Abstract
UDP-glucuronosyltransferase (UGT) is a family of enzymes that catalyze the glucuronidation of various compounds, and thereby has an important role in metabolism and detoxification of a large number of xenobiotic and endogenous compounds. UGTs are present highly in the liver and small intestine, while several investigations on quantification of UGT mRNA reported that UGTs were also expressed in the brain. However, reported expression patterns of UGT isoforms in human brain were often incongruous with each other. In the present study, therefore, we investigated UGT mRNA expressions in brains of humanized UGT1 (hUGT1) mice. We found that among the human UGT1 members, UGT1A1, 1A3, and 1A6 were expressed in the brain. We further observed that nicotine (3 mg/kg) induced the expression of UGT1A3 mRNA in the brain, but not liver. While it was not statistically significant, the nicotine treatment resulted in an increase in the chenodeoxycholic acid glucuronide-formation activity in the brain microsomes. UGT1A3 is involved in metabolism of various antidepressants and non-steroidal antiin-flammatory drugs, which exhibit their pharmacological effects in the brain. Therefore, nicotine-treated hUGT1 mice might be useful to investigate the role of brain UGT1A3 in the regulation of local levels of these drugs and their response.
Keywords: UDP-glucuronosyltransferase (UGT), Nicotine, Brain, Tissue-specific regulation, Humanized animal model
1. Introduction
UDP-glucuronosyltransferase (UGT) is a family of enzymes that catalyze the glucuronidation of various compounds, and thereby has an important role in metabolism and detoxification of a large number of xenobiotic and endogenous compounds. There are 19 functional UGTs in human and they are divided into mainly three subfamilies based on sequence homology [1,2]. Subfamily 1A, 2A, and 2B include 9, 3, and 7 enzymes, respectively. While UGTs are highly expressed in the liver and small intestine, several investigations reported that UGTs were also expressed in brains. Ohno et al. reported that only UGT1A5 and UGT2B17 mRNAs were detected in human brain [3]. Court et al. reported that UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A10, 2A2, 2A3, 2B7, 2B11, and 2B17 mRNAs were detected in human brain [4]. Jones et al. reported that among the UGT2B family members, UGT2B4, 2B10, 2B11, 2B15, and 2B17 mRNAs were detected in the human brain [5]. These data indicate that reported UGT expression patterns in human brain were often incongruous with each other.
Nicotine, which is the major constituent of tobacco that responsible for tobacco dependence, has been reported to induce certain cytochrome P450 (CYP) families. CYP2B is highly expressed in the liver, while the enzyme is slightly expressed in the brains of human, rat, mouse, and monkey [6]. It was reported that nicotine induced CYP2B in the brains but not in the livers in rats and monkeys [7,8]. This tissue-specific induction by nicotine is different from gene induction by classical inducer, such as phenobarbital, carbamazepine, and rifampicin, which usually induce tissue-independently. The higher CYP2B expression was also shown in human brains of smokers [9]. Until recently, however, little has been known about the regulation of brain UGT by nicotine.
Humanized UGT1 (hUGT1) mice were previously developed in a C57BL/6 background. In the liver and small intestine, the expression pattern of UGT1A isoforms in hUGT1 mice was similar to that in humans [10]. It was also reported that the glucuronidation activities in the liver microsomes from hUGT1 mice were also similar to those in human liver microsomes [11]. In the present study, the expression levels of UGT mRNA in the brain were quantitatively evaluated in non-treated and nicotine-treated hUGT1 mice. We further investigated the UGT activity in the brain microsomes that were prepared from the non-treated and nicotine-treated hUGT1 mice.
2. Materials and methods
2.1. Materials
Nicotine was purchased from MP Biomedicals, Inc. (Solon, OH). Diethyl ether was purchased from Nacalai Tesque (Kyoto, Japan). Trizol reagent and primers were obtained from Invitrogen (Carlsbad, CA). ReverTra Ace qPCR RT master Mix and THUNDERBIRD SYBR qPCR Mix were obtained from Toyobo (Tokyo, Japan). Complete mini protease inhibitor was purchased from Roche Applied Science (Vilvoorde, Belgium). Chenodeoxycholic acid (CDCA) was purchased from Wako (Osaka, Japan). UDP-glucuronic acid (UDPGA), alamethicin, and saccharic acid 1,4-lactane were purchased from Sigma—Aldrich (St Louis, MO). Human liver microsomes were obtained from BD Gentest (Woburn, MA). All the other reagents were of the highest grade commercially available.
2.2. Animals
Tg(UGT1A1*28)Ugt1−/−(hUGT1) mice were developed previously in a C57BL/6 background [10]. All animals received food and water ad libitum, and mouse handling and experimental procedures were conducted in accordance with our animal care protocol, which was previously approved by Kitasato University.
Humanized UGT1 mice were injected subcutaneously (s.c.) once daily for 7 days (Group A) or twice daily for 5 days (Group B) with 3.0 mg nicotine ditartrate base per kg body weight in sterile saline. Mice in control groups of Group A and Group B received saline s.c. injections on the same schedule. Twenty-four hours (Group A) or 6 h (Group B) after the last injection, animals were anesthetized with diethyl ether. Animals were decapitated, and brains and livers were removed. Brains and Livers were stored at −80 °C until used for reverse transcription quantitative real-time PCR (RT-qPCR) analysis or preparation of microsomes.
