n = 7 mice for DMSO and 6 mice for Jq1, P = 0.002, t = 3.155). (c) Latency to the return of normal movement after PTZ injection (unpaired two-tailed t test n = 7 mice for DMSO and 6 mice for Jq1, P = 0.036, t = 2.388). (d) Model of seizure induction paradigm. (e) Seizure susceptibility score of mice on day 30 of kindling testing (one sample t test, for DMSO n = 4 mice, for Jq1 n = 4 mice and for non-kindled n = 6 mice, for DMSO P = 0.0473, t = 2.415). (f) Number of mice seizing during each day of testing. (g) Seizure susceptibility of mice seizing during each day of testing (initial n = 8 mice for dmso and Jq1 and N = 6 mice for non-kindled group, for DMSO vs non-kindled unpaired one-tailed t test, day 13 P = 0.00555, t = 2.494, day 15 P = 0.0486, t = 0.928, unpaired two-tailed t test for day 30 P = 0.0265, t = 2.715). #, p<0.05 with univariate analysis. *, p<0.05. ***, p<0.001. min, minutes. Error bars represent standard error.
We also tested the kindling method of seizure induction in which DMSO or Jq1 was given within an hour before mice receive a sub-threshold doses of PTZ37,38. This is repeated every two days for two weeks and experimental mice as well as an additional non-kindled group are tested again two weeks later (Fig. 8d). Mice typically show increased seizure induction over time as PTZ-induced increases in neuronal firing enhance the strength of neuronal connections intensifying the response to future doses37. Mice are considered kindled if they show enhanced susceptibility that is maintained for several weeks. Jq1 had little effect during initial treatments, possibly because mice had only received a small number of doses of Jq1. However, on day 30, only the DMSO-treated mice showed ‘kindling’ compared to the non-kindled group (Fig. 8e–f, Supplementary Fig. 8g, h) These data indicate that Jq1 treatment has similar effects on neuronal function in vivo as we observed in vitro and raise the intriguing possibility of using BET inhibitors for treatment of epilepsy.
Discussion
Our results demonstrate that Brd4 is expressed throughout the brain and plays a critical role in activity-dependent transcription. Neuronal activity acts through CK2 to increase Brd4 association with chromatin. Brd4 then promotes transcription of critical IEGs and synaptic proteins (Supplementary Fig. 9). We found that Jq1 inhibition of Brd4 and its family members blocked novel object preference, indicating impairments in memory consolidation. In addition, consistent with its effects on synaptic proteins, Jq1 treatment also decreased seizures susceptibility in mice. This is the first demonstration that Brd4 has a critical function in neurons and that BET protein inhibition affects memory consolidation.
Implications for the clinical use of BET inhibitors
Our data demonstrate that Brd4 is necessary for rapid activation of genes. As has been demonstrated by numerous groups, the first few minutes following a burst of neuronal activity are of critical importance for a cell to activate the appropriate transcription response3–6. Loss of this rapid response may represent the mechanism through which Brd4 inhibition prevents long-term memory formation after.
BET protein inhibitors have been proposed as a treatment for several types of cancer and are currently in clinical trials. Initial mouse studies reported that Jq1 was well tolerated,17,18,23,33 and we did not find obvious deficits in the health or mobility of mice. However, our study provides new evidence that use of such inhibitors causes memory deficits in mice and thus may also cause neurological problems in patients receiving these drugs. Our results suggest that compounds that do not cross the blood brain barrier may pose less risk of neurological side effects for patients.
The role of CK2 in neurons
Casein Kinase 2 has several established functions in neurons in addition to regulating Brd4. CK2 phosphorylates GluA1 and GluA2 subunits of the AMPA receptor to promote its expression27 and regulates composition of the NMDA receptor28. These synaptic actions of CK2 promote synaptic strength as does the role we propose for CK2 in regulating Brd4. This dual function would allow CK2 to act immediately on the synapse by directly phosphorylating synaptic proteins while also acting through Brd4 to promote expression of these same genes in order to consolidate synaptic changes. The effects of the CK2 inhibitor TBB have also been tested in vivo in an epilepsy model39. TBB treatment blocked recurrent epileptiform discharges in hippocampal slice preparations after magnesium removal. We demonstrated the Jq1 results in decreased seizure susceptibility (Fig. 7) suggesting that some of CK2’s effects on epileptiform discharges may be the result of its action on Brd4 as well as its effects on the synapse.