2.3. RT—qPCR analysis
RNA was extracted from tissues with Trizol reagent. The cDNA was synthesized from total RNA using ReverTra Ace qPCR RT Master Mix according to the manufacturer's protocol. RT-qPCR was performed with THUNDERBIRD SYBR qPCR Mix, and the reactions were run in a CFX96TM Real-Time PCR Detection System (BioRad). The cycling parameters consisted of denaturation at 95 °C for 5 s, annealing temperature for 15 s, and extension at 72 °C for 30 s after one cycle at 95 °C for 1 min. Expression of cyclophilin B (CPH) mRNA was used as an internal control for the cDNA quantity and quality. The used primers and annealing temperature were shown in Table 1. According to a guideline of the Q-PCR analysis [12], the detailed information on the Q-PCR analysis was included in the manuscript according to the (Supplemental Table 1).
Table 1.
Sequence of primers for amplification of cDNA.
| Isoforms | Primer | Sequence | Annealing temperature (°C) |
|---|---|---|---|
| UGT1A1 | Forwarda | 5′-CCT TGC CTC AGA ATT CCT TC-3′ | 60 |
| Reverse | 5′-CAC AGG ACT GTC TGA GGG ATT TT-3′ | ||
| UGT1A3 | Forward | 5′-GAG AGG TGT CAG TGG TGG ATA TTC T-3′ | 60 |
| Reverse | 5′-CAC AGG ACT GTC TGA GGG ATT TT-3′ | ||
| UGT1A4 | Forwardb | 5′-CCT GCT GTG TTT TTT TGG AGG T-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A5 | Forwarda | 5′-ACA ATA TGT CTT TGA TCA TA-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A6 | Forwardb | 5′-CAA CTG TAA GAA GAG GAA AGA C-3′ | 54 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A7 | Forwardb | 5′-CCC CTA TTT TTT CAA AAATGT CTT-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A8 | Forwarda | 5′-GGT CTT CGC CAG GGG AAT AG-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A9 | Forwarda | 5′-GAA CAT TTA TTA TGC CAC CG-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| UGT1A10 | Forwarda | 5′-CTC TTT CCT ATG TCC CCA ATG A-3′ | 60 |
| Reversea | 5′-ATT GAT CCC AAA GAG AAA ACC AC-3′ | ||
| Ugt2b1 | Forward | 5′-TTG GGC CGA GCA ATG AAT CT-3′ | 60 |
| Reverse | 5′-GAT AGC AGC TCA CCA CAG GG-3′ | ||
| Ugt2b5 | Forward | 5′-TTA TGC GCC ACA AAA GAG CC-3′ | 60 |
| Reverse | 5′- AAT CAA GTC AAG TGT TTA GAA GGT G -3′ | ||
| Ugt2b34 | Forward | 5′-TTC CAG TGG TTG GCA TTC CT-3′ | 60 |
| Reverse | 5′-ACA CGA AGA TGC TTG GCT CC-3′ | ||
| Ugt2b35 | Forward | 5′-CTT GTA CAG AGG GGC CAT GAA-3′ | 60 |
| Reverse | 5′- TGG GAC CTC AAA AGT CCA TTC -3′ | ||
| Ugt2b36 | Forward | 5′- AAG GTG TTG GTA TGG CCG GT-3′ | 66 |
| Reverse | 5′-AGT CCA TCT TTC CAC AGC CTT-3′ | ||
| Ugt2b37 | Forward | 5′-ACA GCT TTT GAA TGT GTG GAC T-3′ | 60 |
| Reverse | 5′-GCG ATT AGC TCC CCA CCA AA-3′ | ||
| Cyp2b10 | Forward | 5′-GCA AGC CAT GTT GCT CCT AA-3 | 60 |
| Reverse | 5′-CTT GGA GCC CTG GAG ATT TGG-3′ | ||
| CPHc | Forward | 5′-CAG ACG CCA CTG TCG CTT T-3′ | 60 |
| Reverse | 5′-TGT CTT TGG AAC TTT GTC TGC AA-3′ |
2.4. Preparation of brain microsomes
Brain was homogenized in approximately three volumes of 100 mM phosphate-buffered saline (pH7.4) with protease inhibitor. The homogenate was centrifuged at 9000 g for 15 min at 4 °C. The supernatant was centrifuged at 105,000 g for 60 min at 4 °C, and the pellet was suspended in the same buffer with 20% glycerol and used as the microsomal fraction. Protein concentrations of microsomal fractions were measured by the Bradford method using BSA as a standard [13].
2.5. CDCA glucuronide formation assays
CDCA glucuronide formation was determined as follows. A typical incubation mixture (200 μL of total volume) contained 100 mM phosphate buffer (pH 7.4), 4 mM MgCl2, 5 mM UDPGA, 0.05 mg/mL alamethicin, 10 mM saccharic acid 1,4-lactane, 1.0 mg/mL brain microsomes or 0.05 mg/mL human liver microsomes, and 200 μmM CDCA. The reaction was initiated by the addition of UDPGA after a 9 min preincubation at 37 °C. After incubation at 37 °C for 5 h with the brain microsomes or 30 min with human liver microsomes, the reaction was terminated by addition of 200 μL of cold methanol. The mixtures were then centrifuged at 15,000 g for 10 min.