BET inhibitors as epilepsy drugs
We found that Jq1 decreased the seizure susceptibility, potentially by decreasing levels of the GluA1 subunit of AMPARs which have been linked to epilepsy40–47. Decreased levels of other gene targets that regulate synaptic function may also contribute to the seizure effect though mechanisms such as phosphorylation of GluA148. Although the dose we tested resulted in memory deficits, it is possible that in an overactive epileptic brain Jq1 would restore normal levels of synaptic proteins. Most epilepsy treatments directly target synaptic proteins and receptors. Jq1 treatment represents a novel approach by targeting a protein responsible for the transcriptional regulation of these synaptic receptors instead of modifying proteins already present at the synapse. While many cases of epilepsy respond to available treatments, a significant portion of patients are refractory to current drugs38. It is possible that this novel approach of targeting transcriptional regulators of synaptic proteins rather than targeting synaptic proteins directly may provide a more robust method of dampening the heightened synaptic activity leading to seizures and could provide new avenues of treatments for these patients.
Online Materials and Methods
Cell culture
Neurons were isolated from E16.5 cortices of C57BL/6 mice (Charles River), dissociated in Optimem media with 20mM glucose and plated at 600,000 cells per mL on coverslips or plates coated with poly-D-lysine. One hour later Optimem media was replaced with Neurobasal media supplemented with Pen/Strep, Glutamax and B27 supplement. 3 days after plating, AraC was added to the media to prevent glial cell growth. Neurons were typically used at 12 days in vitro. Glial cells were isolated from P1 cortices, dissociated and grown in DMEM media with 10% fetal bovine serum and Pen/Strep. Media was changed every 3 days and cells were passaged to ensure that no neurons remained in the culture.
Transfections were performed using lipofectamine 2000 (Life Technologies). Neurons were put in a 1mM kynurenic acid solution during transfection to prevent excitotoxicity. Lipofectamine and DNA complexes were left on neurons for 15 minutes. Transfections were performed at 10 DIV for constructs expressing Brd4 and cells were fixed one to two days later. siRNA transfections were performed at 7 DIV and cells were stimulated and fixed 5 to 7 days later. Brd and CK2 siRNAs were from Dharmacon (LG-041493-00-0002 for Brd4 and L-058653-00-0005 for CK2). Target sequences of Brd4 siRNAs are ACAATCAAGTCTAAACTAG, TTACTGGAATGCTCAGGAA, GAGGATAAGTGTAAGCCCA, and GTACAGAGATGCCCAGGAA. Target sequences of CK2 siRNAs are CCGAAGAGCCCTTTAAATA, GGTCAGGGTTTACAGAGTA, CTGAACGAATCATGTCTTA, and TCACCTGGCATCATAGATA. For infections, control or Brd4 lentivirus was added to neurons on day 10 and neurons were stimulated and lysed 3 days later. Brd4 lentivirus contained a pool of 4 siRNAs from Applied Biological Materials (ABM) uses a dual convergent promoter system to express sense and antisense siRNA from different promoters. Target sequences of siRNAs are GGGTGAACTCACGTCAGAA for control virus (ABM LVP015-G) and GTGGATGCCGTCAAGCTGAACCTCCCTGA, GGACTTCAAGCACTATGTTTACAAATTGTT, GGAGATGACATCGTCTTAATGGCAGAAGC, and CCCAGGAATTTGGTGCTGATGTCCGATTG for Brd4 (ABM iV038675). The constitutively expressed Brd4 was from K. Ozato. Stratagene site directed mutagenesis kit was used for creating mutations and deletions. TBB (Tocris 2275) was used at 50μM, BDNF (PeproTech 450-02) was used at 50ng/mL. Tetrodotoxin (Abcam ab120055) was used at. Jq1 was used at 250 nM.
N2A cells were grown in DMEM with 10% serum and tested for mycoplasma infection regularly. N2A transfections were performed in DMEM using lipofectamine 2000 (Life Technologies). Lipfectamine and DNA complexes were left on cells overnight. Cells were harvested for analysis 5 days after transfection.