CDCA glucuronide was quantified using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) [14–16]. The sample of enzyme assay was moved to a vial and 10 μL was separated on a 50 mm × 2.1 mm Acquity 1.6 μm C18 column (Waters Corp, MA, USA) using an Acquity UPLC H-Class system (Waters). The gradient mobile phase consisted of 1 mM ammonium formate (A) and methanol containing 1 mM ammonium formate (B). Initial condition was 40% A and 60% B, followed by a linear gradient to 70% B in 1.5 min. This condition was maintained for 2.7 min, and the column was flushed with 95% B for 2.8 min and re-equilibrated to the initial condition for an additional 8 min. The flow rate was set to 0.2 mL/min. The eluent was introduced by electrospray ionization into the mass spectrometer (Xevo TQD, Waters) operating in negative ionization mode. Multiple reaction monitoring (MRM) mode, using specific precursor/product ion transition, was employed for quantification. The capillary and sampling cone voltages were set to 2900 and 60 V, respectively. Source and des-olvation temperatures were set to 150 and 200 °C, respectively, and the cone and desolvation gas flows were set to 0 and 650.0 L/h, respectively. The collision energy was set to 35 V. Detection of the negative ion was performed by monitoring the transition of m/z 567.4 → 391.4. Data acquisition, instrument control and data handling were performed with MassLynx Software (Waters). The conditions used in the present study were optimized to limit the instability of CDCA glucuronides [16,17].
2.6. Data analysis
In the RT-qPCR analysis, T-test with significance set at 5% was used. Brunner—Munzel test with significance set at 5% was used to examine the significance in the enzyme assays.
3. Results
3.1. Effects of nicotine on the expression level of human UGT1A mRNA in brain and liver
UGT isoforms expressed in the liver have been well examined; however, little is known about UGT isoforms expressed in the brain. In the brain of hUGT1 mouse, the expressions of UGT1A1, 1A3, and 1A6 were detected (Supplemental Tables 2 and 3). In contrast, the expressions of UGT1A4, 1A5, 1A7, 1A8, 1A9, and 1A10 were below the level of quantification limit. Therefore, the effect of nicotine treatment on the regulation of human UGT1A1, 1A3, and 1A6 was investigated in the brain.
In the previous studies, 1.0 mg nicotine per kg body weight was used to induce the expression of CYP2B mRNA in rat and monkey brains [7,8]. Therefore, in our preliminary study, hUGT1 mice were injected subcutaneously (s.c.) once daily for 7 days with 1.0 mg/kg weight nicotine. However, as demonstrated in Supplemental Fig. 1, 1.0 mg/kg nicotine did not regulate the expressions of human UGT1A1, 1A3, 1A6, and mouse Cyp2b10 mRNAs in hUGT1 mice. Therefore, in the present study, 3.0 mg/kg nicotine was used.
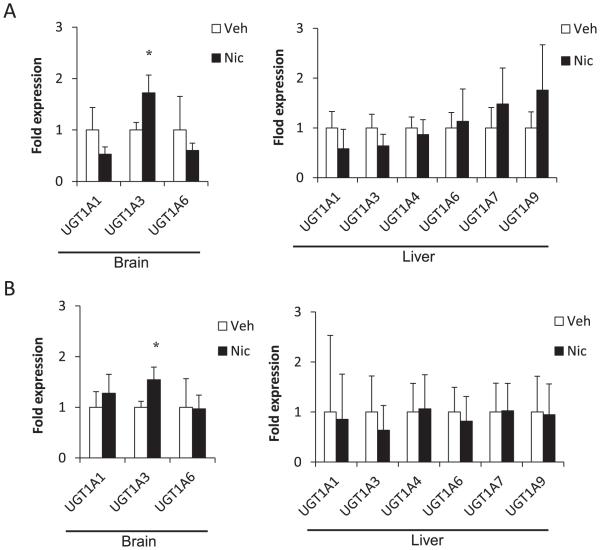
In Group A, hUGT1 mice were treated with 3.0 mg/kg nicotine, once daily for 7 days. Tissues were isolated 24 h after the last injection. The expression of UGT1A3 mRNA was induced by nicotine in the brain (1.7 fold, P < 0.05) (Fig. 1A). The expressions of UGT1A1 and UGT1A6 mRNAs were not changed by nicotine in the brain.
Fig. 1.
Relative expressions of human UGT1A mRNA in brains and livers of hUGT1 mice treated with nicotine or vehicle in Group A (A) and Group B (B). Results are shown as mean ± SD (n = 3). The expression levels in the vehicle treated mice were defined as 1. *P < 0.05. Veh = vehicle, Nic = nicotine.
In Group B, hUGT1 mice were treated with 3.0 mg/kg nicotine, twice daily for 5 days to investigate the effect of higher nicotine treatment on UGT expression. Tissues were isolated 6 h after the last injection. The expression of UGT1A3 mRNA was induced by nicotine in the brain (1.6 fold, P < 0.05) (Fig. 1B), while the expressions of UGT1A1 and UGT1A6 mRNAs were not changed.
The induction of UGT1A1, 1A3, and 1A6 mRNAs by nicotine was not observed in the liver in both groups. These findings indicated that nicotine induced the expression of UGT1A3 mRNA selectively in the brain. Induction of brain UGT1A3 mRNA in the nicotine-treated hUGT1 mice was reproducible (Supplemental Fig. 2).
3.2. Effect of nicotine on expression level of mouse Ugt2b mRNA in brain and liver
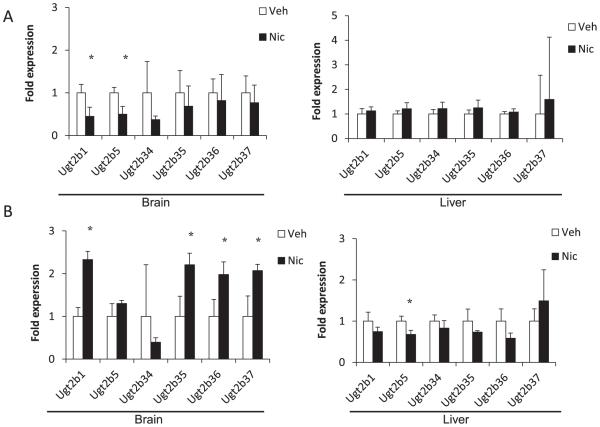
In a previous study, expressions of Ugt2b1, 2b5, 2b34, 2b35, 2b36, and 2b37 mRNAs were detected in mouse brain [18]. In the present study, therefore the expressions of these Ugt2b isoforms were examined in the brain of hUGT1 mice.
In Group A, the expression levels of Ugt2b1 and Ugt2b5 mRNAs were reduced by nicotine in the brain (Ugt2b1, 0.45 fold; Ugt2b5, 0.50 fold; P < 0.05) (Fig. 2A). In the liver, the expressions of the Ugt2b isoforms were not significantly changed by nicotine. Therefore, in Group A, the expressions of Ugt2b1 and Ugt2b5 mRNAs were reduced by nicotine selectivity in the brain.
Fig. 2.
Relative expressions of mouse Ugt2b mRNA in brains and livers of hUGT1 mice treated with nicotine or vehicle in Group A (A) and Group B (B). Results are shown as mean ± SD (n = 3). The expression levels in the vehicle treated mice were defined as 1. *P < 0.05. Veh = vehicle, Nic = nicotine.
In Group B, expression levels of Ugt2b1, 2b35, 2b36, and 2b37 mRNAs were increased by nicotine in the brain (Ugt2b1, 2.3 fold; Ugt2b35, 2.2 fold; Ugt2b36, 2.0 fold; Ugt2b37, 2.1 fold; P < 0.05) (Fig. 2B). In the liver, the expression of Ugt2b5 mRNAs was reduced by nicotine (0.68 fold; P < 0.05). Therefore, in Group B, the expressions of Ugt2b1, Ugt2b35, Ugt2b36, and 2b37 mRNAs were induced selectivity in the brain by nicotine. The expression of Ugt2b34 mRNA was not regulated.
These findings indicated that nicotine regulated Ugt2b expressions dose-independently and tissue-specifically.
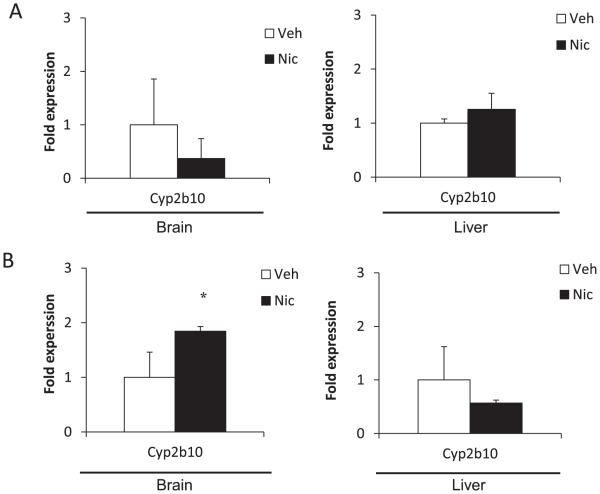
3.3. Effect of nicotine on expression level of mouse Cyp mRNA in brain and liver
It was reported that nicotine induced CYP2B in the brain of rat and monkey, but not in the liver [7,8]. However, it was unknown whether mouse Cyp2b could be regulated by nicotine in the brain. Therefore, we examined the expression levels of mouse Cyp2b10 mRNA in control mouse brain and nicotine-treated mouse brain. In Group A, the expression level of Cyp2b10 mRNA was not statistically changed by nicotine in both brain and liver (Fig. 3A). In Group B, the expression level of Cyp2b10 mRNA was induced by nicotine in the brain (1.7 fold, P < 0.05) (Fig. 3B). However, the expression level of Cyp2b10 mRNA was not changed by nicotine in the liver. Our data indicate that nicotine induced expression of brain Cyp2b10 mRNA selectivity in mouse brain, too.
Fig. 3.