Western blotting
Cells were lysed in RIPA buffer and lysates were separated by SDS-PAGE and transferred to PVDF paper. Antibodies used were Brd4 (Bethyl A301-985A, 1:1000), NeuN (Millipore MAB377, 1:500), GFAP (Abcam ab10062, 1:1000), Gapdh (Abcam ab8245, 1:500), MAPK (Cell Signaling 4695P, 1:3000), phosMAPK (Cell Signaling 4370, 1:3000), H3 (Abcam ab1791, 1:4000), H4 (Abcam ab10158, 1:4000), H3K14ac (Active motif 39697, 1:500), H4K16ac (Active motif 39167, 1:500). Phospho-Brd4 was developed with Millipore. The best bleeds were affinity purified against the phosphorylated target peptide and immuno-depleted against unmodified Brd4. Blots were imaged on an LAS3000 system (FujiFilm).
Reverse transcription, Quantitative PCR, and ChIP
RNA was purified using the QIAGEN RNAeasy kit and reverse transcribed using the applied biosystems kit. qPCR was performed with Power SYR green PCR master mix (applied biosystems) on an applied biosystems quantitative PCR system run using StepOne software. ChIP was done as previously described1. Chromatin shearing was performed with a Bioruptor300 (diagenode) at 4 °C, for 55 cycles of 30 seconds on and off. Immunoprecipitation was performed using 5 μg of antibody bound to 50 μL of magnetic Dynabeads M280 (life sciences). DNA was purified using the QIAGEN Qiaquick PCR purification kit. Antibodies used were Brd4 (Bethyl A301-985A, 5μg/chip), Brd2 (Bethyl A302-583A, 5μg/chip), Brd3 (Bethyl A302-368A, 5μg/chip), H3K9ac (Millipore 07-352, 5μg/chip), H3K14ac (Active motif 39697, 5μg/chip), H4K12ac (Upstate 05-119, 5μg/chip), H4K16ac (Active motif 39167, 5μg/chip), CBP (Santa Cruz 7300, 5μg/chip).
Primers used for qPCR were:
Gapdh forward: AACTCCCTCAAGATTGTCAGCAA
Gapdh reverse: GGCATGGACTGTGGTCATGA
Arc forward: TAACCTGGTGTCCCTCCTAGATC
Arc reverse: GGAAAGACTTCTCAGCAGCTTGA
cFos forward: ACAGCCTTTCCTACTACCATTCCC
cFos reverse: CTGCACAAAGCCAAACTCACCTGT
Nr4A1 forward: ACCAACTCTTCTGGCTTCCCTTAC
Nr4A1 reverse: GGCTGGTTGCTGGTGTTCCATATT
GluA1 forward: TCCTGAAGAACTCCTTAGTG
GluA1 reverse: ATCATGTCCTCATACACAGC
GluA2 forward: AACGGCGTGTAATCCTTGAC
GluA2 reverse: CTCCTGCATTTCCTCTCCTG
Brd4 forward: AAATCAGCTCACCAGGCTGT
Brd4 reverse: TCTTGGGCTTGTTAGGGTTG
Brd2 forward: ACAAGGTAGTGATGAAGGCTCTGTGGAA
Brd2 reverse: CTTGTGGCATTGATGCAACCTTCTGTAGG
Brd3 forward: GGACATCCTCTGGCAGCTTA
Brd3 reverse: CCATCTTCCGAAGGGGACT
Bdnf forward: TGTCTCTGCTTCCTTCCCACAGTT
Bdnf reverse: TGGACGTTTGCTTCTTTCATGGGC
Primers used for ChIP were:
Arc forward: ATAAATAGCCGCTGGTGGCG
Arc reverse: CGGCTCCGAACAGGCTAAG
cFos forward: CGGGTTTCAACGCCGACTA
cFos reverse: TTGGCACTAGAGACGGACAGA
Nr4A1 forward: TGGAATGTCTGCGCGCGTG
Nr4A1 reverse: TATAGATCAAACAATCCGCG
GluA1 forward: ATCTGGCTGTCAGTCGGTGT
GluA1 forward: AAAGAAGCCCTGGTCCAAC
RNA-sequencing sample preparation and analysis
RNA was collected and prepared using a Qiagen RNAeasy kit and the TruSeq RNA Sample Preparation Kit v2 (Illumina). Sequencing was performed with an Illumina HiSeq2500 system. Reads were aligned to the mouse mm9 reference genome using Tophat 2.0.112. Reads with two or fewer mismatches with a maximum of 20 hits for each read were used. Transcript levels were analyzed with Cufflinks 2.2.13. Expressed genes were defined as those with an fpkm of 1 or above. BDNF-induced genes were defined as those with a significant increase in expression with 10 minutes BDNF treatment using a p value with a Bonforroni correction. David ontology cluster analysis was to determine gene groups enriched in Jq1 up and down-regulated groups of genes of fpkm of 1 or above. The RNA-sequencing dataset is available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63809.