Relative expressions of mouse Cyp2b10 mRNA in brains and livers of hUGT1 mice treated with nicotine or vehicle in Group A (A) and Group B (B). Results are shown as mean ± SD (n = 3). The expression levels in the vehicle treated mice were defined as 1. *P < 0.05. Veh = vehicle, Nic = nicotine.
3.4. Effect of nicotine on UGT1A3 activity in brain
CDCA was reported to be mainly glucuronidated by UGT1A3 and moderately by UGT1A1, UGT2B7, and UGT2A [14,17]. CDCA glucuronide was detected in the reaction mixtures containing brain microsomes and human liver microsomes (Supplemental Fig. 3A, B). The glucuronide was not detected when CDCA was incubated without UDPGA (Supplemental Fig. 3C).
The activity of CDCA glucuronidation was determined in the brain microsomes in the control and nicotine-treated hUGT1 mice. We observed that nicotine-treated mice had a 5-fold higher glucuronidation activity compared to the activity in the control mice, although it was not statistically significant (Fig. 4).
Fig. 4.
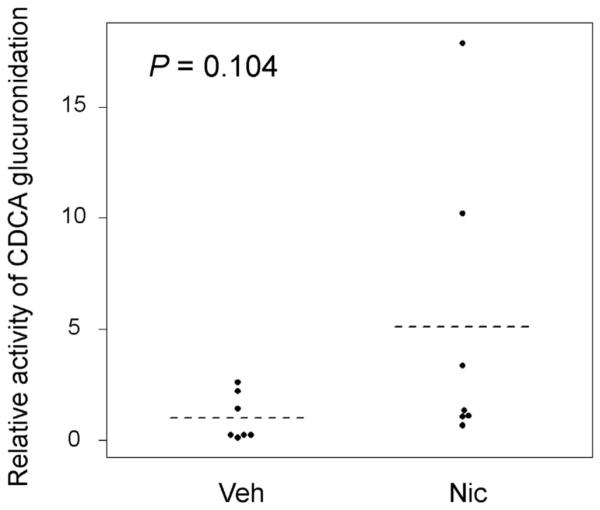
CDCA glucuronidation in brain microsomes. Brain microsomes were prepared from control and nicotine-treated hUGT1 mice. The mean value was shown by dashed lines (n = 7). The relative activity in the vehicle treated mice was defined as 1. *P < 0.05. Veh = vehicle, Nic = nicotine.
4. Discussion
Multiple UGT isoforms have been detected in human brains. However, the reported expression patterns of UGT in human brain were often incongruous with each other [3–5]. In the present study, the expressions of UGT1A1, 1A3, and 1A6 mRNAs were detected in the brain of hUGT1 mouse by RT-qPCR among the UGT1A members, which is relatively in agreement with a previous report by Court et al. [4]. They reported that among the UGT1A members, the expressions of UGT1A1, 1A3, 1A4, 1A6, and 1A7 mRNAs were detected in human brain. The presence of UGT1A pre-mRNA, which contains introns as well, has been suggested [19]. UGT1A family is encoded by unique first exons and common exons 2–5 [20]. In the present study, the forward and reverse primers that were located on exon 1s and common exon 2, respectively, were used to detect UGT1A4 and UGT1A7 mRNAs. When we used forward and reverse primers located on exon 1s of UGT1A4 or UGT1A7 in our preliminary study, those mRNAs were detected in the brain (not data shown). These findings indicate a possibility that not spliced mRNAs, but pre-mRNAs of UGT1A4 and UGT1A7 might be present in the brain. In the previous study by Court et al., the forward and reverse primers located on exon 1s were used to detect UGT1A4 and UGT1A7 mRNAs. Therefore, the detected UGT1A4 and UGT1A7 mRNAs in human brain might have been their pre-mRNAs in their study. To detect spliced mRNAs of UGT1A family members, forward and reverse primers that are crossing introns should be used.
In this study, we found that nicotine (3.0 mg/kg) specifically induced the expression of human UGT1A3 mRNA in the brain (Fig. 1 and Supplementary Fig. 2). While it was not statistically significant, nicotine-treated mice exhibited higher CDCA glucuronidation in the brain microsomes (Fig. 4). UGT1A3 metabolizes various exogenous and endogenous compounds, such as antidepressants (amitriptyline, clomipramine, imipramine), morphine, non-steroidal antiinflammatory drugs (NSAIDs) (ibuprofen, flubiprofen, diclofenac), and benzodiazepine (midazolam, lorazepame, oxazepame), that exhibit their pharmacological effects in brain [21–26]. Therefore, nicotine-treated hUGT1 mice might be useful to investigate the role of brain UGT1A3 in the regulation of local levels of these drugs and their effects on the phenotype.
In the previous studies, 1.0 mg nicotine per kg body weight was used to induce the expression of CYP2B mRNA in rat and monkey brains [7,8]. Therefore, in our preliminary study, hUGT1 mice were injected subcutaneously (s.c.) once daily for 7 days with 1.0 mg/kg weight nicotine. However, as demonstrated in Supplemental Fig. 1, 1.0 mg/kg nicotine did not regulate the expressions of human UGT1A1, 1A3, 1A6, and mouse Cyp2b10 mRNAs in hUGT1 mice. It was reported that the half-lives (t1/2) of nicotine in plasma were 1.3 h and 1.6 h in rats and monkeys, respectively [27,28]. In contrast, the t1/2 of nicotine in blood was reported to be 6 min in mice [29], indicating that mice needed higher nicotine dosages to examine the effect of nicotine on the expression of UGT and CYP mRNA. Therefore, in the current study, hUGT1 mice were treated with 3.0 mg/kg nicotine.