Immunohystochemistry
Adult mice were perfused with 4% paraformaldehyde and brains were removed and kept in paraformaldehyde overnight. Tissue was then washed in PBS and processed for paraffin embedding at the Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using Leica ASP6025 tissue processor. Brains were embedded in paraffin, and paraffin sagittal sections of 5 microns were cut on a Leica RM2155 microtome and collected on superfrost plus slides (Fisher scientific). Slides were baked for 1 hour at 60°C before de-paraffinization and staining.
Staining was performed at the Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using a Discovery XT processor (Ventana Medical Systems). For GFAP and Brd4 co-staining, slices were first stained for Brd4 (Bethyl A301-985A, 2μg/mL). The tissue sections were blocked for 30 minutes in 10% normal goat serum and 2% BSA in PBS. The incubation with the primary antibody was done for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-rabbit IgG (Vector labs PK6101, 1:200). Detection was performed with Streptavidin-HRP D (Ventana Medical Systems) followed by incubation with Tyramide-Alexa Fluor (Invirogen T20948, 1:200). For GFAP, sections were blocked for 30 minutes in 10% normal goat serum and 2% BSA in PBS. Rabbit polyclonal GFAP (Dako Z0334, 1 μg/ml) was incubated for 5 hours at RT, followed by 32 minutes incubation with biotinylated goat anti-rabbit IgG (Vector labs PK6101, 1:200, 6.5 μg/mL). Detection was performed with Blocker D, Streptavidin-HRP and DAB detection. For NeuN, slices were first stained for NeuN and then stained for Brd4 as described above. For NeuN staining, sections were blocked first for 30 min in Mouse IgG Blocking reagent (Vector Labs, MKB-2213) in PBS. Mouse monoclonal NeuN staining (Millipore, MAB377, at 1 μg/mL) was incubated for 3 hours at RT and followed by 60 minutes incubation of biotinylated mouse Secondary (Vector Labs, MOM Kit BMK-2202, 1:200 dilution, 5.75μg/mL). The detection was performed with Secondary Antibody Blocker, Blocker D, Streptavidin-HRP D (Ventana Medical Systems) and DAB Detection Kit (Ventana Medical Systems) according to manufacturer instructions.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 10 minutes, washed with PBS and permeabilized in 0.1% triton in PBS for 10 minutes. Cells were then blocked for 1 hour in 2% serum, 3% BSA and 0.1% triton in PBS and then primary antibody was added in the same solution overnight at 4°C. Cells were washed in PBS for 3 10 minute washes and put in secondary antibody for 1 hour at room temperature. After an additional 3 washes, coverslips were mounted with prolong gold antifade solution (Invitrogen). For surface staining assays, GluA1 antibody was added for 30 minutes to the media live cells at 37°C before fixation. Cells were then blocked without triton and secondary antibody was added immediately following the blocking step. Antibodies used were Brd4 (BethylA301-985A, 1:1000), Arc (Santa Cruz 365736, 1:100), GluA1 (Millipore 2263, 1:300), CK2 (Peirce/Thermo PA5-28686, 1:100), H4K16ac (Abcam 109463, 1:500) and secondary antibodies were AlexaFlour 647 Donkey anti-mouse (Jackson 715-605-150, 1:500) and Rhodamine Red-X goat anti-rabbit (Invitrogen R6394, 1:500).
Microscopy equipment and settings
Slides were imaged at room temperature on an inverted Lieca DMI 6000, TCS SP8 laser scanning confocal confocal microscope with a 405 nm laser and a fully tunable white light laser (470–670 nm) with an acousto-optical beam splitter. The microscope uses 3 gated HyD detectors and one PMT detector and both a conventional scanner and a resonant scanner. Objectives used were a 63x HC PL APO CS2 oil objective with a NA of 1.40 and for whole brain images a 10x HCX PL APO DS dry objective with a NA of 0.4. Type F immersion liquid (Leica) was used for oil objectives. For brain images, the Lieca super-z stage and rapid tiling system was used to compile images. For glua1 surface staining, z-stacks spaced at 0.5 microns were used to image the entire dendrite. For 63x images, images were 184.52 by 184.52 microns, 1052 by 1052 pixels, (5.701 pixels per micron), and 8-bits per pixel. For 10x images, images were 1162.5 by 1162.5 microns, 1052 by 1052 pixels, (0.881 pixels per micron), and 8-bits per pixel.