Propofol, which is metabolized by CYP2B, acts primarily on GABAA receptors and causes anesthetic effects [30,31]. It was reported that selective induction and inhibition of CYP2B in rat brain changed local levels of propofol in the brain and its anesthetic effect [32]. Chlorpyrifos is metabolized by CYP2B to be an active metabolite, chlorpyrifos-oxon, which inhibits acetylcholinesterase (AChE) and causes of neurotoxicity [33]. Neurotoxicity through the metabolism of chlorpyrifos was reduced by the inhibition of brain CYP2B in chlorpyrifos-treated rats [34]. The concentrations of morphine, which is generated from codeine by CYP2D, was reduced in the brain by inhibiting brain CYP2D in codeine-treated rats [35]. These data indicate that the regulation of CYPs in the brain impacts local metabolism of drugs and their response in vivo.
Although the function of UGT in the brain has not been clarified, certain glucuronic acid conjugates were detected in several brain samples. Glucuronic acid conjugates of neurotransmitters, such as serotonin (5-HT) and dopamine (DA), were detected in cerebrospinal fluid (CSF) and microdialysis samples of humans and rats [36–38]. Glucuronic acid conjugates of steroids were also detected in the mouse brain extract [39]. In an in vitro study, King et al. demonstrated that 5-HT glucronide, which is substrate of UGT1A6, was generated in rat brain microsomes [40]. Morphine, which is glucuronidated by UGT2B7 [41], was also glucronidated in human brain homogenates, rat brain homogenates, and rat microglia [42–44]. Furthermore, it was first demonstrated that the brain microsomes had potency to glucuronidate CDCA, which is a selective substrate of UGT1A3, in the previous study. Interestingly, it has been shown that large amounts of CDCA were present in brain [44]. Therefore, it can be possible that UGTs in the brain regulate local levels of these endogenous and exogenous compounds in brain to affect their pharmacological effects in vivo.
In conclusion, we demonstrated that nicotine regulated the expression levels of human UGT1A3, mouse Ugt2b, and mouse Cyp2b10 mRNAs in the brains of hUGT1 mice. Therefore, nicotine-treated hUGT1 mice might be a useful model to investigate the role of brain UGT in the regulation of local levels of these drugs and their responses. Further investigations are needed to elucidate the mechanism of the brain-specific induction of drug metabolizing enzymes by nicotine.
Supplementary Material
Footnotes
This work was supported by a Grant-in-Aid for Encouragement of Young Scientists B [26870562] (R.F.). This work was also supported in part by the National Institutes of Health National Institute of Environmental Health Sciences [Grant P42-ES010337] and National Institute of General Medical Sciences [Grant R01-GM100481].
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dmpk.2015.04.004.
References
- [1].Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro SI, Iyanagi T, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–85. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- [2].Guillemette C, Lévesque E, Harvey M, Bellemare J, Menard V. UGT genomic diversity: beyond gene duplication. Drug Metab Rev. 2010;42:24–44. doi: 10.3109/03602530903210682. [DOI] [PubMed] [Google Scholar]
- [3].Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- [4].Court MH, Zhang X, Ding X, Yee KK, Hesse LM, Finel M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica. 2012;42:266–77. doi: 10.3109/00498254.2011.618954. [DOI] [PubMed] [Google Scholar]
- [5].Jones NR, Lazarus P. UGT2B gene expression analysis in multiple tobacco carcinogen-targeted tissues. Drug Metabolism Dispos. 2014;42:529–36. doi: 10.1124/dmd.113.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Miksys SL, Tyndale RF. Drug-metabolizing cytochrome P450s in the brain. J Psychiatry Neurosci. 2002;27:406. [PMC free article] [PubMed] [Google Scholar]
- [7].Miksys S, Hoffmann E, Tyndale RF. Regional and cellular induction of nicotine-metabolizing CYP2B1 in rat brain by chronic nicotine treatment. Biochem Pharmacol. 2000;59:1501–11. doi: 10.1016/s0006-2952(00)00281-1. [DOI] [PubMed] [Google Scholar]
- [8].Ferguson CS, Miksys S, Palmour RM, Tyndale RF. Differential effects of nicotine treatment and ethanol self-administration on CYP2A6, CYP2B6 and nicotine pharmacokinetics in African green monkeys. J Pharmacol Exp Ther. 2012;343:628–37. doi: 10.1124/jpet.112.198564. [DOI] [PubMed] [Google Scholar]