ImageJ was used to crop images and merge channels into composite RGB images. Photoshop was used to adjust individual channels. In all cases, identical adjustments were applied across all images used in an experiment for each channel. No deconvolution software was used. All image analysis as performed in ImageJ. For Arc staining quantification, a region of interest was selected in the cell body, outside the nucleus, and the average intensity was measured. Regions were selected using dapi and gfp channels and then applied to the Arc channel such that the analysis was performed blind to the Arc staining. For Brd4 staining quantification, the same process was used but inside the nucleus. For surface GluA1 quantification, the z-stacks were summed using ImageJ to create one image per channel. GFP images were converted to binary and used to create a mask surrounding the transfected dendrite. The mask was then applied to the GluA1 image and the average intensity within the dendrite was measured. This was automated using ImageJ macros to prevent user bias. For all image analysis, an average background intensity value was subtracted from each intensity value. To allow for comparisons across experiments, the average control cell value was set to 100 and all conditions were normalized to this value.
Behavior
All experiments were approved by the Institutional Animal Care and Use Committee of the Rockefeller University. C57B/6 male mice (Jackson) were housed up to 5 mice per cage in a 12 hour light-dark cycle. JQ1 (APExBIO) was administered to 2-3 months old mice via intraperitoneal injections. Each mouse was injected daily for 1 week or 3 weeks before testing began with either JQ1 at 50mgs/kg dissolved in DMSO or DMSO alone, diluted into cyclo-dextrin (Sigma). Mice were randomly assigned to groups and groups were then checked to ensure that the average weight per mouse of each group was equivalent at the beginning of the experiment. Injections continued during the week of behavioral testing and testing was performed during the light cycle. Open field testing was performed first and activity was measured for 1 hour. Fusion 3.2 was used to track mice and analyze movement. One day after open field testing, mice were habituated to the novel object recognition box for 10 minutes. One day later mice were habituated for an additional 2 minutes and then 2 identical objects (either a faucet or a lego pyramid) were placed in the box and mice were given 10 minutes to explore. On the following day, mice were returned to the box with one object they had previously seen and one new object in place of the original object and allowed to explore for 10 minutes. All sessions were recorded using ethovision software. Time spent interacting with each object was manually analyzed. Discrimination index was calculated as (% time with novel object − % time with familiar object)/(% time with novel object + % time with familiar object). Fear conditioning tests began 1 day after novel object recognition. Mice were placed in a small box and allowed to explore for 2 minutes. A tone was played for 20 seconds followed by a 0.7 mAmp shock. This was repeated once per minute for 3 shocks total. After an additional 2 minutes, mice were removed from the box. One day later mice were returned to the same box for 7 minutes to measure context dependent freezing. Then the flooring, wall covering, and smell of the box was changed and mice were returned to the box. The tone was then played in the same patter as the original training session without a subsequent shock to measure cued learning. Fear conditioning sessions were run and recorded using FreezeFrame 3 software and scored manually in random order. All experiments were carried out and analyzed with the experimenter blind to the treatment group. One mouse was excluded from analysis because the lights went off in the facility during the discrimination test so the data could not be analyzed.
For novel object ‘learning’ testing, mice were habituated to the novel object recognition box for 10 minutes. One day later mice were given 10 minutes to interact with 2 identical objects. Mice were then removed and one object was replaced with a novel object and mice were returned to the box and again allowed to explore for 10 minutes. For single dose tests, one cohort received a dose of DMSO in the morning approximately 6 hours before testing and a second dose of DMSO within 30 minutes of exposure to objects. One cohort received Jq1 in the morning and DMOS after testing and the final cohort received DMSO in the morning and Jq1 following testing. Mice were tested for novel object preference one day after the first exposure to objects. All sessions were recorded using ethovision software. Time spent interacting with each object was manually analyzed. All experiments were carried out and analyzed with the experimenter blind to the treatment group and which object was considered novel.
Seizure testing
JQ1 (APExBIO) was administered to 3 to 4 month old C57B/6 male or female mice (Jackson) via intraperitoneal injections. For acute seizure testing, each mouse was injected daily for 1 week before testing began with either JQ1 at 50mgs/kg dissolved in DMSO or DMSO alone, diluted into cyclo-dextrin (Sigma). Pentylenetetrazol (PTZ) (Sigma) dissolved in PBS was injected at 50mgs/kg via intraperitoneal injections. For kindling seizure testing, Jq1 was administered 1 hour before PTZ injection. Mice were observed up to one hour after injection or until recovery from seizure (defined by a return to normal movement). The modified Racine scale4 used to measure seizure induction was as follows:
Stage 1: Hypoactivity culminating in behavioral arrest with contact between abdomen and the cage.
Stage 2: Partial clonus (PC) involving the face head or forelimbs.
Stage 3: generalized clonus (GC) including all four limbs and tail, rearing or falling.
Stage 4: Generalized Tonic-Clonic seizure (GTC)
Seizure susceptibility score was calculated as: (0.2) (1/PC latency) + (0.3) (1/GC latency) + (0.5) (1/TC latency).
Statistics
An alpha level of 0.05 was used for all statistical analyses. 2-sided t-tests were performed in excel. A bonforonni correction was applied when comparing multiple groups. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications20,30,37. Data distribution was assumed to be normal but this was not formally tested. For fear conditioning, 2-way ANOVA was performed in R with post-hoc Bonforroni corrections for individual comparisons. For novel object testing, context discrimination was calculated by time spent with objects: (novel-familiar)/(novel+familiar). Univariate analysis was used for each individual group to compare to a context discrimination of zero. Degrees of freedom were calculated as the biological replicates minus one. For all other behavioral testing, t-tests with a bonforroni correction were used to compare between multiple groups. A supplementary methods checklist is available.
Supplementary Material
Acknowledgments
We thank Dr. Keiko Ozato for the Brd4 construct, Dr. Jodi Gresack and the Rockefeller behavioral core for guidance and suggestions, Dr. Kaye Thomas for microscopy expertise, the MSKCC histology core for immunohistochemistry support, Dr. Alexey Soshnev for the schematic model graphic, John Gerace for editing, and members of the Allis lab for feedback. Support for this work was provided by a grant from the Robertson Foundation, a Ruth Kirschstein NRSA fellowship (F32MH103921), the NIH (R01 NS34389, NS081706) and a Simons Foundation Research Award. R.B.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
E.K. wrote the manuscript and designed and carried out experiments. M.H. helped carry out behavioral testing and molecular studies. I.Z. helped with seizure testing. R.D. helped initiate and design the project and provided feedback, and C.D.A provided support, feedback and guidance.
References
- 1.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Gapp K, Woldemichael BT, Bohacek J, Mansuy IM. Epigenetic regulation in neurodevelopment and neurodegenerative diseases. Neuroscience. 2014;264:99–111. doi: 10.1016/j.neuroscience.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 3.Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490(Pt 3):703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey U, Krug M, Brödemann R, Reymann K, Matthies H. Long-term potentiation induced in dendrites separated from rat’s CA1 pyramidal somata does not establish a late phase. Neurosci Lett. 1989;97:135–139. doi: 10.1016/0304-3940(89)90152-3. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 6.Messaoudi E, Ying SW, Kanhema T, Croll SD, Bramham CR. Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation in vivo. J Neurosci Off J Soc Neurosci. 2002;22:7453–7461. doi: 10.1523/JNEUROSCI.22-17-07453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 8.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 9.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan S, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houzelstein D, et al. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Descalzi G, Fukushima H, Suzuki A, Kida S, Zhuo M. Genetic enhancement of neuropathic and inflammatory pain by forebrain upregulation of CREB-mediated transcription. Mol Pain. 2012;8:90. doi: 10.1186/1744-8069-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha RN, et al. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Schael S, et al. Casein kinase 2 phosphorylation of protein kinase C and casein kinase 2 substrate in neurons (PACSIN) 1 protein regulates neuronal spine formation. J Biol Chem. 2013;288:9303–9312. doi: 10.1074/jbc.M113.461293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirguet O, et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56:7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 25.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanno T, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lussier MP, Gu X, Lu W, Roche KW. Casein kinase 2 phosphorylates GluA1 and regulates its surface expression. Eur J Neurosci. 2014;39:1148–1158. doi: 10.1111/ejn.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz-Clemente A, Matta JA, Isaac JTR, Roche KW. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron. 2010;67:984–996. doi: 10.1016/j.neuron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietrich JB. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 2013;352:179–190. doi: 10.1007/s00441-013-1565-2. [DOI] [PubMed] [Google Scholar]