- [9].Miksys S, Lerman C, Shields PG, Mash DC, Tyndale RF. Smoking, alcoholism and genetic polymorphisms alter CYP2B6 levels in human brain. Neurophar-macology. 2003;45:122–32. doi: 10.1016/s0028-3908(03)00136-9. [DOI] [PubMed] [Google Scholar]
- [10].Fujiwara R, Nguyen N, Chen S, Tukey RH. Developmental hyperbilirubinemia and CNS toxicity in mice humanized with the UDP glucuronosyltransferase 1 (UGT1) locus. Proc Natl Acad Sci USA. 2010;107:5024–9. doi: 10.1073/pnas.0913290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kutsuno Y, Sumida K, Itoh T, Tukey RH, Fujiwara R. Glucuronidation of drugs in humanized UDP-glucuronosyltransferase 1 mice: similarity with glucuronidation in human liver microsomes. Pharmacol Res Perspect. 2013;1:00002. doi: 10.1002/prp2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative realtime PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- [13].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- [14].Perreault M, Gauthier-Landry L, Trottier J, Verreault M, Caron P, Finel M, et al. The human UDP-glucuronosyltransferase UGT2A1 and UGT2A2 enzymes are highly active in bile acid glucuronidation. Drug Metab Dispos. 2013;41:1616–20. doi: 10.1124/dmd.113.052613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Griffiths WJ. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. 2003;22:81–152. doi: 10.1002/mas.10046. [DOI] [PubMed] [Google Scholar]
- [16].Caron P, Trottier J, Verreault M, Bélanger J, Kaeding J, Barbier O. Enzymatic production of bile acid glucuronides used as analytical standards for liquid chromatography-mass spectrometry analyses. Mol Pharm. 2006;3:293–302. doi: 10.1021/mp060021l. [DOI] [PubMed] [Google Scholar]
- [17].Trottier J, Verreault M, Grepper S, Monté D, Bélanger J, Kaeding J, et al. Human UDP-glucuronosyltransferase (UGT) 1A3 enzyme conjugates chenodeox-ycholic acid in the liver. Hepatology. 2006;44:1158–70. doi: 10.1002/hep.21362. [DOI] [PubMed] [Google Scholar]
- [18].Buckley DB, Klaassen CD. Tissue-and gender-specific mRNA expression of UDP-glucuronosyltransferases (UGTs) in mice. Drug Metab Dispos. 2007;35:121–7. doi: 10.1124/dmd.106.012070. [DOI] [PubMed] [Google Scholar]
- [19].Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, et al. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos. 2009;37:1759–68. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- [20].Gong QH, Cho JW, Huang T, Potter C, Gholami N, Basu NK, et al. Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics. 2001;11:357–68. doi: 10.1097/00008571-200106000-00011. [DOI] [PubMed] [Google Scholar]
- [21].Stone AN, Mackenzie PI, Galetin A, Houston JB, Miners JO. Isoform selectivity and kinetics of morphine 3-and 6-glucuronidation by human UDPglucuronosyltransferases: evidence for atypical glucuronidation kinetics by UGT2B7. Drug Metab Dispos. 2003;3:1086–9. doi: 10.1124/dmd.31.9.1086. [DOI] [PubMed] [Google Scholar]
- [22].Zhou D, Guo J, Linnenbach AJ, Booth-Genthe CL, Grimm SW. Role of human UGT2B10 in N-glucuronidation of tricyclic antidepressants, amitriptyline, imipramine, clomipramine, and trimipramine. Drug Metab Dispos. 2010;38:863–70. doi: 10.1124/dmd.109.030981. [DOI] [PubMed] [Google Scholar]
- [23].Kato Y, Izukawa T, Oda S, Fukami T, Finel M, Yokoi T, et al. Human UDP-glucuronosyltransferase (UGT) 2B10 in drug N-glucuronidation: substrate screening and comparison with UGT1A3 and UGT1A4. Drug Metab Dispos. 2013;41:1389–97. doi: 10.1124/dmd.113.051565. [DOI] [PubMed] [Google Scholar]
- [24].Sakaguchi K, Green M, Stock N, Reger TS, Zunic J, King C. Glucuronidation of carboxylic acid containing compounds by UDP-glucuronosyltransferase isoforms. Arch Biochem Biophys. 2004;424:219–25. doi: 10.1016/j.abb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- [25].Uchaipichat V, Suthisisang C, Miners JO. The glucuronidation of R-and S-lorazepam: human liver microsomal kinetics, UDP-glucuronosyltransferase enzyme selectivity, and inhibition by drugs. Drug Metab Dispos. 2013;41:1273–84. doi: 10.1124/dmd.113.051656. [DOI] [PubMed] [Google Scholar]
- [26].Duan SX, Guillemette C, Journault K, Krishnaswamy S, von Moltke LL, Greenblatt DJ. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab Dispos. 2002;30:1257–65. doi: 10.1124/dmd.30.11.1257. [DOI] [PubMed] [Google Scholar]
- [27].Kyerematen GA, Owens GF, Chattopadhyay B, Vesell ES. Sexual dimorphism of nicotine metabolism and distribution in the rat. Studies in vivo and in vitro. Drug Metab Dispos. 198;16:823–828. [PubMed] [Google Scholar]
- [28].Seaton M, Kyerematen GA, Morgan M, Jeszenka EV, Vesell ES. Nicotine metabolism in stumptailed macaques, Macaca arctoides. Drug Metab Dispos. 1991;19:946–54. [PubMed] [Google Scholar]
- [29].Petersen DR, Norris KJ, Thompson JA. A comparative study of the disposition of nicotine and its metabolites in three inbred strains of mice. Drug Metab Dispos. 1984;12:725–31. [PubMed] [Google Scholar]
- [30].Duan SX, Hesse LM, Venkatakrishnan K, Greenblatt DJ. Cytochrome P-450 2B6 is responsible for interindividual variability of propofol hydroxylation by human liver microsomes. Anesthesiology. 2001;94:110–9. doi: 10.1097/00000542-200101000-00021. [DOI] [PubMed] [Google Scholar]
- [31].Altomare C, Trapani G, Latrofa A, Serra M, Sanna E, Biggio G, et al. Highly water-soluble derivatives of the anesthetic agent propofol: in vitro and in vivo evaluation of cyclic amino acid esters. Eur J Pharm Sci. 2003;20:17–26. doi: 10.1016/s0928-0987(03)00161-1. [DOI] [PubMed] [Google Scholar]
- [32].Khokhar JY, Tyndale RF. Drug metabolism within the brain changes drug response: selective manipulation of brain CYP2B alters propofol effects. Neuropsychopharmacology. 2010;36:692–700. doi: 10.1038/npp.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sultatos LG. Mammalian toxicology of organophosphorus pesticides. J Toxicol Environ Health. 1994;43:271–89. doi: 10.1080/15287399409531921. [DOI] [PubMed] [Google Scholar]
- [34].Khokhar JY, Tyndale RF. Rat brain CYP2B-enzymatic activation of chlorpyrifos to the oxon mediates cholinergic neurotoxicity. Toxicol Sci. 2012;126:325–35. doi: 10.1093/toxsci/kfs029. [DOI] [PubMed] [Google Scholar]
- [35].Zhou K, Khokhar JY, Zhao B, Tyndale RF. First demonstration that brain CYP2D-mediated opiate metabolic activation alters analgesia in vivo. Biochem Pharmacol. 2013;85:1848–55. doi: 10.1016/j.bcp.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Suominen T, Uutela P, Ketola RA, Bergquist J, Hillered L, Finel M, et al. Determination of serotonin and dopamine metabolites in human brain microdialysis and cerebrospinal fluid samples by UPLC-MS/MS: discovery of intact glucuronide and sulfate conjugates. PloS One. 2013;8:e68007. doi: 10.1371/journal.pone.0068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Uutela P, Reinila R, Harju K, Piepponen P, Ketola RA, Kostiainen R. Analysis of intact glucuronides and sulfates of serotonin, dopamine, and their phase I metabolites in rat brain microdialysates by liquid chromatography– tandem mass spectrometry. Anal Chem. 2009;81:8417–25. doi: 10.1021/ac901320z. [DOI] [PubMed] [Google Scholar]
- [38].Tyce GM, Messick JM, Yaksh TL, Byer DE, Danielson DR, Rorie DK. Amine sulfate formation in the central nervous system. Fed Proc. 1986;45:2247–53. [PubMed] [Google Scholar]
- [39].Kallonen SE, Tammimäki A, Piepponen P, Raattamaa H, Ketola RA, Kostiainen R. Discovery of neurosteroid glucuronides in mouse brain. Anal Chim Acta. 2009;651:69–74. doi: 10.1016/j.aca.2009.07.059. [DOI] [PubMed] [Google Scholar]
- [40].King CD, Rios GR, Assouline JA, Tephly TR. Expression of UDP-glucuronosyltransferases (UGTs) 2B7 and 1A6 in the human brain and identification of 5-hydroxytryptamine as a substrate. Arch Biochem Biophys. 1999;365:156–62. doi: 10.1006/abbi.1999.1155. [DOI] [PubMed] [Google Scholar]
- [41].Yamada H, Ishii K, Ishii Y, Ieiri I, Nishio S, Morioka T, et al. Formation of highly analgesic morphine-6-glucuronide following physiologic concentration of morphine in human brain. J Toxicol Sci. 2003;28:395–401. doi: 10.2131/jts.28.395. [DOI] [PubMed] [Google Scholar]
- [42].Nagano E, Yamada H, Oguri K. Characteristic glucuronidation pattern of physiologic concentration of morphine in rat brain. Life Sci. 2000;67:2453–64. doi: 10.1016/s0024-3205(00)00825-0. [DOI] [PubMed] [Google Scholar]
- [43].Togna AR, Antonilli L, Dovizio M, Salemme A, De Carolis L, Togna GI, et al. In vitro morphine metabolism by rat microglia. Neuropharmacology. 2013;75:391–8. doi: 10.1016/j.neuropharm.2013.08.019. [DOI] [PubMed] [Google Scholar]
- [44].Mano N, Goto T, Uchida M, Nishimura K, Ando M, Kobayashi N, et al. Presenceof protein-bound unconjugated bile acids in the cytoplasmic fraction of rat brain. J Lipid Res. 2004;45:295–300. doi: 10.1194/jlr.M300369-JLR200. [DOI] [PubMed] [Google Scholar]
- [45].Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008;36:1461–4. doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- [46].Fujiwara R, Chen S, Karin M, Tukey RH. Reduced expression of UGT1A1 in intestines of humanized UGT1 mice via Inactivation of NF-κB leads to hyperbilirubinemia. Gastroenterology. 2012;142:109–18. doi: 10.1053/j.gastro.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.