- 30.Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. 2011;34:591–598. doi: 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borges K, Dingledine R. Functional organization of the GluR1 glutamate receptor promoter. J Biol Chem. 2001;276:25929–25938. doi: 10.1074/jbc.M009105200. [DOI] [PubMed] [Google Scholar]
- 32.Myers SJ, et al. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci Off J Soc Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antunes M, Biala G. The novel object recognition memory: neurobiology, test procedure, and its modifications. Cogn Process. 2012;13:93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naydenov AV, et al. ABHD6 blockade exerts antiepileptic activity in PTZ-induced seizures and in spontaneous seizures in R6/2 mice. Neuron. 2014;83:361–371. doi: 10.1016/j.neuron.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferraro TN, et al. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J Neurosci Off J Soc Neurosci. 1999;19:6733–6739. doi: 10.1523/JNEUROSCI.19-16-06733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhir A. Pentylenetetrazol (PTZ) kindling model of epilepsy. In: Board Jacqueline N, Crawley Al, editors. Curr Protoc Neurosci. Unit9.37. Chapter 9. 2012. [DOI] [PubMed] [Google Scholar]
- 38.Bialer M, White HS. Key factors in the discovery and development of new antiepileptic drugs. Nat Rev Drug Discov. 2010;9:68–82. doi: 10.1038/nrd2997. [DOI] [PubMed] [Google Scholar]
- 39.Brehme H, Kirschstein T, Schulz R, Köhling R. In vivo treatment with the casein kinase 2 inhibitor 4,5,6,7- tetrabromotriazole augments the slow afterhyperpolarizing potential and prevents acute epileptiform activity. Epilepsia. 2014;55:175–183. doi: 10.1111/epi.12474. [DOI] [PubMed] [Google Scholar]
- 40.Rogawski MA. AMPA receptors as a molecular target in epilepsy therapy. Acta Neurol Scand Suppl. 2013:9–18. doi: 10.1111/ane.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Abdullah JM. The role of GluA1 in central nervous system disorders. Rev Neurosci. 2013;24:499–505. doi: 10.1515/revneuro-2013-0021. [DOI] [PubMed] [Google Scholar]
- 42.Kato AS, Gill MB, Yu H, Nisenbaum ES, Bredt DS. TARPs differentially decorate AMPA receptors to specify neuropharmacology. Trends Neurosci. 2010;33:241–248. doi: 10.1016/j.tins.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Donevan SD, Rogawski MA. Anticonvulsant activity of AMPA/kainate antagonists: comparison of GYKI 52466 and NBOX in maximal electroshock and chemoconvulsant seizure models. Epilepsy Res. 1993;15:179–184. doi: 10.1016/0920-1211(93)90054-b. [DOI] [PubMed] [Google Scholar]
- 44.Namba T, Morimoto K, Sato K, Yamada N, Kuroda S. Antiepileptogenic and anticonvulsant effects of NBQX, a selective AMPA receptor antagonist, in the rat kindling model of epilepsy. Brain Res. 1994;638:36–44. doi: 10.1016/0006-8993(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 45.Hara H, et al. Effect of YM872, a selective and highly water-soluble AMPA receptor antagonist, in the rat kindling and rekindling model of epilepsy. Eur J Pharmacol. 2006;531:59–65. doi: 10.1016/j.ejphar.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 46.Kodama M, et al. Effects of YM90K, a selective AMPA receptor antagonist, on amygdala-kindling and long-term hippocampal potentiation in the rat. Eur J Pharmacol. 1999;374:11–19. doi: 10.1016/s0014-2999(99)00295-2. [DOI] [PubMed] [Google Scholar]
- 47.Mattes H, Carcache D, Kalkman HO, Koller M. alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) antagonists: from bench to bedside. J Med Chem. 2010;53:5367–5382. doi: 10.1021/jm901688m. [DOI] [PubMed] [Google Scholar]
- 48.Rakhade SN, et al. Glutamate receptor 1 phosphorylation at serine 831 and 845 modulates seizure susceptibility and hippocampal hyperexcitability after early life seizures. J Neurosci Off J Soc Neurosci. 2012;32:17800–17812. doi: 10.1523/JNEUROSCI.6121-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naydenov AV, et al. ABHD6 blockade exerts antiepileptic activity in PTZ-induced seizures and in spontaneous seizures in R6/2 mice. Neuron. 2014;83:361–371. doi: 10.1016/j.neuron.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.